Abstract
Increasing evidence indicates that viruses do not simply propagate as independent virions among cells, organs, and hosts. Instead, viral spread is often mediated by structures that simultaneously vehicle groups of viral genomes, such as polyploid virions, aggregates of virions, virion-containing proteinaceous structures, secreted lipid vesicles, and virus-induced cell-cell contacts. These structures increase the multiplicity of infection, independent of viral population density and transmission bottlenecks. Collective infectious units may contribute to the maintenance of viral genetic diversity, and could have implications for the evolution of social-like virus-virus interactions. These may include various forms of cooperation such as immunity evasion, genetic complementation, division of labor, and relaxation of fitness trade-offs, but also non-cooperative interactions such as negative dominance and interference, potentially leading to conflict.
Keywords: multiplicity of infection, genetic diversity, social evolution, microvesicles, baculoviruses, polyploid virion
Beyond the “one genome – one infectious unit” paradigm
A long-accepted concept in virology is that viruses propagate as free virions in the extracellular milieu, each containing one viral genome. Under this view, the viral genetic program is transmitted in the form of individual copies, each of which constitutes a unit of infection. This implies that, for a cell to become co-infected with two or more viral genomes, several virions should to enter the same cell independently. However, as argued here, this view accounts for only one of several possible ways in which viruses spread inside and among hosts since, in some cases, viral infectious units are constituted by structures containing multiple viral genomes. This conceptual shift has been spurred by several recent findings. For instance, detailed microscopic analysis of enterovirus-infected cultures showed that, in addition to being released to the extracellular milieu as free viral particles, groups of virions inside lipid microvesicles budded from infected cells before lysis, and that these vesicles enabled the co-transmission of multiple viral genomes between cells [1]. In another work, massive parallel sequencing of individual cells infected with the vesicular stomatitis virus (VSV) revealed that, despite receiving a single infectious unit, cells often harbored several viral genetic variants inherited from the parental population, suggesting that infectious units were constituted by multiple, genetically diverse viral genomes [2]. A survey of the literature shows that these are just two examples of possible multi-genome infectious units, and that there exists a variety of structures supporting viral co-transmission, here jointly referred to as collective infectious units. Group infection may have broad implications for the maintenance of viral genetic diversity, the evolution of cooperation and conflict, virus epidemiology, and the design of new antiviral strategies.
Collective infectious units come in all shapes and sizes
Albeit discovered independently, structures such as polyploid virions, virion aggregates, and virion-containing lipid vesicles share the common property of delivering multiple viral genome copies to the same cell/host. In addition, different viruses have evolved the ability to rewire the cytoskeleton and alter the surface of infected cells to promote direct cell-to-cell passage of the viral progeny. The number of genomes within these structures probably varies extensively, from five or less in polyploid virions to potentially thousands in direct cell-cell contacts. Use of different mechanisms and molecular processes to achieve a common goal suggests that this kind of group infection has evolved multiple times because it provides some fitness benefits.
Virions with versatile ploidy
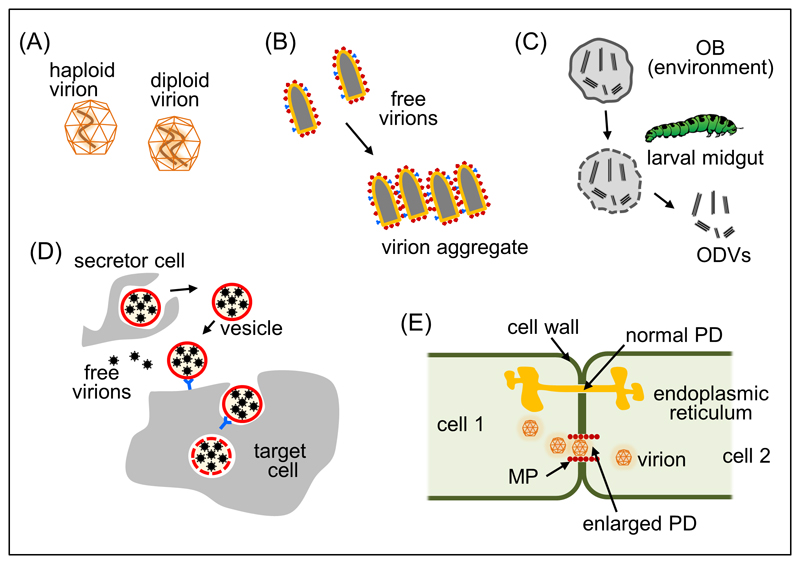
For some viral families, it is well known that each individual virion can contain more than just one copy of the viral genome (Fig. 1A). Probably the first report of a polyploid virion corresponded to the filamentous bacteriophage f1, which occasionally encapsidates multiple genomes in a set of concatenated coat proteins, a structure known as polyphage [3]. Another well-known example is found in retroviruses, which encapsidate two partially annealed RNA genomes per virion. Retroviruses are considered pseudo-diploid because generally only one of the two genomes effectively initiates infection, but true diploidy has also been suggested [4]. True or pseudo-diploidy may be advantageous in terms of increased tolerance to nucleic acid damage, as well as for promoting recombination. For several other families that were initially believed to encapsidate a single genome copy, it was later found that multiple genomes can be co-packaged. This has been shown in paramyxoviruses such as measles virus [5], in filoviruses such as Ebola virus [6], and in birnaviruses such as infectious bursal disease virus [7] and infectious pancreatic necrosis virus [8]. Although the greater modularity of filamentous capsids probably favors co-packaging, clearly the process can also take place in icosahedral virions. Some segmented viruses, such as bursal disease virus and the Rift Valley fever bunyavirus, appear to lack mechanisms for guiding the encapsidation of their different fragments, implying that virions contain random sets of segments [7,9]. In these cases, versatile ploidy can be viewed as a consequence of non-selective encapsidation. Polyploidy can mediate the co-transmission of different genetic variants of a virus and, hence, is expected to promote interactions between virus variants at the intracellular level.
Fig. 1. Structure of collective infectious units in viruses.
(A) Viral capsids can contain one or multiple genome copies, producing n-ploid virions. Filamentous capsids probably have increased tendency to accommodate multiple genomes copies, but polyploidy has also been described in icosahedral capsids. A diploid icosahedral virion is represented. (B) Aggregation is a commonly described process in virion preparations. However, its molecular basis and physiological relevance remain largely unassessed. Aggregation of bullet-shaped virions is shown, indicating hypothetical virion surface molecules that may mediate specific virion-virion contacts. (C) Occlusion bodies (OBs) are virions encased in crystallized polyhedrin and constitute the inter-host transmission structure of baculoviruses. Crystals are dissolved under the alkaline pH of the larval midgut, releasing occlusion-derived virions (ODVs). In turn, these can be single virions (single nucleopolyhedroviruses) or groups of virions (multiple nucleopolyhedroviruses). (D) Virion-containing lipid vesicles include exosome-like vesicles (which can contain full virions or viral RNA) and autophagosome-like vesicles. These are rich in phosphatidylserine, which stimulates phagocytosis and thus potentiates viral entry. (E) Cell-to-cell transmission also ensures that multiple viral genome copies are collectively delivered to the same target cell. Use of plasmodesmata (PD) by plant viruses is depicted as an example. Whereas normal PDs are too small to allow viral passage and contain smooth endoplasmic reticulum, plant viruses encode a movement protein (MP) that promotes cell-to-cell transmission by altering the structure of PDs.
