Abstract
Xenobiotic agonists of constitutive androstane receptor (CAR) induce many hepatic drug metabolizing enzymes, but following prolonged exposure, promote hepatocellular carcinoma, most notably in male mouse liver. Here, we used nuclear RNA-seq to characterize global changes in the mouse liver transcriptome following exposure to the CAR-specific agonist ligand 1,4-bis-[2-(3,5-dichloropyridyloxy)]benzene (TCPOBOP), including changes in novel long noncoding RNAs that may contribute to xenobiotic-induced pathophysiology. Protein-coding genes dysregulated by 3 h TCPOBOP exposure were strongly enriched in KEGG pathways of xenobiotic and drug metabolism, with stronger and more extensive gene responses observed in female than male liver. After 27 h TCPOBOP exposure, the number of responsive genes increased >8-fold in males, where the top enriched pathways and their upstream regulators expanded to include factors implicated in cell cycle dysregulation and hepatocellular carcinoma progression (cyclin-D1, oncogenes E2f, Yap, Rb, Myc, and proto-oncogenes β-catenin, FoxM1, FoxO1, all predicted to be activated by TCPOBOP in male but not female liver; and tumor suppressors p21 and p53, both predicted to be inhibited). Upstream regulators uniquely associated with 3 h TCPOBOP-exposed females include TNF/NFkB pathway members, which negatively regulate CAR-dependent proliferative responses and may contribute to the relative resistance of female liver to TCPOBOP-induced tumor promotion. These responses may be modified by the many long noncoding liver RNAs we show are dysregulated by TCPOBOP or pregnane-X-receptor agonist exposure, including lncRNAs proximal to CAR target genes Cyp2b10, Por, and Alas1. These data provide a comprehensive view of the CAR-regulated transcriptome and give insight into the mechanism of sex-biased susceptibility to CAR-dependent mouse liver tumorigenesis.
Keywords: lincRNA, HCC, PXR, nuclear receptor, cytochrome P450, environmental chemical, environmental toxicology, sex biased gene expression; PCN
Many foreign chemicals, including industrial chemicals, environmental pollutants, and pharmaceuticals alter gene expression through receptor-based mechanisms involving ligand-activated transcription factors, such as the nuclear receptors constitutive androstane receptor (CAR; NR1I3) and pregnane X receptor (PXR; NR1I2) (Chang and Waxman, 2006; Cherian etal., 2015). Constitutive androstane receptor and PXR are expressed at highest levels in mammalian liver, where they act as xenobiotic sensors that detect and respond to foreign chemicals. Some chemicals bind directly to the large and promiscuous ligand-binding pockets of CAR and PXR (Cherian etal., 2015), whereas others activate cell signaling pathways linked to nuclear receptor activation, as exemplified by the activation of CAR by phenobarbital (Hori etal., 2016). Activated CAR and PXR rapidly translocate to the nucleus, where they form heterodimers with retinoid X receptor (NR2B1), bind DNA, and trans-activate many genes (Kobayashi etal., 2015).
Constitutive androstane receptor and PXR induce overlapping sets of genes in liver, including many drug metabolizing enzymes of the cytochrome P450 (Cyp) gene superfamily, as well as phase II conjugation enzymes and drug transporters (Cui and Klaassen, 2016; Timsit and Negishi, 2007). As a consequence, activators of CAR or PXR increase the metabolism and clearance of many xenobiotics. Constitutive androstane receptor also regulates the expression of genes that impact diverse physiological pathways, including lipid metabolism, glucose homeostasis, inflammation, and hepatogenesis (Moreau etal., 2008; Yan etal., 2015). Although some effects of CAR activation are beneficial, others are deleterious. TCPOBOP and other foreign chemical CAR activators induce hepatomegaly, liver tumor promotion, and hepatocarcinogenesis (Kazantseva etal., 2016) and can increase the hepatotoxicity of alcohol and drugs (Chen etal., 2011; Yamazaki etal., 2011). Activators of CAR can also induce nonalcoholic steatohepatitis (Takizawa etal., 2011) and increase serum triglycerides in diabetes and liver disease (Maglich etal., 2009). Constitutive androstane receptor activators can thus dysregulate a wide range of metabolic and other physiological processes in the liver.
Several findings indicate that CAR may impact liver gene expression in a sex-biased manner. Certain CAR-regulated genes show female-bias in expression in liver, eg, Cyp2b10. Furthermore, the greater abundance of CAR in female compared to male mouse liver (Hernandez etal., 2009; Lu etal., 2013) suggests female mice may be more sensitive than males to CAR activators. Although liver sex differences and their impact on expression of drug-metabolizing enzymes and many other genes are well-studied (Waxman and Holloway, 2009), studies of the sex-dependent effects of CAR are limited to CAR-regulated Cyps (Hernandez etal., 2009) and a few other genes (Ledda-Columbano etal., 2003), or use phenobarbital, which gives more complex responses due to its activation of both CAR and PXR (Geter etal., 2014). Furthermore, many prior genome-wide studies of CAR transcriptional responses are limited by their use of microarray technology (Oshida etal., 2015; Ross etal., 2009; Tojima etal., 2012), which often cannot reliably distinguish closely related genes within a family or superfamily. RNA-seq technology addresses this concern, however, to date, RNA-seq analysis of CAR transcriptional responses has been limited to male liver (Cui and Klaassen, 2016; Selwyn etal., 2015). Furthermore, several studies employ exposures lasting days or even weeks (Cui and Klaassen, 2016; Luisier etal., 2014; Ross etal., 2009), which cannot elucidate early, and potentially transient, effects CAR on the transcriptome as they stimulate a mixture of primary and secondary transcriptional responses. A further limitation is that current studies of CAR activation only investigate protein-coding genes; however, it is becoming increasingly clear that noncoding RNAs, in particular long noncoding RNAs (lncRNAs), play vital roles in regulating protein-coding genes both in cis and trans (Goff and Rinn, 2015; Sun and Kraus, 2015). Furthermore, studies of the early effects of CAR on the noncoding transcriptome may be particularly important in view of the emerging role of lncRNAs in hepatocellular carcinoma in both mice and humans (Huang etal., 2014; Klingenberg etal., 2017). Based on our recent work, at least 5000 lncRNAs are expressed in mouse liver, many of which show strong tissue-specific expression, and a subset of which have homologs in other species, including humans (Melia etal., 2016). The responsiveness of such liver-expressed lncRNAs to activators of CAR and other nuclear receptors is unknown.
Here, we use RNA-seq to characterize in a comprehensive manner early responses of the mouse liver transcriptome to a short, 3 h exposure of male and female mice to the CAR agonist ligand TCPOBOP (Tzameli etal., 2000), including changes in expression of protein-coding and noncoding genes. We also assess transcriptional responses 27 h after initiation of TCPOBOP exposure, to identify persistent changes in gene expression, and to capture delayed primary gene responses as well as secondary responses. We report striking sex differences in gene responses to CAR activation, including stronger initial responses in female liver, as well as important qualitative differences after 27 h TCPOBOP exposure, including more extensive dysregulation of many key cell cycle control genes linked to liver tumor promotion in male compared to female mouse liver. We also identify 530 liver-expressed lncRNAs that show strong (>4-fold) changes in expression induced by activators of CAR or PXR, in some cases in association with activation of nearby, co-regulated protein-coding genes, including Cyp2b10 and other drug-metabolizing enzyme genes.
MATERIALS AND METHODS
Animals
All mouse work was carried out in compliance with procedures approved by the Boston University Institutional Animal Care and Use Committee. Male and female CD-1 mice, 7-week-old, were purchased from Charles River Laboratories (Wilmington, Massachusetts) and kept on a 12-hour light cycle (7:30 am to 7:30 pm). 1,4-Bis-[2-(3,5-dichloropyridyloxy)]benzene (TCPOBOP) and pregnenolone 16α-carbonitrile (PCN), purchased from Sigma-Aldrich (St. Louis, Missouri), were dissolved in a 1% DMSO solution in corn oil (vehicle) to give 0.15 mg/ml TCPOBOP or 2.5 mg/ml PCN. Mice were administered each chemical, or vehicle control, by IP injection of 20 μl/g body weight, resulting in a final dose of 3 mg/kg TCPOBOP or 50 mg/kg PCN. Injections were given at a fixed time of day (between 8:00 am and 8:45 am of treatment day 1) to control for circadian effects on liver gene expression (Kettner etal., 2016). Mice were euthanized between 11:00 am and 11:45 am on day 1 (3 h treatment and control groups) or between 11:00 am and 11:45 am on day 2 (27 h treatment and control groups) to give either a 3 h or 27 h exposure (±5 min) for each individual mouse. A small piece of each liver was snap frozen in liquid nitrogen for whole tissue RNA extraction (total RNA). The remainder of the liver was fixed for immunohistochemistry (see below) or was used to isolate nuclei for nuclear RNA analysis.
Tissue fixation, cyrosectioning, and CAR immunostaining
Fresh mouse livers were each placed in approximately 20 ml isopentane in a stainless-steel beaker on dry ice until frozen. Livers were then removed and stored at −80 °C. Each liver was later mounted on metal chucks in a Leica Cryostat (CM1950) and embedded in OCT compound at −20 °C, and 5 μM sections were prepared. Liver sections were adhered to slides and stored at −80 °C. Sections were fixed in 100% methanol at −20 °C for 15 min. Following fixation, tissue was rinsed with cold PBS three times. Hydrogen peroxide (0.3%) was added to the slides for 30 min at room temperature followed by a PBS rinse. Slides were blocked for 30 min with avidin solution (30 μl goat serum + 4 drops avidin block [Vector Avidin/Biotin Blocking Kit, Vector Laboratories, cat. #SP-2001] in 1 ml of PBS) at room temperature, then rinsed with PBS. Slides were then blocked for 30 min with biotin solution (30 μl goat serum + 4 drops biotin block [Vector Avidin/Biotin Blocking Kit] in 1 ml of PBS) at room temperature, then rinsed with PBS. Slides were stained overnight at 4 °C with antibodies against mouse CAR (30 μl goat serum in 1 ml PBS + 1:150 dilution of antibody sc-50462) (Santa-Cruz Biotechnology) or with IgG as a negative control. Slides were rinsed with PBS and incubated for 1 h at 4 °C with secondary antibody (30 μl goat serum in 1 ml PBS + 1:200 dilution of goat-antirabbit secondary antibody). Slides were developed with Vectastain ABC Elite reagent (5 ml PBS + 2 drops of reagent A + 2 drops of reagent B) for 30 min at room temperature and rinsed with PBS. DAB color reagent was prepared by adding 2 drops of buffer stock to 15 ml of distilled water, followed by 4 drops of Vector Peroxidase Substrate Kit DAB (Vector Laboratories, cat. #SK-4100) and 2 drops of hydrogen peroxide solution with thorough mixing. 3,3'-diaminobenzidine tetrahydrochloride (DAB) solution was added to each slide and color allowed to develop for 2–10 min at room temperature. Slides were flushed with distilled water to stop the reaction and photographed.
