Abstract
Background: Bone mineral density (BMD) is a heritable phenotype that predicts fracture risk. We performed fine-mapping by targeted sequencing at WLS, MEF2C, ARHGAP1/F2 and JAG1 loci prioritized by eQTL and bioinformatic approaches among 56 BMD loci from our previous GWAS meta-analysis. Methods and Results: Targeted sequencing was conducted in 1,291 Caucasians from the Framingham Heart Study (n = 925) and Cardiovascular Health Study (n = 366), including 206 women and men with extreme low femoral neck (FN) BMD. A total of 4,964 sequence variants (SNVs) were observed and 80% were rare with MAF <1%. The associations between previously identified SNPs in these loci and BMD, while nominally significant in sequenced participants, were no longer significant after multiple testing corrections. Conditional analyses did not find protein-coding variants that may be responsible for GWAS signals. On the other hand, in the sequenced subjects, we identified novel associations in WLS, ARHGAP1, and 5’ of MEF2C (P-values < 8x10 − 5; false discovery rate (FDR) q-values < 0.01) that were much more strongly associated with BMD compared to the GWAS SNPs. These associated SNVs are less-common; independent from previous GWAS signals in the same loci; and located in gene regulatory elements. Conclusions: Our findings suggest that protein-coding variants in selected GWAS loci did not contribute to GWAS signals. By performing targeted sequencing in GWAS loci, we identified less-common and rare non-coding SNVs associated with BMD independently from GWAS common SNPs, suggesting both common and less-common variants may associate with disease risks and phenotypes in the same loci.
Introduction
Bone mineral density (BMD) is a highly heritable phenotype that is the major predictor for the risk of fracture (1). Over the past few years, genome-wide association studies (GWAS) have identified more than 65 novel genome-wide significant loci for BMD (2). Previously, we published the largest GWAS meta-analysis for BMD involving ∼36,000 Caucasian men and women with 2.5 million HapMap imputed common single nucleotide polymorphisms (SNPs) as part of the Genetic Factors for Osteoporosis (GEFOS) consortium (3). In that study, we identified 56 genome-wide significant loci associated with BMD of the lumbar spine (LS) and/or femoral neck (FN) skeletal sites. Fourteen of these 56 BMD-associated loci were also associated with fracture risk in a case-control meta-analysis involving up to 31,016 fracture cases and 102,444 controls without fracture. As is true for many GWAS efforts, we considered that the identified common SNPs were probably not the causal variants and more likely were in linkage disequilibrium (LD) with underlying un-typed causal variants.
To fine-map GWAS loci and identify potentially causal variants that are responsible for the GWAS signals, we selected four promising GWAS loci from our GEFOS meta-analysis and performed targeted re-sequencing using next generation sequencing techniques as part of the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE)(4) Targeted Sequencing Study.
Results
SNV distribution in sequenced samples
As designed (Table 1), FN BMD as well as LS BMD was significantly lower in the extremely low FN BMD group. The average BMD (for both FN and LS) was similar between the Cohort Random Sample and other Phenotype Groups. Across the four targeted regions, 4,964 variants were observed in the 1,291 sequenced Caucasian samples (average one variant per 80 bp). Most of the common variants (MAF ≥ 5%) have already been captured by HapMap Phase 2 imputation in our previous GWAS meta-analyses (Table 2). However, low-frequency SNVs, especially variants with MAF < 1%, were not observed by HapMap Phase 2 and the 1000G Project. Seventy-five percent of variants with MAF < 1% were newly discovered by our targeted sequencing project. The Supplementary Material, Figure S1 provides a categorization of the types of variants. Many missense variants and variants located in transcription factor binding sites were found. Of note, 76 of 112 (68%) observed missense variants had a MAF < 0.1%.
Table 1.
Sample characteristics of sampling scheme within the Framingham Study (FHS) and the Cardiovascular Health Study (CHS)
| Study Cohort |
FHS (n = 925) |
CHS (n = 366) |
||||
|---|---|---|---|---|---|---|
| Sampling scheme | Extremes with low FN BMD* | Samples from random controls | Samples from other phenotype groups | Extremes with low FN BMD* | Samples from random controls | Samples from other phenotype groups |
| Sample size | 93 | 430 | 402 | 113 | 124 | 129 |
| Percent female | 67% | 51% | 53% | 66% | 50% | 72% |
| Age (yrs)+ | 60 (11 .6) | 62 (9 .0) | 63 (10 .4) | 77 .3 (4 .5) | 77 (4 .8) | 76 .4 (4 .5) |
| Weight (lbs) | 137 (28 .1) | 175 (36 .7) | 177 (40 .3) | 136 (26 .4) | 163 (33 .9) | 163 (37 .8) |
| FN BMD (g/cm2) | 0 .64 (0 .11) | 0 .92 (0 .15) | 0.92 (0 .16) | 0 .50 (0 .06) | 0 .75 (0 .14) | 0 .70 (0 .12) |
| FN BMD range | 0.12 – 0 .80 | 0 .58 – 1 .49 | 0 .51 – 1 .40 | 0 .31-0 .61 | 0 .48-1 .25 | 0 .47-1 .10 |
| FNBMD T-score | −2 .95 (0 .80) | −0 .80 (1 .13) | −0 .81 (1 .20) | −3 .84 (0 .57) | −1 .43 (0 .79) | −1 .62 (0 .53) |
| FN BMD Z-score | −2 .06 (0 .42) | 0 .05 (0 .99) | 0 .15 (1 .01) | −2 .00 (0 .34) | −0 .02 (0 .71) | −0 .07 (0 .75) |
| LS BMD (g/cm2) | 0 .97 (0 .16) | 1 .24 (0 .22) | 1 .25 (0 .22) | 0 .82 (0 .17) | 1 .06 (0 .26) | 1 .04 (0 .22) |
| LS BMD range | 0 .61 – 1 .50 | 0 .69 – 2 .07 | 0 .62 – 1 .98 | 0 .43-1 .37 | 0 .64-1 .93 | 0 .62-1 .81 |
| LS BMD T-score | −1 .86 (1 .29) | 0 .35 (1 .78) | 0.41 (1 .80) | −2 .47 (0 .94) | −1 .35 (1 .76) | −1 .42 (1 .57) |
| LS BMD Z-score | −1 .28 (0 .75) | 0 .02 (1 .01) | 0.13 (0 .97) | −1 .16 (0 .59) | −0 .10 (0 .83) | −0 .17 (0 .79) |
The ‘extreme low FN BMD samples’ were selected based on (1) T-score was < -2; (2) 100 (FHS) and 120 (CHS) individuals with the lowest FN BMD Z-scores in all available samples; and (3) not been selected for ‘The Cohort Random Samples’ . Due to sequencing quality, we have excluded 14 samples (7 FHS and 7 CHS samples) in the association analyses .
