Abstract
Evaluating the potential for methylmercury (MeHg) toxicity relies on accurately predicting the mercury (Hg) body burden that results from eating fish. Hg body burden is directly determined by the slow elimination kinetics of MeHg in the human body (kel = 0.014 days−1 or t1/2 =50 days). Existing studies on MeHg half-life in humans demonstrate a wide range values (t1/2 = 30 to >150 days) and has lead to uncertainty in the derivation of a regulatory standard for acceptable daily oral intake. The causes of variation in MeHg toxicokinetics in humans remain little explored. Here we characterize variation in human MeHg metabolism and elimination rate (kel) in 37 adult volunteers who consumed 3 fish meals. We determined MeHg elimination rates via longitudinal Hg analysis in single hairs using laser ablation inductively coupled plasma mass spectrometry. We also measured MeHg metabolism (biotransformation) via speciation of fecal Hg. We find an average kel = 0.0157 days−1 (t1/2 = 44 days) amongst a more than 2-fold variation in kel across the cohort (0.0248–0.0112 days−1; t1/2 = 28–62 days). Although MeHg biotransformation varied widely between individuals, it showed a positive association with elimination rates across the cohort. A more than 2-fold change in kel over a period of 2 years was seen in some individuals. In 2 individuals, who received antibiotic for unrelated health issues, elimination rate was seen to slow significantly. Associations of kel with age, body mass index, gender, and fish eating habits were not observed. We establish that a measure of methylmercury metabolism and eliminaiton status (MerMES) can reduce uncertainty in determining an individual’s MeHg toxicokinetics subsequent to eating fish.
Keywords: methylmercury, elimination, metabolism, biotransformation, half-life, toxicokinetics, laser ablation ICP-MS
Methylmercury (MeHg) is a persistent environmental contaminant that accumulates in fish and seafood and is best known for its toxic effects on the developing nervous system at doses associated with poisoning episides (Amin-Zaki et al., 1979; Harada, 1995). Despite growing evidence for an overall benefit of fish in the maternal diet, considerable uncertainty remains over the risks of toxicity that come with consumption of fish containing natural background levels of MeHg (Oken et al., 2008; Starling et al., 2015). Strategies to limit MeHg exposure predominantly rely on fish consumption advisories (EPA, 2017), which are built upon the fundamental concept that a risk for toxicity exists when the mercury (Hg) body burden exceeds a defined amount (eg, a Reference Dose [RfD]). Evaluating the potential for MeHg toxicity therefore relies on accurately predicting the Hg body burden that will result from a habit of fish eating. The slow elimination kinetics of MeHg in the human body (kel = 0.014 days−1; t1/2 = 50 days) are what dictate the biological half-life of MeHg and is therefore a direct determinant of the Hg body burden. However, results from existing kinetic studies aimed at resolving a MeHg half-life in humans demonstrate a wide range of values (t1/2 = 30 to >150 days) (Aberg et al., 1969; Al-Shahristani and Shibab, 1974; Birke et al., 1972; Kershaw et al., 1980; Miettinen et al., 1971; Phelps et al., 1980; Rand et al., 2016; Sherlock et al., 1984; Smith et al., 1994; Yaginuma-Sakurai et al., 2012). This wide range of t1/2 values is a source of uncertainty for MeHg toxicokinetics, which is a central component of a 10-fold “uncertainty factor” used in the derivation of the US-EPA RfD (EPA, 2001). Factors contributing to the wide range of reported t1/2 values may reflect methodological differences. However, it may also be due to a naturally occurring biological basis for variation in MeHg toxicokinetics in humans, which remains little explored. One possibility is that genetic polymorphisms, especially in genes involved in MeHg transport and elimination, may be a contributing factor (Engstrom et al., 2016; Llop et al., 2017; Mazzaron Barcelos et al., 2012; Vorojeikina et al., 2017). Nonetheless, we have recently shown that even within an individual MeHg elimination rate can vary almost 2-fold over a period of as little as 6 months (Rand et al., 2016), suggesting that factors in addition to conventional genetics can greatly influence MeHg kinetics. The lingering uncertainty in human MeHg toxicokinetics calls for more rapid and accurate methods to characterize MeHg metabolism and elimination across a broader spectrum of individuals and populations.
Metabolism (biotransformation) of MeHg to inorganic Hg (I-Hg) is proposed to be an essential intermediate step to MeHg elimination (Ishihara, 2000; Rowland et al., 1977). Typically, 90% or more of the Hg derived from a MeHg dose is excreted in feces as I-Hg (Ishihara, 2000; Norseth and Clarkson, 1971). This is consistent with the model whereby, in the course of enterohepatic cycling of MeHg, biotransformation in the gut lumen yields the poorly absorbed I-Hg isoform that is more readily excreted (Norseth and Clarkson, 1971). In rodents, administration of antibiotics has the effect of drastically reducing MeHg elimination while also reducing MeHg biotransformation, highlighting a role for gut microbiota in the process (Rowland et al., 1977). It is therefore reasonable to predict that a faster elimination rate may stem from a greater MeHg biotransformation activity that would also yield a higher proportion of I-Hg in the feces. The relative amount of I-Hg (% of total Hg [T-Hg]) in human feces can vary widely suggesting that biotransformation could be a source of variation in the overall metabolism and elimination of MeHg (Ishihara, 2000; Rand et al., 2016). Although rodent studies point to gut microbiota as mediators of MeHg biotransformation and elimination (Rowland et al., 1977, 1984), an analogous role for gut microbiota in humans has yet to be demonstrated.
