Abstract
In conjunction with the second International Environmental Omics Symposium (iEOS) conference, held at the University of Liverpool (United Kingdom) in September 2014, a workshop was held to bring together experts in toxicology and regulatory science from academia, government and industry. The purpose of the workshop was to review the specific roles that high-content omics datasets (eg, transcriptomics, metabolomics, lipidomics, and proteomics) can hold within the adverse outcome pathway (AOP) framework for supporting ecological and human health risk assessments. In light of the growing number of examples of the application of omics data in the context of ecological risk assessment, we considered how omics datasets might continue to support the AOP framework. In particular, the role of omics in identifying potential AOP molecular initiating events and providing supportive evidence of key events at different levels of biological organization and across taxonomic groups was discussed. Areas with potential for short and medium-term breakthroughs were also discussed, such as providing mechanistic evidence to support chemical read-across, providing weight of evidence information for mode of action assignment, understanding biological networks, and developing robust extrapolations of species-sensitivity. Key challenges that need to be addressed were considered, including the need for a cohesive approach towards experimental design, the lack of a mutually agreed framework to quantitatively link genes and pathways to key events, and the need for better interpretation of chemically induced changes at the molecular level. This article was developed to provide an overview of ecological risk assessment process and a perspective on how high content molecular-level datasets can support the future of assessment procedures through the AOP framework.
Keywords: toxicogenomics, methods, regulatory/policy, risk assessment, predictive toxicology, in vitro and alternatives
Within the field of chemical safety and risk assessment, there is a need to assess toxicity of a continuously growing number of chemicals using finite resources while addressing the ethical concerns surrounding the use of reliable animal alternatives (Krewski, et al., 2010; SCENIHR, 2012). There are currently approximately 140 000 chemicals registered for use under the European Inventory of Existing Commercial chemical Substances (ECHA, 2017) and 85 000 chemicals listed on the U.S. EPA Toxic Substances Control Act Inventory (USEPA, 2016a). Improvements in technologies for measuring molecular-level endpoints (eg, at the gene or protein level) have provided an impetus for evaluating the ability to incorporate these measurements into modern-day risk assessment procedures (Cote et al., 2016; Sturla et al., 2014). Guidance documents such as the European Commission Scientific Committee’s publication “Addressing the New Challenges for Risk Assessment” (SCENIHR, 2012) highlight the advantages and challenges of conducting risk assessments using data from molecular and cellular biological processes (NRC, 2007). However, there is still no cohesive approach to the incorporation of molecular-level measurements into ecological or human health risk assessments. One framework that is seen as a promising solution to address this gap is the Adverse Outcome Pathway (AOP) framework. AOPs represent a valuable framework for the evaluation of biologically plausible and empirically supported links between different levels of biological organization, including molecular and biochemical measurements Ankley et al., 2010; Villeneuve et al., 2014a). An AOP represents the linkage between a chemical perturbation, which leads to a molecular initiating event (MIE). Subsequent changes in the physiology of an organism caused by MIEs subsequently lead to adverse outcomes (AOs). AOs occur at the level of the individual or the population, with the possibility of extrapolating towards impacts on the community and/or ecosystem level (Kramer et al., 2011). Within an AOP, MIEs and AOs are linked by a series of measurable and causally connected key events (KEs); ultimately, the application of an AOP requires a clear definition of MIE and early KEs (Madden et al., 2014).
Omics technologies (which are defined as high-content datasets with measurements of genes, proteins, and/or metabolites for the purpose of this article) enable researchers to assess the responses of tens of thousands of genes and their products in a single sample (Aardema and MacGregor, 2002). In combination with advances in system-level data analysis, omics datasets are used to “learn” the structure of biological pathways from observational data (Mitra et al., 2013), an approach that is also being used more frequently within the field of toxicology (eg, McBride, 2017; Perkins et al., 2011; Quercioli et al, 2017; Thomas et al., 2017). The magnitude and scope of omics datasets (ie, The modENCODE Consortium et al., 2010) have raised our expectations of what these data can do for toxicology and risk assessment, specifically by facilitating the identification of molecular-level changes (eg, MIEs and early KEs) that underlie responses to chemical stressors. In the context of the AOP framework, these data can provide a better understanding of potential molecular toxicity pathways as well as effects that occur at basal biological levels of organization (Ankley et al., 2010; Villeneuve et al., 2014a,b). Although omics have been at the foundation of the application and derivation of some novel AOPs (ie, Antczak et al., 2015), it is still not clear how omics datasets should be used in the context of risk assessment and what further research is needed to facilitate the framework’s implementation and acceptance by the risk assessment community.
Previous reviews and workshops have identified some of the challenges in using omics within risk assessments, including the lack of a standardized approach for interpreting gene expression data or quantitatively connecting omics data to a phenotypic outcome (Ankley and Villeneuve, 2006; ECETOC, 2008, 2010; Garcia-Reyero and Perkins, 2011). In this article, we summarize the findings of a workshop held in conjunction with the second international Environmental Omics Symposium (iEOS) at the University of Liverpool (United Kingdom) in September 2014. The goal of this workshop was to identify how omics data can support chemical risk assessments via the AOP framework. We also discuss future challenges and provide a proposed workflow that can be used to achieve improved incorporation of omics data into the AOP framework and the ecological risk assessment (ERA) process.
THE CURRENT STATE OF ECOLOGICAL RISK ASSESSMENT
The process of ERA is defined as an approach for determining the potential impacts of chemical stressors on ecosystems through the examination of chemical effects (defined as the hazard) in the context of a measured or predicted environmental exposure. The standard approach for conducting an ERA is comprised of 4 components: (1) hazard identification, (2) concentration-response assessment, (3) exposure assessment, and (4) risk characterization (REACH, 2006). Detailed methods for conducting an ERA are outlined most recently in the Registration, Evaluation, Authorization, and Restriction (REACH) chemicals legislation (REACH, 2006) and are also derived from earlier documents (USEPA, 1998).
The current approach for assessing hazard consists of 2 key steps: (1) identification of the hazard of concern, and (2) quantification of the hazard via concentration-response measurements in order to derive a toxicity threshold or reference value, typically based on an apical outcome (REACH, 2006; USEPA, 1998). The European Chemicals Agency (ECHA) also provides guidance for additional types of hazard assessment: (1) to establish a chemical’s safe use in a risk assessment (described earlier), and (2) to screen a set of substances in order to determine groups with particular characteristics, which are indicative of a low or high potential hazard. Once chemicals are grouped based on hazard levels, the investigator may focus development on promising candidates for their application or use the information to support testing strategies for safety evaluation (Tollefsen et al., 2014). Alternatively, a regulator may investigate the necessity of further clarification for substances suspected to be of concern (REACH, 2006).
One major challenge of hazard assessment is the requirement to derive an understanding of an effect in a species of relevance to the environment of concern by extrapolating from a standard laboratory species. The uncertainties of such an approach are then addressed by the use of uncertainty factors to provide a margin of safety (Dorato and Engelhardt, 2005). However, this approach was developed before many modern molecular biology tools were established and, in addition to being associated with very great uncertainty, does not meet the growing demand for managing a large number of synthetic compounds in the environment (SCENIHR, 2012). Because of this, scientists have focused on ways in which high-content datasets could be used to provide knowledge of how chemicals actually interfere with biological processes (Sturla et al., 2014).
OMICS AND CHEMICAL RISK ASSESSMENT
There is a growing agreement that omics approaches offer real potential for informing risk assessments when applied as part of an integrated systems biology approach (Cassman, 2005) and when considered in the context of the AOP framework (Van Aggelen et al., 2010). Although to date, omics datasets cannot provide sufficient evidence to characterize risk within an ERA, these datasets do currently have applications for improving our knowledge of toxicological effects.
