Abstract
The Genome 10K Project was established in 2009 by a consortium of biologists and genome scientists determined to facilitate the sequencing and analysis of the complete genomes of 10,000 vertebrate species. Since then the number of selected and initiated species has risen from ~26 to 277 sequenced or ongoing with funding, an approximately tenfold increase in five years. Here we summarize the advances and commitments that have occurred by mid-2014 and outline the achievements and present challenges of reaching the 10,000-species goal. We summarize the status of known vertebrate genome projects, recommend standards for pronouncing a genome as sequenced or completed, and provide our present and future vision of the landscape of Genome 10K. The endeavor is ambitious, bold, expensive, and uncertain, but together the Genome 10K Consortium of Scientists and the worldwide genomics community are moving toward their goal of delivering to the coming generation the gift of genome empowerment for many vertebrate species.
Keywords: mammal, amphibian, reptile, bird, fish, genome
INTRODUCTION
The advent of low-cost, high-throughput sequencing has ushered in a new age of genome science and has forever changed the landscape of biological research. Projects that could only be dreamt of 10 years ago are now becoming a reality. The Genome 10K Project (hereafter the G10K Project) is one such project (1–3). Sequencing 10,000 vertebrate genomes is an ambitious and worthy goal that will provide a foundation for diverse research and exciting discovery for decades to come. We originally selected a goal of 10,000 species (from a total of over 62,000 named vertebrate species) (Figure 1) as a round number target that was achievable, and which includes nearly every species with even modest biological knowledge available plus several thousand species without much knowledge. A detailed description of the rationale is presented in the original G10K White paper (1).
Figure 1.
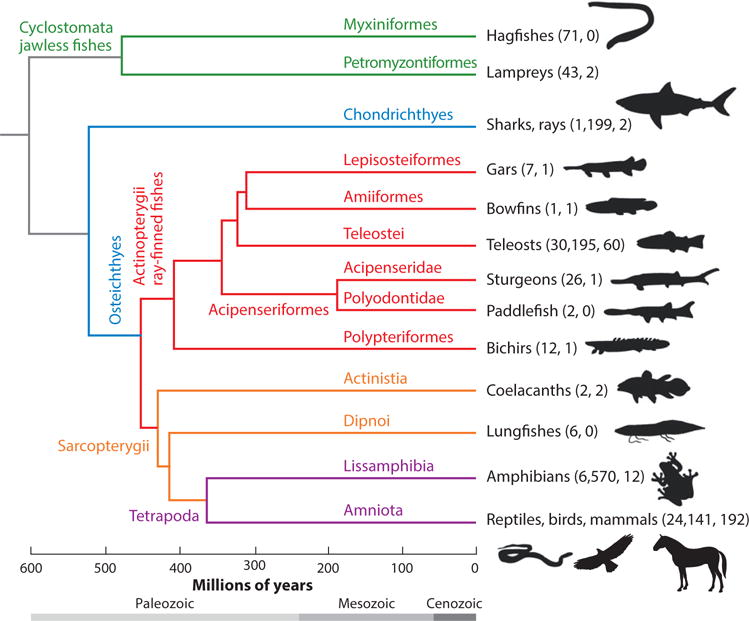
Consensus phylogeny of the major lineages of vertebrates. Topology and divergence dates (Ma) are consensus estimates derived from References 1 and 276 and included citations. Following the common names of taxon groups in parentheses are number of living species for that group and number of species with published and/or pending genomes (see Tables 2 and 3).
The G10K Project was founded in 2009 by bringing together biologists, bioinformaticians, and computational scientists to accumulate and organize specimens, to develop standards for genome assembly and annotation, and to facilitate the release and use of the genome data created through the project. At the first G10K workshop in Santa Cruz, California (April 13–16, 2009), biologists who curated museum or personal frozen collections of biospecimens were convened and asked to develop a list of vertebrate specimens available in collections globally, which then would become the basis of the G10K Project. Amazingly, the group found that 16,203 vertebrate species had already been collected and were housed in existing collections. These were collated into a database (http://genome10k.soe.ucsc.edu) that became the foundation for developing initial plans for whole genome sequencing (WGS) (1).
Since 2009, the G10K Project has grown in membership, in responsibilities, in recognition, and in stewardship. At the most recent G10K workshop (April 24–28, 2013) in Fort Lauderdale, Florida, over 150 scientists gathered to develop plans for future genome sequencing and discuss analytical and computational challenges and the exciting results from the first ~270 vertebrate genomes sequenced to date. Here, we provide an overview of the goals, responsibilities, accomplishments, and insights of the G10K Project, where the project stands today with regard to the vertebrate genomes that have been sequenced thus far, and the remaining challenges involved in reaching the goal of sequencing 10,000 vertebrate genomes.
GENOME 10K RESPONSIBILITIES
The G10K Community of Scientists (G10KCOS) established six primary goals or responsibilities to drive the project forward (Table 1). Our first charge was to accumulate biospecimens that would provide the DNA necessary to develop reference-quality genomes. The 2009 G10K meeting identified over 16,000 species from existing collections in museums, universities, and zoos around the world and cataloged that inventory in an open-access database accessible to the entire community (https://genome10k.soe.ucsc.edu/biospecimen_database). Samples included in this virtual repository ranged from extracted genomic DNA to frozen tissues to cell lines. In addition to compiling this virtual list, we produced an in-depth report of best practices for obtaining and storing vertebrate biospecimens for WGS (4).
Table 1.
Goals of the Genome 10K Project (see text for details)
| 1. Gather and validate voucher biospecimens for whole genome sequencing (WGS) |
| 2. Develop scientific communities around the species, taxonomic groups, and analytical themes (e.g., assembly, annotation, alignment, comparative genomic analyses) |
| 3. Set standards for genome |
| a. Assembly |
| b. Annotation |
| c. Release on browsers |
| d. Rapid data release |
| 4. Monitor progress on vertebrae WGS projects |
| 5. Raise funds |
| 6. Foster and support other genome consortia, such as the following: |
| a. Insect 5K (i5K) http://www.arthropodgenomes.org/wiki/i5K |
| b. Global Invertebrate Genomics Alliance (GIGA) http://www.nova.edu/ocean/giga/ |
| c. Consortium for Snake Genomics http://www.snakegenomics.org/SnakeGenomics/Home.html |
| d. 1000 Fungal Genomes Project (1KFG) http://1000.fungalgenomes.org/home/ |
| e. NSF Plant Genome Research Program (PGRP) http://www.nsf.gov/pubs/2014/nsf14533/nsf14533.htm |
| f. 100K Foodborne Pathogen Genome Project http://100kgenome.vetmed.ucdavis.edu |
A second goal of the G10K Project is to foster the development of research communities centered either around the genomes of species or species groups (e.g., birds) or around bioinformatics themes, namely genome assembly, annotation, alignment, and comparative analyses. Such communities are vital because not only do they help establish criteria for the selection of species to be sequenced but they also ensure interdisciplinary collaboration among scientists with diverse research experiences. For example, whereas one scientist may intend to use a reference genome to analyze genome architecture, another may use the same data to search for evidence of positive selection. Thus, an open-access genome becomes a commodity that drives multifaceted research programs in different fields. Within the G10K Project, communities of scientists are broadly organized around the major classes of vertebrates (fishes, amphibians, nonavian reptiles, birds, and mammals), and these communities strive to identify target species for genome sequencing that benefit the largest group of scientists and fill major genome sampling gaps across the vertebrate tree of life.
A third goal of the G10K Project is to develop a strong and scientifically vetted set of standards concerning specimen selection, DNA preparation, genome assembly, genome feature annotation, whole genome alignment, comparative analyses, and data release. Despite the tremendous progress that has been made in genomics, the field itself is still in an experimental state with no established best practices in the generation and analysis of genome data. Various genomic groups develop their own ideas about sample quality and quantity for de novo sequencing as well as about what constitutes a high-coverage genome. They often use home-brew or unvetted software, even though several groups have established that software programs developed for assembly, annotation, and alignment differ markedly in accuracy and efficiency (5–8). G10K scientists aim to develop a set of consensus-based best practices regarding genomic data generation and analysis. For example, given a shark, frog, or microbat, which tissue(s) would be most useful in producing genomic libraries? How should these biospecimens be preserved? How is DNA derived from them handled? Which sequencing libraries should be prepared? Given the choice of among 20+ genome assembly algorithms and programs, which one produces the most accurate assembly, and what parameters are best for evaluating this? The G10KCOS is developing informed guidelines in addressing issues such as these through collaborations between biologists and bioinformaticians. A preliminary snapshot of G10K endorsed standards is presented in the sidebar, Draft Standards for Genome 10K.
A fourth responsibility for the G10KCOS is to record the progress of vertebrate WGS by maintaining a database of completed and ongoing projects being carried out by genome sequencing centers and by independent research laboratories around the world (http://genome10k.soe.ucsc.edu/species). By doing this, we not only avoid duplication of efforts, given the still relatively high expense of generating and annotating reference-quality genomes, but also help to target the species that will maximize research dividends and increase breadth of phylogenetic coverage in the vertebrate tree of life (9). Table 2 presents a list of 164 vertebrate species with a published genome sequence, and Table 3 lists an additional 113 vertebrate species for which genome sequencing is accomplished or near completion.
Table 2.
List of 164 vertebrate genomes that have been published as of December 9, 20141
| SPECIES | COMMON NAME | ORDER | FAMILY | GENBANK ACCESSION | BIOPROJECT ID | REFERENCE |
|---|---|---|---|---|---|---|
| CYCLOSTOMATA | ||||||
| Lethenteron camtschaticum | Arctic lamprey | Petromyzontiformes | Petromyzontidae | APJL00000000 | PRJNA192554 | 135 |
| Petromyzon marinus | Sea lamprey | Petromyzontiformes | Petromyzontidae | AEFG00000000 | PRJNA12880 | 136 |
| CHONDRICHTHYES | ||||||
| Callorhinchus milii | Elephant shark | Chimaeriformes | Callorhinchidae | AAVX00000000 | PRJNA18361 | 14 |
| ACTINOPTERYGII | ||||||
| Takifugu rubripes | Fugu | Tetraodontiformes | Tetraodontidae | CAAB00000000 | PRJNA1434 | 56, 137 |
| Tetraodon nigroviridis | Freshwater pufferfish | Tetraodontiformes | Tetraodontidae | CAAE00000000 | PRJNA12350 | 57 |
| Oryzias latipes | Japanese medaka | Beloniformes | Adrianichthyidae | BAAF00000000 | PRJNA16702 | 58 |
| Gadus morhua | Atlantic cod | Gadiformes | Gadidae | CAEA00000000 | PRJNA41391 | 138 |
| Anguilla japonica | Japanese eel | Anguilliformes | Anguillidae | AVPY00000000 | PRJNA158309 | 139 |
| Gasterosteus aculeatus | Three-spined stickleback | Gasterosteiformes | Gasterosteidae | AANH00000000 | PRJNA13579 | 59 |
| Danio rerio | Zebrafish | Cypriniformes | Cyprinidae | CABZ00000000 | PRJNA11776 | 60 |
| Thunnus orientalis | Pacific bluefin tuna | Scombriformes | Scombridae | BADN00000000 | PRJDA68701 | 140 |
| Xiphophorus maculatus | Southern platyfish | Cyprinodontiformes | Poeciliidae | AGAJ00000000 | PRJNA72525 | 61 |
| Cynoglossus semilaevis | Tongue sole | Pleuronectiformes | Cynoglossidae | AGRG00000000 | PRJNA73987 | 131 |
| Oncorhynchus mykiss | Rainbow trout | Salmoniformes | Salmonidae | CCAF000000000 | PRJEB4421 | 141 |
| Electrophorus electricus | Electric eel | Gymnotiformes | Gymnotidae | PRJNA249073 | 142 | |
| Cyprinus carpio | Common carp | Cypriniformes | Cyprinidae | PRJNA202478 | 143 | |
| Astyanax mexicanus | Mexican tetra | Characiformes | Characidae | APWO00000000 | PRJNA89115 | 144 |
| Larimichthys crocea | Large yellow croaker | Sciaenidae | JPYK00000000 | PRJNA237858 | 145 | |
| Boleophthalmus pectinirostris | Blue-spotted mudskipper | Gobiiformes | Gobiidae | JACK00000000 | PRJNA232434 | 146 |
| Periophthalmus magnusspinnatus | Giant-fin mudskipper | Gobiiformes | Gobiidae | JACL00000000 | PRJNA232435 | 146 |
| Periophthalmodon schlosseri | Giant mudskipper | Gobiiformes | Gobiidae | JACM00000000 | PRJNA232436 | 146 |
| Scartelaos histophorus | Blue mudskipper | Gobiiformes | Gobiidae | JACN00000000 | PRJNA232437 | 146 |
| SARCOPTERYGII | ||||||
| Latimeria chalumnae | African coelacanth | Coelacanthiformes | Coelacanthidae | AFYH00000000; BAHO00000000 | PRJNA56111; PRJDB500 | 147, 148 |
| Latimeria menadoensis | Indonesian coelacanth | Coelacanthiformes | Coelacanthidae | PRJNA38001 | 148 | |
| AMPHIBIA | ||||||
| Xenopus (Silurana) tropicalis | Western clawed frog | Anura | Pipidae | AAMC00000000 | PRJNA12348 | 65 |
| “REPTILIA” | ||||||
| Anolis carolinensis | Green anole | Squamata | Iguanidae | AAWZ00000000 | PRJNA18787; PRJNA60547 | 87 |
| Python bivittatus | Burmese python | Squamata | Pythonidae | AEQU00000000 | PRJNA61243; PRJNA238085 | 90 |
| Alligator mississippiensis | American alligator | Crocodylia | Alligatoridae | AKHW00000000 | PRJNA159843; PRJNA221578 | 93 |
| Crocodylus porosus | Saltwater crocodile | Crocodylia | Crocodylidae | PRJNA163131 | 93 | |
| Gavialis gangeticus | Indian gharial | Crocodylia | Gavialidae | PRJNA172383 | 93 | |
| Ophiophagus hannah | King cobra | Squamata | Elapidae | AZIM00000000 | PRJNA201683 | 92 |
| Alligator sinensis | Chinese alligator | Crocodylia | Alligatoridae | AVPB00000000 | PRJNA221633 | 94 |
| Chelonia mydas | Green turtle | Testudines | Cheloniidae | AJIM00000000 | PRJNA104937; PRJNA234097 | 96 |
| Pelodiscus sinensis | Chinese softshell turtle | Testudines | Trionychidae | AGCU00000000 | PRJNA68233; PRJNA221645 | 96 |
| Chrysemys picta | Western painted turtle | Testudines | Emydidae | AHGY00000000 | PRJNA78657 | 95 |
| Crotalus mitchellii | Speckled rattlesnake | Serpentes | Viperidae | JPMF01000000 | PRJNA255393 | 149 |
| AVES | ||||||
| Gallus gallus | Red jungle fowl | Galliformes | Phasianidae | AADN00000000 | PRJNA13342 | 103 |
| Meleagris gallopavo | Wild turkey | Galliformes | Phasianidae | ADDD00000000 | PRJNA42129 | 104 |
| Taeniopygia guttata | Zebra finch | Passeriformes | Estrildidae | ABQF00000000 | PRJNA17289 | 105 |
| Amazona vittata | Puerto Rican parrot | Psittaciformes | Psittacidae | AOCU00000000 | PRJNA171587 | 150 |
| Ficedula albicollis | Collared flycatcher | Passeriformes | Muscicapidae | AGTO00000000 | PRJNA208061 | 119 |
| Ficedula hypoleuca | Pied flycatcher | Passeriformes | Muscicapidae | 119 | ||
| Geospiza fortis | Mallard duck | Anseriformes | Anatidae | ADON00000000 | PRJNA46621 | 151 |
| Ara macao | Scarlet macaw | Psittaciformes | Psittacidae | AOUJ00000000 | PRJNA189648 | 152 |
| Columba livia | Rock pigeon | Columbiformes | Columbidae | AKCR00000000 | PRJNA167554; PRJNA170656 | 153 |
| Coturnix japonica | Japanese quail | Galliformes | Phasianidae | BASJ00000000 | PRJDB1146 | 154 |
| Falco cherrug | Saker falcon | Falconiformes | Falconidae | AKMU00000000 | PRJNA217049 | 155 |
| Falco peregrinus | Peregrine falcon | Falconiformes | Falconidae | AKMT00000000 | PRJNA198010 | 155 |
| Geospiza magnirostris | Large ground finch | Passeriformes | Thraupidae | PRJNA178982 | 156 | |
| Melopsittacus undulatus | Australian parakeet (budgerigar) | Psittaciformes | Psittacidae | AGAI00000000 | PRJNA197262 | 157 |
| Pseudopodoces humilis | Ground tit | Passeriformes | Paridae | ANZD00000000 | PRJNA217046 | 158, 159 |
| Aquila chrysaetos | Golden eagle | Accipitriformes | Accipitridae | JDSB00000000 | PRJNA222866 | 160 |
| Colinus virginianus | Northern bobwhite | Galliformes | Odontophoridae | AWGT00000000 | PRJNA188411 | 161 |
| Corvus cornix | Hooded crow | Passeriformes | Corvidae | PRJNA208001 | 162 | |
| Lyrurus (Tetrao) tetrix | Black grouse | Galliformes | Phasianidae | JDSL00000000 | PRJNA179551 | 163 |
| Geospiza fortis | Medium ground finch | Passeriformes | Fringillidae | AKZB00000000 | PRJNA156703 | 112, 113 |
| Aptenodytes forsteri | Emperor penguin | Sphenisciformes | Spheniscidae | JMFQ00000000 | PRJNA235982 | 112, 113 |
| Pygoscelis adeliae | Adelie penguin | Sphenisciformes | Spheniscidae | JMFP00000000 | PRJNA235983 | 112, 113 |
| Acanthisitta chloris | Rifleman | Passeriformes | Acanthisittidae | JJRS00000000 | PRJNA212877 | 112, 113 |
| Antrostomus carolinensis | Chuck-will’s-widow | Caprimulgiformes | Caprimulgidae | JMFU00000000 | PRJNA212888 | 112, 113 |
| Apaloderma vittatum | Bar-tailed trogon | Trogoniformes | Trogonidae | JMFV00000000 | PRJNA212878 | 112, 113 |
| Balearica regulorum | Crowned crane | Gruiformes | Gruidae | JJRR00000000 | PRJNA212879 | 112, 113 |
| Buceros rhinoceros | Javan rhinoceros hornbill | Bucerotiformes | Bucerotidae | JMFK00000000 | PRJNA212887 | 112, 113 |
| Calypte anna | Anna’s hummingbird | Trochiliformes | Trochilidae | JJRV00000000 | PRJNA212866 | 112, 113 |
| Cariama cristata | Red-legged seriema | Gruiformes | Cariamidae | JJRQ00000000 | PRJNA212889 | 112, 113 |
| Cathartes aura | Turkey vulture | Cathartiformes | Cathartidae | JMFT00000000 | PRJNA212890 | 112, 113 |
| Chaetura pelagica | Chimney swift | Apodiformes | Apodidae | PRJNA210808 | 112, 113 | |
| Charadrius vociferus | Killdeer | Charadriiformes | Charadriidae | JMFX00000000 | PRJNA212867 | 112, 113 |
| Chlamydotis macqueenii | MacQueen’s bustard | Gruiformes | Otididae | JMFJ00000000 | PRJNA212891 | 112, 113 |
| Colius striatus | Speckled mousebird | Coliiformes | Coliidae | JJRP00000000 | PRJNA212892 | 112, 113 |
| Corvus brachyrhynchos | American crow | Passeriformes | Corvidae | JMFN01000000 | PRJNA212869 | 112, 113 |
| Cuculus canorus | Common cuckoo | Cuculiformes | Cuculidae | JNOX01000000 | PRJNA212870 | 112, 113 |
| Egretta garzetta | Little egret | Ciconiiformes | Ardeidae | JJRC00000000 | PRJNA232959 | 112, 113 |
| Eurypyga helias | Sunbittern | Gruiformes | Eurypygidae | JJRO00000000 | PRJNA212893 | 112, 113 |
| Fulmarus glacialis | Northern fulmar | Procellariiformes | Procellariidae | JJRN00000000 | PRJNA212894 | 112, 113 |
| Gavia stellata | Red-throated loon | Gaviiformes | Gaviidae | JJRM00000000 | PRJNA212895 | 112, 113 |
| Haliaeetus albicilla | White-tailed eagle | Falconiformes | Accipitridae | JJRL00000000 | PRJNA212896 | 112, 113 |
| Haliaeetus leucocephalus | Bald eagle | Falconiformes | Accipitridae | PRJNA237821 | 112, 113 | |
| Leptosomus discolor | Cuckoo roller | Coraciiformes | Leptosomatidae | JJRK00000000 | PRJNA212897 | 112, 113 |
| Manacus vitellinus | Golden-collared manakin | Passeriformes | Pipridae | JMFM00000000 | PRJNA212872 | 112, 113 |
| Merops nubicus | Northern carmine bee-eater | Coraciiformes | Meropidae | JJRJ00000000 | PRJNA212898 | 112, 113 |
| Mesitornis unicolor | Brown mesite | Gruiformes | Mesitornithidae | JJRI00000000 | PRJNA212899 | 112, 113 |
| Nestor notabilis | Kea | Psittaciformes | Psittacidae | JJRH00000000 | PRJNA212900 | 112, 113 |
| Nipponia nippon | Crested ibis | Ciconiiformes | Threskiornithidae | JMFH00000000 | PRJNA232572 | 112, 113 |
| Opisthocomus hoazin | Hoatzin | Opisthocomiformes | Opisthocomidae | JMFL00000000 | PRJNA212873 | 112, 113 |
| Pelecanus crispus | Dalmatian pelican | Pelicaniformes | Pelicanidae | JJRG00000000 | PRJNA212901 | 112, 113 |
| Phaethon lepturus | White-tailed tropicbird | Phaethontiformes | Phaethontidae | JJRF00000000 | PRJNA212902 | 112, 113 |
| Phalacrocorax carbo | Great black cormorant | Pelicaniformes | Phalacrocoracidae | JMFI00000000 | PRJNA212903 | 112, 113 |