+Age at the time of the bone density measure .
Continuous variables: Mean (SD) .
Table 2.
Distribution of minor allele frequencies (MAF) of 4,964 sequence variants in 1,291 Caucasian samples stratified by variants previously observed in HapMap II, 1000 Genomes Project (1000G), dbSNP (build 135) or newly identified in the current sequencing project (CHARGE novel)
| SNPs References | MAF < 1% | 1% ≤ MAF < 5% | MAF ≥ 5% | Total |
|---|---|---|---|---|
| Hapmap+1000G | 60 (1.5%) | 175 (56.5%) | 757 (98.7%) | 992 (20.0%) |
| dbSNP v135 | 913 (23.5%) | 125 (40.3%) | 9 (1.2%) | 1047 (21.0%) |
| CHARGE novel | 2914 (75.0%) | 10 (3.2%) | 1 (0.1%) | 2925 (59.0%) |
| Total | 3887 (100%) | 310 (100%) | 767 (100%) | 4964 (100%) |
Single variant association analyses
The quantile-quantile plots are shown in the Supplementary Material, Figure S2. The λGC inflation factors were 1.07 and 1.00 for FN BMD and LS BMD, respectively, suggesting that Type I error was well controlled. Table 3 provides the most significant associations between single variants and BMD in the four targeted loci.
Table 3.
List of the significantly associated variants (FDR q-value < 0.01) from single SNP-phenotype association analyses
| BMD | rs number | Chrom position (hg19) | Alleles (reference allele/ effect allele) | MAF | Effect (Beta, g/cm2) | SE | P-values | Functional Roles | ENCODE Regulatory Elements |
|---|---|---|---|---|---|---|---|---|---|
| Chr1p31.3 WLS locus | |||||||||
| FN | rs75170441* | 68567995 | G/T | 0.01 | −0.107 | 0.0271 | 5.07 x 10−5 | ncRNA intron† | Identified as Irf3 motif; Histone modifications |
| LS | rs2566764* | 68583521 | A/G | 0.03 | −0.061 | 0.0152 | 5.55 x 10−5 | ncRNA intron† | Pax motif |
| Chr5q14.3 MEF2C locus | |||||||||
| FN | rs13163005* | 88410884 | C/T | 0.01 | −0.067 | 0.0148 | 6.21 x 10−6 | intergenic | GATA-3 motif; Histone modifications |
| Chr 11p11.2 ARHGAP1/F2 locus | |||||||||
| FN | rs111584437* | 46716912 | C/T | 0.01 | −0.059 | 0.0141 | 2.89 x 10−5 | ARHGAP1 3’UTR | Bound by HNF4A and RFX3 TFs; Open chromatin; Histone modifications |
Statistical significance after multiple testing correction by FDR with q-values < 0.01.
non-coding RNA: These variants are located in the last intron of WLS, whose mRNA sequences are overlapping with a ncRNA (GNG12-AS1).
In the following sections, we used the ‘GWAS SNP’ term to indicate SNPs that were originally found to be genome-wide significant (P < 5x10 − 8) in our previously reported GWAS meta-analyses. Their association with BMD in the relatively small numbers of sequenced samples may not be necessarily genome-wide significant (P < 5x10 − 8) or even marginally significant (P < 0.05). We used the ‘SNVs’ term to indicate sequence variants that were not available in our previously reported GWAS meta-analyses.
The WLS locus
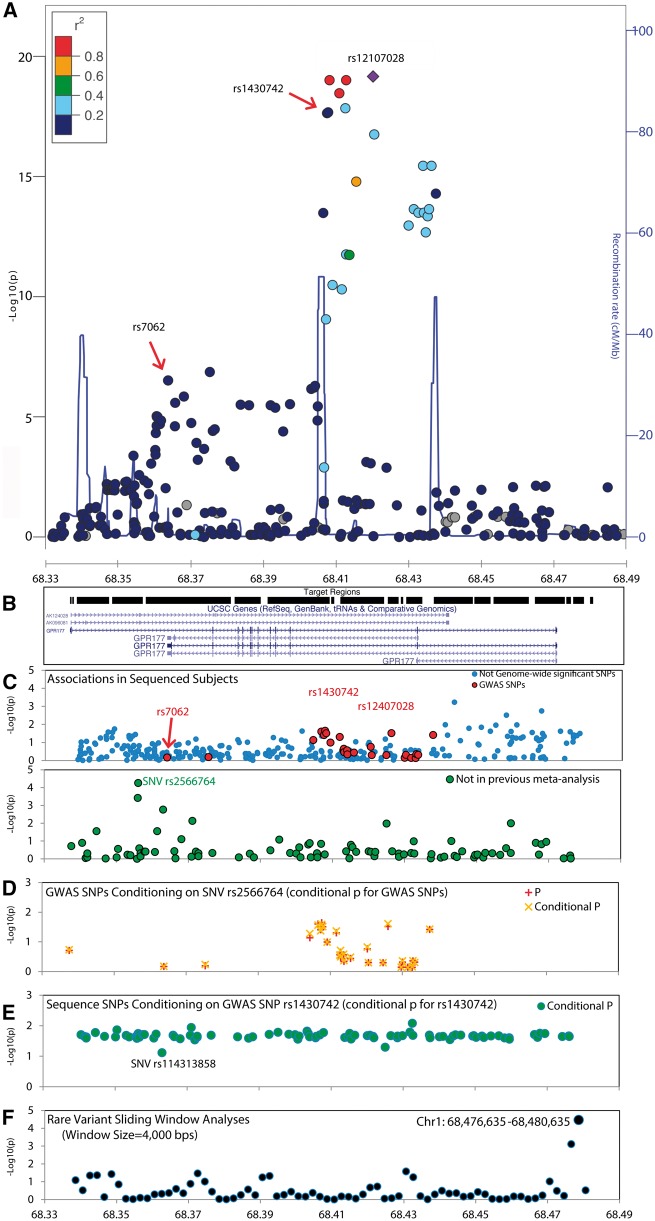
Figure 1C shows the regional association with LS BMD in 1,291 sequenced samples. In Figure 1C, we grouped single nucleotide sequence variants (SNVs) observed in 1,291 sequenced samples into 3 groups, each with different colours. The red dots (GWAS SNPs) indicate SNVs that were previously shown to be genome-wide significantly associated with BMD reported in our previous GWAS meta-analyses (3) in a much larger sample size. Blue dots (Not genome-wide significant SNPs) indicate SNVs that were previously reported as not reaching genome-wide significance in our published meta-analyses (3). Green dots (SNVs) indicate SNVs that were not analysed nor reported in our previously published GWAS meta-analyses (3) that were based on the international HapMap II CEU reference panel.
