Abstract
Polymer lipid nanodiscs have provided an invaluable system for structural and functional studies of membrane proteins in their near-native environment. In spite of the recent advances in the development and usage of polymer lipid nanodisc systems, lack of control over size and poor tolerance to pH and divalent metal ions are major limitations for further applications. Here we report a facile modification of a low molecular weight styrene maleic acid copolymer to form monodispersed lipid bilayer nanodiscs that show ultra-stability towards a pH range of 2.5 to 10 and divalent metal ion concentration. The macro-nanodiscs (>20 nm diameter) show magnetic-alignment properties that can be exploited for high-resolution structural studies of membrane proteins and amyloid proteins using solid-state NMR techniques. As demonstrated in this study, the new polymer, SMA-QA, nanodisc is a robust membrane mimetic tool that offers significant advantages over currently reported nanodisc systems.
Keywords: polymer nanodisc, pH tolerance, lipid bilayer, SMA, magnetic-alignment
Graphical abstract
Lack of control over size and poor tolerance to pH and divalent metal ions are major limitations of polymer nanodiscs. Here, a modified SMA based polymer is demonstrated to form monodispersed nanodiscs showing ultra-stability towards a pH range of 2.5 to 10 and divalent metal ions.
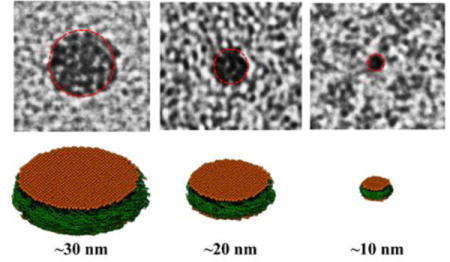
Controlled molecular self-assembly in the formation of soft nanomaterials has been a challenge in bio-nanotechnology.[1–2] Nanodiscs, lipid bilayers surrounded by an amphiphilic belt, are engineered soft-nanomaterials which have been inspired from biological systems such as high-density lipo-particles (HDL).[3] These nanodiscs provide a near native membrane like lipid bilayer environment, and have been used to study the structure and function membrane proteins.[3–5] Recent developments have expanded the formation of nanodiscs using different types of amphiphilic systems such as proteins,[6–10] peptides,[11] and polymers.[12–16] Polymer nanodiscs exhibit significant advantages over conventional protein based nanodiscs, such as detergent free membrane protein extraction[17] and are devoid of interferences from the belt-forming protein or peptide.[18] Currently, no polymer nanodisc systems have been able to demonstrate the precise control of size and morphology over a wide range of sizes, and tolerance towards a broad range of pH and divalent metal ions.[17] These unique properties are needed to greatly expand the applicability of nanodisc technology. Here we report the directed self-assembly of covalently modified styrene maleic acid copolymer with lipid bilayers to form monodispersed nanodiscs that show ultra-stability towards a broad range of pH and divalent metal ion concentration. We also demonstrate the ability to control the size of the self-assembled nanodiscs and size dependent unique magnetic-alignment properties. Lack of control over size and poor tolerance to pH and divalent metal ions are major limitations of polymer nanodiscs. Here, a modified SMA based polymer is demonstrated to form monodispersed nanodiscs showing ultra-stability towards a pH range of 2.5 to 10 and divalent metal ions.
Synthesis of SMA-QA (Styrene Maleimide Quaternary Ammonium) was achieved by the treatment of a low molecular weight SMA (~1.6 kDa) with (2-aminoethyl)trimethylammonium chloride hydrochloride in anhydrous dimethylformamide while heating in the presence of excess triethylamine. Maleimide formation was accomplished by a dehydration reaction using acetic anhydride, sodium acetate, and triethylamine (Figure 1a). The newly synthesized SMA-QA polymer was characterized using FT-IR and 13CPMAS (cross-polarization magic angle spinning) solid-state NMR experiments. FT-IR spectrum of SMA-QA exhibits a shift in carbonyl C=O stretching frequency from 1774 cm−1 to 1693 cm−1 showing the successful formation of maleimide (See supporting information Figure S1). The formation of SMA-QA was further confirmed using 13C CPMAS NMR spectrum (Figure 1b): the peaks appearing ~32 ppm and ~40 ppm are from the CH2 group, and the peaks resonating ~53 ppm and ~62 ppm are from the methyl carbons associated with quaternary ammonium and methylene carbons, respectively. The observed change in the chemical shift of the carbonyl carbon from ~172 (from the reactant SMA polymer) to ~178 ppm (from the product SMA-QA polymer) confirms the presence of maleimide carbonyls and completion of the reaction and product formation (Figure 1b).
