Abstract
Background
We have previously shown that high salt intake suppresses the expression of prolyl hydroxylase domain-containing protein 2 (PHD2), an enzyme promoting the degradation of hypoxia inducible factor (HIF)-1α, and increases HIF-1α along with its target genes in the renal medulla, which promotes sodium excretion and regulate salt sensitivity of blood pressure. However, it remains unknown how high salt inhibits the expression of PHD2.
Method and results
The current study first revealed that high salt-induced PHD2 inhibition was due to the enhanced decay of mRNA. We then found that high salt significantly increased the expression of miR-429, which was subsequently proven to target the 3′-untranslated region of PHD2 and reduce PHD2 levels, in the renal medulla. To define the functional role of renal medullary miR-429 in the regulation of PHD2/HIF-1α-mediated renal adaptation to high salt intake and salt sensitivity of blood pressure, we locally inhibited miR-429 in the renal medulla by LNA anti-miR-429 in uninephrectomized rats. Our results demonstrated that inhibition of miR-429 remarkably increased the levels of PHD2, which disrupted PHD2-associated adaptive activation of HIF-1α-mediated gene expression in response to high salt in the renal medulla, and consequently inhibited urinary sodium excretion, enhanced sodium retention in response to chronic sodium overloading, and as a result, produced a salt-sensitive hypertension.
Conclusion
It is concluded that miR-429 is an important upstream mediator in PHD2/HIF-1α-associated renal adaptation to high salt intake and that deficiency in miR-429-mediated PHD2 inhibition in response to high salt in the renal medulla may represent a pathogenic mechanism for salt-sensitive hypertension.
Keywords: hypertension, hypoxia-inducible factor prolyl hydroxylase, microRNA, mRNA decay, salt balance
INTRODUCTION
Salt-sensitive hypertension accounts for 50% of hypertensive population 1. Importantly, the salt sensitivity of blood pressure is closely associated with a much greater risk to develop organ injuries in hypertension1–3. Mechanism for salt-sensitive hypertension remains not fully understood. It is well documented that renal medullary function plays a pivotal role in the regulation of sodium excretion and arterial blood pressure, and that functional impairment of the renal medulla is involved in salt-sensitive hypertension 4–6. Our previous studies have shown that high salt intake increases the level of the transcription factor hypoxia-inducible factor (HIF)-1α, which activates many antihypertensive genes, such as nitric oxide synthase (NOS) and hemeoxygenase-1 (HO-1) in the renal medulla 7, 8. The products of these HIF-1α target genes increase renal medullary blood flow and inhibit sodium reabsorption, thereby promoting the excretion of extra sodium load and regulating the renal adaptation to high salt intake.
High salt intake significantly increases HIF-1α levels along with many of its target genes such as home oxgenase 1, nitric oxide synthase 2, and endothelin 1 in the renal medulla 4, 9–11. The products of these HIF-1α target genes play critical roles in the maintenance of sodium balance and regulation of arterial blood pressure by increasing blood flow and/or inhibiting tubular reabsorption in the renal medulla 9, 11–13. It has been shown that blockade of HIF-1α to inhibit the expression of its downstream target genes in the renal medulla reduces sodium excretion and induces a salt-sensitive hypertension in response to high salt intake 8. These results suggest that HIF-1α-mediated gene activation in the renal medulla represents an important molecular adaptive mechanism to maintain sodium balance in response to high salt intake.
HIF prolyl-hydroxylases (PHDs) are the major enzymes that regulate the degradation of HIF-1α. PHDs catalyze proline hydroxylation of HIF-1α and that prolyl-hydroxylated HIF-1α is then recognized and targeted for degradation through ubiqutin-proteasome pathway 14, 15. Three isoforms of PHDs, including PHD1, 2 and 3, have been identified 16–18 and present in the kidneys with PHD2 as the predominant isoform 19–23, abundantly expressed in the renal medulla 19, 23. We have shown that inhibition of PHD2 activity increases the levels of HIF-1α in the renal medulla and enhances pressure natriuresis, suggesting a critical role of PHD2 in the regulation of renal medullary function 19. Our data have further demonstrated that high salt intake suppresses the expression of PHD2 in the renal medulla and that this high salt-induced inhibition of PHD2 serves as an important upstream signal to activate HIF-1α-mediated gene regulation facing high salt challenge 24. Moreover, interruption of the high salt-induced suppression of PHD2 and thereafter the HIF-1α-mediated gene activation impairs renal medullary function, blunts pressure natriuresis, attenuates sodium excretion, promotes sodium retention and induces salt-sensitive hypertension in uninephrectomized rats 25. Thus, PHD2-mediated activation of HIF-1α target genes in the renal medulla is an important adaptive mechanism in response to extra salt load and thereby regulates salt sensitivity of blood pressure. However it remains unanswered how high salt intake inhibits PHD2 expression in the renal medulla.
The microRNAs (miRNAs) are crucial regulators affecting gene expression. The miRNAs are short non-coding RNAs, with approximately 17–25 nucleotides in length, which negatively regulate gene expression at the post-transcriptional level by coupling with sites in 3′-untranslated (3′-UTR) in the target mRNAs, thereby altering stability of the target mRNAs 26, 27. The miRNA has been shown to reduce gene expression mostly through mRNA destabilization with a small effect via translational repression 28. It has been recently recognized that miRNAs participate in the regulation of vascular function and renin angiotensin system, implicating a role of miRNAs in the development of hypertension 29. However, little is known about the role of renal miRNAs in the regulation of sodium excretion and long-term control of blood pressure.
In the present study, we found that a high salt diet significantly increased the expression of miR-429 in the renal medulla in rats and further identified that miR-429 potentially targeted PHD2 mRNA. Then, we transfected miR-429 inhibitor into the renal medulla to block the increase of miR-429 levels after high salt challenge and examined its effect on the expression of PHD2 and HIF-1α-mediated gene activation, as well as on the sodium excretion and blood pressure after chronic sodium overloading. Our results for the first time demonstrated that miRNA-429 mediates high salt-induced PHD2 inhibition via enhancing mRNA decay and that inhibition of miRNA-429 to disrupt this PHD2-associated adaptive activation of HIF-1α-mediated gene expression in response to high salt challenge in the renal medulla reduced sodium excretion and produced salt-sensitive hypertension.
