Abstract
Botanical, mycological, zoological, and prokaryotic species names follow the Linnaean format, consisting of an italicized Latinized binomen with a capitalized genus name and a lower case species epithet (e.g., Homo sapiens). Virus species names, however, do not follow a uniform format, and, even when binomial, are not Linnaean in style. In this thought exercise, we attempted to convert all currently official names of species included in the virus family Arenaviridae and the virus order Mononegavirales to Linnaean binomials, and to identify and address associated challenges and concerns. Surprisingly, this endeavor was not as complicated or time-consuming as even the authors of this article expected when conceiving the experiment.
Keywords: Arenaviridae, binomials, ICTV, International Committee on Taxonomy of Viruses, Mononegavirales, virus nomenclature, virus taxonomy
Botanical, mycological, zoological, and prokaryotic species names follow a Latinized binomi(n)al format (i.e., binomial nomenclature) first introduced, formally and systematically, by Carl Linnaeus in 1753 (Linnaeus 1753). This format consists of two italicized words (a binomen or binary combination or scientific/Latin name), with the first capitalized word naming the genus to which the species belongs (“genus name”) and the second lower case word denoting the name/species epithet”) (International Committee on Systematic Bacteriology 1992; International Association for Plant Taxonomy 2011; International Commission on Zoological Nomenclature 2012). Typical examples for binomial species names are Arabidopsis thaliana, Saccharomyces cerevisiae, Homo sapiens, or Escherichia coli. Such species names have several advantages over most alternative species-naming conventions, including: (i) they are internationally recognizable as they do not change in typography even in texts using non-Latin alphabets or other scripts; (ii) they are easier to remember than alphanumerical schemes or “names”; (iii) they may be explicative, that is, each part of the binomial name may relate specific characteristics of species members; (iv) they are in an extinct language that evolves very slowly if at all, ensuring the stability of names and of their etymology; and (v) they frequently differ from the names of species members (e.g., Homo sapiens humans), thereby emphasizing the logical difference between taxon (a concept of the mind) and taxon member (a concrete physical entity) (Drebot et al. 2002; Calisher and Mahy 2003; Van Regenmortel 2003, 2006, 2007, 2016a; Kuhn and Jahrling 2010).
humans), thereby emphasizing the logical difference between taxon (a concept of the mind) and taxon member (a concrete physical entity) (Drebot et al. 2002; Calisher and Mahy 2003; Van Regenmortel 2003, 2006, 2007, 2016a; Kuhn and Jahrling 2010).
In contrast, viral species names do not follow a uniform format, except for a requirement to be italicized and to have the first word capitalized (King et al. 2012; International Committee on Taxonomy of Viruses 2013). The few existing binomial virus species names are not Latinized and typically begin with the capitalized species epithet followed by the lower case name of the genus to which the species belongs (e.g., Lassa mammarenavirus), thus inverting the order of the Linnaean format. The mismatch between the various formats of virus species names and the species name format of all other official taxonomies reflects the complicated history of virus taxonomy and nomenclature.
Importantly, the lack of a uniform virus species-naming scheme impedes the development of comprehensive biological databases. Because all species names currently in use for animals, fungi, plants, prokaryotes, and protozoans follow the Linnaean binomial format, software can easily recognize the genus name and species epithet in a properly formatted database entry: the first word in the < species > entry field will be the genus name, the second, following a space, will be the species epithet. A virus species name that is not following the Linnaean format will be parsed erroneously. For instance, a species name such as the current “Measles virus” will lead the software to assume that the species epithet “Measles” is a genus and that the word “virus” is a species epithet. Likewise, a species name such as the current non-Latinized binomial “Lassa mammarenavirus” will lead the software to assume that “Lassa” and “mammarenavirus” are genus and species epithet, respectively, rather than vice versa (“Mammarenavirus” is the genus in current virus taxonomy). Consequently, virus taxonomy is currently excluded from many bioinformatic projects, such as BioCode (Greuter et al. 2011). Alignment of virus taxonomy with other taxonomies was originally considered by the founders of the International Committee on Nomenclature of Viruses (ICNV), the predecessor of the International Committee on Taxonomy of Viruses (ICTV). Numerous Linnaean species names were published in the first and second ICNV/ICTV Reports (Wildy 1971; Fenner 1976), but this practice was abandoned thereafter. Linnaean virus species names have been commonly used in the past [e.g., “Herpesvirus simiae” (Hummeler et al. 1959; Huemer et al. 2003)] or are currently used [e.g., “Pandoravirus salinus” (Philippe et al. 2013)], but none are officially approved at present. Indeed, Matthews stated in 1985 that “[n]othing releases adrenalin more readily for many virologists than the suggestion that virus species names should be latinized [sic]” (Matthews 1985a), a view that was especially prominent among plant virologists at the time (Matthews 1983, 1985a, 1985b; Milne 1984).
During the most recent, largest-to-date ICTV meeting in February 2016, a straw poll among the attending ICTV Study Group chairs revealed an interest in discussing the advantages and disadvantages of virus species name conversion to the Linnaean format (International Committee on Taxonomy of Viruses 2016). This majority vote to revisit the issue somewhat contrasts with the published view that such a conversion “is unlikely to be a welcome alternative [to current virus species names]” (Van Regenmortel 2016a). Whether such a conversion is even plausible is unclear. Over the years, a number of potential challenges, conceptual and practical, have been postulated. One conceptual debate is whether viruses are alive, that is, considered to be organisms (for contrasting views, see Koonin and Starokadomskyy 2016; Van Regenmortel 2016a, 2016b), and, therefore, whether or not Linnaean virus species names would give the impression that this dispute has been settled in favor of viruses being part of the living world. The most relevant practical concern is the fear that Latinization is so complex that only a few experts, not accessible to typical ICTV Study Groups, could achieve it and that converting the existing virus species names would become a Herculean endeavor (Van Regenmortel 2016a).
Here we set the conceptual questions aside and, as a proof of principle, demonstrate the results of a practical team effort of devising Linnaean binomial species names for all currently accepted species of the family Arenaviridae and the order Mononegavirales. Without attempting to impose the resulting names as a new standard, we seek to provide a more solid basis for consideration of the principle of applying the Linnaean format to virus nomenclature using a case study of how such conversion could be accomplished if the virology community chooses to do so.
The family Arenaviridae currently includes two ICTV-approved genera and 34 species for viruses that infect bats, eulipotyphla, rodents, and snakes (Radoshitzky et al. 2015; Adams et al. 2016). All arenavirus species names are non-Latinized binomials, which led us to believe that these names could be easily converted to the Linnaean format. The order Mononegavirales currently includes eight ICTV-approved families, 34 genera, and 141 species for viruses that infect animals, plants, and fungi (Adams et al. 2016; Afonso et al. 2016). Mononegaviral species naming conventions differ among families and, as a whole, reflect the current nonuniformity of species naming across viral taxa; it was a priori unclear whether a consistent conversion is practically achievable in this case.