Figure 1. Synthesis and characterization of SMA-QA polymer.
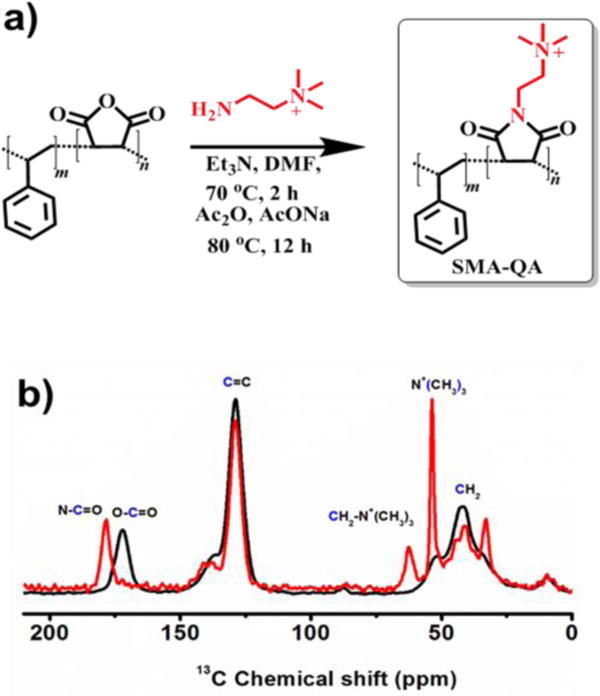
(a) Reaction scheme showing the modifications of SMA polymer to synthesize SMA-QA. (b) 13C CPMAS solid-state NMR spectra of SMA (black) and SMA-QA (red) polymers confirm the formation and successful completion of the chemical reaction. The SMA and SMA-QA polymers were further characterized using FT-IR experiments (Figure S1).
Next, we characterized the ability of SMA-QA to form lipid bilayer nanodiscs. Upon the addition of an aqueous solution of SMA-QA to a turbid solution of DMPC (1,2-dimyristoyl-sn-glycero-3-phosphocholine) multilamellar vesicles (MLVs), the turbid solution spontaneously became clear indicating an efficient solubilization of MLVs by the polymer. The solubilization kinetics was followed by static light scattering (SLS) measurements. Figure 2b shows the SLS profiles of DMPC MLVs for different lipid to polymer weight ratios. As shown in Figure 2b, the large intense scattering observed for DMPC MLVs dramatically decreased upon the addition of SMA-QA polymer demonstrating the solubilization of large DMPC MLVs into small size polymer-lipid nanodiscs. Our results further demonstrate that the kinetics of solubilization of MLVs by the polymer was found to depend on the ratio of DMPC:SMA-QA (Figure 2b). The rate of solubilization of MLVs was accelerated by the increase of the amount of SMA-QA.
Figure 2. Formation and size tunability of SMA-QA lipid nanodiscs.
