Abstract
Background
Campylobacteriosis is a major cause of gastroenteritis in the UK, and although 70% of cases are associated with food sources, the remainder are probably associated with wider environmental exposure.
Methods
In order to investigate wider environmental transmission, we conducted a spatio-temporal analysis of the association of human cases of Campylobacter in the Tyne catchment with weather, climate, hydrology and land use. A hydrological model was used to predict surface-water flow in the Tyne catchment over 5 years. We analysed associations between population-adjusted Campylobacter case rate and environmental factors hypothesized to be important in disease using a two-stage modelling framework. First, we investigated associations between temporal variation in case rate in relation to surface-water flow, temperature, evapotranspiration and rainfall, using linear mixed-effects models. Second, we used the random effects for the first model to quantify how spatial variation in static landscape features of soil and land use impacted on the likely differences between subcatchment associations of case rate with the temporal variables.
Results
Population-adjusted Campylobacter case rates were associated with periods of high predicted surface-water flow, and during above average temperatures. Subcatchments with cattle on stagnogley soils, and to a lesser extent sheep plus cattle grazing, had higher Campylobacter case rates.
Conclusions
Areas of stagnogley soils with mixed livestock grazing may be more vulnerable to both Campylobacter spread and exposure during periods of high rainfall, with resultant increased risk of human cases of the disease.
Keywords: Campylobacter, hydrology, livestock, rainfall, soils
Introduction
In the UK, Campylobacter is a major cause of gastroenteritis, and is thought to result in approximately 700 000 cases per annum, leading to health-associated costs in 2009 of over £50 million.1 The number of human cases of disease is strongly seasonal, with peaks in early summer (June) that vary regionally.2 Infection is mainly caused by consumption of contaminated chicken and beef, and chicken has been identified as a particular problem3 with the majority of samples bought at UK supermarkets found to be contaminated by Campylobacter.4 Although the majority of human Campylobacter cases can be linked to food consumption, between 30% and 50% of cases may be a result of infection from the wider environment.5 The survival and distribution of Campylobacter in the environment change with both space and time, and this will interact with how humans are exposed to the organism. However, given that the numbers of reported Campylobacter cases are dominated by infection through food, and eating behaviour changes seasonally6, this makes it harder to detect cases of infection from the wider environment. Seasonal variation in the prevalence of Campylobacter has not been detected in poultry7 or sheep,8 but the amounts of Campylobacter being shed by dairy cattle does change seasonally.9 To understand the epidemiology of Campylobacter therefore requires analyses that include spatial-temporal patterns, livestock management, meteorology and environmental conditions.
Key Messages
Campylobacter is a major cause of gastroenteritis in the UK, with approximately 30% of cases associated with environmental contamination.
We used hydrological and meteorological data in a temporal model, and livestock and soil maps in a spatial model, to assess potential environmental factors affecting human campylobacteriosis.
Warm wet weather, during periods of high surface-water overland flow, combined with cattle grazing on stagnohumic gley soils, were associated with increased Campylobacter case rates.
To understand Campylobacter case rates it is essential to measure the role of environmental factors such as meteorology, hydrology, livestock grazing and soil type.
More information is needed on human behaviours, especially where and when visits are made to the countryside, that may affect the risk of exposure to Campylobacter.
Molecular epidemiological investigations suggest that the spring\early summer peak of Campylobacter infections may be largely due to environmental exposure.2 Complex pathways of primary and secondary interactions occur between Campylobacter reservoirs in soil, water, wild animals and livestock in the countryside,10,11 and whereas there is evidence for a wide distribution of Campylobacter in the environment, the health risks posed for humans remain unclear. Sequence type (ST) analyses of Campylobacter in Cheshire in North West England12 have indicated the particular importance of dairy cattle, and cattle-derived strains were most often isolated in humans (particularly the ST-61 complex). Cattle appear to have generally higher infection levels than sheep, at about 90% for herds and 55% for flocks.3,13Campylobacter deposited in faeces from individual sheep and cattle has been estimated at 102 to 107 colony-forming units/g,9 although this may be focused on a small number of ‘high-shedding’ animals within a herd.3
Campylobacter survival at any point location will depend on both local soil type and local weather conditions. Soil type is of particular importance14,15 as it affects soil moisture, chemistry and infiltration within the soil;16,17 some studies have indicated that Campylobacter is twice as common in clay compared with non-clay soils.18Campylobacter is microaerophilic and is extremely sensitive to desiccation in warm dry weather and UV radiation,19 and it survives better in the environment at temperatures less than 10°C compared with over 25°C 18,20 Some Campylobacter populations, however, also appear to have adaptive tolerance in the field to some environmental stresses.20 Wet conditions (due to soil type and/or weather) will therefore aid not only Campylobacter survival, but also increase its risk of being spread further during subsequent surface-water flow events. Human Campylobacter infection from environmental sources will be the product of two processes,: the presence of the pathogen and the mechanisms leading to human exposure. There remains considerable uncertainty about the role of different environmental factors in determining Campylobacter survival in the landscape, transmission between hosts and infection in humans. The pathogen is likely to be distributed in large quantities across the landscape through manure spreading, grazing livestock and transmission between domesticated animals and wildlife such as badgers, rabbits and wild birds.21,22
Campylobacter shed by livestock can be transported by surface and subsurface-water flows,23 and there is a large literature on predicting hydrological flows in landscapes and attempts to link this research with the distribution of pathogens.24–26 Most catchment-level landscape models of bacterial movement have focused on overland flow,26 although there have been recent attempts to produce models for small catchments which couple both surface and subsurface movement of bacterial pathogens.25,27 Quantifying the links between human disease and water flow is difficult because of differences in spatial and temporal scales: the cases of disease are comparatively infrequent, with delays between infection and reporting, and high-precision predictions of hydrological flow generally work best for small catchments.28 For large catchments such as that of the River Tyne, where spatio-temporal patterns in Campylobacter cases become more obvious, modelling surface flows becomes challenging, and the problems of integrating disease and environmental models operating at very different spatial and temporal scales increase.29 Storm events are likely to increase the amount of overland flow30,31 and are known to be important in the movement of other bacteria such as Escherichia coli.30 The frequency and intensity of these events are likely to increase with climate change, but their impacts on potential spread of Campylobacter are unclear.32
It is clear that any approach to investigate possible environmental factors associated with increased Campylobacter cases must incorporate the spatio-temporal dynamics of factors that might affect exposure, including weather conditions, catchment hydrology (especially surface-water flows that distribute the pathogen), soil types and livestock grazing patterns, while allowing for the vastly different scales of each process. The primary aim of this study was to improve our understanding of the impact and transmission pathways of environmentally acquired Campylobacter cases in the catchment of the River Tyne, and livestock land use, hydrology, soil conditions and meteorology, using a combined spatio-temporal statistical modelling approach. We hypothesize that the numbers of human cases of Campylobacter will be related to temporal variation in weather and hydrology (rainfall, run-off and temperature) which will impact on the distribution of Campylobacter in the environment, and that this will be moderated by spatial variation in livestock production, soil type and meteorology. The research had the following specific objectives:
to quantify the association between occurrences of human cases of Campylobacter and temperature, rainfall and hydrological responses of the study catchment. We refer to this as our temporal model;
and to investigate the association between the occurrence of Campylobacter cases and land use and soil after adjusting for the temporal variation in weather and hydrology. We refer to this as our spatial model.