Virion aggregates
Early electron microscopy studies suggested that viral particles tend to form aggregates in a variety of viruses, including poxviruses [10], influenzaviruses [11], rhabdoviruses [12], and polioviruses [13]. However, aggregation has been mainly viewed from a technical standpoint as an issue for the purpose of preparing “pure”, monodisperse viral stocks. To what extent aggregation is a natural process has not been investigated in detail, and its potential implications for viral transmission, diversity and evolution remain largely unexplored. Our recent work with VSV shows, however, that each plaque forming unit (PFU) (see Glossary) does not necessarily constitute a biological clone, as it can harbor several genetic variants of the virus [2]. This was demonstrated by sequencing the viral progeny produced by single cells infected with less than one PFU on average. Several of the variants detected in the viral progeny were also present in other cells and could be traced back to the stock used for inoculation. Furthermore, when we co-infected cells with two phenotypically marked genetic variants (wild-type and antibody resistant mutant) and used the progeny viruses for isolating single PFUs by the plaque assay, we found that individual plaques still contained a mixture of the two variants. These results suggest that aggregates of virions are jointly transmitted as part of the same PFU (Fig. 1B). Recently, we have explored the causal link between aggregation and the co-transmission of variants in VSV. We found that free virions can spontaneously bind to other virions in certain media, particularly in saliva, giving rise to collective infectious units [14]. In other viruses, it has been found that salivary mucins [15] as well as some concentrations of non-neutralizing antibodies may sometimes promote aggregation by cross linking virions, without necessarily impeding viral entry [16].
Glossary.
Defective interfering particle (DIP): a viral particle whose genome contains large deletions, that is non-infectious per se but can replicate in the presence of a functional virus (helper).
Effective population size: the relevant population size in terms of random genetic drift (smaller effective population sizes mean stronger drift). Since not all individuals in a population contribute to the next generation with equal probability (as a result of natural selection, population bottlenecks and so on) the effective population size is typically much smaller than the census population size.
Epistasis: non-independent effects of different mutations on a given phenotype. When referring to fitness, negative/positive epistasis means that a combination of mutations is associated to lower/higher fitness than expected from the effects of each mutation alone.
Genetic complementation: the ability of two parental strains with deleterious mutations to yield normal progeny. In viruses, complementation refers to the ability of different deleterious variants co-infecting the same cell to reach similar fitness as the wild-type. Complementation requires that the deleterious mutations map to different genes, or at least to different functional domains of a gene.
Heterotypic cooperation: a mutually beneficial interaction between individuals, in which the benefits originate from specific genetic differences between the interacting partners, and that has evolved in response to selection based on these benefits.
Infectious dose: the minimal number of virus (measured as PFUs, genome copies, or other quantity) that is required to cause infection of a susceptible host.
Multipartite virus: a virus whose genome is divided in two or more segments and in which each segments is encapsidated separately. This is in contrast with narrow-sense segmented viruses, in which all segments are co-packaged. In plants and fungi, most segmented viruses are multipartite, whereas multipartite viruses are extremely rare among animals.
Multiplicity of infection (MOI): sometimes defined as the ratio of infectious viral particles to susceptible cells in a given space. An alternative definition is the number of viral genomes that initiate an infection. These two definitions are similar under the “one genome – one infectious unit” model, but become disconnected otherwise.
Negative dominance: a deleterious mutation that is genetically dominant over the normal allele.
Occlusion body (OB): the inter-host transmission structure of baculoviruses, formed by one or several virions encased in a matrix of crystallized protein.
Plaque forming unit (PFU): an infectious entity capable of forming a plaque, i.e. a defined lysis area in a cell lawn. In lytic viruses, the number of PFUs per mL is used to quantify the infectivity of a viral suspension (viral titer).
Plasmodesmata: plant intercellular channels that mediate molecular transport and communication. Plasmodesmata are used by plant viruses for cell-to-cell movement.
Superinfection exclusion: the process whereby a pre-existing viral infection prevents secondary infections with the same or closely related viruses. This can occur through different mechanisms, including reduced expression of surface receptors, reduced viral uptake, or inhibition of early steps of the infection cycle.
Viral titer / load / population density: these expressions all refer to the concentration of viral infectious units. The viral titer is used in cell culture settings, the viral load for infections in vivo, and viral population density in the epidemiological and ecological contexts
Virological synapse: junctions between cells specifically promoted by viruses to facilitate their cell-to-cell transfer.
Baculovirus multi-virion complexes and occlusion bodies
Baculoviruses are highly virulent insect viruses that are transmitted in nature from larvae to larvae, which typically involves long times of inactivity in the environment, including overwinter periods [17]. As such, environmental stability is an important fitness component for baculoviruses. Whereas intra-host viral dissemination mainly takes place in the form of enveloped virions that bud from the membrane of infected cells, host-to-host transmission is mediated by occlusion bodies (OBs), in which virions are encased inside a proteinaceous matrix made of crystalized polyhedrin (Fig. 1C) [8]. OBs originate from occlusion-derived bodies (ODVs), which are produced in the cell nucleus and are surrounded by a de novo synthesized membrane. ODVs and OBs accumulate during late stages of larval infection and are released to the environment upon host death and liquefaction. In addition to their protective role against UV radiation, OBs are vehicles for the host-to-host co-transmission of multiple viral genomes since, in most baculoviruses, each OB contains tens of virions. In turn, in multiple nucleopolyhedroviruses each ODV can be made of several capsids. The evolutionary advantage of nucleocapsid aggregation is still debated, but it is nevertheless clear that OBs contain non-identical viral genome copies, including genomes with large deletions [19–22].
Virion-containing extracellular vesicles
The well-known distinction between enveloped and non-enveloped viruses has become blurred in recent years after the discovery that some non-enveloped viruses, such as hepatitis A virus [23] and hepatitis E virus [24], can also be released from infected cells inside exosomes, 30-150 nm lipid vesicles with an important role in antiviral immunity among other regulatory functions [25] (Fig. 1D). Exosomes can contain virions and can mediate intercellular transmission of HIV-1 [26] and severe fever with thrombocytopenia syndrome virus [27], but can also transmit naked, infectious RNA genomes such as in hepatitis C virus [28]. Furthermore, work with poliovirus and other enteroviruses such as coxsackievirus and rhinovirus has shown that pools or virions are encapsulated in larger, autophagosome-like lipid vesicles rich in phosphatidylserine [1]. Retrospective analysis of micrographs of bluetongue virus [29] and norovirus [30] also suggested vesicle encapsulation of virions [31]. Given the size of enterovirus vesicles, they could potentially contain hundreds to thousands of virions. However, their virion content has not been precisely defined yet, and an assessment of the actual number of viral genomes per vesicle that effectively initiate the cell infection is lacking. The ability of vesicles to potentially transport large numbers of viral particles was further exemplified in recent work with the Marseille virus (a large DNA virus), which suggested that up to thousands of virions could be encapsulated and transmitted inside giant vesicles [32]. Importantly, vesicle-mediated intercellular transmission of viruses coexists with the classical mode of transmission in the form of free virions, and the evolutionary, genetic, and mechanistic determinants of this duality remain to be elucidated.
