Abstract
The advent of widespread cancer genome sequencing has accelerated our understanding of the molecular aberrations underlying malignant disease at an unprecedented rate. Coupling the large number of bioinformatic methods developed to locate genomic breakpoints with increased sequence read length and a deeper understanding of coding region function has enabled rapid identification of novel actionable oncogenic fusion genes. Using examples of kinase fusions found in liquid and solid tumours, this review highlights major concepts that have arisen in our understanding of cancer pathogenesis through the study of fusion proteins. We provide an overview of recently developed methods to identify potential fusion proteins from next-generation sequencing data, describe the validation of their oncogenic potential and discuss the role of targetted therapies in treating cancers driven by fusion oncoproteins.
Keywords: Cancer, Oncology/cancer, Other receptors, Protein kinases/phosphatases, Targeted therapy
Introduction
Cancer cells exhibit increased genomic instability and deficient DNA repair mechanisms, resulting in an accumulation of genomic aberrations such as mutations and variations in chromosomal structure. Chromosomal rearrangements arise from genomic deletions, duplications, inversions and translocations that can occur with or without additional gain or loss of genetic material. Balanced chromosomal translocations give rise to chimeric fusion proteins by end-to-end joining of coding segments from two normal genes. Based on the functions of the gene-fusion partners, a given gene rearrangement can be classified as: (i) a transcriptional enhancer fused to a transcription factor; (ii) a frame-shifted gene fused to a tumour suppressor; (iii) a constitutive activator fused to a kinase or (iv) a kinase fused to a new localisation signal. Gene fusions promote tumorigenesis by either functioning as a hyper-activated oncogenic driver or by inactivating a tumour suppressor. Here, we will broadly categorise gene fusions into either kinase fusions or transcription-factor fusions, focusing further discussion primarily on kinase gene-fusion discovery, biology and pharmacology.
Oncogenic kinase fusion proteins in cancer
Kinase fusions have played an important role in our ability to understand how cancer develops, how to harness this understanding to inhibit progression or effectively ‘cure’ a cancer, and how cancers develop mechanisms to escape therapeutic target-ting of specific oncogenic lesions. A seminal example is the identification and subsequent characterisation of the BCR-ABL1 fusion protein resulting from a t(9;22)(q34;q11) translocation as an oncogenic driver in chronic myeloid leukaemia. This provided a momentous paradigm shift in our understanding of how cancer can be treated and ushered in the era of precision medicine (Nowell and Hungerford, 1960). The specificity and potency of the ABL1 tyrosine kinase inhibitor imatinib in BCR-ABL1-positive CML resulted in unprecedented anti-leukaemic effects with minimal side effects for patients (Druker et al., 2006; Druker, 2009). The study of BCR-ABL1 also contributed to the field's understanding of how drug-resistant mutations develop upon targetted therapy with kinase inhibitors. For example, the T315I gatekeeper mutation in the BCR-ABL1 kinase domain, which compromises imatinib binding, has been described as a frequent cause of therapeutic resistance (O'Hare et al., 2007a,b; O'Hare et al., 2009; Lu et al., 2011; Gibbons et al., 2012; Mian et al., 2012; Zabriskie et al., 2014). Studies of acquired resistance mutations have led to a fuller understanding of the structure–function relationship of the inhibitor-bound kinase, including binding mode and structural moieties required for inhibitory selectivity and efficacy (Hantschel et al., 2012). These studies have facilitated the rational development of alternative drugs with increased potency and efficacy against such resistance mutations.
A second example is the t(15;17)(q24;q21) translocation that results in the PML-RARα transcription-factor fusion protein, a hallmark of acute promye-locytic leukaemia (Takashima and Misawa, 1992). Fusion proteins of RARα act as potent transcriptional repressors of retinoic acid signalling and contribute to leukaemogenesis by blocking myeloid differentiation at the promyelocyte stage. Elucidation of the resultant biology of this fusion protein informed the application of all-trans retinoid acid and arsenic trioxide, two agents capable of inducing differentiation of promyelocytes, for the treatment of patients with acute promyelocytic leukaemia. The results have been remarkable and represent an extremely effective strategy for managing these patients (Huang et al., 1988; reviewed in De Braekeleer et al., 2014).
In contrast tothe relative exclusivity of BCR-ABL1 and PML-RARα in driving malignant transformation of haematopoietic lineage cells, activating gene fusions of ALK, NTRK and ROS1 are present in diverse subsets of adult and pediatric cancer patients, demonstrating the capability to transform cells from multiple lineages. Similar transformation capacity for multiple cell lineages is observed for oncogenic kinase fusions resulting from chromosomal rearrangement of FGFR1, FGFR2, FGFR3, RET, MET, EGFR, BRAF and RAF1 (Suehara et al., 2012; Drilon et al., 2013; Gainor and Shaw, 2013; Kohno et al., 2013; Parker and Zhang, 2013; Shaw et al., 2013a; Stransky et al., 2014), but will not be discussed here.
The NTRK family comprises three members (NTRK1, NTRK2 and NTRK3) that are primarily expressed in, but not limited to, the central and peripheral nervous system (Huang and Reichardt, 2001). Recent studies have helped to identify multiple NTRK fusions in a broad range of tumour types. NTRK1 fusions include NFASC-NTRK1, BCAN-NTRK1 and TPM3-NTRK1 in glioblastoma multiforme (GBM); MPRIP-NTRK1 and CD74-NTRK1 in non-small cell lung cancer (NSCLC); and TPM3-NTRK1 in intrahepatic cholangiocarcinoma (ICC) (Shah et al., 2013; Vaishnavi et al., 2013; Ardini et al., 2014; Wu et al., 2014). NTRK2 fusions have been identified primarily in pediatric brain tumours, including VCL-NTRK2 and AGBL4-NTRK2 fusions in diffuse intrinsic pontine glioma (DIPG); QKI-NTRK2 and NACC2-NTRK2 in midline pilocytic astrocytoma; and NAV-NTRK2 in low-grade glioma (Jones et al., 2013; Zhang et al., 2013; Wu et al., 2014). To date, the diversity of NTRK3 fusion partners has been limited to BTBD1-NTRK3 in DIPG and ETV6-NTRK3 in DIPG, low-grade glioma, acute myeloid leukaemia, congenital fibrosarcoma (CFS), congenital mesoblastic nephroma (CMN), secretory breast carcinoma (SBC), thyroid cancer and mammary-analogue secretory carcinoma of salivary glands (Knezevich et al., 1998a,b; Tognon et al., 2002; Skalova et al., 2010; Kralik et al., 2011; Leeman-Neill et al., 2014; Wu et al., 2014). Paediatric brain tumours frequently occur during a highly active neural development period that includes processes of neuronal differentiation, maturation, apoptosis and migration. The recurrent expression of NTRK fusion proteins in pediatric neuroglial tumours suggests that aberrant constitutive activation of NTRK signalling in neuroglial cells or progenitors drives unchecked proliferation and survival, leading to cancerous phenotypes (Nakagawara, 2001). NTRK fusion kinases are actionable oncogenes, and their discovery in treatment-refractory cancers like DIPG, GBM and ICC, which pose a formidable clinical challenge, unveils a set of potentially more effective therapeutic targets (Thiele et al., 2009).
ALK is a receptor tyrosine kinase that is mostly undetectable in adult human tissue, with low levels observed in the developing central nervous system, suggesting a role in nervous system maturation (Pulford et al., 1997). Recent studies also implicate ALK signalling in the regulation of vascular morphogenesis (Cunha and Pietras, 2011; McDonald et al., 2011; Larrivee et al., 2012). Chromosomal rearrangement of ALK resulting in the NPM-ALK fusion protein was first identified in anaplastic large-cell lymphoma (ALCL) in 1994 (Morris et al., 1994; Shiota et al., 1994). Since this initial discovery, other ALK fusions that have been discovered in ALCL patients include TFG-ALK, ATIC-ALK, CLTC-ALK, TPM4-ALK, MYH9-ALK and MSN-ALK (Hernandez et al., 1999; Colleoni et al., 2000; Meech et al., 2001; Tort et al., 2001; Cools et al., 2002; Lamant et al., 2003). Despite this presentation in ALCL, it was the discovery of EML4-ALK in a subset of NSCLC patients that propelled aggressive preclinical drug discovery efforts and eventual implementation of crizotinib as the first ALK inhibitor with demonstrated efficacy in ALK fusion-positive patients (Soda et al., 2007; Kwak et al., 2010; Rodig and Shapiro, 2010; Casaluce et al., 2013; Shaw et al., 2013b). Analogous to the clinical experience with ABL1 inhibitors in BCR-ABL1-positive CML, resistance due to acquisition of kinase domain mutations that disrupt drug binding has also emerged in ALK fusion-positive NSCLC patients (Doebele et al., 2012; Awad and Shaw, 2014). Several other ALK fusion proteins have been identified in other cancers, including CLTC-ALK and SQSTM1-ALK in diffuse large B-cell lymphoma and inflammatory myofibroblastic tumour (IMT); TPM3/4-ALK, CARS-ALK, RANBP2-ALK and SEC31L1 ALK in IMT; VCL-ALK in renal-cell carcinoma; C2orf44-ALK in colorectal cancer; FN1-ALK in ovarian cancer; and KIF5B-ALK, KLC1-ALK and ALK-PTPN3 in NSCLC (Lawrence et al., 2000; Bridge et al., 2001; De Paepe et al., 2003; Debelenko et al., 2003; Panagopoulos et al., 2006; Takeuchi et al., 2011; Jung et al., 2012; Lipson et al., 2012; Ren et al., 2012). It is likely that clinical ALK inhibitors will have therapeutic value in patients harbouring these other ALK fusions as well.
The ROS1 fusion protein FIG-ROS1, resulting from an intrachromosomal deletion, was identified initially in the U118MG glioblastoma cell line (Birchmeier et al., 1987; Sharma et al., 1989; Charest et al., 2003a), and subsequently in ICC, ovarian cancer and NSCLC (Birch et al., 2011; Gu et al., 2011; Rimkunas et al., 2012). Similar to the trajectory of ALK fusions in NSCLC, the discovery of SLC34A2-ROS1 and CD74-ROS1 oncoproteins in NSCLC fueled enthusiasm to investigate the occurrence of ROS1 fusions in other cancers and to determine the potential of ROS1-targetted inhibitors to treat patients harbouring these fusions (Rikova et al., 2007). ROS1 and ALK fusion proteins exhibit ∼40% homology in their kinase domains, implying that ALK inhibitors will be functionally effective inhibitors for ROS1. Accordingly, crizotinib efficacy in ROS1 fusion-positive NSCLC has been recently demonstrated (Awad et al., 2013; Davies et al., 2013; Chiari et al., 2014; Shaw et al., 2014). ROS1 fusions have since been identified in several other cancers, including CEP85L-ROS1 in GBM and angiosarcoma; SLC34A2-ROS1 in gastric adenocarcinoma and colorectal cancer; YWHAE-ROS1 and TFG-ROS1 in IMT; and TPM3-ROS1, SDC4-ROS1, EZR-ROS1, LRIG3-ROS1, KDELR2-ROS1, CCDC6-ROS1, LIMA1-ROS1 and MSN-ROS1 in NSCLC (Davies and Doebele, 2013; Gainor and Shaw, 2013; Giacomini et al., 2013; Shah et al., 2013; Shaw et al., 2014).
The identification of multiple ALK, ROS1 and NTRK translocations across many different tumour types helps to support the idea that aberrant tyrosine kinase function contributes to transformative phenotypes. Table 1 summarises the most recent information on NTRK, ALK and ROS1 fusions, identifies their fusion partners, annotates their associated cancers and provides supporting references. To date, all oncogenic tyrosine kinase fusions retain the exon(s) encoding the kinase domain that fuses in frame with an upstream partner. Typically, the fusion partner contains a dimerisation domain resulting in constitutive kinase activity and chronic upregulation of canonical downstream signalling pathways. Notably, kinase fusions are generally observed to be mutually exclusive of one another, consistent with their individual driving roles in oncogenesis. This is illustrated in thyroid cancer, which has one of the highest frequencies of recurrent tyrosine kinase fusions. Approximately 13% of thyroid cancer patients harbour a fusion involving ALK, BRAF, MET, NTRK1, NTRK2, RAF1 or RET, but these never co-occur in the same patient (Stransky et al., 2014). Despite the potential differences in functional properties of this diverse group of somatic kinase fusions, including enzyme kinetics, variable affinity for adaptor proteins to regulate downstream effectors and subcellular localisation, the common denominator is their ability to create a state of ‘oncogenic addiction’, thus making them highly suitable candidates for rational targetted therapy. Furthermore, as patients harbouring a particular oncogenic fusion generally represent a small subset of all patients diagnosed with that malignancy, such targets may appear less attractive when considering the investment necessary to develop robust diagnostic testing and therapeutic compounds. However, it is necessary to consider that a family of fusion proteins harbouring the kinase domain of a specific gene (e.g., NTRK, ALK, ROS1, FGFR, RET) may be present both with multiple different binding partners and across multiple malignancies, elevating the total number of patients eligible for such therapies to a sufficiently compelling level to warrant further development.