FORMAL REQUIREMENTS FOR LINNAEAN BINOMIALS
Linnaean binomials are Latinized, that is, both name components follow Latin grammar, although the components themselves can be derived from languages other than Latin. The genus name must be treated as a Latin singular noun in the nominative case, whereas the species epithet may be an adjective or a noun, in both cases either in the nominative or genitive case. Correct formation of a species name, therefore, requires a basic understanding of Latin grammar. However, the grammatical complexities of virus species names are less intricate than those of nonviral taxonomies; by convention, all virus genus names end in the suffix “-virus.” Consequently, the question of the declension of the cognate species epithet is simplified to only one gender, as the Latin word “virus,” meaning slime or poison, is a noun of the neuter gender (unexpectedly for a word ending in “ us”).
us”).
SPECIES NAME CONVERSION: SPECIFIC CHALLENGES AND POSSIBLE SOLUTIONS
Conversion of True Non-Latinized Binomial Species Names to Linnaean Binomials
Most currently accepted arenaviral and several currently accepted mononegaviral species names follow a true binomial, albeit non-Linnaean, format originally proposed by van Regenmortel et al. (2010). While italicized names are not Latinized, these names consist of only two words, with the first word a capitalized species epithet and the second word a lower case genus name (e.g., Lassa mammarenavirus, Human metapneumovirus). Most viral species epithets in current non-Latinized binomials within and outside of Arenaviridae and Mononegavirales refer to geographic areas (e.g., “Lassa,” “Zaire”), to host taxa (e.g., Sclerotinia), or to vernacular host names (e.g., alethinophid, human). Conversion to the Linnaean format is, therefore, straightforward and achieved by switching the word order and converting the species epithet to a Latinized word in the proper case (e.g., zairense: nominative singular neuter of the suffix “-ensis,” indicating a place of origin, attached to the root “zaire”; sclerotiniae: genitive singular of “sclerotinia,” after the host genus; and hominis: genitive singular of “homo,” also after the host genus). One encountered problem is the presence of diacritical marks in several species names (e.g., Junín mammarenavirus, Sabiá mammarenavirus). Conversion to Linnaean binomials can be achieved, however, by either dropping diacritical marks or by changing the name to contain a geographic location not requiring a diacritic (note also that the ICTV recently ratified a Code rule to disallow the use of diacritics and other special characters in new taxon names). Examples for this type of possible conversion are shown in Tables 1 and 2 in dark green (online version only).
Table 1.
Possible Linnaean binomial names for all currently accepted arenaviral species
| Current ICTV-approved | Possible converted Linnaean | (Unchanged) member virus species name |
|---|---|---|
| species name | species namea | name and abbreviation |
| Genus Mammarenavirus | ||
| Allpahuayo mammarenavirus | Mammarenavirus allpahuayense | Allpahuayo virus (ALLV) |
| AmaparÍ mammarenavirus | Mammarenavirus amapariense | Amapar í virus (AMAV) |
| Bear Canyon mammarenavirus | Mammarenavirus saltusursinenseb | Bear Canyon virus (BCNV) |
| Chapare mammarenavirus | Mammarenavirus chaparense | Chapare virus (CHAPV) |
| Cupixi mammarenavirus | Mammarenavirus cupixense | Cupixi virus (CUPXV) |
| Flexal mammarenavirus | Mammarenavirus flexalense | Flexal virus (FLEV) |
| Gairo mammarenavirus | Mammarenavirus gairense | Gairo virus (GAIV) |
| Guanarito mammarenavirus | Mammarenavirus guanaritense | Guanarito virus (GTOV) |
| Ippy mammarenavirus | Mammarenavirus ippyense | Ippy virus (IPPYV) |
| Jun ín mammarenavirus | Mammarenavirus iuninense | Junín virus (JUNV) |
| Lassa mammarenavirus | Mammarenavirus lassaense | Lassa virus (LASV) |
| Latino mammarenavirus | Mammarenavirus latinum | Latino virus (LATV) |
| Lujo mammarenavirus | Mammarenavirus luioense | Lujo virus (LUJV) |
| Luna mammarenavirus | Mammarenavirus lunaense | Luna virus (LUAV) |
| Lunk mammarenavirus | Mammarenavirus lunkense | Lunk virus (LNKV) |
| Lymphocytic choriomeningitis mammarenavirus | Mammarenavirus choriomeningitidis | lymphocytic choriomeningitis virus (LCMV) |
| Machupo mammarenavirus | Mammarenavirus machupense | Machupo virus (MACV) |
| Mariental mammarenavirus | Mammarenavirus marientalense | Mariental virus (MRLV) |
| Merino Walk mammarenavirus | Mammarenavirus viamerinense | Merino Walk virus (MRWV) |
| Mobala mammarenavirus | Mammarenavirus mobalaense | Mobala virus (MOBV) |
| Mopeia mammarenavirus | Mammarenavirus mopeiense | Mopeia virus (MOPV), Morogoro virus (MORV) |
| Okahandja mammarenavirus | Mammarenavirus okahandiense | Okahandja virus (OKAV) |
| Oliveros mammarenavirus | Mammarenavirus oliverosense | Oliveros virus (OLVV) |
| Paraná mammarenavirus | Mammarenavirus paranaense | Paraná virus (PRAV) |
| Pichindé mammarenavirus | Mammarenavirus pichindense | Pichindé virus (PICHV) |
| Pirital mammarenavirus | Mammarenavirus piritalense | Pirital virus (PIRV) |
| Sabiá mammarenavirus | Mammarenavirus sabiaense | Sabiá virus (SBAV) |
| Tacaribe mammarenavirus | Mammarenavirus tacaribense | Tacaribe virus (TCRV) |
| Tamiami mammarenavirus | Mammarenavirus tamiamense | Tamiami virus (TMMV) |
| Wenzhou mammarenavirus | Mammarenavirus wenzhouense | Wēnzhōu virus (WENV) |
| Whitewater Arroyo mammarenavirus | Mammarenavirus arroyense | Catarina virus (CTNV), Big Brushy Tank virus (BBRTV), Skinner Tank virus (SKTV), Tonto Creek virus (TTCV), Whitewater Arroyo virus (WWAV) |
| Genus Reptarenavirus | ||
| Alethinophid 1 reptarenavirus | Reptarenavirus portaureaec | Golden Gate virus (GOGV) |
| Alethinophid 2 reptarenavirus | Reptarenavirus helsinkii | ROUT virus (ROUTV), University of Helsinki virus (UHV) |
| Alethinophid 3 reptarenavirus | Reptarenavirus californiae | CAS virus (CASV) |
Note: Dark green (online version only) depicts current true binomial names and their Linnaean counterparts; light green (online version only) depicts current binomial-like names and their Linnaean counterparts.
These names are for illustration purposes only and may not be the same as the names that ICTV Study Groups would officially propose if a Linnaean binomial species naming convention were implemented.
saltusursinense “from the Bear Canyon.”
“from the Bear Canyon.”
portaureae “of the Golden Gate.”
“of the Golden Gate.”
Table 2.