MATERIALS AND METHODS
Animals
Experiments were performed using male Sprague-Dawley rats (250–350g, Harlan, Madison, WI). Rats were kept on a low-salt diet (0.4% NaCl, Dyets), with some fed a high-salt diet (8% NaCl) as indicated in the Results section. All animal procedures were approved by the Institutional Animal Care and Use Committee of Virginia Commonwealth University.
In vivo administration of actinomycin D into rats
To investigate whether high salt-reduced PHD2 mRNA level was due to the enhanced decay of PHD2 mRNA, rats were fed with a low or high salt diet for 10 days and then injected with actinomycin D (400ug/100g BW, i.p.) to block the synthesis of mRNA 30. At 3 and 6 hour after actinomycin D injection, kidneys were collected for the measurement of PHD2 mRNA levels in the renal medulla.
Database search
Prediction of miRNA target was retrieved from an online miRNA database miRDB (http://mirdb.org/miRDB/). It was shown that miR-290, miR-143, miR-200b, miR-200c, and miR-429 potentially target PHD2.
RNA extraction and quantitative reverse-transcribed-PCR analysis of individual micro RNAs, prolyl hydroxylase domain-containing protein 2 and heme oxygenase-1 mRNA
Total RNA from renal medullary tissues was extracted using TRIzol solution. For miRNA assay, RNA was reverse-transcribed (RT) and the RT products were then amplified using TaqMan MicroRNA Assays kits (Applied Biosystems) for individual miRNAs (miR-290, miR-143, miR-200b, miR-200c, and miR-429) with a Stratagene Mx3000P Real-Time PCR Detection System (Agilent Technologies) 31, 32. The level of U6 small nuclear RNA (snRNA) was used as endogenous normalizer for the measurement of individual miRNAs. For mRNA assay, RNA was reverse-transcribed (RT) using a cDNA Synthesis Kit (Bio-Rad) and the mRNA levels of PHD2 and HO-1 in the renal medullary were determined by real-time RT-PCR with 18S ribosomal RNA as internal control. The relative gene expressions were calculated using cycle threshold (Ct) values in accordance with the ΔΔCt method.
Assay of 3′-UTR luciferase reporter
To test whether a miRNA directly targets the 3′ UTR of PHD2, luciferase reporter assay was carried out. The constructs of luciferase reporter with rat PHD2 3′-UTR (RmiT) and its control vector (CmiT) were customized from Genecopoeia. Renal medullary interstitial cells (RMIC) were seeded in a 96-well plate (5 ×103 cells/well). After 24 h, cells were co-transfected with RmiT or CmiT together with miRNA-429 precursor or scrambled control. Luciferase activities were measured 24 hours after transfection using the Luc-Pair miR luciferase kit (GeneCopoeia) following the manufacturer’s protocol. The miRNA-429 precursor and scrambled miRNA precursor as a negative control were purchased from Genecopoeia (Maryland, US).
The micro RNA inhibitor
The locked nucleic acid (LNA) miRNA power Inhibitor for rat miR-429 (LNA anti-miR-429) and the scrambled control inhibitor (LNA anti-miR) were purchased from Exiqon (Woburn, MA). The sequence of anti-miR-429 was CGGCATTACCAGACAGTATT. Locked Nucleic Acid (LNA)-modified anti-miR are antisense oligonucleotides with perfect sequence complementary to their target. When introduced into cells, they sequester their target microRNA in highly stable heteroduplexes thereby effectively preventing the microRNA from hybridizing with its normal cellular interaction partners. The pre-designed sequences of the anti-miR and their LNA™ spiking patterns are generated using a patent design algorithm by Exiqon. The resulting inhibitors have uniform high potency regardless of the GC content of their microRNA targets. In addition, the LNA™ microRNA Power Inhibitors have phosphorothioate modified backbones that dramatically improve their biological stability.
Transfection of locked nucleic acid anti-miR-429 into the renal medulla in vivo
LNA modified anti-miR has been shown as an efficient strategy to knock down specific miRNA in vivo and that the inhibitory effect of LNA anti-miR lasts at least for 14 days 32 or 5 weeks 33 after injection. The transfection of LNA anti-miR-429 or LNA scrambled anti-miR into the renal medulla was performed as described previously 25, 33, 34. In brief, uninephrectomized rats were anaesthetized with 2.5% isoflurane and placed on a thermostatic table to maintain a body temperature of 37°C. Then the remaining left kidney was exposed by a flank incision (1–1.5 cm) and placed in a cup to stabilize the organ for implanting a medullary interstitial catheter (4–5 mm long with a tapped tip) into the kidney. The catheter was anchored into place on the kidney surface with Vetbond Tissue Adhesive (3M) and 0.6 mL of mixture containing 50 μg of LNA anti-miR-429 or scrambled LNA anti-miR (Exiqon, MA, USA) and microbubble (Optison) at a ratio of 1:1 was infused into renal medulla at a speed of 10 μl/min. After infusion, the catheter was cut and blocked by a piece of fat tissue with Vetbond Tissue Adhesive. An ultrasound transducer (Sonitron 2000; Rich-Mar, Inola, OK, USA) was directly applied onto the kidneys with a 1-MHz ultrasound at 10% power output, for a total of 60 sec. with 30-sec. intervals on each side of the kidney 35 in the middle and at the end of the infusion. The ultrasound radiation has been shown to significantly improve the efficiency of gene transfection in combination with different transfection reagents 36, 37. Previous studies, including ours, showed that this ultrasound-microbubble technique effectively delivered DNA into cells in the kidneys with a >90% of transfection rate without toxicity to the kidney 8, 35, 38, 39. We also performed in vitro experiments to confirm the inhibition efficiency of LNA anti-miR-429 in cultured RMICs.
Measurement of daily sodium balance
Additional rats were treated the same as above and then housed in metabolic cages. Daily sodium balance was calculated by subtracting sodium excretion from sodium intake. After 1 day of control measurements, rats were fed with 2% NaCl water, and daily sodium balance was measured for 3 more days 33, 40, 41.
Chronic monitoring of arterial blood pressure in conscious rats
Blood pressure was measured using a telemetry system as previously described 42, 43. Three-day baseline mean arterial pressure (MAP) was recorded when the animals remained on low-salt diet. Then rats were fed with a high salt diet (Dyets, Bethlehem, PA), and MAP was recorded for 10 more days. Rats were divided into 3 groups: (i) LNA scrambled anti-miR+ high-salt diet, (ii) LNA anti-miR-429 + high-salt diet, and (iii) LNA anti-miR-429 + low-salt diet. After MAP recording, kidneys were removed and saved for the isolation of protein and RNA later.