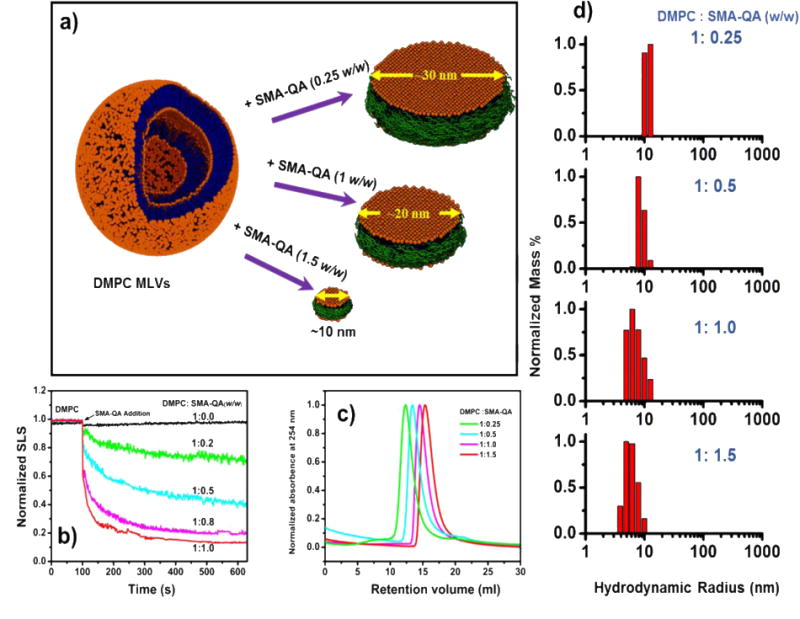
a) Schematic illustrating the formation of SMA-QA nanodiscs of varying size. b) SLS profiles showing the kinetics of DMPC MLVs solubilization. c) Size exclusion profiles of nanodiscs made by varying the DMPC:SMA-QA weight ratio. d) DLS profiles of purified DMPC:SMA-QA nanodiscs demonstrate the formation of different size nanodiscs by varying the weight ratio of DMPC:SMA-QA.
The lipid nanodiscs formed by the SMA-QA polymer were subjected to size exclusion chromatography (SEC) to remove the free polymer. The SEC retention volume of purified nanodiscs was found to be dependent on the lipid:polymer ratio used in the nanodisc formation as shown in Figure 2c. The resulting nanodiscs solutions were further characterized using dynamic light scattering (DLS) experiments. The DLS profiles showed the presence of monodispersed nanodiscs of varying hydrodynamic radii (~5 to ~13 nm) that were dependent on the ratio of DMPC to SMA-QA (Figure 2d). Then, the size and morphology of the resulting nanodiscs were characterized using transmission electron microscopy (TEM) images. The TEM images of DMPC:SMA-QA nanodiscs confirmed the presence of disc shaped, monodispersed particles as shown in Figure 3. The expanded images shown in Figure 3(bottom most row) show the size of individual nanodiscs and their remarkable circular shape.
Figure 3. Remarkably monodispersed and circular shaped polymer nanodiscs revealed by TEM.
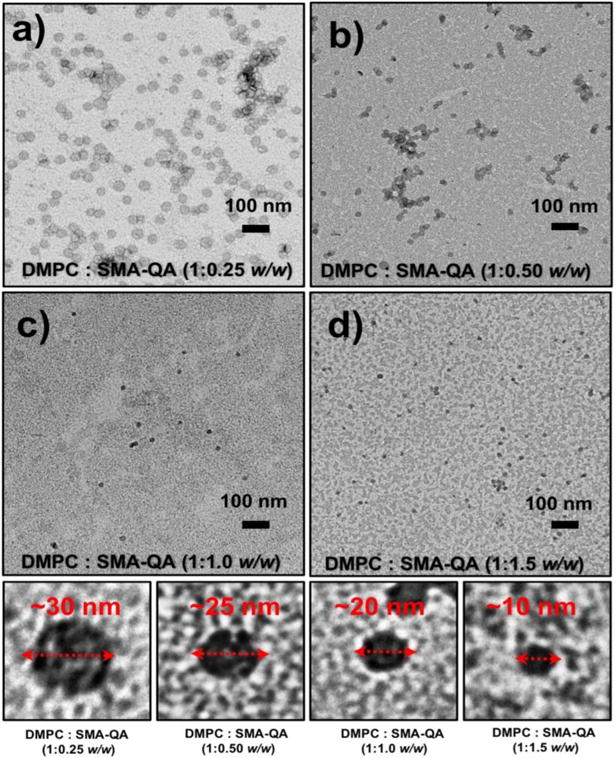
(a-d) TEM images of DMPC:SMA-QA nanodiscs formed with the indicated lipid to polymer ratio. Expanded images of nanodiscs showing the remarkable disc shape of the nanodisc for different sizes (bottom most row).
