Epigenetic changes may contribute to major depressive disorder (MDD). Qi, Yang et al. use microRNA-132 levels in peripheral blood to guide the decomposition of resting-state functional, structural and diffusion MRI in unmedicated patients and healthy controls. Increased microRNA-132 levels in MDD are associated with structural and functional impairment in the fronto-limbic network.
Keywords: major depressive disorder, microRNA132, supervised multimodal fusion, fronto-limbic network, unmedicated
Abstract
There is compelling evidence that epigenetic factors contribute to the manifestation of depression, in which microRNA132 (miR-132) is suggested to play a pivotal role in the pathogenesis and neuronal mechanisms underlying the symptoms of depression. Additionally, several depression-associated genes [MECP2, ARHGAP32 (p250GAP), CREB, and period genes] were experimentally validated as miR-132 targets. However, most studies regarding miR-132 in major depressive disorder are based on post-mortem, animal models or genetic comparisons. This work will be the first attempt to investigate how miR-132 dysregulation may impact covariation of multimodal brain imaging data in 81 unmedicated major depressive patients and 123 demographically-matched healthy controls, as well as in a medication-naïve subset of major depressive patients. MiR-132 values in blood (patients > controls) was used as a prior reference to guide fusion of three MRI features: fractional amplitude of low frequency fluctuations, grey matter volume, and fractional anisotropy. The multimodal components correlated with miR-132 also show significant group difference in loadings. Results indicate that (i) higher miR-132 levels in major depressive disorder are associated with both lower fractional amplitude of low frequency fluctuations and lower grey matter volume in fronto-limbic network; and (ii) the identified brain regions linked with increased miR-132 levels were also associated with poorer cognitive performance in attention and executive function. Using a data-driven, supervised-learning method, we determined that miR-132 dysregulation in major depressive disorder is associated with multi-facets of brain function and structure in fronto-limbic network (the key network for emotional regulation and memory), which deepens our understanding of how miR-132 dysregulation in major depressive disorders contribute to the loss of specific brain areas and is linked to relevant cognitive impairments.
Introduction
Major depressive disorder (MDD) is characterized by affective, cognitive, neuromorphological and molecular abnormalities that may have a neurodevelopmental origin (Gong and He, 2015). According to the World Health Organization, patients with MDD will be the second-leading cause of disability in the world by the year 2020, following cardiovascular disease (Sliz and Hayley, 2012). In contrast to schizophrenia and autism disorder (high hereditary mental illness), depression has a relatively low heritability that has been estimated at 30–40% (Ebmeier et al., 2006), while a growing number of studies have suggested that epigenetics has an important role in MDD (Duman, 2002; Lorenzetti et al., 2009; Urdinguio et al., 2009; Busatto, 2013). MicroRNAs (miRNAs) are important epigenetic regulators that can regulate >50% of human genes (Novina and Sharp, 2004; Lewis et al., 2005; Selbach et al., 2008) and are receiving intensive research interest in psychiatric disease studies. With high levels of potential regulatory influence, certain miRNAs may strongly impact gene expression and play a role in the pathophysiology or aetiology of diseases with an elusive genetic basis.
Previous experimental evidence has demonstrated that miR-132 is a brain-enriched miRNA that plays key roles during both the early and late stages of adult neurogenesis, including the proliferation and differentiation of adult neural stem cells, the regulation of dendritic morphogenesis, and synaptic plasticity (Vo et al., 2005; Wayman et al., 2008; Magill et al., 2010; Im and Kenny, 2012). In addition, miR-132 is predicted to regulate multiple members of neuronal, activity-specific, biological pathways, including long-term depression pathways (Cheng et al., 2007). A recent study demonstrated that serum levels of miR-132 were increased in MDD patients, as compared with healthy control subjects (Li et al., 2013), with a significant positive correlation between Self-Rating Depression Scale scores (Li et al., 2013). Another study also demonstrated that miR-132 expression was increased in the peripheral blood samples of MDD compared to healthy control subjects (Su et al., 2015). In addition, increased expression levels of miR-132 were detected in the hippocampus in an animal model of chronic stress-induced depression (Su et al., 2015). Furthermore, several depression-associated genes [MECP2, ARHGAP32 (p250GAP), CREB, and period genes] were experimentally validated as miR-132 targets (Mouillet-Richard et al., 2012; Su et al., 2015). All of the above evidence suggests that miR-132 may have an important role in the pathogenesis and neuronal mechanisms underlying the symptoms of depression. Based on these findings of abnormal miR-132 expression levels in MDD (Cheng et al., 2007; Zheng et al., 2013), we hypothesized that miR-132 may be associated with alterations in a brain network that takes charge of emotion regulation and is consistently impaired in MDD (Clark et al., 2009), such as the fronto-limbic network. Multiple MDD studies have focused on typical impaired brain regions such as prefrontal cortex, hippocampus and amygdala because of their roles in emotion processing and antidepressant action (Clark et al., 2009). However, most studies used either single-modality analyses or performed a multimodal comparison after separate analyses within each modality. In this case, the cross-information among multiple modalities is often missed (Calhoun and Sui, 2016). A key motivation for this work is to leverage the cross-information in the existing data, thereby revealing important relationships that are specifically related to the measure of interest and cannot be detected by using a single modality (Meda et al., 2014).
Compelling evidence has confirmed that patients with MDD reflect fundamental differences in brain structure and function (Drevets et al., 2008). Moreover, most existing studies about the role of miR-132 in depression are based on post-mortem tissue analyses, animal models or genetic comparisons. To date, no studies have directly linked miR-132 with multimodal brain imaging data together in a joint analysis to investigate the imaging–epigenetic association and the underlying neural mechanisms of major depression. In this work, we aim to apply a novel, supervised, multimodal fusion with the referenced method (Qi et al., 2018) to identify the association between miR-132 levels and three MRI measures for unmedicated patients with MDD under the guidance of one epigenetic reference point (miR-132). Compared to unsupervised fusion, this supervised-learning model can be more goal-directed as it takes advantage of specific prior knowledge to guide the fusion analysis, thus enabling researchers to pinpoint a particular component of interest from a large complex dataset (Qi et al., 2018). Due to the difficulty of procuring miR-132 from brain tissues, here we used miR-132 extracted from peripheral blood as the reference. As previously reported (Walton et al., 2016), epigenetic signatures such as miR-132 measured in blood can serve as a surrogate marker for the brain, especially for mood disorders (Li et al., 2013; Su et al., 2015).