Using acetylcholine esterase inhibition as an example, transcriptomics data were shown to support known MIEs and their connections to KEs within the AOP framework. These KEs include a build-up of acetylcholine in neural synapses as well as uncontrolled excitation within muscular junctions (Hodges et al., in press; Russom et al., 2014). Microarray data from Caenorhabditis elegans, a nematode model organism used for both ecotoxicology and human health studies, identified impacted pathways related to electron carrier activities and lipid metabolism (Vinuela et al., 2010). These findings also correlated with results from studies conducted in vertebrates such as mice (Mus musculus) and zebrafish (Danio rerio) (Garcia-Reyero et al., 2016; Moreira et al., 2010; Tilton et al., 2011). These studies demonstrate that results from omics datasets can be used to confirm and support the known toxicology or pharmacology of classes of chemicals and can provide supporting evidence of KEs at different levels of biological organization and across different taxonomic groups.
Within the AOP framework, omics datasets also enable a more precise definition of the MIE and the selection of biomarkers relevant for assessing effects and/or exposure. Omics data provide both gene- and pathway-level read-outs (eg, changes in individual gene expression levels or statistical indices for the differential regulation of sets of genes within a specific biological pathway) that can be used for selecting measurable and relevant biomarkers (Gatzidou et al., 2007; Song et al., 2016). In addition, by building upon the omics discoveries made by the broader research community (eg, The modENCODE Consortium et al., 2010), a carefully chosen suite of model test species that represent major evolutionary lineages of animal diversity could allow researchers to gain knowledge for accurately predicting chemical susceptibility in a range of organisms.
Using Omics Datasets for Mode/Mechanism of Action Assessment and Chemical Grouping
One of the methods in aquatic toxicology currently used for assigning a specific mode of action (MOA) to an unknown chemical based on its chemical structure is the Verhaar scheme (Ellison et al., 2015; Enoch et al., 2008; Verhaar et al., 1992, 2000). The recently updated approach places chemicals into one of the following categories: (1) inert compounds causing nonpolar narcosis, (2) less inert/more toxic, compounds causing polar narcosis, (3) reactive compounds with increased toxicity, (4) compounds with specific or receptor-mediated toxicity, and (5) chemicals not able to be classified. Although this scheme is freely available on several platforms such as the Organisation for Economic Co-operation and Development (OECD) quantitative structure-activity relationship toolbox (OECD, 2016) and the Joint Research Center Toxtree application (EURL ECVAM, 2016), it is only useful for acute toxicity endpoints and has a limited ability to assign a chemical class, meaning that many chemicals cannot be classified. Further, the biological domain of applicability is somewhat limited relative to the number of species used to derive the categorization scheme.
Other MOA identification approaches that include a broader number of species are also available (eg, Barron et al., 2015; Russom et al., 1997), but these approaches again are based largely on acute toxicity data and therefore are of limited utility relative to chronic and/or sublethal effects. Consequently, current MOA classifying methods have very limited value in understanding population-relevant endpoints as well as extrapolation to other taxa.
Omics datasets offer promise for providing molecular-level evidence indicative of a specific MOA (Antczak et al., 2015; Fabian et al., 2016; Soetaert et al., 2007a,b; Van Aggelen et al., 2010; Van Ravenzwaay et al., 2016). Both the European Centre for Ecotoxicology and Toxicology of Chemicals (ECETOC) and the US EPA have developed specific guidance for these approaches, including how omics datasets might inform ERA by supporting MOA classification (ECETOC, 2008; USEPA, 2004). Although the use of high-content omics datasets has led to numerous publications on chemical MOAs (De Abrew et al., 2015; Massart et al., 2015; Nookaew et al., 2012; Schmeits et al., 2015; Sohm et al., 2015; Zhang et al., 2016), there is still a scientific requirement to validate these results with additional biochemical or physiological studies. The AOP approach provides a platform that aims to meet this scientific requirement while supporting the use of omics datasets in risk assessment.
Computational Biology Approaches and Their Application Within the AOP Framework
One solution to the challenge of using omics datasets for risk assessment procedures is to incorporate computational biology methodologies into the analysis and application of omics data in AOPs. Although these methods and approaches have been utilized in the human health and biomedical research fields (Macarron et al., 2011; Weston and Hood, 2004), there is still a bottleneck in terms of developing the informatics environments for knowledge management and tailoring these methods to support the AOP framework and for assessing chemical risk (Groh et al., 2015).
The objective of systems biology is to “understand network behaviour, and in particular their dynamic aspects, which requires the utilization of mathematical modelling tightly linked to experimental data (Cassman, 2005). Systems biology can be used in the AOP framework to help understand how an MIE or an early KE could lead to an AO. While these datasets can be powerful scientific tools, the generation of large numbers of high-content molecular-level datasets over the past 10 years has not inherently caused a ‘paradigm shift’ in our understanding of mechanistic toxicology. This is partly because of the need for improved integration of large datasets to enable a better identification of the crucial clusters of molecular responses (eg, network hubs) that are at the centre of chemically induced changes (Davidsen et al., 2016). Computational methods designed to learn the structure of an unknown pathway from observational data (eg, Eren et al., 2012; Perkins et al., 2011) can also be used to combine omics data with phenotypic outcomes and to identify genetic pathways that can explain observed phenotypic outcomes.
Davidsen et al. (2016) describe a framework that applies computational biology approaches towards understanding physiology and proposed 3 major categories of application: biomarker discovery, network inference, and computational modelling. Gene, protein, and metabolite biomarkers can be determined using either univariate or multivariate selection approaches, and have previously been used to identify the most relevant explanatory measurements and their application in toxicology (Gatzidou et al., 2007). Network inference can be conducted using reverse engineering (Perkins et al., 2011); however, other statistical methodologies are also available (Davidsen et al., 2016; D’haeseleer et al., 2000). Ecological modelling is also becoming more widely adopted by the AOP community and mechanistic models such as the dynamic energy budget (DEB) are now more prominent in the field (Forbes et al., 2017; Klanjscek et al., 2013). DEB models detail quantitative relationships between energy intake and key apical processes such as growth, maintenance, and reproduction. Upon future model refinement, it may even be possible to develop genomic indicators for these processes.
Data-driven learning of biological mechanisms also relies on integrating available knowledge in the reasoning process. In this respect, an increasing number of toxicology and omics databases relevant for AOP development and risk assessment are now available. Resources such as the comparative toxicogenomics database, ToxNet, PubChem, ChEMBL, and the connectivity map are just some of the online sources that can be used for chemical interaction and toxicology data for both in vivo and in vitro applications (Schroeder et al., 2016; Fay et al., 2017). In addition, coordinated research programs including ToxCast and Tox21 are working to generate pathway-based biological effects data on large panels of chemicals using high-throughput, commercially available tests (Krewski et al., 2010).
Although ToxCast and Tox21 do not currently focus on generating omics datasets, they do represent coordinated efforts for addressing chemical screening and hazard analysis in a high-throughput manner. Another online resource, the AOP Wiki (Adverse Outcome Pathway Wiki, 2017), serves to organize available toxicology knowledge into a clear conceptual framework. Future integration of features allowing for the upload of experimental results or functional genomics data could aid in the development of a novel AOP in cases where other types of data are scarce.
Research conducted in the field of omics applications has already demonstrated the applicability of omics datasets to generate new, or refine existing, AOPs (Rowlands et al., 2013; Thomas et al., 2013a,b). For example, Antczak et al. (2015) were able to identify a putative mechanism for narcosis toxicity in Daphnia magna linked to calcium signalling. Narcosis, or baseline toxicity, is a MOA that is highly prevalent for industrial chemicals but the mechanism underpinning of narcosis remains unclear. Data generated by Antczak et al. (2015) and researchers at the University of Antwerp supported the acceptance of a proposed AOP by the OECD as part of their AOP development program (Project 1.2: The Adverse Outcome Pathways for Nonpolar Narcosis) (OECD, 2015).