Nuclear extraction and RNA purification
Livers were homogenized on ice in a Potter-Elvehjem homogenizer in 8ml of complete homogenization buffer (incomplete homogenization buffer = 10mM HEPES buffer [pH 7.9] containing 10% [v/v] glycerol, 2 M sucrose, 25 mM KCl, 0.15 mM spermine, 0.5 mM spermidine trihydrochloride, and 1 mM EDTA; complete homogenization buffer = incomplete homogenization buffer with 0.05 mM DTT, 1 mM PMSF, 1 mM sodium orthovanadate, 10 mM sodium fluoride, and completeTM Protease Inhibitor Cocktail [1 tablet per 50ml buffer; Roche] added on the day of use). The homogenate was layered onto 3 ml of fresh homogenization buffer and spun in a ThermoFisher TH-641 ultracentrifuge rotor at 25 000 rpm for 35 min. The pellet (intact nuclei) was resuspended in 20 mM Tris–HCl buffer (pH 8.0) containing 75 mM NaCl, 0.5 mM EDTA, 50% (v/v) glycerol, 0.85 mM DTT, and 0.125 mM PMSF, then aliquoted and stored at −80 °C. TRIzol LS reagent was used according to the manufacturer’s protocol (Life Technologies) to purify liver nuclear RNA from approximately 25% of the nuclei in a mouse liver.
qPCR analysis
cDNA synthesis reactions were performed using either liver total RNA or liver nuclear RNA (1 μg) after digestion with DNase I to eliminate genomic DNA contaminants. Reverse transcription and cDNA synthesis were performed using the Applied Biosystems High Fidelity RT kit (ThermoFisher). Primers specific for Cyp2b10, Cyp2c55, and other genes of interest were designed using Primer Express software. Primer pairs were placed in adjacent exons of target genes with amplicons spanning long introns to decrease the possibility of having genomic DNA contamination contribute to the qPCR signal, and are detailed in Supplementary Table 1. Quantitative real time PCR (qPCR) was performed using Power SYBR Green PCR Master Mix (ThermoFisher) in 384-well plates and read on an ABI 7900 Real Time PCR system. Fold-change values were calculated using the ΔΔCt method, using the expression of 18S ribosomal RNA as the background Ct value for normalization between samples.
High-throughput RNA sequencing (RNA-seq) and differential expression analysis
Nuclear RNA-seq was performed using two or three biological replicate pools for each treatment group, with each pool representing liver nuclear RNA from n = 3–4 individual mouse livers. Sequencing libraries were prepared from 1 μg of pooled liver nuclear RNA by poly(A) selection using the NEBNext Poly(A) mRNA Magnetic Isolation Module, followed by processing with the NEBNext Ultra Directional RNA Sequencing for Illumina kit (New England Biolabs). Sequencing was performed at the New York Genome Center (New York, NY) on an Illumina HiSeq2500 instrument and 50 bp paired-end sequence reads were obtained. Data were analyzed using a custom RNA-seq analysis pipeline described elsewhere (Connerney etal., 2017). Briefly, sequence reads were mapped to the mouse genome (release mm9) using TopHat (v2.0.13). Genomic regions that contain exonic sequence in at least one isoform of a gene (exon collapsed regions; Connerney etal., 2017) were defined for each RefSeq gene and for each lncRNA gene. HTSeq (0.6.1p1) was then used to obtain read counts for exon collapsed regions of RefSeq genes, and featureCounts (1.4.6-p5) was used to obtain read counts for exon collapsed regions of lncRNAs. A set of 15 558 liver-expressed lncRNA transcripts was considered for differential expression analysis. These lncRNAs are based on the set of intergenic liver lncRNAs that we defined earlier (Melia etal., 2016), which we updated by including lncRNAs that are antisense or intragenic with respect to RefSeq genes, and by increasing the number of mouse liver RNA-seq datasets used for lncRNA discovery from 45 to 186. RefSeq and lncRNA genes that showed significant differential expression following exposure to TCPOBOP or PCN were identified by EdgeR as outlined elsewhere (Melia etal., 2016). RefSeq genes dysregulated with an expression fold-change (ie, either up regulation or down regulation) >1.5 and a false discovery rate (FDR), ie, an adjusted P-value <.001 were considered significant and are shown in Supplementary Table 2. A total of 530 liver-expressed lncRNAs responded to either TCPOBOP or PCN exposure with an expression fold-change >4 at FDR <0.05 and were considered significant. Two hundred and fifty-two of the 530 lncRNAs (48%) are multiexonic–exonic and 278 are mono-exonic. A fold-change >2 at FDR <0.05 was then applied to the 530 lncRNAs to determine significant regulation in each of the 5 datasets for downstream analysis, as shown in Supplementary Table 3. Of the 530 lncRNAs, 22 have RefSeq NR designations; 18 of these (17 multiexonic and 1 mono-exonic) are also included in the listings of responsive RefSeq genes in Supplementary Table 2. Raw and processed RNA-seq data are available at GEO (https://www.ncbi.nlm.nih.gov/gds) accession number GSE95685.
Pathway and upstream regulator analysis
RefSeq genes showing differential expression following exposure to either TCPOBOP or PCN were submitted to DAVID Bioinformatics Resources (https://david.ncifcrf.gov/; last accessed February 17, 2017) for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. Separate gene lists were submitted for each of the five mouse liver RNA-seq datasets (TCPOBOP exposure of males and females, for both 3 h and at 27 h, and PCN exposure of males for 3 h). Twenty-six unique KEGG pathways were considered significant at FDR <0.05 (as controlled by the Benjamini–Hochberg method) in at least 1 of the 5 datasets (Supplementary Table 4). Pathways were categorized as broadly related to either drug/xenobiotic metabolism or cell cycle/DNA replication/cancer by inspection of the lists of differentially regulated genes associated with each pathway. In separate analyses, data for the full set of 24 197 RefSeq genes for each of the 5 datasets was uploaded to Ingenuity Pathway Analysis (IPA; https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/; last accessed March 6, 2017) and Upstream Regulator analysis was performed on genes showing significant differential expression, as defined above. Upstream regulators with P-value of overlap <.001 and |activation z-score| > 2 and a minimum of 5 target genes were considered significant and are listed in Supplementary Table 5, ranked based on P-value of overlap. Upstream regulators identified by IPA as molecular type “chemical” or “biologic drug” were excluded, as were redundant terms. Upstream regulators were categorized as follows: Nuclear receptor-related, factors broadly related to either drug/xenobiotic or lipid metabolism; factors that are key regulators of CAR-mediated hepatocarcinogenesis (Kazantseva etal., 2016); factors related to Tumor Necrosis Factor (TNF) and NFκB signaling; and factors related to nonliver cancers. Cancer-related regulators were further categorized as protumor or antitumor, based on their activation state, as predicted by IPA upstream regulator analysis, and their known function (eg, activation of a tumor promotor is protumor and activation of a tumor suppressor is antitumor, etc.).
Statistics
Student’s t-test implemented using Prism 6 (Graphpad) was used to assess statistical significance for all qPCR analysis and for comparing RNA-Seq induction values between male and female livers. An exact binomial test implemented in the statistical analysis package R was used to determine statistical significance of liver cancer pathway enrichments in male and female liver, and to evaluate the enrichment of multiexonic lncRNAs in the induced gene sets.
RESULTS
Nuclear RNA Analysis to Assess Short-Term Responses to CAR and PXR Agonists
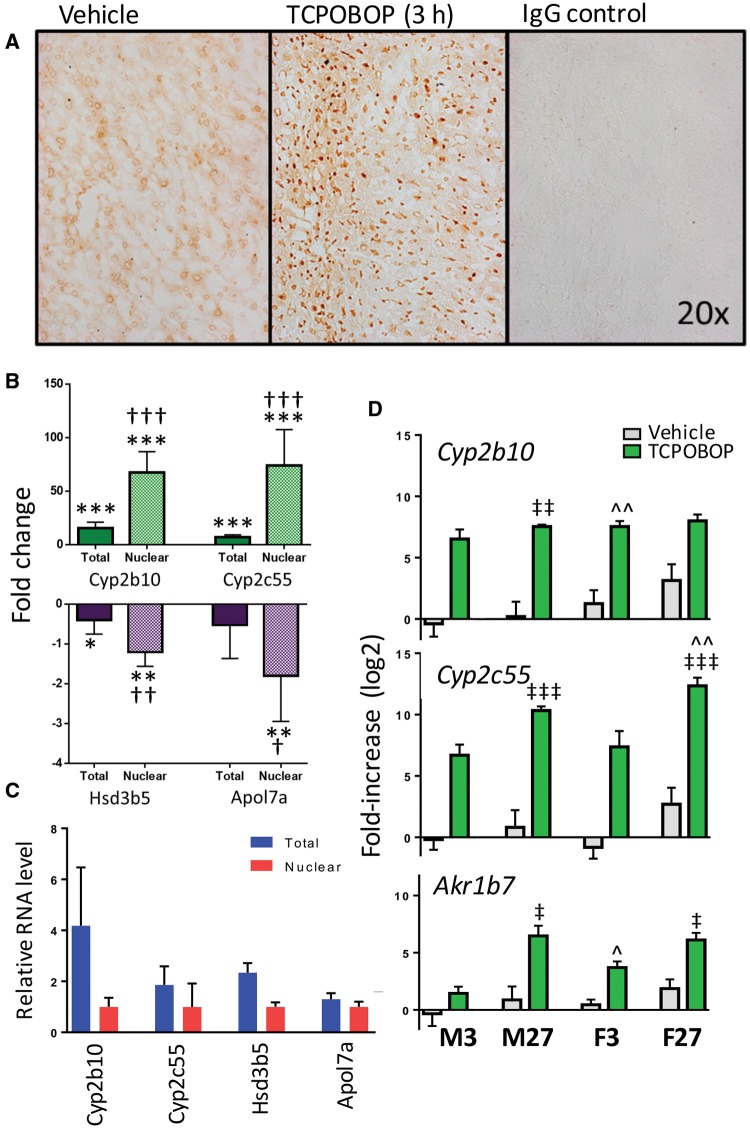
We first verified by immunohistochemistry that CAR protein was present in a large fraction of the hepatocytes in mouse liver and translocated to the nucleus within 3 h of IP injection of the CAR-specific agonist ligand TCPOBOP (Figure 1A). Next, we evaluated the effects of 3 h TCPOBOP exposure on gene expression by qPCR analysis of liver nuclear RNA to capture early transcriptional changes. Results were compared to an analysis of total (unfractionated) liver RNA. Much stronger induction (stronger up regulation) of the CAR target genes Cyp2b10 and Cyp2c55 was observed in the nuclear RNA fraction (65- and 70-fold, increases, respectively; Figure 1B) as compared to total liver RNA (15- and 6-fold increases, respectively). Similarly, the PXR activator PCN, after a 3 h exposure, repressed the expression of Hsd5b and Apol7a to a greater extent in the liver nuclear RNA fraction than in total liver RNA: 2.4- and 4.8-fold repression, respectively, in nuclear RNA versus only 1.4- and 1.5-fold repression, respectively, in total RNA (Figure 1B). All four genes showed lower basal expression in the nuclear than in the total RNA fraction (Figure 1C), which contributes to the greater sensitivity of the nuclear RNA fraction to a change in transcription rate induced by activators of CAR and PXR.
Figure 1.
TCPOBOP stimulates CAR nuclear localization and CAR target gene induction. A, Immunohistochemical analysis of male mouse liver showing CAR localized to the cytoplasm in the absence of TCPOBOP (vehicle control) but translocated to the nucleus after 3 h TCPOBOP exposure. B, Induction of Cyp2b10 and Cyp2c55 by TCPOBOP (3 h) and suppression of Hsd3b5 and Apol7a expression by PCN (3 h) in male mouse liver, as measured by qPCR. RNA isolated from whole liver tissue (“Total”) shows modest induction of Cyp2b10 and Cyp2c55 by TCPOBOP (t-test: *P < .05; **P < .01; ***P < .001 vs vehicle controls), while RNA purified from liver nuclei (“Nuclear”) shows much strong induction (higher fold-change) for both genes (t-test: +P < .05; ++P < .01; +++P < .001 for difference in induction between total and nuclear). Down regulation of Hsd3b5 and Apol7a is also significantly stronger in nuclear RNA compared to total RNA. C, Expression levels measured by qPCR analysis of total liver RNA is higher than for the corresponding nuclear liver RNA samples, all from vehicle control livers, for the four genes shown. D, Induction of Cyp2b10, Cyp2c55, and Akr1b7 in livers of mice treated with TCPOBOP males (M) or females (F) for 3 h (3) or for 27 h (27), as indicated at the bottom. Expression level for the male 3 h vehicle control samples was set to 1. Strong and statistically significant (ANOVA; t-test, P < .05, not marked) induction of all 3 genes was seen in both male and female livers after 3 h or 27 h. A significantly greater increase in expression at 27 h compared to 3 h is seen in males for Cyp2b10, Cyp2c55, and Akr1b7, and in females for Cyp2c55 and Ark1b7 (ANOVA; t-test; ‡P < .05; ‡‡P < .01; ‡‡‡P < .001). A significant sex bias was found for the induction of all three genes, with females showing a stronger induction of Cyp2b10 and Akr1b7 than males at 3 h, and stronger induction of Cyp2c55 at 27 h than males (ANOVA; t-test; ^P < .05; ^^P < .01). For B, C, and D, n = 5–12 individual livers per group.