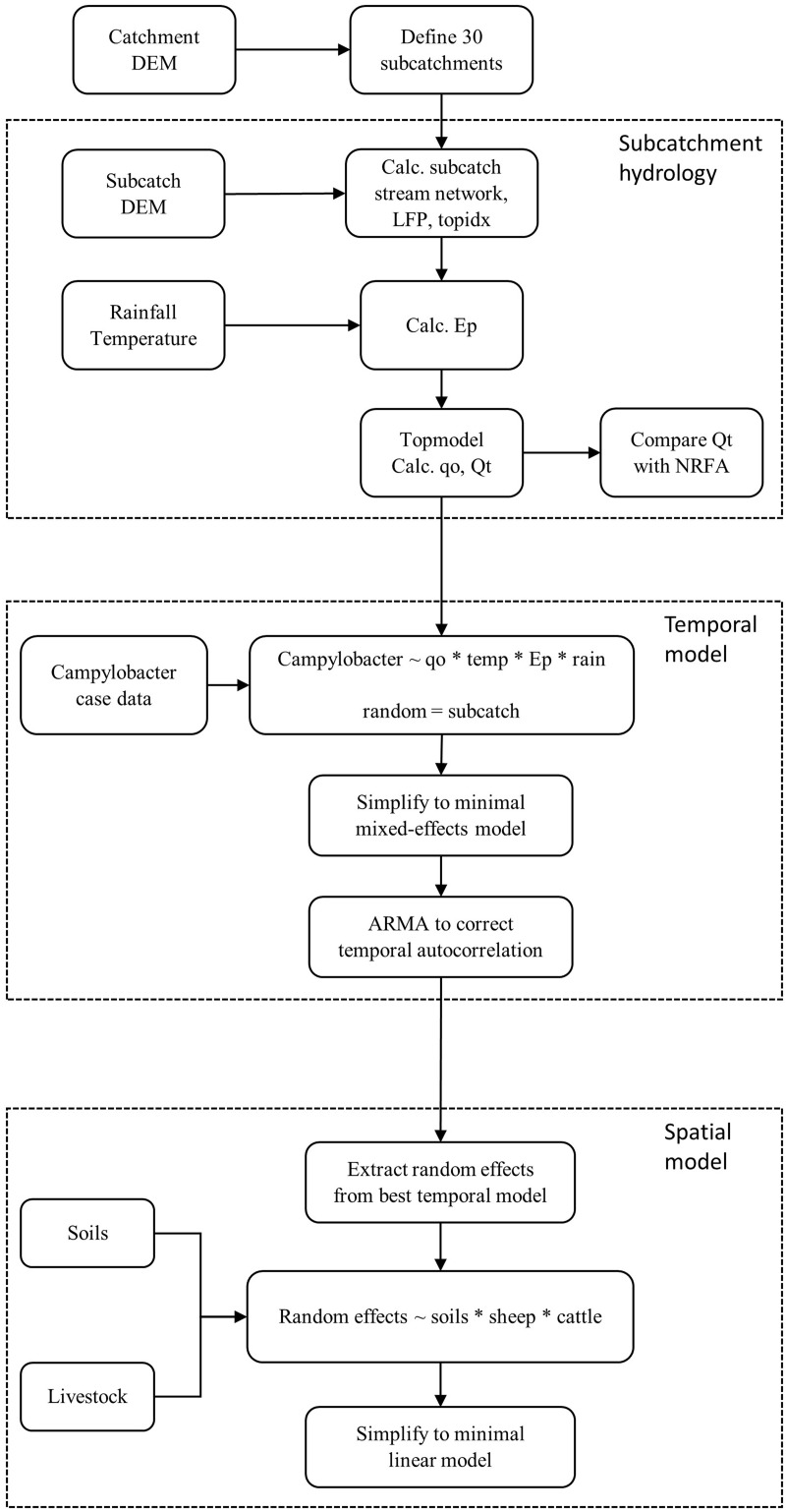
The Topmodel hydrological model33 was initially run for the study catchment and its predictions used as inputs into the temporal model. It investigates Campylobacter case rates in relation to short-term variation in the weather and hydrology across the range of subcatchments. The spatial model quantifies the effects of spatial variation in the underlying landscape (soil and livestock) of these subcatchments with case rates, having already been adjusted for temporal variation in weather and hydrology. The overall structure of the hydrological, temporal and spatial models is summarised in Figure 1.
Figure 1.
Flowchart to summarize procedures used to construct the Topmodel hydrological model, the temporal model of Campylobacter cases and spatial model to relate to soil type, sheep and cattle grazing. (Qt, total flow; qo, overland flow; Ep, evapotranspiration).
Methods
Analyses were undertaken using a combination of Unix shell-scripting to interface with the GRASS geographical information system,34 the Topmodel hydrological model28,35 and the R statistical package.36 One advantage of GRASS, as the geographical information system (GIS) for this type of analysis, is that it integrates well with R.37
Hydrological model
Study area and division into subcatchments
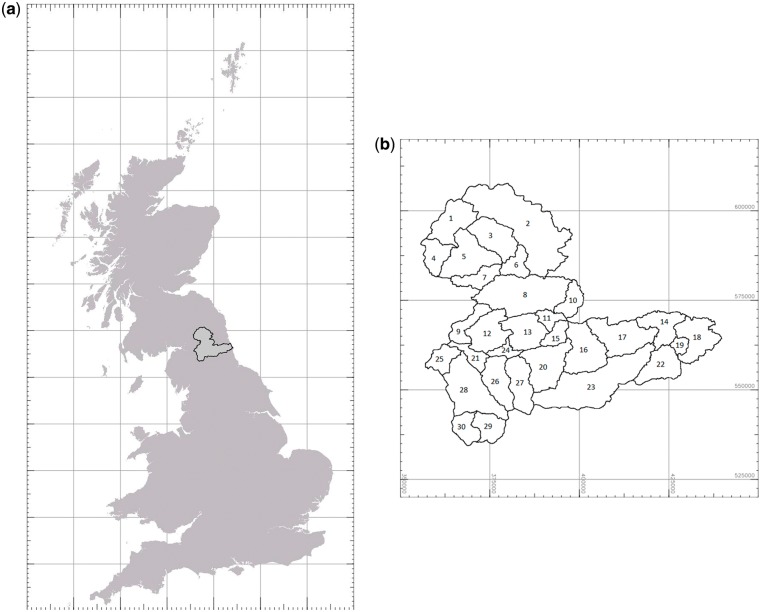
The Tyne catchment is approximately 2944 km2 in North East England (Figure 2). The landscape is highly diverse, ranging from semi-natural wild moorland habitats dominated by upland plant species and sheep grazing (c 1000 km2) through lowland arable (c 275 km2) to the urban sprawl of Tyneside (c 220 km2). Kielder Valley, in the north-west of the catchment, contains Kielder Water, the largest reservoir in the UK at over 10 km2, and part of Kielder Forest (c 380 km2 coniferous plantation within the catchment). The catchment has an altitude range of 0 to 900 m, with average annual rainfall ranging from 630 mm to 1670 mm.
Figure 2.
(a) Location of the Tyne catchment within the UK, with 100-km GB National Grid; and (b) division into 30 subcatchments, with 10-km grid, for use in the analyses.
The Topmodel hydrological model33 was used to predict surface-water overland flow in the Tyne catchment from 1 January 2004 to 31 March 2009, using landscape topology and weather data as inputs. Topmodel breaks down a catchment into a series of subcatchments which are relatively homogeneous hydrologically (it is sometimes described as a ‘semi-distributed’ model). The subcatchments used to divide the main Tyne catchment were derived from the Ordnance Survey 50 -m raster38 map; the number of subcatchments to use is somewhat subjective, but has implications for subsequent analyses. If a large number of small subcatchments is created, there may be insufficient meteorological data within each subcatchment at sufficiently fine resolution to provide inputs for the hydrological models and subsequent analyses. In addition, there may be too few cases of human campylobacteriosis to analyse within small subcatchments. Conversely, if a small number of large subcatchments is created, the hydrological models will become more accurate, but there may be an inadequate number of subcatchments to provide sufficient statistical power to investigate spatial variation in soil type or livestock management effects. We created maps with 20, 30 and 40 subcatchments using the GRASS ‘r.watershed’ command, and undertook identical sets of analyses in each set. In practice, the results of the models were similar for all three sets of subcatchments; for brevity, we only report results from 30 subcatchments in detail in this paper.