| Phoenicopterus ruber ruber | Caribbean flamingo | Phoenicopteriformes | Phoenicopteridae | JJRE00000000 | PRJNA212904 | 112, 113 |
| Picoides pubescens | Downy woodpecker | Piciformes | Picidae | JJRU00000000 | PRJNA212874 | 112, 113 |
| Podiceps cristatus | Great-crested grebe | Podicipediformes | Podicipedidae | JMFS00000000 | PRJNA212905 | 112, 113 |
| Pterocles gutturalis | Yellow-throated sandgrouse | Ciconiiformes | Pteroclidae | JMFR00000000 | PRJNA212906 | 112, 113 |
| Struthio camelus | Ostrich | Struthioniformes | Struthionidae | JJRT00000000 | PRJNA212875 | 112, 113 |
| Tauraco erythrolophus | Angola turaco | Musophagiformes | Musophagidae | JNOY00000000 | PRJNA212908 | 112, 113 |
| Tinamus guttatus | White-throated tinamou | Tinamiformes | Tinamidae | JMFW00000000 | PRJNA212876 | 112, 113 |
| Tyto alba | Barn owl | Strigiformes | Tytonidae | JJRD00000000 | PRJNA212909 | 112, 113 |
| Hemignathus virens | Hawaii amakihi | Passeriformes | Fringillidae | 164 | ||
| MAMMALIA | ||||||
| Homo sapiens | Human | Primates | Hominidae | NCBI36 | 165, 166 | |
| Mus musculus | House mouse | Rodentia | Muridae | 167 | ||
| Rattus norvegicus | Norway rat | Rodentia | Muridae | AABR00000000 | PRJNA10629 | 168 |
| Canis familiaris | Domestic dog | Carnivora | Canidae | AAEX00000000 | PRJNA13179 | 169 |
| Pan troglodytes | Chimpanzee | Primates | Hominidae | AADA01000000 | PRJNA13184 | 170 |
| Felis catus | Domestic cat | Carnivora | Felidae | AANG00000000 | PRJNA16726 | 171 |
| Macaca mulatta | Rhesus macaque | Primates | Cercopithecidae | AANU00000000; AEHK00000000 | PRJNA12537; PRJNA51409 | 172, 173 |
| Monodelphis domestica | Gray short-tailed opossum | Didelphimorphia | Didelphidae | AAFR00000000 | PRJNA12561 | 174 |
| Ornithorhynchus anatinus | Platypus | Monotremata | Ornithorhynchidae | AAPN00000000 | PRJNA12885 | 19 |
| Bos taurus | Cow | Cetartiodactyla | Bovidae | AAFC00000000 | PRJNA12555 | 175, 176 |
| Equus caballus | Horse | Perissodactyla | Equidae | AAWR00000000; ATDM00000000 | PRJNA18661; PRJNA200654 | 177, 178 |
| Ailuropoda melanoleuca | Giant panda | Carnivora | Ursidae | ACTA00000000 | PRJNA38683 | 13 |
| Ovis aries | Domestic sheep | Cetartiodactyla | Bovidae | AMGL00000000 | PRJNA169880 | 179, 180 |
| Cavia porcellus | Guinea pig | Rodentia | Caviidae | AAKN00000000 | PRJNA12583 | 114 |
| Choloepus hoffmanni | Two-toed sloth | Pilosa | Megalonychidae | ABVD00000000 | PRJNA30809 | 114 |
| Cricetulus griseus | Chinese hamster | Rodentia | Cricetidae | AFTD00000000; APMK00000000; AMDS00000000 | PRJNA69991; PRJNA189319; PRJNA167053 | 181–183 |
| Dasypus novemicinctus | Nine-banded armadillo | Cingulata | Dasypodidae | AAGV00000000 | PRJNA12594 | 114 |
| Dipodomys ordii | Ord’s kangaroo rat | Rodentia | Heteromyidae | ABRO00000000 | PRJNA20385 | 114 |
| Echinops telfairi | Lesser hedgehog tenrec | Afrosoricida | Tenrecidae | AAIY00000000 | PRJNA12590 | 114 |
| Erinaceus europaeus | Western European hedgehog | Eulipotyphla | Erinaceidae | AMDU00000000 | PRJNA74585 | 114 |
| Heterocephalus glaber | Naked mole rat | Rodentia | Bathyergidae | AFSB00000000 | PRJNA68323 | 184 |
| Ictidomys tridecemlineatus | Thirteen-lined ground squirrel | Rodentia | Sciuridae | AAQQ01000000; AGTP00000000 | PRJNA13937; PRJNA61725 | 114 |
| Loxodonta africana | African savanna elephant | Proboscidea | Elephantidae | AAGU00000000 | PRJNA12569 | 114 |
| Macaca fascicularis | Crab-eating macaque | Primates | Cercopithecidae | AEHL00000000 | PRJNA51411 | 173, 185 |
| Macropus eugenii | Tammar wallaby | Diprotodontia | Macropodidae | ABQO000000000 | PRJNA12587 | 186 |
| Microcebus murinus | Gray mouse lemur | Primates | Cheirogaleidae | ABDC00000000 | PRJNA19967 | 114 |
| Myotis lucifugus | Little brown bat | Chiroptera | Vespertilionidae | AAPE00000000 | PRJNA16951 | 114 |
| Ochotona princeps | American pika | Lagomorpha | Ochotonidae | AAYZ00000000; ALIT00000000 | PRJNA19235; PRJNA74593 | 114 |
| Odocoileus virginianus | White-tailed deer | Cetartiodactyla | Cervidae | AEGY00000000; AEGZ00000000 | PRJNA52611 | 187 |
| Oryctolagus cuniculus | Rabbit | Lagomorpha | Leporidae | AAGW00000000 | PRJNA12819 | 114 |
| Otolemur garnettii | Bushbaby (small-eared galago) | Primates | Galagidae | AAQR00000000 | PRJNA16955 | 114 |
| Pongo abelii | Sumatran orangutan | Primates | Hominidae | ABGA00000000 | PRJNA20869 | 188 |
| Procavia capensis | Rock hyrax | Hyracoidea | Procaviidae | ABRQ00000000 | PRJNA13972 | 114 |
| Pteropus vampyrus | Large flying fox | Chiroptera | Pteropodidae | ABRP00000000 | PRJNA20325 | 114 |
| Sarcophilus harrisii | Tasmanian devil | Dasyuromorphia | Dasyuridae | AFEY00000000; AEFK00000000 | PRJNA65325; PRJNA51853 | 123, 189 |
| Sorex araneus | European shrew | Eulipotyphla | Soricidae | AALT00000000 | PRJNA13689 | 114 |
| Tarsius syrichta | Philippine tarsier | Primates | Tarsiidae | ABRT000000000 | PRJNA20339 | 114 |
| Tupaia belangeri | Northern tree shrew | Scandentia | Tupaiidae | AAPY00000000 | PRJNA13971 | 114 |
| Tursiops truncatus | Bottle-nosed dolphin | Cetartiodactyla | Delphinidae | ABRN00000000 | PRJNA20367 | 114 |
| Vicugna pacos | Alpaca | Cetartiodactyla | Camelidae | ABRR00000000 JEMW00000000 | PRJNA30567 PRJNA233565 | 114, 190 |
| Bos indicus | Zebu | Cetartiodactyla | Bovidae | AGFL00000000 | PRJNA72827 | 191 |
| Bos grunniens mutus | Yak | Cetartiodactyla | Bovidae | AGSK00000000 | PRJNA74739 | 116 |
| Camelus bactrianus ferus | Bactrian camel | Cetartiodactyla | Camelidae | AGVR00000000 JARL00000000 | PRJNA76177 PRJNA183605 | 190, 192 |
| Capra hircus | Goat | Cetartiodactyla | Bovidae | AJPT00000000 | PRJNA158393 | 37 |
| Daubentonia madagascariensis | Aye-aye | Primates | Daubentoniidae | AGTM000000000 | PRJNA74997 | 193, 194 |
| Gorilla gorilla | Gorilla | Primates | Hominidae | CABD00000000 | PRJEA31265 | 195 |
| Myotis davidii | David’s myotis | Chiroptera | Vespertilionidae | ALWT00000000 | PRJNA171994 | 196 |
| Pteropus alecto | Black flying fox | Chiroptera | Pteropodidae | ALWS00000000 | PRJNA171993 | 196 |
| Pan paniscus | Bonobo | Primates | Hominidae | AJFE00000000 | PRJNA49285 | 197 |
| Sus scrofa | Domestic pig | Cetartiodactyla | Suidae | AJKK00000000 | PRJNA13421; PRJNA144099 | 198, 199 |
| Eidolon helvum | Straw-colored fruit bat | Chiroptera | Pteropodidae | AWHC00000000 | PRJNA209406 | 200 |
| Megaderma lyra | Indian false vampire bat | Chiroptera | Megadermatidae | AWHB00000000 | PRJNA209407 | 200 |
| Pteronotus parnellii | Parnell’s mustached bat | Chiroptera | Mormoopidae | AWGZ00000000 | PRJNA209408 | 200 |
| Rhinolophus ferrumequinum | Greater horseshoe bat | Chiroptera | Rhinolophidae | AWHA00000000 | PRJNA209409 | 200 |
| Myotis brandtii | Brandt’s bat | Chiroptera | Vespertilionidae | ANKR00000000 | PRJNA218631 | 120 |
| Lipotes vexillifer | Yangtze river dolphin | Cetartiodactyla | Lipotidae | AUPI00000000 | PRJNA174066 | 201 |
| Panthera tigris | Amur tiger | Carnivora | Felidae | ATCQ00000000 | PRJNA182708 | 202 |
| Pantholops hodgsonii | Chiru | Cetartiodactyla | Bovidae | AGTT00000000 | PRJNA72465 | 203 |
| Tupaia chinensis | Chinese tree shrew | Scandentia | Tupaiidae | ALAR00000000 | PRJNA169406 | 204 |
| Balaenoptera acutorostrata | Minke whale | Cetartiodactyla | Balaenopteridae | ATDI00000000 | PRJNA72723 | 15 |
| Callithrix jacchus | Common marmoset | Primates | Cebidae | ACFV00000000 | PRJNA20401 | 205 |
| Macaca thibetana | Tibetan macaque | Primates | Cercopithecidae | PRJNA226187 | 206 | |
| Spalax galili | Blind mole rat | Rodentia | Spalacidae | AXCS00000000 | PRJNA213569 | 207 |
| Ursus maritimus | Polar bear | Carnivora | Ursidae | AVOR00000000 | PRJNA210951 | 208 |
| Nomascus leucogenys | White-cheeked gibbon | Primates | Hylobatidae | ADFV00000000 | PRJNA13975 | 209 |
| Camelus dromedarius | Dromedary | Cetartiodactyla | Camelidae | JDVD00000000 | PRJNA234474 | 190 |
| Rhinopithecus roxellana | Golden snub-nosed monkey | Primates | Cercopithecidae | JABR00000000 | PRJNA230020 | 210 |
Species are listed chronologically according to year genome was first published. Species in boldface were sequenced through the BGI-G10K collaborative effort.
Table 3.
List of 113 vertebrate genomes that either are unpublished or have been targeted for de novo sequencing through the BGI-G10K collaborative effort
| SPECIES | COMMON NAME | ORDER | FAMILY | GENBANK ACCESSION or BGI-G10K species |
|---|---|---|---|---|
| CHONDRICHTHYES | ||||
| Sphyrna mokarran | Great hammerhead shark | Carcharhiniformes | Sphyrnidae | BGI-G10K |
| ACTINOPTERYGII | ||||
| Acipenser sinensis | Chinese sturgeon | Acipenseriformes | Acipenseridae | BGI-G10K |
| Amia calva | Bowfin | Amiiformes | Amiidae | BGI-G10K |
| Polypterus senegalus | Bichir | Polypteriformes | Polypteridae | BGI-G10K |
| Hoplostethus atlanticus | Orange roughy | Beryciformes | Trachichthyidae | BGI-G10K |
| Astyanax mexicanus | Blind cave fish | Characiformes | Characidae | BGI-G10K |
| Carassius auratus gibelio | Prussian carp | Cypriniformes | Cyprinidae | BGI-G10K |
| Megalobrama amblycephala | Wuchang bream | Cypriniformes | Cyprinidae | BGI-G10K |
| Hypophthalmichthys molitrix | Silver carp | Cypriniformes | Cyprinidae | BGI-G10K |
| Gobiocypris rarus | Rare gudgeon | Cypriniformes | Cyprinidae | BGI-G10K |
| Hippocampus comes | Tiger tail seahorse | Gasterosteiformes | Syngnathidae | BGI-G10K |
| Scleropages formosus | Golden arowana | Osteoglossiformes | Osteoglossidae | BGI-G10K |
| Chaenocephalus aceratus | Blackfin icefish | Perciformes | Channichthyidae | BGI-G10K |
| Eleginops maclovinus | Patagonian blenny | Perciformes | Eleginopidae | BGI-G10K |
| Boleophthalmus pectinirostris | Mudskipper | Perciformes | Gobiidae | BGI-G10K |
| Periophthalmus magnuspinnatus | Giant-fin mudskipper | Perciformes | Gobiidae | BGI-G10K |
| Sinocyclocheilus grahami | Golden Line fish | Cypriniformes | Cyprinidae | BGI-G10K |
| Dissostichus mawsoni | Antarctic toothfish | Perciformes | Nototheniidae | BGI-G10K |
| Pseudosciaena crocea | Large yellow croaker | Perciformes | Sciaenidae | BGI-G10K |
| Sparus aurata | Gilthead sea bream | Perciformes | Sparidae | BGI-G10K |
| Paralichthys olivaceus | Bastard halibut | Pleuronectiformes | Paralichthyidae | BGI-G10K |
| Thunnus albacares | Yellowfin tuna | Scombriformes | Scombridae | BGI-G10K |
| Epinephelus coioides | Grouper | Perciformes | Serranidae | BGI-G10K |
| Platycephalus bassensis | Sand flathead | Scorpaeniformes | Platycephalidae | BGI-G10K |
| Siganus oramin | Pearl-spotted spinefoot | Perciforms | Siganidae | BGI-G10K |
| Monopterus albus | Finless eel | Synbranchiformes | Synbranchidae | BGI-G10K |
| Mola mola | Ocean sunfish | Tetraodontiformes | Molidae | BGI-G10K |
| Amphilophus citrinellus | Midas cichlid | Cichliformes | Cichlidae | CCOE00000000 |
| Anguilla anguilla | European eel | Anguilliformes | Anguillidae | AZBK00000000 |
| Anoplopoma fimbria | Sablefish | Perciformes | Anoplopomatidae | AWGY00000000 |
| Astyanax mexicanus | Blind cave fish | Characiformes | Characidae | APWO00000000 |
| Cyprinodon nevadensis | Amargosa pupfish | Cyprinodontiformes | Cyprinodontidae | JSUU00000000 |
| Cyprinodon variegatus | Sheepshead minnow | Cyprinodontiformes | Cyprinodontidae | JPKM01000000 |
| Haplochromis burtoni | Burton’s mouthbrooder | Cichliformes | Cichlidae | AFNZ00000000 |
| Lepisosteus oculatus | Spotted gar | Semionotiformes | Lepisosteidae | AHAT00000000 |
| Neolamprologus brichardi | Princess cichlid | Cichliformes | Cichlidae | AFNY00000000 |
| Notothenia coriiceps | Black rockcod | Perciformes | Nototheniidae | AZAD01000000 |
| Oreochromis niloticus | Nile tilapia | Cichliformes | Cichlidae | AERX00000000 |
| Pampus argenteus | Silver pomfret | Scombriformes | Stromateidae | JHEK00000000 |
| Pimephales promelas | Fathead minnow | Cypriniformes | Cyprinidae | JNCD01000000 |
| Poecilia formosa | Amazon molly | Cyprinodontiformes | Poeciliidae | AYCK00000000 |
| Poecilia reticulata | Guppy | Cyprinodontiformes | Poeciliidae | AZHG00000000 |
| Pundamilia nyererei | Flame back cichlid | Cichliformes | Cichlidae | AFNX00000000 |
| Salmo salar | Atlantic salmon | Salmoniformes | Salmonidae | AGKD00000000 (275) |
| Sebastes nigrocinctus | Tiger rockfish | Perciformes | Sebastidae | AUPR00000000 |
| Sebastes rubrivinctus | Flag rockfish | Perciformes | Sebastidae | AUPQ00000000 |
| Stegastes partitus | Bicolor damselfish | Perciformes | Pomacentridae | JMKM00000000 |
| AMPHIBIA | ||||
| Xenopus (Silurana) laevis | African clawed frog | Anura | Pipidae | http://www.xenbase.org/entry/ |
| Ascaphus truei | Coastal tailed frog | Anura | Ascaphidae | BGI-G10K |
| Spea bombifrons | Plains spadefoot toad | Anura | Scaphiopodidae | BGI-G10K |
| Bufo marinus | Cane toad | Anura | Bufonidae | BGI-G10K |
| Limnodynastes dumerilii | Eastern banjo frog | Anura | Limnodynastidae | BGI-G10K |
| Oophaga pumilio | Strawberry dart-poison frog | Anura | Dendrobatidae | BGI-G10K |
| Physalaemus pustulosus | Tungara frog | Anura | Leiuperidae | BGI-G10K |
| Eleutherodactylus coqui | Coqui | Anura | Eleutherodactylidae | BGI-G10K |
| Nanorana parkeri | Tibetan frog | Anura | Dicroglossidae | BGI-G10K |
| Gastrotheca cornuta | Horned marsupial frog | Anura | Hemiphractidae | BGI-G10K |
| Ichthyophis bannanicus | Banna caecilian | Gymnophiona | Ichthyophiidae | BGI-G10K |
| “REPTILIA” | ||||
| Sphenodon punctatus | Tuatara | Sphenodontia | Sphenodontidae | AWC-G10K |
| Eublepharus macularius | Leopard gecko | Squamata | Gekkonidae | BGI-G10K |
| Heloderma suspectum | Gila monster | Squamata | Helodermatidae | BGI-G10K |
| Podarcus muralis | Wall lizard | Squamata | Lacertidae | BGI-G10K |
| Ophisaurus harti | Chinese glass lizard | Squamata | Anguidae | BGI-G10K |
| Aspidoscelis arizonae | Western whiptail | Squamata | Teiidae | BGI-G10K |
| Pogona vitticeps | Central bearded dragon | Squamata | Agamidae | BGI-G10K |
| Shinisaurus crocodilurus | Chinese crocodile lizard | Squamata | Shinisauridae | BGI-G10K |
| Apalone spinifera | Spiny softshell turtle | Testudines | Trionychidae | APJP00000000 |
| AVES | ||||
| Zonotrichia albicollis | White-throated sparrow | Passeriformes | Fringillidae | ARWJ00000000 |
| MAMMALIA | ||||
| Acinonyx jubatus | Cheetah | Carnivora | Felidae | BGI-G10K |
| Panthera leo | Lion | Carnivora | Felidae | BGI-G10K |
| Puma concolor coryi | Puma | Carnivora | Felidae | BGI-G10K |
| Crocuta crocuta | Spotted hyena | Carnivora | Hyaenidae | BGI-G10K |
| Vulpes vulpes | Red fox | Carnivora | Canidae | BGI-G10K |
| Connochaetes taurinus | Blue wildebeest | Cetartiodactyla | Bovidae | BGI-G10K |
| Elaphurus davidianus | Pere David’s deer | Cetartiodactyla | Cervidae | BGI-G10K |
| Sousa chinensis | Chinese white dolphin | Cetartiodactyla | Delphinidae | BGI-G10K |
| Giraffa camelopardalis | Giraffe | Cetartiodactyla | Giraffidae | BGI-G10K |
| Tragulus napu | Greater Malayan chevrotain | Cetartiodactyla | Tragulidae | BGI-G10K |
| Oryx gazella | Gemsbok | Cetartiodactyla | Bovidae | BGI-G10K |
| Muntiacus reevesi | Chinese muntjac | Cetartiodactyla | Cervidae | BGI-G10K |
| Muntiacus muntjak | Indian muntjac | Cetartiodactyla | Cervidae | BGI-G10K |
| Desmodus rotundus | Common vampire bat | Chiroptera | Phyllostomidae | BGI-G10K |
| Dromiciops gliroides | Monito del monte | Microbiotheria | Microbiotheriidae | BGI-G10K |
| Tachyglossus aculeatus | Short-beaked echidna | Monotremata | Tachyglossidae | BGI-G10K |
| Equus przewalskii | Mongolian horse | Perissodactyla | Equidae | BGI-G10K |
| Fukomys damarensis | Damaraland mole rat | Rodentia | Bathyergidae | BGI-G10K |
| Spermophilus dauricus | Daurian souslik ground squirrel | Rodentia | Sciuridae | BGI-G10K |
| Bison bison | American bison | Cetartiodactyla | Bovidae | JPYT00000000 |
| Bubalus bubalis | Water buffalo | Cetartiodactyla | Bovidae | AWWX00000000 |
| Cavia aperea | Brazilian guinea pig | Rodentia | Caviidae | AVPZ00000000 |
| Ceratotherium simum simum | Southern white rhinoceros | Perissodactyla | Rhinocerotidae | AKZM00000000 |
| Chinchilla lanigera | Long-tailed chinchilla | Rodentia | Chinchillidae | AGCD00000000 |
| Chlorocebus sabaeus | Green monkey | Primates | Cercopithecidae | AQIB00000000 |
| Chrysochloris asiatica | Cape golden mole | Afrosoricida | Chrysochloridae | AMDV00000000 |
| Condylura cristata | Star-nosed mole | Eulipotyphla | Talpidae | AJFV00000000 |
| Elephantulus edwardii | Cape elephant shrew | Macroscelidae | Macroscelididae | AMGZ00000000 |
| Eptesicus fuscus | Big brown bat | Chiroptera | Vespertilionidae | ALEH00000000 |
| Galeopterus variegatus | Sunda flying lemur | Dermoptera | Cynocephalidae | JMZW00000000 |
| Jaculus jaculus | Lesser Egyptian jerboa | Rodentia | Dipodidae | AKZC00000000 |
| Leptonychotes weddellii | Weddell seal | Carnivora | Phocidae | APMU00000000 |
| Manis pentadactyla | Chinese pangolin | Pholidota | Manidae | JPTV00000000 |
| Mesocricetus auratus | Golden hamster | Rodentia | Cricetidae | APMT00000000 |
| Microtus ochrogaster | Prairie vole | Rodentia | Cricetidae | AHZW00000000 |
| Mustela putorius furo | Domestic ferret | Carnivora | Mustelidae | AEYP00000000 |
| Octodon degus | Degu | Rodentia | Octodontidae | AJSA00000000 |
| Odobenus rosmarus divergens | Pacific walrus | Carnivora | Odobenidae | ANOP00000000 |
| Orcinus orca | Killer whale | Cetartiodactyla | Delphinidae | ANOL00000000 |
| Orycteropus afer | Aardvark | Tubulidentata | Orycteropodidae | ALYB00000000 |
| Papio anubis | Olive baboon | Primates | Cercopithecidae | AHZZ00000000 |
| Peromyscus maniculatus | North American deer mouse | Rodentia | Cricetidae | AYHN00000000 |
| Physeter catodon | Sperm whale | Cetartiodactyla | Physeteridae | AWZP00000000 |
| Saimiri boliviensis | Bolivian squirrel monkey | Primates | Cebidae | AGCE00000000 |
| Trichechus manatus latirostris | Florida manatee | Sirenia | Trichechidae | AHIN00000000 |
The fifth goal of the G10K Project, raising funds, is an evolving exercise. The G10K Project was initially predicated on the expectation that the costs associated with genome sequencing would decrease rapidly, making it relatively affordable to sequence vertebrate genomes with size scales similar to the human genome (10–12). However, even as sequencing costs decline, the cost of data processing and bioinformatic analysis remains substantive. The G10KCOS is addressing this challenge by fostering training workshops that empower computer-savvy students in analysis of genome data (see below for these bioinformatics challenges). The G10KCOS endorses research development grants and proposals that facilitate local funding of genome projects and encourage investigator-initiated fund development from government, corporate, and entrepreneurial resources. G10K has signed memorandums of understanding with large sequencing centers, such as BGI-Shenzhen and the Broad Institute, to work together to increase the quality and quantity of vertebrate genome sequencing endeavors. For example, in 2010 BGI-Shenzhen agreed to sequence and fund the first ~1% (105 species) of vertebrate genomes in close collaboration with the G10KCOS. At this writing, whole genome sequences have been completed for 70% of these species, and of these, 43 have been published (Tables 2 and 3).