Using Omics Datasets for Species-Sensitivity Extrapolations and Assessing Target Conservation
Current risk assessment approaches rely on extrapolating data from standard laboratory model species to thousands of species of environmental concern. Current species extrapolation approaches tend to be characterized by a large degree of uncertainty due to inherent physiological diversity, life history/trait differences, and a wide range of sensitivities to contaminants (Hecker, 2016; Kramer et al., 2011). AOPs are able to capture information within a defined realm of taxonomic applicability, but this domain may be limited. However, while there are empirically based interspecies correlation estimation tools available (eg, USEPA, 2016b), the general lack of comparative cross-species sensitivity data limits our ability to make robust taxonomic extrapolations in support of ERAs.
One source of inter-species differences in chemical sensitivity arises from changes in molecular targets (ie, potential MIEs) over the course of evolution, in particular macromolecules such as receptors, enzymes, and other functional proteins (Celander et al., 2011; ECETOC, 2008; Gunnarsson et al., 2008; LaLone et al., 2013). Although fundamental biological systems such as reproductive, metabolic, or detoxification pathways are often conserved across species (Hutchinson et al., 2014; Rand-Weaver et al., 2013), small structural or functional variations can lead to substantial differences in chemical sensitivity even in conserved pathways. This was demonstrated in several species of birds and fish, where large differences in the sensitivity to dioxin-like chemicals were driven by seemingly minor amino acid residue differences in the ligand-binding pocket of the arylhydrocarbon receptor (Doering et al., 2014; Karchner et al., 2006).
Information on the sequence and functional homology of a molecular target can provide valuable insights into species sensitivities. However, understanding the impacts of these differences on the MIE and/or KE is necessary to determine differential sensitivity. For example, while the MIE triggered by oestrogen receptor agonists may be highly conserved between oviparous and viviparous animals, the role of KEs such as vitellogenin production represents a critical outcome in one group of species (eg, oviparous vertebrates) but not in another (eg, mammals). In other cases, interspecies differences in sensitivity may arise through changes in less-defined structural features or in physiological processes that are indirectly related to interaction with the target site. This applies in particular to catabolic or metabolic pathways, where differences in toxicokinetics influence the concentrations at biological targets and are the primary drivers in sensitivity (ECETOC, 2008; Escher et al., 2011).
Given the vast inter-species differences present at multiple levels of organization, there is unlikely to be one approach that can effectively characterize species sensitivity on a broad scale. However, the use of omics datasets within well-developed AOPs is a way to develop and validate predictive models for reliable species-sensitivity extrapolations. A tiered approach that includes (1) the use of existing AOP knowledge (eg, available sequence and functional homology information when the MIE and KEs are well-characterized), and (2) the generation of custom de novo gene or protein sequence information can provide a feasible solution to rapidly characterize taxonomic applicability domain of a given toxicity prediction. Expert-curated species similarity maps can then be constructed, with the goal of identifying “forecaster species”, or organisms that can be use as predictors for a range of species in the environment. These similarity maps and forecaster species can then enable a more accurate extrapolation of sensitivity.
This approach, based on the theory of evolution, is similar to the current practice for the risk assessment of dioxin-like contaminants for birds, where species are categorized as chicken-like (highly sensitive), pheasant-like (moderately sensitive), or quail-like (not sensitive) (Karchner et al., 2006). Confirmation of predictors, which can identify drivers of species-specific differences, is proposed using a combination of (1) identification of taxonomic conservation of MIEs based on existing data (eg, LaLone et al., 2016), (2) advanced in silico modeling of MIEs or KEs across species (eg, Madden et al., 2014), (3) the use of targeted high-throughput in vitro assays that follow principles currently used in drug discovery (eg, Doering et al., 2014, 2015), and (4) the development of in ovo tests (eg, Embry et al., 2010). This approach would allow for anchoring to an AOP using more economical and higher-throughput systems while still addressing animal welfare and ethical concerns. Although these sources of information are unable to address all the issues of species differences (eg, life history traits and reproductive strategies), this approach can be considered a means by which species sensitivity can be addressed during a risk assessment.
Additional Experimental Considerations in the AOP Framework: Exposure and Absorption, Distribution, Metabolism, and Excretion
As we move towards pathway-based risk assessments, we need to understand, develop, and utilize tools that can aid in our understanding of the bioavailability of a chemical within an organism (Hubal, 2009). Although considerations of absorption, distribution, metabolism, and excretion (ADME) are commonplace in traditional in vivo toxicity tests, there is currently a void in the derivation and understanding of the role of ADME in omics datasets (Burden et al., 2015). The growing emphasis on the use of in vitro systems raises a key challenge in the need for improved tools to model, measure, and understand the freely available concentration of chemicals in an exposure system (Embry et al., 2014; Pastoor et al., 2014).
Depending on a compound’s physical-chemical properties, there may be variations between the nominal and the freely dissolved concentration that will reach the site of toxic action. These variations, in addition to other factors such as volatility and degradation rates, can arise due to chemical binding to media or in vitro test system constituents (eg, plastics). Having a basic understanding of a chemical’s physicochemical properties, the make-up of an in vitro test system, and how these metrics influence the amount of freely available chemical present in the system are crucial for a more accurate assessment of dose-response relationships (Teeguarden et al., 2016). Although the AOP framework is chemical-independent, the use of exposure information that is dependent on chemical and system properties is an important practical consideration. Approaches such as the Aggregate Exposure Pathway (AEPs), which are aimed at linking exposure and ADME information to that of AOPs, are proposed to overcome some of these obstacles (Teeguarden et al., 2016); however, AEPs are still under development and require further successful demonstration before their widespread use.
CHALLENGES FOR OMICS IN THE AOP FRAMEWORK: REGULATORY ACCEPTANCE
Perhaps one of the biggest challenges facing the application of omics within the AOP concept are the regulatory requirements placed on the generation of data that can be used within hazard and exposure assessment. REACH, for example, sets standards requirements and acceptance criteria for the quality of information and provides guidance on what methods are acceptable for a chemical’s registration (ECHA, 2014a). REACH also actively promotes the use of animal alternative approaches, with an emphasis on chemical read-across, chemical grouping, modelling, and weight of evidence approaches. While there has been an increase in the use of such alternative methods in REACH submission dossiers, current evidence suggests that their application is still limited (ECHA, 2014b).
In the dossiers submitted to ECHA, the use of in vitro assays has been increasing. As an example, the total number of in vitro studies submitted for skin and eye irritation endpoints increased from 442 in 2011 to 1410 in 2013. Currently, the predominant use of in vitro assays is for developing categories and predicting substance properties via read across, which is a consequence of the limited availability of accepted in vitro tests (ECHA, 2014a). The National Academy of Science report “Toxicity in the 21st Century”, provides a vision for how these types of data could be used in the future (NRC, 2007). Although the report states that the current application of in vitro assays cannot be used as direct replacements for the generation of in vivo apical data, recent publications have demonstrated the predictive capabilities of in vitro datasets for apical toxic outcomes, including predictions for toxic mechanisms after binding to nuclear receptors (Drwal et al., 2015) and classifying compounds based on estrogen receptor response data (Norinder and Boyer, 2016). Although there currently is no provision within existing U.S. regulations for how in vitro assays can be used to determine chemical information directly, there is an increasing emphasis on the use of in vitro data in different programs. For example, the USEPA endocrine disruptor screening program has described how a battery of high throughput screening assays can be used to prioritize chemicals for in vivo testing in either mammalian or nonmammalian models (USEPA, 2014, 2016b; Ankley et al., 2016).