We recently described an improved method for the individualized determination of MeHg elimination rates (Rand et al., 2016). Prior prospective studies have been limited in the number of subjects examined since they have relied upon interventions with high level MeHg exposures, long observation periods, repeated blood draws or the use of radioisotopic (203 Hg) MeHg injection or ingestion (Aberg et al., 1969; Al-Shahristani and Shibab, 1974; Birke et al., 1972; Kershaw et al., 1980; Miettinen et al., 1971; Phelps et al., 1980; Sherlock et al., 1984; Smith et al., 1994). Our method is based on longitudinal analysis of Hg in hair of study participants who consume normal dietary levels of fish. Hair is a well-established biomarker that shows a direct relationship with blood Hg levels (Cernichiari et al., 2007; Phelps et al., 1980; Zareba et al., 2008). During hair growth, transport and equilibration of MeHg between the bloodstream and keratinocytes in the follicle occurs, and is subsequently incorporated into the growing hair shaft (Kempson and Lombi, 2011; Zareba et al., 2008). As a result, longitudinal analysis of hair has proven an effective means to obtain a time-resolved profile of MeHg levels in blood (Al-Shahristani and Shibab, 1974; Phelps et al., 1980). Methods of X-ray fluorescence spectrometry and, more recently, laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS) has made Hg analysis of single strands of hair possible (Legrand et al., 2007; Marsh et al., 1987; Stadlbauer et al., 2005). We have previously reported a highly effective method using LA-ICP-MS to resolve changes in Hg levels across single hairs resulting from fish consumption (Rand et al., 2016). Our previous study examined the MeHg elimination rates of 8 individuals over a 60-day period after a controlled tuna steak consumption regimen. We were able to measure MeHg elimination rates and calculate half-lives, which spanned a range consistent with that previously reported (t1/2 = 43–128 days) (Rand et al., 2016). The small sample size of this prior study (n = 8) precluded extensive evaluation of the apparent variation in elimination rates as well as correlation analyses of elimination kinetics with MeHg biotransformation or population characteristics, such as age or gender.
In this study we set out to characterize variation in human MeHg metabolism and elimination. We have applied our methodology of longitudinal hair Hg analysis, together with fecal Hg speciation, to determine a MeHg metabolism and elimination status (MerMES) in 37 individuals subsequent to consumption of just 3 fish meals.
MATERIALS AND METHODS
Study design
Volunteers were recruited from the University of Rochester Medical Center community. Subjects were enrolled if they were between 18 and 80 years of age and in good general health on a self-reported basis. Subjects were excluded if they had known allergies to fish, were pregnant or lactating, used hair dyes or treatments, had known gastrointestinal or renal disorders, or had used systemic antibiotics within the last 2 months. Study procedures were reviewed and approved by the University of Rochester Research Subjects Review Board and consent was obtained from each subject.
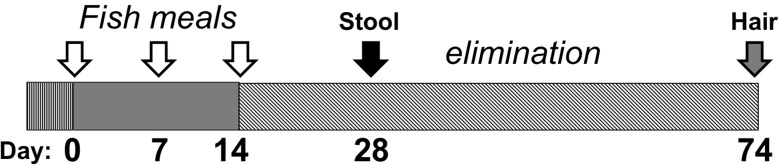
The overall study design is summarized in Figure 1. Each subject agreed to consume 3 fish meals at 7-day intervals. Stool samples were collected at approximately 2 weeks after the last fish meal and hair samples were collected upon completion of the study. Only a small amount of MeHg is eliminated via other routes (eg, <5% via urine in first 70 days; Smith et al., 1994), therefore analyses focused on the biomarkers of hair and feces. Subjects were asked to keep a daily record of types of food consumed at each meal spanning the fish consumption and elimination period. In addition, subjects were asked to record any bouts of illness and new course of medication that was incurred during the study. Concurrent with this study, stool sampling was done at 3 time points (before fish consumption, immediately after the last fish meal, and 2 weeks after the last fish meal). The entire stool was collected for the third time point only, and used for Hg analyses (indicated in Figure 1). Subsampling of all 3 stools was performed for the purpose of nucleic acid extraction, to be used in gut microbiome profiling in a parallel initiative. Results from these microbiome studies will be reported separately.
Figure 1.
Experimental design. Participants ate 3 tuna steaks 7 days apart and refrained from fish and seafood consumption for the elimination period (approximately 60 days). A stool sample collected at 2 weeks after the last fish meal was used to measure the T-Hg and I-Hg levels. On approximately day 74 hair was collected for longitudinal analysis of Hg content.
Study population
Thirty-seven subjects were recruited and retained for the study (23 women, 14 men), coming from the Rochester, NY metropolitan area. In total 22 of the 37 subjects were between the ages of 21–30, 7 were between the ages of 31–40, 3 between the ages of 41–50, 5 over the age of 51, with body weights ranging from 52 to 138 kg (114–304 pounds, Supplementary Table 1). Thirty-five of the subjects were Caucasian, 1 was African American female (S20), and 1 Indian-Asian male (S22). No unusual deviations from each subject’s respective dietary habit was noted prior to and during the course of the trial as determined from entry interviews and the subsequent food diaries kept by the subjects. Subjects were asked to resume and maintain their typical diets, and refrain from any fish or seafood consumption during the 60-day elimination period. Food diaries and personal communication confirmed there was no fish eating during the elimination period by any of the subjects. Two subjects reported illnesses requiring the use of antibiotics during the course of the study, which was recorded in their food diaries.