Possible Linnaean binomial names for all currently accepted mononegaviral species
| Current ICTV-approved | Possible converted Linnaean | (Unchanged) member virus |
|---|---|---|
| species name | species namea | name and abbreviation |
| Family Bornaviridae | ||
| Elapid 1 bornavirus | Bornavirus elapsoideaeb | Loveridge’s garter snake virus 1 (LGSV-1) |
| Mammalian 1 bornavirus | Bornavirus crocidurae | Borna disease viruses 1/2 (BoDV-1/2) |
| Psittaciform 1 bornavirus | Bornavirus alphapsittaciforme | parrot bornaviruses 1/2/3/4/7 (PaBV-1/2/3/4/7) |
| Psittaciform 2 bornavirus | Bornavirus betapsittaciforme | parrot bornavirus 5 (PaBV-5) |
| Passeriform 1 bornavirus | Bornavirus alphapasseriforme | canary bornaviruses 1/2/3 (CnBV-1/2/3) |
| Passeriform 2 bornavirus | Bornavirus betapasseriforme | estrildid finch bornavirus 1 (EsBV-1) |
| Waterbird 1 bornavirus | Bornavirus avisaquaticae | aquatic bird bornaviruses 1/2 (ABBV-1/2) |
| Family Filoviridae | ||
| Lloviu cuevavirus | Cuevavirus lloviense | Lloviu virus (LLOV) |
| Bundibugyo ebolavirus | Ebolavirus bundibugyoense | Bundibugyo virus (BDBV) |
| Reston ebolavirus | Ebolavirus restonense | Reston virus (RESTV) |
| Sudan ebolavirus | Ebolavirus sudanense | Sudan virus (SUDV) |
| Taï Forest ebolavirus | Ebolavirus silvataiense | Taï Forest virus (TAFV) |
| Zaire ebolavirus | Ebolavirus zairense | Ebola virus (EBOV) |
| Marburg marburgvirus | Marburgvirus marburgense | Marburg virus (MARV), Ravn virus (RAVV) |
| Family Mymonaviridae | ||
| Sclerotinia sclerotimonavirus | Sclerotimonavirus sclerotiniae | Sclerotinia sclerotiorum negative-stranded RNA virus 1 (SsNSRV-1) |
| Family Nyamiviridae | ||
| Midway nyavirus | Nyavirus midwayense | Midway virus (MIDWV) |
| Nyamanini nyavirus | Nyavirus nyamaninense | Nyamanini virus (NYMV) |
| Sierra Nevada nyavirus | Nyavirus sierranevadense | Sierra Nevada virus (SNVV) |
| Soybean cyst nematode socyvirus | Socyvirus heteroderae | soybean cyst nematode virus 1 (SbCNV-1) |
| Family Paramyxoviridae | ||
| Atlantic salmon paramyxovirus | Aquaparamyxovirus salmonis | Atlantic salmon paramyxovirus (AsaPV) |
| Newcastle disease virus | Avulavirus avisprimum | avian paramyxovirus 1 (APMV-1) |
| Avian paramyxovirus 2 | Avulavirus avissecundum | avian paramyxovirus 2 (APMV-2) |
| Avian paramyxovirus 3 | Avulavirus avistertium | avian paramyxovirus 3 (APMV-3) |
| Avian paramyxovirus 4 | Avulavirus avisquartum | avian paramyxovirus 4 (APMV-4) |
| Avian paramyxovirus 5 | Avulavirus avisquintum | avian paramyxovirus 5 (APMV-5) |
| Avian paramyxovirus 6 | Avulavirus avissextum | avian paramyxovirus 6 (APMV-6) |
| Avian paramyxovirus 7 | Avulavirus avisseptimum | avian paramyxovirus 7 (APMV-7) |
| Avian paramyxovirus 8 | Avulavirus avisoctavum | avian paramyxovirus 8 (APMV-8) |
| Avian paramyxovirus 9 | Avulavirus avisnonum | avian paramyxovirus 9 (APMV-9) |
| Avian paramyxovirus 10 | Avulavirus avisdecimum | avian paramyxovirus 10 (APMV-10) |
| Avian paramyxovirus 11 | Avulavirus avisundecimum | avian paramyxovirus 11 (APMV-11) |
| Avian paramyxovirus 12 | Avulavirus avisduodecimum | avian paramyxovirus 12 (APMV-12) |
| Fer-de-Lance paramyxovirus | Ferlavirus bothropsi | Fer-de-Lance virus (FDLV) |
| Cedar henipavirus | Henipavirus cedarense | Cedar virus (CedV) |
| Ghanaian bat henipavirus | Henipavirus ghanense | Kumasi virus (KV) |
| Hendra virus | Henipavirus hendrense | Hendra virus (HeV) |
| Mojiang henipavirus | Henipavirus moiangense | Mòjiāng virus (MojV) |
| Nipah virus | Henipavirus nipahense | Nipah virus (NiV) |
| Canine distemper virus | Morbillivirus canis | canine distemper virus (CDV) |
| Cetacean morbillivirus | Morbillivirus cetaceae | cetacean morbillivirus (CeMV) |
| Feline morbillivirus | Morbillivirus felis | feline morbillivirus (FeMV) |
| Measles virus | Morbillivirus hominis | measles virus (MeV) |
| Peste-des-petits-ruminants virus | Morbillivirus caprinae | peste-des-petits-ruminants virus (PPRV) |
| Phocine distemper virus | Morbillivirus phocinae | phocine distemper virus (PDV) |
| Rinderpest virus | Morbillivirus bovinae | rinderpest virus (RPV) |
| Bovine parainfluenza virus 3 | Respirovirus bovistertium | bovine parainfluenza virus 3 (BPIV-3) |
| Human parainfluenza virus 1 | Respirovirus parainfluenzaeprimum | human parainfluenza virus 1 (HPIV-1) |
| Human parainfluenza virus 3 | Respirovirus parainfluenzaetertium | human parainfluenza virus 3 (HPIV-3) |
| Porcine parainfluenza virus 1 | Respirovirus suisprimum | porcine parainfluenza virus 1 (PPIV-1) |
| Sendai virus | Respirovirus muris | Sendai virus (SeV) |
| Human parainfluenza virus 2 | Rubulavirus parainfluenzaesecundum | human parainfluenza virus 2 (HPIV-2) |
| Human parainfluenza virus 4 | Rubulavirus parainfluenzaequartum | human parainfluenza viruses 4a/b (HPIV-4a/b) |
| Mapuera virus | Rubulavirus mapuerense | Mapuera virus (MapV) |
| Mumps virus | Rubulavirus parotitidisc | mumps virus (MuV), bat mumps virus |
| Parainfluenza virus 5 | Rubulavirus parainfluenzaequintum | parainfluenza virus 5 (PIV-5) |
| Porcine rubulavirus | Rubulavirus suis | La Piedad Michoacán Mexico virus (LPMV) |
| Simian virus 41 | Rubulavirus macacae | simian virus 41 |
| Family Pneumoviridae | ||
| Avian metapneumovirus | Metapneumovirus avis | turkey rhinotracheitis virus |
| Human metapneumovirus | Metapneumovirus hominis | human metapneumovirus (HMPV) |
| Bovine respiratory syncytial virus | Orthopneumovirus bovis | bovine respiratory syncytial virus (BRSV) |
| Human respiratory syncytial virus | Orthopneumovirus hominis | human respiratory syncytial viruses A2/B1/S2 (HRSV-A2/B1/S2) |
| Murine pneumonia virus | Orthopneumovirus muris | murine pneumonia virus (MPV) |
| Family Rhabdoviridae | ||
| Alfalfa dwarf cytorhabdovirus | Cytorhabdovirus medicagonis | alfalfa dwarf virus (ADV) |