Topmodel hydrological model
The Topmodel rainfall-runoff model33 was used to predict surface-water overland flow at a daily time-step within each subcatchment. The two main assumptions which Topmodel uses to relate downslope flow from a point to discharge at the catchment outlet are that:
the dynamics of the saturated zone are approximated by successive steady-state representations;
and the hydraulic gradient of the saturated zone is approximated by the local surface topographic slope.
The model uses a relatively simple relationship between catchment storage and local water table depth, which can be related by the topographic index:39
| (1) |
where a = upslope contributing drainage area above a point and β = local slope angle. High values of topidx generally have large upstream contributing areas and\or shallow slopes such as at the base of hillsides or near streams; low values have small upslope-contributing areas and\or steep slopes.
A Linux bash script was stepped through for each subcatchment, using the appropriate digital elevation maps (DEM) and meteorological data. We used this DEM to derive stream networks in each subcatchment, rather than already published stream networks, to ensure maximal agreement between different inputs into Topmodel, especially measures of longest flow path and topidx values (see below). There were a few problems in accurately defining some stream networks, particularly on lowland or relatively flat areas towards the east of the Tyne catchment, or due to minor errors in the DEM, and these were resolved by running the relevant GRASS commands at a coarser 100 -m grid resolution. Daily meteorological data, provided by the UK Meteorological Office, were obtained for the study area from the British Atmospheric Data Centre (BADC) Meteorological Office Integrated Data Service (MIDAS) website, for the period 1 January 2004 to 31 March 2009.40,41 These data are provided in the form of -km grid resolution raster data. We calculated longest flowpath of a stream network within each subcatchment using the GRASS GIS ‘r.stream’ modules, in particular ‘r.stream.distance’ and ‘r.lfp’. The latter calculates the longest flowpath plus the cumulative drainage area upstream at points along the longest flowpath.35 Topmodel also requires input of the potential evapotranspiration (Ep), which we calculated based on daily temperature, rainfall and latitude for each subcatchment using the method of Xu and Singh.42
Topmodel was run via GRASS ‘r.topmodel’ using the default parameter sets, as the model primarily responds to topography and meteorology and is less sensitive to the exact parameter values used.28,43 Topmodel as implemented in GRASS ‘r.topmodel’ required three main inputs: first, a parameter file, which contained the default parameter sets, plus the distance along the longest flowpath and cumulative upstream area ratios in each subcatchment; second, the map in the subcatchment of topographic index values, topidx, calculated according to Eqn. 1; and third, a file containing rainfall and potential evapotranspiration over time. Our aim was to compare saturation overland flow (qo) predicted by Topmodel across different subcatchments, rather than predict exact overland flow values in any one subcatchment. Output files from the separate Topmodel runs in each subcatchment were automatically concatenated into a single file by the Unix bash script, for ease of use in subsequent analyses in R.
Topmodel outputs were compared with daily flow records from the UK National River Flow Archive (NRFA). The NRFA data do not contain overland flow values, so the most suitable comparison variable is total flow (Qt) which is also output from Topmodel. Nine NRFA gauging stations were operational in the Tyne catchment throughout the study period. Water flow in these generally encompassed flows from multiple upstream subcatchments used in our Topmodel runs (Table 1); therefore daily Qt values for upstream subcatchments were aggregated to coincide with the relevant NRFA areas. Strong annual patterns in both Qt and NRFA flow data were observed; therefore to avoid spurious correlation, each set of data was de-seasonalized using a standard sine-cosine harmonic model:
| (2) |
where y = NRFA flow data or Qt Topmodel predictions, α, β, γ = estimated model coefficients, t = day of year/365.25 and ɛ = residual error.
Table 1.
Summary of major soil types in Tyne catchment according to the Soil Survey of England and Wales (SSEW) classification (excluding urban areas)
| Description | SSEW soil ID code | Area (ha) | Comments |
|---|---|---|---|
| Lithomorphic rankers | 3.1 | 173 | Shallow soils over bedrock |
| Brown calcareous earths | 5.1 | 157 | Agricultural soils, <300 m alt. |
| Brown earths | 5.4 | 19325 | Agricultural soils, <300 m alt. |
| Brown sands | 5.5 | 1248 | Agricultural soils, <300 m alt. |
| Brown alluvial soils | 5.6 | 4872 | Agricultural soils, <300 m alt. |
| Podzols | 6.3 | 557 | Well-drained, acidic soils |
| Stagnopodzols | 6.5 | 9427 | Upland, wet, peaty topsoil |
| Stagnogley soils | 7.1 | 99096 | Seasonally waterlogged, lowland |
| Stagnohumic gley soils | 7.2 | 81467 | Seasonally waterlogged, upland |
| Alluvial gley soils | 8.1 | 582 | Riverine-derived gley |
| Cambic gley soils | 8.3 | 2300 | Subsoil not clay-enriched |
| Disturbed soils | 9.2 | 1514 | Deep cultivation/mining/quarries |
| Raw peat soils | 10.1 | 43987 | Undrained, organic, acidic |
Alt, altitude.
Two separate de-seasonalized models were created (one with NRFA flow data as the response, the other Qt Topmodel predictions). Cross-correlation functions (CCF)44 between the residual errors of these two models were then calculated for a range of lag-distances (days) to determine the overall correlation between the observations and predictions, and any time lag that might have occurred as a result of landscape characteristics.
Temporal model of effects of overland flow, temperature, rainfall and evapotranspiration on Campylobacter cases in subcatchments
The aim of the temporal model was to investigate the temporal variation in the population-adjusted Campylobacter case rate in relation to monthly changes in environmental variables associated with weather and hydrology. This was achieved through linear mixed-effects models (LMMs) with population-adjusted Campylobacter case rate as the response, weather and hydrology as fixed effects and subcatchment as the random effect, using the R package ‘nlme’45 which can also account for temporal autocorrelation.
The number of reported cases of Campylobacter per month in each subcatchment was obtained from the Health Protection Agency in 2010 (now Public Health England); they were available for 63 months, from January 2004 to March 2009. The HPA data provide the six-figure UK residential postcode for each Campylobacter case, and these were converted into Ordnance Survey GB National Grid eastings and northings and imported into the GIS. Cases were then overlaid onto the map of the subcatchments. The Campylobacter case data were log-transformed (corrected by log-transformed population size in each sub-catchment), to create a ‘population-adjusted Campylobacter case rate’ in each subcatchment per month. Topmodel outputs were daily; therefore the mean monthly temperature, rainfall, potential evapotranspiration (Ep) and saturated overland flow (qo) were calculated before comparison with the Campylobacter case data.
A standard mixed-effects model45 can be expressed in matrix formulation as:
| (3) |
| (4) |
| (5) |
where yi = ni x 1 vector of observations in i’th group, Xi = ni x 1 model matrix of fixed-effects regressors for observations in group I, β = p x 1 vector of fixed-effects coefficients, Zi = ni x q matrix of regressors for random effects for observations in group I, bi = p x 1 vector of random effects for group I, ɛi = ni x 1 vector of errors for observations in each group, Ψ = q x q covariance matrix for random effects and σ2Λ = ni x ni covariance matrix for errors in group i.