Initial publication of a genome sequencing project frequently generates additional funding, particularly when the published genome of a species stirs excitement and enthusiasm in the public imagination. Whether it is the genome of the giant panda (13), with its revelations about the genetics of its ability to digest bamboo; the elephant shark (14), as a model for the evolution of the vertebrate body plan; or the minke whale (15), providing a glimpse into the adaptations associated with becoming aquatic, many of the opportunities we already have with today’s sequencing technology are too enticing to pass up while waiting for technology to improve.
Lastly, the G10K Project has spread across biology to inspire similar large community initiatives to sequence the genomes of nonvertebrate species (our sixth goal), including insects (i5K), noninsect marine invertebrates (GIGA), plants (NSF Plant Genome Research Program), fungi (1000 Fungal Genomes Project), and microbes (100K Foodborne Pathogen Genome Project) (see Table 1).
BIOINFORMATICS CHALLENGES TO WHOLE GENOME SEQUENCE ANALYSES
The G10KCOS is presently working to identify and prioritize the next set of vertebrate species for genome sequencing (e.g., Reference 16). This process relies on insights from the bioinformaticians who will lead the assembly and analysis of the sequence data (17, 18). A critical first step in genome assembly is to determine what sequence data will be most useful to maximize the potential for de novo and reference-guided genome assembly. Large-insert genomic libraries, long sequence reads, and physical map–based technologies are crucial in assembling longer contiguous sequence fragments. High-quality (undegraded) DNAs in high-microgram quantities are required. Better methods for de novo genome sequencing from smaller (nanogram) amounts of DNA will make sample collection easier for many additional smaller species. Another important consideration for genome assembly is the size and repeat content of the target genome. Larger and more repetitive genomes will be more costly to sequence and assemble. Complex and abundant repeat families present in many species confound genome assembly, especially if the repeating units are long and highly similar to one another. Unfortunately, it is not always possible to determine the repeat content of a genome until some preliminary sequence sampling has been performed.
Another key bioinformatics challenge is sequence heterozygosity and its disposition across the genome. Available assembly algorithms erect a haploid reference genome by merging the information from the two parental genome sequences, often making arbitrary phase assignments, frequently producing chimeric contigs and scaffolds. A highly heterozygous individual can make assembly inaccurate or impossible. This can be assuaged by selecting highly inbred or haploid individuals, but these are unavailable for most species. Abundant segmental duplications, which may appear as additional haplotypes, add to the problem. These may be polymorphic, and hence heterozygous as well. Mixtures of DNA from multiple individuals, undertaken to obtain sufficient input DNA for some sequencing libraries, create an additional layer of complexity.
Given the current challenges in assembling a large (>>3-Gbp), repeat-rich genome with a high level of heterozygosity, many such genome projects are being deferred until the future. Even for typical vertebrate genomes, there is constant awareness that the longer one waits to sequence one’s favorite genome, the cheaper and higher quality it will become. Species for which genomic sequences were generated and assembled relatively early in the large-scale comparative genomics era can be of lower quality, with inaccurate assemblies, missed paralogs, and chimeric chromosomal segments [see, for example, the platypus (19) and giant panda (13) genomes; 20]. Assemblies for certain species that were first to be sequenced (e.g., chicken, chimpanzee) have been validated and improved using complementary mapping and assembly approaches, but they are expensive and time consuming. Prioritizing species for sequencing is a complex process that must balance the needs of individual communities, the overall G10K effort, funding constraints, and emerging technologies.
EVALUATING GENOME ASSEMBLIES
The initial step in making a genome useful to the biological community that studies a species is to produce an assembly of the millions of short DNA reads obtained from next-generation sequencing technology into an ordered and oriented sequence of contigs that resembles the order in which the assembled sequence actually occurs on each chromosome (see References 20–22). Genome assembly begins with homology match detection of reads to build short contigs. Contigs are then joined with mate pair end reads to form scaffolds, which within ideal assemblies span millions of base pairs. The process is completed when scaffolds are assembled into chromosomes using independent physical framework maps. The G10KCOS has evaluated a dozen or more available computational assembly tools, termed assemblers, which have been developed to accomplish this process in the Assemblathon competitions (7, 8). The challenges are detecting and correcting assembly mistakes caused by repeat sequence families, by copy number variation of certain DNA stretches, and by single-nucleotide variants (SNPs), the stuff of evolution and the scourge of a basic assembler (e.g., Reference 21). Assemblathon competitions first compared different assembly tools using a simulated vertebrate genome (7) and then three genome sequences from a cichlid fish, a parakeet, and a snake (8). The Genome Assembly Gold-standard Evaluation consortium and study further evaluated assembly quality of genomes across a broad array of species (5).
Lessons learned from the Assemblathons and other evaluations have led to the development of new assemblers. DISCOVAR de novo (http://www.broadinstitute.org/software/discovar/blog/; 23) is a new assembler developed at the Broad Institute that avoids the need for polymerase chain reaction (PCR) and in fact requires PCR-free libraries. This leads to improvements because compositional biases present in PCR-based approaches confound assemblers by generating nonuniform read depth. Although DISCOVAR is currently being used for resequencing projects, its real promise may be to assemble de novo genomes.
To evaluate assembly quality, new metrics have also been developed beyond N50 (the smallest length N such that at least 50% of the bases in the assembly are in contigs of that length or greater). Probabilistic measures based on likelihood statistics have been used and shown to provide more accurate and objective evaluations of assembly quality, independent of a reference genome (24–26). For example, the program CGAL uses the uniformity of genome coverage to evaluate the likelihood of assembly quality while simultaneously taking into account sequencing errors, insert size distribution, and extent of unassembled data (26). When CGAL was applied to the Assemblathon 1 data set, assemblies with a higher extent of coverage tended to be more accurate. These methods allow researchers to optimize parameters associated with assembly programs to obtain better-quality assemblies (with higher likelihood values) and are likely to become standard tools in obtaining high-quality assemblies (25).
A major finding of the Assemblathon studies is that there is considerable variation among output assemblies. Users cannot simply merge the outputs of many assemblers to arrive at an optimal consensus assembly. One assembly program, Metassembler (M. Schatz, unpublished data; http://schatzlab.cshl.edu/presentations/2011-11-03.Genome%20Informatics.pdf), actually does this, but its accuracy is no better than its best constituents. Assembly is a complex problem with many trade-offs, and there are no easy solutions (25). Has genome assembly with short reads reached a point of diminishing returns? At the G10K 2013 workshop, we learned that though many algorithms are still in development, accuracy is not substantially improved when only short reads are available, suggesting new sequencing approaches are needed to make the next quantum leap.
Large-Insert Sequencing Methods
New methods that improve the outlook for de novo genome assembly by sequencing large inserts with distinctly barcoded short reads are on the horizon. Protocols based on sequencing fosmid pools (~30 kb/fosmid) have gotten less expensive while still achieving long-range order and orientation of contigs (27, 28) (Table 4). Illumina-Moleculo technology, at approximately 10 kb per independently barcoded insert, provides similar benefits at lower cost. Its cost and overall feasibility for G10K have not been well established, though several groups have recently used Illumina-Moleculo reads to haplotype the human genome, with promising results (29).
Table 4.
Long-read and mapping technologies that promise to improve genome assemblies
| Platform or method | Technology | URL/more information |
|---|---|---|
| Pacific Biosciences | Single-molecule, real-time sequencing | http://www.pacificbiosciences.com |
| Illumina Moleculo | Long molecular reads | http://www.illumina.com/technology/next-generation-sequencing/long-read-sequencing-technology.html |
| Oxford Nanopore | Nanopore sensing | https://www.nanoporetech.com/ |
| BGI Complete Genomics | Self-assembling DNA nanoarrays | http://www.completegenomics.com/ |
| OpGen | Whole genome mapping | http://www.opgen.com/ |
| BioNano Genomics | Single-molecule imaging/nanochannel arrays | http://www.bionanogenomics.com/ |
| Nabsys | Single-molecule sequencing with nanodetectors | http://www.nabsys.com/ |
| Stratos Genomics | Single-molecule sequencing by Sequencing by Expansion (SBX) | http://www.stratosgenomics.com/ |
| Electronic BioSciences | Nanopore single-molecule sequencing | http://electronicbio.com/ |
| GenapSys | Gene Electronic Nano-Integrated Ultra-Sensitive (GENIUS) | http://genapsys.com/ |
| Genia | Single-molecule sequencing with nanopores | http://www.geniachip.com/ |
| Lasergen | Lightning terminator technology | http://lasergen.com/ |
| Noblegen | Single-molecule sequencing with nanopores and optical reading | http://www.noblegenbio.com/ |
| QuantuMDx | Single-molecule sequencing (Q-SEQ) | http://www.quantumdx.com/ |
| Sperm haplotyping | Whole genome haplotyping | 34–36 |
| Radiation hybrid maps | Whole chromosome mapping | 279 |
| Trios | Whole genome haplotyping | 280 |
Table 4 lists promising new long-read technologies, although each of these is as yet unproven for very large-scale (~3-Gbp) genome assembly. The single-molecule, real-time sequencing technology (SMRT) manufactured by Pacific Biosciences (PacBio) has been available for several years, but its higher relative cost and higher basic error rate have restricted its use to microbial genomes and eukaryote transcriptomes (30, 31). However, ongoing improvements in SMRT sequencing are beginning to ameliorate these concerns (32), and high-quality assemblies can often be obtained through hybrid approaches in which assemblies are generated using both short reads (e.g., Illumina) and long reads (e.g., PacBio) (33). Oxford Nanopore long reads have evoked considerable hopefulness as genome scientists are piloting genome assembly for accuracy, feasibility, and cost effectiveness. As various long-read technologies improve and their prices fall, it is likely that they will become part of typical genome assembly efforts.
Mapping Methods to Assist in Assembly
Mapping methods can also be used to improve assembly. Richard Durbin from the Wellcome Trust Sanger Institute proposed at the 2013 G10K meeting the sequencing of trios (mother, father, child) to improve genome assembly through a direct haplotype-phase resolved linkage map (280). Using SNP variation as an information source in assembly is a unique and potentially powerful new strategy that would anchor scaffolds to an ad hoc haplotype map. However, this approach does require additional sequencing. These techniques are an addition to single-sperm genome amplification (producing individual genome-wide haplotypes as well as whole genome assemblies) and other sequencing approaches that in theory can build a recombination and/or physical map using bioinformatics analysis (34–36).
Framework physical maps have been a mainstay for anchoring genome assemblies of model species (human, mouse, rat, dog, cat, and others) (20). However, linkage and radiation hybrid physical maps for these genome projects are rather expensive for wider-scale use. Optical mapping, a relatively new tool for building an independent physical map to anchor assembled scaffolds of sequenced genomes (e.g., Reference 37), was evaluated favorably in Assemblathon 2 (8). Map-generating technologies pioneered by BioNano Genomics, the Irys System, use rare-cut genomic DNA subjected to electrophoretic current to produce physical maps as well (38, 39). Physical or optical mapping methods can be used to improve graph navigation (40), to validate chromosomal ordering of contigs, and to detect and break up chimeric contigs. Random fosmid sequencing was also used as a kind of physical map for evaluation in Assemblathon 2. Although laborious and expensive, clone-based sequencing has the advantage of reduced size and no sequence heterozygosity. Genome assemblers can benefit from transcriptome information (41) to guide their algorithms as well as from comparative syntenic similarity employed by the Reference-Assisted Chromosome Assembly algorithm (42). These avenues of research must be explored more thoroughly as genome alignment and comparative genome analyses become more central to the G10K Project.
GENOME ANNOTATION
Genome annotation encompasses the description of a variety of elements that can be identified in a species’ genome, from protein-coding regions and intervening noncoding sequence to repeat families, noncoding RNAs, regulatory motifs, and specific elements (Table 5). For identification of protein-coding genes, transcriptome information via RNA-seq data is invaluable before, during, and after a genome has been assembled (6). Noncoding RNA genes, such as structural RNAs, microRNAs, and long noncoding RNAs, are also identified by RNA-seq in conjunction with bioinformatics sequence analysis and play key roles in the cell (e.g., Reference 43). One of the main reasons to sequence a genome is to investigate its genes, and RNA-seq can provide some of this information at a fraction of the cost of a whole genome assembly. Flanking the genes, one finds a variety of regulatory elements, some of which are highly conserved between species and hence recognized from sequence, whereas others are more rapidly evolving and require experimental assays involving chromatin immunoprecipitation followed by sequencing (ChIP-seq) or DNase I hypersensitive site sequencing.
Table 5.
Example tools used for genome assembly, annotation of genome features, and mapping
Available software programs for discerning genes and other features (Table 5) have been employed to unravel the secrets of new genomes on a regular basis. There are no precise best practices for gene selection, SNP discovery, or repeat annotation, although it has been shown that consistency may be low across different algorithms and methods [e.g., SNP calling (44) or reconstruction from RNA-seq data (45)]. The G10KCOS is considering an annotation-collaborative exercise (such as the Assemblathons and the Alignathon) to develop more explicit guidelines for vertebrate genome annotation.
GENOME ALIGNMENT
A comparative genomics approach between related species is fundamental to the identification and analysis of genes, their regulatory elements, and their adaptive natural history (6, 46–48). As such, comparative analyses of homologous genes in a syntenic context among related well-annotated species is a mainstay of annotation pipelines (49, 50). Such analysis depends heavily upon accurate multiple genome alignments. Exceptions to gene sequence conservation can indicate evolutionary gene changes, chromosome rearrangements, gene family expansion or contraction, and SNP-based signatures of historic selection. Discerning these genome modifications allows critical insights into the events occurring over the course of speciation and divergence of taxa. But comparative analysis of genomes from distantly related species is not simple, rather akin to comparing the assembly blueprints of a Boeing 747 to a Mercedes-Benz sedan, to a Yamaha motor scooter, and to a tricycle. A first step is to design an efficient strategy for aligning the entire gigabase-long genomes of related species.
Genome alignment, the task of aligning all the homologous nucleotides in a set of complete genomes, including those in noncoding regions, is critical if we are to establish the genetic relationships and, by extension, evolutionary history of our shared vertebrate ancestry. Genome alignment can be thought of as a generalized form of the DNA alignment problem, in that all other (classical) forms of alignment are a subclass of this general problem. The Alignathon competition invited participants to submit solutions to constructed or collected data sets (51). Three independent data sets, two simulated from primates and mammals and one a set of 20 Drosophila genomes, were offered for trial of various alignment algorithms. All the data sets involved genomes of approximately 200 Mbp in length, a decision made to create a meaningful challenge that was nonetheless accessible to the broadest possible range of tools. In all, 35 different analytical solutions were submitted by 10 teams using 12 distinct alignment pipelines (51).