Western blot analysis of protein levels HIF-1α and PHD2 in renal medulla
Renal tissue homogenates and nuclear proteins from the renal medulla were extracted and then subjected (50 μg) to Western blot analysis as previously described 19, 44. The primary antibodies against HIF-1α (monoclonal, 1:300) and PHD2 (rabbit polyclonal, 1:300) were from Novus Biologicals (Littleton, CO). The intensities of the blots were analyzed using ImageJ software (http://rsbweb.nih.gov/ij/). The levels of β-actin were used as internal control.
Statistical analysis
Data are presented as means ± SE. The significance of differences in mean values within and between multiple groups was evaluated using an analysis of variance followed by Tukey’s post hoc test. Student t test was used to evaluate statistical significance of differences between 2 groups. P < 0.05 was considered statistically significant.
RESULTS
Effect of high salt intake on the decay of PHD2 mRNA in the renal medulla
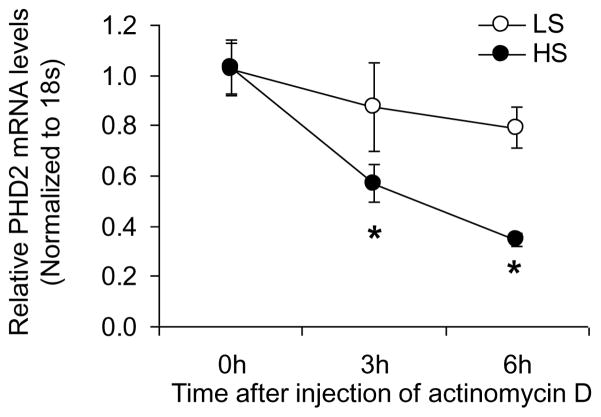
After injection of actinomycin D to block the synthesis of new RNA, PHD2 mRNA levels in the renal medulla were progressively decreased in both low and high salt groups, whereas the reduction of PHD2 mRNA levels was more significant in high salt-treated rats compared with low salt-treated rats (Fig. 1). These results suggest that enhanced decay of PHD2 mRNA pivotally contributes to the high salt-induced reduction of PHD2 mRNA level in the renal medulla.
Figure 1. Effect of high salt intake on the degradation of PHD2 mRNA in the renal medulla.
* P<0.05 vs. LS. LS, low salt; HS, high salt. (n=6)
Effect of high salt intake on the levels of miRNAs potentially targeting PHD2 gene in the renal medulla
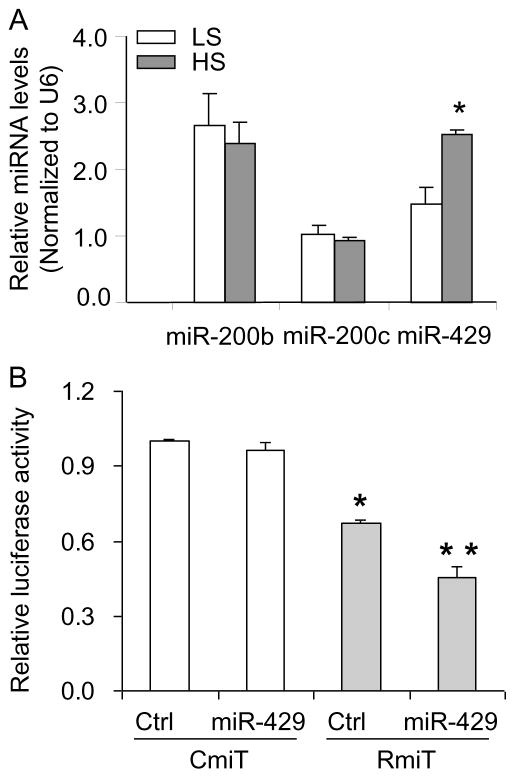
To identify the potential miRNAs that targeted PHD2 mRNA in response to high salt, five miRNAs (miR-290, miR-143, miR-200b, miR-200c, and miR-429), which were predicted to target PHD2 mRNA based on bioinformatic analysis, were selected. The levels of miR-290 and miR-143 were undetectable and the expression patterns of the other three miRNAs in the renal medulla in response to the high salt intake are summarized in Figure 2A. High salt intake significantly increased the level of miR-429, while the expressions of miR-200b and miR-200c remained no change. These data indicates that miR-429 may be the candidate miRNA targeting PHD2 mRNA and directing its decay.
Figure 2. Expression of miRNAs potentially targeting PHD2 mRNA in the renal medulla and interaction of miR-429 with 3′-UTR segment of PHD2 in cultured RMIC cells.
A: High salt intake increased the level of miR-429. * P<0.05 vs. LS. LS, low salt; HS, high salt. (n=5). B: miR-429 decreased luciferase activity when the luciferase gene was constructed with the 3′-UTR segment of PHD2. RMIC cells were co-transfected with rat PHD 2 3′-UTR luciferase reporter constructs RmiT or control 3′-UTR CmiT, and miR-429 precursor (miR-429) or scrambled miRNA precursor (Ctrl). * P<0.05 vs. cells treated with CmiT. ** P<0.05 vs. cells treated with RmiT and Ctrl. RmiT, construct of luciferase reporter with 3′-UTR of rat PHD2; CmiT, construct of luciferase reporter with scrambled 3′-UTR; Ctrl, scrambled miRNA precursor; miR-429, miRNA-429 precursor (n=4–6).
Effect of miR-429 on the luciferase activities of the 3′-UTR of PHD2 construct
To determine whether miR-429 targets PHD2 mRNA, the 3′-UTR segment of rat PHD2 gene was cloned to the downstream of a luciferase reporter gene in the expression construct Rmi-MT01 and that the interactions between miR-429 and the 3′-UTR segment of PHD2 would be expected to decrease luciferase activity. Our results showed that the luciferase activity in RMIC transfected with Rmi-MT01/Ctrl was significantly decreased, which was further decreased in Rmi-MT01/miR-429-treated cells, compared to that in Cmi-MT01/Ctrl and Cmi-MT01/miR-429 treated cells. These data confirmed that miR-429 targets the 3′-UTR of PHD2 mRNA (Fig. 2B).