This formulation gives considerable flexibility in the structure of mixed-effects models. In our study, we used subcatchment as the grouping variable (hence index i varied from 1 to 30). We did not detect evidence of long-term increases or decreases in the population-adjusted Campylobacter case rate during the study period; therefore a random-intercepts, fixed-slopes model was fitted. Initially overland flow, temperature, evapotranspiration and rain were used as predictors, plus all interaction terms, to account for potential collinearity between predictor variables. Non-significant interaction terms were sequentially removed, and where necessary main effects, and each simplified model compared with the previous one. Non-significant main effects were retained in the final model if they were included in a significant interaction term. Model fitting was done via maximum likelihood (ML) rather than the default restricted maximum likelihood (REML), because the fixed effects changed with each model.46 When comparing models, those with a lower Bayesian Information Criterion (BIC) were selected.
After identifying the best linear mixed-effects model, autocorrelation function (ACF) plots were constructed to check for evidence of temporal autocorrelation in the residuals. Initial modelling efforts were focused on identification of the best predictors (i.e. the fixed effects), and then improving these models to account for any temporal autocorrelation.47,48 Autoregressive (AR) moving average (MA) correlation terms were added, testing over a range daily lags for p (autoregressive) and q (moving average) between 0 and 3 to find the optimum values to correct for temporal autocorrelation.49,50 This approach has the advantage of exploring all correction options from pure autoregressive (p > 0 and q = 0), pure moving average (p = 0 and q > 0) or joint autoregressive moving-average (ARMA; p > 0 and q > 0), until the optimal correction is identified, by altering the structure of Λi in the covariance matrix in Eqn. 5 above.
After identification of an appropriate AR, MA or ARMA correlation function (and confirmation of improvement in model fit via BIC and the ACF plots), the random effects (i.e. for each subcatchment) from this model were used as the response variable in the subcatchment analyses as described below.
Spatial model of soil type, sheep and cattle stocking rates on Campylobacter cases
The random effects from the best temporal model quantify differences between the subcatchments in population-adjusted Campylobacter case rates which are not explained by the hydrology, temperature, evapotranspiration or rainfall. These spatial differences between the subcatchments might be due to other environmental factors, in particular soil type and livestock grazing.
Soil data from the Soil Survey of England and Wales (SSEW) maps for northern England51 at 100 -m grid resolution were analysed at the level of the soil group in the SSEW classification. Different soil groups show strong collinearity, and it was not practical to include all the soil groups (plus interactions with livestock) as explanatory variables in the subcatchment models. Therefore, to identify the main patterns of variation in the composition of SSEW soil groups across the subcatchments, the matrix of subcatchments by soil groups was initially analysed using principal components analysis (PCA). The subcatchments by soil groups matrix was Hellinger-transformed52 before the PCA analysis, as the total areas of each soil group within a subcatchment were non-independent. The soil type(s) with the highest correlations with PC1 (and if necessary PC2) were used as predictors in the linear models.
Data on the numbers of livestock derived from the Defra Agricultural Census (2010); these were obtained at 2-km grid resolution.53,54 Total numbers of sheep/km-2 and cattle/km-2 were calculated within GRASS for each subcatchment. There is evidence that sheep and cattle may play slightly different roles in the transmission of Campylobacter to humans;3 therefore they were used as separate predictor variables in the subcatchment analyses.
The linear models used the random effects from the temporal analyses as the response, with explanatory variables of sheep/km-2, cattle/km-2, and the soil type(s) most characteristic of variation between the subcatchments. All main effects and higher-level interactions were initially fitted to the full linear model, which was simplified until a minimal model with lowest BIC was identified.
Results
Hydrological model
Characteristics of subcatchments
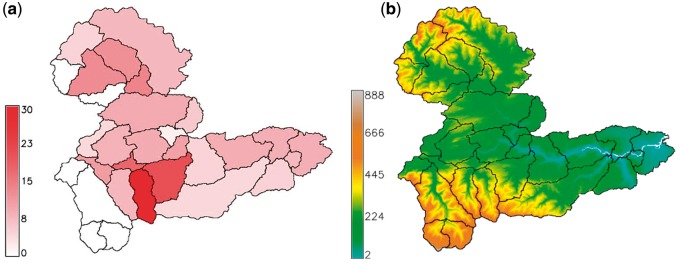
The distributions of the 30 subcatchments are summarized in Figure 2b. The geographical extent of some subcatchments was unchanged irrespective of the number of subcatchments used to subdivide the whole Tyne catchment. These were generally subcatchments in the upland reaches of the Tyne catchment, where the topography was such that there was considerable difference in elevation from river valley to mountain tops, with the result that subcatchments could be clearly defined hydrologically, e.g. the Rede Valley, West and East Allendales: subcatchments 2, 26 and 27, respectively. In contrast, where the topography was flatter, there was more variation in subcatchment area and shape depending on whether 20, 30 or 40 subcatchments were used, e.g. subcatchments 14, 18, 19 and 22 around Tyneside. Highest Campylobacter case rates per head of population were in the Allendales and east of the catchment near Tyneside (Figure 3a). Irrespective of the number of subcatchments used, those in the north-west and south-west of the Tyne catchment were predominantly upland, with higher livestock numbers, compared with the lowland and urbanized eastern end of the catchment (Figure 3b). Soils in the Tyne catchment as a whole are dominated by gley soils with a high clay content, raw peats in the uplands and brown earths on some of the better low-altitude agricultural land (Table 1).
Figure 3.
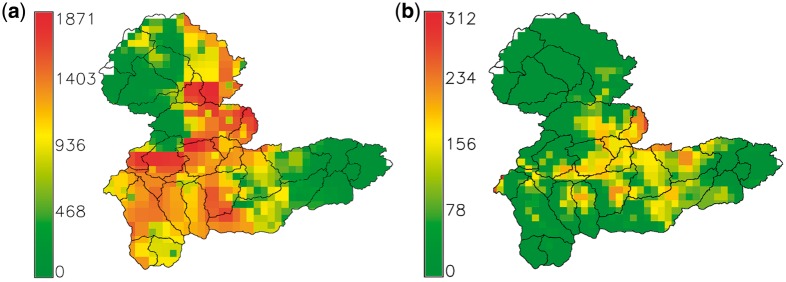
(a) Total number of Campylobacter cases per 1000 population in each subcatchment over whole study period; (b) elevation above sea level in metres.
Topmodel hydrological model
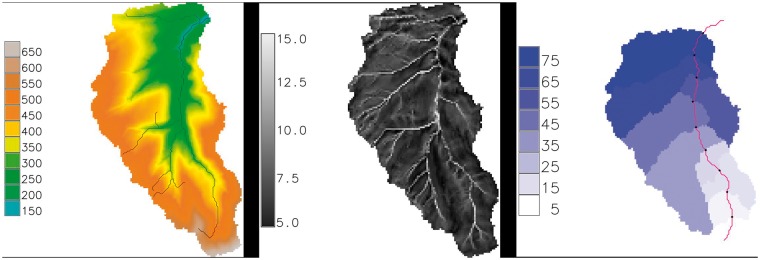
Temperature and rainfall showed strong cyclical patterns on an annual basis, as might be expected, with knock-on effects on predicted evapotranspiration across the whole Tyne region. The saturated overland flow, qo, was less predictable, with large overland flow predicted by Topmodel during the winter of 2006–07. This was associated with a period of intense rainfall, but there were considerable variations between subcatchments in predicted overland flow as a result of between-subcatchment variation in temperature, rainfall and topography. For example, peak overland flow was very high in the south-west of the Tyne catchment in winter 2006–07, particularly around the River West Allen and East Allen, and River South Tyne near Featherstone (subcatchments 26, 27 and 28, respectively, in Figure 2b). In contrast, there was less evidence of any major change in overland flow in the north-west of the catchment during this period, even though this area also experienced higher rainfall. These are subcatchments 1 to 6, around the River Rede, Kielder Valley (including Kielder Water reservoir) and Tarset Burn. Although the north-west is also upland, it has fewer steep-sided valleys than the south-west, which accounts for the lower overland flow. Figure 4 shows more detailed Topmodel outputs for subcatchment 26 (River West Allen): the predicted stream network (Figure 4a); the topidx index, showing higher values near streams (Figure 4b); and the longest flowpath and cumulative drainage basins (Figure 4c).