Although a direct application of omics datasets to risk assessments remains a long-term goal, currently there are a number of knowledge gaps that can be addressed in the immediate future in terms of how omics can be used as part of the current AOP and risk assessment framework. For example, identifying MIEs and supporting KEs at the molecular levels are places in which omics datasets clearly can be used in the current AOP framework (Patlewicz et al., 2015). Within REACH, there are 2 promising scenarios in which omics datasets can be used during the registration process: chemical read-across and weight of evidence. For read-across applications, omics datasets can be used as supporting mechanistic information for the use of read-across when multiple chemical substances have an overlapping chemical property (Fabian et al., 2016; van Ravenzwaay et al., 2016). Endpoints such as overlapping pathway-level changes between the responses to structurally similar chemicals can be submitted as evidence for the use of read-across during the registration process. A quantitative approach is another way in which the expression of biological pathways can be classified as KEs that can be used for the development of a quantitative AOP (Margiotta-Casaluci et al., 2016). In this approach, effect concentrations can be used as quantitative gatekeepers for progression through the pathway in order to aid prediction of AO in the context of a risk assessment. In other words, a known in vitro or in vivo data concentrations that result in KEs can be used to predict progression through a pathway (Margiotta-Casaluci et al., 2016; Conolly et al., 2017).
For weight of evidence, omics datasets can be used to provide scientific support for assessing the relative robustness of data available (De Coen and Versonnen, 2015). There are also several examples of how omics datasets can be applied to current risk assessment paradigms, including the use of gene set analysis to identify major biological responses after chemical exposure (Bourdon-Lacombe et al., 2015). However, there is still a need for a more complete demonstration of the scientific and practical feasibility of applying these methods directly into human health or ERAs (ECHA, 2016; Margiotta-Casaluci et al., 2016; Tralau and Luch, 2015).
DISCUSSION AND CONCLUSION
The increased ability over the last 10 years to generate high-content omics data has provided exciting opportunities to change the way we assess chemicals and provides an opportunity to bring together elements of traditionally disparate disciplines through a systems biology approach (Sturla et al., 2014). Yet even with the opportunities that omics datasets hold to change in the way that risk assessments are conducted, we have yet to see this shift take place from a regulatory perspective. When compared with the applications of omics datasets in other fields such as biomedicine (Macarron et al., 2011; Weston and Hood, 2004), environmental toxicology and risk assessment applications have fallen behind. This is due in part to the increased complexity of the systems in environmental science (eg, multiple species, fewer well-characterized molecular targets and endpoints, abiotic/confounding environmental factors) as well as a lack of central and comprehensive research efforts focused on the application of omics approaches within risk assessment. Although there has been significant progress resulting in over 100 AOPs present in the AOP Wiki, toxicologists are still working to cohesively and rapidly understand mechanisms of toxicity, identify toxicity pathway and MOA, and improve cross-species extrapolations. There are still hurdles in terms of the widespread use and applications of omics datasets in both the AOP and ERA framework, but the increased efficiency and reduced costs of omics data generation and interpretation will further enable omics to play a significant role in identifying toxicity pathways in the near future
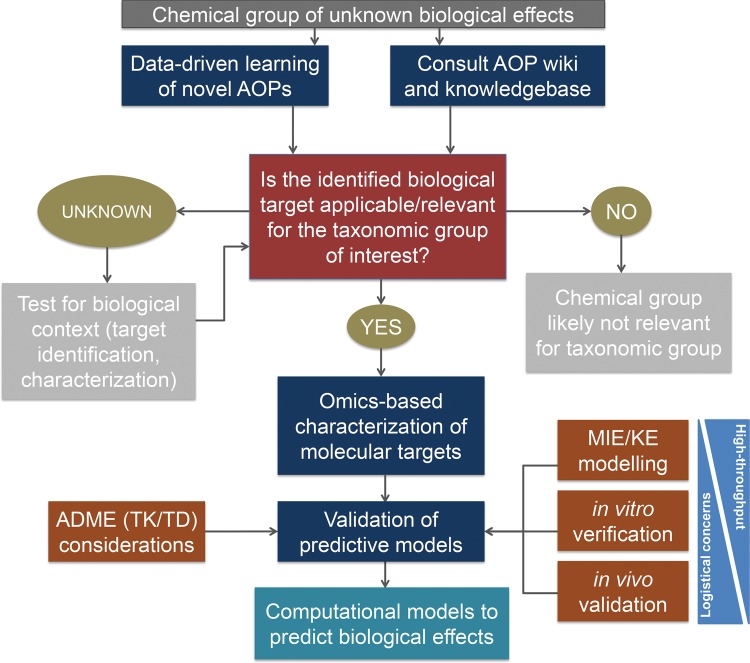
We propose a tiered approach for the coordinated application of omics data and the AOP framework for deriving computational models with the long-term goal of developing data that can support risk assessments (Figure 1). Factors that need to be addressed include (1) the identification of the most relevant pathways for study, (2) the validation of pathway models which can accurately predict biological relevant effects from molecular level perturbations, and (3) strong evidence of a dose-response for the response metric(s) being used. This will require the development of semiquantitative or quantitative relationships between MIEs, intermediate KEs, and AOs (defined as KE relationships) (Wittwehr et al., 2017). In the future, we hope to see omics datasets enhance risk assessment but do not feel that they will completely replace all approaches used in traditional risk assessments.
FIG 1.
Schematic of a proposed tiered approach for the application of omics data and the AOP concept with the ultimate goal to derive computational models that enable assessment of species sensitivity to chemicals of regulatory concern in support of ERA. ADME, absorption, distribution, metabolism, and excretion of a chemical in an organism; KE, key event; MIE, molecular initiating event; TK, toxicokinetics; TD, toxicodynamics.
Another inevitable consequence of the use of large datasets has been the requirement to develop standardized statistical methods for the identification of differentially expressed genes, proteins, or metabolites in response to chemical exposure. This falls in line with suggestions from ECETOC to develop more standardized frameworks for the studies themselves along principles of Good Laboratory Practices, standardized analytical/data processing pipelines and approaches to applying the data for regulatory purposes (ECETOC, 2016). For the field to progress, more complex modelling techniques will be required which include approaches that are able to incorporate complex biological processes such as compensatory homeostatic mechanisms.
Perhaps some of the areas with the strongest potential for short and medium term breakthroughs are providing mechanistic evidence to support chemical read-across, providing weight of evidence in MOA assignment, understanding biological networks, and developing robust species-sensitivity extrapolations. Three key challenges include (1) the need for a cohesive experimental design approach for positively identifying relevant KEs, (2) the integration of a framework for interpreting omics data to identify genes and pathways which are linked to KEs and representative of a pathway or MOA (Ankley et al., 2006), and (3) the need for better interpretation of the causal linkages between chemically induced molecular-level changes to physiological and higher-level effects. In order for omics to have a future within risk assessment, researchers need to demonstrate that the degree of uncertainty generated from omics datasets is at an acceptable level for risk assessment applications, whether it be for screening, prioritizing tests, or for chemical read-across (Perkins et al., 2015).
Despite these challenges, omics datasets can one day be useful in providing supportive evidence for mechanistically driven MOA determination as well as in assessing inter-species sensitivity differences in order to provide the required validation to overcome the regulatory hesitations for accepting these types of evidence. By continuing to focus ongoing research efforts while directly addressing the challenges of applying these omics datasets in the AOP and ERA frameworks, toxicologists can finally enable a data-driven shift in how we assess the safety of a large and growing number of chemicals.
ACKNOWLEDGMENTS
The content of this article is based on the discussions and conclusions made as part of an international expert workshop, “Potential Roles of Omics Data in the use of Adverse Outcome Pathways for Environmental Risk Assessment”, which was held at the University of Liverpool (Liverpool, UK) on September 18th, 2014. Financial support for the workshop was provided by Unilever and was partially supported by a Natural Environment Research Council (NERC) Grant (NE/1028246/2) awarded to Francesco Falciani. The authors would like to acknowledge conference and workshop organising committee member Louise Crompton. All workshop attendees and co-authors approved and contributed to the final version of this manuscript. Opinions, interpretations, conclusions, and recommendations are those of the author(s) and are not necessarily endorsed by the US Environmental Protection Agency or US Army.