Next, we examined the effect of TCPOBOP exposure for either 3 h or 27 h on liver nuclear levels of CAR target genes Cyp2b10, Cyp2c55, and Akr1b7 in both male and female mice. These time points were set 24 h apart to fix the time of day for all treatment groups and thereby control for circadian effects on CAR-dependent gene expression (Lu etal., 2013). TCPOBOP induction of all three genes was not detectable at 30 min and 1 h (data not shown) but was readily apparent after 3 h (Figure 1D). Cyp2b10 induction was near maximal at 3 h in both males and females, whereas Cyp2c55 was submaximally induced at 3 h (50- to 70-fold increases) compared to 27 h (375- to 400-fold increases) in both sexes. Akr1b7 also showed a significantly delayed response to TCPOBOP. This delay was more pronounced in male liver, resulting in a sex difference in induction at 3 h: 3-fold increase in Akr1b7 in male liver versus 14-fold increase in female liver.
Sex-Differential Global Transcriptomic Responses to CAR Activation
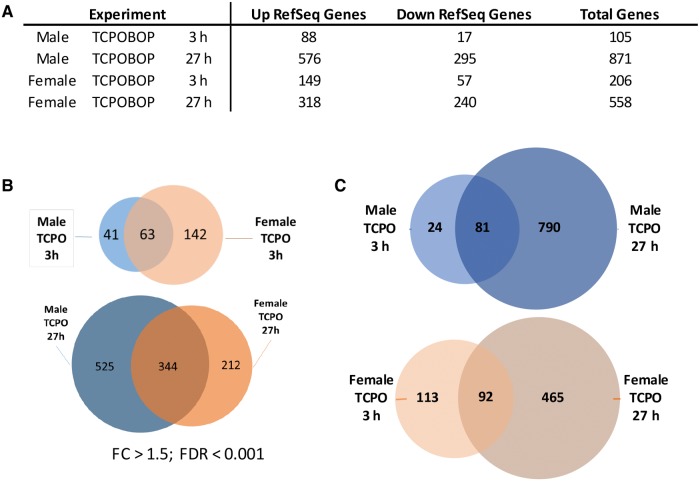
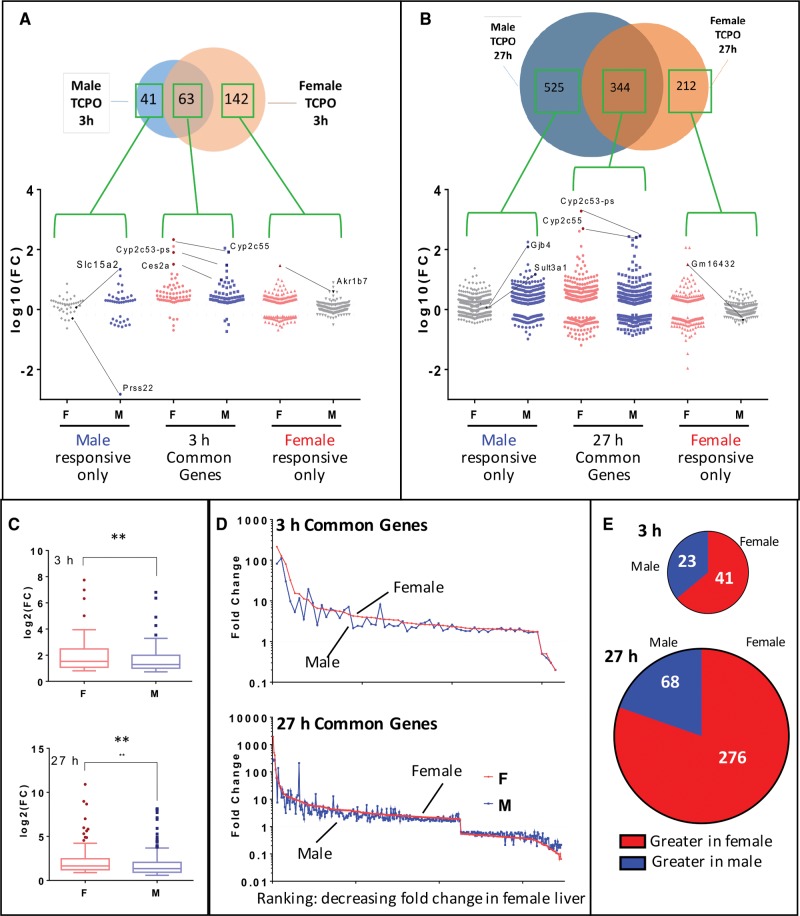
Nuclear RNA-seq analysis was carried out to identify on a global scale TCPOBOP-stimulated RNA responses in both male and female mouse liver. In 3 h TCPOBOP-exposed mice, many more RefSeq genes showed significant changes in expression in female liver (206 genes) than in male liver (105 genes) (Figure 2A), as detailed in Supplementary Table 2. This result may be a consequence of the higher levels of CAR expression reported in female liver (Lu etal., 2013). In all four exposure groups, more genes were up regulated than down regulated (Figure 2A). Although 63 genes showed a common response to 3 h TCPOBOP exposure in males and in females, many other genes were responsive in only one sex: 41 genes were uniquely responsive to 3 h TCPOBOP in male liver and 142 genes were uniquely responsive in female liver (Figure 2B). These sex differences are not an artifact of the threshold values (fold-change and FDR) used to identify significant response genes, as many of the genes showed strong responses to TCPOBOP in the responsive sex but were marginally, if at all, responsive in the opposite sex. Examples include Slc15a2 and Prss22 in male liver and Akr1b7 in female liver (Figure 3A; also see Figure 3B). We also examined the short-term transcriptional responses to the PXR activator PCN, whose gene targets overlap with those of CAR (Supplementary Figure 1 and Supplementary Table 2), consistent with earlier work.
Figure 2.
Nuclear RNA-seq identifies RefSeq genes responsive to TCPOBOP (TCPO). A, Numbers of genes that respond to TCPOBOP treatment in mouse liver at |fold-change|>1.5 and FDR <0.001. B, Overlap between sets of genes responsive to TCPOBOP in males and females at 3 h (top) and 27 h (bottom). C, Overlap of genes responsive to TCPOBOP in males (top) and in females (bottom) at 3 h versus 27 h. Genes that were responsive in multiple datasets, but in opposite directions, were omitted from these analyses. See Supplementary Table 2 for full details on all genes responsive to TCPOBOP or PCN.
Figure 3.
Sex differences in TCPOBOP gene responses identified by nuclear RNA-seq. A and B, Venn diagram show overlap of genes responding to TCPOBOP (TCPO) in male versus female liver after 3 h (A) or after 27 h (B) at |fold-change| >1.5 and FDR <0.001. Omitted are 3 genes that responded in the opposite direction in male versus female liver at 27 h. Graph below Venn diagram shows fold-change values for genes responsive to TCPOBOP either in male liver only, in female liver only, or in both sexes (common genes), as noted, with each individual gene graphed as a separate data point. Genes whose response to TCPOBOP did not meet the fold-change and FDR cutoff values are shown in the two outer datasets as gray dots. Examples of genes showing a strong sex bias in their response include Slc15a1 and Prss22 (3 h male only), Akr1b7 (3 h female only), Gjb4 and Sult3a1 (27 h male only), and Gm16432 (27 h female only), as marked. Many genes that respond to TCPOBOP in common in both sexes are more strongly induced in females, as highlighted by Ces2a (3 h) Cyp2c55 and Cyp2c53-ps (3 and 27 h). C, Box plot of log 2 fold-change values showing that genes up regulated by TCPOBOP in both sexes, after either 3 h (top) or 27 h (bottom), are significantly more strongly induced in female liver (t-test; **P< .01). D, Fold-change values of individual TCPOBOP common response genes in male (blue) and female liver (red), as marked, ranked by decreasing fold-change in female mice. E, Numbers of TCPOBOP common response genes whose |fold-change| is larger in male liver or in female liver, as indicated.
Increasing the time of TCPOBOP exposure from 3 to 27 h resulted in many more genes dysregulated (Figure 2C), in particular in male liver (increase from 105 to 871 responsive in males vs from 206 to 558 genes in females; Figure 2A). Three hundred and forty-four of the genes that responded at 27 h were regulated in common in both sexes (Figure 2B), with 119 genes down regulated and 225 genes up regulated. Although fewer genes responded to TCPOBOP in females than in males at 27 h, the magnitude of the response for the up-regulated male–female common genes was significantly greater in females at both time points (Figure 3C). Similar results were apparent when all of the common response genes were examined individually (Figure 3D). Furthermore, a majority of all genes that showed a common response to TCPOBOP in both sexes were more strongly induced, or were more strongly repressed, in female compared to male liver (Figure 3E). Individual examples are highlighted in Figures 3A and 3B (center panels).
KEGG Pathway Analysis
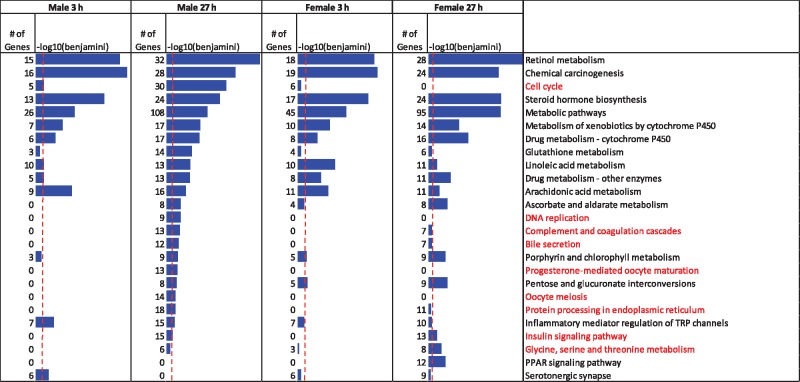
KEGG pathway analysis of the gene sets responsive to 3 h TCPOBOP identified 12 significant pathways in male liver and 11 in female liver (Figure 4; Supplementary Table 4A and C). Nine of these pathways were common between the two 3 h datasets. These pathways all relate to xenobiotic and lipid metabolism and associated metabolic processes, consistent with the known role of CAR as a xenobiotic sensor. The KEGG pathway cell cycle showed significant, albeit weak enrichment in the male but not the female gene set at 3 h. Many more KEGG pathways were identified at 27 h than at 3 h, in both males and females (Figure 4; Supplementary Table 4B and D), consistent with the expanded list of responsive genes at 27 h (Figure 2A). Whereas xenobiotic and lipid metabolism and related terms were prominent on the list of enriched KEGG pathways at 27 h in both males and females, cell cycle and DNA replication showed significant enrichment in male liver only. Furthermore, many more cell cycle pathway genes were dysregulated in male liver at 27 h (30 genes) than at 3 h (5 genes). KEGG pathways enriched in response to the PXR activator PCN are shown in Supplementary Figure 1.