Figure 4.
Example Topmodel outputs for River West Allen, subcatchment 26. (a) Elevation (metres) and predicted stream network; (b) topographic index scores topidx; (c) longest flow path (solid continuous line) and cumulative drainage basins (drained areas upstream km2).
De-seasonalized NRFA flow data were significantly positively cross-correlated with predicted Topmodel Qt estimates at all nine NRFA gauging stations (Table 1), the majority with a CCF of greater than 0.40 and median maximum CCF at a lag of 4 days. CCF values were lowest for the River Derwent (NRFA station 23007) and Team Valley (NRFA station 23017), both of which are lowland catchments that only partially encompass some of the subcatchments we defined for use in Topmodel and include relatively large urban areas. The highest time lag was 7 days recorded for the North Tyne (NRFA station 23022), but the NRFA warn that flow records from this gauging station are strongly affected by Kielder Reservoir, which is important for human-induced water discharge.
Temporal model of effects of overland flow, temperature, rainfall and evapotranspiration on Campylobacter cases
The population-adjusted case rate was most strongly associated with overland surface-water flow, temperature and the interactions between overland flow and temperature, and between evapotranspiration and rain (Table 3), with case rates predicted to be higher with increased overland flow and in warm weather conditions. There was a spring peak in Campylobacter cases every year in the most densely populated subcatchments around Newcastle and Tynemouth (numbers 14, 17, 18, 19 and 22; Figure 2), but seasonal patterns were less consistent across all years of the study in the other subcatchments.
Table 3.
Summary of results of best temporal linear mixed-effects model, with population-adjusted Campylobacter case rates per month as the response, and subcatchment number as the random effect. qo is saturated surface-water overland flow from Topmodel; Ep is potential evapotranspiration. Standard deviation of residual random effects = 0.3104; log-likelihood = −496.95; BIC = 1099.51
| Coefficient | Value | SE | t-value | P-value |
|---|---|---|---|---|
| Intercept | −6.7822 | 0.3669 | −18.484 | <0.0001 |
| qo | 0.0802 | 0.0308 | 2.609 | 0.0092 |
| Temperature | 0.0702 | 0.0223 | 3.151 | 0.0017 |
| Ep | 0.0105 | 0.0217 | 0.484 | 0.6283 |
| Rain | 0.0102 | 0.0095 | 1.077 | 0.2819 |
| qo x temperature | 0.0776 | 0.0294 | 2.638 | 0.0084 |
| Ep x rain | −0.0204 | 0.0091 | −2.239 | 0.0253 |
SE, standard error.
The minimal linear mixed-effects model showed strong residual autocorrelation, evident at lags 2 and 3 in particular, with an optimum ARMA model at lag p = 3 and moving average q = 2. The need for ARMA models could simply reflect time delays between Campylobacter infection and sample date, since the onset of disease post-exposure is variable (see Discussion). Examination of residuals indicated that the model fitted best to the catchments in the north-west of the Tyne catchment, and most poorly in the south and east of the catchment, especially some of the flatter, lowland urban areas, where it is possible that subcatchment area and extents were less accurately defined, or where the epidemiology of the disease was different.
Spatial model of soil type, sheep and cattle stocking rates on Campylobacter cases
Over 60% of the variation in the composition of SSEW soil groups within each subcatchment was explained by the first PCA axis, which represented a trend from stagnohumic gleys and raw peats through to brown earths and stagnogleys. PCA axis 1 was most strongly positively correlated with the stagnogleys (r = 0.906, P < 0.0001); therefore the area of stagnogley in each subcatchment was used as an explanatory variable in subsequent analyses. In contrast, PCA axis 2 only explained 15% of the variation in SSEW soil composition, was not clearly associated with major soil types and was therefore omitted from subsequent analyses.
We assumed that the random effects from the best temporal model described above quantified the unexplained spatial variation across different subcatchments in the population-adjusted Campylobacter case rate. The subcatchment (linear) models used these random effects as response variables, to determine which environmental variables at the subcatchment level were most associated with deviations from the ‘average’ effect explained by the fixed effects of the temporal model. The amount of stagnogley, cattle density and interactions between the stagnogleys x cattle and sheep x cattle, were found to be the most important environmental variables of the random intercepts in the best subcatchment model (F5,24 = 17.39, Adj-R2 = 0.739, P < 0.0001; Table 4). The signs on the estimated coefficient values in Table 4 indicate that deviations from the predicted case rate from the temporal model were negative for individual subcatchments with larger areas of stagnogley (Figure 6b) and areas of higher numbers of cattle (mid-altitude: Figure 5b). However, the positive cattle x stagnogley interaction (Table 4; t = 2.334, P = 0.0283) suggests that in those subcatchments that contained large cattle numbers grazing stagnogley soils, the temporal model under-predicted Campylobacter case rate. Sheep grazing without cattle was mainly associated with the uplands (excluding the north-west around Kielder Forest), but the positive cattle x sheep interaction (Table 4; t = 2.958, P = 0.0069) suggests higher Campylobacter case rates in those (predominantly lower altitude) subcatchments which contained both sheep and cattle grazing.
Table 4.
Summary of results of best spatial (subcatchment) model of the effects of soil type and livestock management, using the random effects from the best linear mixed-effects model as the response. F5,24 = 17.39; P < 0.0001; Adj-R2 = 0.739
| Coefficient | Value | SE | t-value | P-value |
|---|---|---|---|---|
| Intercept | 4.7730 | 0.6876 | 6.942 | <0.0001 |
| Stagnogley | −7.3675 | 1.0718 | −6.874 | <0.0001 |
| Sheep km−2 | −0.0014 | 0.0023 | −0.619 | 0.5416 |
| Cows km−2 | −0.2618 | 0.0794 | −3.299 | 0.0030 |
| Stagnogley x cows km−2 | 0.2089 | 0.0895 | 2.334 | 0.0283 |
| Sheep km−2 x cows km−2 | 0.0004 | 0.0001 | 2.958 | 0.0069 |
SE, standard error.
Figure 6.
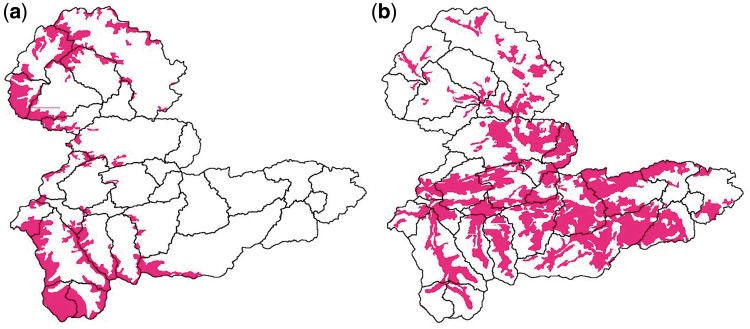
Soil Survey of England and Wales maps of: (a) raw peat soils and (b) stagnogley soils.
Figure 5.
(a) Sheep and (b) cattle grazing in the Tyne catchment based on the DEFRA June 2010 Agricultural Census at 2-km raster scale; units are animals km2.