REFERENCES
- Aardema M. J., MacGregor J. T. (2002). Toxicology and genetic toxicology in the new era of “toxicogenomics”: Impact of “-omics” technologies. Mut. Res. 499, 13–25. [DOI] [PubMed] [Google Scholar]
- Adverse Outcome Pathway Wiki. (2017). Main Page. Available at: https://aopwiki.org/. Accessed June 7, 2017.
- Ankley G. T., Villeneuve D. L. (2006). The fathead minnow in aquatic toxicology: Past, present and future. Aquat. Toxicol. 78, 91–102. [DOI] [PubMed] [Google Scholar]
- Ankley G. T., Bennett R. S., Erickson R. J., Hoff D. J., Hornung M. W., Johnson R. D., Mount D. R., Nichols J. W., Russom C. L., Schmieder P. K., et al. (2010). Adverse outcome pathways: A conceptual framework to support ecotoxicology research and risk assessment. Environ. Toxicol. Chem. 29, 730–741. [DOI] [PubMed] [Google Scholar]
- Ankley G. T., LaLone C. A., Gray L. E., Villeneuve D. L., Hornung M.W. (2016). Evaluation of the scientific underpinnings for identifying estrogenic chemicals in non-mammalian taxa using mammalian test systems. Environ. Toxicol. Chem. 35, 2806–2816. [DOI] [PubMed] [Google Scholar]
- Antczak P., White T. A., Giri A., Michelangeli F., Viant M. R., Cronin M. T., Vulpe C., Falciani F. (2015). Systems biology approach reveals a calcium-dependent mechanism for basal toxicity in Daphnia magna. Environ. Sci. Technol. 49, 11132–11140. [DOI] [PubMed] [Google Scholar]
- Barron M. G., Lilavois C. R., Martin T. M. (2015). MOAtox: A comprehensive mode of action and acute aquatic toxicity database for predictive model development. Aquat. Toxicol. 161, 102–107. [DOI] [PubMed] [Google Scholar]
- Bourdon-Lacombe J. A., Moffat I. D., Deveau M., Husain M., Auerbach S., Krewski D., Thomas R. S., Bushel P. R., Williams A., Yauk C. L. (2015). Technical guide for applications of gene expression profiling in human health risk assessment of environmental chemicals. Regul. Toxicol. Pharmacol. 72, 292–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burden N., Mahony C., Muller B. P., Terry C., Westmoreland C., Kimber I. (2015). Aligning the 3Rs with new paradigms in the safety assessment of chemicals. Toxicology 330, 62–66. [DOI] [PubMed] [Google Scholar]
- Cassman M. (2005). Barriers to progress in systems biology. Nature 438, 1079.. [DOI] [PubMed] [Google Scholar]
- Celander M. C., Goldstone J. V., Denslow N. D., Iguchi T., Kille P., Meyerhoff R. D., Smith B. A., Hutchinson T. H., Wheeler J. R. (2011). Species extrapolation for the 21st century. Environ. Toxicol. Chem. 30, 52–63. [DOI] [PubMed] [Google Scholar]
- Conolly R. B., Ankley G., Cheng W. Y., Mayo M., Miller D. H., Perkins E., Villeneuve D. L., Watanabe K. H. (2017). Quantitative adverse outcome pathways and their application to predictive toxicology. Environ. Sci. Technol. 51, 4661–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote I., Andersen M. E., Ankley G. T., Barone S., Birnbaum L. S., Boekelheide K., Bois F. Y., Burgoon L. D., Chiu W. A., Crawford-Brown D., et al. (2016). The next generation of risk assessment multiyear study- Highlights of findings and future directions. Environ. Health Perspect. 124, 1671–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidsen P. K., Turan N., Egginton S., Falciani F. (2016). Multilevel functional genomics data integration as a tool for understanding physiology: A network biology perspective. J. Appl. Physiol. 120, 297–309. [DOI] [PubMed] [Google Scholar]
- D’haeseleer P., Liang S., Somogyi R. (2000). Genetic network inference: From co-expression clustering to reverse engineering. Bioinformatics 16, 707–726. [DOI] [PubMed] [Google Scholar]
- De Abrew K. N., Overmann G. J., Adams R. L., Tiesman J. P., Dunavent J., Shan Y. K., Carr G. J., Daston G. P., Naciff J. M. (2015). A novel transcriptomics based in vitro method to compare and predict hepatotoxicity based on mode of action. Toxicology 328, 29–39. [DOI] [PubMed] [Google Scholar]
- De Coen W., Versonnen B. (2015). Opportunities for “Omics” under REACH. Paper presented at: 25th Annual European Society of Environmental Toxicology and Chemistry meeting; May 3–7 2015, Barcelona, Spain.
- Doering J. A., Wiseman S., Beitel S. C., Giesy J. P., Hecker M. (2014). Identification and expression of aryl hydrocarbon receptors (AhR1 and AhR2) provide insight in an evolutionary context regarding sensitivity of white sturgeon (Acipenser transmontanus) to dioxin-like compounds. Aquat. Toxicol. 150, 27–35. [DOI] [PubMed] [Google Scholar]
- Doering J., Farmahin R., Wiseman S., Beitel S., Kennedy S., Giesy J., Hecker M. (2015). Differences in activation of aryl hydrocarbon receptors of white sturgeon relative to lake sturgeon are predicted by identities of key amino acids in the ligand binding domain. Environ. Sci. Technol. 49, 4681–4689. [DOI] [PubMed] [Google Scholar]
- Dorato M. A., Engelhardt J. A. (2005). The no-observed-adverse-effect-level in drug safety evaluations: Use, issues, and definition(s). Regul. Toxicol. Pharmacol. 42, 265–274. [DOI] [PubMed] [Google Scholar]
- Drwal M. N., Siramshetty V. B., Banerjee P., Goede A., Preissner R., Dunkel M. (2015). Molecular similarity-based predictions of the Tox21 screening outcome. Front. Environ. Sci. 3, 54. [Google Scholar]
- ECETOC. (2008). Workshop report 11 on the applications of ‘Omic Technologies in Toxicology and Ecotoxicology: Case Studies and Risk Assessments. Available at: www.ecetoc.org/publication/workshop-report-11-workshop-on-the-application-of-omics-in-toxicology-and-ecotoxicology-case-studies-and-risk-assessment. Accessed August 17, 2016.
- ECETOC. (2010). Workshop report 19 on ‘Omics in (Eco)toxicology: Case studies and Risk assessment. Available at: www.ecetoc.org/publication/workshop-report-19-omics-in-ecotoxicology-case-studies-and-risk-assessment/. Accessed August 17, 2016.
- ECETOC. (2016). Workshop on Applying ‘omics technologies in chemicals risk assessment. Available at: http://www.ecetoc.org/event/ecetoc-workshop-applying-omics-technologies-chemicals-risk-assessment/. Accessed May 1, 2017. [DOI] [PMC free article] [PubMed]
- ECHA (European CHemicals Agency). (2014a). Guidance on Information Requirements and Chemical Safety Assessment Chapter R.7b: Endpoint specific guidance.Available at: echa.europa.eu/guidance-documents/guidance-on-information-requirements-and-chemical-safety-assessment. Accessed August 17, 2016.
- ECHA. (2014b) The Use of Alternatives to Testing on Animals for the REACH Regulation Second report under Article 117(3) of the REACH Regulation. Available at: echa.europa.eu/documents/10162/13639/alternatives_test_animals_2014_en.pdf. Accessed August 17, 2016.
- ECHA. (2017). Guidance for identification and naming of substances under REACH and CLP. Version 2.1, May 2017. Available at: https://echa.europa.eu/documents/10162/23036412/substance_id_en.pdf/ee696bad-49f6-4fec-b8b7-2c3706113c7d. Accessed June 7, 2017.
