Abstract
Motivation
Improving the dissemination of information on existing epidemiological studies and facilitating the interoperability of study databases are essential to maximizing the use of resources and accelerating improvements in health. To address this, Maelstrom Research proposes Opal and Mica, two inter-operable open-source software packages providing out-of-the-box solutions for epidemiological data management, harmonization and dissemination.
Implementation
Opal and Mica are two standalone but inter-operable web applications written in Java, JavaScript and PHP. They provide web services and modern user interfaces to access them.
General features
Opal allows users to import, manage, annotate and harmonize study data. Mica is used to build searchable web portals disseminating study and variable metadata. When used conjointly, Mica users can securely query and retrieve summary statistics on geographically dispersed Opal servers in real-time. Integration with the DataSHIELD approach allows conducting more complex federated analyses involving statistical models.
Availability
Opal and Mica are open-source and freely available at [www.obiba.org] under a General Public License (GPL) version 3, and the metadata models and taxonomies that accompany them are available under a Creative Commons licence.
Introduction
To maximize the use of resources and accelerate improvements in health, significant advances are needed in the discoverability and inter-operability of epidemiological study data.1 For one, the seemingly simple task of locating pre-existing data available for research is in fact a significant challenge; information on studies and on the data they collect is often either unavailable or difficult to find. Direct contact with study staff is then necessary to enquire about data availability, which results in a time-intensive process for researchers and study managers alike. At the same time, epidemiological research projects are increasingly drawing on co-analysis across multiple studies to enhance statistical power and improve the precision and robustness of results in a cost-effective manner.2 However, existing studies gather information on individuals using various data collection instruments, data management software and documentation standards, resulting in heterogeneous database systems with disparate formats and models. Large-scale collaborative projects therefore require considerable resource investments to render the information collected by each study compatible and to convert heterogeneous data into compatible formats.
To accelerate the research process while reducing costs associated with it, stakeholders are emphasizing the need for tools that foster the visibility of existing data and enable data compatibility across studies.3–7 In recent years, a number of web portals documenting and disseminating research data in the social,8,9 environmental10 and biomedical sciences11–16 have emerged. Software applications such as ARK,16 REDCap17 and OpenClinica18 support the collection and management of biomedical research data. Other more generic software solutions for research data dissemination have also been developed.19–22 However, little software has been specifically designed to facilitate data harmonization in multi-study projects.23,24 Finally, to our knowledge, no integrated solution exists to address the dual aim of disseminating epidemiological metadata to the research community and supporting data inter-operability across studies.
To address this gap, Maelstrom Research [www.maelstrom-research.org] proposes the OBiBa (Open Source Software for BioBanks) software suite. In this Software Application Profile we present Opal and Mica, two web-based software products that respectively provide out-of-the-box epidemiological data management/harmonization and metadata dissemination solutions for individual epidemiological studies or study consortia. We also show how the full potential of these applications is harnessed when used conjointly; enabling studies and research networks to easily and securely build a federated database system which allows securely querying and analysing data stored on remote servers. Screenshots showing salient parts of each application are presented in Supplementary materials, available as Supplementary data at IJE online.
Implementation: Opal and Mica design
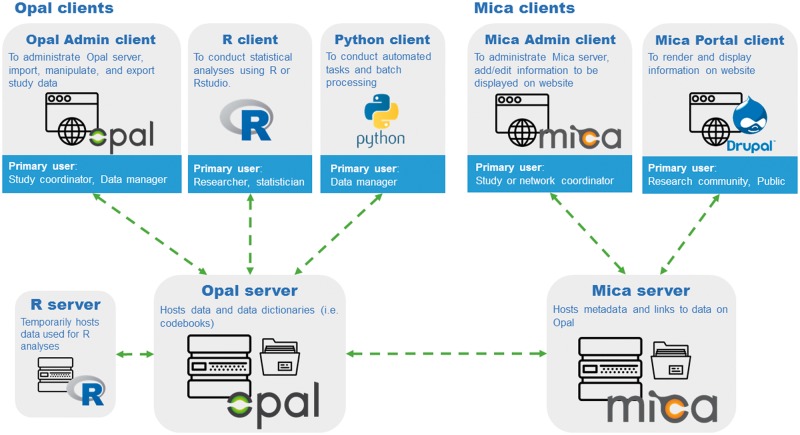
The Opal and Mica software applications are written in Java, JavaScript and PHP languages. Both applications are built upon a RESTful client-server infrastructure, allowing multiple connections between servers and different software web clients using secure HTTPS protocol. As shown in Figure 1, these clients are designed to ensure a broad range of specialized tasks achieved by different types of users. For example, a study/network coordinator or data manager makes use of the Opal and Mica Admin clients’ point-and-click interfaces, to handle day-to-day operations such as server administration, management of user permissions, connections to other applications, data and metadata upload and content editing. Researchers or statisticians use an R client such as RStudio to conduct statistical analyses of study data stored on Opal, and a Python client allows data managers to handle repeatable tasks such as batch processing or periodic data imports. The Mica Portal client renders study metadata and other non-confidential information hosted on the Mica server and publishes this information on a publicly accessible website. The Mica Portal is designed using Drupal, a popular open-source content management system used by websites worldwide. Importantly, individual-level data always remain managed and stored on a secure Opal server, but communication with the Mica server allows extraction of summary statistics to be presented on Mica Portal.