Effect of renal medullary transfection of LNA anti-miR-429 on the levels of PHD2, HIF-1α and its target gene HO-1 in the renal medulla
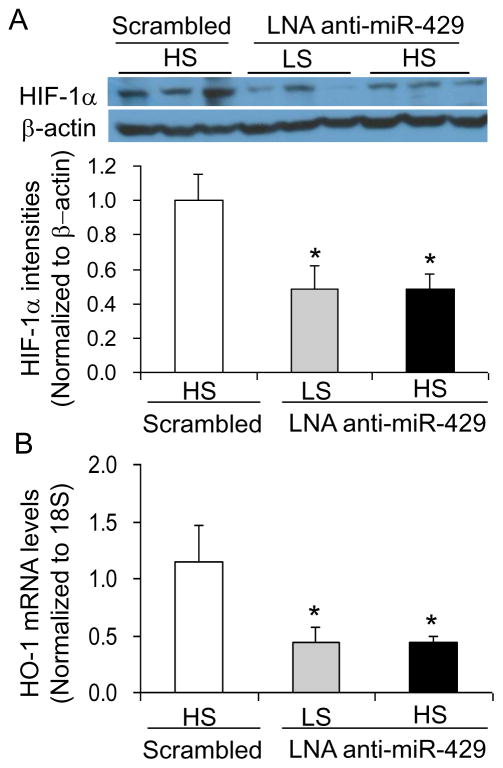
Real-time RT-PCR failed to amplify miR-429 in LNA anti-miR-429-treated rats compared with rats treated with LNA scrambled anti-miR (Fig. 3A) as the RT-PCR system would not be able to amplify the miRNA bound with anti-miR, indicating an inhibition of miR-429 by anti-miR. LNA anti-miR-429-treated rats showed remarkable increases in the levels of PHD2 mRNA and protein in the renal medulla (Fig. 3B&C). As a result, HIF-1α levels were significantly decreased in the renal medulla from rats treated with LNA anti-miR-429 (Fig. 4A). In addition, the mRNA levels of HO-1, one of the important HIF-1α target genes and as a prototype of HIF-1α target genes, in the renal medulla were also similarly decreased in rats treated with LNA anti-miR-429 (Fig. 4B). These results verified the successful inhibition of miR-429 and the consequent increase of PHD2, which inhibited the high salt-induced activation of HIF-1α and its target genes in the renal medulla by LNA anti-miR-429.
Figure 3. Effect of renal medullary transfection of LNA anti-miR-429 on the levels of PHD2 mRNA and PHD2 protein in the renal medulla.
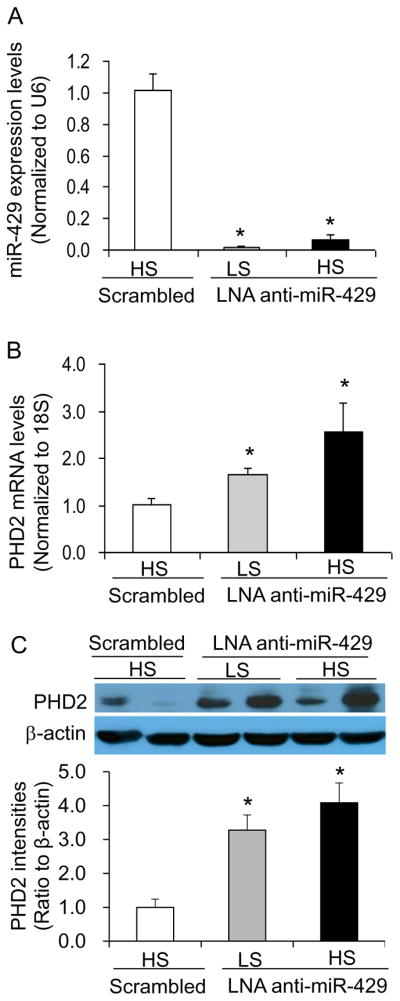
A: Real-time RT-PCR analysis of miR-429. B: Real-time RT-PCR analysis of PHD2 mRNA levels. C: Western blot analyses of PHD2. * P < 0.05 vs. the scramble. n = 5. LS, low salt; HS, high salt; Scrambled, LNA Scrambled anti-miR-429. Renal medullary tissues were obtained at the end of blood pressure recording after 10 days high salt challenge.
Figure 4. Effect of renal medullary transfection of LNA anti-miR-429 on the levels of HIF-1α protein and HO-1 mRNA in the renal medulla.
A: Western blot analyses of HIF-1α. B: Real-time RT-PCR analysis of HO-1 mRNA levels. * P < 0.05 vs. the scramble. n = 5. LS, low salt; HS, high salt; Scrambled, LNA Scrambled anti-miR-429. Renal medullary tissues were obtained at the end of blood pressure recording after 10 days high salt challenge.
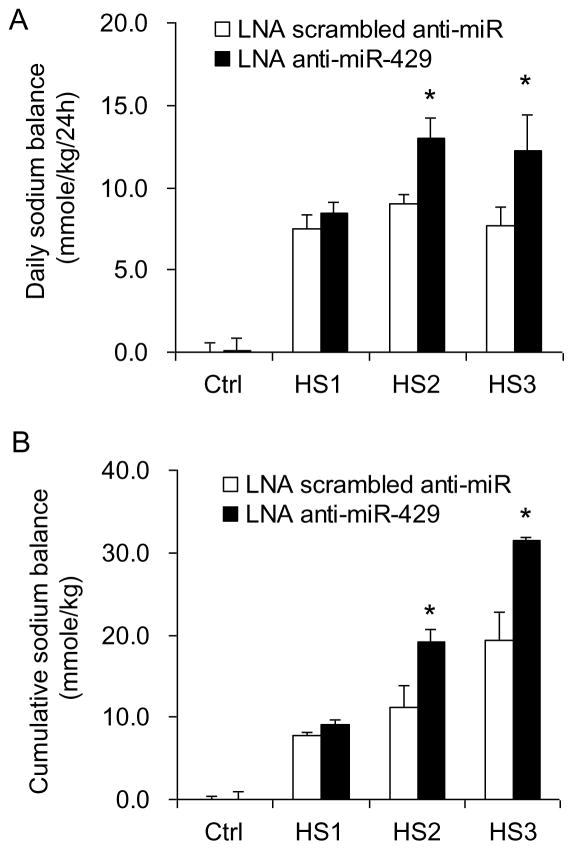
Effect of renal medullary transfection of LNA anti-miR-429 on salt balance after chronic sodium loading. High salt intake produced a positive daily and cumulative salt balance. The daily positive salt balances were progressively increased in the first 2 days and decreased on the third day of high salt intake. The positive salt balances were significantly enhanced in LNA anti-miR-429-treated rats compared with the rats treated with LNA scrambled anti-miR (Fig. 5).