- ECHA. (2016). New Approach Methodologies in Regulatory Science: Proceedings of a Scientific Workshop, April 2016. Available at: https://echa.europa.eu/documents/10162/21838212/scientific_ws_proceedings_en.pdf/a2087434-0407-4705-9057-95d9c2c2cc57. Accessed October 10.
- Ellison C. M., Madden J. C., Cronin M. T., Enoch S. J. (2015). Investigation of the Verhaar scheme for predicting acute aquatic toxicity: Improving predictions obtained from Toxtree ver. 2.6. Chemosphere 139, 146–154. [DOI] [PubMed] [Google Scholar]
- Embry M. R., Bachman A. N., Bell D. R., Boobis A. R., Cohen S. M., Dellarco M., Dewhurst I. C., Doerrer N. G., Hines R. N., Moretto A., et al. (2014). Risk assessment in the 21st century: Roadmap and matrix. Crit. Rev. Toxicol. 44(Suppl 3), 6–16. [DOI] [PubMed] [Google Scholar]
- Embry M. R., Belanger S. E., Braunbeck T. A., Galay-Burgos M., Halder M., Hinton D. E., Leonard M. A., Lillicrap A., Norberg-King T., Whale G. (2010). The fish embryo toxicity test as an animal alternative method in hazard and risk assessment and scientific research. Aquat. Toxicol. 97, 79–87. [DOI] [PubMed] [Google Scholar]
- Enoch S. J., Hewitt M., Cronin M. T., Azam S., Madden J. C. (2008). Classification of chemicals according to mechanism of aquatic toxicity: An evaluation of the implementation of the Verhaar scheme in Toxtree. Chemosphere 73, 243–248. [DOI] [PubMed] [Google Scholar]
- Eren K., Deveci M., Kucuktunc O., Catalyurek U. V. (2012). A comparative analysis of biclustering algorithms for gene expression data. Brief. Bioinformatics 14, 279–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escher B. I., Ashauer R., Dyer S., Hermens J. L., Lee J. H., Leslie H. A., Mayer P., Meador J. P., Warne M. S. (2011). Crucial role of mechanisms and modes of toxic action for understanding tissue residue toxicity and internal effect concentrations of organic chemicals. Integr. Environ. Assess. Manag. 7, 28–49. [DOI] [PubMed] [Google Scholar]
- EURL ECVAM (2016). Toxtree. Available at: https://eurl-ecvam.jrc.ec.europa.eu/laboratories-research/predictive_toxicology/qsar_tools/toxtree. Accessed May 1, 2017.
- Fabian E., Bordag N., Herold M., Kamp H., Krennrich G., Looser R., Ma-Hock L., Mellert W., Montoya G., Peter E., et al. (2016). Metabolite profiles of rats in repeated dose toxicological studies after oral and inhalative exposure. Toxicol. Lett. 255, 11–23. [DOI] [PubMed] [Google Scholar]
- Fay K. A., Villeneuve D. L., LaLone C. A., Song Y., Tollefsen K.-E., Ankley G. T. (2017). Practical approaches to adverse outcome pathway (AOP) development and weight-of-evidence evaluation as illustrated by ecotoxicological case studies. Environ. Toxicol. Chem. In Press, doi: 10.1002/etc.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes V. E., Salice C. J., Birnir B., Bruins R. J. F., Calow P., Ducrot V., Galic N., Garber K., Harvey B. C., Jager H., et al. (2017). A framework for predicting impacts on ecosystem services from (sub)organismal responses to chemicals. Environ. Toxicol. Chem. 36, 845–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Reyero N., Perkins E. J. (2011). Systems biology: Leading the revolution in ecotoxicology. Environ. Toxicol. Chem. 30, 265–273. [DOI] [PubMed] [Google Scholar]
- Garcia-Reyero N., Escalon L., Prats E., Faria M., Soares A. M., Raldúa D. (2016). Targeted Gene Expression in Zebrafish Exposed to Chlorpyrifos-Oxon Confirms Phenotype-Specific Mechanisms Leading to Adverse Outcomes. Bull. Environ. Contam. Toxicol. 96, 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatzidou E. T., Zira A. N., Theocharis S. E. (2007). Toxicogenomics: A pivotal piece in the puzzle of toxicological research. Journal of Applied Toxicology 27, 302–309. [DOI] [PubMed] [Google Scholar]
- Groh K. J., Carvalho R. N., Chipman J. K., Denslow N. D., Halder M., Murphy C. A., Roelofs D., Rolaki A., Shirmer K., Watanabe K. H. (2015). Development and application of the adverse outcome pathway framework for understanding and predicting chronic toxicity: I. Challenges and research needs in ecotoxicology. Chemosphere 120, 764–777. [DOI] [PubMed] [Google Scholar]
- Gunnarsson L., Jauhiainen A., Kristiansson E., Nerman O., Larsson D. G. (2008). Evolutionary conservation of human drug targets in organisms used for environmental risk assessments. Environ. Sci. Technol. 42, 5807–5813. [DOI] [PubMed] [Google Scholar]
- Hecker M. (2016). Non-Model Species in Ecological Risk Assessment In A Systems Biology Approach to Advancing Adverse Outcome Pathways for Risk Assessment (Murphy C., Reyero N., Eds.), Springer, New York, USA. [Google Scholar]
- Hodges G., Gutsell S., Taylor N., Brockmeier E. K., Butler E., Rendal C., Colbourne J. In press. Invertebrate Model species in AOP development In A Systems Biology Approach to Advancing Adverse Outcome Pathways for Risk Assessment (Garcia-Reyero N., Murphy C., Eds.). Springer, New York, NY. [Google Scholar]
- Hubal E. A. (2009). Biologically relevant exposure science for 21st century toxicity testing. Toxicol. Sci. 111, 226–232. [DOI] [PubMed] [Google Scholar]
- Hutchinson T. H., Madden J. C., Naidoo V., Walker C. H. (2014). Comparative metabolism as a key driver of wildlife species sensitivity to human and veterinary pharmaceuticals. Philos. Trans. R Soc. Lond. B Biol. Sci. 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karchner S. I., Franks D. G., Kennedy S. W., Hahn M. E. (2006). The molecular basis for differential dioxin sensitivity in birds: Role of the aryl hydrocarbon receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 6252–6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klanjscek T., Nisbet R. M., Priester J. H., Holden P. A. (2013). Dynamic energy budget approach to modeling mechanisms of CdSe quantum dot toxicity. Ecotoxicology 22, 319–330. [DOI] [PubMed] [Google Scholar]
- Kramer V. J., Etterson M. A., Hecker M., Murphy C. A., Roesijadi G., Spade D. J., Spromberg J. A., Wang M., Ankley G. T. (2011). Adverse outcome pathways and ecological risk assessment: Bridging to population-level effects. Environ. Toxicol. Chem. 30, 64–76. [DOI] [PubMed] [Google Scholar]
- Krewski D., Acosta D. Jr, Andersen M., Anderson H., Bailar J. C. 3rd, Boekelheide K., Brent R., Charnley G., Cheung V. G., Green S. Jr, et al. (2010). Toxicity testing in the 21st century: A vision and a strategy. Journal of Toxicol. Environ. Health B Crit. Rev. 13, 51–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalone C. A., Villeneuve D. L., Burgoon L. D., Russom C. L., Helgen H. W., Berninger J. P., Tietge J. E., Severson M. N., Cavallin J. E., Ankley G. T. (2013). Molecular target sequence similarity as a basis for species extrapolation to assess the ecological risk of chemicals with known modes of action. Aquat. Toxicol. 144–145. [DOI] [PubMed] [Google Scholar]