Table 1. Summary of known NTRK, ALK and ROS1 fusion proteins.
| Gene | Fusion partner | Malignancy | PMID | Reference |
|---|---|---|---|---|
| NTRK1 | NFASC | Glioblastoma Multiforme | 24261984 | Shah et al. (2013) |
| BCAN | Glioblastoma Multiforme | 24261984 | Shah et al. (2013) | |
| TPM3 | Diffuse Intrinsic Pontine Glioma | 24705251 | Wu et al. (2014) | |
| MPRIP | Lung Adenocarcinoma | 24162815 | Vaishnavi et al. (2013) | |
| CD74 | Lung Adenocarcinoma | 24162815 | Vaishnavi et al. (2013) | |
| TPM3 | Colorectal Cancer | 24962792 | Ardini et al. (2014) | |
| RABGAP1L | Cholangiocarcinoma | 24563076 | Ross et al. (2014) | |
| TAF | Papillary Thyroid Carcinoma | 8288244 | Greco et al. (1993) | |
| TFG | Thyroid Carcinoma | 7565764 | Ross et al. (2014) | |
| LMNA | Spitzoid Melanoma | 24445538 | Wiesner et al. (2014) | |
| NTRK2 | VCL | Diffuse Intrinsic Pontine Glioma | 24705251 | Wu et al. (2014) |
| AGBL4 | Diffuse Intrinsic Pontine Glioma | 24705251 | Wu et al. (2014) | |
| QKI | Pilocytic Astrocytoma | 23817572 | Jones et al. (2013) | |
| NACC2 | Pilocytic Astrocytoma | 23817572 | Jones et al. (2013) | |
| PAN3 | Head and Neck Squamous-Cell Carcinoma | 25204415 | Stransky et al. (2014) | |
| TRIM24 | Lung Adenocarcinoma | 25204415 | Stransky et al. (2014) | |
| AFAP1 | Low-Grade Glioma | 25204415 | Stransky et al. (2014) | |
| NAV | Low-Grade Glioma | 23583981 | Zhang et al. (2013) | |
| NTRK3 | BTBD1 | Diffuse Intrinsic Pontine Glioma | 24705251 | Wu et al. (2014) |
| ETV6 | Diffuse Intrinsic Pontine Glioma | 24705251 | Wu et al. (2014) | |
| ETV6 | Low-Grade Glioma | 23583981 | Zhang et al. (2013) | |
| ETV6 | Acute Myeloid Leukaemia | 21401966 | Kralik et al. (2011) | |
| ETV6 | Congenital Fibrosarcoma | 9462753 | Knezevich et al. (1998b) | |
| ETV6 | Congenital Mesoblastic Nephroma | 9823307 | Knezevich et al. (1998a) | |
| ETV6 | Secretory Breast Carcinoma | 12450792 | Tognon et al. (2002) | |
| ETV6 | Thyroid Carcinoma | 24327398 | Leeman-Neill et al. (2014) | |
| ETV6 | Mammary-Analog Secretory Carcinoma of Salivary Gland Origin | 20410810 | Skalova et al. (2010) | |
| ALK | ATIC | Anaplastic Large-Cell Lymphoma | 10702393 | Colleoni et al. (2000) |
| CLTC | Anaplastic Large-Cell Lymphoma | 12112524 | Cools et al. (2002) | |
| MSN | Anaplastic Large-Cell Lymphoma | 11310834 | Tort et al. (2001) | |
| MYH9 | Anaplastic Large-Cell Lymphoma | 12800156 | Lamant et al. (2003) | |
| NPM | Anaplastic Large-Cell Lymphoma | 8122112 | Morris et al. (1994) | |
| RNF213 | Anaplastic Large-Cell Lymphoma | 22570254 | Zhang et al. (2013) | |
| TFG | Anaplastic Large-Cell Lymphoma | 10556217 | Hernandez et al. (1999) | |
| TPM4 | Anaplastic Large-Cell Lymphoma | 11493472 | Meech et al. (2001) | |
| NPM | Anaplastic Large-Cell Lymphoma | 10994999 | Drexler et al. (2000) | |
| NPM | Diffuse Large B-Cell Lymphoma | 21474455 | Grande et al. (2011) | |
| CLTC | Diffuse Large B-Cell Lymphoma | 12750159 | De Paepe et al. (2003) | |
| SQSTM1 | Diffuse Large B-Cell Lymphoma | 21134980 | Takeuchi et al. (2011) | |
| NPM | Follicular Lymphoma | 21474455 | Grande et al. (2011) | |
| NPM | Hodgkin's Disease | 21474455 | Grande et al. (2011) | |
| NPM | Mature B-Cell Neoplasm | 21474455 | Grande et al. (2011) | |
| NPM | Peripheral T-Cell Leukaemia | 21474455 | Grande et al. (2011) | |
| EML4 | Breast Carcinoma | 19737969 | Lin et al. (2009) | |
| C2orf44 | Colorectal Cancer | 22327622 | Lipson et al. (2012) | |
| EML4 | Colorectal Cancer | 19737969 | Lin et al. (2009) | |
| FN1 | Endometrial Stromal Sarcoma | 22570254 | Ren et al. (2012) | |
| VCL | Renal-Cell carcinoma | 21076462 | Debelenko et al. (2011) | |
| EML4 | Lung Adenocarcinoma | 17625570, 18927303 | Soda et al. (2007); Takeuchi et al. (2008) | |
| KIF5B | Lung Adenocarcinoma | 19383809 | Takeuchi et al. (2009) | |
| KLC1 | Lung Adenocarcinoma | 22347464 | Togashi et al. (2012) | |
| PTPN3 | Lung Adenocarcinoma | 22334442 | Jung et al. (2012) | |
| CARS | Inflammatory Myofibroblastic Tumor | 13679433 | Debelenko et al. (2003) | |
| CLTC | Inflammatory Myofibroblastic Tumor | 11485898 | Bridge et al. (2001) | |
| PPFIBP1 | Inflammatory Myofibroblastic Tumor | 23957430 | Yoshida et al. (2013) | |
| RANBP2 | Inflammatory Myofibroblastic Tumor | 12661011 | Ma et al. (2003) | |
| SEC31L1 | Inflammatory Myofibroblastic Tumor | 16161041 | Panagopoulos et al. (2006) | |
| TPM3 | Inflammatory Myofibroblastic Tumor | 10934142 | Lawrence et al. (2000) | |
| TPM4 | Inflammatory Myofibroblastic Tumor | 10934142 | Lawrence et al. (2000) | |
| FN1 | Ovarian Carcinoma | 22570254 | Ren et al. (2012) | |
| DCTN1 | Spitz Nevus/Spitzoid Neoplasms | 24445538 | Wiesner et al. (2014) | |
| TPM3 | Spitz Nevus/Spitzoid Neoplasms | 24445538 | Wiesner et al. (2014) | |
| STRN | Thyroid Carcinoma | 24475247 | Perot et al. (2014) | |
| ROS1 | CEP85L | Angiosarcoma | 23637631 | Giacomini et al. (2013) |
| GOPC | Glioblastoma Multiforme | 12661006 | Charest et al. (2003b) | |
| CEP85L | Glioblastoma Multiforme | 24261984 | Shah et al. (2013) | |
| SLC34A2 | Colorectal Cancer | 23719267 | Davies and Doebele (2013) | |
| CD74 | Lung Adenocarcinoma | 18083107 | Davies and Doebele (2013) | |
| EZR | Lung Adenocarcinoma | 23418494, 22327623 | Takeuchi et al. (2012) | |
| LRIG3 | Lung Adenocarcinoma | 22327623 | Takeuchi et al. (2012) | |
| SDC4 | Lung Adenocarcinoma | 22327623 | Takeuchi et al. (2012) | |
| TPM3 | Lung Adenocarcinoma | 22327623 | Takeuchi et al. (2012) | |
| LIMA1 | Lung Adenocarcinoma | 25264305 | Shaw et al. (2014) | |
| MSN | Lung Adenocarcinoma | 25264305 | Shaw et al. (2014) | |
| GOPC | Lung Adenocarcinoma | 22661537 | Rimkunas et al. (2012) | |
| SLC34A2 | Lung Adenocarcinoma | 18083107 | Rikova et al. (2007) | |
| GOPC | Ovarian Carcinoma | 22163003 | Birch et al. (2011) | |
| MYO5A | Pigmented Spindle Cell Nevus | 24445538 | Wiesner et al. (2014) | |
| PWWP2A | Spitzoid Melanoma | 24445538 | Wiesner et al. (2014) | |
| CLIP1 | Spitz Nevus/Spitzoid Neoplasms | 24445538 | Wiesner et al. (2014) | |
| PPFIBP1 | Spitz Nevus/Spitzoid Neoplasms | 24445538 | Wiesner et al. (2014) | |
| TPM3 | Spitz Nevus/Spitzoid Neoplasms | 24445538 | Wiesner et al. (2014) | |
| ZCCHC8 | Spitz Nevus/Spitzoid Neoplasms | 24445538 | Wiesner et al. (2014) | |
| PPFIBP1 | Spitz Nevus/Spitzoid Neoplasms | 24445538 | Wiesner et al. (2014) | |
| HLAA | Spitz Nevus/Spitzoid Neoplasms | 24445538 | Wiesner et al. (2014) | |
| ERC1 | Spitz Nevus/Spitzoid Neoplasms | 24445538 | Wiesner et al. (2014) | |
| KIAA1598 | Spitz Nevus/Spitzoid Neoplasms | 24445538 | Wiesner et al. (2014) | |
| SLC34A2 | Gastric Adenocarcinoma | 23400546 | Lee et al. (2013) |
Informing fusion protein discovery with cytogenetics and phosphoproteomics
Fusion proteins have been difficult to detect and the process of identifying precise breakpoints has historically been labour intensive. Until recently, the vast majority of fusions were identified through recurrent observations of gross cytogenetic changes in specific cancer subtypes. This approach was previously often restricted to large inter- or intrachromosomal translocations or inversions that affected clear banding patterns. Due to the available resolution at that time, detection was restricted to rearrangements that involved >3 Mb of DNA. Chromosomal banding analysis was also further confounded by cytogenetically complex samples. Paediatric samples have been used in the past to overcome this problem, as they generally possess fewer gross chromosomal changes compared with adult tumours, making it easier to identify recurrent translocations. A number of dominant fusion proteins, such as the ETV6-NTRK3 fusions first described in CFS and CMN, were initially identified in cases of tumours affecting infants or children but have subsequently been found in adult tumours (Knezevich et al., 1998a,b; Tognon et al., 2002; Skalova et al., 2010; Kralik et al., 2011; Leeman-Neill et al., 2014; Wu et al., 2014).
The process of identifying specific fusion partners depended heavily on prior knowledge of the genetic landscape and the availability of probes that could be used to map the area using fluorescence in situ hybridisation (FISH). Locus-specific probes, used in chromosome walking experiments required to identify the breakpoint location, often contained significant amounts of genomic information (80 kb to 1 Mb) in large vectors such as P1-derived artificial chromosomes, yeast artificial chromosomes or bacterial artificial chromosomes. Due to its utility in studying chromosomal aberrations in nondividing cells and its high sensitivity and specificity, FISH has attained broad clinical application for its ability to test for known recurrent translocations. Newer techniques, such as chromosome painting, multicolour FISH and spectral karyotyping, aided in the analysis of samples possessing complex rearrangements and these methods have become invaluable diagnostic and research tools. Using FISH, fusion-gene detection can be theoretically confirmed in less than 24 h, allowing for its use in diagnostic pathology and as a decision-making tool in treatment strategies. FISH-based strategies cannot, however, be used as a screening tool since most FISH methodologies only identify known chromosomal imbalances. This limitation reduces the utility of this technique in biomarker-based patient selection strategies for clinical trials aiming to identify fusion-positive patients across broad tumour types.
Methods other than FISH have successfully identified novel highly recurrent fusion proteins in patient samples. One classic example is the initial identification of the TMPRSS2-ETV1 and TMPRSS2-ERG fusion proteins that were identified in over 50% of prostate cancers using microarray-based techniques and cancer outlier profile analyses (Boehm and Hahn, 2011). Cancer outlier profile analysis ranked ERG and ETV1, two Ets family transcription factors, within the top 10 outlier genes in six independent cancer profiling studies and established mutual exclusivity of expression for the two genes. TMPRSS2 was identified using 5′ rapid amplification of cDNA ends coupled with quantitative PCR-exon walking, and FISH was then used to confirm the fusions. Androgen receptor binding to the TMPRSS2 promoter was shown to drive expression of the Ets family member fusion partner (Tomlins et al., 2005). Although expression of the fusions is restricted to prostate cancer, the prognostic significance of this finding is still controversial: a number of studies suggest that the presence of the fusion is a good prognosticator (Petrovics et al., 2005; Winnes et al., 2007; Saramaki et al., 2008; Taris et al., 2014), whereas others have come to the opposite conclusion (Mehra et al., 2007; Nam et al., 2007a,b; Winnes et al., 2007). Monitoring fusion protein levels in urine samples can now be used to determine response to hormone therapy (Hessels et al., 2007), providing an important example of how fusion proteins can be used as biomarkers for therapy response. How the Ets family fusions contribute to prostate cancer progression and metastasis has been less clear. Recent insights have demonstrated that ERG can block the expression of a number of genes responsible for neuroendocrine and luminal cell differentiation (Mounir et al., 2014), and that it modulates the expression of specific microRNAs with important roles in inducing epithelial-to-mesenchymal transitions (Kim et al., 2014). Clinical targetting of these fusions has been difficult given the uncertainty surrounding their prognostic value and the paucity of mechanistic data, and provides an example of the complexity that can be faced when functionally validating transcription-factor fusion proteins.
Constitutively activated tyrosine-kinase fusions typically drive a rather deterministic set of variations in cellular signalling, including increased tyrosine phosphorylation of substrate proteins and ensuing activation of downstream pathways that govern mitosis, cell survival and metabolism. Cellular protein phosphorylation can be quantitatively assessed using mass spectrometry-based phosphoproteomic techniques, including stable-isotope labelling through chemical modification of peptides, stable-isotope labelling of amino acids in cell culture and label-free methods (Nita-Lazar et al., 2008). Consequently, phosphoproteomics may enable the detection and discovery of functional kinase gene fusions from primary tumour samples or cancer cells. Highly successful implementation of a quantitative phosphotyrosine proteomic strategy was demonstrated by Rikova et al. (2007) with their discovery of ALK and ROS1 fusion proteins in lung cancer. In this study, quantitative phosphoproteomic assessment of 41 NSCLC cell lines and >150 NSCLC tumours and subsequent bioinformatics enrichment analyses revealed expression of oncogenic CD74-ROS1 and SLC34A2-ROS1 in NSCLC patient samples and cell lines. This discovery of ROS1 fusions in the context of NSCLC launched a fervent and productive enquiry into the frequency, relevance and targetability of oncogenic ROS1 fusions in several solid tumours (see Table 1). This finding highlights the potential value of using phosphoproteomics to identify kinase gene fusions and provides insight into cancer biology that is not accessible via genomic or chromosomal analyses.