Figure 5. Effect of renal medullary transfection of LNA anti-miR-429 on salt balance after chronic sodium overloading.
A: Daily sodium balance. B: Cumulative sodium balance. * P< 0.05 vs. the scramble. (n =5)
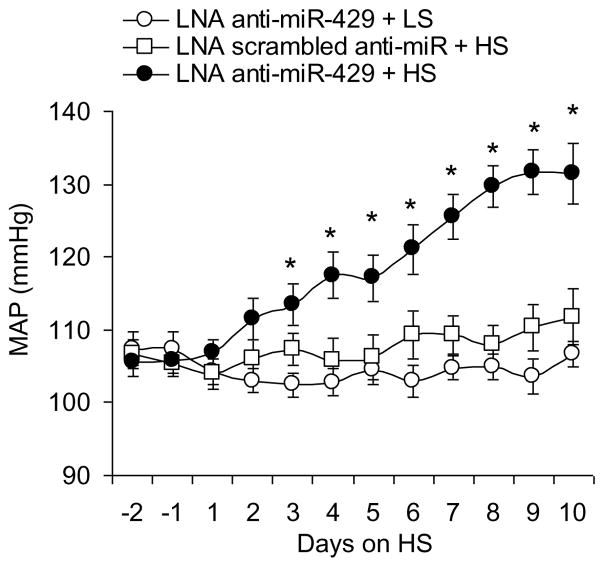
Effect of renal medullary transfection of LNA anti-miR-429 on blood pressure after high salt intake
The MAPs are presented in Figure 6. After the rats were challenged with a high salt diet, the MAP was progressively increased in rats treated with LNA anti-miR-429 compared with that in control rats. However, there was no change in MAP in rats treated with LNA anti-miR-429 when the rats were not challenged with high salt diet (Fig. 6).
Figure 6. Effect of renal medullary transfection of LNA anti-miR-429 on mean arterial pressure (MAP).
* P< 0.05 vs. other groups. (n= 7)
DISCUSSION
In the current study, we found that high salt intake significantly enhanced the decay of PHD2 mRNA, which was accompanied with an increased expression of miR-429, in the renal medulla of SD rats. We confirmed that miR-429 targeted the 3′-UTR of PHD2 mRNA and further demonstrated that inhibition of miR-429 remarkably increased the mRNA levels of PHD2 and decreased the levels of HIF-1α and its target gene HO-1. Our results also demonstrated that blocking the high salt-induced increase of miR-429 to disrupt PHD2-associated adaptive activation of HIF-1α-mediated gene expression in the renal medulla consequently inhibited urinary sodium excretion after chronic sodium overloading, promoted sodium retention, and as a result, produced salt-sensitive hypertension.
Our previous study showed that high salt intake reduced PHD2 mRNA levels in the renal medulla of SD rats 24. To determine whether high salt reduced PHD2 mRNA level was due to the enhanced decay of PHD2 mRNA, rats were treated with low or high salt diet first and then injected with actinomycin D to block the synthesis of new RNA, which enables us to determine the decay of PHD2 mRNA. Our result showed that the degradation rate of PHD2 mRNA in the renal medulla was significantly enhanced by high salt intake, suggesting that enhanced decay of mRNA pivotally contributes to the reduction of renal medullary PHD2 mRNA level in response to high salt intake.
It has been demonstrated that miRNA binding to the 3′-UTR alters the stability of target mRNAs 26, 27 and that miRNA reduces gene expression mostly through mRNA destabilization with a small effect via translational repression 28. To determine whether the enhanced decay of PHD2 mRNA was associated with miRNA, we detected the levels of miRNAs targeting PHD2 mRNA and found that miR429, one member of miR-200 family, was significantly upregulated in the renal medulla by high salt, which led to a further investigation on the interaction between miR-429 and the 3′-UTR of PHD2 mRNA. The interaction between miR-429 and the 3′-UTR of PHD2 mRNA from the results of 3′-UTR luciferase reporter analysis further supported our hypothesis that miR-429 mediates high salt-induced reduction of PHD2 mRNA via enhancing its decay. Our current finding was consistent with a previous report showing that upregulation of miR-200 family miR-200b, miR-200c and miR-429 was responsible for the downregulation of PHD2 mRNA and subsequent increase of HIF-1α during cerebral ischemia 45.
Our results showed that local inhibition of miR-429 by LNA anti-miR-429 increased the levels of PHD2 and subsequently inhibited the activation of HIF-1α in the renal medulla after high salt intake. Given the pivotal role of PHD2 regulation of HIF-1α-mediated gene activation in the renal adaptive mechanisms to high salt challenge and kidney sodium handling 7, 25, manipulation of PHD2/HIF-1α pathway by LNA anti-miR-429 enabled us to evaluate the role of miR-429 in the regulation of renal sodium excretion and long-term control of arterial pressure in response to high salt challenge.
To evaluate the role of miR-429 in the regulation of renal sodium excretion, we examined salt balance after chronic sodium challenge. As high salt-induced inhibition of PHD2 levels is an important molecular adaptive mechanism to maintain salt balance 24, increasing PHD2 expression by blocking miR-429 to reduce HIF-1α levels along with its target genes would be expected to impair normal renal medullary function and disrupt salt balance. Our data showed that inhibition of miR-429 remarkably impaired the capability of the kidneys to remove extra sodium load and promoted sodium retention. These results demonstrated that anti-miR-429 disrupted the renal adaptive response to extra sodium load and additionally indicated that activation of renal medullary miR-429 levels by high salt challenge is an important determinant to promote sodium excretion after high salt intake.
As normal renal medullary function plays a pivotal role in the chronic regulation of blood pressure 6, 46, suppression of the miR-429 in the renal medulla to impair renal sodium excretion would lead to an increase in blood pressure upon chronic high salt challenge. To test this hypothesis, we compared MAPs between animals transfected with LNA anti-miR-429 and LNA scrambled anti-miR into the renal medulla. Our data showed that high salt challenge significantly increased the MAP in LNA anti-miR-429-treated rats, but not in LNA scrambled anti-miR-treated rats, whereas LNA anti-miR-429-treated rats did not develop hypertension when they were maintained on a low salt diet. These results suggested that miR-429 is an important upstream mediator in the regulation of salt sensitivity of blood pressure, probably through governing the renal sodium handling via tuning the PHD2/HIF-1α pathway.