- LaLone C. A., Villeneuve D. L., Lyons D. L., Helgen H. W. D., Robinson S. L. H. W., Swintek J. A. S. L., Saari T.W.J.A., G.,T.W., Ankley G. T. (2016). Sequence alignment to predict across species susceptibility (SeqAPASS): A web-based tool for addressing the challenges of species extrapolation of chemical toxicity. Tox. Sci. 153, 228–245. [DOI] [PubMed] [Google Scholar]
- Macarron R., Banks M. N., Bojanic D., Burns D. J., Cirovic D. A., Garyantes T., Green D. V. S., Hertzberg R. P., Janzen W. P., Paslay J. W., et al. (2011). Impact of high-throughput screening in biomedical research. Nat. Rev. Drug Discov. 10, 188–195. [DOI] [PubMed] [Google Scholar]
- Madden J. C., Rogiers V., Vinken M. (2014). Application of in silico and in vitro methods in the development of adverse outcome pathway constructs in wildlife. Philos. Trans. R Soc. Lond. B Biol. Sci. 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margiotta-Casaluci L., Owen S. F., Huerta B., Rodríguez-Mozaz S., Kugathas S., Barceló D., Rand-Weaver M., Sumpter J. P. (2016). Internal exposure dynamics drive the Adverse Outcome Pathways of synthetic glucocorticoids in fish. Sci. Rep. 6, 21978.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massart S., Perazzolli M., Höfte M., Pertot I., Jijakli M. H. (2015). Impact of the omic technologies for understanding the modes of action of biological control agents against plant pathogens. BioControl 60, 725–746. [Google Scholar]
- McBride M. T. (2017). Future platforms for toxicity testing. International Journal of Risk Assessment and Management 20, 59–87. [Google Scholar]
- Mitra K., Carvunis A.-R., Ramesh S. K., Ideker T. (2013). Integrative approaches for finding modular structure in biological networks. Nat. Rev. Genet. 14, 719–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira E. G., Yu X., Robinson J. F., Griffith W., Hong S. W., Beyer R. P., Bammler T. K., Faustman E. M. (2010). Toxicogenomic profiling in maternal and fetal rodent brains following gestational exposure to chlorpyrifos. Toxicol. Appl. Pharmacol. 245, 310–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council of the National Academies. (2007). Toxicity Testing in the 21st Century: A Vision and a Strategy. Washington, DC 20001: National Academies Press. DOI 10.17226/11970. [Google Scholar]
- Nookaew I., Papini M., Pornputtapong N., Scalcinati G., Fagerberg L., Uhlen M., Nielsen J. (2012). A comprehensive comparison of RNA-Seq-based transcriptome analysis from reads to differential gene expression and cross-comparison with microarrays: A case study in Saccharomyces cerevisiae. Nucleic Acids Res. 40, 10084–10097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norinder U., Boyer S. (2016). Conformal Prediction Classification of a Large Data Set of Environmental Chemicals from ToxCast and Tox21 Estrogen Receptor Assays. Chem. Res. Toxicol. 29, 1003–1010. [DOI] [PubMed] [Google Scholar]
- OECD. (2015). Lists of projects on the AOP development programme workplan. Available at: http://www.oecd.org/chemicalsafety/testing/projects-adverse-outcome-pathways.htm. Accessed June 7, 2017.
- OECD. (2016). The OECD QSAR Toolbox. Available at: http://www.oecd.org/chemicalsafety/risk-assessment/theoecdqsartoolbox.htm. Accessed November 25, 2016.
- Pastoor T. P., Bachman A. N., Bell D. R., Cohen S. M., Dellarco M., Dewhurst I. C., Doe J. E., Doerrer N. G., Embry M. R., Hines R. N., et al. (2014). A 21st century roadmap for human health risk assessment. Crit. Rev. Toxicol. 44(Suppl 3), 1–5. [DOI] [PubMed] [Google Scholar]
- Patlewicz G., Simon T. W., Rowlands J. C., Budinsky R. A., Becker R. A. (2015). Proposing a scientific confidence framework to help support the application of adverse outcome pathways for regulatory purposes. Regul. Toxicol. Pharmacol. 71, 463–477. [DOI] [PubMed] [Google Scholar]
- Perkins E. J., Chipman J. K., Edwards S., Habib T., Falciani F., Taylor R., Van Aggelen G., Vulpe C., Antczak P., Loguinov A. (2011). Reverse engineering adverse outcome pathways. Environ Toxicol Chem 30, 22–38. [DOI] [PubMed] [Google Scholar]
- Perkins E., Garcia-Reyero N., Edwards S., Wittwehr C., Villeneuve D., Lyons D., Ankley G. (2015). The adverse outcome pathway: A conceptual framework to support toxicity testing in the twenty-first century In Computational Systems Toxicology (Hoeng J., Peitsch M. C., Eds.), pp 1–26. Springer, New York, NY. [Google Scholar]
- Quercioli D., Roli A., Morandi E., Perdichizzi S., Polacchini L., Rotondo F., Vaccari M., Villani M., Serra R., Colacci A. (2017). The use of omics-based approaches in regulatory toxicology: An alternative approach to assess the no observed transcriptional effect level. Microchem. J. DOI: https://doi.org/10.1016/j.microc.2017.01.029. [Google Scholar]
- Rand-Weaver M., Margiotta-Casaluci L., Patel A., Panter G. H., Owen S. F., Sumpter J. P. (2013). The read-across hypothesis and environmental risk assessment of pharmaceuticals. Environ. Sci. Technol. 47, 11384–11395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REACH. (2006). Regulation (EC) No 1907/2006 of the European Parliament and of the Council of 18 December 2006 concerning the Registration, Evaluation, Authorisation and Restriction of Chemicals (REACH), establishing a European Chemicals Agency. O. J. Eur. Union 396, 1–849.
- Rowlands J. C., Budinsky R., Gollapudi B., Black M. B., Wolfinger R. D., Cukovic D., Dombkowski A., Thompson C. M., Urban J. D., Thomas R. S. (2013). A genomics-based analysis of relative potencies of dioxin-like compounds in primary rat hepatocytes. Toxicol. Sci. 136, 595–604. [DOI] [PubMed] [Google Scholar]
- Russom C. L., Bradbury S. P., Broderius S. J., Hammermeister D. E., Drummond R. A. (1997). Predicting modes of toxic action from chemical structure: Acute toxicity in the fathead minnow (Pimephales promelas). Environ. Toxicol. Chem. 16, 948–967. [DOI] [PubMed] [Google Scholar]
- Russom C. L., LaLone C. A., Villeneuve D. L., Ankley G. T. (2014). Development of an adverse outcome pathway for acetylcholinesterase inhibition leading to acute mortality. Environ. Toxicol. Chem. 33, 2157–2169. [DOI] [PubMed] [Google Scholar]
- SCENIHR (Scientific Committee on Emerging and Newly Identified Health Risks). (2012). Addressing the New Challenges for Risk Assessment: Discussion paper approved for public consultation in view of receiving feedback from stakeholders for its further development, October 8, 2012. Available at: http://ec.europa.eu/health/scientific_committees/emerging/docs/scenihr_o_037.pdf. Accessed August 17, 2016.