Several important conclusions were reached through the Alignathon competition. First, relatively few groups and very few tools are currently capable of making precise genome alignments even at the scale of the 20-Drosophila-genome data set. For example, 11 of the 35 submitted alignments were computed using variants of the Multiz alignment pipeline (52), which is now over ten years old. Second, many current genome alignment tools have noticeable limitations. In particular, many of the entries were reference-based (genomes aligned to a reference genome as a key step), which produced a noticeable bias in the quality of alignments between nonreference genomes. Notably, only two of the alignment teams attempted to align multiple paralogous sequences. Third, there are few broad metrics for assessing genome alignments of real genomes that can be used to assess the quality of the alignment across the genome, and which do not rely on expert biological information (e.g., the location of annotations), and even fewer that have robust implementations. Fourth, consistent results were found between the simulation study and metrics for assessing the real alignments (53). Lastly, there exists tremendous variability in performance between alignment programs, though there is much less variance when aligning closely related organisms. With increasing evolutionary distance between compared species, all the various whole genome alignment tools get progressively less reliable.
The Alignathon was successful in revealing both the strengths and weaknesses of available whole genome alignment tools, but there remain several important directions for future work that, when pursued, will provide valuable information for the G10K and eukaryotic genomics community as a whole. A proposed second Alignathon competition in the future would address the following topics:
the impact of assembly errors on alignment. Addressing this would ideally be an integrative analysis with the Assemblathon group.
scaling to larger genome sizes with greater complexity and more repeats; i.e., evaluating and comparing results of full-size vertebrate genome alignments.
a comparison of methods for the alignment of genes within genome alignments.
the accuracy of cross-validation methods; one way to assess genome alignments is to set aside the sequence of a target genome and then assess how closely an imputed ancestral genome based upon a genome alignment of the other genomes matches the target genome. Such approaches have been used previously (52, 54) but never for complete genomes and genome alignments.
Computing genome alignments is computationally intense and requires several thousand CPU hours per genome. One of the main problems encountered in the first Alignathon was the lack of groups with sufficient computational power to compete. This is a critical problem that must be addressed by the development of more efficient methods, coupled to an increased commitment to provisioning more powerful computer resources for multiple alignments.
PROGRESS AND FUTURE PLANS FOR WHOLE GENOME SEQUENCING OF 10,000 SPECIES
In the five years since Genome 10K was proposed, the genomes of 277 vertebrate species have been proposed, funded, accomplished at some level, and released; of these, the genomes of 164 species have been reviewed and published (Tables 2 and 3). These achievements reflect efforts from larger sequencing centers, independent projects from individual teams, the BGI-G10K collaboration, and other G10KCOS initiatives, altogether a remarkable accomplishment. An additional 200+ species are named on websites of sequencing centers (BGI, the Broad Institute, the Baylor College of Medicine Human Genome Sequencing Center, The Genome Institute at Washington University, and others) as pending, with a substantial degree of uncertainty about their timetable for completion. The initial G10KCOS selection of species has been discussed (1), and a wealth of vertebrate evolutionary genomic diversity is beginning to be produced. Next, we summarize the challenges, accomplishments, and insights of G10K to date regarding the five principal taxonomic classes of vertebrates (Figure 1).
FISHES
More than half of all vertebrate species are fishes, which include the jawless (Agnatha), cartilaginous (Chondrichthyes), lobe-fin (Sarcopterygii), and ray-fin (Actinopterygii) fishes, with the latter group being the most diverse in number of species (Figure 2). The first nonhuman vertebrate genomes to be sequenced were those of the teleost fishes, a group that contains many species with genomes that are unusually small in size and therefore amenable to whole genome shotgun sequencing (e.g., fugu, Takifugu rubripes; 55, 56). Since then, a draft genome sequence from another pufferfish species, Tetraodon nigroviridis, has been produced (57), along with the genomes of the medaka (Oryzias latipes), three-spined stickleback (Gasterosteus aculeatus), zebrafish (Danio rerio), and platyfish (Xiphophorus maculatus), all of which serve as important model organisms for studies of gene function in development and adaptive evolution (58–61). Annotation of the zebrafish genome revealed over 26,000 protein-coding genes as well as the highest number of species-specific genes yet found for any vertebrate species whole genome sequenced to date. This large gene number is likely due to the whole genome duplication event that occurred early in the history of teleost fishes, resulting in the formation of numerous functional gene duplicates (60). Since these earlier studies, the number of fish WGS projects, both published and ongoing, has increased dramatically, providing many key insights related to physiological adaptations and vertebrate evolution (62, 63).
Figure 2.
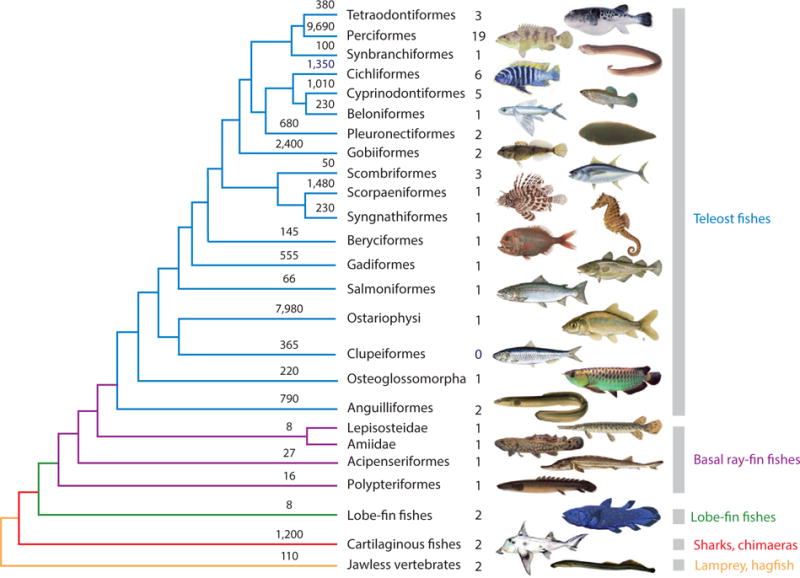
Consensus phylogeny of the major lineages of fishes. Topology and dates (Ma) are derived from combined data tree reported in Reference 1. On the ends of the limbs is the number of living species for that group. Following the common names of taxon groups is number of species with published and/or pending genomes (see Tables 2 and 3).
Given the breadth of vertebrate species diversity represented by the fishes, the majority of species planned to be de novo sequenced by the G10K Project will be fishes, particularly the teleosts (see Reference 16). As a first step toward that goal, 30 of the first 105 species to be selected for WGS through the collaborative efforts of BGI and G10K are fishes, including one cartilaginous fish, the elasmobranch great hammerhead shark (Sphyrna mokarran); two representatives of the early-branching Chondrostei; and 27 species of teleost fishes that encompass 12 orders. At this writing, the genomes of 24 fish species are published and 47 others are near completion (Tables 2 and 3). In anticipation of future genome sequencing efforts, the G10K fish community has identified a global list of 100 fish species that were nominated as gold standards, in that besides WGS, transcriptomes and stable cell lines will be generated for these species (16).
AMPHIBIANS
Amphibians comprise approximately 11% of vertebrate species. New taxa are described and reported for this group every year, implying that total amphibian biodiversity may be greatly underestimated (64). Among 7,300 named amphibian species, we currently have whole genome sequences available for only two species, both being anurans and from the same genus, the western clawed frog [Xenopus (Silurana) tropicalis] and the African clawed frog [Xenopus (Silurana) laevis] (Tables 2 and 3) (65; see also http://www.xenbase.org/entry/). Genome size is extremely variable within amphibians, varying by as much as ~130-fold (66, 67). Further, amphibians harbor some of the largest genomes, which has significantly hampered progress in the sequencing of additional amphibian genomes. The largest tetrapod genomes are found within the salamanders (Caudata), with sizes ranging from ~14 to ~120 Gb (68). Preliminary genomic scans of several salamander taxa indicate that large genome size may be related to the extensive proliferation of long terminal repeat retrotransposons (69). Such large genomes increase the cost of collecting raw data (many more libraries are needed to achieve adequate coverage) and increase the computational complexity of the assembly and analysis of those data. Additionally, the small physical size of most amphibians limits the amount of tissue that is available for making large-insert mate-pair libraries.
Despite the challenges and high costs of obtaining a diversity of amphibian genomes, there are reasons that these costs may be justifiable to some extent, considering how underrepresented this important group is currently among the list of completed vertebrate genomes (Table 2). Future developments in assembly strategies, especially the use of long reads discussed above (Table 4), may enable large genomes to be assembled more readily. Given the remarkable and unique adaptations developed in this vertebrate class, the complete absence of an understanding of the diversity of amphibian genome structure, content, and evolution poses a major gap in our knowledge of living vertebrates (66).
Among the first 105 species nominated for WGS through the BGI-G10K collaborative effort, nine amphibians were chosen to represent a broad level of divergence across the (mostly) anuran tree of life (Table 3). Species targeted for WGS include the coastal tailed frog (Ascaphus truei), a member of the Archaeobatrachia, which includes species showing primitive characteristics not found in other anurans and therefore represents a key lineage in the anuran tree of life. Also included is a member of the amphibian order Gymnophiona (caecilians), represented by the Banna caecilian (Ichthyophis bannanicus). At present, sequencing has been completed for the Tibetan frog (Nanorana parkeri), now in the draft assembly stage. At least one other independent anuran genome project is under way, that of the cane toad (Rhinella marina), a species originally found in Central and South America but later introduced into Hawaii, Australia, and parts of Oceania, where it has become an invasive (70). This species is also in the assembly stage.
WGS has also begun on well-studied frog species with relatively small genome sizes, such as the túngara frog (Physalaemus pustulosus), important in studies of sexual selection (71); the coqui frog (Eleutherodactylus coqui), important in studies of the evolution of direct development (72); and the plains spadefoot toad (Spea bombifrons), important in studies of speciation and adaptive hybridization (73). An additional small-genome species, the eastern banjo frog from Australia (Limnodynastes dumerilii), provides phylogenetic breadth. BGI-G10K is also taking on one large-genome species, the strawberry dart-poison frog (Oophaga pumilio), important for studies of rapid phenotypic evolution under natural and sexual selection (74). WGS data collection should begin soon on the last of the nine amphibian G10K species, including the horned marsupial frog (Gastrotheca cornuta), with its unusual reproductive biology and high conservation concern (75).
Looking toward the future, we see three main priorities for sequencing the genomes of additional amphibian species. First, a high-quality assembly should be provided from at least one member of each of the three extant amphibian orders (Anura, Caudata, and Gymnophiona). The BGI-G10K-selected amphibian species will meet two-thirds of this goal with sequencing the genomes of nine Anura and one Gymnophiona species (Table 3). As for Caudata, independent efforts are currently under way to sequence and assemble the genome of the Mexican axolotl (Ambystoma mexicanum), an important model organism used for research in a variety of fields, including embryogenesis, regenerative biology and medicine, neurology, and sensory biology (see http://www.ambystoma.org/). Amphibians are the sister group to amniotes, and complete genomes from representatives of all three amphibian orders could therefore provide new information about the characteristics of the amniote ancestral genome and how vertebrate lineages have diverged since this ancestor (76).
The second priority would expand WGS and annotation to incorporate species with smaller-sized genomes. Because a reference genome assembly is paramount to genome analyses, frog species with small genomes remain high-priority targets for platinum genome sequencing projects today (see sidebar, Draft Standards for Genome 10K). Furthermore, the availability of high-quality RNA samples for transcriptome sequencing from frozen viable cell cultures opens new opportunities for assisting the advancement of amphibian genomics.
A third priority would target species pairs or larger groups that allow genomic analysis of one of the many biological phenomena that are prominent in amphibian evolution. These include species that produce medically important skin toxins and antimicrobial peptides (77). Genomic data may also be important to many conservation interventions in amphibians and to understanding susceptibility and resistance to chytrid fungal infection and decline, e.g., of Atelopus and Lithobates (78, 79). Finally, the next round of amphibian genome sequencing will certainly need to greatly increase phylogenetic coverage of the amphibian tree of life to facilitate comparative genomic analyses, and in so doing will hopefully provide greater geographical representation as well.
NONAVIAN REPTILES
Living “reptiles” comprise three main lineages: (a) turtles (Testudines); (b) tuatara, lizards, and snakes (Lepidosauria); and (c) alligators and crocodiles (Crocodylia). Reptiles are an ancient group, which is reflected in their extensive diversity; for example, the divergence among major squamate groups (e.g., snakes and lizards) is similar in magnitude to that between humans and kangaroos (~175 My) (80). This diversity manifests across many traits, reflected in appreciable genetic and morphological innovation across reptilian lineages. For example, across reptile species there exists a broad range of life history traits related to reproduction and sex determination. Among the most remarkable are repeated transitions across the phylogeny between environmental and genotypic sex determination (81). Furthermore, species with genotypic sex determination can have sex chromosome systems with either female (ZZ/ZW) or male (XX/XY) heterogamety. These sex chromosomes, and presumably the sex-determining genes they contain, are not conserved across lineages even though the basic syntenic blocks making up the karyotype are conserved (reviewed in Reference 82). Reptiles are therefore excellent models for the study of evolution of sex determination, and of sex chromosomes. Annotation, mapping, and comparison of whole genome sequences from both sexes are invaluable tools for understanding the evolutionary processes governing sex determination (83) and promise to identify, for the first time, a sex-determining gene in a reptile. Squamates (lizards and snakes) are also the only vertebrate group to have true parthenogenesis, or asexual reproduction without any input (genetic or otherwise) from males (84). They are thus excellent systems to investigate the consequences of asexuality in amniotes on a whole genome scale (85).
Despite the extreme variations in genomic content and characteristics present within reptiles (86), they have remained a relatively neglected target of large-scale genome sequencing efforts. A handful of recent nonavian reptile genome sequencing and assembly projects have been motivated by addressing phylogenetic questions and the genomic basis of specific biological questions. The first published nonavian reptile genome, that of the green anole lizard, Anolis carolinensis, revealed a nucleotide organization (isochores) unlike that of any other sequenced vertebrate to date (87–89). Since then, genomes for two snake species, the Burmese python (Python molurus bivittatus) and the king cobra (Ophiophagus hannah), have been published and indicate that snakes may have reevolved GC isochore structure (90–92). Analyses of snake genomes also suggest that the ancestral snake lineage experienced unprecedented levels of positive selection on protein-coding genes, that repeat element content varies widely across snakes, and that snake organ remodeling after feeding is associated with massive shifts in gene expression (90, 91). Draft genome sequences for four species of crocodilians, the American alligator (Alligator mississippiensis), the gharial (Gavialis gangeticus), the saltwater crocodile (Crocodylus porosus), and the Chinese alligator (Alligator sinensis), have been completed and published (93, 94). Together, these crocodilian genomes provide important insights into the ancestral genomes of archosaurs and amniotes and hold potential for understanding characteristics of dinosaur genomes (93). The sister phylogenetic relationship of turtles and archosaurs (birds and crocodiles) was recently affirmed with the complete genome sequence from the western painted turtle, Chrysemys picta (95), which also found that turtles have evolved at a remarkably slow rate at the molecular level. Crocodiles have an even slower rate (93). Thus, current reptilian genomics projects are largely motivated by the specific biological and evolutionary questions that their genomes can address, and ongoing or proposed projects continue to develop among independent research groups or through research consortiums (e.g., the Consortium for Snake Genomics) (Table 1).
Eleven nonavian reptile species were nominated for de novo genome sequencing and assembly through the BGI-G10K collaboration (Tables 2 and 3). Draft assemblies have been completed for eight of these species, of which three have been published (94, 96). Among the species chosen is the tuatara, Sphenodon punctatus, the sole representative of the relictual lineage Rhyncocephalia, which is likely sister to the squamate reptiles. The rarity and significance of this species made obtaining samples for WGS a permitting challenge, and the relatively large genome size (~5 Gb) has also hampered efforts to obtain a reference genome at high coverage.
Other reptilian target species were chosen to address particular questions with regard to key biological characteristics. The Gila monster, Heloderma suspectum, is being sequenced to identify the genes involved in venom evolution (e.g., Reference 97). The Australian central bearded dragon lizard, Pogona vitticeps, is also being targeted because this species provides an ideal model to examine the genomic underpinnings of environmental and genetic sex determination. Gender in this lizard is usually determined by a pair of ZZ or ZW sex microchromosomes (98), but ZZ individuals can be reversed to the female phenotype at high temperatures (99). An annotated genome sequence for the dragon lizard P. vitticeps is currently available online (https://genomics.canberra.edu.au), and a partial physical map for this species is nearing completion. For two turtle species published, the green turtle (Chelonia mydas), a marine species, and the soft-shelled turtle (Pelodiscus sinensis) (96), genome sizes averaged about 2.2 Gb. Comparative genomic analyses indicated dramatic expansion in the olfactory receptor gene family in both species and the loss of several orthologous genes involved in normal development and energy homeostasis (96). Whole-embryo gene expression analysis of both turtle species showed global repatterning of gene regulation following the divergence between the turtle and chicken lineages through which the unique body plan of turtles may have evolved (96).
Future priorities for WGS of additional taxa of nonavian reptiles is collectively based on the number of interesting biological questions such genomes may address, the availability of samples, species having smaller genome sizes and low heterozygosity, and overall vertebrate genome diversity. Species selected for the next round of WGS have been prioritized to address such questions, including the following: (a) the evolution and molecular mechanisms underlying genetic and temperature-dependent sex determination; (b) molecular underpinnings of extreme morphological and molecular convergent evolution; (c) extreme phenotypes (e.g., horns, gliding in lizards, adhesive toe pads, projectile tongues); (d) responses of widely distributed species to past and present climate change; (e) evolution and persistence of parthenogenetic lineages, evolution of deadly venom toxins, and loss of limbs and sight; (f) evolution of viviparity; and (g) the evolutionary placement of debated lineages within the evolutionary tree of nonavian reptiles. Because there are several independent research groups producing moderate-quality genomes of reptiles, the G10KCOS is targeting species that could add value to these other genomes by providing a platinum reference genome of related species.
BIRDS
Modern birds trace their origins to the Jurassic epoch (over 150 Mya), when a theropod lineage of the widespread and successful reptilian dinosaurs spawned a group that would be the only survivors of the Cretaceous-Paleogene dinosaur extinction (~66 Mya) (100). Today, Aves represents the most specious class of terrestrial vertebrates, with some 10,500 bird species occupying a plethora of adaptive niches. One hypothesis is that Neoaves birds and placental mammals, comprising more than 95% of all living bird and mammal species, have captured the ecological niche opportunities that emerged from the cataclysm of the Cretaceous-Paleogene extinction event 66 Mya, which led to the extinction of dinosaurs. An alternative hypothesis is that modern birds radiated 10–80 millions of years before that event (101, 102).
This detailed history, enriched by morphological, behavioral, molecular, and paleontological inference, has produced a fascinating vertebrate group that has informed evolutionary processes, neuroscience, developmental biology, and species conservation. Further, several domestic bird species have significant economic impact (chicken, turkey, ostrich, quail, and others), and many species have been introduced in the pet trade.
During recent decades, the avian systematics community has developed large repositories that house high-quality genetic samples of a substantial number of avian species. These collections provide an essential resource for genomic analyses of avian structural, functional, and behavioral diversity. With representation from 15 natural history collections distributed globally, the G10K biospecimen list (1) includes specimens from 100% of the 32 orders, 91% of the 230 families, 73% of the 2,172 genera, and approximately 50% of the 10,500 species of birds (Figure 3). Each order is represented in multiple biospecimen collections, as are all but 17 families and all but 585 genera, ensuring at least one sample of high quality.
Figure 3.