Figure 1.
Regional association plot for lumbar-spine bone mineral density (LS BMD) on Chr 1p31.3 WLS locus in 1,291 sequenced samples. Panel A: Regional plot from previously published GWAS meta-analysis. P-values were obtained from published GWAS meta-analyses.3Panel B: gene annotation based on human genome reference panel NCBI v36. The dark box represents the sequenced region. Panel C: Regional plots for association results in sequenced subjects. P-values were estimated in sequenced subjects. We grouped single nucleotide sequence variants (SNVs) observed in 1,291 sequenced samples into 3 groups, each with different colours. The red dots (GWAS SNPs) indicate SNVs that were previously showed genome-wide significantly associated with BMD reported in our previous GWAS meta-analyses (3) in much larger sample size. Blue dots (SNPs did not achieved genome-wide significance) indicate SNVs that were previously reported not reaching genome-wide significance in our published meta-analyses (3). Green dots (SNVs) indicate SNVs that were not analysed nor reported in our previously published GWAS meta-analyses (3) that were based on the international HapMap II CEU reference panel. Panel D: Conditional analyses: Association analyses were only performed on GWAS SNPs (SNVs indicated as red dots in Panel C) by conditioning on the most significantly associated SNVs (rs2566764) in sequenced samples. ‘P’ are P-values for GWAS SNPs without conditioning on rs2566764; and ‘Conditional P’ are P-values for the same GWAS SNPs conditioning on rs2566764. Panel E: Conditional analyses: Association analyses were done for SNVs (green dots SNVs in Panel C that were not analysed nor reported in our previously published GWAS meta-analyses) by conditioning on the most significant GWAS SNP (rs1430742, most significant GWAS SNP indicated as red dot in Panel C) in sequenced subjects. ‘Conditional P’ are P-values for GWAS SNP rs1430742 after conditioning on one SNV at time. Panel F: Sliding window (4,000 bps) rare variant association analyses by SKAT. Panel G: Leave-one-out analyses on the significantly associated window (chr1: 68,476,635-68,480,635). There are 11 rare SNVs in the analyses.
Indicated by green dots in Figure 1C, after multiple testing corrections by FDR, SNV rs2566764 (P = 5.55x10 − 5) was significantly associated with LS BMD (FDR q-values < 0.01) in the 1,291 sequenced samples. SNV rs2566764 is less-common (1% ≤ MAF < 5%) and not previously genotyped, imputed or analysed in our published GWAS meta-analysis of BMD (3) based on the HapMap II CEU reference panel. As a comparison (Fig. 1A), we also showed the regional association plot of our previously reported GWAS meta-analyses in a much larger sample size (3). SNV rs2566764 is 7.7kb away from one of the GWAS SNPs (rs7062) and both variants are located in the same LD block within recombination hot-spots (Fig. 1A and C). However, GWAS SNP rs7062 was not associated with LS BMD in the sequenced subjects (P-value = 0.67, Fig. 1C, red dots). In sequenced subjects, the association P-values of our originally reported GWAS SNPs ranged from 0.022 (rs1430742) to 0.719 and none of them achieved statistical significance after multiple testing correction (Red dots in Fig. 1C).
To examine whether SNV rs2566764 was responsible for GWAS signals in the WLS locus, we performed conditional analyses (Fig. 1D) and found none of the association results (P-values and beta-coefficients) of the GWAS SNPs were modified after conditioning on SNV rs2566764, suggesting that the SNV rs2566764 association with LS BMD in sequenced subjects is independent from the originally reported GWAS signals (Fig. 1D). To identify whether the ‘new’ SNVs (identified in the current sequencing study, but not reported in previous GWAS meta-analysis) were responsible for the GWAS signals, we performed association analysis conditioning on the GWAS SNP rs1430742 (the most significant GWAS SNP in sequenced subjects) in the WLS locus (Fig. 1E). Again, we did not find a notable change in the regression coefficients and P-values of GWAS SNP rs1430742 (Fig. 1E), except that the association between GWAS SNP rs1430742 and LS BMD seemed to be eliminated (conditional association P = 0.07 after conditioning on SNV rs114313858).
As for FN BMD, after multiple testing corrections by FDR, SNV rs75170441 (P = 5.07x10 − 5) was significantly associated with FN BMD (FDR q-values < 0.01, indicated by green dots in Supplementary Material, Fig. S3C) in the WLS locus. SNV rs75170441 is less-common (1% ≤ MAF < 5%) and not previously genotyped, imputed or analysed in our published GWAS meta-analysis of BMD based on the HapMap II CEU reference panel. To examine whether SNV rs75170441 was responsible for GWAS signals in the WLS locus, we performed conditional analyses and found none of the association results (P-values and beta-coefficients) of the GWAS SNPs were modified after conditioning on SNV rs75170441, suggesting that the SNV rs75170441 association with FN BMD in sequenced subjects is independent from the originally reported GWAS signals (Supplementary Materials, Figs S3D and E).
The MEF2C locus
SNV rs13163005 was significantly associated with FN BMD (P-value = 6.21 x 10 − 6) at FDR q-values < 0.01. SNV rs13163005 is less common and was not previously genotyped, imputed or analysed in our original GWAS publication (green dots, Supplementary Material, Fig. S4C). The association P-values of the GWAS SNPs ranged from 0.007 (rs10037512) to 0.101 in our sequenced samples and none of them achieved statistical significance after multiple testing corrections (red dots, Supplementary Material, Fig. S4C). SNV rs13163005 is only 56kb away from the most significant GWAS SNP (rs10037512, P = 0.007) in sequenced subjects. However, SNV rs13163005 is not in LD with any GWAS SNPs in this locus. To examine whether SNV rs13163005 is responsible for GWAS signals in MEF2C locus, we performed a conditional analysis on all GWAS SNPs and conditioned on SNV rs13163005. We did not find a notable change in the regression coefficients and P-values for the GWAS SNPs (Supplementary Material, Fig. S4D), suggesting that the SNV rs13163005 association signal is independent from GWAS signals. In addition, to identify any sequence variants that may be responsible for GWAS signals, we performed a conditional analysis of sequence variants and conditioned on GWAS SNP rs10037512. We did not find a notable change in the regression coefficients and P-values for the GWAS SNP rs10037512 (Supplementary Material, Fig. S4E).