- Schmeits P. C., Schaap M. M., Luijten M., van Someren E., Boorsma A., van Loveren H., Peijnenburg A. A., Hendriksen P. J. (2015). Detection of the mechanism of immunotoxicity of cyclosporine A in murine in vitro and in vivo models. Arch. Toxicol. 89, 2325–2337. [DOI] [PubMed] [Google Scholar]
- Schroeder A. L., Ankley G. T., Houck K. A., Villeneuve D. L. (2016). Environmental surveillance and monitoring–The next frontiers for high-throughput toxicology. Environ. Toxicol. Chem. 35, 513–525. [DOI] [PubMed] [Google Scholar]
- Soetaert A., van der Ven K., Moens L. N., Vandenbrouck T., van Remortel P., De Coen W. M. (2007a). Daphnia magna and ecotoxicogenomics: Gene expression profiles of the anti-ecdysteroidal fungicide fenarimol using energy-, molting- and life stage-related cDNA libraries. Chemosphere 67, 60–71. [DOI] [PubMed] [Google Scholar]
- Soetaert A., Vandenbrouck T., van der Ven K., Maras M., van Remortel P., Blust R., De Coen W. M. (2007b). Molecular responses during cadmium-induced stress in Daphnia magna: Integration of differential gene expression with higher-level effects. Aquat. Toxicol. 83, 212–222. [DOI] [PubMed] [Google Scholar]
- Sohm B., Immel F., Bauda P., Pagnout C. (2015). Insight into the primary mode of action of TiO2 nanoparticles on Escherichia coli in the dark. Proteomics 15, 98–113. [DOI] [PubMed] [Google Scholar]
- Song Y., Rundberget J. T., Evenseth L. M., Xie L., Gomes T., Høgåsen T., Iguchi T., Tollefsen K. E. (2016). Whole-organism transcriptomic analysis provides mechanistic insight into the acute toxicity of emamectin benzoate in Daphnia magna. Environ. Sci. Technol. 50, 1194–12003. [DOI] [PubMed] [Google Scholar]
- Sturla S. J., Boobis A. R., FitzGerald R. E., Hoeng J., Kavlock R. J., Schirmer K., Whelan M., Wilks M. F., Peitsch M. C. (2014). Systems toxicology: From basic research to risk assessment. Chem. Res. Toxicol. 27, 314–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeguarden J. G., Tan Y. M., Edwards S. W., Leonard J. A., Anderson K. A., Corley R. A., Kile M. L., Simonich S. M., Stone D., Tanguay R. L., et al. (2016). Completing the link between exposure science and toxicology for improved environmental health decision making: The aggregate exposure pathway framework. Environ. Sci. Technol. 50, 4579–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The modENCODE Consortium., Roy S., Ernst J., Kharchenko P. V., Kheradpour P., Negre N., Eaton M. L., Landolin J. M., Bristow C. A., Ma L., Lin M. F., et al. (2010). Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330, 1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R. S., Wesselkamper S. C., Wang N. C., Zhao Q. J., Petersen D. D., Lambert J. C., Cote I., Yang L., Healy E., Black M. B., et al. (2013a). Temporal concordance between apical and transcriptional points of departure for chemical risk assessment. Toxicol. Sci. 134, 180–194. [DOI] [PubMed] [Google Scholar]
- Thomas R. S., Himmelstein M. W., Clewell H. J., Yang Y., Healy E., Black M. B., Andersen M. E. (2013b). Cross-species transcriptomic analysis of mouse and rat lung exposed to chloroprene. Toxicol. Sci. 131, 629–640. [DOI] [PubMed] [Google Scholar]
- Thomas R. S., Cheung R., Westphal M., Krewski D., Andersen M. E. (2017). Risk science in the 21st century: A data-driven framework for incorporating new technologies into chemical safety assessment. Int. J. Risk Assess. Manage. 20, 88–108. [Google Scholar]
- Tilton F. A., Tilton S. C., Bammler T. K., Beyer R. P., Stapleton P. L., Scholz N. L., Gallagher E. P. (2011). Transcriptional impact of organophosphate and metal mixtures on olfaction: Copper dominates the chlorpyrifos-induced response in adult zebrafish. Aquat. Toxicol. 102, 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollefsen K. E., Scholz S., Cronin M. T., Edwards S. W., de Knecht J., Crofton K., Hartung T., Worth A., Patlewicz G. (2014). Applying Adverse Outcome Pathways (AOPs) to support Integrated Approaches to Testing and Assessment (IATA). Regul. Toxicol. Pharmacol. 70, 629–640. [DOI] [PubMed] [Google Scholar]
- Tralau T., Luch A. (2015). Moving from rats to cellular omics in regulatory toxicology: Great challenge toward sustainability or “up-shit-creek without a paddle”? Arch. Toxicol. 89, 819–821. [DOI] [PubMed] [Google Scholar]
- USEPA (1998). Guidelines for Ecological Risk Assessment, EPA/630/R095/002F, Federal Register 63(93):26846-26924. Available at: https://archive.epa.gov/raf/web/pdf/ecotxtbx.pdf. Accessed August 17, 2016.
- USEPA (2004). Potential Implications of Genomics for Regulatory and Risk Assessment Applications at EPA, EPA 100/B-04/002. Available at: https://cfpub.epa.gov/si/si_public_record_Report.cfm?dirEntryId=101618&CFID=63770336&CFTOKEN=20290707. Accessed August 17, 2016.
- US EPA. (2014). Integrated bioactivity and exposure ranking: A computational approach for the prioritization and screening of chemicals in the endocrine disruptor screening program. US EPA, Office of Chemical Safety and Pollution Prevention, Washington, DC.
- USEPA. (2016a). About the TSCA Chemical Substance Inventory. Available at: https://www.epa.gov/tsca-inventory/about-tsca-chemical-substance-inventory. Accessed October 25, 2016.
- USEPA. (2016b). Endocrine disruptor screening program. Available at: http://www.epa.gov/endo/. Accessed February 12, 2016.
- Van Aggelen G., Ankley G. T., Baldwin W. S., Bearden D. W., Benson W. H., Chipman J. K., Collette T. W., Craft J. A., Denslow N. D., Embry M. R., et al. (2010). Integrating omic technologies into aquatic ecological risk assessment and environmental monitoring: Hurdles, achievements, and future outlook. Environ. Health Perspect. 118, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ravenzwaay B., Sperber S., Lemke O., Fabian E., Faulhammer F., Kamp H., Mellert W., Strauss V., Strigun A., Peter E., et al. (2016). Metabolomics as read-across tool: A case study with phenoxy herbicides. Regul. Toxicol. Pharmacol. 81, 288–304. [DOI] [PubMed] [Google Scholar]
- Verhaar H. J., Solbe J., Speksnijder J., van Leeuwen C. J., Hermens J. L. (2000). Classifying environmental pollutants: Part 3. External validation of the classification system. Chemosphere 40, 875–883. [DOI] [PubMed] [Google Scholar]
- Verhaar H. J. M., van Leeuwen C. J., Hermens J. L. M. (1992). Classifying environmental pollutants. Chemosphere 25, 471–491. [DOI] [PubMed] [Google Scholar]
- Vinuela A., Snoek L. B., Riksen J. A., Kammenga J. E. (2010). Genome-wide gene expression analysis in response to organophosphorus pesticide chlorpyrifos and diazinon in C. elegans. Plos One 5, e12145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve D. L., Crump D., Garcia-Reyero N., Hecker M., Hutchinson T. H., LaLone C. A., Landesmann B., Lettieri T., Munn S., Nepelska M., et al. (2014a). Adverse outcome pathway (AOP) development I: Strategies and principles. Toxicol. Sci. 142, 312–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve D. L., Crump D., Garcia-Reyero N., Hecker M., Hutchinson T. H., LaLone C. A., Landesmann B., Lettieri T., Munn S., Nepelska M., et al. (2014b). Adverse outcome pathway development II: Best practices. Toxicol. Sci. 142, 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston A. D., Hood L. (2004). Systems biology, proteomics, and the future of health care: Toward predictive, preventative, and personalized medicine. J. Proteome Res. 3, 179–196. [DOI] [PubMed] [Google Scholar]
- Wittwehr C., Aladjov H., Ankley G. T., Byrne H. J., de Knecht J., Henzie E., Klambauer G., Landesmann B., Luijten M., MacKay C., et al. (2017). How adverse outcome pathways can aid the development and use of computational prediction models for regulatory toxicology. Toxicol. Sci. 155, 326–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zhang X., Xia P., Zhang R., Wu Y., Xia J., Su G., Zhang J., Giesy J. P., Wang Z., et al. (2016). Activation of AhR-mediated toxicity pathway by emerging pollutants polychlorinated diphenyl sulfides. Chemosphere 144, 1754–1762. [DOI] [PubMed] [Google Scholar]