To the best of our knowledge, this is the first attempt to estimate multimodal biomarkers by jointly mining three types of MRI data under the guidance of a particular miRNA (miR-132) that was used to investigate the imaging-epigenetic covariance in 81 unmedicated patients with MDD and 123 healthy control subjects. Three MRI features [fractional amplitude of low frequency fluctuations (fALFF) from resting-state functional MRI (rs-fMRI), grey matter volume from structural MRI, and fractional anisotropy from diffusion MRI] were jointly decomposed with the subject-wise miR-132 levels as the reference. Furthermore, correlation analyses with multiple cognitive domains [measured by the Cambridge Neuropsychological Test Automated Battery (CANTAB)] and depressive symptoms were also performed to investigate the potential cognitive and symptomatic relationship with the identified brain networks modulated by miR-132.
Materials and methods
Participants
In this study, we recruited 81 unmedicated patients with MDD and 123 demographically-matched healthy control subjects from the Mental Health Center of West China Hospital of Sichuan University that were matched for age and gender. Demographics of all subjects are shown in Table 1. All MDD patients were enrolled based on the Structured Clinical Interview for DSM-IV (SCID; First, 2002) for major depression, with Hamilton Depression Rating Scale (HDRS) score > 17 and age within 18–60 years. Exclusion criteria included patients with concurrent neurological illness, mental retardation, cardiovascular disease, bipolar disorder, schizophrenia, anxiety disorder, alcohol or drug abuse and history of loss of consciousness. To avoid the influence of medication, we ensured all patients enrolled in our study had stopped taking any antidepressants for at least 3 months before scanning (i.e. unmedicated MDD), while a subset of MDD patients (26/81, 32%) were previously treated with antidepressant medications. Detailed clinical information is presented in Table 1. All patients were assessed and scanned as soon as possible (usually within 3 days) to prevent treatment delay. Demographically, similar healthy volunteers were recruited by poster advertisements within the local community. The control subjects were also interviewed by professional psychiatrists using SCID non-patient edition (SCIDI/NP; APA, 2000) to ensure that none of them had a current or past history of depression, other major physical or neurological illnesses, or any substance abuse. All participants signed an informed consent form prior to participation in the study. This study was approved by the Ethics Committee of Sichuan University and conducted according to the Declaration of Helsinki. Tests from CANTAB (Robbins et al., 2010), imaging parameters and preprocessing details are presented in the Supplementary material.
Table 1.
Demographic and clinical information
| MDD (n = 81) | HC (n = 123) | t/χ2-tests | |
|---|---|---|---|
| Demographic characteristics | |||
| Age, mean ± SD | 29 ± 19.8 | 27 ± 18.0 | 0.1 |
| Gender, male/female | 28/53 | 41/82 | 0.9 |
| Education time, years, mean ± SD | 13.5 ± 2.9 | 14.6 ± 3.5 | 0.02 |
| Clinical characteristics | |||
| HDRS, mean ± SD | 22.8 ± 4.1 | NA | NA |
| Total duration of illness, months, mean ± SD | 31.1 ± 48 | NA | NA |
| Current duration of illness, months, mean ± SD | 7.4 ± 12.8 | NA | NA |
| Presence of suicidal thoughts, yes/no | 40/41 | NA | NA |
| Presence of suicidal behaviour, yes/no | 7/74 | NA | NA |
| With/without family history | 6/75 | ||
| Medical characteristics | |||
| No medication (68%) | 55 | 123 | |
| Antidepressants (32%, taken at least 3 months ago) | |||
| SSRIs (selective serotonin reuptake inhibitors) | 20 | NA | NA |
| SNRIs (serotonin–norepinephrine reuptake inhibitors) | 3 | NA | NA |
| NaSSAs (noradrenergic and specific serotonergic antidepressant) | 3 | NA | NA |
NA = not available.
MiR-132 expression measurement in peripheral blood
Whole blood (3 ml) was collected from each subject using Tempus™ Blood RNA Tubes (Applied Biosystems). The blood was immediately mixed vigorously for at least 10 s and frozen at −80°C until use. Total RNAs were isolated using the MagMAX™ for Stabilized Blood Tubes RNA Isolation Kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. The quality of RNAs was assayed using the NanoDrop ND-2000. Reverse transcription reactions were performed using RevertAid™ First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) and miRNA-specific stem-loop primers. The reverse transcription reaction product was immediately stored at −20°C for further analysis. The qPCR (quantitative real-time PCR) reactions were done in real-time cyclers (Analytikjena-Qtower 2.2). The reaction mixture consisted of 5 µl 2×SYBR® Green Supermix (Aidlab), 200 nM miRNA specific primers, and 200 nM universal primers (0.8 µl reverse transcription product); the volume was adjusted to 10 µl with water. PCR reactions were initiated with a 3 min incubation period at 95°C followed by 40 cycles at 95°C for 10 s and at 60°C for 60 s. A melting curve was performed at the end of the PCR run over a range of 66–99°C with the temperature increased stepwise by 0.5°C every 2 s. As described above, all experiments were performed in triplicate. The relative level of miR-132 was calculated with the comparative ΔCt (ΔΔCt) method using small RNA U6 as a normalization control (http://www.mirbase.org/).