New opportunities for detecting fusion proteins using next-generation sequencing
With the advent of massively parallel next-generation sequencing (NGS) technologies, we have entered an era where large-scale mapping of tumour genomes is increasing at an unprecedented rate. Large-scale NGS survey methodologies include whole-genome sequencing (WGS), whole-exome sequencing and whole-transcriptome sequencing (RNA-seq). NGS has also been utilised for more focussed sequencing of a subset of genes using targetted sequencing libraries or sequence capture libraries. Although single-nucleotide variants and small insertions and deletions (indels) can be reliably identified from tumour DNA using current computational analysis pipelines, the ability to uncover gene fusions has only recently become feasible with the development of novel algorithms that enable in silico mining of fusion events. RNA-seq experiments generate a large number of short reads that are either ‘single-end’ or ‘paired-end’, the result of sequencing of only one or both of the two fragment strands, respectively. As listed in Table 2, there are now several bioinformatic tools that utilise these NGS data to identify fusion breakpoints. Most of these computational methods include the following steps: aligning or mapping reads to a designated reference transcriptome library, identifying fusion junctions, filtering, and sequence assembly. The initial step identifies a larger set of gene fusion candidates based on detection of discordant read pairs aligning to two different genes. The precise junction of the fusion product is then mapped, followed by application of a set of filters to cull false positives from the discovery cohort and generate a more accurate set of fusion transcript calls for further validation. Most of the software tools listed in Table 2 require input of paired-end RNA-seq data, with the exception of TopHat, FusionFinder, Fusion-Map and the recently reported Ivy Center fusion discovery tool, each of which has successfully discovered novel gene-fusions from paired-end as well as single-end sequencing reads (Kim and Salzberg, 2011; Shah et al., 2013). Notably, another recently developed pipeline called Pegasus is an automated workflow tool that provides a common interface between several gene-fusion detection tools, permits reconstruction of chimeric transcripts, annotates functional domains based on reading frame and predicts the oncogenic potential of gene fusions based on functionality of cognate partners (Abate et al., 2014). An alternative approach is taken by the Comrad tool, which requires both WGS and RNA-seq data from the same sample; by leveraging both sequencing technologies, Comrad identifies gene fusions only if the fusion event is supported by both sequencing platforms, thereby further reducing false-positive calls (McPherson et al., 2011b).
Table 2. Summary of current bioinformatic tools available to identify fusion breakpoints from NGS data.
| Tool name | PMID | Citation |
|---|---|---|
| Bellerophontes | 22711792 | Abate et al. (2012) |
| BreakFusion | 22563071 | Bradeen et al. (2006) |
| BreakPointer | 22302574 | Sun et al. (2012) |
| ChildSeq-RNA | 24517889 | Qadir et al. (2014) |
| ChimeraScan | 21840877 | Iyer et al. (2011) |
| Comrad | 21478487 | McPherson et al. (2011b) |
| defuse | 21625565 | McPherson et al. (2011a) |
| Dissect | 22689759 | Yorukoglu et al. (2012) |
| EBARDenovo | 23457040 | Chu et al. (2013) |
| EricScript | 23093608 | Benelli et al. (2012) |
| FusionAnalyser | 22570408 | Piazza et al. (2012) |
| FusionCatcher | 21247443 | Edgren et al. (2011) |
| FusionFinder | 22761941 | Francis et al. (2012) |
| FusionHunter | 21546395 | Li et al. (2011) |
| FusionMap | 21593131 | Ge et al. (2011) |
| FusionSeq | 20964841 | Sboner et al. (2010) |
| Ivy Center Fusion discovery tool | 24261984 | Shah et al. (2013) |
| LifeScope | 22496636 | Sakarya et al. (2012) |
| MapSplice | 20802226 | Wang et al. (2010) |
| NFuse | 22745232 | McPherson et al. (2012) |
| Pegasus | 25183062 | Abate et al. (2014) |
| ShortFuse | 21330288 | Kinsella et al. (2011) |
| SnowShoes-FTD | 21622959 | Asmann et al. (2011) |
| SOAPFuse | 23409703 | Jia et al. (2013) |
| TopHat-Fusion | 21835007 | Kim and Salzberg (2011) |
Despite the improvements in gene-fusion calling contributed by these various detection algorithms in the analysis of cancer genomes to date, challenges still remain, including considerable variance in terms of sensitivity of fusion detection, and false discovery rates between different tools. Every tool has its own set of biases pertaining to sensitivity or selectivity parameters. Typically, tools that are designed with a high sensitivity bias tend to also exhibit more false positives, and vice versa. There are several areas of ongoing progress that will further enhance the functionality of these tools, including improving the quality and accuracy of NGS data with increased read depth and length, enhancing the accuracy of mapping tools, and designing even more stringent filtering tools to improve the ratio between true and false positives. Other limitations of current methods include the inability to accurately predict genefusion transcripts present at low levels in the sample, due either to tumour heterogeneity and subclonal expression or instability of fusion transcripts. Most computational tools rely on alignment of RNA-seq reads to a reference genome and mapping to a known transcriptome library. These mapping methods may impede the discovery of novel gene fusions, especially if they harbour novel exons that are not annotated in existing transcriptome libraries, or if they are composed of noncoding genes. Optimisation of in silico fusion-finding tools will likely take these factors into account to maximise the utility of detection and discovery of novel or poorly characterised gene fusions in cancer. An in-depth analysis of the fusion-finding algorithms and a detailed outline for improving these tools have been explored elsewhere (please refer to Beccuti M, 2013; Carrara et al., 2013; Wang et al., 2013).
In contrast to mining for gene fusions from WGS or RNA-seq data sets, targetted sequencing has also been used to identify fusion proteins. Several commercially available platforms use targetted sequencing for capturing gene fusions by designing custom libraries and sequencing at high depths in a smaller selection of genes. One such example is Nu-GEN's Ovation fusion panel target enrichment system, promising detection of most rearrangements using a panel of 500 genes curated by the Wellcome Trust Sanger Institute's Catalog of Somatic Mutations in Cancer as possible fusion genes. In addition, there are Clinical Laboratory Improvement Amendments-certified tests for identifying gene fusions from patient samples, including FoundationOne (Frampton et al., 2013), Caris Life Sciences and UW-OncoPlex platforms. Most of these assays perform RNA-seq using a custom capture primer library that is selectively chosen to amplify genes based on prevalence of aberration in cancer followed by bioinformatic analysis. This approach is very useful to discover rearrangements of known genes. However, targetted sequencing limits the possibilities of discovering novel fusions from genes that are not represented in the capture library.
Overall, the expanding range of tools available to synchronise with NGS data for gene fusion detection and discovery is growing. First-generation tools have revealed a range of possibilities and also shown areas of weakness that can be improved upon. Whether they are used to identify known gene fusions in patients in order to guide treatment decisions, or to discover entirely new fusions for biological characterisation and clinical investigation, the utility of NGS data and accompanying bioinformatic fusion detection tools continues to grow.
The role of validation in translating NGS findings
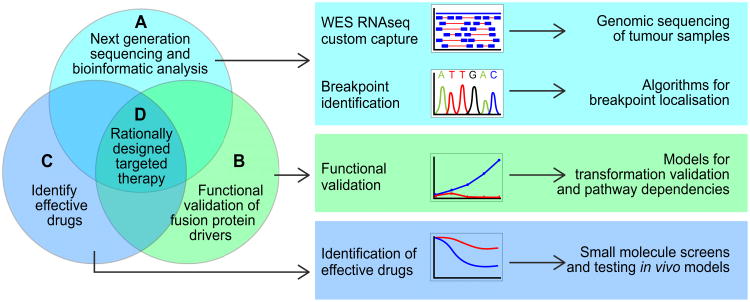
Our newfound ability to identify fusion proteins in a wide variety of cancers has the potential to influence both diagnostic and therapeutic decisions. In our view, there are several important components supporting the rational design of targetted therapies for fusion proteins identified by NGS. First, potential breakpoints, uncovered in genomic sequencing data by bioinformatics tools (Figure 1A), require confirmation by Sanger sequencing or ultra-deep resequencing to rule out false-positive results (Table 2). Second, the translational applicability of confirmed gene fusion discoveries is contingent upon our understanding of the biological significance of these aberrations in the cancer phenotype. Functional validation studies (Figure 1B) can contribute to our mechanistic understanding of a given fusion protein and help to identify common pathways that play important roles in initiation and progression across different cancer types. Third, cell line and animal models, created for functional validation studies, can also be exploited to screen for effective small-molecule inhibitors or drugs (Figure 1C) that block fusion protein-mediated transformation. Lastly, preclinical demonstration of drug efficacy using appropriate in vivo model systems and/or validation of pathway activation in primary human tumour samples is needed to cement the impetus for development of rationally designed clinical trials (Figure 1D).
Figure 1. Methods used to identify, validate and target oncogenic fusion proteins.
(A) A variety of NGS methods generate data from primary patient samples and many different computational methods have been developed to identify fusion proteins (see also: Section – New opportunities for detecting fusion proteins and Table 2). These algorithms vary depending upon the type of genomic sequencing employed and whether the fusion is known or novel. Sanger or deep re-sequencing based confirmation of the fusion breakpoint is important for ruling out false-positive results. (B) Functional validation experiments must be carried out to determine how fusion proteins promote tumorigenesis. Rationally chosen cell line or animal model systems, created to test whether the fusion protein possesses transformative ability, can also be used to identify pathway dependencies. (C) These model systems can also be used to identify small-molecule inhibitors and clinically actionable drugs capable of blocking fusion protein function. (D) Together, these data support the clinical development of rationally designed targetted therapies for fusion protein-driven cancers.
Critical factors to consider for successful functional validation include the current depth of knowledge surrounding the fusion gene partners, as well as the choice of model system and experimental readouts) that are used (reviewed in [75]). The availability of affordable cDNA clones or custom gene synthesis and modern cloning techniques (e.g. In-Fusion® HD from Clontech and Gateway® Cloning from Life Technologies), and the ability to easily shuttle cDNAs into various expression vector systems has enabled the rapid construction and expression of novel fusion proteins in cell-line models. Not surprisingly, selecting and generating model systems to test fusions involving one or more genes of known function is more straightforward than creating model systems to test fusions made from noncoding regions or unknown open reading frames.
A number of classical functional assays, such as the focus formation or growth-factor independence assays, test for the ability of mutations or fusions to drive proliferation or survival, two of the hallmarks of cellular transformation (Hanahan and Weinberg, 2000). Alternatively, model systems used to assess fusion protein-mediated transformation can be created to recapitulate the tumour in which the fusion protein was identified. An example of this approach is the functional validation of TMP3-NTRK1 and BTBD1-NTRK3 fusions identified in non-brainstem high-grade gliomas (Wu et al., 2014). Wu et al. (2014) created an elegant model by stably expressing these NTRK fusions in Tp53-null primary mouse astrocytes. When fusion-expressing cells were transplanted into mouse brains, they rapidly induced high-grade astrocytomas with complete penetrance. The tumours themselves exhibited activation of the PI3K and MAPK pathways, similar to that observed in human HGGs (Wu et al., 2014). This model provided important confirmation of fusion-mediated glial transformation within the context of non-brainstem high-grade gliomas. A similar system was used to validate the transforming capacity of FGFR1 mutations found in pediatric low-grade gliomas that caused duplication of the tyrosine kinase domain (Zhang et al., 2013). Furthermore, the use of analogous cells can provide strong supportive evidence that fusion protein expression plays a causal role in the development of a particular cancer type.
Validation models can extend past cell linestomore complex animal models. One important advantage of in vivo models is the ability to analyse the effects of fusion protein expression on actual tumour formation. One such example was created to study the role of the ETV6-NTRK3 fusion protein in breast cancer progression. ETV6-NTRK3, first discovered in a number of pediatric solid tumours, was subsequently identified in a rare form of breast cancer called SBC. To look at the fusion's potential causal role in breast cancer, it was stably expressed in the human breast epithelial cell line MCF10A and ectopically injected into the mammary fat pads of immunodeficient mice (Tognon et al., 2011). Xenografts possessed a differentiated phenotype and expressed epithelial markers; cells grown in Matrigel cultures formed large polarised mammospheres possessing filled lumens. Cells also expressed high levels of MEK and AKT phosphorylation, contributing to the increase in acinar size and lack of luminal apoptosis, respectively. Li et al. (2007) went further to create a whey acidic protein-inducible promoter-driven knock-in transgenic mouse model to express ETV6-NTRK3 in the mammary epithelial cells of the developing mouse. These animals developed highly penetrant multifocal tumours with characteristics similar to those observed in human SBC. These types of validation studies helped to determine whether similar signalling pathways were activated by this fusion in different cell types and whether similar targetted therapies could be applied across different cancers. Although often thought to be indolent in nature, ETV6-NTRK3-positive cancers can grow rapidly and metastasise, thereby necessitating invasive surgeries and treatment. Drug testing by various groups identified a number of promising drugs for ETV6-NTRK3-driven tumours, including the multi-kinase inhibitor PKC412 (Chi et al., 2012), IGF1R inhibitors (Martin et al., 2006; Tognon et al., 2011; Tognon et al., 2012), and the US Food and Drug Administration (FDA)-approved drug crizotinib (Taipale et al., 2013; Roberts et al., 2014). ETV6-NTRK3 also provided one of the earliest examples of an oncogenic fusion protein that crossed cell boundaries – being capable of transforming mesenchymal (CFS and CMN), epithelial (breast, thyroid and salivary gland carcinoma), haematopoietic (acute myeloid leukaemia and B-cell precursor acute lymphoblastic leukaemia) and neuroglial cells (DIPG and low-grade glioma) (Knezevich et al., 1998a,b; Rubin et al., 1998; Tognon et al., 2002; Skalova et al., 2010; Fehr et al., 2011; Kralik et al., 2011; Leeman-Neill et al., 2014; Wu et al., 2014) – a theme that has reemerged in the context of other fusions (see Table 1).