As for LS BMD, no significant association found (Supplementary Material, Fig. S5C). Since there was no significant association, we did not perform conditional analyses for LS BMD.
The ARHGAP1/F2 locus
SNV rs111584437 was significantly associated with FN BMD (P-value = 2.89 x 10 − 5). SNV rs111584437 is also less common and was not previously genotyped, imputed or analysed in our original GWAS publication. SNV rs111584437 is 6.7kb away from GWAS SNP, rs10769205 (Supplementary Material, Fig. S6C). However, GWAS SNP rs10769205 was not significantly associated with BMD (P-value = 0.22) in the sequenced subjects. The association P-values of other GWAS SNPs in this locus ranged from 0.12 to 0.35 in the sequenced samples (red dots, Supplementary Material, Fig. S6C). To examine whether SNV rs111584437 was responsible for GWAS signals in the ARHGAP1/F2 locus, we performed conditional analysis on GWAS SNPs and conditioned on the SNV rs111584437. We found that SNV rs111584437 signal was independent of GWAS signals (Supplementary Material, Fig. S6D). In addition, to identify any sequence variants that may be responsible for GWAS signals, we performed a conditional analysis of the sequence variants and conditioned on GWAS SNP rs10769205. We did not find notable changes in the regression coefficients and P-values for the GWAS SNP rs10769205 (Supplementary Material, Fig. S6E).
As for LS BMD, no significant association was found after multiple testing corrections (Supplementary Material, Fig. S7).
The JAG1 locus
After correction for multiple testing, no significant associations between any of the single variants and BMD were found (supplementary Materials, Figs S8 and S9). The most significant association in sequenced subjects was found for SNP rs7267595, our originally reported GWAS SNP with nominal P-value equal to 0.005.
Rare variant associations
In addition to single variant-phenotype associations, we also performed rare variant (MAF ≤ 1%) association analyses (Fig. 1F, Supplementary Materials, Figs S3F–9F). The total number of analysed variants with MAF ≤ 1% was 1,167, 272, 830 and 368 variants, respectively for WLS, MEF2C, ARHGAP1/F2 and JAG1 loci. We divided each targeted sequenced locus into several ‘4,000 bp-windows’ with a 2,000 bp-wide overlap between two neighbouring windows. The SKAT rare variant association analysis was performed in each of the 4,000 bp windows. Permutation with smallest P-values approach was performed to correct for multiple testing. As shown in Figure 1F, we found one significant association with LS BMD at the WLS locus in between chromosome position 68,476,635 to 68,480,635 (NCBI human genome reference Build 36/HG18) with association P = 3.36x10 − 5 (permutation with smallest P-value test P = 0.002). There are 11 SNPs with MAF ≤ 1% located inside this region. To identify which rare variant/variants contributed to the signal from this associated genomic region, we performed a leave-one-out analysis, excluding one variant at time and repeating SKAT association analyses. We found only one rare variant (rs71649062) located at chromosome 1 position 68,477,885 (Chr1:68,477,885) that drove the gene-based association signal. As for the MEF2C, ARHGAP1/F2 and JAG1 loci, after multiple testing corrections, we did not find any significant associations by SKAT tests.
Discussion
This is the first attempt using targeted sequencing of selected GWAS loci in over a thousand individuals to follow up GWAS findings from the largest meta-analysis of BMD. Following a widely held view, we hypothesized that GWAS SNPs in non-coding regions were probably in LD with un-genotyped causal variants that were most likely located in the protein-coding regions and were likely functional (5). However, in this current study, we did not find any protein-coding sequence variants that may potentially be responsible for GWAS signals from our previous GWAS meta-analyses (3). Similar to a recent report from the ENCODE project, (6–8) our observations support the possibility that many disease-associated GWAS SNPs in non-coding regions may be themselves causal and functional. On the other hand, via high-resolution targeted sequencing, we were able to identify novel significantly associated SNVs (P-values < 8 x 10 − 5) in the region of the WLS gene, 5’ upstream region of MEF2C and ARHGAP1/F2 loci that were not previously identified in our GWAS meta-analysis (3). One question that our study had to answer is whether these novel significant associations are responsible for the original GWAS signals. Although SNV rs2566764 (WLS gene), rs75170441 (WLS gene), rs13163005 (5’ upstream of MEF2C) and rs111584437 (ARHGAP1/F2 locus) were all located close (300bp to 7kb) to our original GWAS signals, these SNVs are less-common and are not in LD with any of our previously reported GWAS SNPs in these four loci. Furthermore, these novel findings had smaller association P-values compared to the nominal or non-significant association P-values of our original GWAS signals in our sequenced samples. Our conditional analyses suggest these novel associations are independent of previously reported GWAS signals and represent additional newly-discovered associated variants in the same GWAS loci. Less-common variants are predicted to exhibit stronger effect sizes than common variants, consistent with the view that functional allelic variants are subject to purifying selection pressure (9,10). Our novel associated SNVs are relatively less-common in study populations, suggesting common and rare variants in the same locus may be independently associated with disease phenotypes. These novel associated SNVs study have the same effect direction as the effect direction of the reported GWAS common SNPs from our previous GWAS meta-analyses. However, the effect size of our novel associated SNVs is almost similar to the effect size observed from previously reported GWAS common SNPs in the same genomic regions; suggesting that less common SNVs associated with BMD phenotypes may not necessarily have stronger effect size comparing to the effect size of common variants.
SNVs for these novel associations are located in potential regulatory elements with histone protein modification and transcription factor binding. As shown in Table 3, SNVs rs75170441 (in Irf3 motif) and rs2566764 (in Pax motif) are located in transcription factor binding sites and histone protein modification sites of WLS. SNV rs111584437, located in the 3’UTR of the ARHGAP1 gene, has been confirmed experimentally to affect HNF4A and RFX3 transcription factors binding (11,12). Expression quantitative trait loci (eQTLs) have been used to inform the potential perturbation of non-coding SNVs to the expression of potential target genes. We were not able to perform such eQTL analyses using publicly available eQTL databases, since our novel associated SNVs were not genotyped nor imputed due to their low frequencies in general populations. Thus, additional experiments, such as allele-specific expression in human bone tissues or gene-editing experiments in cellular models are needed to further validate our novel association findings.