Our findings on the other hand may suggest that an impaired response of miR-429 to high salt intake could be responsible for the occurrence of salt-sensitive hypertension. In this regard, we have shown before that impairment in PHD2/HIF-1α-mediated renal adaptation to high salt challenge contributes to the mechanism of salt-sensitive hypertension in the Dahl S rats 7, 25. Given the significant role of renal medullary miR-429 in the regulation of PHD2/HIF-1α pathway and sodium excretion shown in the current study, the impairment in miR-429 response to high salt intake would be very possibly the pathogenic mechanism in Dahl S rats, which requires further investigation.
The present study did not attempt to explore the mechanisms by which the high salt intake increased the expression of miR-429. In this regard, the transcriptional regulation may be a candidate mechanism. The transcription of miRNA genes is regulated in a similar manner to that of protein-coding genes and is a major level of control responsible for specific miRNA expression. Therefore, transcription factors remain the major factors that regulate specific miRNA expression 47, 48. Tissue-specific enrichment of certain transcription factors is also one of the mechanisms for tissue-specific miRNA expression 47, 48. It has been shown that p63 and p73 are upstream transcription factors that activate miR-429 transcription 49. To detect the response of p63 and/or p73 to high salt intake and determine their roles in miR-429 expression may shed the light on the mechanism mediating high salt-induced miR-429.
It should be pointed out that the high salt intake in the present study was higher than the high salt intake (5–10 g/day sodium) observed in human population 50, as we focused on the response to a high salt intake. The observation in the present study may or may not be applied to the situation with the usual (normal) range salt intake. Because the salt sensitivity of blood pressure has been observed in normotensive individuals on the usual salt intake 51, whether miR-429-mediated PHD2/HIF-1α pathway participates in the regulation of sodium excretion and salt sensitivity of blood pressure under such a condition with the usual salt intake needs to be further determined.
Another limitation in the present study is that the miR inhibitor, anti-miR, used in the present study is to sequester the target microRNA, which may block the binding of miR-429 to all its targets. Although PHD2/HIF-1α pathway is altered by anti-miR-429 and that PHD2/HIF-1α pathway has been shown to participate in the regulation of sodium excretion and salt sensitivity of blood pressure 7, 25, the effect of anti-miR-429 on renal sodium handling and blood pressure regulation may not necessarily only through PHD2/HIF-1α pathway. The possible unknown roles of other targets of miR-429 cannot be ruled out. Using approaches such as microRNA target site blockers, which bind to the microRNA target site of an mRNA thereby interrupting microRNA interaction with a single mRNA, may help to dissect the role of a specific target of miR-429 vs. miR-429 per se. In addition, there may be unrelated and/or unwanted effects of anti-miR-429 via other targets. Nevertheless, the present study revealed the important role of miR-429 in renal sodium handling and BP regulation. Further investigation is required to clarify which exact target(s) of miR-429 mediates its effects on renal sodium handling.
In summary, the present study for the first time demonstrated that high salt intake enhanced the decay of PHD2 mRNA via inducing the miR-429 level in the renal medulla, and that inhibition of high salt-induced increase of miR-429 level disrupted the PHD2-associated adaptive activation of HIF-1α-mediated gene expression in response to high salt challenge in the renal medulla, which consequently inhibited sodium excretion and promoted sodium retention in response to chronic sodium overloading, thereby producing a salt-sensitive hypertension. These results suggest that miR-429 is an important upstream mediator in PHD2/HIF-1α-associated renal adaptation to high salt intake and that impairment in miR-429-mediated PHD2 inhibition in response to high salt intake in the renal medulla may represent a pathogenic mechanism for salt-sensitive hypertension. These findings provide novel insights into the molecular adaptation to the high salt intake in the kidneys and advance our understanding about the mechanism mediating salt sensitivity of blood pressure. Modulation of this miR-429-associated molecular adaptive pathway to enhance the production of anti-hypertensive factors in the renal medulla in response to high salt challenge may have important therapeutic implications for the prevention and/or treatment of salt-sensitive hypertension.
Acknowledgments
Funding: National Institutes of Health (Grants HL089563 and HL106042)
The Fundamental Research Funds of Sun Yat-sen University (16YKPY47)
Abbreviations
- HIF
hypoxia inducible factor
- HO
heme oxygenase
- LNA
locked nucleic acid
- MAP
mean arterial pressure
- miRNA
microRNA
- PHD
prolyl hydroxylase domain-containing protein
- UTR
untranslated region
Footnotes
Disclosure: the authors declared no conflict of interest.