Figure 4.
KEGG pathways enriched in sets of RefSeq genes responding to TCPOBOP—Shown are gene numbers and Benjamini p-values (blue bars; max value E−17) for KEGG pathways enriched in responsive genes (|fold-change|>1.5 and FDR <0.001) in each of the indicated RNA-Seq datasets, based on DAVID analysis. Pathways not directly related to metabolism are shown in lighter color (red). Pathways related to drug metabolism or other liver metabolic processes are found in all datasets, while pathways related to cell cycle and DNA replication are found exclusively in TCPOBOP exposed males at 27 h. Vertical dashed red lines: Benjamini p-value = .05. See Supplementary Table 4 for full details of KEGG pathway analysis.
Sex Differences in Hepatocarcinogenic Upstream Regulators of CAR Gene Responses
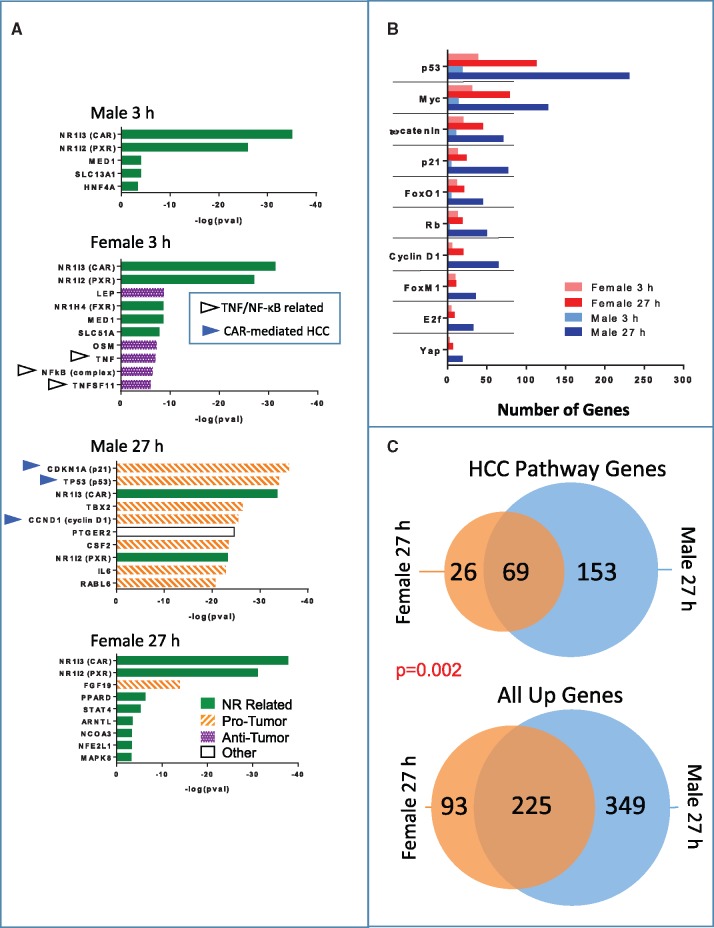
Ingenuity Pathway Analysis upstream regulator analysis identified CAR and PXR as top upstream regulators of the genes that respond to TCPOBOP in all 4 datasets (Figure 5; Supplementary Table 5). This finding reflects the overlap of the gene responses to TCPOBOP, described here, with those reported previously for various activators of CAR and PXR, which populate the gene–gene regulator association database queried by IPA. Furthermore, after 27 h TCPOBOP exposure, many unique upstream regulators apparently distinct from the primary CAR-induced xenobiotic and lipid metabolism responses were identified in male but not female liver. These upstream regulators of male liver 27 h TCPOBOP responses include 10 key factors linked to cell cycle dysregulation and hepatocellular carcinoma progression (Ledda-Columbano etal., 2000; Zhang etal., 2009), all of which have specifically been associated with CAR activation (Kazantseva etal., 2016). Eight of the 10 upstream regulators were protumor factors predicted by IPA to be activated by TCPOBOP (cyclin-dependent kinase inhibitor CCND1/cyclin D1, oncogenes E2f, Yap, and Rb, and proto oncogenes CTNNB1/β-catenin, Myc, FoxM1, and FoxO1) and two were tumor suppressors whose activity was predicted to be inhibited (p21, p53) (Figure 5; Supplementary Table 5C). Mechanistic networks involving these factors, and their prevalence in TCPOBOP-exposed male but not female liver are shown in Supplementary Figure 2. Overall, the predicted activation states of these regulatory factors of CAR-induced hepatocarcinogenesis demonstrate that TCPOBOP stimulates a prominent protumor response in male liver, insofar as oncogenic factors are predicted to be activated and tumor suppressors are predicted to be inhibited by TCPOBOP exposure. Other protumor factors identified as activated upstream regulators for the male 27 h TCPOBOP dataset included TBX2, a cell cycle regulator and direct repressor of p21, RABL6 (RBEL1A), a negative regulator of p53, and growth factors CSF2, VEGF, and HGF (Figure 5; Supplementary Table 5C).
Figure 5.
Upstream regulator analysis of TCPOBOP responsive genes. A, Shown are up to the top 10 upstream regulators identified in each RNA-seq dataset by IPA. Green (solid) bars identify upstream regulators related to activation CAR or other nuclear receptors (NR). Orange (striped) bars identify upstream regulators associated with a protumor response, and purple (dotted) bars identify upstream regulators associated with an antitumor response based on their known biological functions and direction of the activated z-score calculated by IPA. Blue (closed) arrow heads mark upstream regulators known to be involved in CAR-dependent liver tumor promotion, and gray (open) arrow heads mark TNF/NFκB pathway related regulators. B, Gene targets were identified by IPA for each of the 10 indicated protumor upstream regulators identified in male liver at 27 h (see Supplementary Table 5). Shown is the number of gene targets for each upstream regulator that are responsive at |fold-change|>1.5 and FDR <0.001 in each of the four exposure groups. C, A single gene list was obtained from the union of all 27 h TCPOBOP-responsive gene targets of the 10 protumor upstream regulators (HCC pathway gene targets) in male and female liver (Supplementary Table 4). Shown is the overlap of the HCC gene pathway gene targets that are up regulated by TCPOBOP at 27 h in male versus female liver (top), and the overlap of all genes up regulated by TCPOBOP at 27 h in male versus female liver (bottom). These patterns of overlap differ at p = .002 (exact binomial test) (see text).
For each of the 10 hepatocarcinogenesis-associated upstream regulators, the number of target genes that responded to 3 h TCPOBOP exposure was greater in female than in male liver, consistent with the greater hepatoproliferative responses reported for short-term TCPOBOP-treated female mouse liver (Ledda-Columbano etal., 2003); however, by the 27 h time point, many more target genes of these regulators responded to TCPOBOP in male than in female liver (Figure 5B). Furthermore, the number of up-regulated liver cancer pathway-associated genes (ie, genes downstream of these 10 upstream regulators) that were unique to 27 h-TCPOBOP male liver (153 genes) as compared to those unique to 27 h-TCPOBOP female liver (26 genes) (ratio of 153/26 = 5.9) was significantly greater than the corresponding ratio of total genes uniquely up regulated in male versus female liver (ratio = 349 genes in males/93 genes in females = 3.8; p = .002 by exact binomial test) (Figure 5C). Thus, there is a significantly stronger enrichment in male compared to female liver for TCPOBOP-responsive liver cancer pathway genes as compared to for all TCPOBOP-responsive genes. Consistent with these findings, whereas 3 of the 10 factors linked to CAR-dependent hepatocarcinogenesis were identified as upstream regulators of female liver responses to TCPOBOP at the 3 h time point (FoxM1, E2F1, and CTNNB1/β-catenin), the p-values of overlap for their target genes were much weaker in female than in male liver (Supplementary Table 5B vs 5C). FGF19, which was specifically associated with female liver responses to TCPOBOP at 27 h (Figure 5A) induces hepatocellular carcinoma in human liver but not mouse liver (Zhou etal., 2017).
Next, we considered whether any of the upstream regulators activated in female liver might reduce the hepatocarcinogenic potential of TCPOBOP. We identified 18 upstream regulators that were specific to 3 h TCPOBOP-exposed female liver (Figure 5A;Supplementary Table 5B). Strikingly, 5 of the 18 upstream regulators identified only in females at 3 h (TNF, NFkB, RELA, IKBKB, CHUK (IKBKA) are linked to TNF/NFκB signaling, indicating that TCPOBOP preferentially activates this signaling pathway in female liver (Supplementary Table 5B and Supplementary Figure 3). TNF/NFκB signaling exerts negative cross-talk to CAR gene responses (Van Ess etal., 2002) and to the hepatoproliferative response to TCPOBOP (Columbano etal., 2005), suggesting these 5 upstream regulators contribute to the relative resistance of female liver to TCPOBOP hepatocarcinogenesis. Consistent with this proposal, the NFkB inhibitor NFKBIA was a much stronger activated upstream regulator in 27 h-TCPOBOP male liver than in 3 h-TCPOBOP-female liver (Supplementary Table 5C). Finally, these 5 upstream regulators were also identified in our analysis of livers of male mice exposed to the PXR activator PCN (Supplementary Table 5E). Unlike CAR, PXR activation in mouse liver is not associated with hepatic tumor promotion (Shizu etal., 2013).
Impact of TCPOBOP and PCN on Noncoding Transcriptome
Given the increasing recognition that many noncoding RNAs regulate gene expression by epigenetic and other mechanisms, we used the four RNA-Seq datasets described above to investigate the impact of TCPOBOP exposure on 15 558 noncoding RNA transcripts that we identified in mouse liver (see Materials and Methods). These transcripts include long-noncoding RNAs (lncRNAs) that are intergenic, intragenic, or antisense with respect to protein-coding genes. We also identified lncRNAs responsive to the PXR activator PCN (3 h exposure, male liver) to ascertain whether CAR and PXR regulate overlapping sets of lncRNAs, as is seen for their protein-coding gene targets (Supplementary Figure 1B). LncRNAs that respond similarly to both TCPOBOP and PCN are unlikely to be false positive responses. To further mitigate discovery of false positives, we required a fold-change >4 and FDR <0.05 in any of the 5 data sets to identify responsive lncRNAs. A stringent set of 530 lncRNA responding to either TCPOBOP or PCN was thus identified (Supplementary Table 3). Next, to determine the patterns of response to TCPOBOP and PCN, we used a more relaxed set of cutoffs to define differential expression (fold-change >2; FDR <0.05, Table 1). Of 402 TCPOBOP-responsive lncRNAs, 80 also responded to PCN treatment, whereas an additional 118 lncRNAs were uniquely responsive to PCN, a majority of which were down regulated (73 of 118 lncRNAs; Table 1, last line).
Table 1.