The associated variants located in WLS (chr1p31.3) gene and 5’ upstream flanking of the MEF2C (chr5q14.3) gene are of interest because these two genes are related to the Wnt signalling pathway (13,14). This pathway has recently been recognized as a key regulator of bone remodeling and regeneration. WLS may control skeletal development through modulation of autocrine and paracrine Wnt signals in a lineage-specific fashion (15). Wnt signalling has also been found to regulate crucial aspects of cardiovascular physiology (differentiation, morphogenesis and progenitor self-renewal), (16) to increase with aging in the heart, (17) and to drive cardiac progenitor cells to differentiate into a fibrogenic lineage, suggesting its involvement in the pathophysiology of CVD. The MEF2C gene encodes a member of the MADS box transcription enhancer factor 2 (MEF2) family of proteins, which plays a role in myogenesis and is evolutionarily conserved (18). MEF2C fulfils a critical role in the differentiation of vascular smooth muscle cells and regulates myocardin expression (19,20). MEF2C has been found to regulate myocardial gene differential expression in murine and human heart failure (21,22), and is essential for the transcriptional activation of the bone formation inhibitor, SOST (23). Thus, there may be some potential for Wnt signalling pathway’s pleiotropy with both bone and cardiac phenotypes. Similarly, our findings at the chr11p11.2 ARHGAP1/F2 locus are of particular interest, as this region also maps to glucose homeostasis (24), T2DM (25), BMI (26), plasma fibrin fragment D-dimer (27), and HDL lipoproteins (28), suggesting this locus is a potential pleiotropic locus for a variety of metabolic and endocrine functions.
In the JAG1 locus, the most significantly associated variant was one of the genome-wide significantly associated SNPs found in our original GWAS meta-analysis and this nominal association did not reach statistical significance after multiple testing corrections. In addition, SNP rs2273061 in the JAG1 gene was reported to be genome-wide significantly associated with BMD and demonstrated allele-specific expression as a potential functional variant by Kung et al. (29). However, this SNP was only nominally associated with BMD in our sequenced samples (P-value = 0.018); also, in rare variant association analyses we did not observe any association in the JAG1 locus.
One of the limitations of our study is the modest sample size, which was highlighted by the observation that the original GWAS meta-analysis loci were not statistically significantly associated with BMD in the sequenced sample of participants, which made it difficult to perform conditional analyses. Examining novel associations found in this study in additional samples and in other ethnic groups will improve the robustness of our findings. In addition, in-vitro functional experiments will be needed to validate their functional importance.
In summary, we attempted to follow-up previous GWAS loci by targeted sequencing in 1,291 Caucasian samples from the CHARGE Targeted Sequencing Study to fine-map four selected GWAS loci to identify potential causal variants. Despite the modest sample size, we observed an average of one variant every 80 bp and 4,964 novel sequence variants in our four targeted sequencing regions. Our previously discovered GWAS SNPs were either nominally or not statistically associated with BMD in sequenced participants, which may reflect the smaller sample size of sequenced samples compared to genotyped participants in GWAS analyses. From conditional analyses, we did not find any protein-coding variants that may potentially be responsible for GWAS signals, suggesting non-coding GWAS SNPs themselves may be causal and functional. On the other hand, we did identify novel associations that were much more strongly associated with BMD compared to the GWAS SNPs in sequenced subjects. These SNVs are located in potential regulatory elements with evidence from histone protein modification and transcription factor binding sites reported by the ENCODE Project and RoadMap Epigenomics Project. These novel SNVs are less-common and are independent from previously reported GWAS SNPs, suggesting both common and less-common variants may be associated with disease-relevant phenotypes in the same GWAS loci.
Materials and Methods
Bone mineral density
BMD (g/cm2) of the femoral neck (FN) and lumbar spine (LS) were obtained in the Framingham Heart Study (FHS) using a LUNAR DPX-L dual-energy X-ray absorptiometer (Lunar Corp, Madison, WI). In the Cardiovascular Health Study (CHS), FN and LS BMD were measured using a QDR 2000 or 2000+ (Hologic, Inc, Bedford, MA) and were read at the University of California, San Francisco reading centre with Hologic software version 7.10.
Study sample
The detailed information about the CHARGE Targeted Sequencing Study has been described elsewhere (30). In brief, The CHARGE Targeted Sequencing Study used a case-cohort study design and selected samples for targeted sequencing from the three participating cohorts (Framingham Heart Study (FHS), Cardiovascular Health Study (CHS), and the Atherosclerosis Risk in Communities (ARIC)). Since ARIC did not have BMD measures, we only included samples from CHS and FHS. Samples in our analyses included (1) selected extreme low BMD samples; (2) a set of cohort random samples; and (3) samples selected for targeted sequencing from phenotypes other than BMD.
Cohort random sample.
The cohort random sample included 471 participants from CHS and 500 participants from FHS with equal numbers of men and women (30). In the Cohort random sample, 124 CHS participants and 430 FHS participants had BMD measurements and were included in our statistical analyses.
Selection of extreme low FN BMD samples
We selected 100 participants from the FHS and 120 participants from the CHS with the ‘lowest FN BMD’ in each cohort study; and were not selected as the ‘Cohort Random Samples’. The extreme low FN BMD phenotype was selected based on both T-score and Z-score. T-score, the number of standard deviations below the young normal sex-specific average reference population value, is used to diagnose osteoporosis (31). Individuals with T-scores less than -2.5 are considered to have osteoporosis and those with T-scores between -2.5 and -2.0 are considered to have osteopenia. In each study, we first selected participants whose FN BMD T-score was ≤2, and from that subset (not selected as ‘Cohort Random Samples’) we then selected 100 (FHS) to 120 (CHS) individuals with the lowest FN BMD Z-scores (number of standard deviations below the age and sex-matched average value in each study). The sample was selected such that there were two thirds women, as we wanted to represent the distribution of men and women that comprised our GWAS meta-analysis sample (3). After quality control, 206 extreme low FN BMD samples (93 FHS participants and 113 CHS participants) were included in our statistical analyses.