Figure 1.
Opal and Mica software components and relationships.
Client authentication is done through user name and password or by establishing a two-way SSL authentication. Back-end software infrastructure includes Elasticsearch and an R server, which respectively provide a data query engine and an environment to run statistical analyses and reports. MongoDB and/or MySQL are used as database back-end engines for storing study participant data. Opal has no practical limitation related to the number of participants/variables it can store.
Opal: a data management and harmonization tool
Opal provides a centralized web-based data management system allowing study coordinators and data managers to securely import/export a variety of data types (e.g. text, numerical, geolocation, images, videos) and formats (e.g. SPSS, CSV) using a point-and-click interface. Opal then converts, stores and displays these data under a standardized model. Opal can also read data directly from other data source engines such as LimeSurvey or SQL databases. Once data have been imported, the Opal web application facilitates data curation and quality control procedures and allows automated descriptive statistics computation with graphical displays such as bar charts and scatter plots. The application also allows annotating variables with metadata to create rich, searchable data dictionaries. For example, using a variable classification taxonomy developed by Maelstrom Research, each data item collected by a study can be annotated with a standard list of domains of information. This then facilitates metadata browsing and data discoverability using the Mica web portal. Thanks to its integration with the R software and R markdown, statistical analyses and reports on data stored in Opal can also be performed. To ensure privacy, Opal stores participant identifiers in a distinct and secure database and provides administrators with tools to manage them. For example, to de-identify data disseminated to researchers, Opal can create new sets of unique participant identifiers (UIDs) when exporting datasets. Since mapping of UIDs remains in Opal, disseminated UIDs can be re-linked to internal UIDs at import.
When used across multiple studies, Opal is a powerful tool to harmonize epidemiological study data. Opal supports all steps of the Maelstrom Research retrospective data harmonization process.25 As such, Opal includes functionalities to: define variables targeted for harmonization, develop and implement processing algorithms used to derive common-format data, and efficiently document data harmonization decision making. To facilitate algorithm development, Opal also includes a comprehensive JavaScript library of functions commonly used to create harmonized variables. Establishing a secure connection with an R client also allows use of the R programming language to derive common format variables.
Mica: a metadata catalogue and data discovery tool
Mica is used to create websites and metadata portals for individual epidemiological studies or multi-study consortia, with a specific focus on supporting observational cohort studies. The Mica application helps data custodians and study or network coordinators to efficiently organize and disseminate information about their studies and networks without significant technical effort. Mica is made up of a number of different modules developed to add and edit descriptive information pertaining to epidemiological research networks, studies and datasets. The network description module allows Mica users to disseminate information such as network description and the number, design and geographical coverage of participating studies. A study description module is used to assemble and publish information on epidemiological studies such as participant selection criteria, sample size and data collection timelines. A dataset module supports the display and dynamic search of study data dictionaries (i.e. codebooks). For a consortium of studies, a special dataset module also allows documenting and presenting variables targeted for harmonization and harmonization results. A data access module provides customizable web forms and workflows to facilitate online submission and management of data access requests. All Mica modules are customizable, i.e. information fields can be added or modified to meet the specific needs of a study or research network. Further, although the software is preconfigured for English or French, its user interfaces can be translated in any International Organization for Standardization (ISO) language without the need to modify the code. Mica also supports the creation of multilingual websites. A number of other Drupal-based extensions such as events calendars, discussion forums and dissemination tools can be used by studies or research networks to enhance the Mica portal as needed.