Once appropriate cell line or animal model systems have been validated, they can then be used in functional genomic screens or experiments to identify signalling pathways required for fusion protein-mediated transformation. A number of methodologies and approaches have been developed to broadly interrogate transformed cell lines, including but not limited to higher-throughput small-molecule, small interfering RNA or lentiviral short hairpin RNA, and newer clustered regularly interspaced short palin-dromic repeats (CRISPR) technology-based screens (reviewed in Boehm and Hahn, 2011). Results from these high-throughput screens also require additional validation to confirm specific pathway dependencies. Animal model studies can then be used to generate pre-clinical data to confirm drug efficacy and provide support for future clinical trial development.
Targetting fusion proteins: identifying effective drugs and clinical realities
The discovery of gene fusions as oncogenic drivers in cancer spurred the possibility of leveraging such molecular vulnerabilities to develop therapies more selective than cytotoxic chemotherapy or radiation. As previously mentioned, clinical implementation of the ABL1 kinase inhibitor imatinib to inhibit the BCR-ABL1 kinase fusion in CML resulted in 80% decrease in patient mortality (Druker et al., 2006) and a paradigm shift in cancer treatment. Research and development of anti-cancer agents is now largely focussed on identifying pathogenic addictions and developing cognate targetted molecules to enable highly effective and selective therapeutic effects in specific malignancies. Accordingly, the cancer pharmacopeia is exponentially expanding and, coupled with the availability of NGS-based gene fusion discovery methods, we now have the capacity to perform high-throughput pre-clinical testing and nominate effective compounds for clinical investigation at a rapid pace.
The rapid FDA approval of crizotinib (Xalkori) for use as frontline treatment in ALK fusion-positive lung cancer is a recent example of highly efficient translation of laboratory findings to the clinical setting. Specifically, about five years after the EML4-ALK fusion was identified in NSCLC (Soda et al., 2007), the FDA granted accelerated approval to crizotinib, a multi-kinase inhibitor of MET, ALK and ROS1, for the treatment of patients harbouring ALK fusions based upon the demonstration of significantly better progression-free survival and overall response rate compared tochemotherapy (Kwak et al., 2010; Rodig and Shapiro, 2010; Shaw et al., 2013b; Solomon et al., 2014). Similar to the experience with imatinib in CML, many ALK fusion-positive patients relapse while on crizotinib due to adaptive resistance to the inhibitor (Heuckmann et al., 2011; Doebele et al., 2012; Lovly and Pao, 2012; Shaw et al., 2013a). Resistance can be attributed to several escape mechanisms, including acquisition of kinase domain mutations in ALK and amplification or overactivation of bypass pathways that are sufficiently compensatory to enable cell growth and survival in the presence of crizotinib (Camidge et al., 2014). Several structurally distinct, second-generation ALK inhibitors capable of circumventing kinase domain mutation-based resistance are now under development. These include ceritinib (LDK378) and alectinib (CH5424802), which recently received accelerated FDA approval and ‘breakthrough therapy’ designations respectively, that were based on robust inhibitory activity in both crizotinib-naïve and crizotinib-resistant ALK fusion-positive disease (Awad and Shaw, 2014; Katayama et al., 2014). However, alectinib resistance has already been reported from a treated patient (Katayama et al., 2014). This rapid adaptation of cancer cells to selection pressure imposed by inhibitors is a testament to the prolific evolutionary rate of cancer genomes. Research and discovery efforts to anticipate kinase domain mutation driven drug-resistance profiles must therefore be concomitant with development of first-generation inhibitors in order to be prepared with potent and effective second-generation inhibitors. As an example, previously predicted BCR-ABL1 kinase domain mutations recovered from cell-based muta-genesis screening were later identified in patients treated with kinase inhibitors, thus validating the utility of these accelerated methods to ‘stay ahead of the curve’ to combat resistance to targetted therapy (Bradeen et al., 2006; O'Hare et al., 2009; Zabriskie et al., 2014).
In certain cases, the potent dual activity of an FDA-approved drug against another kinase may permit more rapid access to targetted therapeutic strategies in patients harbouring that newly implicated kinase fusion. The kinase domain of ROS1 is homologous to that of ALK, with approximately 38% overall amino acid identity and up to 77% identity in the ATP binding pocket. Given the success of crizotinib in ALK fusion-positive NSCLC and structural homology between ALK and ROS1 kinase domains, crizotinib was quickly repurposed as a ROS1 inhibitor upon the recent discovery of ROS1 fusions in a variety of malignancies including NSCLC. Evaluation of crizotinib in ROS1 fusion-positive NSCLC patients demonstrated an objective response rate of 72% and median response duration of 17.6 months (Shaw et al., 2014), confirming inhibition of ROS1 as an effective clinical strategy in these patients. Also, patients in this study harboured seven different ROS1-fusion partners, but efficacy of treatment was not correlated with any specific type of rearrangement. Akin to the experience with crizotinib in ALK fusion-positive disease, crizotinib resistance due to CD74-ROS1 kinase domain mutation was also recently reported (Awad et al., 2013), reaffirming the need to predict resistance and have a pipeline of second- and third-generation inhibitors available.
Unbiased high-throughput inhibitor screening may also facilitate identification of effective kinase inhibitors that may be missed or unrecognised due to the focussed context of homology studies. We recently reported that foretinib is a potent ROS1 inhibitor (Davare et al., 2013). Unlike all previously reported ALK/ROS1 inhibitors, foretinib does not exhibit substantial inhibitory activity against ALK. This lack of reciprocity was surprising due to the relatively high degree of homology between the ALK and ROS1 kinase domains. Due to its overall potency as a ROS1 inhibitor, foretinib is able to circumvent crizotinib-resistant ROS1 mutants, including the G2032R mutation that emerged in an NSCLC patient. The discovery of foretinib was facilitated by the unbiased nature of the high-throughput screening method. If relying exclusively on ALK homology for ROS1 inhibitor selection or design, this novel activity of foretinib and the possibility of developing similar agents to circumvent crizotinib resistance in ROS1 would not have been possible. Additionally, this differential activity of foretinib for ROS1 but not ALK suggests that subtle but functionally significant structural elements must differentiate the ROS1 and ALK kinase domains, which may be valuable for rational ROS1 inhibitor design in the future. By combining multiple approaches, including pre-emptive mutagenesis screening to predict resistant mutations, generation of patient-derived resistant cell lines, and iteratively challenging these cells with large panels of kinase inhibitors to locate novel vulnerabilities, it may be possible to ‘keep up with resistance’ to targetted therapy.
Successful targetting of non-kinase gene fusions has also been accomplished. The proteasome inhibitor bortezomib (Velcade) was used in MLL-rearranged leukaemia to block degradation of the MLL-fusion protein to induce tumour suppressor programs and ultimately apoptotic cell death (Liu et al., 2014). Momentum is also building to develop agents to target transcription factor fusions with novel methods that include peptidomimetics, synthetic lethality, stapled peptides and RNA interference (reviewed in Redmond and Carroll, 2009).
Conclusions
Given the exponential accumulation of cancer genomic data, we are in the midst of an exciting time of discovery for groups studying how novel fusion proteins contribute to cancer progression. The deluge of genomic data and new algorithm development, coupled with advancements in molecular cloning and expression, have contributed to a significant increase in the number of novel and validated fusion proteins found in various malignancies. Compared with transcription factor fusions, the kinase gene fusions have been more rigorously characterised in terms of their capacity to serve as oncogenic drivers. Development of therapeutically viable kinase inhibitors for translational research and clinical implementation is further along. The molecular mechanisms by which transcription factor fusions function as pathogenic drivers in cancer are, in most cases, likely to be distinct from kinase gene fusions. Overall, kinase fusions seem to activate fairly canonical downstream pathways to regulate cell growth, proliferation, and survival, or migration phenotypes. In contrast, transcription factor fusions likely deregulate gene expression or affect developmental pathways, including arrested differentiation and chronic proliferation of tissue progenitors or stem cells.
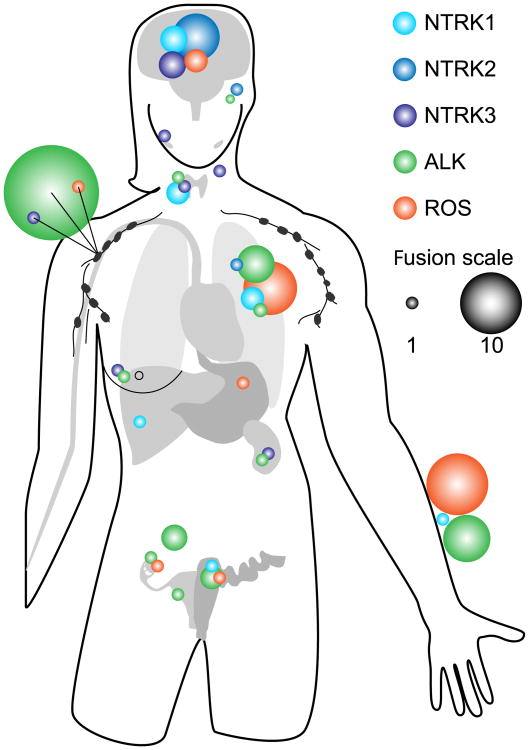
With advancements in NGS, in silico mining tools and phosphoproteomics, it is easier to interrogate large numbers of cancer genomes for the presence of gene fusions en masse. These efforts are beginning to reveal that kinase fusions are more prevalent in cancer than previously estimated. While the overall frequency of a specific kinase fusion in a particular malignancy may be low, the kinase may be frequently partnered with a variety of genes in multiple malignancies. The examples of NTRK, ALK and ROS1 fusions highlighted in this review demonstrate that expression of these tyrosine kinase fusions can transform cells from many lineages (Figure 2), consistent with their detection in a diverse set of malignancies. Receptor tyrosine kinase signalling is frequently coupled to the RAS/MAPK, PI3K/AKT, or JAK/STAT signalling axes that regulate growth, migration, survival and gene transcription. Upregulation of these pathways in dividing cells may be sufficient to drive unchecked proliferation and survival, a key step in oncogenesis.
Figure 2. Prevalence of NTRK, ALK and ROS1 kinase gene fusions across cancer types.
NTRK, ALK and ROS1 fusions, shown as blue/purple, green and orange spheres, respectively, are found partnered with several upstream genes and found expressed in a diverse set of malignancies (see also Table 1). The size of the sphere denotes the number of different fusions found at a given site.
One of the ongoing challenges in cancer genomics is the ability to differentiate the causal oncogenic deviations from the large number of other changes resulting from inherent genomic instability. Often, even if the cancer cell harbours a plethora of genomic alterations, it is only a smaller subset of these changes that are the relevant functional drivers exerting measurable effects upon cell signalling. Thus, the actionable utility of genomics to discover novel cancer drivers is only as good as the efficiency and speed of the functional validation efforts to assign them meaning. Exploring the cancer proteome in parallel with the genome may be a productive exercise to discover cancer drivers by providing functional information on those genetic changes that have discernable proteomic correlates and hence are more likely to be relevant actionable candidates.
Studying fusions has underscored how these unique drivers break though tissue boundaries and provide a context for common therapeutic strategies that may be taken between multiple types of different cancers. Accordingly, we can posit that a malignant lung tumour expressing a ROS1 fusion has more in common from a targetted-therapy standpoint with a ROS1 fusion-driven cholangiocarcinoma tumour than with another lung tumour that may harbour activating mutations in EGFR or fusions of FGFR or NTRK genes. Effective therapeutic approaches, in light of these findings, will require reassessment of current drug testing strategies and clinical trial design aimed at determining the efficacy and benefit of new agents in affected patients. In September 2014, the National Cancer Institute announced that it will be launching a Molecular Analysis for Therapy Choice (MATCH) program, where targetted gene sequencing will be used to match individuals whose tumours harbour specific genetic anomalies to treatment(s) with therapeutic agents selected for activity against the particular anomaly. MATCH will be a complex, first-of-its-kind clinical trial that may have up to 25 unique arms. This will undoubtedly be an iterative process, and many lessons will be learned from the successes and failures of the MATCH program. These studies will shape the future of cancer care and lead us into a streamlined precision medicine program that promises to deliver real-time personalised therapy to improve patient prognosis and long-term outcomes.
Acknowledgments
We apologise to all the authors whose work could not be included in this manuscript owing to space constraints. We would like to thank Nameeta Shah for her valuable insights and constructive feedback that guided several concepts discussed in the review. We thank Christopher Eide for his critical review of this article, Pierrette Lo for useful discussions and editing, and Tiffany Howard for help with the figure illustrations.
Funding: This work was supported in part by a Hyundai Hope on Wheels grant to M.D. C.T. is supported through Howard Hughes Foundation Investigator support awarded to B.J. Druker.
Abbreviations
- ALCL
anaplastic large-cell lymphoma
- CFS
congenital fibrosarcoma
- CML
chronic myeloid leukaemia
- CMN
congenital mesoblastic nephroma
- CRISPR
clustered regularly interspaced short palindromic repeats
- DIPG
diffuse intrinsic pontine glioma
- FDA
Food and Drug Administration
- FISH
fluorescent in situ hybridisation
- GBM
glioblastoma multiforme
- ICC
intrahepatic cholangiocarcnioma
- IMT
inflammatory myofibroblastic tumour
- MATCH
molecular analysis for therapy choice
- NGS
next-generation sequencing
- NSCLC
non-small cell lung cancer
- SBC
secretory breast carcinoma
- WGS
whole-genome sequencing
Footnotes
Definitions: Functional validation: test(s) performed to determine the effects of a mutated protein on the behavior of a cell
Gene fusion: a hybrid gene formed from two previously separate genes resulting from a translocation, inversion or intrachromosomal deletion
Next-generation sequencing: non-Sanger-based high-throughput sequencing technologies
Oncogene: a gene that has the potential to cause cancer
Oncogenic driver: a genomic change found in cancers that is essential to induce cancer progression and metastasis
Targetted therapy: a type of treatment that uses drugs or other substances to identify and attack specific types of cancer cells with less harm to normal cells
Transformation: the change that a normal cell undergoes as it becomes malignant
Tumorigenesis: production or formation of a tumour
Conflict of interest statement: The authors have declared no conflict of interest.