Samples selected for phenotypes other than BMD
To increase sample size, we also included 129 CHS samples and 402 FHS samples with available BMD measurements that were selected for targeted sequencing analyses of phenotypes other than BMD (30). Individuals initially selected for the cohort random sample (of CHARGE Targeted Sequencing Study) or selected from phenotypes other than BMD also included individuals with low BMD.
As shown in Table 1, a total of 1,291 men and women (925 FHS and 366 CHS participants) were included in statistical analyses. All participants were adults and of European ancestry. All participants provided informed consent and Institutional Review Boards approved both FHS and CHS study protocols.
Selection of targeted regions
The four selected targeted loci (WLS, MEF2C, JAG1 and ARHGAP1/F2) emerged from our meta-analysis in 32,961 individuals that identified 56 GWAS loci associated with BMD and replicated in additional 50,933 individuals (3). Regional plots of the GWAS meta-analyses results are shown in Figure 1A and Supplementary Materials, Figures S3A–9A. These four loci were selected for sequencing based on an integrative approach (32): (1) association signals clustered inside a gene or in a regulatory region of a gene; (2) novel candidate genes for BMD or osteoporosis (no sequencing reported previously); (3) involvement in bone and mineral metabolism based on cellular and/or animal models and (4) nominal association with FN BMD in the GWAS analyses in FHS participants selected for targeted sequencing (32). The actual sequencing regions (start and end positions within a locus) were selected based on the location of association signals. For WLS and JAG1 loci, the GWAS SNPs were located inside genes; therefore, the sequencing regions were selected to cover the whole gene (introns and exons) and 5’ flanking regions that were in LD with associated SNPs (r2 ≥ 0.4). For MEF2C and ARHGAP1/F2 loci the GWAS signals were located in the 5’ flanking intergenic regions; therefore, the entire intergenic region with the cluster of association signals (r2 ≥ 0.4) and the exons of nearby genes were sequenced. After excluding genomic regions with high GC content and long repeated sequences, the total sequenced length was ∼ 395 Kbps (116,934 bps for WLS; 122,676 bps for MEF2C loci; 113,955 bps for the ARHGAP1/F2 and 41,464 bps for JAG1 loci).
Target capture and sequencing
Next generation sequencing was performed at the Baylor College of Medicine Human Genome Sequencing Center. For the CHARGE Targeted Sequencing Study, all targeted regions were captured by a customized NimbleGen Capture array, and then sequenced using the ABI SOLiD V4.0 platform. The depth coverage of the targeted bases was 20X on average. The detailed protocols of DNA fragment capture and next-generation sequencing via the SOLID platform can be found on the website, (https://www.hgsc.bcm.edu/sites/default/files/documents/Preparation_of_SOLiD_Multiplex_Capture_Libraries.pdf).
Sequence alignment, variant calling and quality control
Raw short reads were aligned to NCBI human genome reference Build 36 (hg18 at UCSC Browser). Alignments were performed using BFAST (33). The aligned reads were used to call variants with quality filters by SAMtools (34). DNA samples that did not meet the minimum of 65% of the targeted bases at 20X or greater coverage were excluded from subsequent analysis. This project was primarily focused on single nucleotide variants (SNVs), whereas neither small indels nor large copy number variations were investigated.
All Quality Control procedures were carried out in the statistical platform R or Java, in combination with SAMtools (34). The details of the quality controls are described elsewhere (30). In brief, we first filtered any SNVs that mapped more than 100 base pairs from the requested target capture region. The remained SNVs was post-processed to filter variants with apparent strand-bias (variants with alternate allele reads obtained from only positive or negative strand), allelic imbalance for heterozygote genotypes (alternate to reference allele ratio < 0.2 or > 0.8), low allele fraction (< two reads of the alternate alleles), low coverage (a depth of coverage < 10 reads), low quality reads (Phred-scaled base quality score < 30), high missingness across study samples (> 20%), multiple alternative alleles across study samples, overly dense SNV cluster (3 or more variants in a 10 bp window), or deviation from the expectations of Hardy-Weinberg equilibrium (HWE, P < 1 × 10 − 5) in the random cohort samples. After completing the variant-level quality control, we also examined DNA sample quality using several quality scores, including mean fold coverage, mean transition to transversion ratios, mean heterozygote to homozygote ratio, mean non-synonymous to synonymous ratio, number of singletons, number of doubletons and percentage of sites with coverage greater than 20X. None of DNA samples failed these quality matrices.
Sequence annotations
We used the dbSNP (v135) as well as the 1,000G Project (Phase 1 version 3) to classify variants as already known versus novel (35). The functional roles (such as nonsense, missense, splicing, synonymous, 5'UTR, 3'UTR, intronic, intergenic and ncRNA variants) were annotated using the ANNOVAR package (human reference NCBIv36/hg18) (36). To annotate the potential roles of non-coding variants, we used the recently available ENCyclopedia Of DNA Elements (ENCODE) Project (37) via the RegulomeDB (11) and HaploReg (12) databases.
Statistical analyses
We performed SNV-phenotype association analyses in each study and then meta-analyses were conducted to combine association results from FHS and CHS. Quantitative measurement of BMD (g/cm2) at the FN was used as the primary phenotype for association analyses and BMD at the LS as the secondary phenotype. Covariates included in association analyses were age, age2, sex, weight, oestrogen use/menopause status (women only), clinical sites, and principal components for genetic ancestry. Un-weighted linear regression was used to access significance. In addition, a weighted linear regression model to account for the sampling design was applied to obtain effect estimates (http://stattech.wordpress.fos.auckland.ac.nz/files/2012/05/design-paper.pdf).
Single SNV-phenotype association analyses
For sequence variants that had more than 10 copies of the minor allele in the 1,291 participants, we performed single SNV-phenotype association analyses using both weighted linear regression (to obtain beta coefficients) and un-weighted linear regression models (to obtain P-values). We used fixed effects inverse-variance meta-analyses to estimate combined regression results from FHS and CHS association analyses. After performing the meta-analyses, we excluded SNVs with association results in only one of the two studies, heterogeneity P-values of <0.05 and heterogeneity index (Het I2) > 50, or SNVs with an average effect allele frequency of <1%. False discovery rate (FDR) was used for multiple testing correction (FDR q-values < 0.01).