Mono-Exonic and Multiexonic lncRNAs That Respond to TCPOBOP and/or PCN at |Fold-Change|>2 and FDR <0.05
| TCPOBOP Response Profile | Up | Down | Total | Also Responsive to PCN (Same Direction) |
|---|---|---|---|---|
| Number of lncRNAs | ||||
| All TCPOBOP-responsive lncRNAs | 251 | 151 | 402 | 80 |
| Common responses in all 4 TCPOBOP datasets | 30 | 0 | 30 | 15 |
| Early responding, both male and female | 34 | 3 | 37 | 20 |
| Early responding, male only | 7 | 10 | 17 | 7 |
| Early responding, female only | 19 | 20 | 39 | 15 |
| Late responding, both male and female | 50 | 17 | 67 | 7 |
| Late responding, male only | 50 | 44 | 94 | 10 |
| Late responding, female only | 68 | 49 | 117 | 9 |
| Responsive to PCN only | 45 | 73 | 118 | n/a |
Shown are: lncRNAs that respond to TCPOBOP in at least 1 of the 4 TCPOBOP treatment groups (all TCPOBOP-responsive); lncRNAs that respond in all 4 TCPOBOP datasets and in the same direction (ie, up in all 4, or down in all 4) (common responses in all 4); lncRNAs that respond to TCPOBOP at 3 h in males and/or females, as indicated, irrespective of their response to TCPOBOP at 27 h (early responding); lncRNAs that respond to TCPOBOP at 27 h but not at 3 h (late responding); and lncRNAs responsive to PCN in male liver at 3 h but that did not respond in any of the four TCPOBOP datasets (responsive to PCN only). For both early responding and late responding lncRNAs, “both male and female” indicates the lncRNA is either up regulated or down regulated in both sexes. The last column indicates the number of TCPOBOP-responsive lncRNAs in each category that responded in the same direction to 3 h PCN treatment in male liver. Ten lncRNAs whose response to TCPOBOP was discrepant across the 4 conditions were excluded from this analysis. n/a, not applicable.
Time Course of lncRNA Induction
Thirty lncRNAs were induced by TCPOBOP early (3 h) and persisted at 27 h in both sexes (Table 1), with inductions as high as approximately 400-fold seen for lncRNA_5998, which is upstream of Cyp2b10 (see below). Fifteen of these 30 lncRNAs were also induced by PCN. Early gene responses—both induction and repression—characterized many other lncRNAs, a subset of which showed sex-specific responses to TCPOBOP (Table 1). Some of these early responding lncRNAs could contribute to the late (secondary) changes in expression of TCPOBOP-responsive protein-coding genes. Other lncRNAs showed a delayed response to TCPOBOP (ie, lncRNA induction not seen until 27 h): 67 such late lncRNAs responded to TCPOBOP in both male and female liver, 94 responded in male liver only, and 117 responded in female liver only (Table 1, Supplementary Figure 4). The sex-specificity of the late lncRNA responses is intriguing, given the striking sex differences in late protein-coding gene responses in male liver and their association with liver tumor promotion, presented above.
The overall set of 530 responsive lncRNAs was significantly enriched for multiexonic lncRNA genes, which are more likely to be functional than mono-exonic lncRNAs (Liu etal., 2017). Thus, 252 of the 530 responsive lncRNAs (47.5%) are multiexonic as compared to only 20.2% of the overall set of 15 558 liver-expressed lncRNAs (2.35-fold enrichment; p < E−15, exact binomial test). Considering only multiexonic lncRNAs, 3 h TCPOBOP exposure altered the expression of 52 and 62 lncRNAs in male and female liver, respectively, a majority of which were up regulated (Table 2). After 27 h TCPOBOP exposure, 112 multiexonic lncRNAs were dysregulated in male liver and 131 multiexonic lncRNAs were dysregulated in female liver; again, a majority (72%−76%) were up regulated. Examples of multiexonic lncRNAs responsive to TCPOBOP exposure are shown in Figures 6 and 7; Supplementary Figure 5. A total of 126 multiexonic lncRNAs were responsive to PCN, 57% of which were down regulated (Table 2). This is consistent with the greater proportion of protein-coding genes down regulated by PCN as compared to TCPOBOP (Figure 2A).
Table 2.
TCPOBOP and PCN Responsive Multiexonic lncRNAs and Their Relationship to Coregulated RefSeq Genes
| Multi-exonic lncRNA gene sets | Male TCPOBOP 3 h | Male TCPOBOP 27 h | Female TCPOBOP 3 h | Female TCPOBOP 27 h | MalePCN 3 h |
|---|---|---|---|---|---|
| Number of lncRNAs | |||||
| Responsive lncRNAs | 52 | 112 | 62 | 131 | 126 |
| Up regulated | 40 | 83 | 39 | 93 | 53 |
| Down regulated | 12 | 29 | 23 | 38 | 73 |
| Proximity to RefSeq | |||||
| Overlaps RefSeq gene | 19 | 31 | 17 | 27 | 26 |
| Same TAD as RefSeq gene | 7 | 42 | 14 | 37 | 54 |
| Cisversusdistal | |||||
| Cis co-expressed lncRNA-RefSeq pairs (%) | 26 (50%) | 64 (58%) | 30 (48%) | 59 (45%) | 73 (58%) |
| Cis opposite lncRNA-RefSeq pairs (%) | 0 | 9 (8%) | 1 (2%) | 5 (4%) | 7 (6%) |
| Distal lncRNAs (%) | 26 (50%) | 39 (35%) | 31 (50%) | 67 (51%) | 46 (37%) |
Shown are responsive multiexonic lncRNAs (|FC|>2, FDR <0.05) in each dataset and their genomic location relative to the nearest responsive RefSeq gene (|FC|>1.5, FDR <0.001). Cis co-expression and cis opposite expression refer to lncRNA–RefSeq gene pairs where both genes respond to TCPOBOP or PCN and either overlap in their genomic coordinates by at least 1 base pair, or are within the same topologically associating domain (TAD), as indicated under Proximity to RefSeq. The lncRNA and RefSeq genes in each pair can either be co-expressed or show opposite expression, as indicated. Distal lncRNAs are those whose nearest regulated RefSeq gene is in a different TAD on the same chromosome.
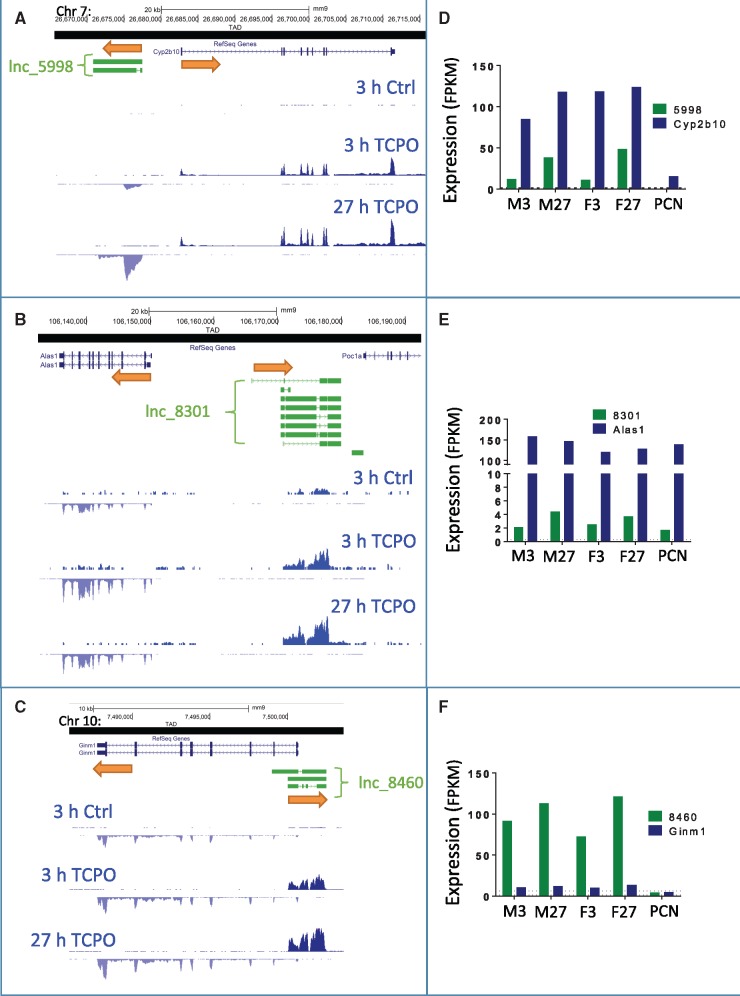
Figure 6.
Liver-expressed lncRNAs responsive to TCPOBOP or PCN that are proximal to regulated RefSeq genes. A–C, Structure and relative position of select regulated multiexonic lncRNAs that are in close proximity to, or overlapping, a responsive RefSeq gene. Shown are RNA-seq read densities for male liver after 3 and 27 h TCPOBOP treatment, as well as vehicle control tracks. Bars with arrows indicate direction of transcription. D–F, Relative expression (FPKM, fragments per kilo base per million sequence reads) for each lncRNA and its nearby responsive RefSeq gene, as measured by RNA-Seq. The mean expression level (FPKM) of each transcript in the vehicle control samples was as follows: Cyp2b10, 1.2; lncRNA_5998, 0.1; Ginm1, 6.3; lncRNA_8460, 1.4; Alas1, 32.1; and lncRNA_8301, 0.3, and is marked by a horizontal dashed line in each figure. Y-axis scale for panel A: positive strand, 350; negative strand, -200; panel B: positive strand, 10; negative strand, -500; panel C: positive strand, 400; negative strand, -50. RNA-seq tracks are shown for male livers with vehicle control (Ctrl) or TCPOBOP treatment (TCPO).
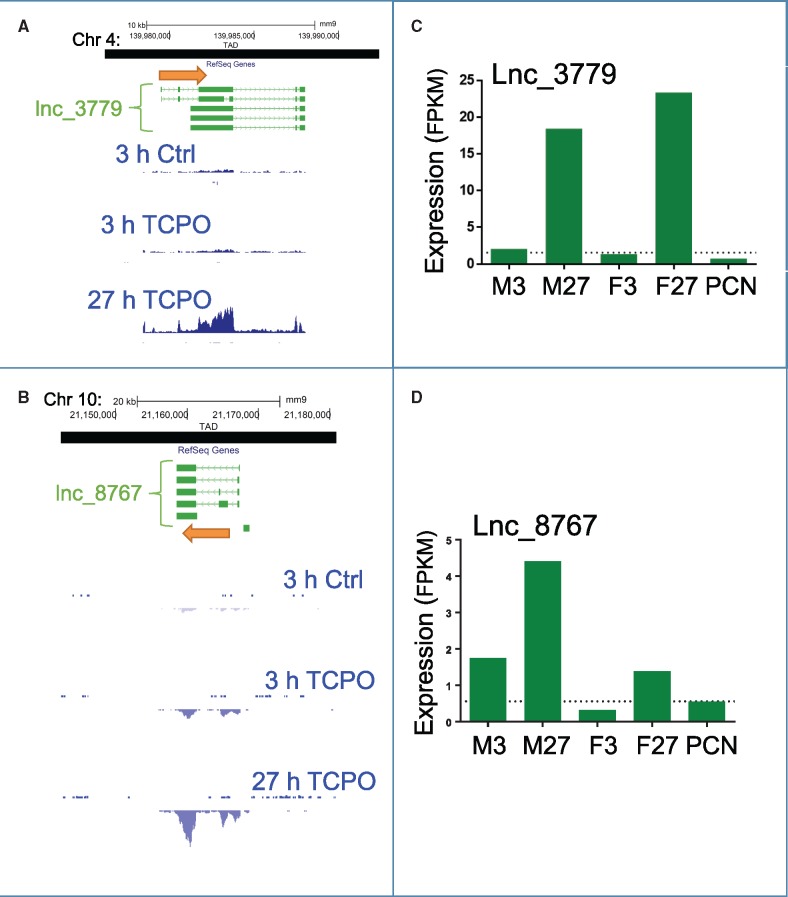
Figure 7.