The precise control over the size of nanodiscs for a wide range of sizes (ranging from 10 nm to 30 nm) inspired us to test their magnetic-alignment properties using solid-state NMR experiments under static conditions. The 31P (spin=1/2) and 14N (spin=1) spectra obtained from polymer nanodiscs are shown in Figure 4. The 31P NMR spectrum of DMPC:SMA-QA (1:0.25 w/w) shows a single narrow peak ~ -16 ppm (Figure 4a) demonstrating the magnetic-alignment of polymer nanodiscs with the bilayer normal perpendicular to the magnetic field direction (Figure 4a).[14] Due to the large size (~30 nm diameter), the slow tumbling of nanodiscs (the large nanodiscs are also called as macro-nanodiscs[14]) allows for the magnetic-alignment in an external magnetic field. On the other hand, a narrow peak was observed at the isotropic chemical shift frequency (~−2 ppm) for small nanodiscs (~10 nm) demonstrating their fast tumbling in the NMR time scale (Figure 4c).
Figure 4. SMA-QA nanodiscs exhibit magnetic-alignment and remarkable tolerance to pH and divalent metal ions.
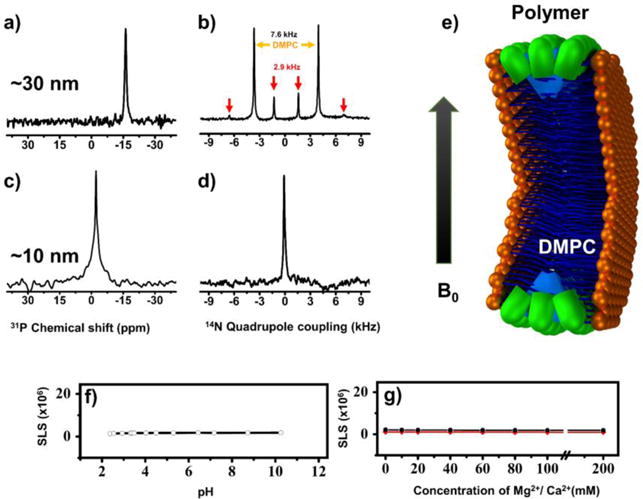
31P (a) and 14N (b) NMR spectra of magnetically-aligned large-size (~30 nm diameter) nanodiscs made from DMPC: SMA-QA (1:0.25 w/w). 31P (c) and 14N (d) NMR spectra of isotropic nanodiscs (~10 nm diameter) made from DMPC:SMA-QA (1:1.5 w/w). e) Schematic of a nanodisc illustrating the orientations of the lipid headgroup and polymer in magnetically-aligned nanodiscs. SLS profiles of DMPC:SMAQA (1:1 w/w) nanodiscs showing remarkable stability towards pH (f) and the presence of Mg2+ and Ca2+ (g) ions.
The 14N NMR spectra of SMA-QA nanodiscs (~30 nm) show three doublets corresponding to quadrupolar coupling values ~14, ~7.6 and ~2.9 kHz (Figure 4b). The magnitude of the observed quadrupolar coupling is dependent on the orientation of the quaternary ammonium Cβ-N bond vector with respect to the magnetic field direction (see the supporting information). The observed quadrupolar coupling magnitude of ~7.6 kHz arise from the choline group of DMPC and confirms the uniaxial orientation of nanodiscs with the lipid bilayer normal perpendicular to the magnetic field direction in agreement with 31P NMR data, as reported for other polymer nanodiscs in the literature.[19] Whereas the observed quadrupolar couplings of magnitude ~2.9 kHz and ~14 kHz are from the quaternary ammonium group of the SMA-QA polymer. The presence of these two distinct doublets suggests that SMA-QA polymer belt completely surrounds the lipid bilayer in a highly ordered fashion and with orientations as shown in Figure 4e, which is similar to the orientations of detergent molecules in aligned bicelles.[20] Additional experiments using paramagnetic lanthanide ions to tilt the orientation of lipid macro-nanodiscs would reveal the exact orientations of the polymer molecules. As expected, the 14N NMR spectrum of small nanodiscs (~10 nm diameter) showed a single narrow peak at the isotropic chemical shift value (0 ppm) suggesting the fast tumbling nature of isotropic nanodiscs in agreement with 31P NMR data.