Conditional analyses
We performed a conditional analysis to estimate whether the SNVs associations with BMD phenotypes identified from the targeted sequencing explained the associations between SNPs previously identified from the GWAS meta-analysis and BMD phenotypes. In following sections, we used the ‘GWAS SNP’ term to indicate SNPs that were found to be genome-wide significant (P < 5x10 − 8) in our previously reported GWAS meta-analyses, as opposing to the ‘new’ sequence variants (SNVs, identified in the current study, but not reported in previous GWAS meta-analysis). For previously reported GWAS SNPs, their association with BMD in sequenced samples may not necessarily be genome-widely significant (P < 5x10 − 8) or even marginally significant (P < 0.05).
Using WLS locus and the LS BMD phenotype as an example, Following are two conditional models that we analysed:
Model 1
BMD = α + β(covariates) + β[the most significant SNV in sequenced samples (chr1:68356109, rs2566764)] + β[GWAS SNP in the sequenced locus]
Model 2
BMD = α + β(covariates) + β[the most significant GWAS SNP in sequenced samples (chr1:68407663, rs1430742)] + β[other SNV in the locus]
For Model 1, the SNVs that were added into the conditional analyses were rs2566764 for LS BMD and rs75170441 for FN BMD in the WLS locus; rs13163005 for FN BMD in the MEF2C locus; and rs111584437 for FN BMD in the ARHGAP1/F2 locus. As for Model 2, the GWAS SNPs that were added into conditional analyses were rs1430742 for both FN and LS BMD in the WLS locus; rs10037512 for FN BMD in the MEF2C locus; and rs10769205 for FN BMD in the ARHGAP1/F2 locus. When there was no significant association between SNVs and BMD phenotypes, we did not perform conditional analyses.
Rare variant association analyses (the sliding window approach)
For rare variants with minor allele frequency (MAF) ≤ 1%, we performed Sequence Kernel Association Test (SKAT)(38) in FHS and CHS, separately, using the sliding window approach. We divided each targeted sequenced locus into several ‘4,000 bp-wide’ windows with a 2,000 bp overlap between two neighbouring windows. The SKAT was performed in each of the 4,000bp windows. We used SKATmeta R package (http://cran.r-project.org/web/packages/skatMeta/vignettes/skatMeta.pdf), which meta-analyses the score statistics obtained from SKAT analyses from each of FHS and CHS studies, instead of combining P-values of SKAT analyses. Exclusion criteria after meta-analysis consisted of windows with heterogeneity P-values of <0.05 and Het I2 > 50.
We accessed significance (P-values) with unweighted linear regression analyses, since the P-values are not biased due to sample selection. We did not use weighted linear regression for the SKAT analyses, since we only estimated P-values, but not the effect size from the SKAT analyes.
Multiple testing corrections for rare variant association analyses
To correct for multiple testing, we performed a permutation test (the minimum P-value selected from each of 1,000 permutations to form the null distribution) to identify windows that achieve statistical significance.
Leave-one-out analyses for rare variants
In those windows that showed significant association with BMD, we performed leave-one-out analyses to identify rare variant or variants that contribute to the association signal within a window. In a window, we excluded one variant (MAF ≤ 1%) at time and repeated SKAT tests. The variant or variants that drove association signals were considered to contribute to the associated tests.
Supplementary Material
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
Funding
‘Building on GWAS for NHLBI-diseases: the U.S. CHARGE Consortium’ was funded by the NIH (the American Recovery and Reinvestment Act of 2009, ARRA, 5RC2HL102419). Data for ‘Building on GWAS for NHLBI-diseases: the U.S. CHARGE Consortium’ was provided by Eric Boerwinkle on behalf of the Atherosclerosis Risk in Communities (ARIC) Study, L. Adrienne Cupples, PI for the Framingham Heart Study (FHS) and Bruce Psaty, PI for the Cardiovascular Health Study (CHS). The FHS - Conducted and supported by the NHLBI in collaboration with Boston University (BU) (N01-HC-25195), and its contract with Affymetrix, Inc., for genome-wide genotyping services (N02-HL-6-4278), for quality control by FHS investigators using genotypes in the SNP Health Association Resource (SHARe) project. A portion of this research was conducted using the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at BU School of Medicine and Boston Medical Center. The Framingham Osteoporosis Study is supported by NIAMS (R01 AR 41398, R01 AR 061162 and R21 AR056405). CHS: This CHS research was supported by NHLBI contracts HHSN268201200036C, HHSN268200800007C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086; and NHLBI grants HL080295, HL087652, HL105756 with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided through AG023629 from the National Institute on Aging (NIA). David Karasik was supported by ERC FP7-PEOPLE-(CIG). A full list of CHS investigators and institutions can be found at http://www.chs-nhlbi.org/pi.htm.
