Abstract
Background
Hereditary hypophosphatemia is a group of rare renal phosphate wasting disorders. The diagnosis is based on clinical, radiological, and biochemical features, and may require genetic testing to be confirmed.
Methodology
Clinical features and mutation spectrum were investigated in patients with hereditary hypophosphatemia. Genomic DNA of 23 patients from 15 unrelated families were screened sequentially by PCR-sequencing analysis for mutations in the following genes: PHEX, FGF23, DMP1, ENPP1, CLCN5, SLC34A3 and SLC34A1. CytoScan HD Array was used to identify large deletions.
Results
Genetic evaluation resulted in the identification of an additional asymptomatic but intermittent hypophosphatemic subject. Mutations were detected in 21 patients and an asymptomatic sibling from 13 families (86.6%, 13/15). PHEX mutations were identified in 20 patients from 12 families. Six of them were novel mutations present in 9 patients: c.983_987dupCTACC, c.1586+2T>G, c.1206delA, c.436+1G>T, c.1217G>T, and g.22,215,887–22,395,767del (179880 bp deletion including exon 16–22 and ZNF645). Six previously reported mutations were found in 11 patients. Among 12 different PHEX mutations, 6 were de novo mutations. Patients with de novo PHEX mutations often had delayed diagnosis and significantly shorter in height than those who had inherited PHEX mutations. Novel compound heterozygous mutations in SLC34A3 were found in one patient and his asymptomatic sister: c.1335+2T>A and c.1639_1652del14. No mutation was detected in two families.
Conclusions
This is the largest familial study on Turkish patients with hereditary hypophosphatemia. PHEX mutations, including various novel and de novo variants, are the most common genetic defect. More attention should be paid to hypophosphatemia by clinicians since some cases remain undiagnosed both during childhood and adulthood.
Introduction
Hereditary hypophosphatemia or hypophosphatemic rickets (HR) when it occurs in children is a group of rare renal phosphate-wasting disorders with a prevalence of 3.9 per 100,000 live births [1]. It is characterized by hypophosphatemia and bone mineralization defects such as rickets and osteomalacia [2–4]. The patients usually present with a spectrum of clinical features such as growth retardation with disproportioned short stature, enlargement of growth plates of rapidly growing bones, lower limb deformities, bone pain, loss of teeth, and dental abscess.
Various genetic defects are known to cause the disease [5–7]. Among them, inactivating mutations in the PHEX (phosphate regulating gene with homologies to endopeptidases on the X chromosome, MIM 300550) lead to X-linked HR (XLHR, MIM 307800), the most common form of hereditary hypophosphatemia [8]. More than 424 different mutations have been listed in the Human Gene Mutation Database (HGMD, http://www.hgmd.cf.ac.uk/ac/index.php, accessed on Jan 17, 2018). Gain-of-function mutations at codon R176 or R179 in the proteolytic cleavage domain of FGF23 (fibroblast growth factor 23, MIM 605380) cause autosomal dominant HR (ADHR, MIM 193100) [9, 10]. Five different mutations are currently listed in the HGMD (accessed on Jan 17, 2018). Inactivating mutations in the DMP1 (dentin matrix acidic phosphoprotein 1, MIM 600980) or ENPP1 (ectonucleotide pyrophosphatase/phosphodiesterase 1, MIM 173335) result in autosomal recessive HR type 1 (ARHR1, MIM 241520) [11, 12] or autosomal recessive HR type 2 (ARHR2, MIM 613312) [13], respectively. The frequency of ARHR1 and ARHR2 is about the same. Currently, there are 9 different DMP1 and 8 ENPP1 mutations listed in the HGMD (accessed on Jan 17, 2018). Interestingly, ENPP1 mutations are more frequently reported in patients with idiopathic infantile arterial calcification or generalized arterial calcification of infancy (49 different mutations are listed in the HGMD), suggesting that a different pathway is involved in the generation of ARHR2 [14]. Inactivating mutations in the SLC34A1 (sodium-dependent phosphate cotransporter 2a, MIM 182309) give rise to either autosomal recessive hypophosphatemic nephrolithiasis/osteoporosis-1 (MIM 612286) [15, 16] or Fanconi renotubular syndrome-2 (MIM 613388) [17], whereas a loss-of-function mutation in the SLC34A3 (sodium-dependent phosphate cotransporter 2c, MIM 609826) results in hereditary HR with hypercalciuria (HHRH, MIM 241530) [18, 19]. SLC34A1 mutation also causes autosomal-recessive infantile hypercalcemia-2 (HCINF2, MIM 616963) due to renal phosphate wasting-induced overproduction of 1,25(OH)2D3 [20]. Unlike inactivation mutation in CYP24A1 (MIM 126065) which causes infantile hypercalcemia-1 (HCINF1) [21], patients with HCINF2 respond rapidly to phosphate supplementation [20]. Currently, there are 25 different SLC34A1 and 33 SLC34A3 mutations listed in the HGMD (accessed on Jan 17, 2018). In addition, inactivating mutations in the CLCN5 (chloride voltage-gated channel 5, MIM 300008) lead to X-linked recessive HR (XLRHR, MIM 300554) and may be part of clinical presentations in Dent disease 1 (MIM 300009) characterized by low molecular weight proteinuria, hypercalciuria, nephrocalcinosis, nephrolithiasis, and renal failure [22–24]. More than 250 different CLCN5 mutations are listed in the HGMD (accessed on Jan 17, 2018).
PHEX and DMP1 negatively regulate the expression of FGF23, a novel phosphaturic hormone produced in osteocytes, which is secreted into the circulation after undergoing O-glycosylation by GALNT3 [25, 26]. FGF23 is a key regulator of phosphate reabsorption and 1,25(OH)2D3 synthesis in the proximal renal tubules [27, 28]. FGF23 levels are increased in patients with XLHR, ADHR, ARHR1, and ARHR2 leading to phosphaturia, low serum phosphate, and low serum 1,25(OH)2D3 level due to its inhibition of vitamin D 1-α hydroxylase. However, in patients with HCINF2, HHRH, and XLRHR, FGF23 levels are normal or low-normal, suggesting an FGF23-independent renal tubular defect [29]. In contrast to patients with XLHR, ADHR or ARHP, who are usually treated with high doses of alphacalcidol or calcitriol and multiple daily doses of oral phosphate, oral phosphate supplementation alone is sufficient for patients with HHRH.
In our previous studies, 27 patients from 16 unrelated Turkish families were investigated [30–32]. In the present study, we analyzed clinical and genetic characteristics of additional patients from 15 unrelated families from a different region of Turkey.
Subjects and methods
Patients
Parents and available siblings of index patients (n = 51) from 15 unrelated Turkish families were enrolled to the study. Hereditary hypophosphatemia was diagnosed based on clinical and laboratory assessment. The clinical and laboratory data at the time of diagnosis were summarized in Table 1. The cases suspected of having renal failure, malabsorption, renal tubular acidosis and other tubulopathies that cause secondary hypophosphatemia (e.g., Fanconi’s syndrome, glycogen storage disease, and galactosemia), vitamin D-dependent rickets type 1 and 2 were excluded from the study. This study was approved by the institutional review board of the Dokuz Eylul University Faculty of Medicine. Written informed consent was obtained from the children and their parents.