Feature extraction and correction
Three representative MRI features (fALFF, grey matter volume and fractional anisotropy) were extracted and then each modality was reduced into a feature matrix with each row representing one subject (Calhoun and Adali, 2009) (see Supplementary material for further details). These three matrices were then normalized to have the same average sum of squares (computed across all subjects and all voxels for each modality) to ensure all modalities had the same ranges. Multivariate analysis of covariance (MANCOVA) was next performed on each of the normalized feature matrices. Gender, age and education were set as covariates along with their interactions, and all were regressed out to minimize their potential impact on the imaging data. The statistical power of the three MRI features before fusion was 0.89 for fALFF, 0.85 for fractional anisotropy and 0.94 for grey matter, respectively, and 0.90 for miR-132 levels, which are all high enough to ensure accurate and robust conclusions about the group effects detected. Detailed statistical analysis is presented in the Supplementary material.
Fusion with reference
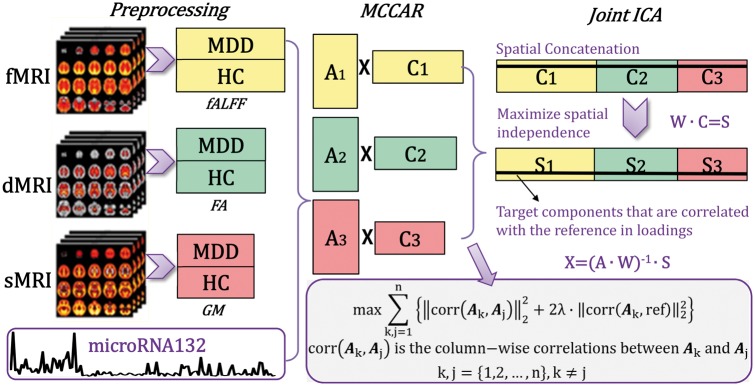
The resulting data were jointly analysed by a fusion-with-reference model called ‘MCCAR+jICA’ (multisite canonical correlation analysis with reference + joint independent component analysis) (Qi et al., 2018), as shown in Fig. 1, in which miR-132 was set as the reference to guide the joint decomposition of three MRI features based on supervised learning. The code of MCCAR+jICA has been incorporated into the Fusion ICA Toolbox (FIT, https://www.nitrc.org/projects/fit), which has been released for public use.
Figure 1.
Flowchart of the supervised data fusion strategy (MCCAR+jICA). The strategy simultaneously maximizes the inter-modality covariation and correlations between loadings of certain components and the reference (i.e. the miR-132 levels).
This model can simultaneously maximize the inter-modality covariation and correlations of certain imaging components with the miR-132 levels. As a result, it enables identification of a joint multimodal component(s) that has robust correlations with both miR-132 and amongst themselves (inter-modality correlations), which may not be detected by a blind (unsupervised) N-way multimodal fusion approach (Supplementary Fig. 3). For more details on the supervised fusion model as well as its effectiveness, please refer to our methodology paper on MCCAR+jICA (Qi et al., 2018). A cross-validation method was used to determine the parameter in miR-132 directed fusion analysis; details are presented in Supplementary Fig. 1, using λ = 0.95. Twenty-four components were estimated for each feature according to an improved minimum description length criterion (Li et al., 2007). Two-sample t-tests were further performed on mixing coefficients of each component for each modality. We aimed to investigate the joint independent component (ICs), which are significantly correlated with miR-132 levels, covarying among modalities and group discriminative.
To avoid the medication influence, we also conducted the same miR-132 directed fusion on a subset of medication-naïve patients with MDD (n = 55, excluding 32% of MDD patients who had been treated with antidepressants >3 months ago) and demographically-matched healthy control subjects (n = 65) with the demographic information listed Supplementary Table 1.
Results
Expression levels of miR-132 in peripheral blood
Because it is difficult to determine miR-132 levels directly from the brain of patients, increasing efforts have focused on detecting miR-132 levels in the blood; specifically, miR-132 levels in the brain of patients with mood disorders can be reflected by levels measured in their blood (Li et al., 2013; Su et al., 2015). We found that the expression levels of miR-132 in unmedicated patients with MDD were higher than those in healthy control subjects (P = 0.0136), in accordance with previous studies (Li et al., 2013; Su et al., 2015). Such a group difference still exists between a medication-naïve MDD subset and demographically-matched healthy control subjects (P = 0.0167). Furthermore, there is no group difference of the expression levels of miR-132 between the unmedicated patients with MDD and medication naïve patients with MDD (P = 0.803). For more details, see the between subgroups boxplots in Supplementary Fig. 2.
MiR-132 associated multimodal covarying imaging patterns
One joint component (denoted as ICref, with the same IC order in all three modalities) was found to be both correlated with miR-132 (r = −0.342*, −0.335*, −0.376* (*FDR-corrected for multiple comparison) for functional MRI, diffusion MRI and structural MRI, respectively; (see Supplementary material for details) and significantly group-discriminating (P = 0.0003*, 0.001*, 0.036 for functional MRI, diffusion MRI and structural MRI, respectively). The spatial maps were transformed into Z scores, visualized at |Z| > 2 as in Fig. 2A and adjusted as healthy control subjects > MDD for all modalities on the mean of loading parameters (boxplot in Fig. 2B). So the positive Z-values (red brain regions) indicate higher contribution in healthy control subjects than patients with MDD and the negative Z-values (blue brain regions) indicate higher contribution in patients with MDD than healthy control subjects. The identified regions in ICref are summarized in Supplementary Table 2 for fALFF (Talairach labels), fractional anisotropy (white matter tracts, from John Hopkins Atlas), and grey matter (Montreal Neurological Institute labels), respectively. Figure 2C shows the correlations between ICref loadings and miR-132 levels (healthy control subjects: red dots, MDD: blue dots); the lower IC loadings correspond to higher miR-132 levels in patients with MDD. Note that even excluding two MDD subjects whose miR-132 levels are much higher than others (as shown in Fig. 2C), the correlations between miR-132 and joint components still remains significant (functional MRI_ICref: r = 0.269, P = 0.002*; DTI_ICref: r = 0.283, P = 0.001*; structural MRI_ICref: r = 0.287, P = 0.001*). Moreover, the partial correlations between miR-132 levels and the brain component loadings remain significant even after regressing out group effects (Supplementary Table 3).
Figure 2.