Absence of dental amalgam was initially a requirement for recruitment of subjects in order to avoid confounding effects on speciation of fecal Hg (Rand et al., 2016). At the end of the recruitment period, to meet recruitment goals, 4 subjects were admitted who carried dental amalgams. The number of tooth surfaces filled with dental amalgam for each subject was recorded by visual inspection at the time of hair collection on the final visit. These subjects had amalgam surfaces numbering from 1 to 11 (Supplementary Table 1).
Fish meal consumption
Wild caught yellowfin tuna steaks of various sizes ranging from 120 to 300 g, were prepared by and purchased from a local supermarket. Steaks were cut from 2 fish of similar age and size. A small sample (approximately 15 g) from each of 3 body regions (nape, body, and tail) of each fish was obtained for Hg analysis, which was completed prior to cutting of the fish steaks. Steaks were cut, weighed, individually wrapped and frozen by the market staff. Portion sizes varied between subjects; however, the maximum portion size provided to any 1 subject was previously determined according to that subject’s body weight so as to adhere to recommendations the FDA/EPA of 8–12 oz/week of fish (227–340 g/week) and to target a maximum predicted blood level that did not exceed the EPA RfD level of 5.8 µg/l in Hg in blood. The weight of fish consumed at each meal was recorded by the subjects. The maximum individual portion size consumed was 300 g (10.6 oz) for a 138 kg (304 lb) subject (S23). Hg levels in the fish were determined using cold vapor atomic absorption (CVAA) spectroscopy according to the methods of Magos and Clarkson (Magos and Clarkson, 1972). Hg levels in the 2 fish were 1.22 ppm Hg (nape = 1.24 ppm, body = 1.19 ppm, and tail = 1.24 ppm) and 1.12 ppm Hg (nape = 0.98 ppm, body = 0.933 ppm, and tail = 1.27 ppm), respectively. All Hg values were within the range of those reported previously for yellowfin tuna by the FDA (1991–2010). Subjects were provided with 3 portions of frozen fish steaks in a cooler for transport home. Preparation of fish for eating was at the discretion of each subject. Previous studies show that cooking methods do have an appreciable effect on MeHg bioavailability, commonly reducing bioavailability relative to consumption of raw fish (reviewed in Bradley et al., 2017). Incomplete absorption of MeHg provided in the fish portions was therefore anticipated, yet acceptable since we were confident that detectable levels of Hg would result in hair from this feeding paradigm (Rand et al., 2016). Furthermore, with the assumption of a single-compartment first order kinetic model (see “MeHg elimination rate determination” section below), the elimination rate constant is a parameter that is independent of absorption rate and dose level.
Hair and stool sampling
Hair samples were taken approximately 60 days after the final fish meal (Figure 1). Hairs were manually extracted using a brisk pull of a tuft of 4–8 strands from the crown region of the scalp using a gloved hand. This resulted in abstraction of follicle/root remnants on the hair shaft. Hairs were stored in sealed plastic bags until mounting for Hg analysis. No additional steps were taken to clean the hair prior to analysis, as previous studies show that washing and chelating steps are ineffective for removal of potentially contaminating hair surface Hg from exogenous exposures (Ali Aldroobi et al., 2013; Li et al., 2008). Hairs were mounted on double-sided clear tape (Scotch brand) previously adhered to a clean glass microscope slide. Single hair strands were aligned straight and adhered to the tape using watch-maker forceps, and were then pressed into the adhesive surface using a clean microscope slide. These steps allowed for the immobilization of the hair with approximately two-thirds of the shaft diameter sitting above the surface of the adhesive. One-centimeter increments were marked on the tape adjacent to the hair strand as reference points to orient the hair for laser ablation.
Stool samples were collected by the subjects using a Fisherbrand Commode Specimen Collection System (Fisher Scientific No. 02-544-208). Stool samples were either frozen immediately at home by the subjects or brought directly to the laboratory where they were frozen at −20 °C until preparation for Hg analyses.
Longitudinal analysis of Hg in hair
Longitudinal analysis of Hg in hair was performed with LA-ICP-MS at the Dartmouth Trace Element Analysis Lab. Laser ablation was carried out with a UP213 laser (ESI, Bozeman, Montana) equipped with a programmable X-Y stage for predetermining coordinates for ablation of a series of spots on the hair shaft. Hair growth occurs at approximately 1.1 cm per month (Harkey, 1993; Myers and Hamilton, 1951), and 3.0 cm of the initial segment, including the root portion of the shaft, was expected to cover the 60-day elimination period, the 14-day fish meal period and a period prior to the fish consumption to assess background Hg levels. Accordingly, 90 consecutive spots were ablated at a 333 µm interval (approximately 1-day growth) over a 3.0 cm segment of the hair segment closest to the root. Elemental detection was performed with an Agilent 8800 QQQ ICP-MS (Santa Clara, California) set to monitor 202Hg and 34 S isotopes in no gas mode. Sulfur concentration has been shown to be stable across the length of hair (Legrand et al., 2004; Rodushkin and Axelsson, 2003), therefore, the 34 S isotope was used as an internal standard for the relative mass of hair captured with each ablation in accord with previously published methods (Legrand et al., 2007). Sampling parameters included laser settings of 55 µm spot size with 3.0-s dwell time at 10 Hz and laser power set at 40% equating to a fluence of 4.00 J/cm2. With this method we typically achieve detection limits for Hg of 10–50 ng/g. Data were collected as time resolved intensity of 202Hg and 34 S and exported as Excel data files. Hg values were expressed as a ratio of the peak areas of the transient signals for 202Hg and 34 S multiplied by a factor of 1000 since the signal intensity for 202Hg was approximately 3 orders of magnitude lower than 34 S, consistent with the low abundance of Hg relative to S in hair. We refer to the relative concentration of Hg in hair as a ratio denoted as Hg:S (×1000 in arbitrary units). For the bulk of analyses Hg values were not quantified with respect to a standard reference material (SRM). As we previously reported in Rand et al. (2016), absolute Hg quantification using this method was not necessary since the goal of this study was to determine a kinetic parameter of Hg elimination, which can be derived from the change in relative concentration of Hg over time.