Table 1. Summary of clinical and laboratory findings in patients with hereditary hypophosphatemia.
| Family | Subjects | Clinical features | Age (years) | Height SDS | Ca mg/dL | P mg/dL | ALP IU/L | 25OHD3 ng/ml | 1,25(OH)2 D3 pg/mL | PTH ng/L | TPR | TmP/GFR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | I-1 Father | normal | 38 | -1.37 | ND | ND | ND | ND | ND | ND | ND | ND |
| I-2 Mother | normal | 38 | 0.56 | ND | ND | ND | ND | ND | ND | ND | ND | |
| I-3 Daughter | Inability to walk, genu varum deformity | 2.4 | -3.76 | 9.2 | 2.5 | 829 | 21.2 | 50.8 | 94 | 56.9 | 1.42 | |
| II | II-1 Father | normal | 38 | -0.62 | ND | ND | ND | ND | ND | ND | ND | ND |
| II-2 Mother | Short stature, genu varum deformity | 37 | -3.95 | 9.8 | 2.1 | 111 | 10.5 | ND | 73 | 87 | 1.70 | |
| II-3 Daughter | Short stature,genu varum deformity | 2 | -2.09 | 10.2 | 2.1 | 531 | 31.1 | ND | 52.5 | 62.3 | 1.31 | |
| III | III-1 Father | normal | 42 | -0.26 | ND | ND | ND | ND | ND | ND | ND | ND |
| III-2 Mother | Genu varum,short stature | 40 | -3.28 | 10.1 | 1.7 | 106 | ND | ND | ND | ND | ND | |
| III.3 Daughter | Inability to walk,genu varum deformity | 5 | -2.13 | 8.5 | 2.2 | 667 | 20 | 30 | 319 | 67.8 | 1.49 | |
| III-4 Son | Inability to walk, genu varum deformity | 3.2 | -1.58 | 9.2 | 3.1 | 600 | 80 | ND | 107 | 69.1 | 2.1 | |
| IV | IV-1 Father | normal | 49 | -1.65 | ND | ND | ND | ND | ND | ND | ND | ND |
| IV-2 Mother | normal | 40 | 0.26 | ND | ND | ND | ND | ND | ND | ND | ND | |
| IV-3 Daughter | Short stature, genu varum deformity | 5 | -3.33 | 9.4 | 2.0 | 559 | 28.3 | ND | 138 | 79.6 | 1.59 | |
| V | V-1 Father | Fractures, genu varum deformity | 30 | -3.71 | 9.6 | 2.1 | 143 | 23.7 | ND | 40 | 69.9 | 1.46 |
| V-2 Mother | normal | 32 | -1.28 | ND | ND | ND | ND | ND | ND | ND | ND | |
| V-3 Daughter | Short stature, widening of wrist | 1.3 | -2.80 | 10 | 3.4 | 465 | >70 | ND | 48.1 | 60.6 | 2.06 | |
| VI | VI-1 Father | normal | 34 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| VI-2 Mother | normal | 37 | 2.05 | ND | ND | ND | ND | ND | ND | ND | ND | |
| VI3 Daughter | normal | 8 | ND | ND | ND | ND | ND | ND | ND | ND | ND | |
| VI-4 Son | Inability to walk,genu varum deformity | 3.5 | -3.76 | 9.8 | 2.8 | 925 | 35 | ND | 62.2 | 69.7 | 1.95 | |
| VII | VII-1 Father | normal | 35 | 0.03 | ND | ND | ND | ND | ND | ND | ND | ND |
| VII-2 Mother | normal | 27 | 0.26 | ND | ND | ND | ND | ND | ND | ND | ND | |
| VII-3 Son | Inability to walk,genu varum deformity | 4 | -3.44 | 9.0 | 3.1 | 393 | 4.9 | ND | 154.7 | 58.7 | 1.82 | |
| VIII | VIII-1 Father | normal | 35 | -0.51 | ND | ND | ND | ND | ND | ND | ND | ND |
| VIII-2 Mother | normal | 33 | -0.26 | ND | ND | ND | ND | ND | ND | ND | ND | |
| VIII-3 Daughter | Short stature | 4.5 | -3.89 | 9.8 | 2.4 | 581 | 39.1 | 130 | 71.5 | 78.5 | 1.88 | |
| IX | IX-1 Father | normal | 36 | -0.95 | ND | ND | ND | ND | ND | ND | ND | ND |
| IX-2 Mother | Short stature, genu varum | 36 | -2.35 | 9.65 | 1.83 | 81 | 15.7 | ND | 130 | 95.5 | 1.69 | |
| IX-3 Son | Genu varum deformity | 2.1 | -0.75 | 10.1 | 2.6 | 827 | 32.3 | ND | 107.1 | 74 | 1.92 | |
| X | X-1 Father | normal | 39 | -0.54 | ND | ND | ND | ND | ND | ND | ND | ND |
| X-2 Mother | normal | 33 | -2.05 | ND | ND | ND | ND | ND | ND | ND | ND | |
| X-3 Son | Nephrolithiasis | 5 | -0.54 | 9.8 | 2.5 | 548 | 12.5 | 27.5 | 33.9 | 85 | 2.1 | |
| X-4 Daughter | Normal | 6.9 | -0.23 | 10.3 | 3.6 | 325 | 27.5 | ND | 13 | 82.5 | 2.97 | |
| XI | XI-1 Father | Normal | 31 | -0.25 | ND | ND | ND | ND | ND | ND | ND | ND |
| XI-2 Mother | Normal | 28 | -0.56 | ND | ND | ND | ND | ND | ND | ND | ND | |
| XI-3 Son | Genu varum deformity | 2.5 | -0.4 | 10.3 | 3.2 | 434 | 6.8 | ND | 16.9 | 70 | 1.1 | |
| XI-3 Son | Normal | 6.2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | |
| XII* | XII-1 Father | normal | 41 | 0.72 | 9.59 | 3.73 | 78 | ND | ND | 30.1 | ND | ND |
| XII-2 Mother | normal | 44 | -1.46 | 9.52 | 2.12 | 114 | ND | ND | 61.2 | ND | ND | |
| XII-3 Son | Genu varum deformity | 2.9 | -0.90 | 9.6 | 2.9 | 623 | 52 | ND | 62 | 74.9 | 2.29 | |
| XII-4 Daughter | Genu varum deformity | 2.9 | 1.18 | 9.8 | 2.9 | 401 | 35.1 | ND | 71.4 | 77 | 2.48 | |
| XIII | XIII-1 Father | Normal | 56 | 9.89 | 2.76 | 61 | 22.2 | ND | 50.8 | 88 | 2.43 | |
| XIII-2 Mother | Normal | 53 | 9.73 | 3.23 | 98 | 10.9 | ND | 72 | 93.4 | 3.01 | ||
| XIII-3 Son | Inability to walk, rickets | 7.5 | -0.75 | 9.9 | 2.1 | 2533 | ND | 36.3 | 58 | 82 | 1.74 | |
| XIV | XIV-1 Father | Normal | 36 | -0.2 | 8.7 | 3.7 | 77 | 39.9 | ND | 63 | 93 | 3.4 |
| XIV-2 Mother | Shortstature, genu varum deformities | 35 | -2.8 | 8.9 | 2.3 | 44 | < 8 | ND | 95 | 97 | 2.2 | |
| XIV-3 Daughter | Short stature, genu varum deformities | 2.7 | -2.01 | 9.4 | 2.2 | 364 | 27 | ND | 49 | 75 | 1.65 | |
| XV | XV-1 Father | Normal | 25 | -1.13 | ND | ND | ND | ND | ND | ND | ND | ND |
| XV-2 Mother | Normal | 28 | -0.40 | ND | ND | ND | ND | ND | ND | ND | ND | |
| XV-3 Daughter | Short stature, genu varum deformities | 3.3 | -3.04 | 9.4 | 2.9 | 677 | 107 | 54.2 | 79.3 | 83.4 | 2.8 | |
| XV-4 Sister | Normal | 6.9 | -0.02 | 10.2 | 4.96 | 176 | 21.5 | ND | 11.4 | 92.3 | 4.5 | |
| Normal range | 8.5–10.5 | 3.7–5.61 | 90–3252 | 20–100 | 17–53 | 15–65 | >85% | 2.9–6.13 | ||||
*from a consanguineous family
ND: not done; SDS: standard deviation score or Z-score
SI unit conversions: to convert the values for 25OHD to nmol/L, multiply by 2.5; to convert the values for 1,25(OH)2D to pmol/L, multiply by 2.4
to convert the value for calcium to mmol/L, divide by 4; to convert the values for phosphate to mmol/L, divide by 3.1.
1 Normal range of serum phosphate is 4.8–8.2 mg/dL for 0–5 days of age, 3.8–6.5 mg/dL for 1–3 years of age, 3.7–5.6 mg/dL for 4–11 years of age, 2.9–5.4 mg/dL for 12–18 years of age and 2.7–4.5 mg/dL for adults.