LncRNAs responsive to TCPOBOP that are not proximal to the nearest regulated RefSeq gene. A, Structure of lncRNA_3779 and a subset of its isoforms (also see Supplementary Figure 5G), which is in the same TAD but >300 kb away from the closest TCPOBOP responsive transcript (Supplementary Table 6). B, Structure of lncRNA_8767, whose nearest TCPOBOP responsive RefSeq gene is in a different TAD (ie, is distal). Also shown are RNA-seq read densities for male liver after 3 and 27 h TCPOBOP treatment, as well as vehicle control tracks. C and D, Expression levels (FPKM values) of lncRNA_3779 and lncRNA_8767 across the 5 datasets, as measured by RNA-Seq. The mean expression level (FPKM) of each transcript in the vehicle control samples was as follows: lncRNA_3779, 1.6; and lncRNA_8767, 0.6, and is indicated by a horizontal dashed line. Y-Axis scale for panel A: positive strand, 50; negative strand, -50; panel B: positive strand, 10; negative strand, -10. RNA-seq tracks are shown for male livers with vehicle control (Ctrl) or TCPOBOP treatment (TCPO).
LncRNAs Proximal to Regulated RefSeq Protein-Coding Genes are Responsive to TCPOBOP
LncRNAs are frequently co-expressed with nearby protein-coding genes, and often regulate expression of protein-coding genes in cis (Engreitz etal., 2016). Accordingly, we examined the proximity of the lncRNAs responsive to TCPOBOP or PCN to the nearest regulated protein-coding gene. Across the five RNA-seq data sets, a total of 129 responsive multiexonic lncRNAs overlapped with, or were within the same genomic topologically associating domain (TAD) (Vietri Rudan etal., 2015), as a co-responsive RefSeq gene (cis co-expression, 45%−58% of the responsive multiexonic lncRNAs; Table 2). Twenty-one other multiexonic lncRNAs (2%−8%) were cis to RefSeq genes that showed the opposite response to TCPOBOP or PCN in one or more datasets (Table 2; see Supplementary Table 6 for listing of all cis co-expressed and cis opposite-regulated multiexonic lncRNA-RefSeq pairs). Because gene regulatory elements are largely contained within TADs, whose boundaries form looped, insulated chromatin domains (Hnisz etal., 2016), these lncRNAs have the potential to regulate their nearest responsive RefSeq gene through a cis mechanism, one that involves either positive regulation (129 cis co-expressed gene pairs) or negative regulation (21 cis opposite gene pairs).
The cis co-expressed lncRNAs comprise four classes of orientation relative to their nearest co-regulated RefSeq genes: (1) lncRNAs upstream of and transcribed from the opposite strand as the RefSeq gene. Two examples are lncRNA_5998, which is 5.2 kb upstream of Cyp2b10 (Figure 6A;Supplementary Figure 5A), and lncRNA_8301, which is 15.6 kb upstream of Alas1 (Figure 6B;Supplementary Figure 5B). Cyp2b10 and Alas1 are well known CAR and PXR targets and are highly induced in all 5 datasets, as are their adjacent co-regulated lncRNAs (Figs. 6D and E). (2) lncRNAs characterized by antisense transcription starting within the first intron of the co-regulated RefSeq gene. Examples include lncRNA_8460, which is antisense to Ginm1 (Figure 6C;Supplementary Figure 5C), and lncRNA_4655, which is antisense to Por (Supplementary Figure 5D). lncRNA_4655 is expressed at a low level, but is induced in all 5 datasets (Supplementary Table 3), as is Por (Supplementary Table 2). Conversely, lncRNA_8460 is highly expressed and rapidly induced in all 5 datasets, whereas the overlapping RefSeq gene, Ginm1, is expressed at a low level and only weakly induced by TCPOBOP at 27 h in both males and females (Figs. 6C and F). (3) lncRNAs found within an intron of a highly expressed RefSeq gene and often spanning multiple RefSeq genes. Examples include two lncRNAs in the Cyp2c locus that span the genomic region from Cyp2c37 through Cyp2c50. lncRNA_15011 originates in intron 5 of Cyp2c37 and ends in intron 4 of Cyp2c50 (isoform 2), whereas lncRNA_15014 originates in intron 7 of Cyp2c37 and ends in intron 6 of Cyp2c50 (isoform 2) (Supplementary Figure 5E). Both lncRNAs are highly expressed and are induced in all 5 datasets (Supplementary Table 3), as are Cyp2c37 and Cyp2c50 (Supplementary Table 2). (4) lncRNAs that are within the same TAD as a regulated RefSeq gene, but are distant in terms of linear chromosome distance. The DNA loop that anchors the TAD structure has been shown to bring linearly distant sequences into close three-dimensional proximity and allow for cis regulation (Hnisz etal., 2016). An example is lncRNA_3779, which is highly induced in TCPOBOP-exposed male and female liver at 27 h (Figs. 7A and C). In male liver, the TCPOBOP-responsive RefSeq protein-coding gene closest to lncRNA_3779 is Klhdc7a, which is approximately 455 kb away, whereas in female liver the nearest responsive RefSeq gene is Padi4, which is approximately 313 kb away (Supplementary Figure 5G). LncRNA_3779, Klhdc7a, and Padi4 are all located within the same TAD. Interestingly, lncRNA_3779 is upregulated in both 27 h datasets, whereas Klhdc7a is down regulated in both and Padi4 is upregulated in TCPOBOP-treated females at 27 h but not males (Figure 7C, Supplementary Tables 2 and 3).
LncRNAs Distal to Regulated RefSeq Genes Also Respond to TCPOBOP
In addition to the above lncRNAs, which are cis to TCPOBOP or PCN responsive protein-coding genes, we identified approximately 100 other lncRNAs—some highly expressed—whose nearest regulated RefSeq gene is not in the same TAD. These distal lncRNAs comprise 35%−51% of the responsive multiexonic lncRNAs in each dataset (Table 2). One example is lncRNA_8767 (Figs. 7B and D;Supplementary Figure 5H). The distant location of this TCPOBOP-induced lncRNA relative to the nearest responsive protein-coding gene, and its complex gene structure (4 multiexonic isoforms; Figure 7B) indicates this lncRNA—and many others in this class—is specifically targeted for transcription and is not a spurious by-product of the very active transcription of a highly expressed nearby protein-coding gene. Because cis gene–gene regulatory element interactions are largely contained within TADs (Hnisz etal., 2016), these lncRNAs are unlikely to regulate TCPOBOP or responsive protein-coding genes by a cis mechanism, but rather, may utilize one of the trans regulatory mechanisms described for lncRNAs (Sun and Kraus, 2015).
DISCUSSION
The impact of the CAR-specific agonist ligand TCPOBOP on the mouse transcriptome has been investigated after exposures lasting several days or weeks (Cui and Klaassen, 2016; Luisier etal., 2014; Ross etal., 2009), which gives insights into the long-term effects of CAR activation, but does not inform about early transcriptional responses, including early events associated with hepatocyte proliferation linked to tumor promotion. Here, we identify genes induced or repressed after exposure to TCPOBOP for either 3 h (presumed primary response genes) or 27 h (primary + secondary response genes). The set of genes dysregulated in 3 h TCPOBOP-treated mouse liver is enriched for xenobiotic and lipid metabolism, and includes Cyp2b10 and other established primary targets of CAR. Many more genes were dysregulated after 27 h exposure to TCPOBOP, likely reflecting secondary transcriptional responses. Moreover, at 27 h, CAR responses in male but not female liver showed strong enrichment for genes functionally linked to CAR-dependent hepatocyte proliferation and hepatic tumor promotion (Kazantseva etal., 2016) and for their upstream regulators. These regulators include cyclin D1, which was predicted to be activated, and p53 and p21, which were predicted to be inhibited (Kazantseva etal., 2014). These findings are consistent with the greater susceptibility of male mice to CAR-dependent tumor promotion (Li etal., 2012) and suggest that the molecular events that lead to hepatic tumor promotion are initiated early (within 27 h) following CAR activation. Finally, we identify more than 500 liver-expressed lncRNAs that respond to either TCPOBP and PCN exposure, many of which are novel genes and likely to contribute to the biological and pathophysiological responses to CAR and PXR activation.
We detected TCPOBOP-induced changes in gene expression with increased sensitivity by RNA-seq analysis of liver nuclear RNA, rather than by analysis of total (unfractionated) liver RNA. The increased sensitivity provided by the nuclear fraction likely reflects a buffering effect of the induction response by the comparatively stable pool of transcripts found in the cytoplasm. Nuclear RNA was also more sensitive for detection of gene down regulation, where the buffering effect of preexisting mature cytoplasmic mRNA can mask a decrease in gene transcription, especially for mature mRNAs that are relatively stable and when the analysis is carried out at an early time point, ie, before a new steady state level of cytoplasmic mRNA is established. The use of nuclear RNA-seq also increased the sensitivity for detecting changes in expression of liver-expressed lncRNAs, a large majority of which show strong nuclear localization (Melia etal., 2016).
It is unclear what triggers the expansion of TCPOBOP gene responses in male liver from the narrow focus on genes of xenobiotic and lipid metabolism seen after 3 h, to genes linked to cell cycle dysregulation and liver hepatocellular carcinoma promotion seen at 27 h. This striking change in gene responses at 27 h to encompass many genes that contribute to liver tumor promotion was not seen in female liver. This limitation of liver tumor promotion response in female liver may in part be due to the robust early activation of TNF and NFκB signaling pathways that we observed in TCPOBOP-treated females but not males. Importantly, TNF/NFκB signaling exerts negative cross-talk on CAR-dependent gene responses (Van Ess etal., 2002) and suppresses the hepatoproliferative effects of TCPOBOP exposure (Columbano etal., 2005), suggesting that TNF-activated NFκB contributes to the partial resistance of female liver to the hepato-proliferative response to TCPOBOP. TNF/NFκB signaling was also predicted to be activated in 3 h PCN-exposed male liver (Supplementary Figure 3), which also does not exhibit the strong hepatocarcinogenic responses associated with male liver TCPOBOP exposure (Shizu etal., 2013). Additional mechanisms for male-biased liver tumor promotion may involve the protumor actions of androgen receptor-FoxA1 complexes and the antitumor effects of estrogen receptor-FoxA1 complexes, via the inhibition of Myc (Li etal., 2012). Of note, Foxa1 was induced 2-fold in our 27 h TCPOBOP datasets, in both male and female liver.
Female mice were more responsive than males to TCPOBOP-induced changes in gene expression, as indicated by the approximately 2-fold greater number of responsive genes at 3 h in females than in males, and by the larger magnitude of gene responses in females both 3 and 27 h after initiating TCPOBOP exposure. This is consistent with earlier reports that CAR is more highly expressed and more active in female than male mouse liver (Hernandez etal., 2009; Lu etal., 2013), and with the finding that the androgens androstanol and androstenedione are inverse agonists of CAR activity (Kawamoto etal., 2000; Kohalmy etal., 2007) that can compete with exogenous CAR agonists for CAR binding in male liver. In addition, estradiol and estrone act as agonists of CAR (Kawamoto etal., 2000) and stimulate basal CAR activation in female liver (Koh etal., 2012). Given this strong intrinsic female bias in CAR responsiveness, it is striking that a male-biased gene response pattern emerged 27 h after TCPOBOP exposure, when many genes involved in cell cycle control and hepatocellular carcinoma progression were preferentially dysregulated in male liver, as discussed above.
Female mice are more resistant to hepatocellular carcinoma induced by the tumor initiator diethylnitrosamine when combined with a nongenotoxic tumor promotor, such TCPOBOP or phenobarbital (Kalra etal., 2008). However, exposure to TCPOBOP alone induces hepatocyte proliferation more frequently in female than in male liver (Ledda-Columbano etal., 2003). There are several possible explanations for the apparent disparity between those findings and our findings here. First, the reported female bias in hepatocyte proliferation is limited to TCPOBOP exposures up to 36 h, as there is no apparent sex bias after 48 h (Ledda-Columbano etal., 2003). This is consistent with our finding of an early female bias in cell proliferation gene expression (Figure 5B). Conceivably, the reversal of the sex bias in expression of cell cycle control genes by 27 h in our dataset may account for the loss of sex bias in hepatic proliferation at 48 h (Ledda-Columbano etal., 2003). Second, given our finding of a strong enrichment for key mediators of CAR-dependent liver tumor promotion in 27 h-TCPOBOP-treated male but not female liver, as well as the early activation in female liver of TNF/NFκB signaling, which is associated with inhibition of hepatoproliferative response to TCPOBOP (Columbano etal., 2005), the transcriptional environment in male liver may be poised for stronger and more rapid liver tumor progression than female liver, despite lower overall cell proliferation rates. Indeed, acute activation of CAR can lead to increased cell proliferation but not liver tumor formation (Dong etal., 2015). Thus, early measurements of cell proliferation alone may not be sufficient to predict the hepatocellular promotion effects of TCPOBOP and perhaps other CAR agonists.