The identified joint components that are not only correlated with miR-132 levels, but also show significant group differences in all modalities. (A) The spatial maps visualized at |Z| > 2, where the positive Z-values (red regions) means healthy control subjects > MDD and the negative Z-values (blue regions) means healthy control subjects < MDD. (B) Boxplot of the loading parameters of ICref that were adjusted as healthy control subjects > MDD on the mean of loadings. (C) Loadings of ICref and miR-132 levels were negatively correlated (healthy control subjects: red dots, MDD: blue dots); thus MDD correspond to higher miR-132 expression levels and lower loading weights compared to healthy control subjects. (D) Correlation between loadings of ICref and CANTAB cognitive domain scores. The higher Rapid Visual Information Processing score (RVP attention test) corresponds to better attention, which is linked with higher fractional anisotropy and fALFF in the identified red regions. Similarly, a higher IED score (Intra-Extra Dimensional Set Shift, executive function test) is associated with higher grey matter volume in the identified grey matter regions, suggesting better executive function. *Significance passed FDR correction for multiple comparisons. The grey regions in C and D indicate a 95% confidence interval. dMRI = diffusion MRI; FA = fractional anisotropy; fMRI = functional MRI; GM = grey matter; sMRI = structural MRI.
As shown in Fig. 2, higher expression levels of miR-132 in MDD are linked with decreased values in both fALFF and grey matter in hippocampus, parahippocampus, amygdala, dorsolateral prefrontal cortex (DLPFC) [Brodmann areas (BA) 9, 46] and insula, which consist of a fronto–limbic neural circuit. For functional MRI only, higher miR-132 levels are associated with lower fALFF in anterior cingulate cortex and inferior parietal lobule, but higher fALFF in visual cortex. For grey matter only, increased miR-132 associates with decreased grey matter volume in thalamus, caudate, and anterior cingulate cortex. MiR-132 is also associated with the co-occurring reduced fractional anisotropy in anterior thalamic radiation, superior longitudinal fasciculus, forceps major, forceps minor, etc.
MiR-132 association with cognitive measures and symptoms
Correlation analysis with multiple cognitive domains such as attention and executive function (measured by CANTAB) were performed, as these two domains are generally impaired in patients with MDD. As shown in Fig. 2D, increased miR-132 levels were linked with lower fALFF in the attention network, corresponding to a poorer attention performance [r = 0.181, P = 0.014, Rapid Visual Information Processing (RVP) (Rock et al., 2014) attention test]. In addition, lower fractional anisotropy values in major white matter tracts are also linked with worse performance on attention tests (r = 0.252, P = 6.1 × 10−4*). Grey matter reductions in the prefrontal and hippocampus regions are linked with poorer executive function as measured by Intra-Extra Dimensional Set Shift (IED) (Rock et al., 2014) where r = 0.194 and P = 0.009*. Finally, as to the association with depressive symptoms, only fractional anisotropy_ICref show significant correlation with HDRS in our results (r = −0.223, P = 0.04), suggesting that the lower fractional anisotropy values in corpus callosum are associated with the more severe depressive symptoms (Li et al., 2011), as shown in Supplementary Fig. 4; grey matter_ICref and fALFF_ICref are not correlated with HDRS. This might be due to structural changes in the corpus callosum that have been shown to induce altered hemispheric connectivity and impaired emotional and cognitive control, which may predispose individuals to more severe depressive symptoms (Xu et al., 2013). Another reason may lie with the original correlations between miR-132 levels and HDRS, which are not significant due to limited sample size of MDD patients (n = 81); consequently, the identified miR-132 associated components are not necessarily correlated with depressive symptoms since the reference-guided fusion will lead to joint imaging components that are significantly correlated with miR-132 levels. Instead, correlation analyses between HDRS with other components (we estimated 24 components for each modality in the supervised fusion) identified another grey matter component (GM_12) that was significantly anti-correlated with HDRS (r = −0.242, P = 0.029), as displayed in Supplementary Fig. 5. The lower the grey matter volume in highlighted regions (including hippocampus, anterior cingulate cortex and the left thalamus), the more severe depressive symptoms are exhibited in unmedicated patients with MDD, which is consistent with previous findings (Campbell et al., 2004; Busatto, 2013).
Subset replication
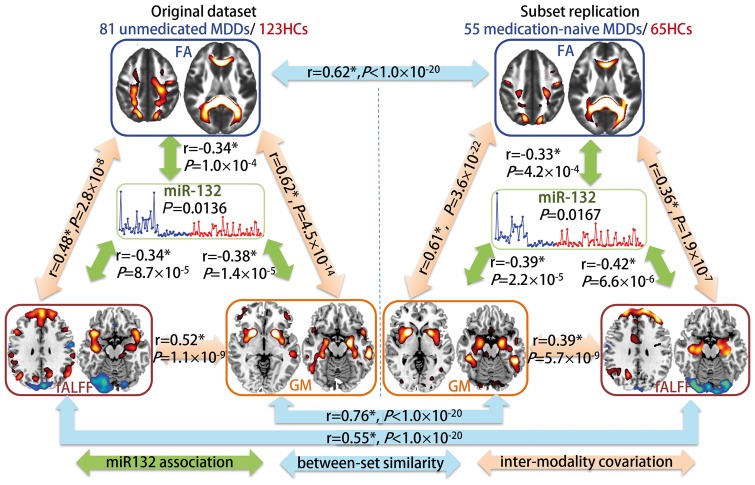
To test the influence of previous use of medication, we also conduct the same miR-132 directed fusion analysis on a subset, including 55 medication-naïve patients with MDD and 65 matched healthy control subjects (Supplementary Fig. 6). Figure 3 compared the miR-132 associated multimodal covarying patterns between the whole unmedicated patients with MDD group and the medication-naïve subset. Evidently, high spatial similarity was demonstrated between the two datasets with r = 0.76, 0.62, and 0.55 (Fig. 3, blue arrows) for grey matter, fractional anisotropy and fALFF respectively, suggesting little impact from the previous use of medication on the brain patterns associated with the dysregulation of miR-132. Furthermore, the miR-132 associated components (Fig. 3, green arrows) are also pair-wisely correlated between modalities (Fig. 3, orange arrows), implicating significant multimodal covariation across subjects.