Quantification of hair Hg was attempted on a limited number of subject samples. A standard of hair SRM (IAEA 86, 0.573 ppm Hg total [IAEA, 2017]) was prepared in a pressed pellet with 5% cellulose binder added. Cellulose binder independently demonstrated no Hg or S by ICP-MS (T.P, B.J. unpublished observation). The matrix demonstrated surface heterogeneity due to the varied size of hair fragments in the SRM powder preparation (Supplementary Figure 1A). To determine an Hg:S ratio that corresponds to the 0.573 ppm Hg value of the SRM, ablation of the pellet was done by rastering across the pellet surface with laser intensity and spot size equivalent to that used for the subject hair ablation. A mean value of 20.8 (Hg:S ×1000) was determined (Supplementary Figure 1B) and used to convert Hg:S determinations to ppm values for the 3 subjects’ hairs that were analyzed.
MeHg elimination rate determination
Data were plotted in terms of Hg:S versus time (days). Growth rate of each individual hair was calculated by determining the distance between the Hg:S spikes that correspond to the first and last fish meal and then dividing by the number of days between these meals. Elimination data spanned the interval beginning 2 spots (approximately 48 h) after the last fish meal to the margin of root material at the follicle end of the hair. Elimination rates were calculated based on a 1-compartment model with the assumption of a first-order process according to:
and its natural log transformed expression:
where Ct is the relative concentration of Hg (ie, Hg:S) at time t and C0 is the concentration of Hg at t = 0. The elimination rate constant (kel [days−1]) was determined from the slope of a linear regression line fit to a plot of ln(Hg:S) versus time created using Prism 6 (Graphpad Software, Inc. La Jolla, California) as previously reported in Rand et al., 2016). For most subjects in this study the premeal Hg:S level in hair was low with respect to the Hg:S resulting from the fish intake. However, some subjects (eg, S14; Figure 2) demonstrated an elevated premeal hair Hg:S relative to the other participants. In several instances the premeal Hg:S in hair demonstrated a declining, and in some case inclining, slope (see Figure 2, S14 and Figure 5, S2) indicating this was not representative of a stable baseline Hg level in these subjects, and may even represent an elimination of MeHg from a previous fish meal (see Figure 4). Therefore, calculations of elimination rates were performed assuming a baseline Hg:S value of zero in all subjects. In this study, where elimination periods spanned only a single half-life for each subject, the data were interpreted to reflect the elimination rate for the approximately 60-day period subsequent to the fish meals. For purpose of discussion of the variation in elimination rates the kel was used to estimate a half-life (t1/2 = 0.693/kel) in units of days.
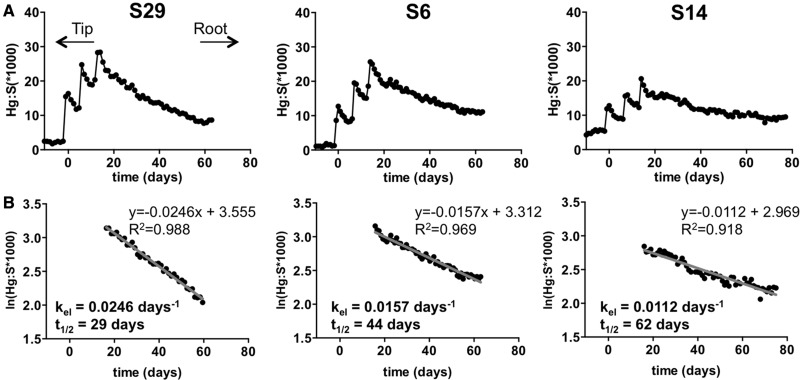
Figure 2.
Longitudinal hair Hg levels and elimination rates. Longitudinal hair Hg analyses leading to determinations of MeHg elimination rates are illustrated for 3 subjects: S29, S6, and S14. A, Hg levels were determined relative to sulfur (Hg:S × 1000 in arbitrary units) in laser ablated spots sampled at 333 µm intervals along a single hair for each subject. Orientation of the hair is indicated from tip to root (left to right). Growth rates for each hair were determined from the interval of the fish meal Hg spikes (see “Materials and methods” section) and were used to derive the time scale (in days) on the X-axis. B, Semi-log plots of the relative Hg concentration (ln(Hg:S*1000)) versus time) for the elimination period following the fish meals was fit by linear regression. The line equation and coefficient of determination (R2) are indicated. Slope values were used in calculations of the elimination rate constants (kel in days−1). kel was used to calculate an estimated half life (t1/2 in days) for each subject.