Cyclin D1 (Ccnd1 gene) is induced in TCPOBOP-exposed mouse liver and is a key mediator of CAR-dependent increases in hepatocyte proliferation (Ledda-Columbano etal., 2003; Kazantseva etal., 2014). Overexpression or dysregulation of cyclin D1 drives tumor formation in many tissues, including liver (Bartek and Lukas, 2011; Deane etal., 2001; Jirawatnotai etal., 2011; Kazantseva etal., 2014). We observed early induction of cyclin D1 in female but not male liver, consistent with prior reports (Ledda-Columbano etal., 2003), followed by strong up regulation of cyclin D1 in both sexes at 27 h. However, we also found that cyclin D1 is a highly significant upstream regulator of downstream TCPOBOP responses in male but not in female liver, with 52 cyclin D1 target genes being responsive to TCPOBOP in males compared to only 11 target genes in females at 27 h. The increase in hepatocyte proliferation within 24 h of TCPOBOP treatment (Ledda-Columbano etal., 2000; Li etal., 2012) is reduced in cyclin D1-deficient mice (Ledda-Columbano etal., 2000, 2002), consistent with our finding that liver cyclin D1 induction is an early response to TCPOBOP exposure. However, longer exposures to TCPOBOP (≥36 h) in the same cyclin D1-deficient mice showed no difference in hepatocyte proliferation, apparently due to compensatory effects of cyclin E (Ledda-Columbano etal., 2002). Thus, while cyclin D1 is not strictly necessary for long-term TCPOBOP mediated hepatocyte proliferation, it is required for the initial response.
We also found that the tumor suppressor p53 and its target gene p21 are upstream regulators of TCPOBOP gene responses whose activity is inhibited by TCPOBOP in male but not female liver. In healthy cells, p53 induces p21, and p21, in turn, inhibits cyclin D1 signaling to trigger cell cycle arrest (Xiong etal., 1993). Disruption of either p53 or p21 signaling is linked to liver cancer progression (Deane etal., 2001; Zhang etal., 2009). Interestingly, 151 of the 217 male liver TCPOBOP-responsive genes that are downstream of either p53, p21, or cyclin D1 are unique to only one of these three regulators, suggesting these regulators may in part act independently of the canonical p53 → p21 → cyclin D1 pathway that connects them.
Human liver CAR (hCAR), which is highly homologous to mouse CAR (mCAR) on a protein level (Omiecinski etal., 2011), functions as a xenobiotic sensor in much the same way as mCAR. However, as discussed elsewhere, activation of hCAR by phenobarbital or by hCAR-specific activators, may or may not elicit the same tumor promotion response in human liver, or in transgenic mouse models, as occurs in mouse liver following activation of mCAR (Braeuning etal., 2015; Yamada etal., 2015). As our studies focus on the biology of mCAR when activated by the mCAR-specific agonist ligand TCPOBOP, any extrapolation of data to the biology to human liver tumors must be done with caution. Nevertheless, our finding that β-catenin is rapidly activated by TCPOBOP in mouse liver is of much interest, insofar as activating mutations in CTNNB1 (β-catenin) have been reported in as many as 33% of human hepatocellular carcinomas (Dong etal., 2015) and β-catenin is proposed to be critical in driving phenobarbital-mediated tumor promotion in mouse (Braeuning etal., 2015).
Our comprehensive characterization of the noncoding liver transcriptome led to the discovery of 530 liver-expressed lncRNAs, including 252 multiexonic lncRNAs, whose expression in mouse liver is either induced or repressed following exposure to TCPOBOP or PCN. LncRNAs have diverse actions, which include tethering to their site of transcription (Werner and Ruthenburg, 2015), which enables them to recruit activating or repressive histone mark writers to nearby target genes in cis, as well as distal actions by epigenetic and other mechanisms that operate in trans (Goff and Rinn, 2015; Sun and Kraus, 2015).
One hundred fifty of the 252 multiexonic lncRNAs discovered here are encoded by genes that are either proximal to, or are found within the same genomic TAD (Hnisz etal., 2016), ie, are cis to, one or more TCPOBOP- or PCN-responsive protein-coding genes. Furthermore, 129 of these lncRNAs show the same direction of response to TCPOBOP or PCN as their cis protein-coding genes, while 21 lncRNAs show the opposite responses. Examples include lncRNA_5998, which is upstream and antisense to Cyp2b10, is proximal to an enhancer that binds CAR and contributes to Cyp2b10 induction (Honkakoski and Negishi, 1997), and is induced to a similar level as Cyp2b10 (Figure 6). Other examples include lncRNAs transcribed antisense to an overlapping, responsive protein-coding gene that they may regulate. This regulation may involve antisense mechanisms (Pelechano and Steinmetz, 2013) that lead to decreased sense transcription by RNA polymerase II (Hobson etal., 2012), recruitment of histone modifiers to silence the genomic region (Rinn etal., 2007), or other mechanisms. Examples include lncRNA_8450, which is induced by TCPOBOP to very high levels (approximately 100 FPKM) and is transcribed antisense to the weakly induced overlapping protein-coding gene Ginm1 (Figure 6D), suggesting lncRNA_8450 may actively interfere with Ginm1 expression. Many other TCPOBOP or PCN responsive lncRNAs are distant from other TCPOBOP or PCN responsive genes, and some of these may operate through trans regulatory mechanisms. Further study is required to elucidate the functions of the CAR and PXR-responsive lncRNAs described here and their impact on CAR- and PXR-dependent physiological and pathophysiological processes.
Several lncRNAs have been associated with hepatocellular carcinoma formation and progression in mouse and human liver. Examples include the lncRNAs HOTAIR and MALAT-1, which show increased expression in human liver cancer and are involved in epigenetic regulation and splicing, respectively (Huang etal., 2014). Other lncRNAs stabilize β-catenin at the transcript level (lncRNAs DANCR and lncRNA-UFC1) or at the protein level (lnc-beta-catm), and thereby promote human hepatocellular carcinoma progression (Klingenberg etal., 2017). In mice, lncRNAs involved in liver tumor progression include Dreh and LALR1 (Huang etal., 2014). The corresponding mouse lncRNAs, Malat1 and Dancr, are highly expressed in mouse liver, but were not significantly dysregulated by short-term activation of CAR in our experiments (Supplementary Table 2). The set of lncRNAs reported here to be strongly induced or repressed by activated CAR may provide novel insight into mouse liver tumor progression, particularly those lncRNAs that show a strong male bias in their response to TCPOBOP.
In conclusion, we have found marked sex differences in the responses of the mouse liver transcriptome to the mouse CAR-specific agonist ligand TCPOBOP. We have presented evidence for a male-biased tumor-promotion response that involves several factors known to contribute to CAR-dependent liver tumor promotion, including β-catenin, p53, and cyclin D1. Our findings also reveal a significant female-bias in the activation of TNF/NFκB signaling, which is known to negatively regulate hepatoproliferative responses following CAR activation. Further study will be required to validate these findings, based on RNA-seq transcriptomic data, at the protein level. In addition, we have presented the discovery of a large number of long noncoding RNA genes, many of them novel, which are significantly dysregulated following the activation of CAR or PXR, some of which may contribute to the regulation of CAR and PXR responses that impact hepatic metabolism and carcinogenesis.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
AUTHORS’ CONTRIBUTIONS
N.J.L. and D.J.W. conceived of the study, and jointly wrote and edited the manuscript. N.J.L. carried out the experimental work and data analysis. T.M. carried out lncRNA gene structure discovery and T.M. and A.R. jointly developed the pipeline used for RNA-seq data analysis. N.J.L. performed downstream lncRNA analysis, including differential expression, and relative distance and orientation to RefSeq genes. D.J.W. supervised the work and contributed to data analysis.
FUNDING
The study was supported in part by NIEHS grant ES024421 from the National Institutes of Health (to D.J.W.).