Discussion
This study has identified associations between population-adjusted Campylobacter cases rates and landscape hydrology, land use and meteorology. Meteorological conditions were important over the 63-month study period, with higher Campylobacter case rates associated with higher temperatures periods of high surface-water overland flow. Subcatchment hydrology was affected by both the weather conditions and topology of each subcatchment. Periods of high surface-water overland flow, in those subcatchments where the topography resulted in increased flow rates, were also associated with higher population-adjusted Campylobacter case rates. Within each subcatchment, stagnogley soils were associated with lower risks of Campylobacter, except in those areas where overall livestock density was high on stagnogleys, when the converse was true. When farm animal grazing type was broken down separately for sheep and cattle; the density of cows grazing on stagnogley soils was an important risk factor, as were larger densities of both sheep and cattle within a subcatchment.
The temporal model indicates that Campylobacter cases were higher during periods of high overland flow. Storm events are known to result in more rapid surface-water transport of bacterial pathogens,55,56 particularly when soils are already saturated. The amount of transport from livestock sources onto adjacent fields, paths and roads will depend partly on the land management, drainage ditches, small scale topography etc. Mole drains will increase the amount of subsurface transport.23 Runoff is generally lower on soils with a large soil pore structure, such as peats\stagnohumic gleys than more compacted clay soils, such as stagnogleys. Our use of Topmodel does not explicitly attempt to model the effects of soil type, land management or livestock management, but our results nevertheless indicate that over the 63-month study period, high overland flow was associated with increased Campylobacter cases. Note that the accuracy of the Topmodel predictions when compared with the NRFA data was unaffected by the predominant soil type in the relevant subcatchments (Table 2); predictions were least accurate in urbanized or flat lowland subcatchments.
Table 2.
Comparison of National Rivers Authority (NRFA) daily flow rates and predicted Topmodel Qt (saturated and overland flow). Cross-correlation functions were calculated on the residuals from the annually de-seasonalized models for NRFA and Qt estimates described in Eqn. 1. The table indicates the maximum CCF values calculated, and the lag (in days) for this maximum. NRFA gauging stations usually measured rivers that encompassed multiple upstream Topmodel subcatchments, and data from the latter were aggregated before comparison
| NRFA station name | NRFA code | Max. CCF | Lag (days) | Upstream Topmodel subcatchments |
|---|---|---|---|---|
| River Tyne, Benwell | 23001 | 0.528 | 4 | 1–13, 15, 20, 21, 24–30 |
| North Tyne, Reaverhill | 23003 | 0.522 | 4 | 1–8 |
| South Tyne, Haydon Bridge | 23004 | 0.463 | 4 | 9, 12, 21, 24, 25, 26–30 |
| South Tyne, Featherstone | 23006 | 0.353 | 4 | 25, 28–30 |
| River Derwent | 23007 | 0.184 | 4 | 23 |
| River Rede | 23008 | 0.429 | 2 | 2 |
| Kielder Burn | 23011 | 0.409 | 3 | 1 |
| Team Valley | 23017 | 0.177 | 1 | 14, 19, 22 |
| North Tyne, Uglydubb | 23022 | 0.365 | 7 | 1, 4, 5 |
Campylobacter does not survive well with drying, and we originally assumed that there would be a negative effect of temperature on Campylobacter case rates; in practice the reverse was found (Table 3), with temperature and the temperature x evapotranspiration interaction associated with higher number of cases. There are several possible explanations for this apparent anomaly. First, some strains of C. jejuni are more resistant to oxidative stresses associated with drought and higher temperatures.57 The markers that encode for the genes thought to be associated with oxidative stress resistance are more common in strains from grazing livestock5 and have been detected at higher frequencies in grazed areas. Second, increased human Campylobacter case rates during the summer may simply reflect greater outdoor activity and exposure of local residents, transmission by wildlife and use of barbeques with a knock-on increased risk of consumption of partially cooked meat (although widespread barbeque use in remote, sheep-rearing, high rainfall areas in the south-west of the Tyne catchment where cases were highest, seems unlikely!). A spring peak in Campylobacter cases was observed across all years in the urban conurbation to the east of the catchment, and to a lesser extent in some, but not all, years in the more rural parts of the west and south-west of the catchment. Again, human behaviour may play a role in this peak, with larger number of visits to the countryside in the spring.
The subcatchment analyses provide insights into spatial, i.e. subcatchment-level, processes that affect Campylobacter case rates, after having adjusted for the weather-related temporal changes over the study period. Stagnogleys are clay soils which have a relatively impervious subsurface horizon and a distinct topsoil, and are prone to waterlogging during periods of heavy rain.58 The negative main-effect coefficient for stagnogleys (Table 4; coefficient = -7.3675; P < 0.0001) indicates that Campylobacter cases were generally lower in those subcatchments, but only if the effects of livestock grazing are ignored (see later). Conversely, raw peats and stagnohumic gley were strongly negatively correlated with soil PCA Axis 1; if either are used instead of stagnogley as the soil predictor in the subcatchment analyses, they are associated with increased Campylobacter cases. Soil-borne pathogens survive better in a more open soil matrix that allows increased penetration into the soil and that contains more organic matter, with reduced desiccation during dry weather,15 such as raw peats and stagnohumic gley. Whereas published data on Campylobacter survival in stagnogleys are not available, survival would be expected to be lower due to the greater tendency of stagnogleys to surface desiccation in dry weather and the lower organic matter content. Stagnogleys are also more productive than stagnohumic gley and peat soils, and probably not subject to human access as frequently because they are more intensively managed.
Subcatchments with high numbers of cattle were associated with decreased Campylobacter case rates, in contrast to the findings of most previous studies 3. Most cattle grazing in the Tyne catchment occurs in lowland areas immediately to the west of the main Newcastle\Gateshead conurbation (Figure 5b; subcatchments 10, 11, 13, 15, 16, 17 and 23). The decrease in Campylobacter case rates with cattle may result from people being less likely to cross fields stocked by cattle, or avoidance of slurry-treated areas and dung-pats. In addition, the risk of human infection within cattle-grazed fields may depend on the number of ‘high-shedding’ animals within the herd, rather than the average proportion of individual cows infected.3
Stagnogley soils are vulnerable to poaching by livestock, especially in wet weather,59 and this can be exacerbated by spatial clustering of livestock within fields, especially around water troughs, feeding areas etc. Poaching will lead to reduced infiltration of water into the soil, but will lead to increased surface-water flow.60 Higher rates of Campylobacter contamination have been recorded on clay soils such as stagnogleys.18 Our results accord with these earlier studies,59,60 with cattle grazing on stagnogley soils being associated with higher Campylobacter case rates (Table 4; stagnogley by cattle interaction). Higher Campylobacter case rates also occurred in subcatchments with both sheep and cattle (Table 4; sheep x cattle interaction). Whereas both sheep and cattle are known to be excretors of Campylobacter,61 our results suggest that cattle are more important (Table 4). Run-off water from sheep and cattle-grazed agricultural land, eventually entering the groundwater drinking supply, has also been identified as a source of human Campylobacter infections in France.62
There have been few attempts to investigate pathogen spread at the catchment scale, with some of the most detailed single studies being undertaken in Australia and New Zealand25,27 under different climatic and agricultural systems from those in the UK. We have used a standard hydrological model that is not excessive in either its input data requirements or CPU run time, to understand temporal changes in meteorological conditions, in combination with surface-water flow predicted from the hydrological model, to understand the changes in Campylobacter cases between 2004 and 2009. Although we did not have sufficient data to undertake more advanced fully integrated spatio-temporal analyses,63 the use of outputs from separate temporal analyses as inputs into subcatchment analyses has allowed us to investigate the relationships between livestock management, soil type and human Campylobacter cases. It is nevertheless clear that the environment has a major role, but the main limitation of this study was that it was not possible to quantify the relative importance of environmental versus food-borne sources of infection. Another challenge is the delay between infection and reporting of cases, which may partly explain the need to use ARMA models; delays from onset of infection to receipt of specimen are on average 5 days in England and Wales6 but is typically 16 days in Scotland,6 based on data collected from 1989 to 1999. Nevertheless, our calculated lag times are slightly less than might be expected a priori. In Campylobacter the incubation period is usually 2–5 days before symptoms arise; and in our data where a doctor could provide an onset date, the median was 4 days, giving an overall delay between infection and specimen of 6 to 9 days. This compares lags of to 2 to 3 days for our ARMA model. The types of analyses we describe would become much more powerful, and useful for public health, if we had had data on the strain types (ST) of the individual human Campylobacter infections, plus samples of Campylobacter taken from the countryside and livestock. Powerful Bayesian methods have recently been developed to source-attribute models, using ST data.64 This would permit a formal link of source to humans, particularly as our results suggest the infection sources change over time and that the risks to human health also depend on human behaviour and activity in the wider countryside.