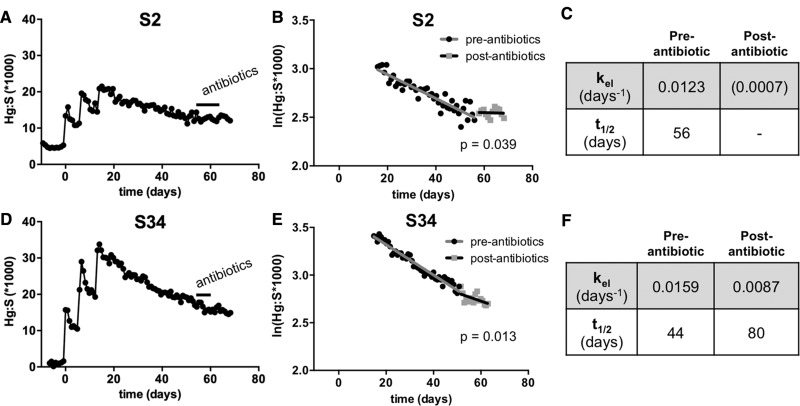
Figure 5.
Elimination rate changes in response to antibiotics. Study participant S2 (A–C) and S34 (D–F) reported taking antibiotics 42 days after the last fish meal during the elimination period of the study. (A, D) Profile of Hg:S levels in S2 and S34. (B, E) Semi-log plots of the Hg concentration (ln[Hg:S]) vs. time) for the elimination period following the fish meals in S2 and S34. Linear regression fits were performed for the preantibiotic and the postantibiotic periods. Analysis of covariance demonstrated a statistical difference in slopes (kel) within each subject (p values for the difference in the pre- and postantibiotic slopes are shown). (C, F) Summary of the kel and t1/2 for MeHg elimination from the pre- and postantibiotic elimination periods. Note, the postantibiotic slope for S2 was not significantly different from zero, however, was significantly different from the preantibiotic slope.
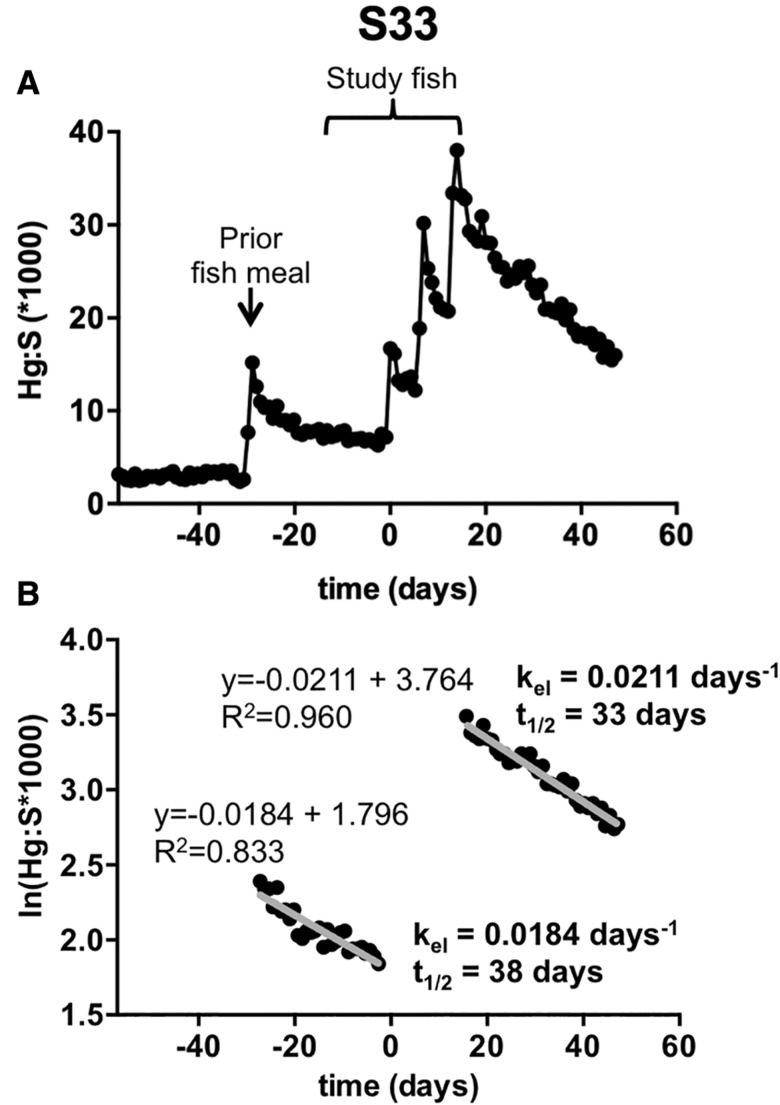
Figure 4.
Sensitivity in detection of Hg from fish meals. A, A spike in the Hg:S ratio prior to the first fish meal of the study revealed a prior fish meal consumed by participant S33. B, Semi-log plot of ln(Hg:S) versus time for the elimination period following the prior fish meal was fit by linear regression. Slope values were used in calculations of the kel (in days−1). kel was used to calculate an estimated half life (t1/2 in days). Statistical analysis of covariance demonstrated no differences in slopes (kel) between the 2 elimination evaluations.