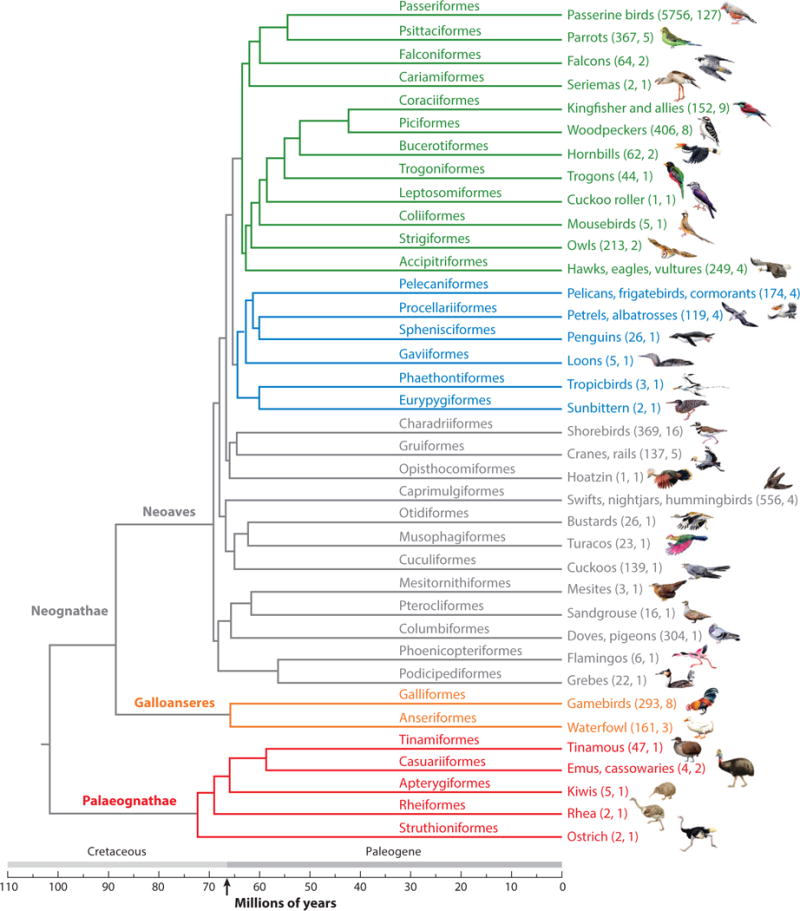
Consensus phylogeny of the major lineages of birds. In parentheses are the number of living species as defined by Howard and Moore (277), with the exception of the Passerine species count, which is taken from (1)/number of species with published and/or pending genomes (Tables 2 and 3). Data include both those genomes published to date as listed in Table 2 and those currently undergoing final assembly and annotation as part of the Avian Phylogenomic Consortium (Table 3). The underlying time-calibrated phylogenetic tree is a composite of the Neognath phylogeny published by Jarvis et al. (112) and Palaeognath phylogeny published by Mitchell et al. (278). Illustrations courtesy of Jon Fjeldå.
Until recently, whole genome sequence assessment was limited to three species, the chicken (Gallus gallus), domestic turkey (Meleagris gallopavo), and zebra finch (Taeniopygia guttata) (103–105). Further, the phylogenetic relationships among many bird taxa were unresolved or controversial except for the most coarse-grained divergences (106–108). The smaller genome size of birds relative to other vertebrates (68) and reduced sequencing costs made it possible to expand WGS efforts into nonmodel species to expand our understanding of the structure and function of avian genomes (109).
The avian genomics community has achieved a seminal realization of the vision outlined by Genome 10K for comparative genomic analyses. With unparalleled collaborative interaction, a comprehensive multifactorial WGS approach has been mounted by an international team (led by investigators from BGI, Duke University, and the University of Copenhagen) for 48 avian species representing each order of the Neognathae infraclass (Table 2) and two Palaeognath orders (110–112), and complemented by a group of reptilian outgroup species genomes, the American alligator (A. mississippiensis) (93) green sea turtle (C. mydas) (96), and green anole lizard (A. carolinensis) (87). In a December 2014 release of some 28 papers published in Science, Genome Biology, and other outlets, the richest comparative genomics analysis of any vertebrate group has appeared.
The findings of the collaborative Avian Phylogenomics Group address a wide variety of inquiries that we shall mention here only briefly, referring the reader to the more detailed reports for added substance (111–113; see also http://www.sciencemag.org/content/346/6215/1308.short and http://www.sciencemag.org/content/346/6215/1308/suppl/DC1). For starters, the studies provided a robust redrawing of the phylogenetic history of avian orders and a genomics inquiry into the making of a bird, or rather a bird genome (Figure 3). The findings help resolve the debate on the timing of the Neoaves divergence, dating it to around 66 Mya in a nearly starlike, big bang radiation of species. Targeted genomic screens for association were offered for special adaptations that are unique to birds, including vocal learning, skeletal adaptations to flight, feather development, dietary and developmental components to endentulism (toothlessness), wide-wavelength visual capacity, sex determination, sexual adaptations, behaviors, plumage color varieties, endogenous retroviral footprints, genome contraction relative to reptiles and mammals, genome exchange breakpoints, and ecological accommodation. Inspired by their own success, the G10K example, and the vast biospecimen collections already inventoried, the Avian Phylogenomics Group and an international consortium of scientists are pursuing a Bird 10K initiative to capture whole genome sequences for every living bird species.
The avian phylogenomic efforts have also addressed and informed many of the bioinformatics challenges listed here that in turn inform all envisioned interspecies comparative genomic efforts. Better ad hoc phylogenetic algorithms were developed and more robust and comparable assemblies and alignment stipulations were tested with real species by the bird exercise. In many ways, the genomes generated from the 48 bird species offer a refreshing preview to the hopes and perils of the coming adventures for the G10K Project.
MAMMALS
Mammals comprise approximately 9% of the total diversity of vertebrates, but they have received a disproportionate focus from WGS studies. This no doubt stems from the fact that humans are nested among the eutherian mammals and that understanding the genomes of our closest mammalian relatives will provide insights into our own biology. A recent comparative genomic analysis of the functional elements among 29 eutherian genomes showed that up to 5.5% of the human genome has evolved through purifying selection and also allowed identification of ~4.2% of the genome that is comprised of constrained bases (i.e., nucleotide positions that show conservation across most or all 29 eutherian genomes) (114). Moreover, this analysis provided strong evidence for the dispersal of transposable elements across mammalian genomes and the accelerated evolution of specific elements along the primate lineage. The mammal and bird genome studies illustrate a timely glimpse of the profound insights gained when a large number of phylogenetically diverse genomes are analyzed in a comparative context.
Many but not all projects for sequencing mammalian genomes were initiated at major genome sequencing centers (the Broad Institute, the Genome Institute at Washington University, Baylor College of Medicine Human Genome Sequencing Center, and BGI-Shenzhen). Species targeted for de novo sequencing by these research centers and independent groups have been sampled from across the mammalian supra-ordinal groups (Monotremata, Marsupialia, Afrotheria, Laurasiatheria, Euarchontoglires, and Xenarthra). Emphases have concentrated among four orders of mammals: Carnivora (cats, dogs, bears, and their allies), Cetartiodactyla (ungulates, dolphins, and whales), Primates (great apes and monkeys), and Rodentia (mice, rats, and allies) (Tables 2 and 3) (Figure 4). Eutherian mammalian outgroups for which there are published genome sequences include two marsupials and one monotreme mammal (Table 2). The number of mammals sequenced has risen to 111 (66 published and 45 near completion) (Tables 2 and 3). Indeed, 41% of accomplished vertebrate genome sequence analyses involve mammals.
Figure 4.
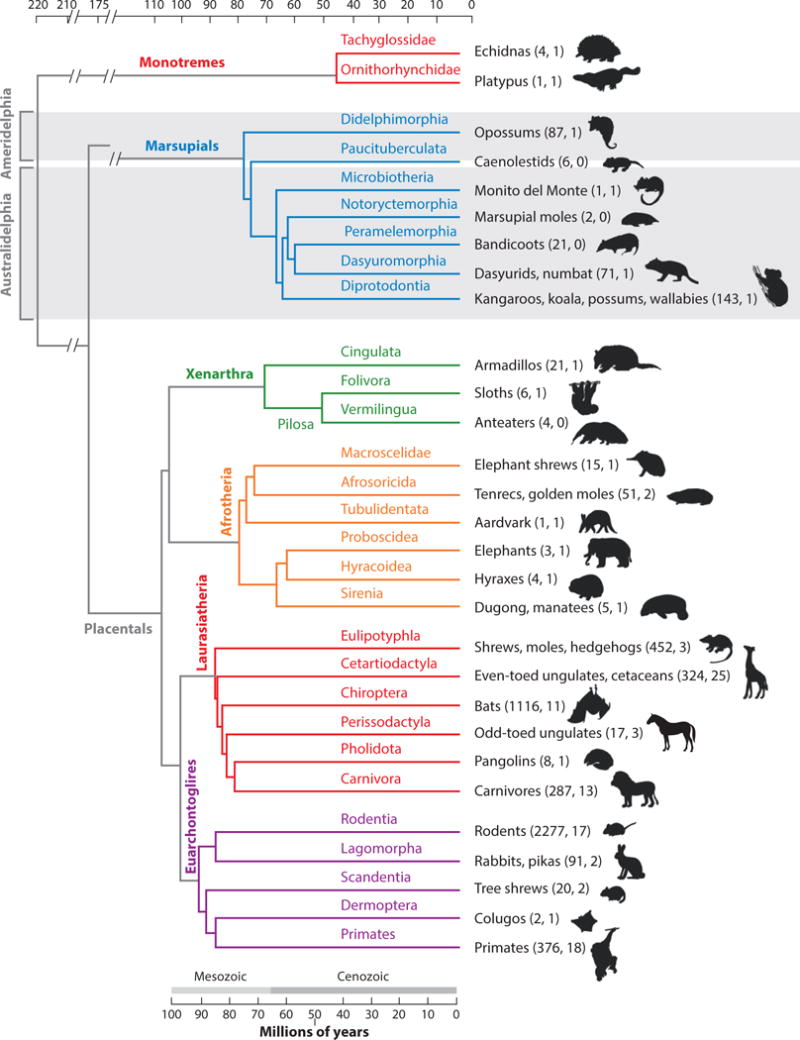
Consensus phylogeny of the major lineages of mammals. Topology and dates (Ma) are consensus estimates derived from References 1 and 276 and included citations. Following the common names of taxon groups in parentheses are the number of living species for that group and number of species with published and/or pending genomes (see Tables 2 and 3).
The mammal species selected for WGS by the initial BGI-G10K collaboration were chosen for reasons described previously (1) with attention to avoiding competitive overlap between the different genome sequencing centers. This has provided an opportunity to begin filling in the branches of the mammal tree of life by focusing on family-level representatives and/or closely related species (Figure 4). Our selection from Carnivora includes four species of large cats (tiger, Panthera tigris; African lion, Panthera leo; cheetah, Acinonyx jubatus; and American puma, Puma concolor). Combined with the felid genome projects being carried out by other research groups, this means that reference genomes will be available for four of the eight major lineages of the Felidae (115). The largest focus of the BGI-G10K collaborative project is in the Cetartiodactyla, with 16 species targeted for de novo sequencing, for which draft assemblies have been completed for 11 species, with a dozen more in progress. Species in this group were chosen not only to address questions related to domestication and understanding of the genetic basis of particular adaptations [e.g., high-altitude adaptation in the domestic yak (116)] but also with an emphasis on understanding the role of genomic architecture and chromosomal rearrangement in genome and organismal evolution (e.g., Reference 42). Primate studies have received focused efforts owing to interest in organization, evolution, and adaptation of the human genome (117). Studies of great apes, including chimpanzees, bonobos, gorillas, and orangutans, have contributed insights into population expansions and reductions as well as phylogeography of our closest relatives, all of which are endangered species (e.g., Reference 118).
The G10KCOS identified several broad research themes that will be used to choose the next round of mammal species for WGS. Species were chosen not only based on their phylogenetic distribution but also with regard to addressing fundamental questions in evolution, behavior, ecology, physiology, and conservation. For example, pairs or groups of species from canids to Old World primates were identified that could be used to address fundamental questions on the genomics of speciation, such as the identification of regions (or islands) of high divergence that may be involved in reproductive isolation that change in size and dimension over time (see, e.g., Reference 119). Another theme revolved around comparing species, particularly within bats and marsupials, that differ dramatically in metabolic rate and how this relates to differences in body size and longevity (e.g., Reference 120). Many mammals, such as bears, squirrels, bats, and opossums, undergo hibernation as part of their life history, and therefore, pairs of species within each of these groups were identified for WGS to explore the genomic basis of hibernation and the ability to deal with deleterious effects of hibernation (e.g., Reference 121). Finally, given the potential revolutionary impact of genomics on conservation genetics and management (122), several species will be targeted for WGS that are amenable to addressing fundamental questions related to inbreeding and outbreeding depression, disease resistance, and use of genomic information to guide and inform deextinction efforts.
ANCIENT VERTEBRATE PALEOGENOMES
Although de novo genome sequencing of extant species exploits high-quality DNA extracted from purposefully collected tissues, another topic that fires the public imagination is paleogenomics— the sequencing and analysis of genome-scale information from historic or ancient samples, particularly those representing extinct species. Until recently, the sequencing of paleogenomes would have been inconceivable, owing to the sheer number of PCR-based Sanger sequencing reactions required to recover the gigabases of information within a preserved eukaryotic cell. Following publication of draft genomes of ancient humans, horses, and extinct species of Neandertals, Denisovans, the woolly mammoth, and passenger pigeons, popular perception has moved from asking if paleogenomes can be sequenced to when it will happen (124–128).
Considerable challenges to paleogenomic sequencing remain, however. Firstly, although the achievements thus far are undeniably impressive, the financial and physical resource requirements for paleogenomic sequencing remain beyond the capabilities of most research programs. Secondly, although experimental protocols for isolating paleogenomic data have improved considerably within the past several years, different preservation contexts clearly require different experimental approaches, and the field remains in the early stages of fully understanding how and why DNA is sometimes preserved. Thirdly, even if specimens are identified that contain high concentrations of target DNA relative to DNA from exogenous sources that colonize the sample postmortem, this target DNA will be heavily fragmented and damaged, precluding the generation of large-insert libraries or ultra-long reads that are critical for scaffolding de novo genome assemblies. As a result, most extinct genomes will, at best, be assembled via mapping to high-quality genomes of extant relative species—the success of which is limited by evolutionary distance. For example, extrapolation from in silico and experimental data sets based around mapping ancient sequencing reads to various mammal genomes suggests that at 5–6 My divergence (e.g., elephant-mammoth), 60–80% of the genome will map, whereas at >60 My (e.g., moa-extant ratite), success could fall below 20% (129, 130).
THE GENOMIC ROAD AHEAD
The G10K Project has fostered and witnessed many accomplishments and discoveries since its inception in 2009. The number of vertebrate species for which whole genomes are being produced or have been published has increased dramatically and will likely continue to rise exponentially in the future. By bringing together biologists, bioinformaticians, and computational scientists, the G10KCOS has tried to lead the way in establishing best practices in biospecimen collection and preparation as well as in genome assembly and alignment. As we have shown in this review, such efforts will need to be applied to other areas of analysis, especially for genomes of large size. The successes so far provide optimism for the future. Genome science continues to be a dynamic field with advancing technologies. Although the vast majority of genome sequencing performed today is on the Illumina platform, and assembly algorithms are dominated by de Bruijn graphs, this may not be true in five years. It is difficult to estimate how genome science will change in the next decade. There are a variety of exciting new technologies, but it is impossible to perform cost-benefit analyses without the products themselves and the algorithms designed to use them. These advances afford new opportunities for elucidating the changes in genome structure and sequence that have resulted in the diversity of vertebrate life. The generation of reference genomes is finding application in health and well-being of humans and other vertebrates and is being applied to efforts for stewardship of our planetary biodiversity and efforts to conserve species threatened with extinction.
DRAFT STANDARDS FOR GENOME 10K.
The G10KCOS continued an ongoing process of setting standards for “doing a vertebrate genome” that actually began in 2009 with the first G10K workshop. The groups recognized nine important areas for discussion and recommendations that all bear on what a G10K species genome project should encompass. Detailed reports about each of these areas have been or will be published separately and deposited on the G10K website for guidance in nomination and sequence analyses of present and future selected species. Similar recommendations for standards have appeared for other genome consortia (Table 1). The areas of consideration, discussed throughout this article but summarized here, include
Standards for biospecimen collections and DNA provision. In general, approximately 100 μg of high–smolecular weight DNA (>50 Kbp) are ideal for construction of high–molecular weight mate-pair libraries. These should be from a single individual selected for minimal heterozygosity to optimize assembly. A detailed description of standards for DNA collection, storage, and processing for G10K has appeared (4).
Recommendations for WGS of males and females. G10K recommends that sequencing of both a male and female be considered for each new species. Comparisons of male to female genomes implicate specific (Y or W) sequences, including dosage-dependent gene regions critical for pinpointing the sex-determining gene (s) (e.g., Reference 131). If sequencing both sexes is not possible, the heterogametic sex should be chosen, because XY males and ZW females represent both sex chromosomes and comprise unabridged sex chromosome genes useful for quality control, population, and forensic applications.
Sequencing standards. These standards involve optimal quality control standards for current generation sequencing, including >60× coverage to assure that >98% of the species’ euchromatic genome is represented.
Assembly standards. G10K standards for assembly encourage large contig and scaffold N50 (on the order of megabases), while minimizing (to a very few) the number of false joins that create chimeric scaffolds using an independent physical map–based framework. There is a cost-benefit consideration here, as some physical maps are very accurate but impractical owing to expense in many species (e.g., a pedigree linkage map in a humpback whale). Physical maps can be generated by various methods (Table 4), and a promising but as-yet-unfulfilled hope is the connecting of contigs to scaffolds using long-read technologies that are not yet optimized or scaled to larger vertebrate genomes (Table 4). Nonetheless, every good genome sequence seems to benefit from high-resolution physical maps (20).
Genome annotation standards. A G10K genome should have genes, SNPs, indels, repetitive elements, and other genome features annotated so the noncomputational user can access the genomic features and aspects readily. Table5 gives a listing of some standard genome features and publically available software that help annotate them.
Standards for archiving and placing a genome in a browser. It is essential that a final genome assembly be submitted to the International Nucleotide Sequence Database Collaboration (INSDC, http://www.insdc.org/) so that it is available in a standard repository to all scientists. Submission to the INSDC can occur through the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/genbank/), European Bioinformatics Institute (http://www.ebi.ac.uk/ena), or DNA Databank of Japan (http://www.ddbj.nig.ac.jp/). The G10KCOS also encourages that all new genomes should be loaded into a genome browser, such as Gbrowse (132), JBrowse (133), track data hubs on the UCSC Genome Browser (134), NCBI, or Ensembl, for viewing and downloading. This format for viewing genomes is convenient and familiar and very much more useful to biological researchers than a trace archive or raw reads.
Standards for genome alignment. Every species’ genome has an evolutionary context and is indisputably connected to all others in a deep evolutionary genealogy that must be better understood. The first step in comparative genomics is to align homologous segments across related genomes so that comparative analyses can be achieved. No perfect algorithm for genome alignment has been developed or claimed, especially for the large vertebrate genomes we discuss here. The Alignathon community of G10K has endeavored to maximize consensus experience in the Alignathon competition discussed elsewhere in this article (51). Achievement of best practices and transfer of these alignment methods to the next generation of genome scientists are goals that the G10KCOS embraces.
G10K data release. The G10KCOS endorses rapid publication and release of genome sequences in the spirit of facilitating wide uses and application. All species’ genome sequences, assembly, and annotation shall be released freely with public access upon publication or within two years of delivery of a sample to a sequencing facility, whichever comes first. The latter clause is intended to handle cases of delayed publication.
Platinum Genome 10K species. Owing to cost limitation, not all species will enjoy the scientific rigor demanded by the standards outlined above; indeed, some light-coverage sequences will be assessed, e.g., for SNP discovery, with no attention to de novo assembly and annotation. To facilitate genomic studies of such genomes, selected reference genomes called platinum genomes should be nominated for major taxonomic groups (e.g., orders or large families that differ by 30–50 My of evolutionary time). The G10KCOS will nominate reference species for which high-resolution physical maps or a long-insert sequencing equivalent will be generated and monitor the progress of such projects to maximize genome opportunities for these platinum species.
Acknowledgments
S.J.O., K.P.K., G.T., and A.K. were supported as PI by Russian Ministry of Science Mega-grant no.11.G34.31.0068; S.J.O. is Principal Investigator.