Funding
This work was supported by the Medical Research Council Grant, the Natural Environment Research Council, the Economic & Social Research Council, the Biotechnology & Biological Sciences Research Council and the Food Standards Agency through the Environmental & Social Ecology of Human Infectious Diseases Initiative (ENIGMA Consortium—study of Campylobacter project: grant reference G1100799/1).
Acknowledgements
We thank other ENIGMA Consortium investigators for advice on this research, including Rob Christley, Christiane Hertz-Fowler, Paul Wigley, Nicola Williams and Craig Winstanley (University of Liverpool), Peter Diggle (Lancaster University), Iain Lake (University of East Anglia), Ken Forbes and Norval Strachan (University of Aberdeen), Rachel Grith and Dan Rigby (University of Manchester), Paul Cross (Bangor University), Tom Humphrey (Swansea University), Malcolm Bennett (University of Nottingham), David Howard (Centre for Ecology and Hydrology) and Brendan Wren (London School of Hygiene and Tropical Medicine).
Conflict of interest: None declared.
References
- 1. Tam CC, O’Brien SJ. Economic cost of Campylobacter, norovirus and rotavirus disease in the United Kingdom. Plos One 2016;11:e0138526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Strachan NJC, Rotariu O, Smith-Palmer A, et al. Identifying the seasonal origins of human campylobacteriosis. Epidemiol Infect 2013;141:1267–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stanley K, Jones K. Cattle and sheep farms as reservoirs of Campylobacter. J Appl Microbiol 2003;94:104–13s. [DOI] [PubMed] [Google Scholar]
- 4. FSA. Food Standards Agency:Campylobacter Survey:Cumulative Results from the Full 12 Months (Q1 - Q4). 2015. https://www.food.gov.uk/news-updates/news/2015/14003/campylobacter-survey-results-12months (9 October 2017, date last accessed). [Google Scholar]
- 5. Champion OL, Gaunt MW, Gundogdu O, et al. Comparative phylogenomics of the food-borne pathogen Campylobacter jejuni reveals genetic markers predictive of infection source. Proc Natl Acad Sci U S A 2005;102:16043–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kovats RS, Edwards SJ, Charron D, et al. Climate variability and campylobacter infection: an international study. Int J Biometeorol 2005;49:207–14. [DOI] [PubMed] [Google Scholar]
- 7. Humphery TJ, Henley A, Lanning DG. The colonization of broiler chickens with Campylobacter jejuni:some epidemiological investigations. Epidemiol Infect 1993;110:601–07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stanley KN, Wallace JS, Currie JE, Diggle PJ, Jones K. Seasonal variation of thermophilic campylobacters in lambs at slaughter. J Appl Microbiol 1998;84:1111–16. [DOI] [PubMed] [Google Scholar]
- 9. Stanley KN, Wallace JS, Currie JE, Diggle PJ, Jones K. The seasonal variation of thermophilic campylobacters in beef cattle, dairy cattle and calves. J Appl Microbiol 1998;85:472–80. [DOI] [PubMed] [Google Scholar]
- 10. Cabrita J, Rodrigues J, Braganca F, Morgado C, Pires I, Gonçalves AP. Prevalence, biotypes, plasmid profile and antimicrobial resistance of Campylobacter isolated from wild and domestic animals from northeast Portugal. J Appl Bacteriol 1992;73:279–85. [DOI] [PubMed] [Google Scholar]
- 11. Bronowski C, James CE, Winstanley C. Role of environmental survival in transmission of Campylobacter jejuni. FEMS Microbiol Lett 2014;356:8–19. [DOI] [PubMed] [Google Scholar]
- 12. Kwan PSL, Barrigas M, Bolton FJ, et al. Molecular epidemiology of Campylobacter jejuni populations in dairy cattle, wildlife, and the environment in a farmland area. Appl Environ Microb 2008;74:5130–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oporto B, Esteban JI, Aduriz G, Juste RA, Hurtado A. Prevalence and strain diversity of thermophilic campylobacters in cattle, sheep and swine farms. J Appl Microbiol 2007;103:977–84. [DOI] [PubMed] [Google Scholar]
- 14. Donnison A, Ross C. Survival and retention of Eschenchia coli O157:H7 and Campylobacter in contrasting soils from the Toenepi catchment. N Z J Agricult Res 2009;52:133–44. [Google Scholar]
- 15. Jamieson RC, Gordon RJ, Sharples KE, Stratton GW, Madani A. Movement and persistence of fecal bacteria in agricultural soils and subsurface drainage water: A review. Canadian Biosystems Engineering 2002;44:1–9. [Google Scholar]
- 16. Sjogren RE. Prolonged survival of an environmental Escherchia coli in laboratory soil microcosms. Water, Air, and Soil Pollution 1994;75:389–403. [Google Scholar]
- 17. Aislabie J, McLeod M, Ryburn J, McGill A, Thornburrow D. Soil type influences the leaching of microbial indicators under natural rainfall following application of dairy shed effluent. Soil Research 2011;49:270–79. [Google Scholar]
- 18. Kemp R, Leatherbarrow AJ, Williams NJ, et al. Prevalence and genetic diversity of Campylobacter spp. in environmental water samples from a 100-square-kilometer predominantly dairy farming area. Appl Environ Microbiol 2005;71:1876–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Butler RC, Lund V, Carlson DA. Susceptibility of Campylobacter jejuni and Yersinia enterocolitica to UV radiation. Appl Environ Microbiol 1987;53:375–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Murphy C, Carroll C, Jordan KN. Environmental survival mechanisms of the foodborne pathogen Campylobacter jejuni. J Appl Microbiol 2006;100:623–32. [DOI] [PubMed] [Google Scholar]
- 21. French N, Barrigas M, Brown P, et al. Spatial epidemiology and natural population structure of Campylobacter jejuni colonizing a farmland ecosystem. Environ Microbiol 2005;7:1116–26. [DOI] [PubMed] [Google Scholar]
- 22. Kwan PSL, Xavier C, Santovenia M, et al. Multilocus sequence typing confirms wild birds as the source of a Campylobacter outbreak associated with the consumption of raw peas. Appl Environ Microb 2014;80:4540–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Oliver DM, Heathwaite L, Haygarth PM, Clegg CD. Transfer of Escherichia coli to Water from Drained and Undrained Grassland after Grazing. J Environ Qual 2005;34:918–25. [DOI] [PubMed] [Google Scholar]
- 24. Dorner SM, Anderson WB, Slawson RM, Kouwen N, Huck PM. Hydrologic Modeling of Pathogen Fate and Transport. Environ Sci Technol 2006;40:4746–53. [DOI] [PubMed] [Google Scholar]
- 25. Haydon S, Deletic A. Development of a coupled pathogen-hydrologic catchment model. J Hydrol 2006;328:467–80. [Google Scholar]
- 26. Jamieson R, Gordon R, Joy D, Lee H. Assessing microbial pollution of rural surface waters: A review of current watershed scale modeling approaches. Agricultural Water Management 2004;70:1–17. [Google Scholar]