Hg analysis and speciation in fecal samples
Determination of T-Hg and I-Hg in fecal samples was performed by CVAA (Magos and Clarkson, 1972). Fecal samples were prepared in triplicate by homogenization of approximately 1.0 g of feces (wet weight) in 1 ml of 1% cysteine and 2 ml of 0.9% saline solution. Homogenization was performed in 5 ml tubes (Eppendorf No. 0030119460) preloaded with 2.8 g of 1-mm zirconium beads using a Bullet Blender 5E homogenizer (Next Advance, Avril Park, New York). Homogenate and beads were transferred to a 20 ml glass scintillation vial containing 2 ml of 45% NaOH, mixed and heated for 10 min at 90 °C. Samples were cooled and 5 ml of 0.9% saline was added and vortexed for 15 s. Samples were stored at 4 °C until analysis. T-Hg was determined by CVAA and I-Hg was determined by omission of CdCl2 prior to the final reduction step, which effectively restricts organic Hg (ie, MeHg in the feces) from contributing to the Hg analysis (Magos and Clarkson, 1972). Hg determinations were made on a wet weight basis and expressed in parts per billion. Triplicate determinations were made for each sample (T-Hg and I-Hg) to obtain an average value. Fecal I-Hg is expressed as the % of T-Hg (I-Hg/T-Hg ×100).
Statistical analyses
Statistical differences in kel between subjects was evaluated using a method of analysis of covariance within the Prism software package (Graphpad Software, Inc. La Jolla, California) and expressed as a p-value for difference in slopes, with significance occurring at p < .05. As an additional point of reference, a 95% CI for the resulting kel and t1/2 was determined from the built in linear regression tool within the Prism software. Correlations of kel and biotransformation (I-Hg, % of T-Hg in feces) were done with Spearman rank analysis within the Prism software to obtain a coefficient (ρ) and a p-value with significance occurring at p < .05. Additional correlations with age, body mass index, gender and fish eating frequency were done with ANOVA and t-tests using the Prism software.
RESULTS
Determination of MeHg Elimination Rates
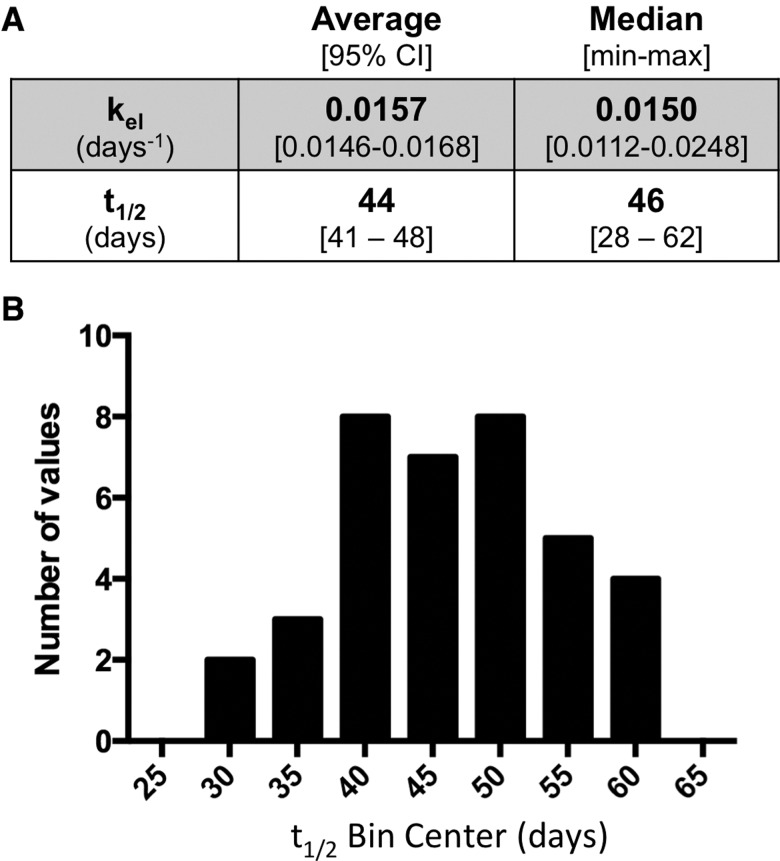
The longitudinal profile of the Hg levels (Hg:S) in the hair that grew over the course of the experimental period is shown in Figure 2A for 3 individuals, S29, S6, and S14, that demonstrate the range of kinetic variation found in the cohort. Spikes in Hg:S were seen that correlate with the day that each fish meal was consumed. A decline in Hg:S followed the final Hg:S spike, which was used as an indicator of the corresponding decrease in blood MeHg concentration during the MeHg elimination process. Elimination rate constants were determined from the slope of linear regression analysis of ln(Hg:S) versus time (Figure 2B). Elimination rates for the 37 subjects ranged from kel = 0.0248 to 0.0112 days−1 (t1/2 = 28–62 days), giving and average of kel = 0.0157 days−1 (t1/2 = 44 days) (Figure 3A). The half-life values across the subjects showed an approximate normal distribution (Figure 3B), with median values of t1/2 = 46 days (kel = 0.0150 days−1) (Figure 3A). Elimination rate constants and estimated half-life values for each subject are summarized in Supplementary Table 1.
Figure 3.
Average value and variation in elimination rates. A, Average and median values for the elimination rates and half-life values for the 37 subjects. B, Distribution of the half-life values binned in 5-day intervals.