Figure 3.
Comparison of miR-132 associated multimodal covarying patterns between unmedicated patients with MDD and medication-naïve patients with MDD. The orange arrows represent inter-modality covariation between different modalities; the blue arrows represent intra-modality similarity cross different datasets and the green arrows denote the correlations between miR-132 and the target joint components. *Denotes the correlations passing FDR correction for multiple comparisons.
Discussion
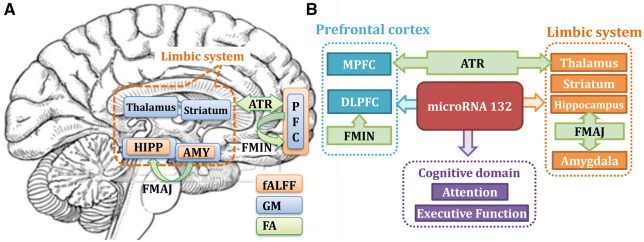
To the best of our knowledge, this is the first study to link miR-132 with multimodal human brain imaging in depression. We attempt to reveal how abnormal epigenetic factors such as miR-132 levels contribute to the multimodal neuroimaging impairment, thus providing insight on the pathophysiology of depression. Results indicate that miR-132 impacts multiple functional and structural regions in unmedicated patients with MDD, especially in the fronto-limbic circuits (prefrontal cortex and the limbic system), which is the key network for emotion regulation, memory and learning (Clark et al., 2009). Figure 4 presents a summary of the identified multimodal covarying patterns closely associated with the miR-132 levels in unmedicated patients with MDD and healthy control subjects.
Figure 4.
Summary of our findings about miR-132 dysregulation in unmedicated depression. (A) Mapping of three MRI components in the same brain with three colours. Note that prefrontal cortex, hippocampus and amygdala are all identified in both fALFF and grey matter, constituting the fronto-limbic system that has been implicated in mediating emotional processing (Phillips et al., 2003) and the pathophysiology of MDD in adults (Drevets, 1999; Sinclair, 2013). Note that these regions were also touched or linked by the identified white matter tracts such as the anterior thalamic radiation (ATR), superior longitudinal fasciculus (SLF), forceps minor (FMIN) and forceps major (FMAJ) (green arrows); while this is all data-driven, it further validates the effectiveness of multimodal fusion. (B) Inference and summary based on our results. Here higher miR-132 expression levels are related with lower fALFF and grey matter in the fronto-limbic circuits and result in poorer attention and executive functions in MDD. Note that forceps minor interconnects the left and right prefrontal cortex across hemispheres (detected in both fALFF and grey matter), anterior thalamic radiation links prefrontal cortex (PFC) with the thalamus, providing evidence for disrupted anatomical connections in fronto-limbic circuitry. Our results clearly support that key depression-associated brain areas, previously implicated in functional, anatomical and structural brain-imaging studies, may be affected by miR-132 dysregulation. AMY = amygdala; DLPFC = dorsolateral prefrontal cortex; FA = fractional anisotropy; GM = grey matter; HIPP = hippocampus; MPFC = medial prefrontal cortex.
As summarized in Fig. 4A, we successfully extracted DLPFC and medial prefrontal cortex, as well as the limbic system both in fALFF and grey matter. These two compartments constitute the fronto-limbic system, which is the most commonly identified network across the existing analyses in MDD (Vasic et al., 2008; Zhang et al., 2012). Particularly, fronto-limbic neural network mediate emotional processing (Phillips et al., 2003) and this system has been implicated in the pathophysiology of MDD in adults (Drevets, 1999; Sinclair, 2013) and in youth (MacMaster et al., 2008). This pattern of baseline and change-related activation appears consistent with the previously proposed involvement of limbic-thalamo-cortical circuits in depression (Drevets, 1998). As summarized in Fig. 4B, the limbic system that is connected by forceps major (Cullen et al., 2010) in fractional anisotropy is connecting with PFC (both detected in fALFF and grey matter), which is traversed by tracts of forceps minor in fractional anisotropy, along with the thalamus-prefrontal circuit that is connected by anterior thalamic radiation in fractional anisotropy. Note that all of the above identified white matter tracts linked the limbic system and prefrontal areas (Fig. 4, green arrows). This is all data-driven, which further validates the effectiveness of multimodal fusion and supports that miR-132 dysregulation may affect both function and structure of the fronto-limbic circuits. Furthermore, this miR-132 associated fronto-limbic network was replicated in a medication naïve MDD subset (Fig. 3), indicating that such an association is not affected by taking antidepressants 3 months prior.
Regarding the expression patterns across different tissues, miR-132 gene expression is detected in normal human tissues at baseline conditions in the brain, pituitary, ovary and tibial nerve according to the human gene database (http://www.genecards.org/cgi-bin/carddisp.pl?gene=MIR132#expression). Note that miR-132 is regulated by CREB and its levels increase significantly after neuronal activity (Nudelman et al., 2010). With respect to miR-132 expression in different brain regions, previous experimental results have demonstrated that miR-132 is a brain-enriched miRNA that plays a key role during both the early and late stages of adult neurogenesis (Vo et al., 2005; Wayman et al., 2008; Magill et al., 2010; Im and Kenny, 2012). In addition, according to animal model studies, the expression levels of miR-132 are higher in prefrontal areas compared with the other brain regions (Vasic et al., 2008; Miller et al., 2012). Specifically, as in our results, increased miR-132 expression profiles lead to decreased fALFF and reduced grey matter volume in prefrontal areas (including DLPFC and medial PFC). Further, increased expression levels of miR-132 were also detected in hippocampus in a rat model of chronic stress-induced depression (Su et al., 2015). MiR-132 has also been shown to control the development of dendrites and spines, as well as synaptic integration in cultured hippocampal neurons and newborn hippocampal neurons (Magill et al., 2010). Correspondingly, both brain regions (PFC and hippocampus) were identified in our results.