| Barley yellow striate mosaic cytorhabdovirus | Cytorhabdovirus hordei | barley yellow striate mosaic virus (BYSMV), maize sterile stunt virus (MSSV), wheat chlorotic streak virus (WCSV) |
| Broccoli necrotic yellows cytorhabdovirus | Cytorhabdovirus brassicae | broccoli necrotic yellows virus (BNYV) |
| Festuca leaf streak cytorhabdovirus | Cytorhabdovirus festucae | festuca leaf streak virus (FLSV) |
| Lettuce necrotic yellows cytorhabdovirus | Cytorhabdovirus lactucanecanted | lettuce necrotic yellows virus (LNYV) |
| Lettuce yellow mottle cytorhabdovirus | Cytorhabdovirus lactucamaculantee | lettuce yellow mottle virus (LYMoV) |
| Northern cereal mosaic cytorhabdovirus | Cytorhabdovirus cerealisboreif | northern cereal mosaic virus (NCMV) |
| Sonchus cytorhabdovirus 1 | Cytorhabdovirus sonchi | sonchus virus (SonV) |
| Strawberry crinkle cytorhabdovirus | Cytorhabdovirus fragariae | strawberry crinkle virus (SCV) |
| Wheat American striate mosaic cytorhabdovirus | Cytorhabdovirus tritici | wheat American striate mosaic virus (WASMV) |
| Coffee ringspot dichorhavirus | Dichorhavirus coffeae | coffee ringspot virus (CoRSV) |
| Orchid fleck dichorhavirus | Dichorhavirus orchidaceae | orchid fleck virus (OFV) |
| Adelaide River ephemorovirus | Ephemerovirus flumenadelaidense | Adelaide River virus (ARV) |
| Berrimah ephemerovirus | Ephemerovirus berrimahense | Berrimah virus (BRMV) |
| Bovine fever ephemerovirus | Ephemerovirus bubulifebrisg | bovine ephemeral fever virus (BEFV) |
| Kotonkan ephemerovirus | Ephemerovirus kotonkani | kotonkan virus (KOTV) |
| Obodhiang ephemerovirus | Ephemerovirus obodhiangense | Obodhiang virus (OBOV) |
| Aravan lyssavirus | Lyssavirus aravanense | Aravan virus (ARAV) |
| Australian bat lyssavirus | Lyssavirus ballinense | Australian bat lyssavirus (ABLV) |
| Bokeloh bat lyssavirus | Lyssavirus bokelohense | Bokeloh bat lyssavirus (BBLV) |
| Duvenhage lyssavirus | Lyssavirus duvenhagei | Duvenhage virus (DUVV) |
| European bat 1 lyssavirus | Lyssavirus alphaeuropense | European bat lyssavirus 1 (EBLV-1) |
| European bat 2 lyssavirus | Lyssavirus betaeuropense | European bat lyssavirus 2 (EBLV-2) |
| Ikoma lyssavirus | Lyssavirus ikomense | Ikoma lyssavirus (IKOV) |
| Irkut lyssavirus | Lyssavirus irkutense | Irkut virus (IRKV) |
| Khujand lyssavirus | Lyssavirus khuiandense | Khujand virus (KHUV) |
| Lagos bat lyssavirus | Lyssavirus lagosense | Lagos bat virus (LBV) |
| Mokola lyssavirus | Lyssavirus mokolense | Mokola virus (MOKV) |
| Rabies lyssavirus | Lyssavirus rabies | rabies virus (RABV) |
| Shimoni bat lyssavirus | Lyssavirus shimonense | Shimoni bat virus (SHIBV) |
| West Caucasian bat lyssavirus | Lyssavirus occidenscaucasense | West Caucasian bat virus (WCBV) |
| Hirame novirhabdovirus | Novirhabdovirus paralichthyos | Hirame rhabdovirus (HIRV) |
| Oncorhynchus 1 novirhabdovirus | Novirhabdovirus salmonidae | infectious hematopoietic necrosis virus (IHNV) |
| Snakehead novirhabdovirus | Novirhabdovirus channae | snakehead rhabdovirus (SHRV) |
| Oncorhynchus 2 novirhabdovirus | Novirhabdovirus piscicidanteh | viral hemorrhagic septicemia virus (VHSV) |
| Datura yellow vein nucleorhabdovirus | Nucleorhabdovirus daturae | datura yellow vein virus (DYVV) |
| Eggplant mottled nucleorhabdovirus | Nucleorhabdovirus solanimelongenae | eggplant mottled dwarf virus (EMDV) |
| Maize fine streak nucleorhabdovirus | Nucleorhabdovirus zealineante | maize fine streak virus (MSFV) |
| Maize Iranian mosaic nucleorhabdovirus | Nucleorhabdovirus zeairanense | maize Iranian mosaic virus (MIMV) |
| Maize mosaic nucleorhabdovirus | Nucleorhabdovirus zeamosaicante | maize mosaic virus (MMV) |
| Potato yellow dwarf nucleorhabdovirus | Nucleorhabdovirus solanituberosi | potato yellow dwarf virus (PYDV) |
| Rice yellow stunt nucleorhabdovirus | Nucleorhabdovirus oryzae | rice yellow stunt virus (RYSV), rice transitory yellowing virus (RTYV) |
| Sonchus yellow net nucleorhabdovirus | Nucleorhabdovirus retesonchi | sonchus yellow net virus (SYNV) |
| Sowthistle yellow vein nucleorhabdovirus | Nucleorhabdovirus venasonchi | sowthistle yellow vein virus (SYVV) |
| Taro vein chlorosis nucleorhabdovirus | Nucleorhabdovirus colocasiae | taro vein chlorosis virus (TaVCV) |
| Anguillid perhabdovirus | Perhabdovirus anguillae | eel virus European X (EVEX), eel virus American (EVA) |
| Perch perhabdovirus | Perhabdovirus percae | perch rhabdovirus (PRV) |
| Sea trout perhabdovirus | Perhabdovirus truttae | lake trout rhabdovirus (LTRV), Swedish sea trout rhabdovirus (SSTV) |
| Drosophila affinis sigmavirus | Sigmavirus drosophilaeaffinis | Drosophila affinis sigmavirus (DAffSV) |
| Drosophila ananassae sigmavirus | Sigmavirus drosophilaeananassae | Drosophila ananassae sigmavirus (DAnaSV) |
| Drosophila immigrans sigmavirus | Sigmavirus drosophilaeimmigrantis | Drosophila immigrans sigmavirus (DImmSV) |
| Drosophila melanogaster sigmavirus | Sigmavirus drosophilaemelanogastris | Drosophila melanogaster sigmavirus (DMelSV) |
| Drosophila obscura sigmavirus | Sigmavirus drosophilaeobscurae | Drosophila obscura sigmavirus (DObsSV) |
| Drosophila tristis sigmavirus | Sigmavirus drosophilaetristis | Drosophila tristis sigmavirus (DTriSV) |
| Muscina stabulans sigmavirus | Sigmavirus muscinaestabulantis | Muscina stabulans sigmavirus (MStaSV) |
| Pike fry sprivivirus | Sprivivirus esocis | pike fry rhabdovirus (PFRV), grass carp rhabdovirus (GrCRV), Tench rhabdovirus (TenRV) |
| Carp sprivivirus | Sprivivirus cyprinidae | spring viremia of carp virus (SVCV) |
| Coastal Plains tibrovirus | Tibrovirus planioraei | Coastal Plains virus (CPV) |
| Tibrogargan tibrovirus | Tibrovirus tibrogarganense | Bivens Arm virus (BAV), Tibrogargan virus (TIBV) |
| Durham tupavirus | Tupavirus durhamense | Durham virus (DURV) |