2 Normal range of serum ALP is 80–380 IU/L for 0–1 year, 90–325 IU/L for 1–9 years, 82–380 IU/L for 9–18 years, 41–120 IU/L for over 18 years.
3 The normal ranges of TmP/GFR (mg/dL) vary with age: Birth, 3.6–8.6; 3 months of age, 3.7–8.25; 6 months of age, 2.9–6.5; 2–15 years of age, 2.9–6.1; adult, 2.2 to 3.6 mg/dL.
Genomic DNA isolation
Genomic DNA from peripheral blood leukocytes was isolated using Gentra Blood Kit (Qiagen Corp, CA).
DNA amplification and sequencing
DNA samples were first analyzed for mutations in all the coding exons and intron-exon boundaries of PHEX. If no mutations were identified, we then screened for mutations in the FGF23 followed by consecutive analysis of DMP1, ENPP1, CLCN5, SLC34A3 and SLC34A1. PCR primers and conditions were described previously[11, 13, 22, 30, 33, 34]. The resulting PCR products were directly sequenced using an automated ABI PRISM 3700 sequencer (Foster City, CA) or cloned into TA vector (Invitrogen, CA). Individual clones were subsequently sequenced.
Identification of a large PHEX deletion
Copy number variation in genomic DNA was analyzed by CytoScan HD Array according to the manufacturer’s procedure (Affymetrix, Santa Clara, CA). The closest undeleted probes to the boundary of deleted region (copy number loss) in the PHEX gene were used as a starting point to map the breaking point. A total of 6 walking primers were used to perform PCR until a specific DNA fragment was amplified. The PCR products were directly sequenced.
RNA isolation and RT-PCR
Total RNA from peripheral blood leukocytes was extracted as described previously [35]. Two μg of total RNA were reverse-transcribed into cDNA using Promega reverse transcription system (Promega, Madison, WI). To determine the effect of a splice site mutation at c.1586+2T>G, RT-PCR was used to amplify PHEX transcripts using the following two primers: 5’- CTATCCAGAGTTTATAATGAA-3’ (forward primer located in exon 13), and 5’- CTGTTCCCCAAAAGAAAGGCTT-3’ (reverse primer located in exon 16). The PCR conditions were 95°C for 5 minutes followed by 35 cycles of amplification (95°C for 1 min, 54°C for min, and 72°C for 1 min). The resulting PCR products were analyzed by gel electrophoresis and subsequently sequenced.
In silico analysis of variants
The significance of single nucleotide variant (SNV) was predicted using the following four web-based programs: Mutation Taster (http://www.mutationtaster.org), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/), SIFT (http://sift.jcvi.org/), and PROVEAN (http://provean.jcvi.org/index.php). SNV was considered to be mutation if they had: low allelic frequency (<0.01) in the normal population database; predicted to be damaging or disease-causing by at least three of four prediction programs; and present only in the patient or in strict segregation with phenotype in familial cases. Biallelic or compound heterozygous mutations were considered as disease-causing mutations. Monoallelic mutations in genes with dominant inheritance were also considered as disease-causing mutations.
Statistical analysis
Student’s t test or Chi-square test (two-tailed) was used to compare two groups. A p value of 0.05 or less was considered significant.
Results
Hereditary hypophosphatemia were diagnosed in 23 patients from 15 families: 17 children (mean age at diagnosis 4.9±1.9 years, age range 1.3–8.9 years), and 6 adults (mean age at diagnosis 34.2±4.5 years, age range 28–40 years). All the children had clinical and laboratory features of rickets such as genu varum deformity, metaphyseal X-ray findings, and/or fractures. The X-ray images of metaphyseal widening and irregularity, cupping and fraying of the metaphyseal region, haziness of cortical margins and epiphysis, and enlargement of the space between the epiphysis and metaphysis were present in all HR children except for X-3 and X-4. Loss of teeth and gum abscess were observed in case IX-3. Cases III-3, VI-4, and VII-3 had undergone a surgical operation for correction of severe lower limb deformities. Bone pain was not recorded in any of the cases.
All the adults were diagnosed based upon relevant clinical (bone pain, walking difficulty, muscle weakness) and laboratory findings (hypophosphatemia, mild-moderate elevated PTH, increased renal phosphate loss). Ten children (58.8%, 10/17) and five adults (83.3%, 5/6) had short stature. Although affected parents had short stature, genu varum, and/or bone fractures after minor trauma starting from childhood, none was diagnosed previously. Genetic analysis identified disease-causing gene mutations in 22 subjects (21 patients and 1 asymptomatic sibling) from 13 families: 12 different PHEX mutations in 20 patients from 12 families, and two different SLC34A3 mutations in two compound heterozygous siblings, one of whom was asymptomatic. No mutation was identified in two patients from two different families.
PHEX mutations
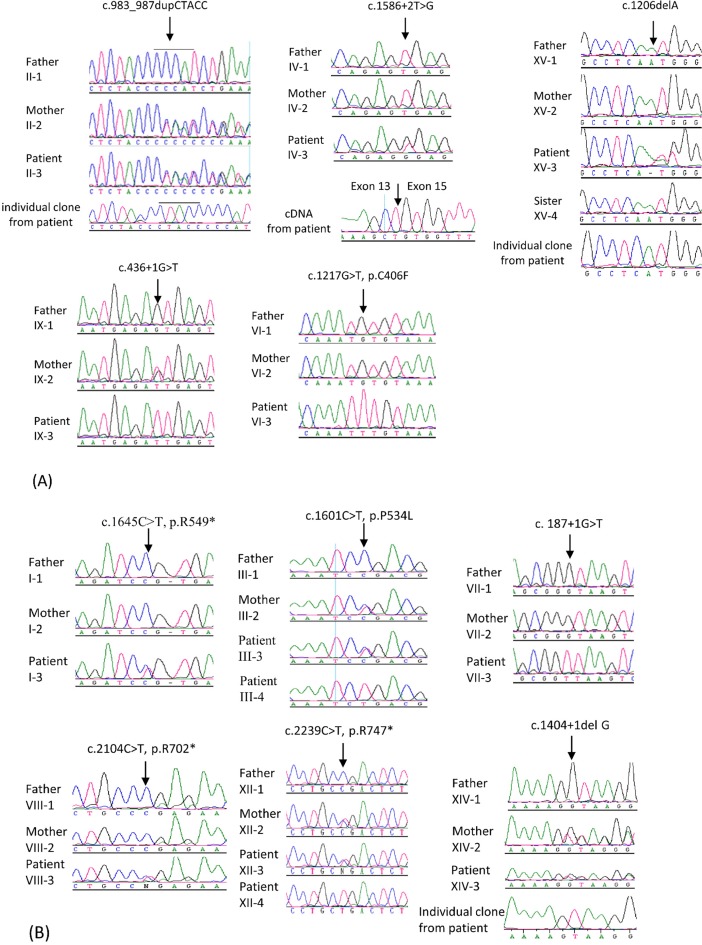
Six novel PHEX mutations were identified in 9 patients from 6 families: c.983_987dupCTACC, c.1586+2T>G, c.1206delA, c.436+1G>T, c.1217G>T (p.C406F), and a large deletion missing exon 16–22 (Fig 1A and Table 2). Although c.436+1G>T is a novel mutation, c.436+1G>A and c.436+1G>C have been reported previously in a Chinese and US patient, respectively [36, 37]. The c.1217G>T (p.C406F) is located at the same position as our previously identified mutation c.1217G>A (p.C406Y) in a Turkish patient [30]. Six previously reported PHEX mutations were found in 11 patients from 6 families: c.1645C>T (p.R549*) [38], c.1601C>T (p.P534L) [38], c. 187+1G>T [39], c.2104C>T (p.R702*) [38], c.2239C>T (p.R747*) [40], and c.1404+1del G (direct submission to PHEX mutation database: http://www.phexdb.mcgill.ca/) (Fig 1B and Table 2). The c.1586+2T>G, c.1206delA, c.1217G>T, c.1645C>T, c. 187+1G>T, and c.2104C>T were not present in the parents and therefore were de novo mutations, accounting for 50% of identified PHEX mutations. The effect of splice donor site mutation c.1586+2T>G on mRNA splicing was investigated by RT-PCR analysis of cDNA from patient’s peripheral lymphocytes. As shown in Fig 1A, the mutation resulted in exon 14 skipping. The c.1586+1G>A was reported previously without annotation of splicing consequence [2]. Based on our current study, the mutation is likely result in exon 14 skipping as well. Most of the mutations were splice site mutations, deletions or duplication, or nonsense mutations. These mutations are predicted to cause frameshift, resulting in either truncated protein or nonsense-mediated decay. Only two missense mutations were identified: c.1601C>T (reported) and c.1217G>T (novel). We performed in silico analysis of these two missense mutations as well as two previously reported mutations that were located in the same codon as c.1217G>T (c.1217G>A, p.C406Y and c.1216T>C, p.C406R) by four web-based programs. As shown in Table 3, all of them were predicted to be disease-causing mutations.