Direct cell-to-cell transfer of viruses
The co-delivery of multiple viral genomes to the same target cell can be further facilitated by direct contacts between cells. Viruses have been long known to exploit pre-existing cell contacts for cell-to-cell movement, such as plamodesmata in plant cells [33–35], neurological synapses in herpes virus [36] and measles virus [37], and immunological synapses in HIV-1 [38] and other immunotropic viruses (Fig. 1E). In solid tissues, effective cell-to-cell transfer can also occur in the absence of specific cell signalling structures if viral budding is concomitant with attachment/entry into a neighbor cell [39]. In addition, it subsequently became clear that direct cell-to-cell viral spread is not restricted to use of physiological cell contacts [40,41]. Contacts induced specifically by viruses are referred to as virological synapses. In HIV-1 and other retroviruses, viral glycoproteins located in the cell surface stimulate the establishment of filopodes, which are used as bridges for cell-to-cell spread [42]. Several lines of evidence indicate that cell-to-cell virion passage through virological synapses is a major route of intra-host viral dissemination and has implications for antiviral therapy in HIV-1 [43–45], human T cell leukaemia virus 1 (HTLV-1) [46,47], and hepatitis C virus [48,49]. Furthermore, HTLV-1 is capable of inducing the formation of a modified extracellular matrix that is colonized by a high density of infectious particles. This so-called “viral biofilm” allows spread of virions between neighbor cells in a protected environment [46]. In vaccinia virus and other poxviruses, newly assembled viral particles remain associated to the surface of the infected cell, where they induce actin polymerization and the production of cellular projections that enhance viral transmission to neighboring cells [50]. However, the actual number of genomes that initiate a new cell infection following cell-to-cell transfer could be much lower than the total number of genomes transferred [51]. Cell-to-cell viral transfer is still an incompletely understood process, and its implications for viral spread, diversity, and evolution are only beginning to be investigated [52].
Implications of collective infectivity for viral fitness and evolution
The fact that collective infectious units drive the simultaneous spread of multiple viral genomes to the same target cell/host may have implications for viral genetic diversity and evolution. Possible ways in which collective infectious units may determine viral fitness are discussed below. So far, most of the ideas presented are untested hypotheses, and they often constitute alternative, non-exclusive scenarios.
Collective infectious units and viral transmissibility
Although each infected multicellular host typically hosts billions of infectious units, only a minuscule fraction, generally on the order of one to tens or less, will colonize new individuals, meaning that severe population bottlenecks take place during viral host-to-host transmission [53,54]. Similarly, intra-host population structure in the form of compartmentalized organs with restricted viral trafficking has been demonstrated in animal and plant viruses [35,54,55]. Finally, yet another fundamental driver of population structure in viruses is the fact that replication is confined to discrete cells, or even to subcellular compartments. Under the “one genome – one infectious unit” paradigm, stringent population bottlenecks make it unlikely that two or more viral genome copies can be co-transmitted to the same target cells. In contrast, a major consequence of collective infectious units is that they increase the cellular multiplicity of infection (MOI) regardless of viral population density or transmission bottlenecks (Box 1). The increased MOI afforded by collective infectious units might thus be beneficial for viral transmissibility. For instance, in segmented viruses lacking regulatory mechanisms for ensuring encapsidation of each segment, polyploidy could increase the chances that at all segments are transmitted. Consistent with this view, birnavirus polyploid capsids showed greater infectivity than haploid capsids [7,8]. Furthermore, even if most virions contain a full set of segments, stochastic segment loss during early steps of the cell infection cycle can lead to non-productive infections, as has been shown for influenza virus [56,57]. Collective infectious units may also contribute to the transmissibility of multipartite viruses. However, the theoretical MOI required for successful transmission becomes prohibitively high as the number of independently encapsidated genome segments increases [58,59]. To what extent collective transmission, for instance through plasmodesmata, fulfils this requirement is unclear, but it is likely that other mechanisms are at play to ensure the transmission of all segments in multipartite viruses. It has been suggested that the multipartite beet necrotic yellow vein virus tends to disseminate inside the plant in the form of RNA-protein complexes rather than mature virions, and that RNA-RNA interactions between segments ensure their joint cell-to-cell transfer and systemic spread [60].
Box 1. Collective infectious units and the MOI concept.
The MOI is a central concept in virology, which has been traditionally defined as the ratio of infectious viral particles to susceptible cells in a given space. Based on this definition and under the “one viral genome – one infectious unit” view, the number of viral genome copies (n) that will enter any given cells is calculated assuming that viral particles infect cells independently. This is modeled using a Poisson probability distribution, with parameter equal to the MOI, such that According to this, low MOI values mean that the vast majority of cell infections will be initiated from n = 1 viral genome copy. However, this will be a wrong prediction if viruses aggregate into collective infectious units. In the latter case, most infected cells will contain several viral genome copies regardless of the MOI. In other words, collective infectious units imply that the commonly used definition of MOI does not capture adequately what happens at the individual cell level. An alternative definition of the MOI is the average number of viral genomes that initiate an infection, regardless of the number of infectious particles per susceptible cell. The “given space” used in the traditional definition of MOI should thus be the individual infected cell, or it may also be the individual multicellular host, depending of the scale of analysis. If one follows this redefinition, the cellular MOI becomes disentangled from the fraction of cells infected.