References
- Abate F, Acquaviva A, Paciello G, Foti C, Ficarra E, Ferrarini A, Delledonne M, Iacobucci I, Soverini S, Martinelli G, Macii E. Bellerophontes: An RNA-Seq data analysis framework for chimeric transcripts discovery based on accurate fusion model. Bioinformatics. 2012;28:2114–2121. doi: 10.1093/bioinformatics/bts334. [DOI] [PubMed] [Google Scholar]
- Abate F, Zairis S, Ficarra E, Acquaviva A, Wiggins CH, Frattini V, Lasorella A, Iavarone A, Inghirami G, Rabadan R. Pegasus: A comprehensive annotation and prediction tool for detection of driver gene fusions in cancer. BMC Syst Biol. 2014;8:97. doi: 10.1186/s12918-014-0097-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardini E, Bosotti R, Borgia AL, DePonti C, Somaschini A, Cammarota R, Amboldi N, Raddrizzani L, Milani A, Magnaghi P, Ballinari D, Casero D, Gasparri F, Banfi P, Avanzi N, Saccardo MB, Alzani R, Bandiera T, Felder E, Donati D, Pesenti E, Sartore-Bianchi A, Gambacorta M, Pierotti MA, Siena S, Veronese S, Galvani A, Isacchi A. The TPM3-NTRK1 rearrangement is a recurring event in colorectal carcinoma and is associated with tumor sensitivity to TRKA kinase inhibition. Mol Oncol. 2014;8(8):1495–1507. doi: 10.1016/j.molonc.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asmann YW, Hossain A, Necela BM, Middha S, Kalari KR, Sun Z, Chai HS, Williamson DW, Radisky D, Schroth GP, Kocher JP, Perez EA, Thompson EA. A novel bioinformatics pipeline for identification and characterization of fusion transcripts in breast cancer and normal cell lines. Nucleic Acids Res. 2011;39:e100. doi: 10.1093/nar/gkr362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awad MM, Katayama R, McTigue M, Liu W, Deng YL, Brooun A, Friboulet L, Huang D, Falk MD, Timofeevski S, Wilner KD, Lockerman EL, Khan TM, Mahmood S, Gainor JF, Digumarthy SR, Stone JR, Mino-Kenudson M, Christensen JG, Iafrate AJ, Engelman JA, Shaw AT. Acquired resistance to crizotinib from a mutation in CD74-ROS1. New Engl J Med. 2013;368:2395–2401. doi: 10.1056/NEJMoa1215530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awad MM, Shaw AT. ALK inhibitors in non-small cell lung cancer: Crizotinib and beyond. Clin Adv Hematol Oncol. 2014;12:429–439. [PMC free article] [PubMed] [Google Scholar]
- Beccuti M, Carrara M, Cordero F, Donatelli S, Calogero RA. The structure of state-of-art gene fusion-finder algorithms. Genome Bioinformatics. 2013;1(1):2. [Google Scholar]
- Benelli M, Pescucci C, Marseglia G, Severgnini M, Torricelli F, Magi A. Discovering chimeric transcripts in paired-end RNA-seq data by using EricScript. Bioinformatics. 2012;28:3232–3239. doi: 10.1093/bioinformatics/bts617. [DOI] [PubMed] [Google Scholar]
- Birch AH, Arcand SL, Oros KK, Rahimi K, Watters AK, Provencher D, Greenwood CM, Mes-Masson AM, Tonin PN. Chromosome 3 anomalies investigated by genome wide SNP analysis of benign, low malignant potential and low grade ovarian serous tumours. PloS One. 2011;6:e28250. doi: 10.1371/journal.pone.0028250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchmeier C, Sharma S, Wigler M. Expression and rearrangement of the ROS1 gene in human glioblastoma cells. Proc Natl Acad Sci U S A. 1987;84:9270–9274. doi: 10.1073/pnas.84.24.9270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm JS, Hahn WC. Towards systematic functional characterization of cancer genomes. Nat Rev Genet. 2011;12:487–498. doi: 10.1038/nrg3013. [DOI] [PubMed] [Google Scholar]
- Bradeen HA, Eide CA, O'Hare T, Johnson KJ, Willis SG, Lee FY, Druker BJ, Deininger MW. Comparison of imatinib mesylate, dasatinib (BMS-354825), and nilotinib (AMN107) in an N-ethyl-N-nitrosourea (ENU)-based mutagenesis screen: High efficacy of drug combinations. Blood. 2006;108:2332–2338. doi: 10.1182/blood-2006-02-004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge JA, Kanamori M, Ma Z, Pickering D, Hill DA, Lydiatt W, Lui MY, Colleoni GW, Antonescu CR, Ladanyi M, Morris SW. Fusion of the ALK gene to the clathrin heavy chain gene, CLTC, in inflammatory myofibroblastic tumor. Am J Pathol. 2001;159:411–415. doi: 10.1016/S0002-9440(10)61711-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camidge DR, Pao W, Sequist LV. Acquired resistance to TKIs in solid tumours: Learning from lung cancer. Nat Rev Clin Oncol. 2014;11:473–481. doi: 10.1038/nrclinonc.2014.104. [DOI] [PubMed] [Google Scholar]
- Carrara M, Beccuti M, Lazzarato F, Cavallo F, Cordero F, Donatelli S, Calogero RA. State-of-the-art fusion-finder algorithms sensitivity and specificity. BioMed Res Int. 2013;2013:340620. doi: 10.1155/2013/340620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaluce F, Sgambato A, Maione P, Rossi A, Ferrara C, Napolitano A, Palazzolo G, Ciardiello F, Gridelli C. ALK inhibitors: A new targeted therapy in the treatment of advanced NSCLC. Targeted Oncol. 2013;8:55–67. doi: 10.1007/s11523-012-0250-9. [DOI] [PubMed] [Google Scholar]
- Charest A, Kheifets V, Park J, Lane K, McMahon K, Nutt CL, Housman D. Oncogenic targeting of an activated tyrosine kinase to the Golgi apparatus in a glioblastoma. Proc Natl Acad Sci U S A. 2003a;100:916–921. doi: 10.1073/pnas.242741799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charest A, Lane K, McMahon K, Park J, Preisinger E, Conroy H, Housman D. Fusion of FIG to the receptor tyrosine kinase ROS in a glioblastoma with an interstitial del(6)(q21q21) Genes Chromosomes Cancer. 2003b;37:58–71. doi: 10.1002/gcc.10207. [DOI] [PubMed] [Google Scholar]
- Chi HT, Ly BT, Kano Y, Tojo A, Watanabe T, Sato Y. ETV6-NTRK3 as a therapeutic target of small molecule inhibitor PKC412. Biochem Biophys Res Commun. 2012;429:87–92. doi: 10.1016/j.bbrc.2012.10.087. [DOI] [PubMed] [Google Scholar]
- Chiari R, Buttitta F, Iacono D, Bennati C, Metro G, Di Lorito A, Iezzi M, Tiseo M, Mazzoni F, Cappuzzo F, Marchetti A, Crino L. Dramatic response to crizotinib in ROS1 fluorescent in situ hybridization- and immunohistochemistry-positive lung adenocarcinoma: A case series. Clin Lung Cancer. 2014;15:470–474. doi: 10.1016/j.cllc.2014.06.004. [DOI] [PubMed] [Google Scholar]
- Chu HT, Hsiao WW, Chen JC, Yeh TJ, Tsai MH, Lin H, Liu YW, Lee SA, Chen CC, Tsao TT, Kao CY. EBARDenovo: Highly accurate de novo assembly of RNA-Seq with efficient chimera-detection. Bioinformatics. 2013;29:1004–1010. doi: 10.1093/bioinformatics/btt092. [DOI] [PubMed] [Google Scholar]
- Colleoni GW, Bridge JA, Garicochea B, Liu J, Filippa DA, Ladanyi M. ATIC-ALK: A novel variant ALK gene fusion in anaplastic large cell lymphoma resulting from the recurrent cryptic chromosomal inversion, inv(2)(p23q35) Am J Pathol. 2000;156:781–789. doi: 10.1016/S0002-9440(10)64945-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cools J, Wlodarska I, Somers R, Mentens N, Pedeutour F, Maes B, DeWolf-Peeters C, Pauwels P, Hagemeijer A, Marynen P. Identification of novel fusion partners of ALK, the anaplastic lymphoma kinase, in anaplastic large-cell lymphoma and inflammatory myofibroblastic tumor. Genes Chromosomes Cancer. 2002;34:354–362. doi: 10.1002/gcc.10033. [DOI] [PubMed] [Google Scholar]
- Cunha SI, Pietras K. ALK1 as an emerging target for antiangiogenic therapy of cancer. Blood. 2011;117:6999–7006. doi: 10.1182/blood-2011-01-330142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davare MA, Saborowski A, Eide CA, Tognon C, Smith RL, Elferich J, Agarwal A, Tyner JW, Shinde UP, Lowe SW, Druker BJ. Foretinib is a potent inhibitor of oncogenic ROS1 fusion proteins. Proc Natl Acad Sci U S A. 2013;110:19519–19524. doi: 10.1073/pnas.1319583110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies KD, Doebele RC. Molecular pathways: ROS1 fusion proteins in cancer. Clin Cancer Res. 2013;19:4040–4045. doi: 10.1158/1078-0432.CCR-12-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies KD, Mahale S, Astling DP, Aisner DL, Le AT, Hinz TK, Vaishnavi A, Bunn PA, Jr, Heasley LE, Tan AC, Camidge DR, Varella-Garcia M, Doebele RC. Resistance to ROS1 inhibition mediated by EGFR pathway activation in non-small cell lung cancer. PloS One. 2013;8:e82236. doi: 10.1371/journal.pone.0082236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBraekeleer E, Douet-Guilbert N, DeBraekeleer M. RARA fusion genes in acute promyelocytic leukemia: A review. Expert Rev Hematol. 2014;7:347–357. doi: 10.1586/17474086.2014.903794. [DOI] [PubMed] [Google Scholar]
- DePaepe P, Baens M, vanKrieken H, Verhasselt B, Stul M, Simons A, Poppe B, Laureys G, Brons P, Vandenberghe P, Speleman F, Praet M, DeWolf-Peeters C, Marynen P, Wlodarska I. ALK activation by the CLTC-ALK fusion is a recurrent event in large B-cell lymphoma. Blood. 2003;102:2638–2641. doi: 10.1182/blood-2003-04-1050. [DOI] [PubMed] [Google Scholar]
- Debelenko LV, Arthur DC, Pack SD, Helman LJ, Schrump DS, Tsokos M. Identification of CARS-ALK fusion in primary and metastatic lesions of an inflammatory myofibroblastic tumor. Lab Invest. 2003;83:1255–1265. doi: 10.1097/01.lab.0000088856.49388.ea. [DOI] [PubMed] [Google Scholar]
- Debelenko LV, Raimondi SC, Daw N, Shivakumar BR, Huang D, Nelson M, Bridge JA. Renal cell carcinoma with novel VCL-ALK fusion: New representative of ALK-associated tumor spectrum. Modern Pathol. 2011;24:430–442. doi: 10.1038/modpathol.2010.213. [DOI] [PubMed] [Google Scholar]
- Doebele RC, Pilling AB, Aisner DL, Kutateladze TG, Le AT, Weickhardt AJ, Kondo KL, Linderman DJ, Heasley LE, Franklin WA, Varella-Garcia M, Camidge DR. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin Cancer Res. 2012;18:1472–1482. doi: 10.1158/1078-0432.CCR-11-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler HG, Gignac SM, vonWasielewski R, Werner M, Dirks WG. Pathobiology of NPM-ALK and variant fusion genes in anaplastic large cell lymphoma and other lymphomas. Leukemia. 2000;14:1533–1559. doi: 10.1038/sj.leu.2401878. [DOI] [PubMed] [Google Scholar]
- Drilon A, Wang L, Hasanovic A, Suehara Y, Lipson D, Stephens P, Ross J, Miller V, Ginsberg M, Zakowski MF, Kris MG, Ladanyi M, Rizvi N. Response to Cabozantinib in patients with RET fusion-positive lung adenocarcinomas. Cancer Discov. 2013;3:630–635. doi: 10.1158/2159-8290.CD-13-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druker BJ. Perspectives on the development of imatinib and the future of cancer research. Nat Med. 2009;15:1149–1152. doi: 10.1038/nm1009-1149. [DOI] [PubMed] [Google Scholar]
- Druker BJ, Guilhot F, O'Brien SG, Gathmann I, Kantarjian H, Gattermann N, Deininger MW, Silver RT, Goldman JM, Stone RM, Cervantes F, Hochhaus A, Powell BL, Gabrilove JL, Rousselot P, Reiffers J, Cornelissen JJ, Hughes T, Agis H, Fischer T, Verhoef G, Shepherd J, Saglio G, Gratwohl A, Nielsen JL, Radich JP, Simonsson B, Taylor K, Baccarani M, So C, Letvak L, Larson RA, Investigators I. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. New Engl J Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- Edgren H, Murumagi A, Kangaspeska S, Nicorici D, Hongisto V, Kleivi K, Rye IH, Nyberg S, Wolf M, Borresen-Dale AL, Kallioniemi O. Identification of fusion genes in breast cancer by paired-end RNA-sequencing. Genome Biol. 2011;12:R6. doi: 10.1186/gb-2011-12-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A, Loning T, Stenman G. Mammary analogue secretory carcinoma of the salivary glands with ETV6-NTRK3 gene fusion. Am J Surg Pathol. 2011;35:1600–1602. doi: 10.1097/PAS.0b013e31822832c7. [DOI] [PubMed] [Google Scholar]
- Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, Schnall-Levin M, White J, Sanford EM, An P, Sun J, Juhn F, Brennan K, Iwanik K, Maillet A, Buell J, White E, Zhao M, Balasubramanian S, Terzic S, Richards T, Banning V, Garcia L, Mahoney K, Zwirko Z, Donahue A, Beltran H, Mosquera JM, Rubin MA, Dogan S, Hedvat CV, Berger MF, Pusztai L, Lechner M, Boshoff C, Jarosz M, Vietz C, Parker A, Miller VA, Ross JS, Curran J, Cronin MT, Stephens PJ, Lipson D, Yelensky R. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis RW, Thompson-Wicking K, Carter KW, Anderson D, Kees UR, Beesley AH. FusionFinder: A software tool to identify expressed gene fusion candidates from RNA-Seq data. PloS One. 2012;7:e39987. doi: 10.1371/journal.pone.0039987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gainor JF, Shaw AT. Novel targets in non-small cell lung cancer: ROS1 and RET fusions. Oncologist. 2013;18:865–875. doi: 10.1634/theoncologist.2013-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H, Liu K, Juan T, Fang F, Newman M, Hoeck W. FusionMap: Detecting fusion genes from next-generation sequencing data at base-pair resolution. Bioinformatics. 2011;27:1922–1928. doi: 10.1093/bioinformatics/btr310. [DOI] [PubMed] [Google Scholar]
- Giacomini CP, Sun S, Varma S, Shain AH, Giacomini MM, Balagtas J, Sweeney RT, Lai E, Del Vecchio CA, Forster AD, Clarke N, Montgomery KD, Zhu S, Wong AJ, vande Rijn M, West RB, Pollack JR. Breakpoint analysis of transcriptional and genomic profiles uncovers novel gene fusions spanning multiple human cancer types. PLoS Genet. 2013;9:e1003464. doi: 10.1371/journal.pgen.1003464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons DL, Pricl S, Kantarjian H, Cortes J, Quintas-Cardama A. The rise and fall of gatekeeper mutations? The BCR-ABL1 T315I paradigm. Cancer. 2012;118:293–299. doi: 10.1002/cncr.26225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande E, Bolos MV, Arriola E. Targeting oncogenic ALK: A promising strategy for cancer treatment. Mol Cancer Ther. 2011;10:569–579. doi: 10.1158/1535-7163.MCT-10-0615. [DOI] [PubMed] [Google Scholar]
- Greco A, Mariani C, Miranda C, Pagliardini S, Pierotti MA. Characterization of the NTRK1 genomic region involved in chromosomal rearrangements generating TRK oncogenes. Genomics. 1993;18:397–400. doi: 10.1006/geno.1993.1482. [DOI] [PubMed] [Google Scholar]
- Gu TL, Deng X, Huang F, Tucker M, Crosby K, Rimkunas V, Wang Y, Deng G, Zhu L, Tan Z, Hu Y, Wu C, Nardone J, MacNeill J, Ren J, Reeves C, Innocenti G, Norris B, Yuan J, Yu J, Haack H, Shen B, Peng C, Li H, Zhou X, Liu X, Rush J, Comb MJ. Survey of tyrosine kinase signaling reveals ROS kinase fusions in human cholangiocarcinoma. PloS One. 2011;6:e15640. doi: 10.1371/journal.pone.0015640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hantschel O, Grebien F, Superti-Furga G. The growing arsenal of ATP-competitive and allosteric inhibitors of BCR-ABL. Cancer Res. 2012;72:4890–4895. doi: 10.1158/0008-5472.CAN-12-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez L, Pinyol M, Hernandez S, Bea S, Pulford K, Rosenwald A, Lamant L, Falini B, Ott G, Mason DY, Delsol G, Campo E. TRK-fused gene (TFG) is a new partner of ALK in anaplastic large cell lymphoma producing two structurally different TFG-ALK translocations. Blood. 1999;94:3265–3268. [PubMed] [Google Scholar]
- Hessels D, Smit FP, Verhaegh GW, Witjes JA, Cornel EB, Schalken JA. Detection of TMPRSS2-ERG fusion transcripts and prostate cancer antigen 3 in urinary sediments may improve diagnosis of prostate cancer. Clin Cancer Res. 2007;13:5103–5108. doi: 10.1158/1078-0432.CCR-07-0700. [DOI] [PubMed] [Google Scholar]
- Heuckmann JM, Holzel M, Sos ML, Heynck S, Balke-Want H, Koker M, Peifer M, Weiss J, Lovly CM, Grutter C, Rauh D, Pao W, Thomas RK. ALK mutations conferring differential resistance to structurally diverse ALK inhibitors. Clin Cancer Res. 2011;17:7394–7401. doi: 10.1158/1078-0432.CCR-11-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang EJ, Reichardt LF. Neurotrophins: Roles in neuronal development and function. Ann Rev Neurosci. 2001;24:677–736. doi: 10.1146/annurev.neuro.24.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ME, Ye YC, Chen SR, Chai JR, Lu JX, Zhoa L, Gu LJ, Wang ZY. Use of all-trans retinoic acid in the treatment of acute promyelocytic leukemia. Blood. 1988;72:567–572. [PubMed] [Google Scholar]
- Iyer MK, Chinnaiyan AM, Maher CA. ChimeraScan: A tool for identifying chimeric transcription in sequencing data. Bioinformatics. 2011;27:2903–2904. doi: 10.1093/bioinformatics/btr467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W, Qiu K, He M, Song P, Zhou Q, Zhou F, Yu Y, Zhu D, Nickerson ML, Wan S, Liao X, Zhu X, Peng S, Li Y, Wang J, Guo G. SOAPfuse: An algorithm for identifying fusion transcripts from paired-end RNA-Seq data. Genome Biol. 2013;14:R12. doi: 10.1186/gb-2013-14-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Hutter B, Jager N, Korshunov A, Kool M, Warnatz HJ, Zichner T, Lambert SR, Ryzhova M, Quang DA, Fontebasso AM, Stutz AM, Hutter S, Zuckermann M, Sturm D, Gronych J, Lasitschka B, Schmidt S, Seker-Cin H, Witt H, Sultan M, Ralser M, Northcott PA, Hovestadt V, Bender S, Pfaff E, Stark S, Faury D, Schwartzentruber J, Majewski J, Weber UD, Zapatka M, Raeder B, Schlesner M, Worth CL, Bartholomae CC, vonKalle C, Imbusch CD, Radomski S, Lawerenz C, vanSluis P, Koster J, Volckmann R, Versteeg R, Lehrach H, Monoranu C, Winkler B, Unterberg A, Herold-Mende C, Milde T, Kulozik AE, Ebinger M, Schuhmann MU, Cho YJ, Pomeroy SL, vonDeimling A, Witt O, Taylor MD, Wolf S, Karajannis MA, Eberhart CG, Scheurlen W, Hasselblatt M, Ligon KL, Kieran MW, Korbel JO, Yaspo ML, Brors B, Felsberg J, Reifenberger G, Collins VP, Jabado N, Eils R, Lichter P, Pfister SM International Cancer Genome Consortium PedBrain Tumor, P. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet. 2013;45:927–932. doi: 10.1038/ng.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y, Kim P, Jung Y, Keum J, Kim SN, Choi YS, Do IG, Lee J, Choi SJ, Kim S, Lee JE, Kim J, Lee S, Kim J. Discovery of ALK-PTPN3 gene fusion from human non-small cell lung carcinoma cell line using next generation RNA sequencing. Genes Chromosomes Cancer. 2012;51:590–597. doi: 10.1002/gcc.21945. [DOI] [PubMed] [Google Scholar]
- Katayama R, Friboulet L, Koike S, Lockerman EL, Khan TM, Gainor JF, Iafrate AJ, Takeuchi K, Taiji M, Okuno Y, Fujita N, Engelman JA, Shaw AT. Two novel ALK mutations mediate acquired resistance to the next-generation ALK inhibitor alectinib. Clin Cancer Res. 2014;20:5686–5696. doi: 10.1158/1078-0432.CCR-14-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Salzberg SL. TopHat-Fusion: An algorithm for discovery of novel fusion transcripts. Genome Biol. 2011;12:R72. doi: 10.1186/gb-2011-12-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Wu L, Zhao JC, Jin HJ, Yu J. TMPRSS2-ERG gene fusions induce prostate tumorigenesis by modulating microRNA miR-200c. Oncogene. 2014;33:5183–5192. doi: 10.1038/onc.2013.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsella M, Harismendy O, Nakano M, Frazer KA, Bafna V. Sensitive gene fusion detection using ambiguously mapping RNA-Seq read pairs. Bioinformatics. 2011;27:1068–1075. doi: 10.1093/bioinformatics/btr085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knezevich SR, Garnett MJ, Pysher TJ, Beckwith JB, Grundy PE, Sorensen PH. ETV6-NTRK3 gene fusions and trisomy 11 establish a histogenetic link between mesoblastic nephroma and congenital fibrosarcoma. Cancer Res. 1998a;58:5046–5048. [PubMed] [Google Scholar]
- Knezevich SR, McFadden DE, Tao W, Lim JF, Sorensen PH. A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nat Genet. 1998b;18:184–187. doi: 10.1038/ng0298-184. [DOI] [PubMed] [Google Scholar]
- Kohno T, Tsuta K, Tsuchihara K, Nakaoku T, Yoh K, Goto K. RET fusion gene: Translation to personalized lung cancer therapy. Cancer Sci. 2013;104:1396–1400. doi: 10.1111/cas.12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralik JM, Kranewitter W, Boesmueller H, Marschon R, Tschurtschenthaler G, Rumpold H, Wiesinger K, Erdel M, Petzer AL, Webersinke G. Characterization of a newly identified ETV6-NTRK3 fusion transcript in acute myeloid leukemia. Diagnostic Pathol. 2011;6:19. doi: 10.1186/1746-1596-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, Ou SH, Dezube BJ, Janne PA, Costa DB, Varella-Garcia M, Kim WH, Lynch TJ, Fidias P, Stubbs H, Engelman JA, Sequist LV, Tan W, Gandhi L, Mino-Kenudson M, Wei GC, Shreeve SM, Ratain MJ, Settleman J, Christensen JG, Haber DA, Wilner K, Salgia R, Shapiro GI, Clark JW, Iafrate AJ. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. New Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamant L, Gascoyne RD, Duplantier MM, Armstrong F, Raghab A, Chhanabhai M, Rajcan-Separovic E, Raghab J, Delsol G, Espinos E. Non-muscle myosin heavy chain (MYH9): A new partner fused to ALK in anaplastic large cell lymphoma. Genes Chromosomes Cancer. 2003;37:427–432. doi: 10.1002/gcc.10232. [DOI] [PubMed] [Google Scholar]
- Larrivee B, Prahst C, Gordon E, del Toro R, Mathivet T, Duarte A, Simons M, Eichmann A. ALK1 signaling inhibits angiogenesis by cooperating with the Notch pathway. Dev Cell. 2012;22:489–500. doi: 10.1016/j.devcel.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence B, Perez-Atayde A, Hibbard MK, Rubin BP, Dal Cin P, Pinkus JL, Pinkus GS, Xiao S, Yi ES, Fletcher CD, Fletcher JA. TPM3-ALK and TPM4-ALK oncogenes in inflammatory myofibroblastic tumors. Am J Pathol. 2000;157:377–384. doi: 10.1016/S0002-9440(10)64550-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Lee SE, Kang SY, Do IG, Lee S, Ha SY, Cho J, Kang WK, Jang J, Ou SH, Kim KM. Identification of ROS1 rearrangement in gastric adenocarcinoma. Cancer. 2013;119:1627–1635. doi: 10.1002/cncr.27967. [DOI] [PubMed] [Google Scholar]
- Leeman-Neill RJ, Kelly LM, Liu P, Brenner AV, Little MP, Bogdanova TI, Evdokimova VN, Hatch M, Zurnadzy LY, Nikiforova MN, Yue NJ, Zhang M, Mabuchi K, Tronko MD, Nikiforov YE. ETV6-NTRK3 is a common chromosomal rearrangement in radiation-associated thyroid cancer. Cancer. 2014;120:799–807. doi: 10.1002/cncr.28484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chien J, Smith DI, Ma J. FusionHunter: Identifying fusion transcripts in cancer using paired-end RNA-seq. Bioinformatics. 2011;27:1708–1710. doi: 10.1093/bioinformatics/btr265. [DOI] [PubMed] [Google Scholar]
- Li Z, Tognon CE, Godinho FJ, Yasaitis L, Hock H, Herschkowitz JI, Lannon CL, Cho E, Kim SJ, Bronson RT, Perou CM, Sorensen PH, Orkin SH. ETV6-NTRK3 fusion oncogene initiates breast cancer from committed mammary progenitors via activation of AP1 complex. Cancer Cell. 2007;12:542–558. doi: 10.1016/j.ccr.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin E, Li L, Guan Y, Soriano R, Rivers CS, Mohan S, Pandita A, Tang J, Modrusan Z. Exon array profiling detects EML4-ALK fusion in breast, colorectal, and non-small cell lung cancers. Mol Cancer Res. 2009;7:1466–1476. doi: 10.1158/1541-7786.MCR-08-0522. [DOI] [PubMed] [Google Scholar]
- Lipson D, Capelletti M, Yelensky R, Otto G, Parker A, Jarosz M, Curran JA, Balasubramanian S, Bloom T, Brennan KW, Donahue A, Downing SR, Frampton GM, Garcia L, Juhn F, Mitchell KC, White E, White J, Zwirko Z, Peretz T, Nechushtan H, Soussan-Gutman L, Kim J, Sasaki H, Kim HR, Park SI, Ercan D, Sheehan CE, Ross JS, Cronin MT, Janne PA, Stephens PJ. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18:382–384. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Westergard TD, Cashen A, Piwnica-Worms DR, Kunkle L, Vij R, Pham CG, DiPersio J, Cheng EH, Hsieh JJ. Proteasome inhibitors evoke latent tumor suppression programs in pro-B MLL leukemias through MLL-AF4. Cancer Cell. 2014;25:530–542. doi: 10.1016/j.ccr.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovly CM, Pao W. Escaping ALK inhibition: Mechanisms of and strategies to overcome resistance. Sci Transl Med. 2012;4:120ps122. doi: 10.1126/scitranslmed.3003728. [DOI] [PubMed] [Google Scholar]