Fig 1. Sequence analysis of PHEX in patients with hereditary hypophosphatemia.
(A) Five novel PHEX mutations. c.983_987dupCTACC (patient II-3 and her mother) and c.436+1G>T (patient IX-3 and his mother) are inherited mutations from mother; c.1586+2T>G (patinet IV-3), c.1206delA (patient XV-3), and c.1217G>T (patientVI-3) are de novo mutations not present in parents. The c.1586+2T>G results in exon 14 skipping. (B) Six previously reported PHEX mutations. c.1645C>T, c. 187+1G>T, and c.2104C>T are de novo mutations. c.1601C>T, c.2239C>T, and c.1404+1del G are inherited mutations transmitted from mother. Mutation is indicated by an arrow.
Table 2. Mutational analysis of patients with hereditary hypophosphatemia.
| Family | Subjects | Gene | Location | Nucleotide change | Amino acid change | Zygocity | Mutation |
|---|---|---|---|---|---|---|---|
| I | I-1 Father | PHEX | Wild-type | ||||
| I-2 Mother | PHEX | Wild-type | |||||
| I-3 Patient | PHEX | Exon 15 | c.1645C>T | p.R549* | Het | De novo, reported | |
| II | II-1 Father | PHEX | |||||
| II-2 Mother | PHEX | Exon 9 | c.978_982dupCTACC | H329Tfs*3 | Het | Novel | |
| II-3 Patient | PHEX | Exon 9 | c.978_982dupCTACC | H329Tfs*3 | Het | Novel | |
| III | III-1 Father | PHEX | Wild-type | ||||
| III-2 Mother | PHEX | Exon 15 | c.1601C>T | p.P534L | Het | Reported | |
| III-3 Patient | PHEX | Exon 15 | c.1601C>T | p.P534L | Het | Reported | |
| III-4 Patient | PHEX | Exon 15 | c.1601C>T | p.P534L | Hemi-zygous | Reported | |
| IV | IV-1 Father | PHEX | Wild-type | ||||
| IV-2 Mother | PHEX | Wild-type | |||||
| IV-3 Patient | PHEX | Splice donor site at intron 14, Exon 14 skipping | c.1586+2T>G | Frameshift | Het | De novo, novel (c.1586+1G>A reported previously) | |
| V | V-1 Father | PHEX | Exon 16–22 | g.22,215,887–22,395,767del(179880 bp deletion) | Hemi-zygous | Reported but not characterized previously | |
| V-2 Mother | PHEX | Wild-type | |||||
| V-3 Patient | PHEX | Exon 16–22 | g.22,215,887–22,395,767del(179880 bp deletion) | Truncated | Het | Reported but not characterized previously | |
| VI | VI-1 Father | PHEX | Wild-type | ||||
| VI-2 Mother | PHEX | Wild-type | |||||
| VI-3 Sister | PHEX | Wild-type | |||||
| VI-4 Patient | PHEX | Exon 11 | c.1217G>T | p.C406F | Hemi-zygous | De novo, novel (c.1217G>A, p.C406Y; and c.1216T>C, p.C406R reported previously) | |
| VII* | VII-1 Father | PHEX | Wild-type | ||||
| VII-2 Mother | PHEX | Wild-type | |||||
| VII-3 Patient | PHEX | Splice donor site at intron 2 | c. 187+1G>T | Frameshift | Hemi-zygous | De novo, reported | |
| VIII | VIII-1 Father | PHEX | Wild-type | ||||
| VIII-2 Mother | PHEX | Wild-type | |||||
| VIII-3 Patient | PHEX | Exon 21 | c.2104C>T | p.R702* | Het | De novo, reported | |
| IX | IX-1 Father | PHEX | Wild-type | ||||
| IX-2 Mother | PHEX | Splice donor site at intron 4 | c.436+1G>T | Frameshift | Het | Novel (c.436+1G>A and c.436+1G>C reported previously) | |
| IX-3 Patient | PHEX | Splice donor site at intron 4 | c.436+1G>T | Frameshift | Hemi-zygous | Novel (c.436+1G>A and c.436+1G>C reported previously) | |
| X | X-1 Father | SLC34A3 | Splice donor site at intron 12 | c.1335+2T>A | Frameshift | Het | Novel |
| X-2 Mother | SLC34A3 | Exon 13 | c.1639_1652delCGCTCCTGGGCCTG | R547Afs*41 | Het | Novel | |
| X-3 Patient | SLC34A3 | Splice donor site at intron 12 and Exon 13 | c.1335+2T>A and c.1639_1652delCGCTCCTGGGCCTG | Frameshift | Compound Het | Novel | |
| X-4 Sister | SLC34A3 | Splice donor site at intron 12 and Exon 13 | c.1335+2T>A and c.1639_1652delCGCTCCTGGGCCTG | Frameshift | Compound Het | Novel | |
| XI | XI-1 Father | Wild-type | |||||
| XI-2 Mother | Wild-type | ||||||
| XI-3 Patient | No mutation detected | ||||||
| XII | XII-1 Father | PHEX | Wild-type | ||||
| XII-2 Mother | PHEX | Exon 22 | c.2239C>T | p.R747* | Het | Reported | |
| XII-3 Patient | PHEX | Exon 22 | c.2239C>T | p.R747* | Hemi-zygous | Reported | |
| XII-4 Patient | PHEX | Exon 22 | c.2239C>T | p.R747* | Het | Reported | |
| XIII | XIII-1 Father | Wild-type | |||||
| XIII-2 Mother | Wild-type | ||||||
| XIII-3 Patient | No mutation detected | ||||||
| XIV | XIV-1 Father | PHEX | Wild-type | ||||
| XIV-2 Mother | PHEX | Splice donor site at intron 12 | c.1404+1delG | Frameshift | Het | Reported | |
| XIV-3 Patient | PHEX | Splice donor site at intron 12 | c.1404+1delG | Frameshift | Het | Reported | |
| XV | XV-1 Father | PHEX | Wild-type | ||||
| XV-2 Mother | PHEX | Wild-type | |||||
| XV-3 Patient | PHEX | Exon 11 | c.1206del A | Q402Hfs*6 | Het | De novo, novel | |
| XV-4 Sister | PHEX | Wild-type |
*from a consanguineous family
Table 3. In silico prediction of single nucleotide variations in the PHEX gene.