Contributing Authors for the G10KCOS
Klaus-Peter Koepfli,1* Benedict Paten,2* Agostinho Antunes,3,4 Kathy Belov,5 Carlos Bustamante,6 Todd A. Castoe,7 Hiram Clawson,2 Andrew J. Crawford,8,9 Mark Diekhans,2 Dan Distel,10 Richard Durbin,11 Dent Earl,2 Matthew K. Fujita,7 Tony Gamble,12,13 Arthur Georges,14 Neil Gemmell,15 M. Thomas P. Gilbert,16 Jennifer Marshall Graves,17 Richard E. Green,18 Glenn Hickey,18 Erich D. Jarvis,19 Warren Johnson,20 Aleksey Komissarov,1 Ian Korf,21 Robert Kuhn,2 Denis M. Larkin,22 Harris Lewin,23 Jose V. Lopez,24 Jian Ma,25 Tomas Marques-Bonet,26,27 Webb Miller,28 Robert Murphy,29 Pavel Pevzner,30,31 Beth Shapiro,32 Cynthia Steiner,33 Gaik Tamazian,1 Byrappa Venkatesh,34 Jun Wang,35,36,37 Robert Wayne,38 Edward Wiley,39 Huanming Yang,35,40 Guojie Zhang,35,41 David Haussler,2 Oliver Ryder,33 and Stephen J. O’Brien1, 24
1Theodosius Dobzhansky Center for Genome Bioinformatics, St. Petersburg State University, 199034 St. Petersburg, Russian Federation
2UC Santa Cruz Genomics Institute, University of California, Santa Cruz, California 95064
3CIMAR/CIIMAR, Centro Interdisciplinar de Investigação Marinha e Ambiental, Universidade do Porto, 4050-123 Porto, Portugal
4Departamento de Biologia, Faculdade de Ciências, Universidade do Porto, 4169-007 Porto, Portugal
5Faculty of Veterinary Science, University of Sydney, Sydney, NSW 2006, Australia
6Department of Genetics, Stanford University School of Medicine, Stanford, California 94305
7Department of Biology, University of Texas at Arlington, Arlington, Texas 76019
8Departamento de Ciencias Biológicas, Universidad de los Andes, A.A. 4976, Bogotá, Colombia
9Smithsonian Tropical Research Institute, Apartado Postal 0843-03092, Panamá, República de Panamá
10Ocean Genome Legacy Foundation, Center for Marine Genomic Research, New England Biolabs, Beverly, Massachusetts 01915
11Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1RQ, United Kingdom
12Department of Genetics, Cell Biology and Development, University of Minnesota, Minneapolis, Minnesota 55455
13The Bell Museum of Natural History, University of Minnesota, Minneapolis, Minnesota 55455
14Institute for Applied Ecology, University of Canberra, ACT 2601, Australia
15Allan Wilson Centre, Department of Anatomy, University of Otago, Dunedin 9054, New Zealand
16Centre for GeoGenetics, Natural History Museum of Denmark, Øster Voldgade 5-7, 1350 Copenhagen, Denmark
17La Trobe Institute of Molecular Science, La Trobe University, Melbourne, VIC 3086, Australia
18Baskin School of Engineering, University of California, Santa Cruz, California 95064
19Department of Neurobiology, Howard Hughes Medical Institute, Duke University Medical Center, Durham, North Carolina 27710
20Smithsonian Conservation Biology Institute, National Zoological Park, Washington, DC 20008
21Department of Molecular and Cell Biology and Genome Center, University of California, Davis, California 95616
22Royal Veterinary College, University of London, London NW1 0TU, United Kingdom
23Department of Evolution and Ecology and UC Davis Genome Center, University of California, Davis, California 95616
24Oceanographic Center, Nova Southeastern University, Dania Beach, Florida 33004
25Department of Bioengineering, University of Illinois at Urbana-Champaign, Urbana, Illinois 61801
26ICREA at Institut Biologia Evolutiva, CSIC-Universitat Pompeu Fabra, Barcelona 08003, Spain
27Centro Nacional de Analisis Genomico, Barcelona 08028, Spain
28Center for Comparative Genomics and Bioinformatics, Pennsylvania State University, University Park, Pennsylvania 16802
29Centre for Biodiversity and Conservation Biology, Department of Natural History, Royal Ontario Museum, Toronto M5S 2C6, Canada
30Department of Computer Science and Engineering, University of California at San Diego, La Jolla, California 92093
31Algorithmic Biology Lab, St. Petersburg Academic University, St. Petersburg 194021, Russian Federation
32Department of Ecology and Evolutionary Biology, University of California, Santa Cruz, California 95064
33San Diego Zoo Institute for Conservation Research, Escondido, California 92027
34Institute of Molecular and Cell Biology, Agency for Science, Technology and Research, Biopolis, Singapore, Republic of Singapore
35BGI-Shenzhen, Shenzhen 518083, China
36Department of Biology, University of Copenhagen, DK-2200 Copenhagen N, Denmark
37Princess Al Jawhara Center of Excellence in the Research of Hereditary Disorders, King Abdulaziz University, Jeddah 21441, Saudi Arabia
38Department of Ecology and Evolutionary Biology, University of California, Los Angeles, California 90095
39Department of Ecology and Evolutionary Biology, University of Kansas, Natural History Museum and Biodiversity Research Center, Lawrence, Kansas 66045
40James D. Watson Institute of Genome Sciences, Hangzhou 310008, China
41Center for Social Evolution, Department of Biology, University of Copenhagen, DK-2100 Copenhagen, Denmark
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
The Annual Review of Animal Biosciences is online at animal.annualreviews.org
LITERATURE CITED
- 1.Genome 10K Community Sci. Genome 10K: a proposal to obtain whole-genome sequence for 10,000 vertebrate species. J Hered. 2009;100(6):659–74. doi: 10.1093/jhered/esp086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayden EC. 10,000 genomes to come. Nature. 2009;462(7269):21. doi: 10.1038/462021a. [DOI] [PubMed] [Google Scholar]
- 3.Pennisi E. No genome left behind. Science. 2009;326(5954):794–95. doi: 10.1126/science.326_794. [DOI] [PubMed] [Google Scholar]
- 4.Wong PB, Wiley EO, Johnson WE, Ryder OA, O’Brien SJ, et al. Tissue sampling methods and standards for vertebrate genomics. GigaScience. 2012;1:8. doi: 10.1186/2047-217X-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salzberg SL, Phillippy AM, Zimin A, Puiu D, Magoc T, et al. GAGE: a critical evaluation of genome assemblies and assembly algorithms. Genome Res. 2012;22(3):557–67. doi: 10.1101/gr.131383.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yandell M, Ence D. A beginner’s guide to eukaryotic genome annotation. Nat Rev Genet. 2012;13(5):329–42. doi: 10.1038/nrg3174. [DOI] [PubMed] [Google Scholar]
- 7.Earl D, Bradnam K, John J, Darling A, Lin D, et al. Assemblathon 1: a competitive assessment of de novo short read assembly methods. Genome Res. 2011;21(12):2224–41. doi: 10.1101/gr.126599.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradnam KR, Fass JN, Alexandrov A, Baranay P, Bechner M, et al. Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. GigaScience. 2013;2(1):10. doi: 10.1186/2047-217X-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azvolinsky A. Sequencing the tree of life. The Scientist. 2014 Apr 24; [Google Scholar]
- 10.Mardis ER. A decade’s perspective on DNA sequencing technology. Nature. 2011;470(7333):198–203. doi: 10.1038/nature09796. [DOI] [PubMed] [Google Scholar]
- 11.Sboner A, Mu XJ, Greenbaum D, Auerbach RK, Gerstein MB. The real cost of sequencing: higher than you think! Genome Biol. 2011;12(8):125. doi: 10.1186/gb-2011-12-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayden EC. The $1000 genome. Nature. 2014;507(7492):294–95. doi: 10.1038/507294a. [DOI] [PubMed] [Google Scholar]
- 13.Li R, Fan W, Tian G, Zhu H, He L, et al. The sequence and de novo assembly of the giant panda genome. Nature. 2010;463(7279):311–17. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venkatesh B, Lee AP, Ravi V, Maurya AK, Lian MM, et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature. 2014;505(7482):174–79. doi: 10.1038/nature12826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yim H-S, Cho YS, Guang X, Kang SG, Jeong J-Y, et al. Minke whale genome and aquatic adaptation in cetaceans. Nat Genet. 2014;46(1):88–92. doi: 10.1038/ng.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernardi G, Wiley EO, Mansour H, Miller MR, Orti G, et al. The fishes of Genome 10K. Mar Genomics. 2012;7:3–6. doi: 10.1016/j.margen.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Schatz MC, Delcher AL, Salzberg SL. Assembly of large genomes using second-generation sequencing. Genome Res. 2010;20:1165–73. doi: 10.1101/gr.101360.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schatz BMC, Langmead B. The DNA data deluge. IEEE Spectrum. 2013 Jun 27; doi: 10.1109/MSPEC.2013.6545119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warren WC, Hillier LW, Marshall Graves JA, Birney E, Ponting CP, et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008;453(7192):175–83. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewin HA, Larkin DM, Pontius J, O’Brien SJ. Every genome sequence needs a good map. Genome Res. 2009;19(11):1925–28. doi: 10.1101/gr.094557.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagarajan N, Pop M. Sequence assembly demystified. Nat Rev Genet. 2013;14(3):157–67. doi: 10.1038/nrg3367. [DOI] [PubMed] [Google Scholar]
- 22.Salzberg SL, Yorke JA. Beware of mis-assembled genomes. Bioinformatics. 2005;21(24):4320–21. doi: 10.1093/bioinformatics/bti769. [DOI] [PubMed] [Google Scholar]
- 23.Weisenfeld NI, Yin S, Sharpe T, Lau B, Hegarty R, et al. Comprehensive variation discovery in single human genomes. Nat Genet. 2014;46:1350–55. doi: 10.1038/ng.3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clark SC, Egan R, Frazier PI, Wang Z. ALE: a generic assembly likelihood evaluation framework for assessing the accuracy of genome and metagenome assemblies. Bioinformatics. 2013;29(4):435–43. doi: 10.1093/bioinformatics/bts723. [DOI] [PubMed] [Google Scholar]
- 25.Ghodsi M, Hill CM, Astrovskaya I, Lin H, Sommer DD, et al. De novo likelihood-based measures for comparing genome assemblies. BMC Res Notes. 2013;6:334. doi: 10.1186/1756-0500-6-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rahman A, Pachter L. CGAL: computing genome assembly likelihoods. Genome Biol. 2013;14(1):R8. doi: 10.1186/gb-2013-14-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alexeyenko A, Nystedt B, Vezzi F, Sherwood E, Ye R, et al. Efficient de novo assembly of large and complex genomes by massively parallel sequencing of fosmid pools. BMC Genomics. 2014;15(1):439. doi: 10.1186/1471-2164-15-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huddleston J, Ranade S, Malig M, Antonacci F, Chaisson M, et al. Reconstructing complex regions of genomes using long-read sequencing technology. Genome Res. 2014;24(4):688–96. doi: 10.1101/gr.168450.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuleshov V, Xie D, Chen R, Pushkarev D, Ma Z, et al. Whole-genome haplotyping using long reads and statistical methods. Nat Biotechnol. 2014;32(3):261–66. doi: 10.1038/nbt.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chin C-S, Alexander DH, Marks P, Klammer AA, Drake J, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–69. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 31.Koren S, Harhay GP, Smith TP, Bono JL, Harhay DM, et al. Reducing assembly complexity of microbial genomes with single-molecule sequencing. Genome Biol. 2013;14(9):R101. doi: 10.1186/gb-2013-14-9-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts RJ, Carneiro MO, Schatz MC. The advantages of SMRT sequencing. Genome Biol. 2013;14(6):405. doi: 10.1186/gb-2013-14-7-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koren S, Schatz MC, Walenz BP, Martin J, Howard JT, et al. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol. 2012;30(7):693–700. doi: 10.1038/nbt.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu S, Zong C, Fan W, Yang M, Li J, et al. Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science. 2012;338(6114):1627–30. doi: 10.1126/science.1229112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, Fan HC, Behr B, Quake SR. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell. 2012;150(2):402–12. doi: 10.1016/j.cell.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirkness EF, Grindberg RV, Yee-Greenbaum J, Marshall CR, Scherer SW, et al. Sequencing of isolated sperm cells for direct haplotyping of a human genome. Genome Res. 2013;23:826–32. doi: 10.1101/gr.144600.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong Y, Xie M, Jiang Y, Xiao N, Du X, et al. Sequencing and automated whole-genome optical mapping of the genome of a domestic goat (Capra hircus) Nat Biotechnol. 2013;31(2):135–41. doi: 10.1038/nbt.2478. [DOI] [PubMed] [Google Scholar]
- 38.Lam ET, Hastie A, Lin C, Ehrlich D, Das SK, et al. Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat Biotechnol. 2012;30(8):771–76. doi: 10.1038/nbt.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hastie AR, Dong L, Smith A, Finklestein J, Lam ET, et al. Rapid genome mapping in nanochannel arrays for highly complete and accurate de novo sequence assembly of the complex Aegilops tauschii genome. PLOS ONE. 2013;8(2):e55864. doi: 10.1371/journal.pone.0055864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin HC, Goldstein S, Mendelowitz L, Zhou S, Wetzel J, et al. AGORA: assembly guided by optical restriction alignment. BMC Bioinform. 2012;13(1):189. doi: 10.1186/1471-2105-13-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xue W, Li J-T, Zhu Y-P, Hou G-Y, Kong X-F, et al. L_RNA_Scaffolder: scaffolding genomes with transcripts. BMC Genomics. 2013;14(1):604. doi: 10.1186/1471-2164-14-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim J, Larkin DM, Cai Q, Asan, Zhang Y, et al. Reference-assisted chromosome assembly. PNAS. 2013;110(5):1785–90. doi: 10.1073/pnas.1220349110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu X, Sun S. Comparing a few SNP calling algorithms using low-coverage sequencing data. BMC Bioinform. 2013;14(1):274. doi: 10.1186/1471-2105-14-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steijger T, Abril JF, Engström PG, Kokocinski F, Akerman M, et al. Assessment of transcript reconstruction methods for RNA-seq. Nat Methods. 2013;10(12):1177–84. doi: 10.1038/nmeth.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siepel A, Diekhans M, Brejová B, Langton L, Stevens M, et al. Targeted discovery of novel human exons by comparative genomics. Genome Res. 2007;17(12):1763–73. doi: 10.1101/gr.7128207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alföldi J, Lindblad-Toh K. Comparative genomics as a tool to understand evolution and disease. Genome Res. 2013;23(7):1063–68. doi: 10.1101/gr.157503.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kellis M, Wold B, Snyder MP, Bernstein BE, Kundaje A, et al. Defining functional DNA elements in the human genome. PNAS. 2014;111(17):6131–38. doi: 10.1073/pnas.1318948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Flicek P, Amode MR, Barrell D, Beal K, Billis K, et al. Ensembl 2014. Nucleic Acids Res. 2014;42:D749–55. doi: 10.1093/nar/gkt1196. Database Issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karolchik D, Barber GP, Casper J, Clawson H, Cline MS, et al. The UCSC genome browser database: 2014 update. Nucleic Acids Res. 2014;42:D764–70. doi: 10.1093/nar/gkt1168. Database Issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Earl D, Nguyen NK, Hickey G, Nguyen N, Harris RS, et al. Alignathon : a competitive assessment of whole genome alignment methods. bioRxiv. 2014 doi: 10.1101/gr.174920.114. doi: http://dx.doi.org/10.1101/003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14(4):708–15. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim J, Ma J. PSAR: measuring multiple sequence alignment reliability by probabilistic sampling. Nucleic Acids Res. 2011;39(15):6359–68. doi: 10.1093/nar/gkr334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paten B, Herrero J, Beal K, Birney E. Sequence progressive alignment, a framework for practical large-scale probabilistic consistency alignment. Bioinformatics. 2009;25(3):295–301. doi: 10.1093/bioinformatics/btn630. [DOI] [PubMed] [Google Scholar]
- 55.Brenner S, Elgar G, Sandford R, MacRae A, Venkatesh B, Aparicio S. Characterization of the pufferfish (Fugu) genome as a compact model vertebrate genome. Nature. 1993;366(6452):265–68. doi: 10.1038/366265a0. [DOI] [PubMed] [Google Scholar]
- 56.Aparicio S, Chapman J, Stupka E, Putnam N, Chia J-M, et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297(5585):1301–10. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- 57.Jaillon O, Aury J-M, Brunet F, Petit J-L, Stange-Thomann N, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431(7011):946–57. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- 58.Kasahara M, Naruse K, Sasaki S, Nakatani Y, Qu W, et al. The medaka draft genome and insights into vertebrate genome evolution. Nature. 2007;447(7145):714–19. doi: 10.1038/nature05846. [DOI] [PubMed] [Google Scholar]
- 59.Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484(7392):55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496(7446):498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schartl M, Walter RB, Shen Y, Garcia T, Catchen J, et al. The genome of the platyfish, Xiphophorus maculatus, provides insights into evolutionary adaptation and several complex traits. Nat Genet. 2013;45(5):567–72. doi: 10.1038/ng.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Philip S, Machado JP, Maldonado E, Vasconcelos V, O’Brien SJ, et al. Fish lateral line innovation: insights into the evolutionary genomic dynamics of a unique mechanosensory organ. Mol Biol Evol. 2012;29(12):3887–98. doi: 10.1093/molbev/mss194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spaink HP, Jansen HJ, Dirks RP. Advances in genomics of bony fish. Brief Funct Genomics. 2014;13(2):144–56. doi: 10.1093/bfgp/elt046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Köhler J, Vieites DR, Bonett RM, García FH, Glaw F, et al. New amphibians and global conservation: a boost in species discoveries in a highly endangered vertebrate group. Bioscience. 2005;55(8):693–96. [Google Scholar]
- 65.Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, et al. The genome of the western clawed frog Xenopus tropicalis. Science. 2010;328(5978):633–36. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gregory TR. Variation across amphibian species in the size of the nuclear genome supports a pluralistic, hierarchical approach to the C-value enigma. Biol J Linn Soc Lond. 2003;79(2):329–39. [Google Scholar]
- 67.Dufresne F, Jeffery N. A guided tour of large genome size in animals: what we know and where we are heading. Chromosome Res. 2011;19(7):925–38. doi: 10.1007/s10577-011-9248-x. [DOI] [PubMed] [Google Scholar]
- 68.Gregory TR. The C-value enigma in plants and animals: a review of parallels and an appeal for partnership. Ann Bot. 2005;95(1):133–46. doi: 10.1093/aob/mci009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun C, Shepard DB, Chong RA, López Arriaza J, Hall K, et al. LTR retrotransposons contribute to genomic gigantism in plethodontid salamanders. Genome Biol Evol. 2012;4(2):168–83. doi: 10.1093/gbe/evr139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shine R. The ecological impact of invasive cane toads (Bufo marinus) in Australia. Q Rev Biol. 2010;85(3):253–91. doi: 10.1086/655116. [DOI] [PubMed] [Google Scholar]
- 71.Ryan MJ. The Túngara Frog: A Study in Sexual Selection and Communication. Chicago: Univ. Chicago Press; 1985. [Google Scholar]
- 72.Callery EM, Fang H, Elinson RP. Frogs without polliwogs: evolution of anuran direct development. BioEssays. 2001;23(3):233–41. doi: 10.1002/1521-1878(200103)23:3<233::AID-BIES1033>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 73.Pfennig KS. Facultative mate choice drives adaptive hybridization. Science. 2007;318(5852):965–67. doi: 10.1126/science.1146035. [DOI] [PubMed] [Google Scholar]
- 74.Richards-Zawacki CL, Wang IJ, Summers K. Mate choice and the genetic basis for colour variation in a polymorphic dart frog: inferences from a wild pedigree. Mol Ecol. 2012;21(15):3879–92. doi: 10.1111/j.1365-294X.2012.05644.x. [DOI] [PubMed] [Google Scholar]
- 75.Gagliardo R, Crump P, Griffith E, Mendelson J, Ross H, Zippel K. The principles of rapid response for amphibian conservation, using the programmes in Panama as an example. Int Zoo Yearb. 2008;42(1):125–35. [Google Scholar]
- 76.Vandebergh W, Bossuyt F. Radiation and functional diversification of alpha keratins during early vertebrate evolution. Mol Biol Evol. 2012;29(3):995–1004. doi: 10.1093/molbev/msr269. [DOI] [PubMed] [Google Scholar]
- 77.Clarke BT. The natural history of amphibian skin secretions, their normal functioning and potential medical applications. Biol Rev Camb Philos Soc. 1997;72(3):365–79. doi: 10.1017/s0006323197005045. [DOI] [PubMed] [Google Scholar]