| Tupaia tupavirus | Tupavirus tupaiae | tupaia virus (TUPV) |
| Lettuce big-vein associated varicosavirus | Varicosavirus lactucavenamagna | lettuce big-vein associated virus (LBVaV) |
| Alagoas vesiculovirus | Vesiculovirus alagoasense | vesicular stomatitis Alagoas virus (VSAV) |
| Carajas vesiculovirus | Vesiculovirus caraiasense | Carajás virus (CJSV) |
| Chandipura vesiculovirus | Vesiculovirus chandipurense | Chandipura virus (CHPV) |
| Cocal vesiculovirus | Vesiculovirus cocalense | Cocal virus (COCV) |
| Indiana vesiculovirus | Vesiculovirus indianense | vesicular stomatitis Indiana virus (VSIV) |
| Isfahan vesiculovirus | Vesiculovirus isfahanense | Isfahan virus (ISFV) |
| Maraba vesiculovirus | Vesiculovirus marabense | Maraba virus (MARAV) |
| New Jersey vesiculovirus | Vesiculovirus newierseyense | vesicular stomatitis New Jersey virus (VSNJV) |
| Piry vesiculovirus | Vesiculovirus piryense | Piry virus (PIRYV) |
| Flanders virus | Conversion not possible because not assigned to a genus | Flanders virus (FLAV) |
| Ngaingan virus | Conversion not possible because not assigned to a genus | Ngaingan virus (NGAV) |
| Wongabel virus | Conversion not possible because not assigned to a genus | Wongabel virus (WONV) |
| Family Sunviridae | ||
| Reptile sunshinevirus 1 | Sunshinevirus reptilis | Sunshine Coast virus (SunCV) |
| Unassigned | ||
| Xincheng anphevirus | Anphevirus xinchengense | Xīnchéng mosquito virus (XcMV) |
| Lishi arlivirus | Arlivirus lishiense | Lĭshì spider virus 2 (LsSV-2) |
| Sanxia wastrivirus | Wastrivirus sanxiense | Sānxiá water strider virus 4 (SxWSV-4) |
| Tacheng chengtivirus | Chengtivirus tachengense | T chéng tick virus 6 (TcTV-6) chéng tick virus 6 (TcTV-6) |
| Wenzhou crustavirus | Crustavirus wenzhouense | Wēnzhōu crab virus 1 (WzCV-1) |
Note: Dark green (online version only) depicts current true binomial names and their Linnaean counterparts; light green (only version only) depicts current binomial-like names and their Linnaean counterparts; purple (online version only) depicts current non-binomial names and their Linnaean counterparts.
aThese names are for illustration purposes only and may not be the same as the names that ICTV Study Groups would officially propose if a Linnaean binomial species naming convention were implemented.
bFor species names referring to host names of their virus members, we used the singular possessive of the lowest taxon these hosts belong to.
cFor species names referring to disease caused by their virus members, we used Latin translations for the disease names.
dlactucanecante “lettuce-killing”.
“lettuce-killing”.
elactucamaculante “lettuce-sullying”.
“lettuce-sullying”.
fcerealisborei “of the northern cereal”.
“of the northern cereal”.
gbubulifebris “of the bovine fever”.
“of the bovine fever”.
hpiscicidante “fish-killing”.
“fish-killing”.
iplaniorae “of the plain of the coast”.
“of the plain of the coast”.
Conversion of Non-Latinized Binomial-like Species Names to Linnaean Binomials
Several currently accepted arenaviral and mononegaviral species names are similar to binomials in structure, with a species epithet and a genus name as outlined above, but consist of more than two words (e.g., in the names Soybean cyst nematode socyvirus and Lymphocytic choriomeningitis mammarenavirus, “Soybean cyst nematode” and “Lymphocytic choriomeningitis” serve as the respective species epithets). Conversion to true binomials, therefore, requires reduction, contraction, or replacement of words in addition to Latinization. Conversion of names that contain virus host information could be achieved by, for example, referring to the host taxa: soybean cyst nematodes are members of the genus Heterodera (Soybean cyst nematode socyvirus Socyvirus heteroderae), alfalfa is the member of the species Medicago sativa (Alfalfa dwarf cytorhabdovirus
Socyvirus heteroderae), alfalfa is the member of the species Medicago sativa (Alfalfa dwarf cytorhabdovirus Cytorhabdovirus medicagonis), and barley is a member of the species Hordeum vulgare (Barley yellow striate mosaic cytorhabdovirus
Cytorhabdovirus medicagonis), and barley is a member of the species Hordeum vulgare (Barley yellow striate mosaic cytorhabdovirus Cytorhabdovirus hordei). Geographical locations that consist of multiple words may be contracted using Latin words (e.g., Taï Forest ebolavirus, referring to Taï Forest in Côte d’Ivoire, could be converted to Ebolavirus silvataiense: “silva” is forest in Latin).
Cytorhabdovirus hordei). Geographical locations that consist of multiple words may be contracted using Latin words (e.g., Taï Forest ebolavirus, referring to Taï Forest in Côte d’Ivoire, could be converted to Ebolavirus silvataiense: “silva” is forest in Latin).
Other current non-Latinized binomial-like species names include numbers. These numbers are located either between species epithet and genus name (e.g., Alethinophid 1 reptarenavirus, Psittaciform 1 bornavirus) or after the genus name (e.g., Sonchus cytorhabdovirus 1). Conversion of these names would be more challenging, especially for sets of species names that differ in numbers but are otherwise identical. One straightforward way would be to convert the numbers into Latinized alphanumericals (Psittaciform 1 bornavirus Bornavirus alphapsittaciforme). This conversion approach may be problematic as long alphanumerical lists (e.g., alphapsittaciforme, betapsittaciforme, gammapsittaciforme, etc.) are nondescript and, therefore, difficult to memorize. A more elegant conversion could be achieved if the numbers were omitted entirely and new species names formed were based on the known properties of the associated member viruses. Examples for these types of possible conversion are shown in Tables 1 and 2 in light green (online version only).