In addition, as shown in Figs 2 and 4, increased miR-132 in MDD is also associated with both reduced fALFF and grey matter values in the limbic system, especially hippocampus and amygdala. In an animal model of chronic stress-induced depression, miR-132 expression was found to be upregulated in the hippocampus of CUS (chronic unpredictable stressors)-exposed rats (Su et al., 2015). MiR-132 has also been shown to control the development of dendrites and spines and spur synaptic integration in cultured hippocampal neurons and newborn hippocampal neurons (Magill et al., 2010). Furthermore, BDNF, CREB, and glucocorticoids are also the key components for hippocampal neurogenesis, all of which are directly related to miR-132 (Zheng et al., 2013). The above findings support our speculation that increased miR-132 may play an important role in the aetiology of depression in the hippocampus, which is also consistent with two meta-analysis studies that indicate grey matter volume of hippocampus was markedly reduced in MDD patients (MacMaster et al., 2008). The amygdala, another component of limbic system, is the most notable brain structure involved in emotional responses and emotion-related memories (Richter-Levin and Akirav, 2000). The decreased amygdala volume observed in our study is consistent with the evidences resulted from region of interest-based analysis of amygdala in unmedicated MDD patients (Hamilton et al., 2008), and complemented by findings in post-mortem brains of patients who had suffered from unipolar depression (Bowley et al., 2002).
Note that the interactions between amygdala and hippocampus are also key players in emotional learning. The hippocampus and amygdala are the core regions in the limbic system and have widespread connections to diverse cortical areas, such as the frontal cortex, anterior thalamic nuclei, basal ganglia and hypothalamus, all of which are regions known to constitute the neuroanatomical network of mood regulation (Dolcos et al., 2014). The amygdala modifies hippocampal responses and vice versa. This dual interaction of amygdala and hippocampus and the dynamics between them may support emotionally based memory uniqueness, as previously reported (Richter-Levin and Akirav, 2000). This is also in line with our results that higher miR-132 expression levels are associated with lower fALFF and grey matter volume in hippocampus-amygdala nuclei and poorer executive function in MDD. The above extracted fronto-limbic system is the key network for emotional regulation and memory, which is potentially linked with cognitive impairment in unmedicated patients with MDD. These alterations implicate a distributed neural circuit composed of multiple sectors of the prefrontal cortex in interaction with limbic regions (hippocampus, amygdala, and thalamus) (Clark et al., 2009).
Another interesting finding of the present study was that individual miR-132 is found to affect cognitive performance mainly focused on domains of attention (RVP) and executive function (IED), while recent meta-analysis also suggest that these two domains are the most severely affected cognitive domains in patients with MDD (Rock et al., 2014). As in our results, the expression levels of miR-132 is higher in patients with MDD than healthy control subjects, which is consistent with reports that overexpression of miR-132 leads to profound cognitive deficits (Hansen et al., 2013). The higher expression levels of miR-132 lead to decreased fALFF within PFC, insular and anterior cingulate cortex (key parts of the salience network), but increased fALFF in the visual cortex (BA17, BA18, BA19). There is evidence that the inhibition of miR-132 alters dendritic spine maturation and prevents ocular dominance plasticity in neurons within the visual cortex (Mellios et al., 2011; Tognini et al., 2011), supporting our findings. Hence, all of the fALFF regions mentioned have previously been implicated in depression and are associated with attention deficits (Seeley et al., 2007), which is in accordance with fMRI_ICref correlated with attention domain (RVP). In addition, dysregulation of miR-132 has also been suggested to be important in hippocampal-dependent learning and memory (Hansen et al., 2013) in a number of neurocognitive disorders. These miR-132 expression levels are found tightly regulated within a narrow range in order to ensure normal learning and memory formation (Hansen et al., 2013) that correspond to executive function domain (IED) correlated with structural MRI_ICref. According to our results, we can speculate that increased peripheral blood miR-132 levels in patients with MDD may be closely relevant to their dysfunction in attention and executive function, which provides promising evidence that miR-132 dysregulation may influence the neurocognitive impairment in patients with MDD.
A possible limitation of this work is that we only tested one miRNA that was consistently implicated in depression studies, while there may exist a large library of MDD-related microRNAs and their target genes; these will be left for future investigations. Second, we measured the miR-132 levels in peripheral blood instead of directly extracted from the brain. Given the difficulty of procuring miR-132 from brain tissues, epigenetic signatures measured in more accessible tissues such as blood can serve as a surrogate marker for the brain (Zeng, 2011; Walton et al., 2016). Based on previous observations (Li et al., 2013; Su et al., 2015), the overall levels of miR-132 in the brain in mood disorders can be reflected by its levels in blood. It has therefore been suggested that epigenetic processes (increased miR-132 expression levels in unmedicated MDD) might impact both functional and structural changes (fronto-limbic system) in the brain through modulating gene expressions. Another limitation is that current study is mainly descriptive. The intrinsic causality between miR-132 and brain alterations awaits further dedicated investigation on pathology of depression, although it has been reported that miR-132 has direct associations with neuronal activity in brain (Im and Kenny, 2012). In addition, the reason we choose these unmedicated patient samples is to minimize the medication impact on both miR-132 expression and brain imaging data. However, in future studies, we may conduct similar comparisons between drug naive patients with MDD and recurrent depressive disorders. Further, we can include temporal features, such as dynamic states or functional network connectivity matrices, as fusion input for functional MRI to capture links between both temporal and spatial changes.
In summary, using a data-driven, supervised-learning method, we revealed that miR-132 dysregulation in MDD may affect multi-facets of both brain function and structure on the fronto-limbic circuits, which is the first step in determining the miR-132 variants on multimodal neuroimaging in unmedicated depression. This work deepens our understanding of how miR-132 dysregulation in MDD contribute to the loss of specific brain areas and provides insight into how plausible epigenetic factors may influence the pathophysiology of depression. Collectively, as summarized in Fig. 4, our results suggest (i) higher miR-132 levels in MDD are associated with both lower fALFF and lower grey matter in fronto-limbic network (Soares and Mann, 1997); and (ii) the identified brain regions linked with increased miR-132 levels were also associated with poorer cognitive performance in domains of attention and executive function. All of these associations are consistent with either unmedicated or medication-naïve patients. These findings support that the fronto-limbic system as previously implicated in major depression in functional, anatomical and structural brain-imaging studies were associated with miR-132 dysregulation, while the intrinsic causality between miR-132 and brain alterations requires further investigation and verification.