Collective infection and viral growth under adverse conditions
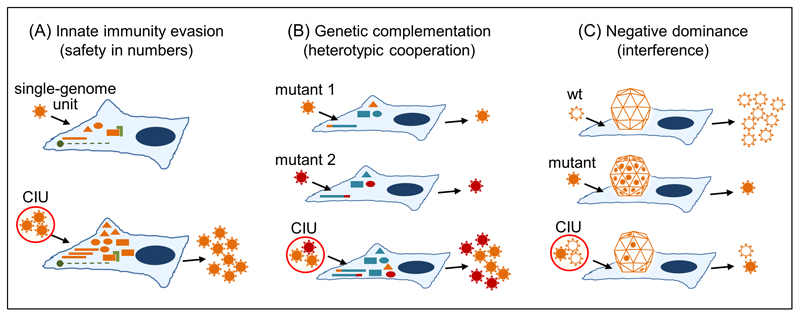
In principle, a disadvantage of collective infectious units is that aggregation is costly for dispersal and increases competition. A possible factor that may compensate these costs is the Allee effect, i.e. a positive correlation between the founder group size and fitness. Several lines of evidence suggest that, under adverse conditions for the virus, the chances of completing the infection cycle successfully increase with the cellular MOI (Fig. 2A). For instance, by increasing the MOI regardless of viral load, cell-to-cell transmission could help HIV-1 replicate in the presence of antiretroviral therapy [44,61,62], as well as of natural viral restriction factors such as TRIM5α [63]. Also, many attenuated viruses can only infect cells with crippled antiviral innate immunity responses, such as cancer cells [64]. However, these attenuated viruses can sometimes successfully grow in normal cells, provided that the MOI at inoculation is high enough [65]. This suggests that initiating the infection with a higher number of genome copies could help the virus overcome innate immunity responses [66]. As a result, disrupting structures of collective infection might promote viral attenuation (Box 2). Analogous arguments can be made for inter-host transmission, since transmission in the form of multi-genome units may allow the virus to increase the chances of reaching the infectious dose, which should be strongly determined by innate immunity. Notice, however, that for collective infection to confer a net benefit, the cost of reduced dispersal should be outbalanced, and whether this condition is met remains untested. In addition to overcoming innate immunity, some collective infectious units may further confer the virus the ability to evade antibody neutralization. This has been shown for cell-to-cell transmission in HIV-1 [67], although HIV-infected cells can also be targeted by neutralizing antibodies [68]. Relatedly, viral biofilms can protect HTLV-1 from antibody neutralization [46]. Lastly, the high phosphatidylserine content of virion-containing vesicles stimulates phagocytosis, because phosphatidylserine is a physiological “eat me” signal normally used for recycling apoptotic cells [69]. This strategy for enhancing viral entry is known as apoptotic mimicry [70]. However, these benefits are not intrinsically associated with the collective nature of infectious units, because encapsulation of single virions in lipid vesicles is expected to have similar effects on immune evasion and mimicry.
Fig. 2. Possible social-like interactions mediated by collective infectious units in viruses.
(A) Co-infection of cells with multiple viral genomes may increase their ability to evade innate immunity responses. Upon viral entry, pathogen-associated molecular patterns (proteins, DNA/RNA, etc.) are sensed by the cell and trigger an antiviral state controlled by immunity effectors. By increasing the cellular MOI, the virus may overwhelm these responses and more successfully complete the infection cycle. Orange bars: genome copies; orange triangles, ellipses and rectangles: viral products; green: cellular responses to infection. CIU: collective infectious unit. (B) Different genetic variants of a virus may complement each other when present in the same cell. Two deleterious mutants (orange and red) mapping to different viral genes are represented. Normal viral genes and proteins are represented in blue. Whereas genetic complementation requires that different loci are involved, other forms of heterotypic cooperation can be envisaged that could involve the same locus, including synergistic interactions between beneficial mutations, or division of labor. For instance, in multifunctional proteins, co-infecting variants may specialize in subsets of these functions. (C) Negative dominance as an example of interference, a non-cooperative virus-virus interaction. Negative dominance is particularly likely in oligomeric structures (here, a capsid). By forming mixed oligomers, a deleterious variant (orange filled) interferes with the wild-type variant (wt, white filled). Other forms of non-cooperative interactions have been described, as exemplified by DIPs, which act as social cheats.
Box 2. Possible practical implications of collective infectious units.
Virus-virus interactions among members of multi-genome infectious structures may have implications for antiviral research. For instance, it has been proposed that transmissible DIP-like viruses could be engineered as an epidemiological-level strategy for combating viruses [102]. Also, use of DIPs to combat influenza virus is under investigation [93]. Clearly, the success of these strategies depends on whether DIPs are co-transmitted with the wild-type virus, which in turn should depend on whether the virus propagates as groups or individually. Another interesting suggestion is using negative dominance to retard the emergence of drug resistance [87,103]. In drugs targeting oligomeric structures such as viral capsids, even if a drug-resistant variant appears, it may be suppressed by the presence of wild-type capsids in the same cell, since mosaic capsids would also be drug-sensitive [87]. Again, this evolution-proof approach critically relies on high MOIs, which means that its feasibility may depend on whether the virus propagates using multi-genome structures. Yet another possible application of collective infectious units is in the field of biopesticides. Baculovirus OBs containing batteries of variants may allow targeting larvae that are resistant to the wild-type virus, or even targeting different species of plagues with a single product. Collective infectious units may also be used in the field of cancer virotherapy. If co-transmission of groups of genomes enhances viral replication in fully immunocompetent cells but is not required in cells with innate immunity defects, mutants lacking the ability to form collective infectious units might exhibit selective replication in cancerous cells, which typically show innate immunity defects. Finally, some types of collective infectious units such as virion aggregates or virion-containing vesicles that evade pre-existing humoral immunity may provide an efficient vehicle for the delivery of viruses for therapeutic purposes.
Collective infectious units and the maintenance of genetic diversity
Bottlenecks and population structure should strongly reduce the effective population size and promote the action of random genetic drift. Drift allows the fixation of slightly deleterious mutations which, in the absence of recombination, will produce irreversible fitness losses in a process known as Muller´s ratchet [71,72]. However, transmission of viruses as collective infectious units among cells, organ, and hosts may alleviate population bottlenecks and mitigate their effects on viral fitness and diversity. Furthermore, by bringing multiple genomes into the same cells, collective infectious units should promote recombination between different genetic variants of a virus, or even between different viral species. This appears to be a likely scenario in viruses that propagate within vesicles. If two such species infect the same cell type, extracellular vesicles containing virions of both species could be produced. For instance, enterovirus 71 and coxsackievirus A16, both of which are causative agents of hand, foot, and mouth disease (HFMD) in humans, use the scavenger receptor class B2, which is expressed in a broad range of cell types [73,74]. Interestingly, these viruses have been reported to co-circulate at the epidemiological level in recent HFMD outbreaks, and co-infection has been associated with an increased risk of clinical complications [75]. As a note of caution, the number of genomes that successfully initiate infection may be much lower than the total number of genomes in collective infectious units. For cell-to-cell spread, the estimated number of founders per cell appears to be lower than six in widely different viruses, including tobacco mosaic virus [76], tomato mosaic virus [51], alphaherpesviruses [77], and HIV-1 [61]. In contrast, this number was estimated to be 20-40 during cell-to-cell spread of turnip mosaic virus throughout the leaf mesophyll, although superinfection exclusion limited the ability of progeny from different cells to get transferred to the same recipient cells, implying that the co-transmitted genomes shared a high degree of genetic relatedness [35]. In sum, to what extent collective infectious units modulate population structure remains an open question.