Bornavirus alphapsittaciforme). This conversion approach may be problematic as long alphanumerical lists (e.g., alphapsittaciforme, betapsittaciforme, gammapsittaciforme, etc.) are nondescript and, therefore, difficult to memorize. A more elegant conversion could be achieved if the numbers were omitted entirely and new species names formed were based on the known properties of the associated member viruses. Examples for these types of possible conversion are shown in Tables 1 and 2 in light green (online version only).
In addition, some current non-Latinized binomial-like species names contain genus names that do not refer to existing genera. For instance, the two species names Atlantic salmon paramyxovirus and Avian paramyxovirus 2 imply that these species belong to a genus “Paramyxovirus,” which does not exist (there is only a family Paramyxoviridae). These names could easily be converted to Linnaean binomials, in analogy to other non-Latinized binomials, for instance by simply replacing the genus names with the correct ones (e.g., Atlantic salmon paramyxovirus Atlantic salmon aquaparamyxovirus
Atlantic salmon aquaparamyxovirus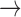 Aquaparamyxovirus salmonis). Examples for these types of possible conversion are shown in Tables 1 and 2 in purple (online version only).
Aquaparamyxovirus salmonis). Examples for these types of possible conversion are shown in Tables 1 and 2 in purple (online version only).
Conversion of Non-binomial Species Names to Linnaean Binomials
Numerous mononegaviral species names are not yet in any binomial or binomial-like format and often differ from the names of their member viruses only in italicization (e.g., measles virus is a member of the species Measles virus). Conversion of these names to Linnaean binomials would simply require the replacement of the word “virus” with a genus name and then following the steps outlined for other names above (Measles virus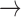 Measles morbillivirus
Measles morbillivirus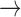 Morbillivirus rubeolae: “rubeola” is measles in Latin). Examples for this type of possible conversion are shown in Table 2 in purple (online version only).
Morbillivirus rubeolae: “rubeola” is measles in Latin). Examples for this type of possible conversion are shown in Table 2 in purple (online version only).
Conversion of Free-Floating Species Names to Linnaean Binomials
Three mononegaviral species names (Flanders virus, Ngaingan virus, and Wongabel virus) are “free-floating” within the family Rhabdoviridae, that is, they have not been assigned to any of the current genera (purple in Table 2). Consequently, these names cannot be converted to Linnaean binomials until genera for them are established (a proposal to include these species in genera is currently in preparation). The implementation of Linnaean binomial virus species names by the ICTV would, therefore, require a change in the International Code of Virus Classification and Nomenclature (ICVCN) mandating genus assignment or establishment concomitant with species establishment.
DISCUSSION
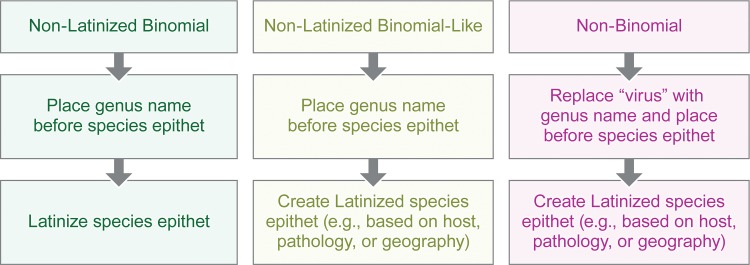
The results of the exercise described here suggest that conversion of current virus species names to Linnaean binomials is a practical task that could be achieved rapidly by individual ICTV Study Groups. The draft conversion of the 34 arenaviral and 141 mononegaviral species names (Tables 1 and 2) was achieved within three work days by a single individual (T.S.P.) following a simple flow chart (Fig. 1).
Figure 1.
Stepwise flow chart for conversion of current virus species names to the Linnaean binomial format. Colors correspond to those used in Tables 1 and 2 (online version only).
The main logistical challenge turned out to be the identification of the etymological origins of some words in current species names (e.g., “Piry” in Piry vesiculovirus; “Aravan” in Aravan lyssavirus) for proper Latinization. Subsequent discussion between the members of all relevant ICTV Study Groups to refine or replace the draft preliminary names by correcting and devising alternatives took an additional 2–3 weeks.
Clearly, the Linnaean names listed in Tables 1 and 2 are not without controversy and could be easily ameliorated. As often is the case in nomenclature, a number of questions remains, such as whether meaning should be closely attached to names. If future scientific discoveries contradict the species name (e.g., if a virus species is named after the presumed host of the member virus and later research demonstrates a wider host spectrum or the presumed host not to be the host), how concerned should scientists be with the mismatch? Another question is whether certain within-taxa consistencies are preferable (e.g., if most species within a genus are named after the hosts of their members, then perhaps all species names within that genus should be devised that way). Actual ICTV-supported conversion of current virus species names to the Linnaean format would likely result in more extensive ICTV Study Group discussions on which types of names should be chosen. Nevertheless, the overall process described herein can most likely be extrapolated to virus species beyond the family Arenaviridae and order Mononegavirales, albeit with some virus taxon-specific complexities and concerns. Thus, the current Master List of ICTV-approved species names (3704 in 2016) could likely be processed within a few months or even in a much shorter time, if a concerted effort is undertaken by the relevant ICTV Study Groups.
ICTV Study Group members either already possess or can acquire the basic knowledge of Latin needed for most name conversions, and the guidance of experts with a more extensive background in taxonomy-oriented Latin would be advantageous. Some creativity would be required in instances of sets of virus species names that differ from each other by only a number or an alphanumerical and species name epithets that consist of merely a number or a letter–-a situation that barely exists in other, nonviral, taxonomies.
Linnaean-style binomials (genus names preceding the species epithet) but without Latinization have been proposed as a practical alternative to the more radical complete binomial nomenclature of viruses by some authors of this article. Following those proposals, most current species names could be converted into new names that would not look as unfamiliar to the virologists as properly declined Latinized names. Importantly, uniform grammar of these names would also allow incorporation of viral taxa into comprehensive taxonomic databases. However, Latinized binomials may have important advantages over non-Latinized binomials. Throughout the history of science, different languages have dominated scientific publications, from Arabic and Greek to Italian, French, and German. While today English is today considered the unofficial language of international science, this preference may quite possibly change in the future. Moreover, many researchers publish findings in their native languages in specialty journals.
In contrast to English or other spoken languages, Latin is static, that is, the meaning of Latin words will not change over time or depend on geographic areas. Therefore, the meaning of Latin words is quite precise no matter when they are used. In zoology and botany, this Latin name stability allows easy translation of an animal or plant name in a manuscript written in a foreign language because of the common Latinized species name. For instance, the German “Meerschweinchen (Cavia porcellus),” Chinese “ . (Cavia porcellus),” and English “guinea pig (Cavia porcellus)” instantly clarify that “Meerschweinchen,” “
. (Cavia porcellus),” and English “guinea pig (Cavia porcellus)” instantly clarify that “Meerschweinchen,” “ ,” and “guinea pig” refer to the same animal belonging to the species Cavia porcellus. This advantage may even apply within a given language. For instance, in American English, “catamount,” “cougar,” “mountain lion,” and “puma” are all synonymous names for the felid belonging to the species Puma concolor. The four names map to a unique Latin name of a species, but they do not necessarily map uniquely to specific words in every other extant language. Such name diversity is also present in virology. For instance, in Chinese, both “
,” and “guinea pig” refer to the same animal belonging to the species Cavia porcellus. This advantage may even apply within a given language. For instance, in American English, “catamount,” “cougar,” “mountain lion,” and “puma” are all synonymous names for the felid belonging to the species Puma concolor. The four names map to a unique Latin name of a species, but they do not necessarily map uniquely to specific words in every other extant language. Such name diversity is also present in virology. For instance, in Chinese, both “ ” and “
” and “ ” are in common use for the English “Ebola virus,” but both Chinese names could immediately be interrelated via the associated virus species name. Using a universal, “dead” language such as Latin for species names may not only keep biological taxonomy consistent, but also keep it neutral with respect to emergent globally dominant languages. As an additional advantage, Latinized species names will appear foreign to most readers. The distinct appearance of these names could make it easier for nonspecialists to easily distinguish names of viruses (physical entities that are typically labeled in a spoken language, such as English) and the names of species (concepts of the mind).