| SNV NM_000444 | MAF (minor allele frequency) | SIFT (score) | PROVEAN (score) | PolypPhen-2 (score) | Mutation Taster |
|---|---|---|---|---|---|
| c.1601C>T, p.P534L | neither found in ExAC nor 1000G | damaging (0.003) | deleterious (-6.89) | probably damaging (1.00) | disease causing |
| c.1217G>T, p.C406F | neither found in ExAC nor 1000G | damaging (0.000) | deleterious (-10.87) | probably damaging (1.00) | disease causing |
| c.1217G>A, p.C406Y | neither found in ExAC nor 1000G | damaging (0.000) | deleterious (-10.87) | probably damaging (1.00) | disease causing |
| c.1216T>C, p.C406R | neither found in ExAC nor 1000G | damaging (0.000) | deleterious (-11.86) | probably damaging (1.00) | disease causing |
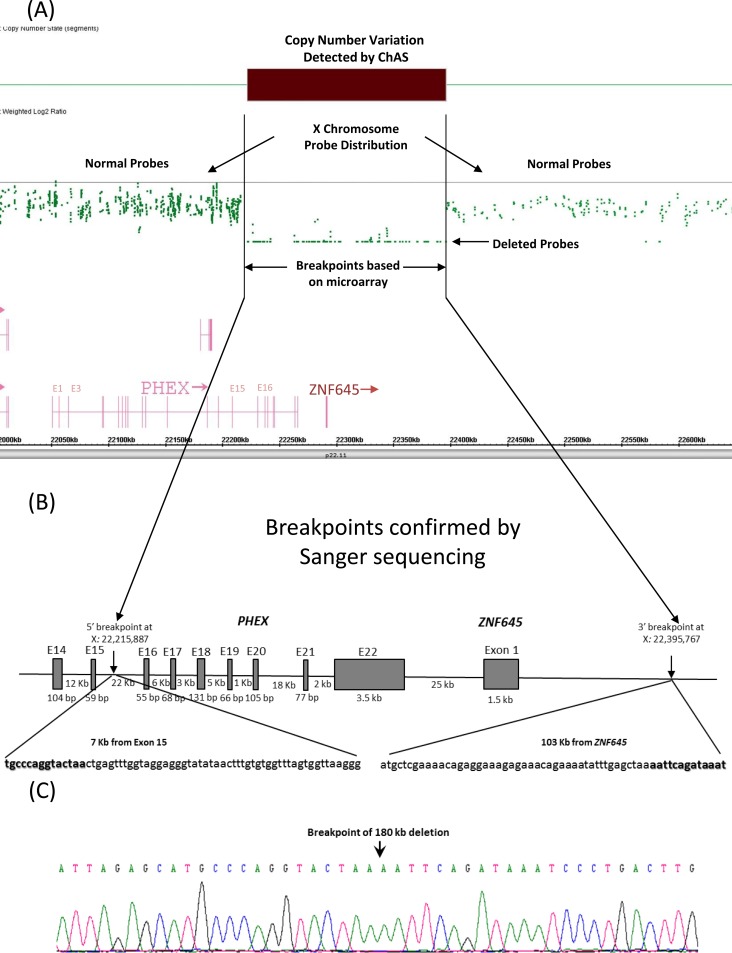
We could not amplify by PCR the region containing PHEX exon 16–22 from affected father of index patient (V-3) from family V whereas the same region was amplified from the female patient and her mother’s DNA, suggesting the possible presence of a large deletion. We next performed a copy number analysis and identified the deleted region (Fig 2A). Based on the 5’ and 3’ undeleted probe sequences closest to the deleted region, we performed chromosome walking to identify the breakpoint. As shown in Fig 3B and 3C, the 5’ breakpoint was located in the intron 15 and 7 kb from the exon 15 of the PHEX gene (X: 22,215,887); the 3’ breakpoint was 103 kb from the ZNF645 gene and 130 kb from the PHEX gene (X: 22,395,767), resulting in the deletion of 179,880 bp DNA fragment. Single TAA is present at the 5’ breakpoint and immediately before the 3’ breakpoint, which may be used for homologous recombination (Fig 2B and 2C).
Fig 2. Characterization of a large deletion of PHEX in patient V-3.
(A) Genechip cytogenetics array results. The top is showing weighted log2 ratios plot showing a normal copy number state for the X chromosome. The deletion is indicated by a dark bar and enclosed in the black lines. More than 70 SNP probes are present in the deleted region and indicated by an arrow. (B) Schematic representation of 179,880 bp deletion of PHEX and ZNF645. The 5’ breakpoint is located in the intron 15 of PHEX and 7 kb from exon 15. The 3’ breakpoint point is 103 kb from ZNF645, resulting in the deletion of exon 16–22 of PHEX and entire ZNF645. The 5’ and 3’ undeleted nucleotide sequences are highlightted in bold. Deletion analysis is based on GRCh37/hg19. (C) Electropherogram of the breaking point. The DNA fragment was amplified by PCR using primers flanking the 5’ and 3’ deletion point. The breakpoint is indicated by an arrow.
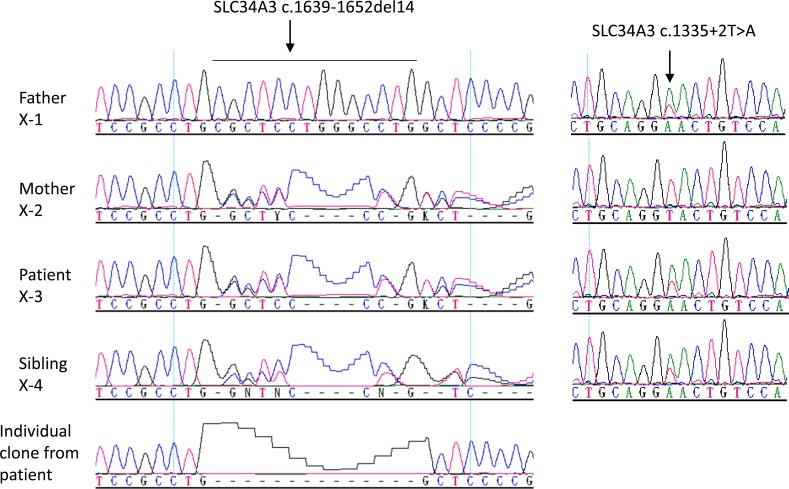
Fig 3. Sequence analysis of SLC34A3 in a patient with hereditary hypophosphatemia.
A novel heterozygous mutation c.1639-1652del14 is present in the mother, patient, and his sister. A novel heterozygous splice donor site mutation c.1335+2T>A is present in the father, patient, and his sister. Compound heterozygous SLC34A3 mutations are carried by both patient and his sister. Mutation is indicated by an arrow.
The mean age at diagnosis of 14 children with PHEX mutations (6 males, 8 females) were 3.3±1.1 (range, 1.3–5.0) years. The mean SD score of height was -2.26±1.46 (-3.89–1.18) and 71.4% of them (10/14) had short stature. All had clinical findings compatible with rickets. Laboratory tests of all subjects revealed typical features of XLHR: normal levels of calcium (9.5±0.5 mg/dL; range 8.5–10.3 mg/dL), low levels of serum phosphate (2.7±0.5 mg/dL; range, 2.0–3.6 mg/dL), TRP (69.8±8.0%, range, 59.9–79.6%), and TmP/GFR (1.88±0.36; range, 1.31–2.48), increased levels of ALP (598±175 IU/L; range, 364–925), and normal/mildly increased levels of PTH (84.5±34.8 pg/mL; range, 31.9–154.7).
The mean age at diagnosis of adults with PHEX mutations (1 male, 5 females) was 34.2±4.5 years (range, 28–40 years). Mean SD score of height was -2.92±0.93 (range, -3.95– -1.46) and 5 of them (83.3%) were short. All of these patients had normal calcium (10.1±0.4 mg/dL; range, 8.9–10.1 mg/dL, normal: 8.5–10.5 mg/dL), hypophosphatemia (2.0±0.2 mg/dL; range, 1.7–2.3 mg/dL, normal: 2.7–4.5 mg/dL), normal urine calcium/creatinine ratio (range, 0.04–0.15 mg/mg; normal: <0.2), and decreased TmP/GFR (1.76±0.31; range, 1.46–2.2, normal: 2.5–3.6). ALP values were in normal range in all except one (111 ± 22 IU/L, range, 81–143; normal: 41–120). Moreover, varying levels were found for PTH (79.8±34.4 pg/mL; range, 40–130 pg/mL; normal: 11.1–79.5 pg/mL) and TRP (87.4±14.4%; range 69.9–97%; normal: >85%) (Table 1).