Collective infectious units as potential drivers of heterotypic cooperation
High MOIs associated with multi-genome infectious structures may promote mutually beneficial interactions between different variants of a virus (heterotypic cooperation) at the intracellular level. A frequently cited type of such interaction is genetic complementation, whereby the fitness of two or more mutants with different defects is restored as a result of the ability of viral products to function in trans, meaning that functional proteins provided by the co-infecting genomes are used as public goods inside the cell [78,79] (Fig. 2B). At very high mutation rates such as those experienced by RNA viruses [80], most genomes will be loaded with deleterious mutations and, hence, may significantly benefit from complementation. However, it should also be noticed that, in principle, complementation achieves the same fitness as the wild-type at best. In contrast, other forms of genetic interaction may lead to more fruitful cooperation. Recent work with coxsackievirus (a vesicle-forming enterovirus) found that some beneficial mutations showed marked negative epistasis when combined in the same genome, but that this antagonism disappeared when the mutations were present in different genomes co-infecting the same cell [81]. More generally, the fact that most viral genomes are highly compact and encode multifunctional proteins imposes fitness tradeoffs, whereby the various functions of a given protein cannot be simultaneously optimized [82–84]. It is possible that the high MOI conferred by group infection could favor a division of labor whereby each co-infecting variant specializes in a given sub-function, therefore relaxing the fitness tradeoffs experienced by a single genome. Heterotypic cooperation was suggested for measles virus, where a deleterious mutant partially restored fitness by co-packaging with a second-site mutant in diploid capsids [85]. Such mosaic capsids showed a phenotype not exhibited by the wild-type, since they were capable of infecting cells that lacked the canonical measles virus receptor. A suggested molecular basis for this interaction was related to the oligomeric structure of capsids. The hetero-oligomers formed in cells infected with both mutants appeared to exhibit a more favorable stability/flexibility balance than homo-oligomers. In baculoviruses, it has been suggested that the genetic diversity of occlusion bodies correlates with viral transmissibility [19] and long-term viral evolvability [86]. Despite the above lines of work, how heterotypic cooperation occurs and how it is maintained by selection remains unclear in most cases.
Possible negative interactions among members of collective infectious units
In contrast to the mutually beneficial interactions outlined above, a deleterious mutation encoding a malfunctioning viral product can interfere with normal products present in the same cell, a process known as negative dominance. This is particularly likely in oligomeric structures such as viral capsids, and has been demonstrated in poliovirus using wild-type and drug-resistant variants [87]. As suggested by this work, negative dominance could be exploited for the design of evolution-proof antiviral strategies (Box 2). By ensuring the maintenance of high MOIs, multi-genome structures could allow these virus-virus interactions to take place regardless of viral load or transmission bottlenecks. Inasmuch high mutation rates have been argued to increase opportunities for genetic complementation they could favor the emergence of cheater-like viruses that exploit the wild-type products, severely impairing population fitness [88–90] (Fig. 2C). In many cases, the mechanisms whereby cheaters are selectively favored remain to be elucidated. Baculovirus occlusion bodies and intra-host populations typically contain multiple genetic variants, including genomes with large deletions [19–22]. An cost has been suggested for factors involved in host-to-host baculovirus transmission, such as the polyhedrin gene [91] and, therefore, mutants defective for these genes should be selected at the intra-host level, whereas they could cheat on wild-type viruses for inter-host transmission. An extreme case of such non-cooperative interactions is defective interfering particles (DIPs), whose genomes have large, lethal deletions and can only replicate in the presence of a functional variant, called helper virus [92,93]. At high cellular MOI, DIPs tend to be superior to the wild-type because their shorter genomes are replicated faster and, hence, DIPs can reach very high population frequencies. Increasing evidence suggest that DIPs occur in vivo and, in some cases, they may even undergo host-to-host transmission [94–101]. From the perspective of the “one genome – one infectious unit” paradigm, inter-host transmission of DIPs may seem paradoxical because, soon after transmission, the viral load should be too low to enable co-infection of cells with DIPs and functional variants at any appreciable rate. In contrast, collective infectious units are compatible with this scenario. Interestingly, viruses appear to have evolved a series of mechanisms that could limit the spread of DIPs and invasion of cheaters in general, including superinfection exclusion and subcellular compartmentalization.
Concluding remarks
Results obtained with widely different viruses, including plant and animal viruses, RNA and DNA viruses, and enveloped and non-enveloped viruses, indicate that collective infectious units are widespread in nature, but further work is required to better characterize these structures and to gauge their importance for viral dissemination at the intra-host and inter-host levels (see Outstanding Questions). These multi-genome structures may promote virus-virus interactions and the evolution of social-like traits, may contribute to explaining the co-circulation of genetic variants of a virus and of different viral species at the epidemiological level, and may influence immune evasion and pathogenesis. Importantly, the fact that collective infectious units deliver groups of genetically diverse genomes to the same cells should not be interpreted a priori as evidence for cooperation, or as an adaptive trait. Finally, cooperative structures, but also viral cheating, might inspire new antiviral strategies.
Outstanding Questions.
In addition to polyploid virions, virion aggregates, occlusion bodies, lipid vesicles, cell-cell contacts, and viral biofilms, are there other structures for the co-transmission of groups of genomes in the same infectious unit?
What is the molecular basis of virion aggregation and how frequent is it under physiological conditions?
What are the viral genetic determinants, if any, of virion polyploidy, aggregation, virion encapsulation in lipid vesicles, and cell-to-cell transmission?
Are some types of viruses more prone to using collective infectious units than others?
Why do multi-genome infectious structures generally coexist with free virions and how this affects viral spread at the intra-host and inter-host levels?
What specific fitness effects are associated with collective infectious units? In particular, are these structures mainly used for viral cooperation or do they serve as vehicles for the spread of cheater-like viruses?
What are the specific public goods shared by members of collective infectious units?
Can co-infecting viral genomes cooperate through division of labor at the molecular level or by relaxing fitness trade-offs that operate at the level of individual genomes?
How many viral genomes effectively initiate infection within each collective infectious unit?
How do social evolution concepts such as kin selection apply to viral collective infectious units?
Can we draw parallels between viral collective infectious units and microbial social networks such as biofilms?
Can we use collective infectious units to develop transmissible defective viruses as an antiviral strategy?
Is it possible to target collective infectious structures for attenuating viruses?
Trends Box.
The spread of viruses among cells, organs, and hosts is often mediated by structures that carry multiple viral genome copies, such as polyploid virions, virion aggregates, occlusion bodies, virus-containing lipid vesicles, and virological synapses.
These structures increase the multiplicity of infection, defined as the number of viral genomes that initiate infection.
High multiplicities of infection may promote the emergence of social-like virus-virus interactions, such as cooperation to evade immunity and/or antiviral treatments and division of labor, but also of non-cooperative interactions such as negative dominance and interference.
Collective infectious units may be exploited for retarding drug resistance evolution, for producing attenuated viruses, or for co-delivering different genetic variants of a virus to target cells/hosts.