” are in common use for the English “Ebola virus,” but both Chinese names could immediately be interrelated via the associated virus species name. Using a universal, “dead” language such as Latin for species names may not only keep biological taxonomy consistent, but also keep it neutral with respect to emergent globally dominant languages. As an additional advantage, Latinized species names will appear foreign to most readers. The distinct appearance of these names could make it easier for nonspecialists to easily distinguish names of viruses (physical entities that are typically labeled in a spoken language, such as English) and the names of species (concepts of the mind).
Different taxonomic codes for different kingdoms utilize additional rules helping to avoid various kinds of complication and confusion. For instance, the International Code of Botanical Nomenclature prohibits the use of tautonymic species names, that is, names in which the genus name is identical in spelling with the species epithet; such names, however, are allowed in zoological taxonomy (e.g., Gorilla gorilla, Rattus rattus). Tautonymic species names currently do exist in mononegavirus taxonomy (e.g., Marburg marburgvirus).
Botanical and zoological, but not prokaryotic, species names are typically followed by the “authority,” that is, information on who first published the description and name of the species (in botany, for instance, “Arabidopsis thaliana (L.) Heynh.”; in zoology, for instance, “Mus musculus Linnaeus, 1758”). In the case of virus species names, considerations may be put forward to omit any authoritative information, as is done in prokaryotic taxonomy.
Another source of contention in nonviral taxonomies is “priority,” that is, the problem of choosing the official name between two distinct species names after it is determined that both names refer to the same member organism. Priority would unlikely be a significant issue for virus taxonomy at this time as the number of ICTV-accepted species is still relatively low. The history of establishing virus species spans only a few decades, rather than hundreds of years, and such names are generally better documented than in older, nonviral, taxonomies. An opportunity exists to update the ICVCN with new rules on resolving potential naming conflicts once they arise. In contrast to other taxonomies, a mechanism is already in place for virus taxonomists to modify the ICVCN annually if necessary. It should also be noted that “authority” and “priority” are concepts that apply in taxonomies in which the first valid publication of a taxon determines the application of taxon names. Viruses are explicitly excluded from such a process at the moment, because the use of taxon names is determined by the ICTV through its approval process.
We conclude that conversion of current virus species names to the Linnaean format would not be as complicated or time-consuming as even the authors of this article had expected when this thought experiment was initiated. The current ICVCN rules also do not prevent the adoption of Linnaean binomials, and such names can be proposed officially by virologists or ICTV Study Groups. However, the ICVCN would have to be amended if uniform application of Linnaean binomials was desired. Whether such an amendment should be pursued will require extensive consultation across the virology community and will be the decision of the ICTV. The actual Linnaean species names to be chosen for each virus would ultimately be the purview of the various ICTV Study Groups and the ICTV membership.
FUNDING
This work was supported in part through Battelle Memorial Institute’s prime contract with the US National Institute of Allergy and Infectious Diseases (NIAID) under Contract No. HHSN272200700016I. A subcontractor to Battelle Memorial Institute who performed this work is J.H.K., an employee of Tunnell Government Services, Inc. This work was also funded in part under National Institutes of Health (NIH) contract HHSN272201000040I/HHSN27200004/D04 and R24AI120942 (N.V., R.B.T.), and the National Science Foundation (NSF) Individual Research and Development (IR/D) program (A.R.M.).
The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the US Department of the Army, the US Department of Defense, the US Department of Health and Human Services, the Department of Homeland Security Science and Technology Directorate, the US NSF, or of the institutions and companies affiliated with the authors. In no event shall any of these entities have any responsibility or liability for any use, misuse, inability to use, or reliance upon the information contained herein. The US departments do not endorse any products or commercial services mentioned in this publication.
ACKNOWLEDGMENTS
We thank Laura Bollinger (NIH/NIAID Integrated Research Facility at Fort Detrick, Frederick, MD, USA) for critically editing the manuscript, Andrew J. Davison (MRC – University of Glasgow Centre for Virus Research, Glasgow, UK) and Michael J. Adams (Department of Plant Pathology and Microbiology, Rothamsted Research, Harpenden, Herts, UK) of the ICTV Executive Committee for suggestions on manuscript improvement, and Arya Ariël Kuhn for sustained vocal support and (dia)critical remarks.