The major disadvantages of SMA and other polymers used to form nanodiscs is their poor stability towards pH and metal ions.[21] This instability is attributed to the presence of carboxylic/carboxylate groups that form the hydrophilic region of the amphiphilic polymer.[17] SMA-QA was specifically engineered and synthesized with a quaternary ammonium group as the hydrophilic portion of the polymer to increase the tolerance to pH and metal ions. The stability of the SMA-QA nanodiscs against pH and metal ion concentration was examined using SLS measurements (Figure 4(f,g)). The SLS profiles of the DMPC:SMA-QA (1:1 w/w) nanodiscs showed no change in the scattering intensity over a wide range of pH (from ~2.5 to 10) and in the presence of metal ion concentrations up to 200 mM. These results signify the ultra-stability of DMPC:SMA-QA nanodiscs, further expanding the applicability of nanodisc technology to a wider range of biological and biomedical applications.
In conclusion, the newly developed SMA-QA polymer allows for the formation of monodispersed lipid nanodiscs with a precise size control and ultra-stability against pH (2.5–10) and metal ion concentrations up to 200 mM. The formation and stability of DMPC:SMA-QA nanodiscs were characterized using various biophysical experiments including solid-state NMR. The macro-nanodiscs (>20 nm diameter) showed magnetic-alignment properties which can be utilized in the structural studies of membrane proteins by solid-state NMR techniques.[22–26] Because of these unique properties of SMA-QA polymer nanodiscs, SMA-QA is a robust membrane mimetic tool that offers significant advantages over all currently reported nanodisc systems, and therefore we foresee a significant expansion in the applicability of nanodiscs technology. We expect direct and immediate impacts in the structural biology studies of those membrane proteins that are sensitive to pH and divalent metal ions[27–31] and amyloid proteins that self-assemble at the membrane interface.[32–33]
Supplementary Material
Acknowledgments
This study was supported by the NIH (GM084018 to A.R.).
Footnotes
Supporting Information: Materials and methods, FTIR spectra of SMA-QA, orientation dependence of nitrogen-14 quadrupole coupling are included in the supporting information.
References
- 1.Whitesides G, Mathias J, Seto C. Science. 1991;254:1312–1319. doi: 10.1126/science.1962191. [DOI] [PubMed] [Google Scholar]
- 2.Sarikaya M, Tamerler C, Jen AKY, Schulten K, Baneyx F. Nat Mater. 2003;2:577–585. doi: 10.1038/nmat964. [DOI] [PubMed] [Google Scholar]
- 3.Denisov IG, Sligar SG. Nat Struct Mol Biol. 2016;23:481–486. doi: 10.1038/nsmb.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Denisov IG, Sligar SG. Chemical Reviews. 2017;117:4669–4713. doi: 10.1021/acs.chemrev.6b00690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee SC, Pollock NL. Biochem Soc Trans. 2016;44:1011–1018. doi: 10.1042/BST20160015. [DOI] [PubMed] [Google Scholar]
- 6.Bayburt TH, Grinkova YV, Sligar SG. Nano Letters. 2002;2:853–856. [Google Scholar]
- 7.Nasr ML, Baptista D, Strauss M, Sun ZJ, Grigoriu S, Huser S, Pluckthun A, Hagn F, Walz T, Hogle JM, Wagner G. Nat Methods. 2017;14:49–52. doi: 10.1038/nmeth.4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hagn F, Etzkorn M, Raschle T, Wagner G. J Am Chem Soc. 2013;135:1919–1925. doi: 10.1021/ja310901f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hagn F, Wagner G. J Biomol NMR. 2015;61:249–260. doi: 10.1007/s10858-014-9883-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bayburt TH, Sligar SG. FEBS Lett. 2010;584:1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang M, Huang R, Ackermann R, Im SC, Waskell L, Schwendeman A, Ramamoorthy A. Angew Chem Int Ed Engl. 2016;55:4497–4499. doi: 10.1002/anie.201600073. [DOI] [PubMed] [Google Scholar]