References
- 1.Chrysant GS, Bakir S, Oparil S. Dietary salt reduction in hypertension--what is the evidence and why is it still controversial? Prog Cardiovasc Dis. 1999;42:23–38. doi: 10.1016/s0033-0620(99)70007-1. [DOI] [PubMed] [Google Scholar]
- 2.Campese V. Salt sensitivity in hypertension. Renal and cardiovascular implications [clinical conference] Hypertension. 1994;23:531–550. doi: 10.1161/01.hyp.23.4.531. [DOI] [PubMed] [Google Scholar]
- 3.Weinberger MH, Fineberg NS, Fineberg SE, Weinberger M. Salt Sensitivity, Pulse Pressure, and Death in Normal and Hypertensive Humans. Hypertension. 2001;37:429–432. doi: 10.1161/01.hyp.37.2.429. [DOI] [PubMed] [Google Scholar]
- 4.Li N, Yi F, dos Santos EA, Donley DK, Li PL. Role of renal medullary heme oxygenase in the regulation of pressure natriuresis and arterial blood pressure. Hypertension. 2007;49:148–54. doi: 10.1161/01.HYP.0000250086.06137.fb. [DOI] [PubMed] [Google Scholar]
- 5.Cowley AW, Jr, Mattson DL, Lu S, Roman RJ. The renal medulla and hypertension. Hypertension. 1995;25:663–73. doi: 10.1161/01.hyp.25.4.663. [DOI] [PubMed] [Google Scholar]
- 6.Mattson DL. Importance of the renal medullary circulation in the control of sodium excretion and blood pressure. Am J Physiol Regul Integr Comp Physiol. 2003;284:R13–27. doi: 10.1152/ajpregu.00321.2002. [DOI] [PubMed] [Google Scholar]
- 7.Li N. Hypoxia inducible factor-1alpha-mediated gene activation in the regulation of renal medullary function and salt sensitivity of blood pressure. Am J Cardiovasc Dis. 2012;2:208–15. [PMC free article] [PubMed] [Google Scholar]
- 8.Li N, Chen L, Yi F, Xia M, Li P-L. Salt-Sensitive Hypertension Induced by Decoy of Transcription Factor Hypoxia-Inducible Factor-1{alpha} in the Renal Medulla. Circ Res. 2008;102:1101–1108. doi: 10.1161/CIRCRESAHA.107.169201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mattson DL, Higgins DJ. Influence of dietary sodium intake on renal medullary nitric oxide synthase. Hypertension. 1996;27:688–92. doi: 10.1161/01.hyp.27.3.688. [DOI] [PubMed] [Google Scholar]
- 10.Nakano D, Pollock D. New concepts in endothelin control of sodium balance. Clin Exp Pharmacol Physiol. 2012;39:104–10. doi: 10.1111/j.1440-1681.2011.05517.x. [DOI] [PubMed] [Google Scholar]
- 11.Tan DY, Meng S, Cason GW, Manning RD., Jr Mechanisms of salt-sensitive hypertension: role of inducible nitric oxide synthase. Am J Physiol Regul Integr Comp Physiol. 2000;279:R2297–303. doi: 10.1152/ajpregu.2000.279.6.R2297. [DOI] [PubMed] [Google Scholar]
- 12.Nakanishi K, Hara N, Nagai Y. Salt-sensitive hypertension in conscious rats induced by chronic nitric oxide blockade. Am J Hypertens. 2002;15:150–6. doi: 10.1016/s0895-7061(01)02267-1. [DOI] [PubMed] [Google Scholar]
- 13.Mattson DL, Maeda CY, Bachman TD, Cowley AW., Jr Inducible nitric oxide synthase and blood pressure. Hypertension. 1998;31:15–20. doi: 10.1161/01.hyp.31.1.15. [DOI] [PubMed] [Google Scholar]
- 14.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–8. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 15.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, Ratcliffe PJ. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–72. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 16.Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O’Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, Tian YM, Masson N, Hamilton DL, Jaakkola P, Barstead R, Hodgkin J, Maxwell PH, Pugh CW, Schofield CJ, Ratcliffe PJ. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 17.Page EL, Chan DA, Giaccia AJ, Levine M, Richard DE. Hypoxia-inducible factor-1alpha stabilization in nonhypoxic conditions: role of oxidation and intracellular ascorbate depletion. Mol Biol Cell. 2008;19:86–94. doi: 10.1091/mbc.E07-06-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McMahon S, Charbonneau M, Grandmont S, Richard DE, Dubois CM. Transforming growth factor beta1 induces hypoxia-inducible factor-1 stabilization through selective inhibition of PHD2 expression. J Biol Chem. 2006;281:24171–81. doi: 10.1074/jbc.M604507200. [DOI] [PubMed] [Google Scholar]
- 19.Li N, Yi F, Sundy CM, Chen L, Hilliker ML, Donley DK, Muldoon DB, Li PL. Expression and actions of HIF prolyl-4-hydroxylase in the rat kidneys. Am J Physiol Renal Physiol. 2007;292:F207–16. doi: 10.1152/ajprenal.00457.2005. [DOI] [PubMed] [Google Scholar]
- 20.Takeda K, Cowan A, Fong GH. Essential role for prolyl hydroxylase domain protein 2 in oxygen homeostasis of the adult vascular system. Circulation. 2007;116:774–81. doi: 10.1161/CIRCULATIONAHA.107.701516. [DOI] [PubMed] [Google Scholar]
- 21.Rosenberger C, Rosen S, Shina A, Frei U, Eckardt KU, Flippin LA, Arend M, Klaus SJ, Heyman SN. Activation of hypoxia-inducible factors ameliorates hypoxic distal tubular injury in the isolated perfused rat kidney. Nephrol Dial Transplant. 2008;23:3472–8. doi: 10.1093/ndt/gfn276. [DOI] [PubMed] [Google Scholar]
- 22.Takeda K, Aguila HL, Parikh NS, Li X, Lamothe K, Duan LJ, Takeda H, Lee FS, Fong GH. Regulation of adult erythropoiesis by prolyl hydroxylase domain proteins. Blood. 2008;111:3229–35. doi: 10.1182/blood-2007-09-114561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schodel J, Klanke B, Weidemann A, Buchholz B, Bernhardt W, Bertog M, Amann K, Korbmacher C, Wiesener M, Warnecke C, Kurtz A, Eckardt KU, Willam C. HIF-prolyl hydroxylases in the rat kidney: physiologic expression patterns and regulation in acute kidney injury. Am J Pathol. 2009;174:1663–74. doi: 10.2353/ajpath.2009.080687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Zhu Q, Xia M, Li PL, Hinton SJ, Li N. Hypoxia-inducible factor prolyl-hydroxylase 2 senses high-salt intake to increase hypoxia inducible factor 1alpha levels in the renal medulla. Hypertension. 2010;55:1129–36. doi: 10.1161/HYPERTENSIONAHA.109.145896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu Q, Liu M, Han WQ, Li PL, Wang Z, Li N. Overexpression of HIF prolyl-hydoxylase-2 transgene in the renal medulla induced a salt sensitive hypertension. J Cell Mol Med. 2012;16:2701–7. doi: 10.1111/j.1582-4934.2012.01590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 27.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 28.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–40. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Batkai S, Thum T. MicroRNAs in hypertension: mechanisms and therapeutic targets. Curr Hypertens Rep. 2012;14:79–87. doi: 10.1007/s11906-011-0235-6. [DOI] [PubMed] [Google Scholar]