Acknowledgments
I thank Stu West and Stéphane Blanc for insightful comments. This work is financially supported by a European Research Council (ERC) Consolidator Grant (724519, Vis-à-Vis).
References
- 1.Chen YH, et al. Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell. 2015;160:619–630. doi: 10.1016/j.cell.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Combe M, et al. Single-cell analysis of RNA virus infection identifies multiple genetically diverse viral genomes within single infectious units. Cell Host Microbe. 2015;18:424–432. doi: 10.1016/j.chom.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.López J, Webster RE. Morphogenesis of filamentous bacteriophage f1: orientation of extrusion and production of polyphage. Virology. 1983;127:177–193. doi: 10.1016/0042-6822(83)90382-3. [DOI] [PubMed] [Google Scholar]
- 4.Iglesias-Sánchez MJ, López-Galíndez C. Each genomic RNA in HIV-1 heterozygous virus generate new virions. Virology. 2005;333:316–323. doi: 10.1016/j.virol.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 5.Rager M, et al. Polyploid measles virus with hexameric genome length. EMBO J. 2002;21:2364–2372. doi: 10.1093/emboj/21.10.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beniac DR, et al. The organisation of Ebola virus reveals a capacity for extensive, modular polyploidy. PLoS ONE. 2012;7:e29608. doi: 10.1371/journal.pone.0029608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luque D, et al. Infectious bursal disease virus is an icosahedral polyploid dsRNA virus. Proc Natl Acad Sci USA. 2009;106:2148–2152. doi: 10.1073/pnas.0808498106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lago M, et al. Aquabirnavirus polyploidy: A new strategy to modulate virulence? J Gen Virol. 2016;10 doi: 10.1099/jgv.0.000434. [DOI] [PubMed] [Google Scholar]
- 9.Wichgers Schreur PJ, Kortekaas J. Single-molecule FISH reveals non-selective packaging of Rift Valley fever virus genome segments. PLoS Pathog. 2016;12:e1005800. doi: 10.1371/journal.ppat.1005800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galasso GJ, Sharp DG. Virus particle aggregation and the plaque-forming unit. J Immunol. 1962;88:339–347. [PubMed] [Google Scholar]
- 11.Wallis C, Melnick JL. Virus aggregation as the cause of the non-neutralizable persistent fraction. J Virol. 1967;1:478–488. doi: 10.1128/jvi.1.3.478-488.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galasso GJ. Quantitative studies on the quality, effects of aggregation and thermal inactivation of vesicular stomatitis virus. Arch Gesamte Virusforsch. 1967;21:437–446. doi: 10.1007/BF01241742. [DOI] [PubMed] [Google Scholar]
- 13.Floyd R. Viral aggregation: mixed suspensions of poliovirus and reovirus. Appl Environ Microbiol. 1979;38:980–986. doi: 10.1128/aem.38.5.980-986.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cuevas JM, et al. Emergence of supra-virion infectious units from free viral particles in an enveloped virus. Nat Microbiol. 2017 doi: 10.1038/nmicrobiol.2017.78. Under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bergey EJ, et al. Interaction of HIV-1 and human salivary mucins. J Acquir Immune Defic Syndr. 1994;7:995–1002. [PubMed] [Google Scholar]
- 16.Dimmock NJ. Neutralization of animal viruses. Curr Top Microbiol Immunol. 1993;183:1–149. doi: 10.1007/978-3-642-77849-0. [DOI] [PubMed] [Google Scholar]
- 17.Fleming-Davies AE, Dwyer G. Phenotypic variation in overwinter environmental transmission of a baculovirus and the cost of virulence. Am Nat. 2015;186:797–806. doi: 10.1086/683798. [DOI] [PubMed] [Google Scholar]
- 18.Slack J, Arif BM. The baculoviruses occlusion-derived virus: virion structure and function. Adv Virus Res. 2007;69:99–165. doi: 10.1016/S0065-3527(06)69003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simon O, et al. Dynamics of deletion genotypes in an experimental insect virus population. Proc Biol Sci. 2006;273:783–790. doi: 10.1098/rspb.2005.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zwart MP, et al. Complex dynamics of defective interfering baculoviruses during serial passage in insect cells. J Biol Phys. 2013;39:327–342. doi: 10.1007/s10867-013-9317-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rohrmann GF. Baculovirus nucleocapsid aggregation (MNPV vs SNPV): an evolutionary strategy, or a product of replication conditions? Virus Genes. 2014;49:351–357. doi: 10.1007/s11262-014-1113-5. [DOI] [PubMed] [Google Scholar]
- 22.Chateigner A, et al. Ultra deep sequencing of a baculovirus population reveals widespread genomic variations. Viruses. 2015;7:3625–3646. doi: 10.3390/v7072788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng Z, et al. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature. 2013;496:367–371. doi: 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagashima S, et al. Hepatitis E virus egress depends on the exosomal pathway, with secretory exosomes derived from multivesicular bodies. J Gen Virol. 2014;95:2166–2175. doi: 10.1099/vir.0.066910-0. [DOI] [PubMed] [Google Scholar]
- 25.Schorey JS, et al. Exosomes and other extracellular vesicles in host-pathogen interactions. EMBO Rep. 2015;16:24–43. doi: 10.15252/embr.201439363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kadiu I, et al. Biochemical and biologic characterization of exosomes and microvesicles as facilitators of HIV-1 infection in macrophages. J Immunol. 2012;189:744–754. doi: 10.4049/jimmunol.1102244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silvas JA, et al. Extracellular vesicles mediate receptor-independent transmission of novel tick-borne bunyavirus. J Virol. 2015;90:873–886. doi: 10.1128/JVI.02490-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramakrishnaiah V, et al. Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc Natl Acad Sci USA. 2013;110:13109–13113. doi: 10.1073/pnas.1221899110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hyatt AD, et al. Release of bluetongue virus-like particles from insect cells is mediated by BTV nonstructural protein NS3/NS3A. Virology. 1993;193:592–603. doi: 10.1006/viro.1993.1167. [DOI] [PubMed] [Google Scholar]
- 30.Lew JF, et al. Identification of minireovirus as a Norwalk-like virus in pediatric patients with gastroenteritis. J Virol. 1994;68:3391–3396. doi: 10.1128/jvi.68.5.3391-3396.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altan-Bonnet N, Chen YH. Intercellular transmission of viral populations with vesicles. J Virol. 2015;89:12242–12244. doi: 10.1128/JVI.01452-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arantes TS, et al. The large Marseillevirus explores different entry pathways by forming giant infectious vesicles. J Virol. 2016;90:5246–5255. doi: 10.1128/JVI.00177-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar D, et al. Cell-to-cell movement of viruses via plasmodesmata. J Plant Res. 2015;128:37–47. doi: 10.1007/s10265-014-0683-6. [DOI] [PubMed] [Google Scholar]