Once populated with study and variable metadata, Mica includes a powerful search engine which allows investigators to quickly find the information they need for their research projects. For example, in a Mica instance with only one study, users may use the metadata search features to easily browse the variables it collects. In a Mica instance with multiple studies, users can quickly identify a list of studies with a given profile (e.g. cohorts recruiting middle aged participants), which collect data on a specific health outcome (e.g. depression), risk factors of interest (e.g. physical activity) and confounding factors (e.g. age, sex, income, education, work status). As shown in the subsequent section, connecting Mica to one or more Opal database(s) allows users to search beyond the metadata by securely querying the actual study data hosted on remote servers.
Connecting Opal and Mica: a federated database infrastructure for collaborative epidemiology
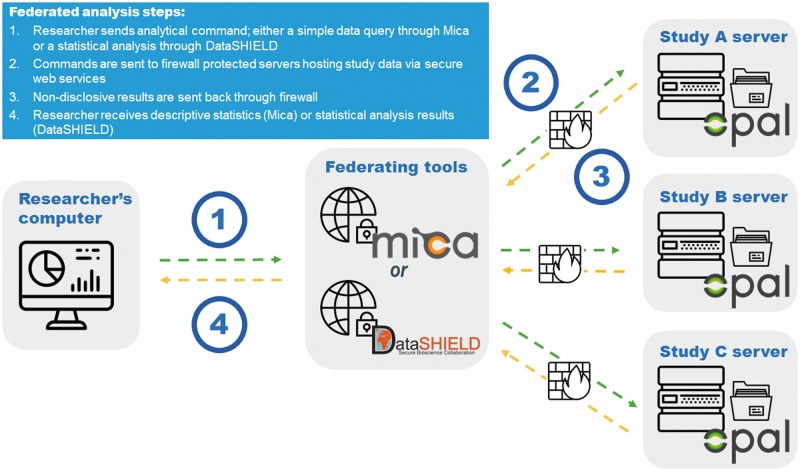
Opal and Mica were developed in parallel as inter-operable and complementary applications. When used conjointly, they allow study networks to create a federated database infrastructure via secure web services. Under such a framework, individual-level data are securely stored in study-specific Opal instances, but can be remotely queried using a Mica-powered web portal. This then gives the research community the ability to retrieve descriptive statistics (e.g. minimum, maximum, mean, standard deviation, counts) and produce contingency tables in real-time across multiple geographically dispersed study databases. Since all potentially disclosive data remain behind the originating institution's firewall, and only aggregate descriptive statistics are sent to the Mica web portal, the privacy and confidentiality of study participants are ensured. More complex analyses involving statistical models or specialized disclosure controls (e.g. for scatter plots) can be applied to harmonized datasets thanks to Opal’s integration with the DataSHIELD statistical analysis approach.26,27 Under DataSHIELD, analysis requests are sent from a central analysis computer to the harmonized data held on several Opal instances. Computation is done simultaneously but in parallel on each Opal server linked by non-disclosive summary statistics. As in the federated Opal-Mica configuration, individual-level data thereby stay at source, under the governance structure and control of the originating study/institution. Figure 2 outlines the steps of a federated data analysis using Opal, Mica and DataSHIELD. Use of these software applications as a federated database infrastructure was first piloted in the BioSHaRE consortium.28 Below are some of the key epidemiological research initiatives that have made use of Opal and Mica.
Figure 2.
Federated database infrastructure using Opal, Mica and DataSHIELD.
Usage case 1: Canadian Longitudinal Study on Aging (CLSA)]
The CLSA29 is a cohort study which is following 50 000 men and women between the ages of 45 and 85 for at least 20 years. To support its data platform, the Opal and Mica software respectively serve as CLSA's data management system and data dissemination portal [https://datapreview.clsa-elcv.ca/datasets]. To date, 10 terabytes of data including questionnaires, magnetic resonance images (MRI), videos and sound tracks collected from 11 assessment centres across Canada are stored, curated and analysed with Opal. Users of the Mica-powered CLSA portal can search variables by domains of information or standardized scales/questionnaires and securely interrogate the data-holding Opal server to retrieve variable distributions in real-time.
Usage case 2: Canadian Partnership for Tomorrow Project (CPTP)
The CPTP30 is a cohort made up of five regional studies following 300 000 Canadians aged 30 to 74. Ensuring compatible data across participating studies was the keystone of the CPTP when it was formed in 2008.30 To facilitate this objective, the Opal software was used to harmonize questionnaire items, physical measurements and biospecimens-related data across regions. A single point of access to these harmonized datasets is provided via CPTP’s Mica-powered data portal [https://portal.partnershipfortomorrow.ca/]. In addition to real-time retrieval of descriptive statistics, the CPTP portal also gives researchers an overview of the harmonization potential across participating studies and data transformation algorithms used to derive common-format variables. Data access requests are also submitted and managed using this online tool.