Funding
This study was funded by the National High-Tech Development Program (863 plan, No. 2015AA020513), ‘100 Talents Plan’ of Chinese Academy of Sciences, the Chinese National Natural Science Foundation No. 81471367, No. 81671344 and No. 61773380, the Strategic Priority Research Program of the Chinese Academy of Sciences (grant No. XDB02060005), the National Science and Technology Support plan (2015BAI13B02), and NIH grants R01EB005846, 1R01MH094524 and P20GM103472.
Supplementary material
Supplementary material is available at Brain online.
Supplementary Material
Glossary
Abbreviations
- fALFF
fractional amplitude of low frequency fluctuations
- HDRS
Hamilton Depression Rating Scale
- IC
independent component
- MDD
major depressive disorder
References
- APA. Diagnostic and statistical manual of mental disorders: DSM-IV-TR. Washington, DC: American Psychiatric Publishing Inc.; 2000. [Google Scholar]
- Bowley MP, Drevets WC, Ongur D, Price JL. Low glial numbers in the amygdala in major depressive disorder. Biol Psychiatry 2002; 52: 404–12. [DOI] [PubMed] [Google Scholar]
- Busatto GF. Structural and functional neuroimaging studies in major depressive disorder with psychotic features: a critical review. Schizophr Bull 2013; 39: 776–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Adali T. Feature-based fusion of medical imaging data. IEEE Trans Inf Technol Biomed 2009; 13: 711–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Sui J. Multimodal fusion of brain imaging data: a key to finding the missing link(s) in complex mental illness. Biol Psychiatry 2016; 1: 230–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S, Marriott M, Nahmias C, MacQueen GM. Lower hippocampal volume in patients suffering from depression: a meta-analysis. Am J Psychiatry 2004; 161: 598–607. [DOI] [PubMed] [Google Scholar]
- Cheng HY, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, et al. microRNA modulation of circadian-clock period and entrainment. Neuron 2007; 54: 813–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark L, Chamberlain SR, Sahakian BJ. Neurocognitive mechanisms in depression: implications for treatment. Annu Rev of Neurosci 2009; 32: 57–74. [DOI] [PubMed] [Google Scholar]
- Cullen KR, Klimes-Dougan B, Muetzel R, Mueller BA, Camchong J, Houri A, et al. Altered white matter microstructure in adolescents with major depression: a preliminary study. J Am Acad Child Adolesc Psychiatry 2010; 49: 173–83.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolcos F, Wang L, Mather M. Current research and emerging directions in emotion-cognition interactions. Front Integr Neurosci 2014; 8: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drevets WC. Functional neuroimaging studies of depression: the anatomy of melancholia. Annu Rev Med 1998; 49: 341–61. [DOI] [PubMed] [Google Scholar]
- Drevets WC. Prefrontal cortical-amygdalar metabolism in major depression. J Neuropsychiatry Clin Neurosci 1999; 877: 614–37. [DOI] [PubMed] [Google Scholar]
- Drevets WC, Price JL, Furey ML. Brain structural and functional abnormalities in mood disorders: implications for neurocircuitry models of depression. Brain Struct Funct 2008; 213: 93–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman RS. Pathophysiology of depression: the concept of synaptic plasticity. Eur Psychiatry 2002; 19: 306–10. [DOI] [PubMed] [Google Scholar]
- Ebmeier KP, Donaghey C, Steele JD. Recent developments and current controversies in depression. Lancet 2006; 367: 153–67. [DOI] [PubMed] [Google Scholar]
- First MB, Spitzer RL, Gibbon ML, Williams JBW. Structured clinical interview for DSM-IV-TR axis I disorders, research version, patient edition. New York, NY: Biometrics Research; 2002. [Google Scholar]
- Gong Q, He Y. Depression, neuroimaging and connectomics: a selective overview. Biol Psychiatry 2015; 77: 223–35. [DOI] [PubMed] [Google Scholar]
- Hamilton JP, Siemer M, Gotlib IH. Amygdala volume in major depressive disorder: a meta-analysis of magnetic resonance imaging studies. Mol Psychiatry 2008; 13: 993–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KF, Karelina K, Sakamoto K, Wayman GA, Impey S, Obrietan K. miRNA-132: a dynamic regulator of cognitive capacity. Brain Struct Funct 2013; 218: 817–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im HI, Kenny PJ. MicroRNAs in neuronal function and dysfunction. Trends Neurosci 2012; 35: 325–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are MicroRNA targets. Cell 2005; 120: 15–20. [DOI] [PubMed] [Google Scholar]
- Li XL, Fang YN, Gao QC, Lin EJ, Hu SH, Ren L, et al. A diffusion tensor magnetic resonance imaging study of corpus callosum from adult patients with migraine complicated with depressive/anxious disorder. Headache 2011; 51: 237–45. [DOI] [PubMed] [Google Scholar]
- Li YJ, Xu M, Gao ZH, Wang YQ, Yue Z, Zhang YX, et al. Alterations of serum levels of BDNF-related miRNAs in patients with depression. PLoS One 2013; 8: e63648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YO, Adali T, Calhoun VD. Estimating the number of independent components for functional magnetic resonance imaging data. Hum Brain Mapp 2007; 28: 1251–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzetti V, Allen NB, Fornito A, Yucel M. Structural brain abnormalities in major depressive disorder: a selective review of recent MRI studies. J Affect Disord 2009; 117: 1–17. [DOI] [PubMed] [Google Scholar]