When the patients with PHEX mutation were evaluated according to the type of inheritance (de novo or inherited), height SD scores and the frequency of short stature were significantly lower in the inherited group (Table 4). Although patients with an inherited mutation was younger at the time of diagnosis when compared to the patients with a de novo mutation, this difference was not statistically significant. The patients were successfully treated with calcitriol and phosphate.
Table 4. Clinical and laboratory characteristics of children with de novo or inherited PHEX mutations.
| Parameters | De novo (n = 6) | Inherited (n = 8) | p Value |
|---|---|---|---|
| Age (years) | 3.8 (3.1 / 4.6) | 2.9 (2.2 / 4.1) | 0.228 |
| Height SDS | -3.60 (-3.79 /-3.30) | -1.51 (-2.10 / -0.79) | 0.001* |
| Short stature | 6 | 4 | 0.035* |
| Ca (mg/dL) | 9.6 (9.4 / 9.9) | 9.5 (8,5 / 9.9) | 0.755 |
| P (mg/dL) | 2.6 (2.3 / 3.1) | 2.8 (2.2 / 3.0) | 0.983 |
| ALP (IU/L) | 629 (526 / 853) | 532 (406 / 656) | 0.228 |
| 25-(OH) vitamin D (ng/mL) | 31.6 (17.1 / 56.0) | 33.7 (24.5 / 67.0) | 0.755 |
| PTH (ng/mL) | 86.5 (69.2 / 142.1) | 66.7 (49.2 / 106) | 0.181 |
| TRP | 73.7 (58.2 / 78.8) | 71.6 (63.7 / 74.9) | 0.662 |
| TmP/GFR | 1.85 (1.54 / 2.05) | 1.99 (1.53 / 2.42) | 0.662 |
*Statistically significant
SLC34A3 mutations
Two novel SLC34A3 compound heterozygous mutations were found in the index case from Family X (X-3) and his asymptomatic sister (X-4): a splice donor site mutation c.1335+2T>A and a 14 bp deletion c.1639_1652delCGCTCCTGGGCCTG (Fig 3 and Table 2). A heterozygous c.1335+2T>A and c.1639_1652delCGCTCCTGGGCCTG were carried by his non-consanguineous father and mother, respectively. The index case, a 5-year-old boy was first evaluated by a nephrologist (SK) due to nephrocalcinosis. Hypophosphatemia and elevated ALP were also observed in addition to hypercalciuria (0.23 mg/mg urine calcium/creatinine ratio, normal <0.20) but were not considered clinically significant initially due to lack of other rachitic findings (Table 1). He had no additional symptoms or findings consistent with rickets. Oral phosphate treatment was started at the age of 14 years following the genetic diagnosis. Consequently, ALP level decreased from 483 U/L to 258 U/L (normal, 74–390). His 6.9-year-old asymptomatic sister had the same mutation. Detailed investigation revealed normal clinical and laboratory variables except for the intermittent mild hypophosphatemia (3.6–4.4 mg/dL, normal 3.7–5.6) and increased urine calcium/creatinine ratio (0.7 mg/mg). She did not require phosphate treatment during 2 years of follow-up. Their father (Case X-1) had recurrent nephrolithiasis but mother (Case X-2) was free of symptoms. Biochemical profiles were normal in both parents.
Families without known mutations
No mutation was detected in the patients from families XI and XIII. Copy number analysis of whole genome did not reveal any large deletions as well.
Case XI-3, a 2.5-year-old boy, was admitted due to skeletal deformities of his hands and feet. His parents were not relatives. Physical examination revealed a normal height (SD score -0.4), prominent widening of wrists, and genu varum. Hypophosphatemia, increased ALP level and urine phosphate excretion, normal calcium and PTH levels, and rachitic X-ray findings were found (Table 1). Vitamin D supplement due to low 25-hydroxyvitamin D3 level did not result in improvement. Calcitriol and neutral phosphate treatment with the diagnosis of HR resulted in resolution of rickets and normalization of laboratory variables.
The other patient (XIII-3) was a 7.5-year-old boy who was evaluated due to inability to walk during the past 1 year. Physical examination was normal other than widening of wrists and genu varum deformities. Laboratory and skeletal X-ray findings were consistent with HR (Table 1). Calcitriol and neutral phosphate were started, and clinical and laboratory improvement were achieved within the first year. His treatment was gradually reduced and stopped when he was 13 years old. The most recent follow-up without treatment was made when he was 21 years old. Physical examination revealed a height of 175.5 cm (-0.19 SDS) and no skeletal abnormalities. Laboratory studies showed normal calcium (9.3 mg/dL, normal 8.8–10.5), ALP (192 IU/L, normal 93–309), and 25- hydroxyvitamin D3 (20.6 ng/mL, normal >20), but low serum phosphate (1.96 mg/dL, normal 2.7–4.5), mildly increased PTH (96 pg/mL, normal 11.1–79.9), normal TRP (91.0%, normal <85%), decreased TmP/GFR (1.78, normal 2.5–4.5), and a normal urine calcium/creatinine ratio (0.01 mg/mg creatinine, normal <0.20).
Discussion
In the present study, we investigated 23 patients from 15 unrelated families with hereditary hypophosphatemia. Disease-causing mutations were found in 21 patients and one asymptomatic sibling (carrying compound heterozygous SLC34A3 mutations) including 8 novel mutations: 6 in PHEX, and 2 in SLC34A3. De novo PHEX mutations were found in 6 out of 12 families. Together with our previous studies [30–32], we have investigated 50 patients from 31 unrelated families with hereditary hypophosphatemia, representing the largest series of patients from Turkish population. Among the 31 families, PHEX, FGF23, CLCN5, and SLC34A3 mutations was found in 26 (83.9%), 1 (3.2%), 1 (3.2%), and 1 (3.2%) families, respectively. The overall mutation detection rate is about 94% (29/31) and PHEX mutation is the most common genetic defect in the Turkish patients. Other mutations are rare such as FGF23, DMP1, ENPP1, SLC34A1, SLC34A3, and CLCN5. In fact, we have not detected DMP1, ENPP1, or SLC34A1 mutation in our patients. Our data are similar to those from other parts of the would such as Italy [2], Denmark [5] and US [6].
The PHEX gene is located on the X chromosome and consists of 22 short exons encoding a protein of 749 amino acids [41]. It is primarily expressed in osteoblasts and osteocytes, and plays an important role in phosphate metabolism [41]. An inactivating mutation of PHEX leads to increased serum FGF23 level and thus renal phosphate loss. Most PHEX mutations found in the current study are either splice site or insertion/deletion mutations, resulting in either truncated proteins devoid of normal function or truncated mRNA likely going through nonsense-mediated decay [42]. The delay in diagnosis of our patients with de novo PHEX mutations is likely due to lack of family history and awareness by their physicians, which results in treatment delay and more severe growth retardation. We could not detect mutation in two patients from 2 families, which may indicate possible existence of a promoter mutation in the screened genes or a novel disease-causing gene, which needs to be investigated further.
We have characterized another PHEX deletion with identification of the breakpoint (g.22,215,887–22,395,767del). The large PHEX deletion missing exon 16–22 has been reported previously but the breakpoint has never been investigated [5, 43]. It is not clear whether previously reported cases have the same breakpoint or not. Interestingly, TAA repeats are also present in the 5’ breakpoint of our previously characterized 40 kb PHEX deletion [31]. Unlike the 40 kb PHEX deletion which has 13 TAA tandem repeats present before the 3’ breakpoint, only one TAA repeat is present before the 3’ breakpoint in the current deletion, suggest that the deletion mediated by homologous recombination may not need multiple tandem repeats. The entire ZNF645 is also deleted together with exon 16–22 of the PHEX gene. ZNF645 is a RING finger protein and function as an E3 ubiquitin-protein ligase. It may play a role in human sperm production and quality control [44].