Supplementary Material
References
- 1. Marshall D., Johnell O., Wedel H. (1996) Meta-analysis of how well measures of bone mineral density predict occurrence of osteoporotic fractures [see comments]. BMJ, 312, 1254–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hsu Y.H., Kiel D.P. (2012) Clinical review: Genome-wide association studies of skeletal phenotypes: what we have learned and where we are headed. J. Clin. Endocrinol. Metab., 97, E1958–E1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Estrada K., Styrkarsdottir U., Evangelou E., Hsu Y.H., Duncan E.L., Ntzani E.E., Oei L., Albagha O.M., Amin N., Kemp J.P., et al. (2012) Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat. Genet., 44, 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Psaty B.M., O'Donnell C.J., Gudnason V., Lunetta K.L., Folsom A.R., Rotter J.I., Uitterlinden A.G., Harris T.B., Witteman J.C., Boerwinkle E. (2009) Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ. Cardiovasc. Genet., 2, 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bush W.S., Moore J.H. (2012) Chapter 11: Genome-wide association studies. PLoS Comput. Biol., 8, e1002822.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gerstein M.B., Kundaje A., Hariharan M., Landt S.G., Yan K.K., Cheng C., Mu X.J., Khurana E., Rozowsky J., Alexander R., et al. (2012) Architecture of the human regulatory network derived from ENCODE data. Nature, 489, 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ecker J.R., Bickmore W.A., Barroso I., Pritchard J.K., Gilad Y., Segal E. (2012) Genomics: ENCODE explained. Nature, 489, 52–55. [DOI] [PubMed] [Google Scholar]
- 8. Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J., et al. (2012) Systematic localization of common disease-associated variation in regulatory DNA. Science, 337, 1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kryukov G.V., Shpunt A., Stamatoyannopoulos J.A., Sunyaev S.R. (2009) Power of deep, all-exon resequencing for discovery of human trait genes. Proc. Natl Acad. Sci. U S A, 106, 3871–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tennessen J.A., Bigham A.W., O'Connor T.D., Fu W., Kenny E.E., Gravel S., McGee S., Do R., Liu X., Jun G., et al. (2012) Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science, 337, 64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S., et al. (2012) Annotation of functional variation in personal genomes using RegulomeDB. Genome Res., 22, 1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ward L.D., Kellis M. (2012) HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res., 40, D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Logan C.Y., Nusse R. (2004) The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol., 20, 781–810. [DOI] [PubMed] [Google Scholar]
- 14. Goldring S.R., Goldring M.B. (2007) Eating bone or adding it: the Wnt pathway decides. Nat. Med., 13, 133–134. [DOI] [PubMed] [Google Scholar]
- 15. Maruyama T., Jiang M., Hsu W. (2012) Gpr177, a novel locus for bone-mineral-density and osteoporosis, regulates osteogenesis and chondrogenesis in skeletal development. J. Bone Miner. Res., 28, 1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marinou K., Christodoulides C., Antoniades C., Koutsilieris M. (2012) Wnt signaling in cardiovascular physiology. Trends Endocrinol. Metab., 23, 628–636. [DOI] [PubMed] [Google Scholar]
- 17. Naito A.T., Shiojima I., Komuro I. (2010) Wnt signaling and aging-related heart disorders. Circ. Res., 107, 1295–1303. [DOI] [PubMed] [Google Scholar]
- 18. Krainc D., Haas M., Ward D.C., Lipton S.A., Bruns G., Leifer D. (1995) Assignment of human myocyte-specific enhancer binding factor 2C (hMEF2C) to human chromosome 5q14 and evidence that MEF2C is evolutionarily conserved. Genomics, 29, 809–811. [DOI] [PubMed] [Google Scholar]
- 19. Creemers E.E., Sutherland L.B., McAnally J., Richardson J.A., Olson E.N. (2006) Myocardin is a direct transcriptional target of Mef2, Tead and Foxo proteins during cardiovascular development. Development, 133, 4245–4256. [DOI] [PubMed] [Google Scholar]
- 20. Gordon J.W., Pagiatakis C., Salma J., Du M., Andreucci J.J., Zhao J., Hou G., Perry R.L., Dan Q., Courtman D., et al. (2009) Protein kinase A-regulated assembly of a MEF2{middle dot}HDAC4 repressor complex controls c-Jun expression in vascular smooth muscle cells. J Biol Chem, 284, 19027–19042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lin Q., Schwarz J., Bucana C., Olson E.N. (1997) Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science, 276, 1404–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Putt M.E., Hannenhalli S., Lu Y., Haines P., Chandrupatla H.R., Morrisey E.E., Margulies K.B., Cappola T.P. (2009) Evidence for coregulation of myocardial gene expression by MEF2 and NFAT in human heart failure. Circ Cardiovasc Genet, 2, 212–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Leupin O., Kramer I., Collette N.M., Loots G.G., Natt F., Kneissel M., Keller H. (2007) Control of the SOST bone enhancer by PTH using MEF2 transcription factors. J. Bone Miner. Res., 22, 1957–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dupuis J., Langenberg C., Prokopenko I., Saxena R., Soranzo N., Jackson A.U., Wheeler E., Glazer N.L., Bouatia-Naji N., Gloyn A.L., et al. (2010) New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet., 42, 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Strawbridge R.J., Dupuis J., Prokopenko I., Barker A., Ahlqvist E., Rybin D., Petrie J.R., Travers M.E., Bouatia-Naji N., Dimas A.S., et al. (2011) Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes, 60, 2624–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Speliotes E.K., Willer C.J., Berndt S.I., Monda K.L., Thorleifsson G., Jackson A.U., Lango Allen H., Lindgren C.M., Luan J., Magi R., et al. (2010) Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet., 42, 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Smith N.L., Huffman J.E., Strachan D.P., Huang J., Dehghan A., Trompet S., Lopez L.M., Shin S.Y., Baumert J., Vitart V., et al. (2011) Genetic predictors of fibrin D-dimer levels in healthy adults. Circulation, 123, 1864–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Asselbergs F.W., Guo Y., van Iperen E.P., Sivapalaratnam S., Tragante V., Lanktree M.B., Lange L.A., Almoguera B., Appelman Y.E., Barnard J., et al. (2012) Large-Scale Gene-Centric Meta-analysis across 32 Studies Identifies Multiple Lipid Loci. Am. J. Hum. Genet., 91, 823–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kung A.W., Xiao S.M., Cherny S., Li G.H., Gao Y., Tso G., Lau K.S., Luk K.D., Liu J.M., Cui B., et al. (2010) Association of JAG1 with bone mineral density and osteoporotic fractures: a genome-wide association study and follow-up replication studies. Am. J. Hum. Genet., 86, 229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lin H., Wang M., Brody J.A., Bis J.C., Dupuis J., Lumley T., McKnight B., Rice K.M., Sitlani C.M., Reid J.G., et al. (2014) Strategies to design and analyze targeted sequencing data: cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium Targeted Sequencing Study. Circ. Cardiovasc. Genet., 7, 335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kanis J.A., Melton L.J., 3rd, Christiansen C., Johnston C.C., Khaltaev N. (1994) The diagnosis of osteoporosis. J. Bone Miner. Res., 9, 1137–1141. [DOI] [PubMed] [Google Scholar]
- 32. Hsu Y.H., Zillikens M.C., Wilson S.G., Farber C.R., Demissie S., Soranzo N., Bianchi E.N., Grundberg E., Liang L., Richards J.B., et al. (2010) An integration of genome-wide association study and gene expression profiling to prioritize the discovery of novel susceptibility Loci for osteoporosis-related traits. PLoS Genet., 6, e1000977.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Homer N., Merriman B., Nelson S.F. (2009) BFAST: an alignment tool for large scale genome resequencing. PLoS One, 4, e7767.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A. (2012) An integrated map of genetic variation from 1,092 human genomes. Nature, 491, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang K., Li M., Hakonarson H. (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res., 38, e164.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dunham I., Kundaje A., Aldred S.F., Collins P.J., Davis C.A., Doyle F., Epstein C.B., Frietze S., Harrow J., Kaul R., et al. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature, 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu M.C., Lee S., Cai T., Li Y., Boehnke M., Lin X. (2011) Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet., 89, 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.