- MacMaster FP, Mirza Y, Szeszko PR, Kmiecik LE, Easter PC, Taormina SP, et al. Amygdala and hippocampal volumes in familial early onset major depressive disorder. Biol Psychiatry 2008; 63: 385–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magill ST, Cambronne XA, Luikart BW, Lioy DT, Leighton BH, Westbrook GL, et al. microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc Natl Acad Sci USA, 2010; 107: 20382–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meda SA, Ruano G, Windemuth A, O'Neil K, Berwise C, Dunn SM, et al. Multivariate analysis reveals genetic associations of the resting default mode network in psychotic bipolar disorder and schizophrenia. Proc Natl Acad Sci USA 2014; 111: E2066–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellios N, Sugihara H, Castro J, Banerjee A, Le C, Kumar A, et al. miR-132, an experience-dependent microRNA, is essential for visual cortex plasticity. Nat Neurosci 2011; 14: 1240–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BH, Zeier Z, Xi L, Lanz TA, Deng S, Strathmann J, et al. MicroRNA-132 dysregulation in schizophrenia has implications for both neurodevelopment and adult brain function. Proc Natl Acad Sci USA 2012; 109: 3125–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouillet-Richard S, Baudry A, Launay JM, Kellermann O. MicroRNAs and depression. Neurobiol Dis 2012; 46: 272–8. [DOI] [PubMed] [Google Scholar]
- Novina CD, Sharp PA. The RNAi revolution. Nat Rev Neurosci 2004; 430: 161–4. [DOI] [PubMed] [Google Scholar]
- Nudelman AS, DiRocco DP, Lambert TJ, Garelick MG, Le J, Nathanson NM, et al. Neuronal activity rapidly induces transcription of the CREB-regulated microRNA-132, in vivo. Hippocampus 2010; 20: 492–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips ML, Drevets WC, Rauch SL, Lane R. Neurobiology of emotion perception I: the neural basis of normal emotion perception. Biol Psychiatry 2003; 54: 504–14. [DOI] [PubMed] [Google Scholar]
- Qi S, Calhoun VD, van Erp TGM, Bustillo J, Damaraju E, Turner JA, et al. Multimodal fusion with reference: searching for joint neuromarkers of working memory deficits in schizophrenia. IEEE Trans Med Imaging 2018; 37: 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter-Levin G, Akirav I. Amygdala-hippocampus dynamic interaction in relation to memory. Mol Neurobiol 2000; 22: 11–20. [DOI] [PubMed] [Google Scholar]
- Robbins TW, James M, Owen AM, Sahakian BJ, McInnes L, Rabbitt P. Cambridge Neuropsychological Test Automated Battery (CANTAB): a factor analytic study of a large sample of normal elderly volunteers. Dement Geriatr Cogn Disord 2010; 5: 266–81. [DOI] [PubMed] [Google Scholar]
- Rock PL, Roiser JP, Riedel WJ, Blackwell AD. Cognitive impairment in depression: a systematic review and meta-analysis. Psychol Med 2014; 44: 2029–40. [DOI] [PubMed] [Google Scholar]
- Seeley WW, Menon V, Schatzberg AF, Keller J, Glover GH, Kenna H, et al. Dissociable intrinsic connectivity networks for salience processing and executive control. J Neurosci 2007; 27: 2349–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature 2008; 455: 58–63. [DOI] [PubMed] [Google Scholar]
- Sinclair L. Screening of postpartum women shows high rates of depression. Psychiatr News 2013; 48: 22–1. [Google Scholar]
- Sliz D, Hayley S. Major depressive disorder and alterations in insular cortical activity: a review of current functional magnetic imaging research. Front Hum Neurosci 2012; 6: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares J, Mann JJ. The anatomy of mood disorders–review of structural neuroimaging studies. Biol Psychiatry 1997; 41: 86–106. [DOI] [PubMed] [Google Scholar]
- Su M, Hong J, Zhao Y, Liu S, Xue X. MeCP2 controls hippocampal brain-derived neurotrophic factor expression via homeostatic interactions with microRNA132 in rats with depression. Mol Med Rep 2015; 12: 5399–406. [DOI] [PubMed] [Google Scholar]
- Tognini P, Putignano E, Coatti A, Pizzorusso T. Experience-dependent expression of miR-132 regulates ocular dominance plasticity. Nat Neurosci 2011; 14: 1237–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urdinguio RG, Sanchez-Mut JV, Esteller M. Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancet Neurol 2009; 8: 1056–72. [DOI] [PubMed] [Google Scholar]
- Vasic N, Walter H, Hose A, Wolf RC. Gray matter reduction associated with psychopathology and cognitive dysfunction in unipolar depression: a voxel-based morphometry study. J Affect Disord 2008; 109: 107–16. [DOI] [PubMed] [Google Scholar]
- Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, et al. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci USA 2005; 102: 16426–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton E, Hass J, Liu J, Roffman JL, Bernardoni F, Roessner V, et al. Correspondence of DNA methylation between blood and brain tissue and its application to schizophrenia research. Schizophr Bull 2016; 42: 406–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HY, et al. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci USA 2008; 105: 9093–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Jiang W, Ren L, Ouyang X, Jiang Y, Wu F, et al. Impaired interhemispheric connectivity in medication-naive patients with major depressive disorder. J Psychiatry Neurosci, 2013; 38: 43–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Liu J, Wang Y, Wang L, Weng S, Tang Y, et al. MicroRNA-210 as a novel blood biomarker in acute cerebral ischemia. Front Biosci 2011; 3: 1265–72. [DOI] [PubMed] [Google Scholar]
- Zhang X, Yao S, Zhu X, Wang X, Zhu X, Zhong M. Gray matter volume abnormalities in individuals with cognitive vulnerability to depression: a voxel-based morphometry study. J Affect Disord 2012; 136: 443–52. [DOI] [PubMed] [Google Scholar]
- Zheng Z, Zeng Y, Huang H, Xu F. MicroRNA-132 may play a role in coexistence of depression and cardiovascular disease: a hypothesis. Med Sci Monit 2013; 19: 438–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.