Supplementary Material
REFERENCES
- Bartek J., Lukas J. (2011). DNA repair: Cyclin D1 multitasks. Nature 474, 171–172. [DOI] [PubMed] [Google Scholar]
- Braeuning A., Henderson C. J., Wolf C. R., Schwarz M. (2015). Model systems for understanding mechanisms of nongenotoxic carcinogenesis: Response. Toxicol. Sci. 147, 299–300. [PubMed] [Google Scholar]
- Chang T. K., Waxman D. J. (2006). Synthetic drugs and natural products as modulators of constitutive androstane receptor (CAR) and pregnane X receptor (PXR). Drug Metab. Rev. 38, 51–73. [DOI] [PubMed] [Google Scholar]
- Chen X., Meng Z., Wang X., Zeng S., Huang W. (2011). The nuclear receptor CAR modulates alcohol-induced liver injury. Lab. Invest. 91, 1136–1145. [DOI] [PubMed] [Google Scholar]
- Cherian M. T., Chai S. C., Chen T. (2015). Small-molecule modulators of the constitutive androstane receptor. Expert. Opin. Drug Metab. Toxicol. 11, 1099–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Columbano A., Ledda-Columbano G. M., Pibiri M., Cossu C., Menegazzi M., Moore D. D., Huang W., Tian J., Locker J. (2005). Gadd45beta is induced through a CAR-dependent, TNF-independent pathway in murine liver hyperplasia. Hepatology 42, 1118–1126. [DOI] [PubMed] [Google Scholar]
- Connerney J., Lau-Corona D., Rampersaud A., Waxman D. J. (2017). Activation of male liver chromatin accessibility and STAT5-dependent gene transcription by plasma growth hormone pulses. Endocrinology 158, 1386–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J. Y., Klaassen C. D. (2016). RNA-Seq reveals common and unique PXR- and CAR-target gene signatures in the mouse liver transcriptome. Biochim. Biophys. Acta 1859, 1198–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane N. G., Parker M. A., Aramandla R., Diehl L., Lee W. J., Washington M. K., Nanney L. B., Shyr Y., Beauchamp R. D. (2001). Hepatocellular carcinoma results from chronic cyclin D1 overexpression in transgenic mice. Cancer Res. 61, 5389–5395. [PubMed] [Google Scholar]
- Dong B., Lee J. S., Park Y. Y., Yang F., Xu G., Huang W., Finegold M. J., Moore D. D. (2015). Activating CAR and beta-catenin induces uncontrolled liver growth and tumorigenesis. Nat. Commun. 6, 5944.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz J. M., Haines J. E., Perez E. M., Munson G., Chen J., Kane M., McDonel P. E., Guttman M., Lander E. S. (2016). Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geter D. R., Bhat V. S., Gollapudi B. B., Sura R., Hester S. D. (2014). Dose-response modeling of early molecular and cellular key events in the CAR-mediated hepatocarcinogenesis pathway. Toxicol. Sci. 138, 425–445. [DOI] [PubMed] [Google Scholar]
- Goff L. A., Rinn J. L. (2015). Linking RNA biology to lncRNAs. Genome Res 25, 1456–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez J. P., Mota L. C., Huang W., Moore D. D., Baldwin W. S. (2009). Sexually dimorphic regulation and induction of P450s by the constitutive androstane receptor (CAR). Toxicology 256, 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D., Day D. S., Young R. A. (2016). Insulated neighborhoods: Structural and functional units of mammalian gene control. Cell 167, 1188–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobson D. J., Wei W., Steinmetz L. M., Svejstrup J. Q. (2012). RNA polymerase II collision interrupts convergent transcription. Mol. Cell 48, 365–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkakoski P., Negishi M. (1997). Characterization of a phenobarbital-responsive enhancer module in mouse P450 Cyp2b10 gene. J. Biol. Chem. 272, 14943–14949. [DOI] [PubMed] [Google Scholar]
- Hori T., Moore R., Negishi M. (2016). p38 MAP kinase links CAR activation and inactivation in the nucleus via phosphorylation at threonine 38. Drug Metab. Dispos. 44, 871–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. L., Zheng L., Hu Y. W., Wang Q. (2014). Characteristics of long non-coding RNA and its relation to hepatocellular carcinoma. Carcinogenesis 35, 507–514. [DOI] [PubMed] [Google Scholar]
- Jirawatnotai S., Hu Y., Michowski W., Elias J. E., Becks L., Bienvenu F., Zagozdzon A., Goswami T., Wang Y. E., Clark A. B., et al. (2011). A function for cyclin D1 in DNA repair uncovered by protein interactome analyses in human cancers. Nature 474, 230–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra M., Mayes J., Assefa S., Kaul A. K., Kaul R. (2008). Role of sex steroid receptors in pathobiology of hepatocellular carcinoma. World J Gastroenterol. 14, 5945–5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto T., Kakizaki S., Yoshinari K., Negishi M. (2000). Estrogen activation of the nuclear orphan receptor CAR (constitutive active receptor) in induction of the mouse Cyp2b10 gene. Mol. Endocrinol. 14, 1897–1905. [DOI] [PubMed] [Google Scholar]
- Kazantseva Y. A., Pustylnyak Y. A., Pustylnyak V. O. (2016). Role of nuclear constitutive androstane receptor in regulation of hepatocyte proliferation and hepatocarcinogenesis. Biochemistry (Mosc) 81, 338–347. [DOI] [PubMed] [Google Scholar]
- Kazantseva Y. A., Yarushkin A. A., Pustylnyak V. O. (2014). CAR-mediated repression of Foxo1 transcriptional activity regulates the cell cycle inhibitor p21 in mouse livers. Toxicology 321, 73–79. [DOI] [PubMed] [Google Scholar]
- Kettner N. M., Voicu H., Finegold M. J., Coarfa C., Sreekumar A., Putluri N., Katchy C. A., Lee C., Moore D. D., Fu L. (2016). Circadian homeostasis of liver metabolism suppresses hepatocarcinogenesis. Cancer Cell 30, 909–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg M., Matsuda A., Diederichs S., Patel T. (2017). Non-coding RNA in hepatocellular carcinoma: Mechanisms, biomarkers and therapeutic targets. J. Hepatol. doi:10.1016/j.jhep.2017.04.009. [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Hashimoto M., Honkakoski P., Negishi M. (2015). Regulation of gene expression by CAR: An update. Arch. Toxicol. 89, 1045–1055. [DOI] [PubMed] [Google Scholar]
- Koh K. H., Jurkovic S., Yang K., Choi S. Y., Jung J. W., Kim K. P., Zhang W., Jeong H. (2012). Estradiol induces cytochrome P450 2B6 expression at high concentrations: Implication in estrogen-mediated gene regulation in pregnancy. Biochem. Pharmacol. 84, 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohalmy K., Tamasi V., Kobori L., Sarvary E., Pascussi J. M., Porrogi P., Rozman D., Prough R. A., Meyer U. A., Monostory K. (2007). Dehydroepiandrosterone induces human CYP2B6 through the constitutive androstane receptor. Drug Metab. Dispos. 35, 1495–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledda-Columbano G. M., Pibiri M., Concas D., Cossu C., Tripodi M., Columbano A. (2002). Loss of cyclin D1 does not inhibit the proliferative response of mouse liver to mitogenic stimuli. Hepatology 36, 1098–1105. [DOI] [PubMed] [Google Scholar]
- Ledda-Columbano G. M., Pibiri M., Concas D., Molotzu F., Simbula G., Cossu C., Columbano A. (2003). Sex difference in the proliferative response of mouse hepatocytes to treatment with the CAR ligand, TCPOBOP. Carcinogenesis 24, 1059–1065. [DOI] [PubMed] [Google Scholar]
- Ledda-Columbano G. M., Pibiri M., Loi R., Perra A., Shinozuka H., Columbano A. (2000). Early increase in cyclin-D1 expression and accelerated entry of mouse hepatocytes into S phase after administration of the mitogen 1, 4-Bis[2-(3,5-Dichloropyridyloxy)] benzene. Am. J. Pathol. 156, 91–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Tuteja G., Schug J., Kaestner K. H. (2012). Foxa1 and Foxa2 are essential for sexual dimorphism in liver cancer. Cell 148, 72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. J., Horlbeck M. A., Cho S. W., Birk H. S., Malatesta M., He D., Attenello F. J., Villalta J. E., Cho M. Y., Chen Y., et al. (2017). CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355, 6320.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y. F., Jin T., Xu Y., Zhang D., Wu Q., Zhang Y. K., Liu J. (2013). Sex differences in the circadian variation of cytochrome p450 genes and corresponding nuclear receptors in mouse liver. Chronobiol. Int. 30, 1135–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luisier R., Lempiainen H., Scherbichler N., Braeuning A., Geissler M., Dubost V., Muller A., Scheer N., Chibout S. D., Hara H., et al. (2014). Phenobarbital induces cell cycle transcriptional responses in mouse liver humanized for constitutive androstane and pregnane x receptors. Toxicol. Sci. 139, 501–511. [DOI] [PubMed] [Google Scholar]
- Maglich J. M., Lobe D. C., Moore J. T. (2009). The nuclear receptor CAR (NR1I3) regulates serum triglyceride levels under conditions of metabolic stress. J. Lipid Res. 50, 439–445. [DOI] [PubMed] [Google Scholar]
- Melia T., Hao P., Yilmaz F., Waxman D. J. (2016). Hepatic long intergenic noncoding RNAs: High promoter conservation and dynamic, sex-dependent transcriptional regulation by growth hormone. Mol. Cell Biol. 36, 50–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau A., Vilarem M. J., Maurel P., Pascussi J. M. (2008). Xenoreceptors CAR and PXR activation and consequences on lipid metabolism, glucose homeostasis, and inflammatory response. Mol. Pharm. 5, 35–41. [DOI] [PubMed] [Google Scholar]
- Omiecinski C. J., Coslo D. M., Chen T., Laurenzana E. M., Peffer R. C. (2011). Multi-species analyses of direct activators of the constitutive androstane receptor. Toxicol. Sci. 123, 550–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshida K., Vasani N., Jones C., Moore T., Hester S., Nesnow S., Auerbach S., Geter D. R., Aleksunes L. M., Thomas R. S., et al. (2015). Identification of chemical modulators of the constitutive activated receptor (CAR) in a gene expression compendium. Nucl. Recept. Signal 13, e002.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V., Steinmetz L. M. (2013). Gene regulation by antisense transcription. Nat. Rev. Genet. 14, 880–893. [DOI] [PubMed] [Google Scholar]
- Rinn J. L., Kertesz M., Wang J. K., Squazzo S. L., Xu X., Brugmann S. A., Goodnough L. H., Helms J. A., Farnham P. J., Segal E., et al. (2007). Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P. K., Woods C. G., Bradford B. U., Kosyk O., Gatti D. M., Cunningham M. L., Rusyn I. (2009). Time-course comparison of xenobiotic activators of CAR and PPARalpha in mouse liver. Toxicol. Appl. Pharmacol. 235, 199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selwyn F. P., Cui J. Y., Klaassen C. D. (2015). RNA-Seq quantification of hepatic drug processing genes in germ-free mice. Drug Metab. Dispos. 43, 1572–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shizu R., Benoki S., Numakura Y., Kodama S., Miyata M., Yamazoe Y., Yoshinari K. (2013). Xenobiotic-induced hepatocyte proliferation associated with constitutive active/androstane receptor (CAR) or peroxisome proliferator-activated receptor alpha (PPARalpha) is enhanced by pregnane X receptor (PXR) activation in mice. PLoS One 8, e61802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M., Kraus W. L. (2015). From discovery to function: The expanding roles of long noncoding RNAs in physiology and disease. Endocr. Rev. 36, 25–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa D., Kakizaki S., Horiguchi N., Yamazaki Y., Tojima H., Mori M. (2011). Constitutive active/androstane receptor promotes hepatocarcinogenesis in a mouse model of non-alcoholic steatohepatitis. Carcinogenesis 32, 576–583. [DOI] [PubMed] [Google Scholar]
- Timsit Y. E., Negishi M. (2007). CAR and PXR: The xenobiotic-sensing receptors. Steroids 72, 231–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tojima H., Kakizaki S., Yamazaki Y., Takizawa D., Horiguchi N., Sato K., Mori M. (2012). Ligand dependent hepatic gene expression profiles of nuclear receptors CAR and PXR. Toxicol. Lett. 212, 288–297. [DOI] [PubMed] [Google Scholar]
- Tzameli I., Pissios P., Schuetz E. G., Moore D. D. (2000). The xenobiotic compound 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene is an agonist ligand for the nuclear receptor CAR. Mol. Cell Biol. 20, 2951–2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ess P. J., Mattson M. P., Blouin R. A. (2002). Enhanced induction of cytochrome P450 enzymes and CAR binding in TNF (p55(-/-)/p75(-/-)) double receptor knockout mice following phenobarbital treatment. J Pharmacol Exp Ther 300, 824–830. [DOI] [PubMed] [Google Scholar]
- Vietri Rudan M., Barrington C., Henderson S., Ernst C., Odom D. T., Tanay A., Hadjur S. (2015). Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 10, 1297–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman D. J., Holloway M. G. (2009). Sex differences in the expression of hepatic drug metabolizing enzymes. Mol. Pharmacol. 76, 215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M. S., Ruthenburg A. J. (2015). Nuclear fractionation reveals thousands of chromatin-tethered noncoding RNAs adjacent to active genes. Cell Rep. 12, 1089–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Hannon G. J., Zhang H., Casso D., Kobayashi R., Beach D. (1993). p21 is a universal inhibitor of cyclin kinases. Nature 366, 701–704. [DOI] [PubMed] [Google Scholar]
- Yamada T., Cohen S. M., Lake B. G. (2015). The mode of action for phenobarbital-induced rodent liver tumor formation is not relevant for humans: Recent studies with humanized mice. Toxicol. Sci. 147, 298–299. [DOI] [PubMed] [Google Scholar]
- Yamazaki Y., Moore R., Negishi M. (2011). Nuclear receptor CAR (NR1I3) is essential for DDC-induced liver injury and oval cell proliferation in mouse liver. Lab. Invest. 91, 1624–1633 (Research Support, N.I.H., Extramural Research Support, N.I.H., Intramural). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Chen B., Lu J., Xie W. (2015). Deciphering the roles of the constitutive androstane receptor in energy metabolism. Acta Pharmacol. Sin. 36, 62–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M. F., Zhang Z. Y., Fu J., Yang Y. F., Yun J. P. (2009). Correlation between expression of p53, p21/WAF1, and MDM2 proteins and their prognostic significance in primary hepatocellular carcinoma. J. Transl. Med. 7, 110.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Luo J., Chen M., Yang H., Learned R. M., DePaoli A. M., Tian H., Ling L. (2017). Mouse species-specific control of hepatocarcinogenesis and metabolism by FGF19/FGF15. J. Hepatol. doi:10.1016/j.jhep.2017.01.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.