- 78.La Marca E, Lips KR, Lötters S, Puschendorf R, Ibáñez R, et al. Catastrophic population declines and extinctions in neotropical harlequin frogs (Bufonidae: Atelopus) Biotropica. 2005;37(2):190–201. [Google Scholar]
- 79.Savage AE, Zamudio KR. MHC genotypes associate with resistance to a frog-killing fungus. PNAS. 2011;108(40):16705–10. doi: 10.1073/pnas.1106893108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hedges SB, Vidal N. Lizards, snakes, and amphisbaenians (Squamata) In: Hedges SB, Kumar S, editors. The Timetree of Life. Oxford: Oxford Univ. Press; 2009. pp. 383–89. [Google Scholar]
- 81.Sarre SD, Ezaz T, Georges A. Transitions between sex-determining systems in reptiles and amphibians. Annu Rev Genomics Hum Genet. 2011;12:391–406. doi: 10.1146/annurev-genom-082410-101518. [DOI] [PubMed] [Google Scholar]
- 82.O’Meally D, Ezaz T, Georges A, Sarre SD, Graves JAM. Are some chromosomes particularly good at sex? Insights from amniotes. Chromosome Res. 2012;20(1):7–19. doi: 10.1007/s10577-011-9266-8. [DOI] [PubMed] [Google Scholar]
- 83.Vicoso B, Emerson JJ, Zektser Y, Mahajan S, Bachtrog D. Comparative sex chromosome genomics in snakes: differentiation, evolutionary strata, and lack of global dosage compensation. PLOS Biol. 2013;11(8):e1001643. doi: 10.1371/journal.pbio.1001643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kearney M, Fujita MK, Ridenour J. Lost sex in the reptiles: constraints and correlations. In: Schön I, Martens K, Dijk P van, editors. Lost Sex: The Evolutionary Biology of Parthogenesis. Dordrecht, Neth: Springer; Sci: 2009. pp. 447–74. [Google Scholar]
- 85.Fujita MK, Moritz C. Origin and evolution of parthenogenetic genomes in lizards: current state and future directions. Cytogenet Genome Res. 2009;127:2–4. 261–72. doi: 10.1159/000295177. [DOI] [PubMed] [Google Scholar]
- 86.Organ CL, Moreno RG, Edwards SV. Three tiers of genome evolution in reptiles. Integr Comp Biol. 2008;48(4):494–504. doi: 10.1093/icb/icn046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Alföldi J, Di Palma F, Grabherr M, Williams C, Kong L, et al. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature. 2011;477(7366):587–91. doi: 10.1038/nature10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fujita MK, Edwards SV, Ponting CP. The Anolis lizard genome: an amniote genome without isochores. Genome Biol Evol. 2011;3:974–84. doi: 10.1093/gbe/evr072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Eckalbar WL, Hutchins ED, Markov GJ, Allen AN, Corneveaux JJ, et al. Genome reannotation of the lizard Anolis carolinensis based on 14 adult and embryonic deep transcriptomes. BMC Genomics. 2013;14:49. doi: 10.1186/1471-2164-14-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Castoe TA, de Koning APJ, Hall KT, Yokoyama KD, Gu W, et al. Sequencing the genome of the Burmese python (Python molurus bivittatus) as a model for studying extreme adaptations in snakes. Genome Biol. 2011;12(7):406. doi: 10.1186/gb-2011-12-7-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Castoe TA, de Koning APJ, Hall KT, Card DC, Schield DR, et al. The Burmese python genome reveals the molecular basis for extreme adaptation in snakes. PNAS. 2013;110(51):20645–50. doi: 10.1073/pnas.1314475110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vonk FJ, Casewell NR, Henkel CV, Heimberg AM, Jansen HJ, et al. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. PNAS. 2013;110(51):20651–56. doi: 10.1073/pnas.1314702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Green RE, Braun EL, Armstrong J, Earl D, Nguyen N, et al. Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science. 2014;346(6215):1254449. doi: 10.1126/science.1254449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wan Q-H, Pan S-K, Hu L, Zhu Y, Xu P-W, et al. Genome analysis and signature discovery for diving and sensory properties of the endangered Chinese alligator. Cell Res. 2013;23(9):1091–105. doi: 10.1038/cr.2013.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shaffer HB, Minx P, Warren DE, Shedlock AM, Thomson RC, et al. The western painted turtle genome, a model for the evolution of extreme physiological adaptations in a slowly evolving lineage. Genome Biol. 2013;14(3):R28. doi: 10.1186/gb-2013-14-3-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang Z, Pascual-Anaya J, Zadissa A, Li W, Niimura Y, et al. The draft genomes of soft-shell turtle and green sea turtle yield insights into the development and evolution of the turtle-specific body plan. Nat Genet. 2013;45(6):701–6. doi: 10.1038/ng.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fry BG, Roelants K, Champagne DE, Scheib H, Tyndall JDA, et al. The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annu Rev Genomics Hum Genet. 2009;10:483–511. doi: 10.1146/annurev.genom.9.081307.164356. [DOI] [PubMed] [Google Scholar]
- 98.Ezaz T, Quinn AE, Miura I, Sarre SD, Georges A, Marshall Graves JA. The dragon lizard Pogona vitticeps has ZZ/ZW micro-sex chromosomes. Chromosome Res. 2005;13(8):763–76. doi: 10.1007/s10577-005-1010-9. [DOI] [PubMed] [Google Scholar]
- 99.Quinn AE, Georges A, Sarre SD, Guarino F, Ezaz T, Marshall Graves JA. Temperature sex reversal implies sex gene dosage in a reptile. Science. 2007;316(5823):411. doi: 10.1126/science.1135925. [DOI] [PubMed] [Google Scholar]
- 100.Feduccia A. The Origin and Evolution of Birds. New Haven, CT: Yale Univ. Press; 1999. [Google Scholar]
- 101.Jetz W, Thomas GH, Joy JB, Hartmann K, Mooers AO. The global diversity of birds in space and time. Nature. 2012;491:444–48. doi: 10.1038/nature11631. [DOI] [PubMed] [Google Scholar]
- 102.Lee MSY, Cau A, Naish D, Dyke GJ. Morphological clocks in paleontology, and a mid-Cretaceous origin of crown. Aves Syst Biol. 2014;63:442–49. doi: 10.1093/sysbio/syt110. [DOI] [PubMed] [Google Scholar]
- 103.Int. Chicken Genome Seq. Consort. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432(7018):695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 104.Dalloul RA, Long JA, Zimin AV, Aslam L, Beal K, et al. Multi-platform next-generation sequencing of the domestic turkey (Meleagris gallopavo): genome assembly and analysis. PLOS Biol. 2010;8(9):e1000475. doi: 10.1371/journal.pbio.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Warren WC, Clayton DF, Ellegren H, Arnold AP, Hillier LW, et al. The genome of a songbird. Nature. 2010;464(7289):757–62. doi: 10.1038/nature08819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hackett SJ, Kimball RT, Reddy S, Bowie RCK, Braun EL, et al. A phylogenomic study of birds reveals their evolutionary history. Science. 2008;320(5884):1763–68. doi: 10.1126/science.1157704. [DOI] [PubMed] [Google Scholar]
- 107.Pacheco MA, Battistuzzi FU, Lentino M, Aguilar RF, Kumar S, Escalante AA. Evolution of modern birds revealed by mitogenomics: timing the radiation and origin of major orders. Mol Biol Evol. 2011;28(6):1927–42. doi: 10.1093/molbev/msr014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.McCormack JE, Harvey MG, Faircloth BC, Crawford NG, Glenn TC, Brumfield RT. A phylogeny of birds based on over 1,500 loci collected by target enrichment and high-throughput sequencing. PLOS ONE. 2013;8(1):e54848. doi: 10.1371/journal.pone.0054848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bonneaud C, Burnside J, Edwards SV. High-speed developments in avian genomics. Bioscience. 2008;58(7):587. [Google Scholar]
- 110.Zhang G, Li B, Li C, Gilbert MTP, Jarvis E, et al. The avian phylogenomics project data. GigaScience Database. 2014 doi: 10.1186/2047-217X-3-26. http://dx.doi.org/10.5524/101000. [DOI] [PMC free article] [PubMed]
- 111.O’Brien SJ, Haussler D, Ryder O. The birds of Genome10K. GigaScience. 2014;3(1):32. doi: 10.1186/2047-217X-3-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jarvis ED, Mirarab S, Aberer AJ, Li B, Houde P, et al. Whole genome analyses resolve early branches in the tree of life of modern birds. Science. 2014;346(6215):1320–31. doi: 10.1126/science.1253451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang G, Li C, Li Q, Li B, Larkin DM, et al. Comparative genomics reveals insights into avian genome evolution and adaptation. Science. 2014;346(6215):1311–20. doi: 10.1126/science.1251385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478(7370):476–82. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Johnson WE, Eizirik E, Pecon-Slattery J, Murphy WJ, Antunes A, et al. The late miocene radiation of modern Felidae: a genetic assessment. Science. 2006;311(5757):73–77. doi: 10.1126/science.1122277. [DOI] [PubMed] [Google Scholar]
- 116.Qiu Q, Zhang G, Ma T, Qian W, Wang J, et al. The yak genome and adaptation to life at high altitude. Nat Genet. 2012;44(8):946–49. doi: 10.1038/ng.2343. [DOI] [PubMed] [Google Scholar]
- 117.Rogers J, Gibbs RA. Comparative primate genomics: emerging patterns of genome content and dynamics. Nat Rev Genet. 2014;15(5):347–59. doi: 10.1038/nrg3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Prado-Martinez J, Sudmant PH, Kidd JM, Li H, Kelley JL, et al. Great ape genetic diversity and population history. Nature. 2013;499(7459):471–75. doi: 10.1038/nature12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ellegren H, Smeds L, Burri R, Olason PI, Backström N, et al. The genomic landscape of species divergence in Ficedula flycatchers. Nature. 2012;491(7426):756–60. doi: 10.1038/nature11584. [DOI] [PubMed] [Google Scholar]
- 120.Seim I, Fang X, Xiong Z, Lobanov AV, Huang Z, et al. Genome analysis reveals insights into physiology and longevity of the Brandt’s bat. Myotis brandtii Nat Commun. 2013;4:2212. doi: 10.1038/ncomms3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xu Y, Shao C, Fedorov VB, Goropashnaya AV, Barnes BM, Yan J. Molecular signatures of mammalian hibernation: comparisons with alternative phenotypes. BMC Genomics. 2013;14(1):567. doi: 10.1186/1471-2164-14-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Steiner CC, Putnam AS, Hoeck PEA, Ryder OA. Conservation genomics of threatened animal species. Annu Rev Anim Biosci. 2013;1(1):261–81. doi: 10.1146/annurev-animal-031412-103636. [DOI] [PubMed] [Google Scholar]
- 123.Miller W, Hayes VM, Ratan A, Petersen C, Wittekindt NE, et al. Genetic diversity and population structure of the endangered marsupial Sarcophilus harrisii (Tasmanian devil) PNAS. 2012;108(30):12348–53. doi: 10.1073/pnas.1102838108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, et al. A draft sequence of the Neandertal genome. Science. 2010;328(5979):710–22. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rasmussen M, Li Y, Lindgreen S, Pedersen JS, Albrechtsen A, et al. Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature. 2010;463(7282):757–62. doi: 10.1038/nature08835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Meyer M, Kircher M, Gansauge M-T, Li H, Racimo F, et al. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338(6104):222–26. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Orlando L, Ginolhac A, Zhang G, Froese D, Albrechtsen A, et al. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature. 2013;499(7456):74–78. doi: 10.1038/nature12323. [DOI] [PubMed] [Google Scholar]
- 128.Hung C-M, Shaner P-JL, Zink RM, Liu W-C, Chu T-C, et al. Drastic population fluctuations explain the rapid extinction of the passenger pigeon. PNAS. 2014;111:10636–41. doi: 10.1073/pnas.1401526111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Miller W, Drautz DI, Ratan A, Pusey B, Qi J, et al. Sequencing the nuclear genome of the extinct woolly mammoth. Nature. 2008;456(7220):387–90. doi: 10.1038/nature07446. [DOI] [PubMed] [Google Scholar]
- 130.Shapiro B, Hofreiter M. A paleogenomic perspective on evolution and gene function: new insights from ancient DNA. Science. 2014;343(6169):1236573. doi: 10.1126/science.1236573. [DOI] [PubMed] [Google Scholar]
- 131.Chen S, Zhang G, Shao C, Huang Q, Liu G, et al. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat Genet. 2014;46(3):253–60. doi: 10.1038/ng.2890. [DOI] [PubMed] [Google Scholar]
- 132.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, et al. The Generic Genome Browser: a building block for a model organism system database. Genome Res. 2002;12(10):1599–610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Skinner ME, Uzilov AV, Stein LD, Mungall CJ, Holmes IH. JBrowse: a next-generation genome browser. Genome Res. 2009;19(9):1630–38. doi: 10.1101/gr.094607.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Raney BJ, Dreszer TR, Barber GP, Clawson H, Fujita PA, et al. Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC genome browser. Bioinformatics. 2014;30(7):1003–5. doi: 10.1093/bioinformatics/btt637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mehta TK, Ravi V, Yamasaki S, Lee AP, Lian MM, et al. Evidence for at least six hox clusters in the Japanese lamprey (Lethenteron japonicum) PNAS. 2013;110(40):16044–49. doi: 10.1073/pnas.1315760110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Smith JJ, Kuraku S, Holt C, Sauka-Spengler T, Jiang N, et al. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet. 2013;45(4):415–21. doi: 10.1038/ng.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kai W, Kikuchi K, Tohari S, Chew AK, Tay A, et al. Integration of the genetic map and genome assembly of fugu facilitates insights into distinct features of genome evolution in teleosts and mammals. Genome Biol Evol. 2011;3:424–42. doi: 10.1093/gbe/evr041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Star B, Nederbragt AJ, Jentoft S, Grimholt U, Malmstrøm M, et al. The genome sequence of Atlantic cod reveals a unique immune system. Nature. 2011;477(7363):207–10. doi: 10.1038/nature10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Henkel CV, Dirks RP, de Wijze DL, Minegishi Y, Aoyama J, et al. First draft genome sequence of the Japanese eel, Anguilla japonica. Gene. 2012;511(2):195–201. doi: 10.1016/j.gene.2012.09.064. [DOI] [PubMed] [Google Scholar]
- 140.Nakamura Y, Mori K, Saitoh K, Oshima K, Mekuchi M, et al. Evolutionary changes of multiple visual pigment genes in the complete genome of Pacific bluefin tuna. PNAS. 2013;110(27):11061–66. doi: 10.1073/pnas.1302051110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Berthelot C, Brunet F, Chalopin D, Juanchich A, Bernard M, et al. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat Commun. 2014;5:3657. doi: 10.1038/ncomms4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gallant JR, Traeger LL, Volkening JD, Moffett H, Chen P-H, et al. Genomic basis for the convergent evolution of electric organs. Science. 2014;344(6191):1522–25. doi: 10.1126/science.1254432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Xu P, Zhang X, Wang X, Li J, Liu G, et al. Genome sequence and genetic diversity of the common carp. Cyprinus carpio Nat Genet. 2014;46(11):1212–19. doi: 10.1038/ng.3098. [DOI] [PubMed] [Google Scholar]
- 144.McGaugh SE, Gross JB, Aken B, Blin M, Borowsky R, et al. The cavefish genome reveals candidate genes for eye loss. Nat Commun. 2014;5:5307. doi: 10.1038/ncomms6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wu C, Zhang D, Kan M, Lv Z, Zhu A, et al. The draft genome of the large yellow croaker reveals well-developed innate immunity. Nat Commun. 2014;5:5227. doi: 10.1038/ncomms6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.You X, Bian C, Zan Q, Xu X, Liu X, et al. Mudskipper genomes provide insights into the terrestrial adaptation of amphibious fishes. Nat Commun. 2014;5:5594. doi: 10.1038/ncomms6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Amemiya CT, Alföldi J, Lee AP, Fan S, Philippe H, et al. The African coelacanth genome provides insights into tetrapod evolution. Nature. 2013;496(7445):311–16. doi: 10.1038/nature12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nikaido M, Noguchi H, Nishihara H, Toyoda A, Suzuki Y, et al. Coelacanth genomes reveal signatures for evolutionary transition from water to land. Genome Res. 2013;23(10):1740–48. doi: 10.1101/gr.158105.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gilbert C, Meik JM, Dashevsky D, Card DC, Castoe TA, Schaack S. Endogenous hepadna viruses, borna viruses and circoviruses in snakes. Proc Biol Sci. 2014;281(1791):20141122. doi: 10.1098/rspb.2014.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Oleksyk TK, Pombert J-F, Siu D, Mazo-Vargas A, Ramos B, et al. A locally funded Puerto Rican parrot (Amazona vittata) genome sequencing project increases avian data and advances young researcher education. GigaScience. 2012;1(1):14. doi: 10.1186/2047-217X-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Huang Y, Li Y, Burt DW, Chen H, Zhang Y, et al. The duck genome and transcriptome provide insight into an avian influenza virus reservoir species. Nat Genet. 2013;45(7):776–83. doi: 10.1038/ng.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Seabury CM, Dowd SE, Seabury PM, Raudsepp T, Brightsmith DJ, et al. A multi-platform draft de novo genome assembly and comparative analysis for the scarlet macaw (Ara macao) PLOS ONE. 2013;8(5):e62415. doi: 10.1371/journal.pone.0062415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Shapiro MD, Kronenberg Z, Li C, Domyan ET, Pan H, et al. Genomic diversity and evolution of the head crest in the rock pigeon. Science. 2013;339(6123):1063–67. doi: 10.1126/science.1230422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kawahara-Miki R, Sano S, Nunome M, Shimmura T, Kuwayama T, et al. Next-generation sequencing reveals genomic features in the Japanese quail. Genomics. 2013;101(6):345–53. doi: 10.1016/j.ygeno.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 155.Zhan X, Pan S, Wang J, Dixon A, He J, et al. Peregrine and saker falcon genome sequences provide insights into evolution of a predatory lifestyle. Nat Genet. 2013;45(5):563–66. doi: 10.1038/ng.2588. [DOI] [PubMed] [Google Scholar]
- 156.Rands CM, Darling A, Fujita M, Kong L, Webster MT, et al. Insights into the evolution of Darwin’s finches from comparative analysis of the Geospiza magnirostris genome sequence. BMC Genomics. 2013;14:95. doi: 10.1186/1471-2164-14-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ganapathy G, Howard JT, Ward JM, Li J, Li B, et al. High-coverage sequencing and annotated assemblies of the budgerigar genome. GigaScience. 2014;3:11. doi: 10.1186/2047-217X-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Cai Q, Qian X, Lang Y, Luo Y, Xu J, et al. Genome sequence of ground tit Pseudopodoces humilis and its adaptation to high altitude. Genome Biol. 2013;14(3):R29. doi: 10.1186/gb-2013-14-3-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Qu Y, Zhao H, Han N, Zhou G, Song G, et al. Ground tit genome reveals avian adaptation to living at high altitudes in the Tibetan plateau. Nat Commun. 2013 May;4:2071. doi: 10.1038/ncomms3071. [DOI] [PubMed] [Google Scholar]
- 160.Doyle JM, Katzner TE, Bloom PH, Ji Y, Wijayawardena BK, Dewoody JA. The genome sequence of a widespread apex predator, the golden eagle (Aquila chrysaetos) PLOS ONE. 2014;9(4):e95599. doi: 10.1371/journal.pone.0095599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Halley YA, Dowd SE, Decker JE, Seabury PM, Bhattarai E, et al. A draft de novo genome assembly for the northern bobwhite (Colinus virginianus) reveals evidence for a rapid decline in effective population size beginning in the late Pleistocene. PLOS ONE. 2014;9(3):e90240. doi: 10.1371/journal.pone.0090240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Poelstra JW, Vijay N, Bossu CM, Lantz H, Ryll B, et al. The genomic landscape underlying phenotypic integrity in the face of gene flow in crows. Science. 2014;344(6190):1410–14. doi: 10.1126/science.1253226. [DOI] [PubMed] [Google Scholar]
- 163.Wang B, Ekblom R, Bunikis I, Siitari H, Höglund J. Whole genome sequencing of the black grouse (Tetrao tetrix): Reference guided assembly suggests faster-Z and MHC evolution. BMC Genomics. 2014;15:180. doi: 10.1186/1471-2164-15-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Callicrate T, Dikow R, Thomas JW, Mullikin JC, NISC Comp. Seq. Progr et al. Genomic resources for the endangered Hawaiian honeycreepers. BMC Genomics. 2014;15:1098. doi: 10.1186/1471-2164-15-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, et al. The sequence of the human genome. Science. 2001;291(5507):1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 166.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 167.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420(6915):520–62. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 168.Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, et al. Genome sequence of the brown Norway rat yields insights into mammalian evolution. Nature. 2004;428(6982):493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 169.Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438(7069):803–19. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 170.Chimpanzee Seq. Anal. Consort. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437(7055):69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 171.Pontius JU, Mullikin JC, Smith DR, Lindblad-Toh K, Gnerre S, et al. Initial sequence and comparative analysis of the cat genome. Genome Res. 2007;17(11):1675–89. doi: 10.1101/gr.6380007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316(5822):222–34. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 173.Yan G, Zhang G, Fang X, Zhang Y, Li C, et al. Genome sequencing and comparison of two nonhuman primate animal models, the cynomolgus and Chinese rhesus macaques. Nat Biotechnol. 2011;29(11):1019–23. doi: 10.1038/nbt.1992. [DOI] [PubMed] [Google Scholar]