The c.1335+2T>A and c.1639_1652del14 mutations in SLC34A3 are predicted to cause frameshift and result in loss-of-function of the gene. Given the autosomal recessive inheritance of HHRH, both patient and his sister should have the disease. The apparent phenotype-genotype discordance in the patient’s sister is not clearly known. Various genetic and/or epigenetic factors may contribute to the phenotype variation. A recent meta-analysis have demonstrated that nearly 25% of individuals with homozygous or compound heterozygous SLC34A3 mutations have no rickets/osteomalacia, and 50% of individuals without renal calcifications (nephrolithiasis or nephrocalcinosis), reflecting the heterogeneity in clinical presentations caused by SLC34A3 mutations [45].
In summary, we have systemically investigated genetic defects in Turkish patients with hereditary hypophosphatemia. PHEX mutation is the most common genetic defect accounting for 84% of the disease. De novo PHEX mutations are common and often contribute to the delay in diagnosis and treatment. The early diagnosis of new-born family members may be possible by screening asymptomatic children who have family history of disease-causing mutations or the knowledge of frequent de novo PHEX mutations in the population. This will enable early treatment and a better growth outcome.
Acknowledgments
The authors would like to thank the families involved in this study. This study is supported by a KACST grant 13-MED1765-20.
Data Availability
All relevant data are within the paper.
Funding Statement
The study is supported by a KACST grant 13-MED1765-20 to YS. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Beck-Nielsen SS, Brock-Jacobsen B, Gram J, Brixen K, Jensen TK. Incidence and prevalence of nutritional and hereditary rickets in southern Denmark. Eur J Endocrinol. 2009;160(3):491–7. doi: 10.1530/EJE-08-0818 Epub 2008 Dec 18. [DOI] [PubMed] [Google Scholar]
- 2.Capelli S, Donghi V, Maruca K, Vezzoli G, Corbetta S, Brandi ML, et al. Clinical and molecular heterogeneity in a large series of patients with hypophosphatemic rickets. Bone. 2015;79:143–9. doi: 10.1016/j.bone.2015.05.040 Epub Jun 5. [DOI] [PubMed] [Google Scholar]
- 3.Razali NN, Hwu TT, Thilakavathy K. Phosphate homeostasis and genetic mutations of familial hypophosphatemic rickets. J Pediatr Endocrinol Metab. 2015;28(9–10):1009–17. doi: 10.1515/jpem-2014-0366 [DOI] [PubMed] [Google Scholar]
- 4.Bastepe M, Juppner H. Inherited hypophosphatemic disorders in children and the evolving mechanisms of phosphate regulation. Rev Endocr Metab Disord. 2008;9(2):171–80. doi: 10.1007/s11154-008-9075-3 Epub 2008 Mar 26. [DOI] [PubMed] [Google Scholar]
- 5.Beck-Nielsen SS, Brixen K, Gram J, Brusgaard K. Mutational analysis of PHEX, FGF23, DMP1, SLC34A3 and CLCN5 in patients with hypophosphatemic rickets. J Hum Genet. 2012;57(7):453–8. doi: 10.1038/jhg.2012.56 Epub Jun 14. [DOI] [PubMed] [Google Scholar]
- 6.Carpenter TO. New perspectives on the biology and treatment of X-linked hypophosphatemic rickets. Pediatr Clin North Am. 1997;44(2):443–66. [DOI] [PubMed] [Google Scholar]
- 7.de Menezes Filho H, de Castro LC, Damiani D. Hypophosphatemic rickets and osteomalacia. Arq Bras Endocrinol Metabol. 2006;50(4):802–13. [DOI] [PubMed] [Google Scholar]
- 8.A gene (PEX) with homologies to endopeptidases is mutated in patients with X-linked hypophosphatemic rickets. The HYP Consortium. Nat Genet. 1995;11(2):130–6. doi: 10.1038/ng1095-130 . [DOI] [PubMed] [Google Scholar]
- 9.Strom TM, Juppner H. PHEX, FGF23, DMP1 and beyond. Curr Opin Nephrol Hypertens. 2008;17(4):357–62. doi: 10.1097/MNH.0b013e3282fd6e5b [DOI] [PubMed] [Google Scholar]
- 10.Autosomal dominant hypophosphataemic rickets is associated with mutations in FGF23. Nat Genet. 2000;26(3):345–8. doi: 10.1038/81664 [DOI] [PubMed] [Google Scholar]
- 11.Lorenz-Depiereux B, Bastepe M, Benet-Pages A, Amyere M, Wagenstaller J, Muller-Barth U, et al. DMP1 mutations in autosomal recessive hypophosphatemia implicate a bone matrix protein in the regulation of phosphate homeostasis. Nat Genet. 2006;38(11):1248–50. Epub 2006 Oct 8. doi: 10.1038/ng1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng JQ, Ward LM, Liu S, Lu Y, Xie Y, Yuan B, et al. Loss of DMP1 causes rickets and osteomalacia and identifies a role for osteocytes in mineral metabolism. Nat Genet. 2006;38(11):1310–5. doi: 10.1038/ng1905 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levy-Litan V, Hershkovitz E, Avizov L, Leventhal N, Bercovich D, Chalifa-Caspi V, et al. Autosomal-recessive hypophosphatemic rickets is associated with an inactivation mutation in the ENPP1 gene. Am J Hum Genet. 2010;86(2):273–8. doi: 10.1016/j.ajhg.2010.01.010 Epub Feb 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lorenz-Depiereux B, Schnabel D, Tiosano D, Hausler G, Strom TM. Loss-of-function ENPP1 mutations cause both generalized arterial calcification of infancy and autosomal-recessive hypophosphatemic rickets. Am J Hum Genet. 2010;86(2):267–72. doi: 10.1016/j.ajhg.2010.01.006 Epub Feb 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prie D, Huart V, Bakouh N, Planelles G, Dellis O, Gerard B, et al. Nephrolithiasis and osteoporosis associated with hypophosphatemia caused by mutations in the type 2a sodium-phosphate cotransporter. N Engl J Med. 2002;347(13):983–91. doi: 10.1056/NEJMoa020028 [DOI] [PubMed] [Google Scholar]
- 16.Beck L, Karaplis AC, Amizuka N, Hewson AS, Ozawa H, Tenenhouse HS. Targeted inactivation of Npt2 in mice leads to severe renal phosphate wasting, hypercalciuria, and skeletal abnormalities. Proc Natl Acad Sci U S A. 1998;95(9):5372–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magen D, Berger L, Coady MJ, Ilivitzki A, Militianu D, Tieder M, et al. A loss-of-function mutation in NaPi-IIa and renal Fanconi's syndrome. N Engl J Med. 2010;362(12):1102–9. doi: 10.1056/NEJMoa0905647 [DOI] [PubMed] [Google Scholar]
- 18.Bergwitz C, Roslin NM, Tieder M, Loredo-Osti JC, Bastepe M, Abu-Zahra H, et al. SLC34A3 mutations in patients with hereditary hypophosphatemic rickets with hypercalciuria predict a key role for the sodium-phosphate cotransporter NaPi-IIc in maintaining phosphate homeostasis. Am J Hum Genet. 2006;78(2):179–92. Epub 2005 Dec 9. doi: 10.1086/499409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lorenz-Depiereux B, Benet-Pages A, Eckstein G, Tenenbaum-Rakover Y, Wagenstaller J, Tiosano D, et al. Hereditary hypophosphatemic rickets with hypercalciuria is caused by mutations in the sodium-phosphate cotransporter gene SLC34A3. Am J Hum Genet. 2006;78(2):193–201. Epub 2005 Dec 9. doi: 10.1086/499410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlingmann KP, Ruminska J, Kaufmann M, Dursun I, Patti M, Kranz B, et al. Autosomal-Recessive Mutations in SLC34A1 Encoding Sodium-Phosphate Cotransporter 2A Cause Idiopathic Infantile Hypercalcemia. J Am Soc Nephrol. 2016;27(2):604–14. doi: 10.1681/ASN.2014101025 Epub 2015 Jun 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Streeten EA, Zarbalian K, Damcott CM. CYP24A1 mutations in idiopathic infantile hypercalcemia. N Engl J Med. 2011;365(18):1741–2; author reply 2–3. doi: 10.056/NEJMc1110226#SA2 [DOI] [PubMed] [Google Scholar]