- 34.Ritzenthaler C. Parallels and distinctions in the direct cell-to-cell spread of the plant and animal viruses. Curr Opin Virol. 2011;1:403–409. doi: 10.1016/j.coviro.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 35.Gutiérrez S, et al. The multiplicity of cellular infection changes depending on the route of cell infection in a plant virus. J Virol. 2015;89:9665–9675. doi: 10.1128/JVI.00537-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zaichick SV, et al. Alphaherpesviruses and the cytoskeleton in neuronal infections. Viruses. 2011;3:941–981. doi: 10.3390/v3070941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawrence DM, et al. Measles virus spread between neurons requires cell contact but not CD46 expression, syncytium formation, or extracellular virus production. J Virol. 2000;74:1908–1918. doi: 10.1128/jvi.74.4.1908-1918.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDonald D, et al. Recruitment of HIV and its receptors to dendritic cell-T cell junctions. Science. 2003;300:1295–1297. doi: 10.1126/science.1084238. [DOI] [PubMed] [Google Scholar]
- 39.Vassalli JD, et al. Direct cell-to-cell transmission of vesicular stomatitis virus. J Cell Sci. 1986;85:125–131. doi: 10.1242/jcs.85.1.125. [DOI] [PubMed] [Google Scholar]
- 40.Sattentau QJ. The direct passage of animal viruses between cells. Curr Opin Virol. 2011;1:396–402. doi: 10.1016/j.coviro.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 41.Zhong P, et al. Cell-to-cell transmission of viruses. Curr Opin Virol. 2013;3:44–50. doi: 10.1016/j.coviro.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sherer NM, et al. Retroviruses can establish filopodial bridges for efficient cell-to-cell transmission. Nat Cell Biol. 2007;9:310–315. doi: 10.1038/ncb1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Álvarez RA, et al. Unique features of HIV-1 spread through T cell virological synapses. PLoS Pathog. 2014;10:e1004513. doi: 10.1371/journal.ppat.1004513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agosto LM, et al. HIV cell-to-cell transmission: effects on pathogenesis and antiretroviral therapy. Trends Microbiol. 2015;23:289–295. doi: 10.1016/j.tim.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Komarova NL, et al. Synaptic transmission and the susceptibility of HIV infection to anti-viral drugs. Sci Rep. 2013;3:2103. doi: 10.1038/srep02103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gross C, Thoma-Kress AK. Molecular mechanisms of HTLV-1 cell-to-cell transmission. Viruses. 2016;8:74. doi: 10.3390/v8030074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pais-Correia AM, et al. Biofilm-like extracellular viral assemblies mediate HTLV-1 cell-to-cell transmission at virological synapses. Nat Med. 2010;16:83–89. doi: 10.1038/nm.2065. [DOI] [PubMed] [Google Scholar]
- 48.Xiao F, et al. Hepatitis C virus cell-cell transmission and resistance to direct-acting antiviral agents. PLoS Pathog. 2014;10:e1004128. doi: 10.1371/journal.ppat.1004128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Graw F, et al. Quantification of hepatitis C virus cell-to-cell spread using a stochastic modeling approach. J Virol. 2015;89:6551–6561. doi: 10.1128/JVI.00016-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dodding MP, Way M. Nck- and N-WASP-dependent actin-based motility is conserved in divergent vertebrate poxviruses. Cell Host Microbe. 2009;6:536–550. doi: 10.1016/j.chom.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 51.Miyashita S, et al. Viruses roll the dice: the stochastic behavior of viral genome molecules accelerates viral adaptation at the cell and tissue levels. PLoS Biol. 2015;13:e1002094. doi: 10.1371/journal.pbio.1002094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Graw F, Perelson AS. Modeling viral spread. Annu Rev Virol. 2016;3:555–572. doi: 10.1146/annurev-virology-110615-042249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zwart MP, Elena SF. Matters of size: genetic bottlenecks in virus infection and their potential impact on evolution. Annu Rev Virol. 2015;2:161–179. doi: 10.1146/annurev-virology-100114-055135. [DOI] [PubMed] [Google Scholar]
- 54.Gutiérrez S, et al. Virus population bottlenecks during within-host progression and host-to-host transmission. Curr Opin Virol. 2012;2:546–555. doi: 10.1016/j.coviro.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 55.Kuss SK, et al. Multiple host barriers restrict poliovirus trafficking in mice. PLoS Pathog. 2008;4:e1000082. doi: 10.1371/journal.ppat.1000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brooke CB, et al. Most influenza a virions fail to express at least one essential viral protein. J Virol. 2013;87:3155–3162. doi: 10.1128/JVI.02284-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heldt FS, et al. Single-cell analysis and stochastic modelling unveil large cell-to-cell variability in influenza A virus infection. Nat Commun. 2015;6:8938. doi: 10.1038/ncomms9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iranzo J, Manrubia SC. Evolutionary dynamics of genome segmentation in multipartite viruses. Proc Biol Sci. 2012;279:3812–3819. doi: 10.1098/rspb.2012.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sicard A, et al. The strange lifestyle of multipartite viruses. PLoS Pathog. 2016;12:e1005819. doi: 10.1371/journal.ppat.1005819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dall'Ara M, et al. Ins and outs of multipartite positive-strand RNA plant viruses: packaging versus systemic spread. Viruses. 2016;8:E228. doi: 10.3390/v8080228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Del Portillo A, et al. Multiploid inheritance of HIV-1 during cell-to-cell infection. J Virol. 2011;85:7169–7176. doi: 10.1128/JVI.00231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sigal A, et al. Cell-to-cell spread of HIV permits ongoing replication despite antiretroviral therapy. Nature. 2011;477:95–98. doi: 10.1038/nature10347. [DOI] [PubMed] [Google Scholar]
- 63.Richardson MW, et al. Mode of transmission affects the sensitivity of human immunodeficiency virus type 1 to restriction by rhesus TRIM5alpha. J Virol. 2008;82:11117–11128. doi: 10.1128/JVI.01046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Russell SJ, et al. Oncolytic virotherapy. Nat Biotechnol. 2012;30:658–670. doi: 10.1038/nbt.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paglino JC, et al. LuIII parvovirus selectively and efficiently targets, replicates in, and kills human glioma cells. J Virol. 2012;86:7280–7291. doi: 10.1128/JVI.00227-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stitz L, Schellekens H. Influence of input multiplicity of infection on the antiviral activity of interferon. J Gen Virol. 1980;46:205–210. doi: 10.1099/0022-1317-46-1-205. [DOI] [PubMed] [Google Scholar]
- 67.Chen P, et al. Predominant mode of human immunodeficiency virus transfer between T cells is mediated by sustained Env-dependent neutralization-resistant virological synapses. J Virol. 2007;81:12582–12595. doi: 10.1128/JVI.00381-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bruel T, et al. Elimination of HIV-1-infected cells by broadly neutralizing antibodies. Nat Commun. 2016;7 doi: 10.1038/ncomms10844. 10844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nagata S, et al. Autoimmunity and the clearance of dead cells. Cell. 2010;140:619–630. doi: 10.1016/j.cell.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 70.Moller-Tank S, Maury W. Phosphatidylserine receptors: enhancers of enveloped virus entry and infection. Virology. 2014;468–470:565–580. doi: 10.1016/j.virol.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chao L. Fitness of RNA virus decreased by Muller´s ratchet. Nature. 1990;348:454–455. doi: 10.1038/348454a0. [DOI] [PubMed] [Google Scholar]