- 174.Mikkelsen TS, Wakefield MJ, Aken B, Amemiya CT, Chang JL, et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447(7141):167–77. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- 175.Bovine Genome Seq. Anal. Consort. Elsik CG, Tellam RL, Worley KC. The genome sequence of Taurine cattle: a window to ruminant biology and evolution. Science. 2009;324(5926):522–28. doi: 10.1126/science.1169588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zimin AV, Delcher AL, Florea L, Kelley DR, Schatz MC, et al. A whole-genome assembly of the domestic cow. Bos taurus Genome Biol. 2009;10(4):R42. doi: 10.1186/gb-2009-10-4-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wade CM, Giulotto E, Sigurdsson S, Zoli M, Gnerre S, et al. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science. 2009;326(5954):865–67. doi: 10.1126/science.1178158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Huang J, Zhao Y, Shiraigol W, Li B, Bai D, et al. Analysis of horse genomes provides insight into the diversification and adaptive evolution of karyotype. Sci Rep. 2014;4:4958. doi: 10.1038/srep04958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Archibald AL, Cockett NE, Dalrymple BP, Faraut T, Kijas JW, et al. The sheep genome reference sequence: a work in progress. Anim Genet. 2010;41(5):449–53. doi: 10.1111/j.1365-2052.2010.02100.x. [DOI] [PubMed] [Google Scholar]
- 180.Jiang Y, Xie M, Chen W, Talbot R, Maddox JF, et al. The sheep genome illuminates biology of the rumen and lipid metabolism. Science. 2014;344(6188):1168–73. doi: 10.1126/science.1252806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Xu X, Nagarajan H, Lewis NE, Pan S, Cai Z, et al. The genomic sequence of the Chinese hamster ovary (CHO)-K1 cell line. Nat Biotechnol. 2011;29(8):735–41. doi: 10.1038/nbt.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Brinkrolf K, Rupp O, Laux H, Kollin F, Ernst W, et al. Chinese hamster genome sequenced from sorted chromosomes. Nat Biotechnol. 2013;31(8):694–95. doi: 10.1038/nbt.2645. [DOI] [PubMed] [Google Scholar]
- 183.Lewis NE, Liu X, Li Y, Nagarajan H, Yerganian G, et al. Genomic landscapes of Chinese hamster ovary cell lines as revealed by the Cricetulus griseus draft genome. Nat Biotechnol. 2013;31(8):759–65. doi: 10.1038/nbt.2624. [DOI] [PubMed] [Google Scholar]
- 184.Kim EB, Fang X, Fushan AA, Huang Z, Lobanov AV, et al. Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature. 2011;479(7372):223–27. doi: 10.1038/nature10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Higashino A, Sakate R, Kameoka Y, Takahashi I, Hirata M, et al. Whole-genome sequencing and analysis of the Malaysian cynomolgus macaque (Macaca fascicularis) genome. Genome Biol. 2012;13(7):R58. doi: 10.1186/gb-2012-13-7-r58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Renfree MB, Papenfuss AT, Deakin JE, Lindsay J, Heider T, et al. Genome sequence of an Australian kangaroo, Macropus eugenii, provides insight into the evolution of mammalian reproduction and development. Genome Biol. 2011;12(8):R81. doi: 10.1186/gb-2011-12-8-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Seabury CM, Bhattarai EK, Taylor JF, Viswanathan GG, Cooper SM, et al. Genome-wide polymorphism and comparative analyses in the white-tailed deer (Odocoileus virginianus): a model for conservation genomics. PLOS ONE. 2011;6(1):e15811. doi: 10.1371/journal.pone.0015811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Locke DP, Hillier LW, Warren WC, Worley KC, Nazareth LV, et al. Comparative and demographic analysis of orang-utan genomes. Nature. 2011;469(7331):529–33. doi: 10.1038/nature09687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Murchison EP, Schulz-Trieglaff OB, Ning Z, Alexandrov LB, Bauer MJ, et al. Genome sequencing and analysis of the Tasmanian devil and its transmissible cancer. Cell. 2012;148(4):780–91. doi: 10.1016/j.cell.2011.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wu H, Guang X, Al-Fageeh MB, Cao J, Pan S, et al. Camelid genomes reveal evolution and adaptation to desert environments. Nat Commun. 2014;5:5188. doi: 10.1038/ncomms6188. [DOI] [PubMed] [Google Scholar]
- 191.Canavez FC, Luche DD, Stothard P, Leite KRM, Sousa-Canavez JM, et al. Genome sequence and assembly of Bos indicus. J Hered. 2012;103(3):342–48. doi: 10.1093/jhered/esr153. [DOI] [PubMed] [Google Scholar]
- 192.Bactrian Camels Genome Seq. Anal. Consort. Genome sequences of wild and domestic Bactrian camels. Nat Commun. 2012;3:1202. doi: 10.1038/ncomms2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Perry GH, Reeves D, Melsted P, Ratan A, Miller W, et al. A genome sequence resource for the aye-aye (Daubentonia madagascariensis), a nocturnal lemur from Madagascar. Genome Biol Evol. 2012;4(2):126–35. doi: 10.1093/gbe/evr132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Perry GH, Louis EE, Ratan A, Bedoya-Reina OC, Burhans RC, et al. Aye-aye population genomic analyses highlight an important center of endemism in northern Madagascar. PNAS. 2013;110(15):5823–28. doi: 10.1073/pnas.1211990110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Scally A, Dutheil JY, Hillier LW, Jordan GE, Goodhead I, et al. Insights into hominid evolution from the gorilla genome sequence. Nature. 2012;483(7388):169–75. doi: 10.1038/nature10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zhang G, Cowled C, Shi Z, Huang Z, Bishop-Lilly KA, et al. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science. 2013;339(6118):456–60. doi: 10.1126/science.1230835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Prüfer K, Munch K, Hellmann I, Akagi K, Miller JR, et al. The bonobo genome compared with the chimpanzee and human genomes. Nature. 2012;486(7404):527–31. doi: 10.1038/nature11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Fang X, Mou Y, Huang Z, Li Y, Han L, et al. The sequence and analysis of a Chinese pig genome. GigaScience. 2012;1(1):16. doi: 10.1186/2047-217X-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Groenen MAM, Archibald AL, Uenishi H, Tuggle CK, Takeuchi Y, et al. Analyses of pig genomes provide insight into porcine demography and evolution. Nature. 2012;491(7424):393–98. doi: 10.1038/nature11622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Parker J, Tsagkogeorga G, Cotton JA, Liu Y, Provero P, et al. Genome-wide signatures of convergent evolution in echolocating mammals. Nature. 2013;502(7470):228–31. doi: 10.1038/nature12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Zhou X, Sun F, Xu S, Fan G, Zhu K, et al. Baiji genomes reveal low genetic variability and new insights into secondary aquatic adaptations. Nat Commun. 2013;4:2708. doi: 10.1038/ncomms3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Cho YS, Hu L, Hou H, Lee H, Xu J, et al. The tiger genome and comparative analysis with lion and snow leopard genomes. Nat Commun. 2013 May;4:2433. doi: 10.1038/ncomms3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Ge R-L, Cai Q, Shen Y-Y, San A, Ma L, et al. Draft genome sequence of the Tibetan antelope. Nat Commun. 2013 May;4:1858. doi: 10.1038/ncomms2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Fan Y, Huang Z-Y, Cao C-C, Chen C-S, Chen Y-X, et al. Genome of the Chinese tree shrew. Nat Commun. 2013;4:1426. doi: 10.1038/ncomms2416. [DOI] [PubMed] [Google Scholar]
- 205.Marmoset Genome Seq. Anal. Consort. The common marmoset genome provides insight into primate biology and evolution. Nat Genet. 2014;46:850–57. doi: 10.1038/ng.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Fan Z, Zhao G, Li P, Osada N, Xing J, et al. Whole-genome sequencing of Tibetan macaque (Macaca thibetana) provides new insight into the macaque evolutionary history. Mol Biol Evol. 2014;31(6):1475–89. doi: 10.1093/molbev/msu104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Fang X, Nevo E, Han L, Levanon EY, Zhao J, et al. Genome-wide adaptive complexes to underground stresses in blind mole rats. Spalax Nat Commun. 2014;5:3966. doi: 10.1038/ncomms4966. [DOI] [PubMed] [Google Scholar]
- 208.Liu S, Lorenzen ED, Fumagalli M, Li B, Harris K, et al. Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears. Cell. 2014;157(4):785–94. doi: 10.1016/j.cell.2014.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Carbone L, Harris RA, Gnerre S, Veeramah KR, Lorente-Galdos B, et al. Gibbon genome and the fast karyotype evolution of small apes. Nature. 2014;513(7517):195–201. doi: 10.1038/nature13679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Zhou X, Wang B, Pan Q, Zhang J, Kumar S, et al. Whole-genome sequencing of the snub-nosed monkey provides insights into folivory and evolutionary history. Nat Genet. 2014;46(12):1303–10. doi: 10.1038/ng.3137. [DOI] [PubMed] [Google Scholar]
- 211.Gnerre S, MacCallum I, Przybylski D, Ribeiro FJ, Burton JN, et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. PNAS. 2011;108(4):1513–18. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Luo R, Liu B, Xie Y, Li Z, Huang W, et al. Soapdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1(1):18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268(1):78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 214.Stanke M, Keller O, Gunduz I, Hayes A, Waack S, Morgenstern B. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006;34:W435–39. doi: 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Deleted in proof.
- 216.Birney E, Clamp M, Durbin R. Genewise and Genomewise. Genome Res. 2004;14(5):988–95. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Slater GSC, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinform. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Kapustin Y, Souvorov A, Tatusova T, Lipman D. Splign: algorithms for computing spliced alignments with identification of paralogs. Biol Direct. 2008;3:20. doi: 10.1186/1745-6150-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–79. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, et al. The variant call format and VCFtools. Bioinformatics. 2011;27(15):2156–58. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, et al. The genome analysis toolkit : a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2013;6(9):677–81. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Hormozdiari F, Hajirasouliha I, Dao P, Hach F, Yorukoglu D, et al. Next-generation VariationHunter: combinatorial algorithms for transposon insertion discovery. Bioinformatics. 2010;26(12):i350–57. doi: 10.1093/bioinformatics/btq216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Iqbal Z, Caccamo M, Turner I, Flicek P, McVean G. De novo assembly and genotyping of variants using colored de Bruijn graphs. Nat Genet. 2012;44(2):226–32. doi: 10.1038/ng.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Nijkamp JF, van den Broek MA, Geertman J-MA, Reinders MJT, Daran J-MG, de Ridder D. De novo detection of copy number variation by co-assembly. Bioinformatics. 2012;28(24):3195–202. doi: 10.1093/bioinformatics/bts601. [DOI] [PubMed] [Google Scholar]
- 226.Alkan C, Kidd JM, Marques-Bonet T, Aksay G, Antonacci F, et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet. 2009;41(10):1061–67. doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Klambauer G, Schwarzbauer K, Mayr A, Clevert DA, Mitterecker A, et al. Cn.MOPS: mixture of Poissons for discovering copy number variations in next-generation sequencing data with a low false discovery rate. Nucleic Acids Res. 2012;40(9):e69. doi: 10.1093/nar/gks003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Morgulis A, Gertz EM, Schäffer AA, Agarwala R. WindowMasker: window-based masker for sequenced genomes. Bioinformatics. 2006;22(2):134–41. doi: 10.1093/bioinformatics/bti774. [DOI] [PubMed] [Google Scholar]
- 229.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27(2):573–80. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Morgulis A, Gertz EM, Schäffer AA, Agarwala R. A fast and symmetric dust implementation to mask low-complexity DNA sequences. J Comput Biol. 2006;13(5):1028–40. doi: 10.1089/cmb.2006.13.1028. [DOI] [PubMed] [Google Scholar]
- 231.Deleted in proof.
- 232.Thiel T, Michalek W, Varshney RK, Graner A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare l.) Theor Appl Genet. 2003;106(3):411–22. doi: 10.1007/s00122-002-1031-0. [DOI] [PubMed] [Google Scholar]
- 233.Wang X, Lu P, Luo Z. GMATo: a novel tool for the identification and analysis of microsatellites in large genomes. Bioinformation. 2013;9(10):541–44. doi: 10.6026/97320630009541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Sperber GO, Airola T, Jern P, Blomberg J. Automated recognition of retroviral sequences in genomic data–RetroTector©. Nucleic Acids Res. 2007;35(15):4964–76. doi: 10.1093/nar/gkm515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.McCarthy EM, McDonald JF. LTR_STRUC: a novel search and identification program for LTR retrotransposons. Bioinformatics. 2003;19(3):362–67. doi: 10.1093/bioinformatics/btf878. [DOI] [PubMed] [Google Scholar]
- 236.Xu Z, Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35:W265–68. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Ellinghaus D, Kurtz S, Willhoeft U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinform. 2008;9(1):18. doi: 10.1186/1471-2105-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Jiang Z, Hubley R, Smit A, Eichler EE. DupMasker: a tool for annotating primate segmental duplications. Genome Res. 2008;18(8):1362–68. doi: 10.1101/gr.078477.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Deleted in proof.
- 240.Huang T-H, Fan B, Rothschild MF, Hu Z-L, Li K, Zhao SH. MiRFinder: an improved approach and software implementation for genome-wide fast microRNA precursor scans. BMC Bioinform. 2007;8:341. doi: 10.1186/1471-2105-8-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. MiRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–58. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Lorenz R, Bernhart SH, Höner zu Siederdissen C, Tafer H, Flamm C, et al. ViennaRNA package 2.0. Algorithms Mol Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Deleted in proof.
- 244.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27(11):1571–72. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Chen P, Cokus SJ, Pellegrini M. BS seeker: precise mapping for bisulfite sequencing software. BMC Bioinform. 2010;11:203. doi: 10.1186/1471-2105-11-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Guo W, Fiziev P, Yan W, Cokus S, Sun X, et al. BS-Seeker2: a versatile aligning pipeline for bisulfite sequencing data. BMC Genomics. 2013;14(1):774. doi: 10.1186/1471-2164-14-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Souaiaia T, Zhang Z, Chen T. FadE: whole genome methylation analysis for multiple sequencing platforms. Nucleic Acids Res. 2013;41(1):e14. doi: 10.1093/nar/gks830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Deleted in proof.
- 249.Deleted in proof.
- 250.De Bie T, Cristianini N, Demuth JP, Hahn MW. CAFÉ: a computational tool for the study of gene family evolution. Bioinformatics. 2006;22(10):1269–71. doi: 10.1093/bioinformatics/btl097. [DOI] [PubMed] [Google Scholar]
- 251.Han MV, Thomas GWC, Lugo-Martinez J, Hahn MW. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFÉ 3. Mol Biol Evol. 2013;30(8):1987–97. doi: 10.1093/molbev/mst100. [DOI] [PubMed] [Google Scholar]
- 252.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15(8):1034–50. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Deleted in proof.
- 254.Garber M, Guttman M, Clamp M, Zody MC, Friedman N, Xie X. Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics. 2009;25(12):i54–62. doi: 10.1093/bioinformatics/btp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–91. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 256.Antao T, Lopes A, Lopes RJ, Beja-Pereira A, Luikart G. LOSITAN: a workbench to detect molecular adaptation based on a Fst-outlier method. BMC Bioinform. 2008;9:323. doi: 10.1186/1471-2105-9-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Gautier M, Vitalis R. rehh: an R package to detect footprints of selection in genome-wide SNP data from haplotype structure. Bioinformatics. 2012;28(8):1176–77. doi: 10.1093/bioinformatics/bts115. [DOI] [PubMed] [Google Scholar]
- 259.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–52. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-seq. Bioinformatics. 2009;25(9):1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Murphy WJ, Larkin DM, Everts-van der Wind A, Bourque G, Tesler G, et al. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science. 2005;309(5734):613–17. doi: 10.1126/science.1111387. [DOI] [PubMed] [Google Scholar]
- 263.Larkin DM, Pape G, Donthu R, Auvil L, Welge M, Lewin HA. Breakpoint regions and homologous synteny blocks in chromosomes have different evolutionary histories. Genome Res. 2009;19(5):770–77. doi: 10.1101/gr.086546.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Grabherr MG, Russell P, Meyer M, Mauceli E, Alföldi J, et al. Genome-wide synteny through highly sensitive sequence alignment: Satsuma. Bioinformatics. 2010;26(9):1145–51. doi: 10.1093/bioinformatics/btq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Soderlund C, Nelson W, Shoemaker A, Paterson A. SyMAP: a system for discovering and viewing syntenic regions of FPC maps. Genome Res. 2006;16(9):1159–68. doi: 10.1101/gr.5396706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Harris RS. PhD Thesis. Pa. State Univ.; 2007. Improved pairwise alignment of genomic DNA. [Google Scholar]
- 267.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–92. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Zhao M, Wang Q, Wang Q, Jia P, Zhao Z. Computational tools for copy number variation (CNV) detection using next-generation sequencing data: features and perspectives. BMC Bioinform. 2013;14(Suppl 11):S1. doi: 10.1186/1471-2105-14-S11-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Merkel A, Gemmell N. Detecting short tandem repeats from genome data: opening the software black box. Brief Bioinform. 2008;9(5):355–66. doi: 10.1093/bib/bbn028. [DOI] [PubMed] [Google Scholar]
- 272.Lim KG, Kwoh CK, Hsu LY, Wirawan A. Review of tandem repeat search tools: a systematic approach to evaluating algorithmic performance. Brief Bioinform. 2013;14(1):67–81. doi: 10.1093/bib/bbs023. [DOI] [PubMed] [Google Scholar]
- 273.Oleksyk TK, Smith MW, O’Brien SJ. Genome-wide scans for footprints of natural selection. Philos Trans R Soc Lond B Biol Sci. 2010;365(1537):185–205. doi: 10.1098/rstb.2009.0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Scheinfeldt LB, Tishkoff SA. Recent human adaptation: genomic approaches, interpretation and insights. Nat Rev Genet. 2013;14(10):692–702. doi: 10.1038/nrg3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Davidson WS, Koop BF, Jones SJM, Iturra P, Vidal R, et al. Sequencing the genome of the Atlantic salmon (Salmo salar) Genome Biol. 2010;11(9):403. doi: 10.1186/gb-2010-11-9-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Hedges SB, Kumar S. The Timetree of Life. New York: Oxford Univ. Press; 2009. [Google Scholar]
- 277.Dickinson EC, Remsen JV Jr, editors. The Howard and Moore Complete Checklist of the Birds of the World. 4th Eastbourne, UK: Aves Press; 2013. [Google Scholar]
- 278.Mitchell KJ, Llamas B, Soubrier J, Rawlence NJ, Worthy TH, et al. Ancient DNA reveals elephant birds and kiwi are sister taxa and clarifies ratite bird evolution. Science. 2014;344:898–900. doi: 10.1126/science.1251981. [DOI] [PubMed] [Google Scholar]
- 279.Cox DR, Burmeister M, Price ER, Kim S, Myers RM. Radiation hybrid mapping: a somatic cell genetic method for constructing high-resolution maps of mammalian chromosomes. Science. 1990;250(4978):245–50. doi: 10.1126/science.2218528. [DOI] [PubMed] [Google Scholar]
- 280.Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]