- Lu XY, Cai Q, Ding K. Recent developments in the third generation inhibitors of Bcr-Abl for overriding T315I mutation. Curr Med Chem. 2011;18:2146–2157. doi: 10.2174/092986711795656135. [DOI] [PubMed] [Google Scholar]
- Ma Z, Hill DA, Collins MH, Morris SW, Sumegi J, Zhou M, Zuppan C, Bridge JA. Fusion of ALK to the Ran-binding protein 2 (RANBP2) gene in inflammatory myofibroblastic tumor. Genes Chromosomes Cancer. 2003;37:98–105. doi: 10.1002/gcc.10177. [DOI] [PubMed] [Google Scholar]
- Martin MJ, Melnyk N, Pollard M, Bowden M, Leong H, Podor TJ, Gleave M, Sorensen PH. The insulin-like growth factor I receptor is required for Akt activation and suppression of anoikis in cells transformed by the ETV6-NTRK3 chimeric tyrosine kinase. Mol Cell Biol. 2006;26:1754–1769. doi: 10.1128/MCB.26.5.1754-1769.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J, Bayrak-Toydemir P, Pyeritz RE. Hereditary hemorrhagic telangiectasia: An overview of diagnosis, management, and pathogenesis. Genet Med. 2011;13:607–616. doi: 10.1097/GIM.0b013e3182136d32. [DOI] [PubMed] [Google Scholar]
- McPherson A, Hormozdiari F, Zayed A, Giuliany R, Ha G, Sun MG, Griffith M, Heravi Moussavi A, Senz J, Melnyk N, Pacheco M, Marra MA, Hirst M, Nielsen TO, Sahinalp SC, Huntsman D, Shah SP. deFuse: An algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol. 2011a;7:e1001138. doi: 10.1371/journal.pcbi.1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson A, Wu C, Hajirasouliha I, Hormozdiari F, Hach F, Lapuk A, Volik S, Shah S, Collins C, Sahinalp SC. Comrad: Detection of expressed rearrangements by integrated analysis of RNA-Seq and low coverage genome sequence data. Bioinformatics. 2011b;27:1481–1488. doi: 10.1093/bioinformatics/btr184. [DOI] [PubMed] [Google Scholar]
- McPherson A, Wu C, Wyatt AW, Shah S, Collins C, Sahinalp SC. nFuse: Discovery of complex genomic rearrangements in cancer using high-throughput sequencing. Genome Res. 2012;22:2250–2261. doi: 10.1101/gr.136572.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meech SJ, McGavran L, Odom LF, Liang X, Meltesen L, Gump J, Wei Q, Carlsen S, Hunger SP. Unusual childhood extramedullary hematologic malignancy with natural killer cell properties that contains tropomyosin 4–anaplastic lymphoma kinase gene fusion. Blood. 2001;98:1209–1216. doi: 10.1182/blood.v98.4.1209. [DOI] [PubMed] [Google Scholar]
- Mehra R, Tomlins SA, Shen R, Nadeem O, Wang L, Wei JT, Pienta KJ, Ghosh D, Rubin MA, Chinnaiyan AM, Shah RB. Comprehensive assessment of TMPRSS2 and ETS family gene aberrations in clinically localized prostate cancer. Mod Pathol. 2007;20:538–544. doi: 10.1038/modpathol.3800769. [DOI] [PubMed] [Google Scholar]
- Mian AA, Metodieva A, Badura S, Khateb M, Ruimi N, Najajreh Y, Ottmann OG, Mahajna J, Ruthardt M. Allosteric inhibition enhances the efficacy of ABL kinase inhibitors to target unmutated BCR-ABL and BCR-ABL-T315I. BMC Cancer. 2012;12:411. doi: 10.1186/1471-2407-12-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SW, Kirstein MN, Valentine MB, Dittmer KG, Shapiro DN, Saltman DL, Look AT. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science. 1994;263:1281–1284. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- Mounir Z, Lin F, Lin VG, Korn JM, Yu Y, Valdez R, Aina OH, Buchwalter G, Jaffe AB, Korpal M, Zhu P, Brown M, Cardiff RD, Rocnik JL, Yang Y, Pagliarini R. TMPRSS2:ERG blocks neuroendocrine and luminal cell differentiation to maintain prostate cancer proliferation. Oncogene. 2014 doi: 10.1038/onc.2014.308. [DOI] [PubMed] [Google Scholar]
- Nakagawara A. Trk receptor tyrosine kinases: A bridge between cancer and neural development. Cancer Lett. 2001;169:107–114. doi: 10.1016/s0304-3835(01)00530-4. [DOI] [PubMed] [Google Scholar]
- Nam RK, Sugar L, Wang Z, Yang W, Kitching R, Klotz LH, Venkateswaran V, Narod SA, Seth A. Expression of TMPRSS2:ERG gene fusion in prostate cancer cells is an important prognostic factor for cancer progression. Cancer Biol Therapy. 2007a;6:40–45. doi: 10.4161/cbt.6.1.3489. [DOI] [PubMed] [Google Scholar]
- Nam RK, Sugar L, Yang W, Srivastava S, Klotz LH, Yang LY, Stanimirovic A, Encioiu E, Neill M, Loblaw DA, Trachtenberg J, Narod SA, Seth A. Expression of the TMPRSS2:ERG fusion gene predicts cancer recurrence after surgery for localised prostate cancer. Br J Cancer. 2007b;97:1690–1695. doi: 10.1038/sj.bjc.6604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nita-Lazar A, Saito-Benz H, White FM. Quantitative phosphoproteomics by mass spectrometry: Past, present, and future. Proteomics. 2008;8:4433–4443. doi: 10.1002/pmic.200800231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell PC, Hungerford DA. Chromosome studies on normal and leukemic human leukocytes. J Natl Cancer Institute. 1960;25:85–109. [PubMed] [Google Scholar]
- O'Hare T, Eide CA, Deininger MW. Bcr-Abl kinase domain mutations and the unsettled problem of Bcr-AblT315I: Looking into the future of controlling drug resistance in chronic myeloid leukemia. Clin Lymphoma Myeloma. 2007a;7(Suppl 3):S120–130. doi: 10.3816/clm.2007.s.012. [DOI] [PubMed] [Google Scholar]
- O'Hare T, Eide CA, Deininger MW. Bcr-Abl kinase domain mutations, drug resistance, and the road to a cure for chronic myeloid leukemia. Blood. 2007b;110:2242–2249. doi: 10.1182/blood-2007-03-066936. [DOI] [PubMed] [Google Scholar]
- O'Hare T, Shakespeare WC, Zhu X, Eide CA, Rivera VM, Wang F, Adrian LT, Zhou T, Huang WS, Xu Q, Metcalf CA, 3rd, Tyner JW, Loriaux MM, Corbin AS, Wardwell S, Ning Y, Keats JA, Wang Y, Sundaramoorthi R, Thomas M, Zhou D, Snodgrass J, Commodore L, Sawyer TK, Dalgarno DC, Deininger MW, Druker BJ, Clackson T. AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell. 2009;16:401–412. doi: 10.1016/j.ccr.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagopoulos I, Nilsson T, Domanski HA, Isaksson M, Lindblom P, Mertens F, Mandahl N. Fusion of the SEC31L1 and ALK genes in an inflammatory myofibroblastic tumor. Int J Cancer. 2006;118:1181–1186. doi: 10.1002/ijc.21490. [DOI] [PubMed] [Google Scholar]
- Parker BC, Zhang W. Fusion genes in solid tumors: An emerging target for cancer diagnosis and treatment. Chin J Cancer. 2013;32:594–603. doi: 10.5732/cjc.013.10178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perot G, Soubeyran I, Ribeiro A, Bonhomme B, Savagner F, Boutet-Bouzamondo N, Hostein I, Bonichon F, Godbert Y, Chibon F. Identification of a recurrent STRN/ALK fusion in thyroid carcinomas. PloS One. 2014;9:e87170. doi: 10.1371/journal.pone.0087170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovics G, Liu A, Shaheduzzaman S, Furusato B, Sun C, Chen Y, Nau M, Ravindranath L, Chen Y, Dobi A, Srikantan V, Sesterhenn IA, McLeod DG, Vahey M, Moul JW, Srivastava S. Frequent overexpression of ETS-related gene-1 (ERG1) in prostate cancer transcriptome. Oncogene. 2005;24:3847–3852. doi: 10.1038/sj.onc.1208518. [DOI] [PubMed] [Google Scholar]
- Piazza R, Pirola A, Spinelli R, Valletta S, Redaelli S, Magistroni V, Gambacorti-Passerini C. FusionAnalyser: A new graphical, event-driven tool for fusion rearrangements discovery. Nucleic Acids Res. 2012;40:e123. doi: 10.1093/nar/gks394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulford K, Lamant L, Morris SW, Butler LH, Wood KM, Stroud D, Delsol G, Mason DY. Detection of anaplastic lymphoma kinase (ALK) and nucleolar protein nucleophosmin (NPM)-ALK proteins in normal and neoplastic cells with the monoclonal antibody ALK1. Blood. 1997;89:1394–1404. [PubMed] [Google Scholar]
- Qadir MA, Zhan SH, Kwok B, Bruestle J, Drees B, Popescu OE, Sorensen PH. ChildSeq-RNA: A next-generation sequencing-based diagnostic assay to identify known fusion transcripts in childhood sarcomas. J Mol Diagn. 2014;16:361–370. doi: 10.1016/j.jmoldx.2014.01.002. [DOI] [PubMed] [Google Scholar]
- Redmond AM, Carroll JS. Defining and targeting transcription factors in cancer. Genome Biol. 2009;10:311. doi: 10.1186/gb-2009-10-7-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Tan ZP, Zhu X, Crosby K, Haack H, Ren JM, Beausoleil S, Moritz A, Innocenti G, Rush J, Zhang Y, Zhou XM, Gu TL, Yang YF, Comb MJ. Identification of anaplastic lymphoma kinase as a potential therapeutic target in ovarian cancer. Cancer Res. 2012;72:3312–3323. doi: 10.1158/0008-5472.CAN-11-3931. [DOI] [PubMed] [Google Scholar]
- Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, Hu Y, Tan Z, Stokes M, Sullivan L, Mitchell J, Wetzel R, Macneill J, Ren JM, Yuan J, Bakalarski CE, Villen J, Kornhauser JM, Smith B, Li D, Zhou X, Gygi SP, Gu TL, Polakiewicz RD, Rush J, Comb MJ. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- Rimkunas VM, Crosby KE, Li D, Hu Y, Kelly ME, Gu TL, Mack JS, Silver MR, Zhou X, Haack H. Analysis of receptor tyrosine kinase ROS1-positive tumors in non-small cell lung cancer: Identification of a FIG-ROS1 fusion. Clin Cancer Res. 2012;18:4449–4457. doi: 10.1158/1078-0432.CCR-11-3351. [DOI] [PubMed] [Google Scholar]
- Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang YL, Pei D, McCastlain K, Ding L, Lu C, Song G, Ma J, Becksfort J, Rusch M, Chen SC, Easton J, Cheng J, Boggs K, Santiago-Morales N, Iacobucci I, Fulton RS, Wen J, Valentine M, Cheng C, Paugh SW, Devidas M, Chen IM, Reshmi S, Smith A, Hedlund E, Gupta P, Nagahawatte P, Wu G, Chen X, Yergeau D, Vadodaria B, Mulder H, Winick NJ, Larsen EC, Carroll WL, Heerema NA, Carroll AJ, Grayson G, Tasian SK, Moore AS, Keller F, Frei-Jones M, Whitlock JA, Raetz EA, White DL, Hughes TP, Guidry Auvil JM, Smith MA, Marcucci G, Bloomfield CD, Mrozek K, Kohlschmidt J, Stock W, Kornblau SM, Konopleva M, Paietta E, Pui CH, Jeha S, Relling MV, Evans WE, Gerhard DS, Gastier-Foster JM, Mardis E, Wilson RK, Loh ML, Downing JR, Hunger SP, Willman CL, Zhang J, Mullighan CG. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. New Engl J Med. 2014;371:1005–1015. doi: 10.1056/NEJMoa1403088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodig SJ, Shapiro GI. Crizotinib, a small-molecule dual inhibitor of the c-Met and ALK receptor tyrosine kinases. Curr Opin Invest Drugs. 2010;11:1477–1490. [PubMed] [Google Scholar]
- Ross JS, Wang K, Gay L, Al-Rohil R, Rand JV, Jones DM, Lee HJ, Sheehan CE, Otto GA, Palmer G, Yelensky R, Lipson D, Morosini D, Hawryluk M, Catenacci DV, Miller VA, Churi C, Ali S, Stephens PJ. New routes to targeted therapy of intrahepatic cholangiocarcinomas revealed by next-generation sequencing. Oncologist. 2014;19:235–242. doi: 10.1634/theoncologist.2013-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin BP, Chen CJ, Morgan TW, Xiao S, Grier HE, Kozakewich HP, Perez-Atayde AR, Fletcher JA. Congenital mesoblastic nephroma t(12;15) is associated with ETV6-NTRK3 gene fusion: Cytogenetic and molecular relationship to congenital (infantile) fibrosarcoma. Am J Pathol. 1998;153:1451–1458. doi: 10.1016/S0002-9440(10)65732-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakarya O, Breu H, Radovich M, Chen Y, Wang YN, Barbacioru C, Utiramerur S, Whitley PP, Brockman JP, Vatta P, Zhang Z, Popescu L, Muller MW, Kudlingar V, Garg N, Li CY, Kong BS, Bodeau JP, Nutter RC, Gu J, Bramlett KS, Ichikawa JK, Hyland FC, Siddiqui AS. RNA-Seq mapping and detection of gene fusions with a suffix array algorithm. PLoS Comput Biol. 2012;8:e1002464. doi: 10.1371/journal.pcbi.1002464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saramaki OR, Harjula AE, Martikainen PM, Vessella RL, Tammela TL, Visakorpi T. TMPRSS2:ERG fusion identifies a subgroup of prostate cancers with a favorable prognosis. Clin Cancer Res. 2008;14:3395–3400. doi: 10.1158/1078-0432.CCR-07-2051. [DOI] [PubMed] [Google Scholar]