- 22.Fisher SE, van Bakel I, Lloyd SE, Pearce SH, Thakker RV, Craig IW. Cloning and characterization of CLCN5, the human kidney chloride channel gene implicated in Dent disease (an X-linked hereditary nephrolithiasis). Genomics. 1995;29(3):598–606. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto K, Cox JP, Friedrich T, Christie PT, Bald M, Houtman PN, et al. Characterization of renal chloride channel (CLCN5) mutations in Dent's disease. J Am Soc Nephrol. 2000;11(8):1460–8. [DOI] [PubMed] [Google Scholar]
- 24.Hoopes RR Jr., Raja KM, Koich A, Hueber P, Reid R, Knohl SJ, et al. Evidence for genetic heterogeneity in Dent's disease. Kidney Int. 2004;65(5):1615–20. doi: 10.1111/j.1523-1755.2004.00571.x [DOI] [PubMed] [Google Scholar]
- 25.Kato K, Jeanneau C, Tarp MA, Benet-Pages A, Lorenz-Depiereux B, Bennett EP, et al. Polypeptide GalNAc-transferase T3 and familial tumoral calcinosis. Secretion of fibroblast growth factor 23 requires O-glycosylation. J Biol Chem. 2006;281(27):18370–7. Epub 2006 Apr 25. doi: 10.1074/jbc.M602469200 [DOI] [PubMed] [Google Scholar]
- 26.Liu S, Guo R, Simpson LG, Xiao ZS, Burnham CE, Quarles LD. Regulation of fibroblastic growth factor 23 expression but not degradation by PHEX. J Biol Chem. 2003;278(39):37419–26. doi: 10.1074/jbc.M304544200 Epub 2003 Jul 21. [DOI] [PubMed] [Google Scholar]
- 27.Quarles LD. Skeletal secretion of FGF-23 regulates phosphate and vitamin D metabolism. Nat Rev Endocrinol. 2012;8(5):276–86. Epub 2012/01/18. doi: nrendo.2011.218 [pii] doi: 10.1038/nrendo.2011.218 [doi]. PubMed . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amatschek S, Haller M, Oberbauer R. Renal phosphate handling in human—what can we learn from hereditary hypophosphataemias? Eur J Clin Invest. 2010;40(6):552–60. doi: 10.1111/j.1365-2362.2010.02286.x Epub 2010 Apr 14. [DOI] [PubMed] [Google Scholar]
- 29.Bergwitz C, Juppner H. Disorders of phosphate homeostasis and tissue mineralisation. Endocr Dev. 2009;16:133–56. doi: 10.1159/000223693 Epub 2009 Jun 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Durmaz E, Zou M, Al-Rijjal RA, Baitei EY, Hammami S, Bircan I, et al. Novel and de novo PHEX mutations in patients with hypophosphatemic rickets. Bone. 2013;52(1):286–91. doi: 10.1016/j.bone.2012.10.012 Epub Oct 16. [DOI] [PubMed] [Google Scholar]
- 31.Guven A, Al-Rijjal RA, BinEssa HA, Dogan D, Kor Y, Zou M, et al. Mutational analysis of PHEX, FGF23 and CLCN5 in patients with hypophosphataemic rickets. Clin Endocrinol (Oxf). 2017;87(1):103–12. doi: 10.1111/cen.13347 Epub 2017 May 11. [DOI] [PubMed] [Google Scholar]
- 32.Zou M, Bulus D, Al-Rijjal RA, Andiran N, BinEssa H, Kattan WE, et al. Hypophosphatemic rickets caused by a novel splice donor site mutation and activation of two cryptic splice donor sites in the PHEX gene. J Pediatr Endocrinol Metab. 2015;28(1–2):211–6. doi: 10.1515/jpem-2014-0103 [DOI] [PubMed] [Google Scholar]
- 33.Larsson T, Yu X, Davis SI, Draman MS, Mooney SD, Cullen MJ, et al. A novel recessive mutation in fibroblast growth factor-23 causes familial tumoral calcinosis. J Clin Endocrinol Metab. 2005;90(4):2424–7. Epub 005 Feb 1. doi: 10.1210/jc.2004-2238 [DOI] [PubMed] [Google Scholar]
- 34.Ichikawa S, Tuchman S, Padgett LR, Gray AK, Baluarte HJ, Econs MJ. Intronic deletions in the SLC34A3 gene: a cautionary tale for mutation analysis of hereditary hypophosphatemic rickets with hypercalciuria. Bone. 2014;59:53–6. doi: 10.1016/j.bone.2013.10.018 Epub Oct 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zou M, Baitei EY, Al-Rijjal RA, Parhar RS, Al-Mohanna FA, Kimura S, et al. KRAS(G12D)-mediated oncogenic transformation of thyroid follicular cells requires long-term TSH stimulation and is regulated by SPRY1. Lab Invest. 2015;95(11):1269–77. doi: 10.1038/labinvest.2015.90 Epub Jul 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu S, Wei M, Xiao J, Wang CY, Qiu ZQ. [Three PHEX gene mutations in Chinese subjects with hypophosphatemic rickets and literature review]. Zhongguo Dang Dai Er Ke Za Zhi. 2014;16(5):518–23. [PubMed] [Google Scholar]
- 37.Ichikawa S, Traxler EA, Estwick SA, Curry LR, Johnson ML, Sorenson AH, et al. Mutational survey of the PHEX gene in patients with X-linked hypophosphatemic rickets. Bone. 2008;43(4):663–6. doi: 10.1016/j.bone.2008.06.002 Epub Jun 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rowe PS, Oudet CL, Francis F, Sinding C, Pannetier S, Econs MJ, et al. Distribution of mutations in the PEX gene in families with X-linked hypophosphataemic rickets (HYP). Hum Mol Genet. 1997;6(4):539–49. [DOI] [PubMed] [Google Scholar]
- 39.Filisetti D, Ostermann G, von Bredow M, Strom T, Filler G, Ehrich J, et al. Non-random distribution of mutations in the PHEX gene, and under-detected missense mutations at non-conserved residues. Eur J Hum Genet. 1999;7(5):615–9. doi: 10.1038/sj.ejhg.5200341 [DOI] [PubMed] [Google Scholar]
- 40.Francis F, Strom TM, Hennig S, Boddrich A, Lorenz B, Brandau O, et al. Genomic organization of the human PEX gene mutated in X-linked dominant hypophosphatemic rickets. Genome Res. 1997;7(6):573–85. [DOI] [PubMed] [Google Scholar]
- 41.Du L, Desbarats M, Viel J, Glorieux FH, Cawthorn C, Ecarot B. cDNA cloning of the murine Pex gene implicated in X-linked hypophosphatemia and evidence for expression in bone. Genomics. 1996;36(1):22–8. doi: 10.1006/geno.1996.0421 [DOI] [PubMed] [Google Scholar]
- 42.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909 [DOI] [PubMed] [Google Scholar]
- 43.Morey M, Castro-Feijoo L, Barreiro J, Cabanas P, Pombo M, Gil M, et al. Genetic diagnosis of X-linked dominant Hypophosphatemic Rickets in a cohort study: tubular reabsorption of phosphate and 1,25(OH)2D serum levels are associated with PHEX mutation type. BMC Med Genet. 2011;12:116 doi: 10.1186/1471-2350-12-116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu YQ, Bai G, Zhang H, Su D, Tao DC, Yang Y, et al. Human RING finger protein ZNF645 is a novel testis-specific E3 ubiquitin ligase. Asian J Androl. 2010;12(5):658–66. doi: 10.1038/aja.2010.54 Epub Jul 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dasgupta D, Wee MJ, Reyes M, Li Y, Simm PJ, Sharma A, et al. Mutations in SLC34A3/NPT2c are associated with kidney stones and nephrocalcinosis. J Am Soc Nephrol. 2014;25(10):2366–75. doi: 10.1681/ASN.2013101085 Epub 2014 Apr 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.