- 12.Oluwole A, Danielczak B, Meister A, Babalola J, Vargas C, Keller S. Angew Chem Int Ed Engl. 2017;56:1919–1924. doi: 10.1002/anie.201610778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orwick MC, Judge PJ, Procek J, Lindholm L, Graziadei A, Engel A, Gröbner G, Watts A. Angew Chem Int Ed Engl. 2012;51:4653–4657. doi: 10.1002/anie.201201355. [DOI] [PubMed] [Google Scholar]
- 14.Ravula T, Ramadugu SK, Di Mauro G, Ramamoorthy A. Angew Chem Int Ed Engl. 2017;56:11466–11470. doi: 10.1002/anie.201705569. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 15.Sahu ID, Zhang R, Dunagan MM, Craig AF, Lorigan GA. J Phys Chem B. 2017;121:5312–5321. doi: 10.1021/acs.jpcb.7b01705. [DOI] [PubMed] [Google Scholar]
- 16.Oluwole AO, Danielczak B, Meister A, Babalola JO, Vargas C, Keller S. Angew Chem Int Ed Engl. 2017;56:1919–1924. doi: 10.1002/anie.201610778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee SC, Knowles TJ, Postis VLG, Jamshad M, Parslow RA, Lin Y-P, Goldman A, Sridhar P, Overduin M, Muench SP, Dafforn TR. Nature Protocols. 2016;11:1149–1162. doi: 10.1038/nprot.2016.070. [DOI] [PubMed] [Google Scholar]
- 18.Dorr JM, Koorengevel MC, Schafer M, Prokofyev AV, Scheidelaar S, van der Cruijsen EA, Dafforn TR, Baldus M, Killian JA. Proc Natl Acad Sci U S A. 2014;111:18607–18612. doi: 10.1073/pnas.1416205112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramadugu VSK, Di Mauro GM, Ravula T, Ramamoorthy A. Chem Commun (Camb) 2017;53:10824–10826. doi: 10.1039/c7cc06409h. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Dvinskikh SV, Yamamoto K, Durr UH, Ramamoorthy A. J Magn Reson. 2007;184:228–235. doi: 10.1016/j.jmr.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ravula T, Hardin NZ, Ramadugu SK, Ramamoorthy A. Langmuir. 2017;33:10655–10662. doi: 10.1021/acs.langmuir.7b02887. [DOI] [PubMed] [Google Scholar]
- 22.Ravula T, Ramadugu SK, Di Mauro G, Ramamoorthy A. Angew Chem Int Ed Engl. 2017;56:11466–11470. doi: 10.1002/anie.201705569. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 23.Park SH, Berkamp S, Cook GA, Chan MK, Viadiu H, Opella SJ. Biochemistry. 2011;50:8983–8985. doi: 10.1021/bi201289c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durr UH, Gildenberg M, Ramamoorthy A. Chem Rev. 2012;112:6054–6074. doi: 10.1021/cr300061w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qin H, Miao Y, Cross TA, Fu R. J Phys Chem B. 2017;121:4799–4809. doi: 10.1021/acs.jpcb.7b02468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salnikov ES, Aisenbrey C, Aussenac F, Ouari O, Sarrouj H, Reiter C, Tordo P, Engelke F, Bechinger B. Sci Rep. 2016;6:20895. doi: 10.1038/srep20895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Postis V, Rawson S, Mitchell JK, Lee SC, Parslow RA, Dafforn TR, Baldwin SA, Muench SP. Biochim Biophys Acta. 2015;1848:496–501. doi: 10.1016/j.bbamem.2014.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown LS, Ladizhansky V. Protein Sci. 2015;24:1333–1346. doi: 10.1002/pro.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Staus DP, Strachan RT, Manglik A, Pani B, Kahsai AW, Kim TH, Wingler LM, Ahn S, Chatterjee A, Masoudi A, Kruse AC, Pardon E, Steyaert J, Weis WI, Prosser RS, Kobilka BK, Costa T, Lefkowitz RJ. Nature. 2016;535:448–452. doi: 10.1038/nature18636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao Y, Cao E, Julius D, Cheng Y. Nature. 2016;534:347–351. doi: 10.1038/nature17964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das N, Murray DT, Cross TA. Nat Protoc. 2013;8:2256–2270. doi: 10.1038/nprot.2013.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez Camargo DC, Korshavn KJ, Jussupow A, Raltchev K, Goricanec D, Fleisch M, Sarkar R, Xue K, Aichler M, Mettenleiter G, Walch AK, Camilloni C, Hagn F, Reif B, Ramamoorthy A. eLife. 2017;6:e31226. doi: 10.7554/eLife.31226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahmed R, VanSchouwen B, Jafari N, Ni X, Ortega J, Melacini G. J Am Chem Soc. 2017;139:13720–13734. doi: 10.1021/jacs.7b05012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