- 27. McBride G, Ball A, French N, et al. Campylobacter in Food and the Environment; Examining the Link to Public Health. Wellongton: Ministry of Agriculture and Forestry, 2011. [Google Scholar]
- 28. Beven K. TOPMODEL: A critique. Hydrol Process 1997;11:1069–85. [Google Scholar]
- 29. Oliver DM, Heathwaite AL, Fish RD, et al. Scale appropriate modelling of diffuse microbial pollution from agriculture. Progress in Physical Geography 2009;33:358–77. [Google Scholar]
- 30. McKergow LA, Davies-Colley RJ. Stormflow dynamics and loads of Escherichia coli in a large mixed land use catchment. Hydrol Process 2010;24:276–89. [Google Scholar]
- 31. Sharpley AN, Kleinman PJA, Heathwaite AL, Gburek WJ, Folmar GJ, Schmidt JP. Phosphorus loss from an agricultural watershed as a function of storm size. J Environ Qual 2008;37:362–68. [DOI] [PubMed] [Google Scholar]
- 32. Sterk A, Schijven J, de Roda Husman AM, de Nijs T. Effect of climate change on runoff of Campylobacter and Cryptosporidium from land to surface water. Water Res 2016;95:90–102. [DOI] [PubMed] [Google Scholar]
- 33. Beven K, Lamb R, Quinn P, Romanowicz R, Freer J, Singh VP. Topmodel. Computer models of watershed hydrology 1995:627–68. [Google Scholar]
- 34. Neteler M, Bowman MH, Landa M, Metz M. GRASS GIS: A multi-purpose open source GIS. Environ Modell Softw 2012;31:124–30. [Google Scholar]
- 35. Cho H. GIS hydrological modelling system using programming interface of GRASS. Hydro Laboratory, Kyungpook National University, 2000. [Google Scholar]
- 36. R Core Development Team. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing, 2015. [Google Scholar]
- 37. Bivand RS. Using the R statistical data analysis language on GRASS 5.0 GIS database files. Comput Geosci-UK 2000;26:1043–52. [Google Scholar]
- 38. OS Ordnance Survey MasterMap Topography Layer 1:50, 000.2016. Edina Digimap Ordnance Survey Service: https://digimap.edina.ac.uk/. [Google Scholar]
- 39. Quinn PF, Beven KJ, Lamb R. The ln(a/tan/β) index: How to calculate it and how to use it within the Topmodel framework. Hydrol Process 1995;9:161–82. [Google Scholar]
- 40. BADC: Natural Environment Research Council. British Atmospheric Data Centre. 2016. http://badc.nerc.ac.uk/home/index.html (9 October 2017, date last accessed). [Google Scholar]
- 41. NCAS British Atmospheric Data Centre. MIDAS: Met Office Integrated Data Archive System (MIDAS) Land and Marine Surface Stations Data (1853-current). Exeter, UK: Met Office, 2012. [Google Scholar]
- 42. Xu CY, Singh VP. Evaluation and generalization of temperature-based methods for calculating evaporation. Hydrol Process 2001;15:30–19. [Google Scholar]
- 43. Beven K. Prophecy, reality and uncertainty in distributed hydrological modelling. Advances in Water Resources 1993;16:41–51. [Google Scholar]
- 44. Cryer JD, Chan K-S. Time Series Analysis :With Applications in R. 2nd edn New York, NY: Springer, 2008. [Google Scholar]
- 45. Pinheiro J, Bates D. Mixed-effects Models in S and S-PLUS. New York, NY: Springer Science & Business Media,,2006. [Google Scholar]
- 46. Faraway JJ. Extending the Linear Model With R (Texts in Statistical Science). Boca Raton, FL: Chapman & Hall/CRC, 2005. [Google Scholar]
- 47. Diggle P, Heagerty P, Liang K-Y, Zeger SL. Analysis of Longitudinal Data. Oxford, UK: Oxford University Press, 2013. [Google Scholar]
- 48. Verbeke G, Molenberghs G. Linear Mixed Models for Longitudinal Data. New York, NY: Springer, 2000. [Google Scholar]
- 49. Zuur AF, Ieno EN, Walker NJ, Saveliev AA, Smith GM. Mixed Effects Models and Extensions in Ecology With R. New York, NY: Springer, 2009. [Google Scholar]
- 50. Sanderson RA, Goffe LA, Leifert C. Time-series models to quantify short-term effects of meteorological conditions on bumblebee forager activity in agricultural landscapes. Agr Forest Entomol 2015;17:270–76. [Google Scholar]
- 51. Avery BW. Soil Classification for England and Wales (Higher Categories). Harpenden, UK: Soil Survey, 1980. [Google Scholar]
- 52. Legendre P, Gallagher E. Ecologically meaningful transformations for ordination of species data. Oecologia 2001;129:271–80. [DOI] [PubMed] [Google Scholar]
- 53. DEFRA. June Survey of Agriculture. London: DEFRA, 2010. [Google Scholar]
- 54. Edina Edina Agricultural Census Data. 2017. https://agcensus.edina.ac.uk (9 October 2017, date last accessed). [Google Scholar]
- 55. Haydon S, Deletic A. Sensitivity testing of a coupled Escherichia coli–Hydrologic catchment model. J Hydrol 2007;338:161–73. [Google Scholar]
- 56. Krometis L-AH, Characklis GW, Simmons OD, Dilts MJ, Likirdopulos CA, Sobsey MD. Intra-storm variability in microbial partitioning and microbial loading rates. Water Res 2007;41:506–16. [DOI] [PubMed] [Google Scholar]
- 57. Gundogdu O, Mills DC, Elmi A, Martin MJ, Wren BW, Dorrell N. The Campylobacter jejuni transcriptional regulator CJ1556 plays a role in the oxidative and aerobic stress response and is important for bacterial survival in vivo. J Bacteriol 2011;193:4238–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jarvis RA, Bendelow VC, Bradley RI, et al. Soils and Their Use in Northern England Rothamsted, UK: Lawes Agricultural Trust (Soil Survey of England and Wales), 1984. [Google Scholar]
- 59. Curran Cournane F, McDowell RW, Condron LM. Effects of cattle treading and soil moisture on phosphorus and sediment losses in surface runoff from pasture. N Z J Agric Res 2010;53:365–76. [Google Scholar]
- 60. Trimble SW, Mendel AC. The cow as a geomorphic agent—a critical review. Geomorphology 1995;13:233–53. [Google Scholar]
- 61. Ogden ID, Dallas JF, MacRae M, et al. Campylobacter excreted into the environment by animal sources: prevalence, concentration shed, and host association. Foodborne Pathogens and Disease 2009;6:1161–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gallay A, De Valk H, Cournot M, et al. A large multi-pathogen waterborne community outbreak linked to faecal contamination of a groundwater system, France, 2000. Clin Microbiol Infect 2006;12:561–70. [DOI] [PubMed] [Google Scholar]
- 63. Blangiardo M, Cameletti M, Baio G, Rue H. Spatial and spatio-temporal models with R-INLA. Spat Spatiotemporal Epidemiol 2013;4:3–49. [DOI] [PubMed] [Google Scholar]
- 64. Miller P, Marshall J, French N, Jewell C. Source Attribution: the R package sourceR. 2016. https://cran.r-project.org/web/packages/sourceR/index.html (9 October 2017, date last accessed). [DOI] [PMC free article] [PubMed] [Google Scholar]