Sensitivity in Elimination Rate Determinations With Longitudinal Hg Analysis by LA-ICP-MS
As discussed earlier, and in our prior study (Rand et al., 2016), the Hg:S ratio at baseline in some samples appeared unstable, in some cases trending towards a decreasing slope. For subject S33 examination of hair in the region that corresponds to growth occurring before the study revealed the presence of a spike in Hg:S (Figure 4A). This spike is consistent with this subject consuming a fish meal prior to initiating the study, which had been confirmed earlier from this subject’s recall at recruitment. For this subject, decay of Hg from the study fish meals gave kel = 0.0211 days−1, with an estimated t1/2 of 33 days (Figure 4B). The rate determined from the Hg decay of the prior fish meal gave a slightly slower elimination (kel = 0.0184 days−1, t1/2 = 38 days) (Figure 4B). Multiple regression analysis of these two ln(Hg:S) versus time plots showed an insignificant difference in slope. These data demonstrate the sensitivity of our method to detect fish meals consumed by subjects on their own accord, which in turn could be used to determine elimination kinetics.
Variation in Elimination Rate Constants With Antibiotic Use
Over the course of the study, 2 individuals were prescribed antibiotics by their physicians to address a separate medical condition. Subject S2 began a 10-day regimen of Augmentin (amoxicillin) 42 days after the last fish meal and subject S34 was prescribed Azithromycin for 6 days, also starting 42 days after the last fish meal. Their Hg:S versus time plots showed a discernable change in the elimination of MeHg (Figs. 5A, B, D, and E). Elimination rate constants were calculated from the slope of the ln(Hg:S) versus time plots for periods spanning the pre- and postantibiotic administration. For both subject S2 and S34 significant slowing in the elimination rate was seen for the period subsequent to the antibiotic administration compared with the preantibiotic period (Figs. 5C and F). Multiple linear regression analysis showed that the slopes of the ln(Hg:S) versus time plots for the preantibiotic and postantibiotic period for both S2 and S34 were significantly different (p = .039 and .013, respectively). These data suggests that the antibiotic administration significantly decreased the ability of these study participants to eliminate the MeHg.
Variation in Individual Elimination Rate Constants Over Time
Our prior pilot study demonstrated a significant change in elimination rate for 4 of the 8 subjects between 2 trials conducted approximately 6 months apart (Rand et al., 2016). To evaluate this intraindividual variability further, we took advantage of the fact that 4 of the subjects in the previous pilot study also participated in this study. As seen in Table 1, subjects S3 and S22 demonstrated a significant increase in elimination rates (decreased t1/2) over the 2 trials of the prior pilot study. This trend continued into the present study resulting in a greater than 2-fold change in half-life for these individuals over the more than 2-years separating these studies. In contrast, S34 and S38, who showed no significant change in elimination rate between trials in the pilot study (Rand et al., 2016); exhibited only a slightly (although significant) faster elimination rates in this study (Table 1).
Table 1.
Change in MeHg Half-Life Values Over Time
|
t1/2 (Days) |
|||||
|---|---|---|---|---|---|
| Subject | Gender | Age | Trial 1 (Spring 2014) | Trial 2 (Fall 2014) | Present study (Fall 2016) |
| S3 | F | 23 | 128 | 85* | 59* |
| S22 | M | 35 | 116 | 66* | 38* |
| S34 | M | 52 | 43 | 54 | 44* |
| S38 | F | 53 | 50 | 52 | 45* |
Significant difference in kel (p < .05 for difference in slopes) observed between this determination the one immediately prior to it within the same subject.
Relationship of Hair Hg to Fish Consumed
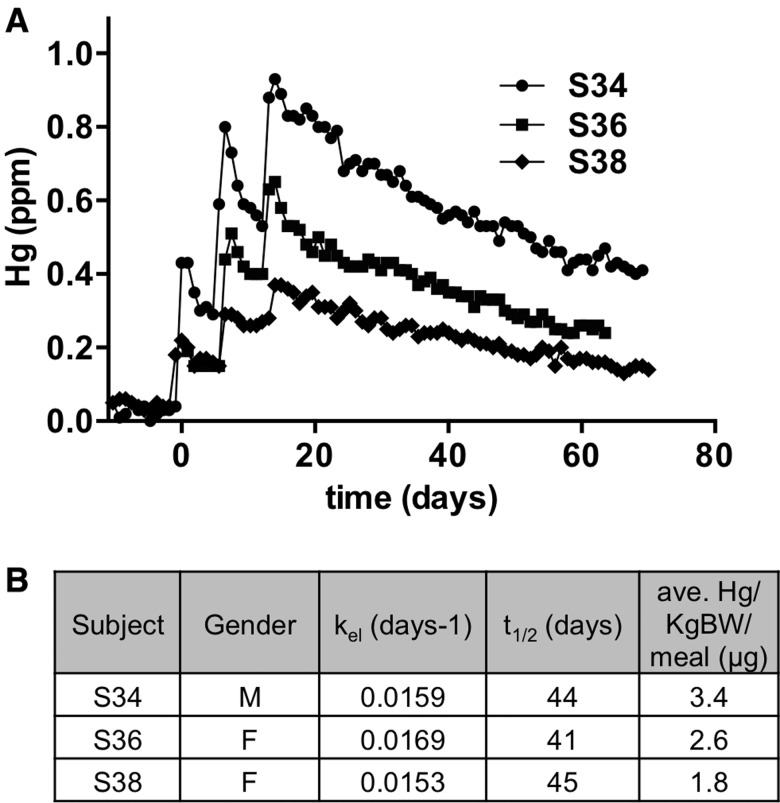