- 72.Silander OK, et al. Understanding the evolutionary fate of finite populations: the dynamics of mutational effects. PLoS Biol. 2007;5:e94. doi: 10.1371/journal.pbio.0050094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamayoshi S, et al. Scavenger receptor B2 is a cellular receptor for enterovirus 71. Nat Med. 2009;15:798–801. doi: 10.1038/nm.1992. [DOI] [PubMed] [Google Scholar]
- 74.Yamayoshi S, et al. Human SCARB2-dependent infection by coxsackievirus A7, A14, and A16 and enterovirus 71. J Virol. 2012;86:5686–5696. doi: 10.1128/JVI.00020-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Han JF, et al. Human enterovirus co-infection in severe HFMD patients in China. J Clin Virol. 2014;61:621–622. doi: 10.1016/j.jcv.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 76.González-Jara P, et al. The multiplicity of infection of a plant virus varies during colonization of its eukaryotic host. J Virol. 2009;83:7487–7494. doi: 10.1128/JVI.00636-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Taylor MP, et al. Alphaherpesvirus axon-to-cell spread involves limited virion transmission. Proc Natl Acad Sci USA. 2012;109:17046–17051. doi: 10.1073/pnas.1212926109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Andino R, Domingo E. Viral quasispecies. Virology. 2015;479–480C:46–51. doi: 10.1016/j.virol.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ciota AT, et al. Cooperative interactions in the West Nile virus mutant swarm. BMC Evol Biol. 2012;12:58–12. doi: 10.1186/1471-2148-12-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sanjuán R, Domingo-Calap P. Mechanisms of viral mutation. Cell Mol Life Sci. 2016;73:4433–4448. doi: 10.1007/s00018-016-2299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bordería AV, et al. Group selection and contribution of minority variants during virus adaptation determines virus fitness and phenotype. PLoS Pathog. 2015;11:e1004838. doi: 10.1371/journal.ppat.1004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Belshaw R, et al. Pacing a small cage: mutation and RNA viruses. Trends in Ecology & Evolution. 2008;23:188–193. doi: 10.1016/j.tree.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Holmes EC. Error thresholds and the constraints to RNA virus evolution. Trends Microbiol. 2003;11:543–546. doi: 10.1016/j.tim.2003.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Woo J, et al. Constraints on HIV-1 diversity from protein structure. J Virol. 2010;84:12995–13003. doi: 10.1128/JVI.00702-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shirogane Y, et al. Cooperation between different RNA virus genomes produces a new phenotype. Nat Commun. 2012;3 doi: 10.1038/ncomms2252. 1235. [DOI] [PubMed] [Google Scholar]
- 86.Clavijo G, et al. Mixed genotype transmission bodies and virions contribute to the maintenance of diversity in an insect virus. Proc Biol Sci. 2010;277:943–951. doi: 10.1098/rspb.2009.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tanner EJ, et al. Dominant drug targets suppress the emergence of antiviral resistance. Elife. 2014:3. doi: 10.7554/eLife.03830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Grande-Pérez A, et al. Suppression of viral infectivity through lethal defection. Proc Natl Acad Sci USA. 2005;102:4448–4452. doi: 10.1073/pnas.0408871102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Manrubia SC, et al. Pathways to extinction: beyond the error threshold. Philos Trans R Soc Lond B Biol Sci. 2010;365:1943–1952. doi: 10.1098/rstb.2010.0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Perales C, et al. The impact of quasispecies dynamics on the use of therapeutics. Trends Microbiol. 2012;20:595–603. doi: 10.1016/j.tim.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 91.Harrison RL, Summers MD. Mutations in the Autographa californica multinucleocapsid nuclear polyhedrosis virus 25 kDa protein gene result in reduced virion occlusion, altered intranuclear envelopment and enhanced virus production. J Gen Virol. 1995;76:1451–1459. doi: 10.1099/0022-1317-76-6-1451. [DOI] [PubMed] [Google Scholar]
- 92.Huang AS, Baltimore D. Defective viral particles and viral disease processes. Nature. 1970;226:325–327. doi: 10.1038/226325a0. [DOI] [PubMed] [Google Scholar]
- 93.Marriott AC, Dimmock NJ. Defective interfering viruses and their potential as antiviral agents. Rev Med Virol. 2010;20:51–62. doi: 10.1002/rmv.641. [DOI] [PubMed] [Google Scholar]
- 94.Aaskov J, et al. Long-term transmission of defective RNA viruses in humans and Aedes mosquitoes. Science. 2006;311:236–238. doi: 10.1126/science.1115030. [DOI] [PubMed] [Google Scholar]
- 95.Baum A, et al. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc Natl Acad Sci USA. 2010;107:16303–16308. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brackney DE, et al. West Nile virus genetic diversity is maintained during transmission by Culex pipiens quinquefasciatus mosquitoes. PLoS ONE. 2011;6:e24466. doi: 10.1371/journal.pone.0024466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ke R, et al. Phylodynamic analysis of the emergence and epidemiological impact of transmissible defective dengue viruses. PLoS Pathog. 2013;9:e1003193. doi: 10.1371/journal.ppat.1003193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Noppornpanth S, et al. Characterization of hepatitis C virus deletion mutants circulating in chronically infected patients. J Virol. 2007;81:12496–12503. doi: 10.1128/JVI.01059-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pfaller CK, et al. Measles virus defective interfering RNAs are generated frequently and early in the absence of C protein and can be destabilized by adenosine deaminase acting on RNA-1-like hypermutations. J Virol. 2015;89:7735–7747. doi: 10.1128/JVI.01017-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Serrano A, et al. Analagous population structures for two alphabaculoviruses highlight a functional role for deletion mutants. Appl Environ Microbiol. 2013;79:1118–1125. doi: 10.1128/AEM.03021-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tapia K, et al. Defective viral genomes arising in vivo provide critical danger signals for the triggering of lung antiviral immunity. PLoS Pathog. 2013;9:e1003703. doi: 10.1371/journal.ppat.1003703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Notton T, et al. The case for transmissible antivirals to control population-wide infectious disease. Trends Biotechnol. 2014;32:400–405. doi: 10.1016/j.tibtech.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 103.Tanner EJ, et al. Exploiting genetic interference for antiviral therapy. PLoS Genet. 2016;12:e1005986. doi: 10.1371/journal.pgen.1005986. [DOI] [PMC free article] [PubMed] [Google Scholar]