Usage case 3: InterConnect
InterConnect is an international research network aiming to help explain the difference in the risk of diabetes and obesity between high- and low-risk populations. To facilitate research collaborations, over 100 diabetes-related studies have been documented on InterConnect’s Mica-powered web portal to date. Through a number of exemplar projects addressing research questions of aetiological and public health interest, InterConnect is also making use of the Opal software and DataSHIELD approach to set up a federated database infrastructure and securely co-analyse harmonized datasets across participating studies from around the world, without the need to physically share individual-level data.
Usage case 5: Maelstrom Research
In addition to developing the Mica application, Maelstrom Research and its partner networks have put this software to use by developing the Maelstrom Research Catalogue, a dynamic compendium of epidemiological studies worldwide that provides researchers with a free, user-friendly, web-based solution for data discovery. To date, 174 studies from more than 55 different countries are documented in the Catalogue, 93 of which include fully annotated and searchable metadata on over 660 000 variables. New studies and searchable variable metadata are added to the Catalogue on a weekly basis.
Discussion
The scientific potential of the wealth of data being collected by epidemiological studies is considerable. A higher volume and variety of data can lead to better precision and interpretation of research results. However, much like other domains of scientific enquiry, unlocking the potential of these data requires improving the organization, compatibility and accessibility of research data.31 The Opal and Mica software packages attempt to address some of these challenges by creating a comprehensive data framework fostering and enhancing the use of epidemiological study data.
New Opal and Mica software features are continuously being developed based on user feedback. Example of current developments include support for genotyping data management and dissemination and for handling STATA and SAS data formats and data query improvements (e.g. number of study participants fulfilling multiple criteria). By incorporating new functionalities in software release cycles, they are made available to all current and future users. The Opal and Mica applications are open-source and freely available for download at [www.obiba.org] under a GNU General Public Licence (GPL) version 3. User guides, detailed software/hardware requirements and links to pre-built packages are available at [wiki.obiba.org]. Study and variable metadata models and classification taxonomies developed by Maelstrom Research to accompany the software are available under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International Public License. We encourage new epidemiological studies and networks to make use of these applications to help them meet their data management, harmonization and dissemination needs.
Supplementary Data
Supplementary data are available at IJE online.
Funding
This work was supported by: the Ontario Institute for Cancer Research; the Canadian Partnership Against Cancer; the Canadian Longitudinal Study on Aging; the province of Quebec’s ‘Ministère de l'Économie, de la Science et de l'Innovation’; Génome Québec; the National Institute on Aging [grant agreement P01AG043362]; and the European Union’s Seventh Framework Program through the BioSHaRE [grant agreement 261433], InterConnect [grant agreement 602068] and BBMRI-LPC [grant agreement 313010] projects.
Supplementary Material
Acknowledgements
We would like to thank software developers and scientific staff who have worked on these applications over the past 10 years. We also acknowledge the invaluable feedback we have received from Opal and Mica users over the years. We are also grateful to the referees and the IJE editorial office for their comments and suggestions on this manuscript. Finally, we would like to thank the Canadian Longitudinal Study on Aging for accepting publication of screenshots of their Opal application in the Supplementary materials, available at IJE online.
Conflict of interest: YM owns Epigeny, a company that offers services based on the Opal and Mica software described in this article.