- 30.Yao J, Kathpalia P, Bushinsky DA, Favus MJ. Hyperresponsiveness of vitamin D receptor gene expression to 1,25-dihydroxyvitamin D3. A new characteristic of genetic hypercalciuric stone-forming rats. J Clin Invest. 1998;101:2223–32. doi: 10.1172/JCI1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmittgen TD, Lee EJ, Jiang J, Sarkar A, Yang L, Elton TS, Chen C. Real-time PCR quantification of precursor and mature microRNA. Methods. 2008;44:31–8. doi: 10.1016/j.ymeth.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, Taylor NE, Lu L, Usa K, Cowley AW, Jr, Ferreri NR, Yeo NC, Liang M. Renal medullary microRNAs in Dahl salt-sensitive rats: miR-29b regulates several collagens and related genes. Hypertension. 2010;55:974–82. doi: 10.1161/HYPERTENSIONAHA.109.144428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 34.Zhu Q, Wang Z, Xia M, Li PL, Zhang F, Li N. Overexpression of HIF-1alpha transgene in the renal medulla attenuated salt sensitive hypertension in Dahl S rats. Biochim Biophys Acta. 2012;1822:936–41. doi: 10.1016/j.bbadis.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lan HY, Mu W, Tomita N, Huang XR, Li JH, Zhu HJ, Morishita R, Johnson RJ. Inhibition of renal fibrosis by gene transfer of inducible Smad7 using ultrasound-microbubble system in rat UUO model. J Am Soc Nephrol. 2003;14:1535–48. doi: 10.1097/01.asn.0000067632.04658.b8. [DOI] [PubMed] [Google Scholar]
- 36.Hosseinkhani H, Aoyama T, Ogawa O, Tabata Y. Ultrasound enhances the transfection of plasmid DNA by non-viral vectors. Curr Pharm Biotechnol. 2003;4:109–122. doi: 10.2174/1389201033489883. [DOI] [PubMed] [Google Scholar]
- 37.Newman CMH, Bettinger T. Gene therapy progress and prospects: Ultrasound for gene transfer. Gene Ther. 2007;14:465–475. doi: 10.1038/sj.gt.3302925. [DOI] [PubMed] [Google Scholar]
- 38.Hou C-C, Wang W, Huang XR, Fu P, Chen T-H, Sheikh-Hamad D, Lan HY. Ultrasound-Microbubble-Mediated Gene Transfer of Inducible Smad7 Blocks Transforming Growth Factor-{beta} Signaling and Fibrosis in Rat Remnant Kidney. Am J Pathol. 2005;166:761–771. doi: 10.1016/s0002-9440(10)62297-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koike H, Tomita N, Azuma H, Taniyama Y, Yamasaki K, Kunugiza Y, Tachibana K, Ogihara T, Morishita R. An efficient gene transfer method mediated by ultrasound and microbubbles into the kidney. J Gene Med. 2005;7:108–16. doi: 10.1002/jgm.632. [DOI] [PubMed] [Google Scholar]
- 40.Li P, Morris M, Ferrario CM, Barrett C, Ganten D, Callahan MF. Cardiovascular, endocrine, and body fluid-electrolyte responses to salt loading in mRen-2 transgenic rats. Am J Physiol. 1998;275:H1130–7. doi: 10.1152/ajpheart.1998.275.4.H1130. [DOI] [PubMed] [Google Scholar]
- 41.DiBona GF, Jones SY, Sawin LL. Angiotensin receptor antagonist improves cardiac reflex control of renal sodium handling in heart failure. Am J Physiol. 1998;274:H636–41. doi: 10.1152/ajpheart.1998.274.2.H636. [DOI] [PubMed] [Google Scholar]
- 42.Zhu Q, Wang Z, Xia M, Li PL, Van Tassell BW, Abbate A, Dhaduk R, Li N. Silencing of hypoxia-inducible factor-1alpha gene attenuated angiotensin II-induced renal injury in Sprague-Dawley rats. Hypertension. 2011;58:657–64. doi: 10.1161/HYPERTENSIONAHA.111.177626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu J, Zhu Q, Xia M, Guo TL, Wang Z, Li PL, Han WQ, Yi F, Li N. Transplantation of mesenchymal stem cells into the renal medulla attenuated salt-sensitive hypertension in Dahl S rat. J Mol Med (Berl) 2014;92:1139–45. doi: 10.1007/s00109-014-1199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu Q, Hu J, Han WQ, Zhang F, Li PL, Wang Z, Li N. Silencing of HIF prolyl-hydroxylase 2 gene in the renal medulla attenuates salt-sensitive hypertension in Dahl S rats. Am J Hypertens. 2014;27:107–13. doi: 10.1093/ajh/hpt207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee ST, Chu K, Jung KH, Yoon HJ, Jeon D, Kang KM, Park KH, Bae EK, Kim M, Lee SK, Roh JK. MicroRNAs induced during ischemic preconditioning. Stroke. 2010;41:1646–51. doi: 10.1161/STROKEAHA.110.579649. [DOI] [PubMed] [Google Scholar]
- 46.Bergstrom G, Evans RG. Mechanisms underlying the antihypertensive functions of the renal medulla. Acta Physiol Scand. 2004;181:475–86. doi: 10.1111/j.1365-201X.2004.01321.x. [DOI] [PubMed] [Google Scholar]
- 47.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 48.Zhang X, Zeng Y. Regulation of mammalian microRNA expression. J Cardiovasc Transl Res. 2010;3:197–203. doi: 10.1007/s12265-010-9166-x. [DOI] [PubMed] [Google Scholar]
- 49.Knouf EC, Garg K, Arroyo JD, Correa Y, Sarkar D, Parkin RK, Wurz K, O’Briant KC, Godwin AK, Urban ND, Ruzzo WL, Gentleman R, Drescher CW, Swisher EM, Tewari M. An integrative genomic approach identifies p73 and p63 as activators of miR-200 microRNA family transcription. Nucleic Acids Res. 2012;40:499–510. doi: 10.1093/nar/gkr731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Graudal N, Jürgens G, Baslund B, Alderman MH. Compared With Usual Sodium Intake, Low- and Excessive-Sodium Diets Are Associated With Increased Mortality: A Meta-Analysis. American Journal of Hypertension. 2014;27:1129–1137. doi: 10.1093/ajh/hpu028. [DOI] [PubMed] [Google Scholar]
- 51.Kawasaki T, Delea CS, Bartter FC, Smith H. The effect of high-sodium and low-sodium intakes on blood pressure and other related variables in human subjects with idiopathic hypertension. The American journal of medicine. 1978;64:193–8. doi: 10.1016/0002-9343(78)90045-1. [DOI] [PubMed] [Google Scholar]