- Sboner A, Habegger L, Pflueger D, Terry S, Chen DZ, Rozowsky JS, Tewari AK, Kitabayashi N, Moss BJ, Chee MS, Demichelis F, Rubin MA, Gerstein MB. FusionSeq: A modular framework for finding gene fusions by analyzing paired-end RNA-sequencing data. Genome Biol. 2010;11:R104. doi: 10.1186/gb-2010-11-10-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah N, Lankerovich M, Lee H, Yoon JG, Schroeder B, Foltz G. Exploration of the gene fusion landscape of glioblastoma using transcriptome sequencing and copy number data. BMC Genomics. 2013;14:818. doi: 10.1186/1471-2164-14-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Birchmeier C, Nikawa J, O'Neill K, Rodgers L, Wigler M. Characterization of the ros1-gene products expressed in human glioblastoma cell lines. Oncogene Res. 1989;5:91–100. [PubMed] [Google Scholar]
- Shaw AT, Hsu PP, Awad MM, Engelman JA. Tyrosine kinase gene rearrangements in epithelial malignancies. Nat Rev Cancer. 2013a;13:772–787. doi: 10.1038/nrc3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AT, Kim DW, Nakagawa K, Seto T, Crino L, Ahn MJ, DePas T, Besse B, Solomon BJ, Blackhall F, Wu YL, Thomas M, O'Byrne KJ, Moro-Sibilot D, Camidge DR, Mok T, Hirsh V, Riely GJ, Iyer S, Tassell V, Polli A, Wilner KD, Janne PA. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. New Engl J Med. 2013b;368:2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- Shaw AT, Ou SH, Bang YJ, Camidge DR, Solomon BJ, Salgia R, Riely GJ, Varella-Garcia M, Shapiro GI, Costa DB, Doebele RC, Le LP, Zheng Z, Tan W, Stephenson P, Shreeve SM, Tye LM, Christensen JG, Wilner KD, Clark JW, Iafrate AJ. Crizotinib in ROS1-rearranged non-small-cell lung cancer. New Engl J Med. 2014;371:1963–1971. doi: 10.1056/NEJMoa1406766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota M, Fujimoto J, Semba T, Satoh H, Yamamoto T, Mori S. Hyperphosphorylation of a novel 80 kDa protein-tyrosine kinase similar to Ltk in a human Ki-1 lymphoma cell line, AMS3. Oncogene. 1994;9:1567–1574. [PubMed] [Google Scholar]
- Skalova A, Vanecek T, Sima R, Laco J, Weinreb I, Perez-Ordonez B, Starek I, Geierova M, Simpson RH, Passador-Santos F, Ryska A, Leivo I, Kinkor Z, Michal M. Mammary analogue secretory carcinoma of salivary glands, containing the ETV6-NTRK3 fusion gene: A hitherto undescribed salivary gland tumor entity. Am J Surg Pathol. 2010;34:599–608. doi: 10.1097/PAS.0b013e3181d9efcc. [DOI] [PubMed] [Google Scholar]
- Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, Bando M, Ohno S, Ishikawa Y, Aburatani H, Niki T, Sohara Y, Sugiyama Y, Mano H. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- Solomon BJ, Mok T, Kim DW, Wu YL, Nakagawa K, Mekhail T, Felip E, Cappuzzo F, Paolini J, Usari T, Iyer S, Reisman A, Wilner KD, Tursi J, Blackhall F, Investigators P. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. New Engl J Med. 2014;371:2167–2177. doi: 10.1056/NEJMoa1408440. [DOI] [PubMed] [Google Scholar]
- Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat Commun. 2014;5:4846. doi: 10.1038/ncomms5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suehara Y, Arcila M, Wang L, Hasanovic A, Ang D, Ito T, Kimura Y, Drilon A, Guha U, Rusch V, Kris MG, Zakowski MF, Rizvi N, Khanin R, Ladanyi M. Identification of KIF5B-RET and GOPC-ROS1 fusions in lung adenocarcinomas through a comprehensive mRNA-based screen for tyrosine kinase fusions. Clin Cancer Res. 2012;18:6599–6608. doi: 10.1158/1078-0432.CCR-12-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R, Love MI, Zemojtel T, Emde AK, Chung HR, Vingron M, Haas SA. Breakpointer: Using local mapping artifacts to support sequence breakpoint discovery from single-end reads. Bioinformatics. 2012;28:1024–1025. doi: 10.1093/bioinformatics/bts064. [DOI] [PubMed] [Google Scholar]
- Taipale M, Krykbaeva I, Whitesell L, Santagata S, Zhang J, Liu Q, Gray NS, Lindquist S. Chaperones as thermodynamic sensors of drug-target interactions reveal kinase inhibitor specificities in living cells. Nat Biotechnol. 2013;31:630–637. doi: 10.1038/nbt.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima T, Misawa S. Retinoic acid receptor alpha gene in t (15; 17) APL. Nihon rinsho Jap J Clin Med. 1992;50:1363–1368. [PubMed] [Google Scholar]
- Takeuchi K, Choi YL, Soda M, Inamura K, Togashi Y, Hatano S, Enomoto M, Takada S, Yamashita Y, Satoh Y, Okumura S, Nakagawa K, Ishikawa Y, Mano H. Multiplex reverse transcription-PCR screening for EML4-ALK fusion transcripts. Clin Cancer Res. 2008;14:6618–6624. doi: 10.1158/1078-0432.CCR-08-1018. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Choi YL, Togashi Y, Soda M, Hatano S, Inamura K, Takada S, Ueno T, Yamashita Y, Satoh Y, Okumura S, Nakagawa K, Ishikawa Y, Mano H. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res. 2009;15:3143–3149. doi: 10.1158/1078-0432.CCR-08-3248. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Soda M, Togashi Y, Ota Y, Sekiguchi Y, Hatano S, Asaka R, Noguchi M, Mano H. Identification of a novel fusion, SQSTM1-ALK, in ALK-positive large B-cell lymphoma. Haematologica. 2011;96:464–467. doi: 10.3324/haematol.2010.033514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K, Soda M, Togashi Y, Suzuki R, Sakata S, Hatano S, Asaka R, Hamanaka W, Ninomiya H, Uehara H, Lim Choi Y, Satoh Y, Okumura S, Nakagawa K, Mano H, Ishikawa Y. RET, ROS1 and ALK fusions in lung cancer. Nat Med. 2012;18:378–381. doi: 10.1038/nm.2658. [DOI] [PubMed] [Google Scholar]
- Taris M, Irani J, Blanchet P, Multigner L, Cathelineau X, Fromont G. ERG expression in prostate cancer: The prognostic paradox. Prostate. 2014;74:1481–1487. doi: 10.1002/pros.22863. [DOI] [PubMed] [Google Scholar]
- Thiele CJ, Li Z, McKee AE. On Trk–the TrkB signal transduction pathway is an increasingly important target in cancer biology. Clin Cancer Res. 2009;15:5962–5967. doi: 10.1158/1078-0432.CCR-08-0651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togashi Y, Soda M, Sakata S, Sugawara E, Hatano S, Asaka R, Nakajima T, Mano H, Takeuchi K. KLC1-ALK: A novel fusion in lung cancer identified using a formalin-fixed paraffin-embedded tissue only. PloS One. 2012;7:e31323. doi: 10.1371/journal.pone.0031323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognon C, Knezevich SR, Huntsman D, Roskelley CD, Melnyk N, Mathers JA, Becker L, Carneiro F, MacPherson N, Horsman D, Poremba C, Sorensen PH. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2:367–376. doi: 10.1016/s1535-6108(02)00180-0. [DOI] [PubMed] [Google Scholar]
- Tognon CE, Martin MJ, Moradian A, Trigo G, Rotblat B, Cheng SW, Pollard M, Uy E, Chow C, Carboni JM, Gottardis MM, Pollak M, Morin GB, Sorensen PH. A tripartite complex composed of ETV6-NTRK3, IRS1 and IGF1R is required for ETV6-NTRK3-mediated membrane localization and transformation. Oncogene. 2012;31:1334–1340. doi: 10.1038/onc.2011.323. [DOI] [PubMed] [Google Scholar]
- Tognon CE, Somasiri AM, Evdokimova VE, Trigo G, Uy EE, Melnyk N, Carboni JM, Gottardis MM, Roskelley CD, Pollak M, Sorensen PH. ETV6-NTRK3-mediated breast epithelial cell transformation is blocked by targeting the IGF1R signaling pathway. Cancer Res. 2011;71:1060–1070. doi: 10.1158/0008-5472.CAN-10-3096. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- Tort F, Pinyol M, Pulford K, Roncador G, Hernandez L, Nayach I, Kluin-Nelemans HC, Kluin P, Touriol C, Delsol G, Mason D, Campo E. Molecular characterization of a new ALK translocation involving moesin (MSN-ALK) in anaplastic large cell lymphoma. Lab Invest. 2001;81:419–426. doi: 10.1038/labinvest.3780249. [DOI] [PubMed] [Google Scholar]
- Vaishnavi A, Capelletti M, Le AT, Kako S, Butaney M, Ercan D, Mahale S, Davies KD, Aisner DL, Pilling AB, Berge EM, Kim J, Sasaki H, Park SI, Kryukov G, Garraway LA, Hammerman PS, Haas J, Andrews SW, Lipson D, Stephens PJ, Miller VA, Varella-Garcia M, Janne PA, Doebele RC. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med. 2013;19:1469–1472. doi: 10.1038/nm.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, He X, Mieczkowski P, Grimm SA, Perou CM, MacLeod JN, Chiang DY, Prins JF, Liu J. MapSplice: Accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010;38:e178. doi: 10.1093/nar/gkq622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Xia J, Jia P, Pao W, Zhao Z. Application of next generation sequencing to human gene fusion detection: Computational tools, features and perspectives. Brief Bioinformatics. 2013;14:506–519. doi: 10.1093/bib/bbs044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner T, He J, Yelensky R, Esteve-Puig R, Botton T, Yeh I, Lipson D, Otto G, Brennan K, Murali R, Garrido M, Miller VA, Ross JS, Berger MF, Sparatta A, Palmedo G, Cerroni L, Busam KJ, Kutzner H, Cronin MT, Stephens PJ, Bastian BC. Kinase fusions are frequent in Spitz tumours and spitzoid melanomas. Nat Commun. 2014;5:3116. doi: 10.1038/ncomms4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnes M, Lissbrant E, Damber JE, Stenman G. Molecular genetic analyses of the TMPRSS2-ERG and TMPRSS2-ETV1 gene fusions in 50 cases of prostate cancer. Oncol Rep. 2007;17:1033–1036. [PubMed] [Google Scholar]
- Wu G, Diaz AK, Paugh BS, Rankin SL, Ju B, Li Y, Zhu X, Qu C, Chen X, Zhang J, Easton J, Edmonson M, Ma X, Lu C, Nagahawatte P, Hedlund E, Rusch M, Pounds S, Lin T, Onar-Thomas A, Huether R, Kriwacki R, Parker M, Gupta P, Becksfort J, Wei L, Mulder HL, Boggs K, Vadodaria B, Yergeau D, Russell JC, Ochoa K, Fulton RS, Fulton LL, Jones C, Boop FA, Broniscer A, Wetmore C, Gajjar A, Ding L, Mardis ER, Wilson RK, Taylor MR, Downing JR, Ellison DW, Zhang J, Baker SJ St. Jude Children's Research Hospital-Washington University Pediatric Cancer Genome, P. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet. 2014;46:444–450. doi: 10.1038/ng.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorukoglu D, Hach F, Swanson L, Collins CC, Birol I, Sahinalp SC. Dissect: Detection and characterization of novel structural alterations in transcribed sequences. Bioinformatics. 2012;28:i179–187. doi: 10.1093/bioinformatics/bts214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A, Shibata T, Tsuta K, Watanabe S, Tsuda H. Inflammatory myofibroblastic tumour of the lung with a novel PPFIBP1-ALK fusion variant. Histopathology. 2013;63:881–883. doi: 10.1111/his.12218. [DOI] [PubMed] [Google Scholar]
- Zabriskie MS, Eide CA, Tantravahi SK, Vellore NA, Estrada J, Nicolini FE, Khoury HJ, Larson RA, Konopleva M, Cortes JE, Kantarjian H, Jabbour EJ, Kornblau SM, Lipton JH, Rea D, Stenke L, Barbany G, Lange T, Hernandez-Boluda JC, Ossenkoppele GJ, Press RD, Chuah C, Goldberg SL, Wetzler M, Mahon FX, Etienne G, Baccarani M, Soverini S, Rosti G, Rousselot P, Friedman R, Deininger M, Reynolds KR, Heaton WL, Eiring AM, Pomicter AD, Khorashad JS, Kelley TW, Baron R, Druker BJ, Deininger MW, O'Hare T. BCR-ABL1 compound mutations combining key kinase domain positions confer clinical resistance to ponatinib in Ph chromosome-positive leukemia. Cancer Cell. 2014;26:428–442. doi: 10.1016/j.ccr.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wu G, Miller CP, Tatevossian RG, Dalton JD, Tang B, Orisme W, Punchihewa C, Parker M, Qaddoumi I, Boop FA, Lu C, Kandoth C, Ding L, Lee R, Huether R, Chen X, Hedlund E, Nagahawatte P, Rusch M, Boggs K, Cheng J, Becksfort J, Ma J, Song G, Li Y, Wei L, Wang J, Shurtleff S, Easton J, Zhao D, Fulton RS, Fulton LL, Dooling DJ, Vadodaria B, Mulder HL, Tang C, Ochoa K, Mullighan CG, Gajjar A, Kriwacki R, Sheer D, Gilbertson RJ, Mardis ER, Wilson RK, Downing JR, Baker SJ, Ellison DW St. Jude Children's Research Hospital-Washington University Pediatric Cancer Genome, P. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet. 2013;45:602–612. doi: 10.1038/ng.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]