References
- 1. Castillo T, Gregory A, Moore S. et al. Enhancing Discoverability of Public Health and Epidemiology Research Data. London: Wellcome Trust, 2014. [Google Scholar]
- 2. Thompson A. Thinking big: large-scale collaborative research in observational epidemiology. Eur J Epidemiol 2009;24:727–31. [DOI] [PubMed] [Google Scholar]
- 3. Piwowar HA, Becich MJ, Bilofsky H, Crowley RS. Towards a data sharing culture: recommendations for leadership from academic health centers. PLoS Med 2008;5:1315–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Walport M, Brest P. Sharing research data to improve public health. Lancet 2011;377:537–39. [DOI] [PubMed] [Google Scholar]
- 5. Roger VL, Boerwinkle E, Crapo JD. et al. Strategic transformation of population studies: recommendations of the working group on epidemiology and population sciences from the National Heart, Lung, and Blood Advisory Council and Board of External Experts. Am J Epidemiol 2015;181:363–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Murtagh MJ, Turner A, Minion JT, Fay M, Burton PR. International data sharing in practice: new technologies meet old governance. Biopreserv Biobank 2016;14:231–40. [DOI] [PubMed] [Google Scholar]
- 7. Wilkinson MD, Dumontier M, Aalbersberg IJJ. et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 2016;3:160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Interuniversity Consortium for Political and Social Research (ICPSR). ICPSR. 2016. https://www.icpsr.umich.edu/ (19 December 2016, date last accessed). [Google Scholar]
- 9. Consortium of European Social Science Data Archives (CESSDA). CESSDA ERIC. 2017. https://www.cessda.eu (17 August 2017, date last accessed). [Google Scholar]
- 10. Michener WK, Allard S, Budden A. et al. Participatory design of DataONE—Enabling cyberinfrastructure for the biological and environmental sciences. Ecol Inform 2012;11:5–15. [Google Scholar]
- 11. Wichmann H-E, Kuhn KA, Waldenberger M. et al. Comprehensive catalog of European biobanks. Nat Biotechnol 2011;29:795–97. [DOI] [PubMed] [Google Scholar]
- 12. CLOSER. CLOSER - Promoting Excellence in Longitudinal Research. 2016. http://www.closer.ac.uk/ (21 December 2016, date last accessed). [Google Scholar]
- 13. Gehring U, Casas M, Brunekreef B. et al. Environmental exposure assessment in European birth cohorts: results from the ENRIECO project. Environ Health 2013;12:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Centre for Longitudinal Studies. CLS - Centre for Longitudinal Studies. 2016. http://www.cls.ioe.ac.uk/ (21 December 2016, date last accessed). [Google Scholar]
- 15. UK Biobank. UK Biobank Data Showcase. 2016. http://www.ukbiobank.ac.uk/data-showcase/ (2 February 2017, date last accessed). [Google Scholar]
- 16. Bickerstaffe A, Ranaweera T, Endersby T. et al. The Ark: a customizable web-based data management tool for health and medical research. Bioinformatics 2017;33:624–26. [DOI] [PubMed] [Google Scholar]
- 17. Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap) - a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009;42:377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. OpenClinica. Electronic Data Capture, Randomization and Patient Engagement Made Easy. 2017. https://www.openclinica.com/ (25 May 2017, date last accessed). [Google Scholar]
- 19. Crosas M. The Dataverse Network®: An open-source application for sharing, discovering and preserving data. d-lib magazine 2011;17:1.–. [Google Scholar]
- 20. Norwegian Centre For Research Data . Nesstar. 2016. http://www.nesstar.com/ (21 December 2016, date last accessed). [Google Scholar]
- 21. CKAN. CKAN – The Open Source Data Portal Software. 2016. http://ckan.org/ (20 December 2016, date last accessed). [Google Scholar]
- 22. International Household Survey Network (IHSN). NADA Microdata Cataloguing Tool. 2015. http://www.ihsn.org/home/software/nada (19 December 2016, date last accessed). [Google Scholar]
- 23. Pang C, van Enckevort D, de Haan M. et al. MOLGENIS/connect: a system for semi-automatic integration of heterogeneous phenotype data with applications in biobanks. Bioinformatics 2016;32:2176–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Winters K, Netscher S. Proposed standards for variable harmonization documentation and referencing: a case study using QuickCharmStats 1.1. PLoS One 2016;11:e0147795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fortier I, Raina P, Van den Heuvel ER. et al. Maelstrom Research guidelines for rigorous retrospective data harmonization. Int J Epidemiol 2017;46:103–05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wolfson M, Wallace SE, Masca N. et al. DataSHIELD: resolving a conflict in contemporary bioscience - performing a pooled analysis of individual-level data without sharing the data. Int J Epidemiol 2010;39:1372–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gaye A, Marcon Y, Isaeva J. et al. DataSHIELD: taking the analysis to the data, not the data to the analysis. Int J Epidemiol 2014;43:1929–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Doiron D, Burton P, Marcon Y. et al. Data harmonization and federated analysis of population-based studies: the BioSHaRE project. Emerg Themes Epidemiol 2013;10:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Raina PS, Wolfson C, Kirkland SA. et al. The Canadian longitudinal study on aging (CLSA). Can J Aging 2009;28:221–29. [DOI] [PubMed] [Google Scholar]
- 30. Borugian MJ, Robson P, Fortier I. et al. The Canadian Partnership for Tomorrow Project: building a pan-Canadian research platform for disease prevention. CMAJ 2010;182:1197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Staff S. Challenges and opportunities. Science 2011;331:692–93. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.