References
- Adams M.J., Lefkowitz E.J., King A.M.Q., Gorbalenya A.E., Harrach B., Harrison R.L., Knowles N.J., Kropinski A.M., Krupovic M., Kuhn J.H., Mushegian A.R., Nibert M., Sabanadzovic S., Sanfaçon H., Siddell S.G., Simmonds P., Zerbini F.M., Davison A.J. 2016. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2016). Arch. Virol. 161:2921–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonso C.L., Amarasinghe G.K., Bányai K., Báo Y., Basler C.F., Bavari S., Bejerman N., Blasdell K.R., Briand F.-X., Briese T., Bukreyev A., Calisher C.H., Chandran K., Chéng J., Clawson A.N., Collins P.L., Dietzgen R.G., Dolnik O., Domier L.L., Dürrwald R., Dye J.M., Easton A.J., Ebihara H., Farkas S.L., Freitas-Astúa J., Formenty P., Fouchier R.A.M., Fù Y., Ghedin E., Goodin M.M., Hewson R., Horie M., Hyndman T.H., Ji?ng D., Kitajima E.W., Kobinger G.P., Kondo H., Kurath G., Lamb R.A., Lenardon S., Leroy E.M., Li C.-X., Lin X.-D., Liú L., Longdon B., Marton S., Maisner A., Mühlberger E., Netesov S.V., Nowotny N., Patterson J.L., Payne S.L., Paweska J.T., Randall R.E., Rima B.K., Rota P., Rubbenstroth D., Schwemmle M., Shi M., Smither S.J., Stenglein M.D., Stone D.M., Takada A., Terregino C., Tesh R.B., Tian J.-H., Tomonaga K., Tordo N., Towner J.S., Vasilakis N., Verbeek M., Volchkov V.E., Wahl-Jensen V., Walsh J.A., Walker P.J., Wang D., Wang L.-F., Wetzel T., Whitfield A.E., Xiè J., Yuen K.-Y., Zhang Y.-Z., Kuhn J.H. 2016. Taxonomy of the order Mononegavirales: update 2016.Arch. Virol. 161:2351–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calisher C.H., Mahy B. W. 2003. Taxonomy: get it right or leave it alone. Am. J. Trop. Med. Hyg. 68:505–506. [DOI] [PubMed] [Google Scholar]
- Drebot M.A., Henchal E., Hjelle B., LeDuc J.W., Repik P.M., Roehrig J.T., Schmaljohn C.S., Shope R.E., Tesh R.B., Weaver S.C., Calisher C. H. 2002. Improved clarity of meaning from the use of both formal species names and common (vernacular) virus names in virological literature. Arch. Virol. 147:2465–2472. [DOI] [PubMed] [Google Scholar]
- Fenner F. 1976. Classification and nomenclature of viruses - second report of the International Committee on Taxonomy of Viruses. Intervirology 7:1–115. [DOI] [PubMed] [Google Scholar]
- Greuter W., Garrity G., Hawksworth D.L., Jahn R., Kirk P.M., Knapp S., McNeill J., Michel E., Patterson D.J., Pyle R., Tindall. B.J. 2011. Draft BioCode (2011): principles and rules regulating the naming of organisms. Taxon 60:201–212. [Google Scholar]
- Huemer H.P., Wechselberger C., Bennett A.M., Falke D., Harrington L. 2003. Cloning and expression of the complement receptor glycoprotein C from Herpesvirus simiae (herpes B virus): protection from complement-mediated cell lysis. J. Gen. Virol. 84:1091–1100. [DOI] [PubMed] [Google Scholar]
- Hummeler K., Davidson W.L., Henle W., Laboccetta A.C., Ruch H. G. 1959. Encephalomyelitis due to infection with Herpesvirus simiae (herpes B virus); a report of two fatal, laboratory-acquired cases. N. Engl. J. Med. 261:64–68. [DOI] [PubMed] [Google Scholar]
- International Association for Plant Taxonomy. 2011. International Code of Nomenclature for algae, fungi, and plants (Melbourne Code). Oberreifenberg, Germany: Koeltz Scientific Books (Regnum Vegetabile; vol. 154).Available from: URL http://www.iapt-taxon.org/nomen/main.php. [Google Scholar]
- International Commission on Zoological Nomenclature. 2012. International Code of Zoological Nomenclature. 4th ed.London, UK: The International Trust for Zoological Nomenclature.Available from: URL http://www.iczn.org/iczn/index.jsp#. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Committee on Systematic Bacteriology. 1992. International Code of Nomenclature of Bacteria: Bacteriological Code, 1990 Revision. Washington, DC: ASM Press. [PubMed] [Google Scholar]
- International Committee on Taxonomy of Viruses. 2013. The International Code of Virus Classification and Nomenclature, February 2013. Available from: URL http://www.ictvonline.org/codeOfVirusClassification.asp. [Google Scholar]
- International Committee on Taxonomy of Viruses. 2016. Brief notes of the meeting of the ICTV Executive Committee (EC) and Study Group (SG) chairs held on 1–2 February 2016 at Hinxton Hall, Wellcome Trust Genome Campus, Cambridgeshire, UK. Available from: URL http://talk.ictvonline.org/meetings/sg_hinxton/w/sg_meeting. [Google Scholar]
- King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E. J. 2012. The International Code of Virus Classification and Nomenclature. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J. editors. Virus taxonomy—ninth report of the International Committee on Taxonomy of Viruses. London, UK: Elsevier/Academic Press. p. 1273–1277. [Google Scholar]
- Koonin E.V., Starokadomskyy P. 2016. Are viruses alive? The replicator paradigm sheds decisive light on an old but misguided question. Stud. Hist. Philos. Biol. Biomed. Sci. 59:125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J.H., Jahrling P. B. 2010. Clarification and guidance on the proper usage of virus and virus species names. Arch. Virol. 155:445–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnaeus C. 1753. Species plantarum. Stockholm, Sweden [Latin]: Laurentius Salvius. [Google Scholar]
- Matthews R.E.F. 1983. A critical appraisal of viral taxonomy. Boca Raton (FL): CRC Press. [Google Scholar]
- Matthews R.E.F. 1985a. Viral taxonomy. Microbiol. Sci. 2:74–76. [PubMed] [Google Scholar]
- Matthews R.E.F. 1985b. Viral taxonomy for the nonvirologist. Annu. Rev. Microbiol. 39:451–474. [DOI] [PubMed] [Google Scholar]
- Milne R. G. 1984. The species problem in plant virology. Microbiol. Sci. 1:113–117. [PubMed] [Google Scholar]
- Philippe N., Legendre M., Doutre G., Coute Y., Poirot O., Lescot M., Arslan D., Seltzer V., Bertaux L., Bruley C., Garin J., Claverie J.M., Abergel C. 2013. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science 341:281–286. [DOI] [PubMed] [Google Scholar]
- Radoshitzky S.R. Báo Y. Buchmeier M.J. Charrel R.N. Clawson A.N. Clegg C.S. DeRisi J.L. Emonet S. Gonzalez J.-P. Kuhn J.H. Lukashevich I.S. Peters C.J. Romanowski V. Salvato M.S. Stenglein M.D. de la Torre J. C.. 2015. Past, present, and future of arenavirus taxonomy. Arch. Virol. 160:1851–1874. [DOI] [PubMed] [Google Scholar]
- Van Regenmortel M. H. 2003. Viruses are real, virus species are man-made, taxonomic constructions. Arch. Virol. 148:2481–2488. [DOI] [PubMed] [Google Scholar]
- Van Regenmortel M. H. 2006. Virologists, taxonomy and the demands of logic. Arch. Virol. 151:1251–1255. [DOI] [PubMed] [Google Scholar]
- Van Regenmortel M. H. 2007. Virus species and virus identification: past and current controversies. Infect. Genet. Evol. 7:133–144. [DOI] [PubMed] [Google Scholar]
- Van Regenmortel M.H., Burke D.S., Calisher C.H., Dietzgen R.G., Fauquet C.M., Ghabrial S.A., Jahrling P.B., Johnson K.M., Holbrook M.R., Horzinek M.C., Keil G.M., Kuhn J.H., Mahy B.W., Martelli G.P., Pringle C., Rybicki E.P., Skern T., Tesh R.B., Wahl-Jensen V., Walker P.J., Weaver S. C. 2010. A proposal to change existing virus species names to non-Latinized binomials. Arch. Virol. 155:1909–1919. [DOI] [PubMed] [Google Scholar]
- Van Regenmortel M. H. V. Classes, taxa and categories in hierarchical virus classification: a review of current debates on definitions and names of virus species. Bionomina 10. 2016a Forthcoming. [Google Scholar]
- van Regenmortel M.H.V. 2016b. The metaphor that viruses are living is alive and well, but it is no more than a metaphor. Stud. Hist. Philos. Biol. Biomed. Sci. 59:117–124. [DOI] [PubMed] [Google Scholar]
- Wildy P. 1971. Classification and nomenclature of viruses - first report of the International Committee on Nomenclature of Viruses. Basel, Switzerland: S. Karger. [Google Scholar]