The amount of fish eaten by the subjects varied somewhat due to the variety of portion sizes made available from the preparation of the tuna steaks (see methods). We observed that the Hg:S peak values in hair also varied substantially between subjects (see S2 vs S34 in Figs. 5A and D), suggesting that MeHg levels in hair could reflect the different amounts of fish that individual subjects consumed. However, comparisons of Hg:S ratios between the bulk of the hair samples was not feasible since the hair sample analyses by LA-ICP-MS required more than 10 separate sessions, which were carried out over the course of a 6-month period, and since Hg quantification with respect to a SRM was not employed as it was not needed for the kinetic analyses (see methods). Therefore, we attempted to quantify Hg levels in the analysis of 3 hairs from subjects who ate a range of fish amounts. Analyses were performed within the final session of LA-ICP-MS such that interindividual Hg levels could be compared accurately. Quantifying elements on a mass basis with LA-ICP-MS poses a challenge in matching the matrix composition of SRM with that of the test sample. To accommodate this we fabricated a compressed pellet of SRM hair powder (Supplementary Figure 1). Ablation of this standard gave an Hg:S (×1000) ratio that was representative of the known value of T-Hg (in ppm) in this SRM. Hairs from subjects S34, S36, and S38 were then analyzed in sequence with the SRM pellet using the same laser and ICP-MS parameters (Figure 6). The Hg concentration (in ppm) versus time plots showed that the 3 subjects attained different peak Hg levels in their hair subsequent to the 3 fish meals. Subjects S34 reached peak Hg levels of approximately 0.95 ppm with the 3rd meal, while S36 peaked at approximately 0.65 ppm and S38 peaked at approximately 0.37 ppm Hg. The amount of fish consumed by each of these subjects was then translated to the average amount of MeHg exposure with each meal, expressed as µg Hg/kg body weight/meal (Figure 6B). Across the 3 subjects it was seen that the levels of Hg measured in the hair corresponded very well with the amount of MeHg exposure derived from the fish meals.
Figure 6.
Relationship of Hair Hg to fish consumed. A, Hg:S determinations were converted to Hg in ppm based on mean value of 20.8 (Hg:S × 1000) = 0.573 ppm for SRM hair (see methods and Supplementary Figure 1). Subject hair Hg profiles were aligned on the day scale to compare relative Hg levels. B, Subject gender, elimination rates, half-life values and average µg Hg/kg body weight/meal are summarized.
Correlation of Elimination Rate With Biotransformation in Feces
With a goal of determining the I-Hg (% of T-Hg) as a proxy for MeHg biotransformation in feces, I-Hg and T-Hg were determined on a wet weight basis using CVAA methods. Measurements were made for 33 subjects. Four subjects, who carried dental amalgam surfaces, were excluded since we have previously shown the presence of amalgams substantially increases the I-Hg fraction in feces (Rand et al., 2016). It is also to be noted that the CVAA method used here has an accuracy of ±5% for I-Hg determination in mixed MeHg/I-Hg samples due to a low rate of intrinsic demethylation that can occur in the reaction vessel (M.R., S.C., T.S., G.W. personal observation, also reported in Lind et al., 1994). We therefore used 95% I-Hg as a cut-off to group samples which cannot effectively be discerned from 100% biotransformation from those with <95% I-Hg (% of T-Hg). The majority of subjects showed >95% I-Hg whereas 12 subjects showed <95% I-Hg in their feces (Supplementary Table 2).
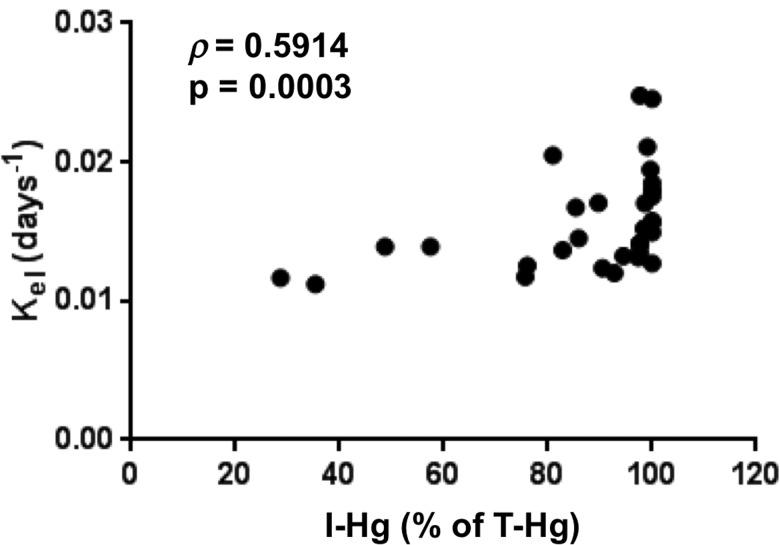
We next evaluated a possible correlation of biotransformation with MeHg elimination rates within these 33 subjects lacking dental amalgam. The plot of kel versus I-Hg (% of T-Hg) demonstrated a positive trend of I-Hg with the increase in kel (Figure 7). Due to the large grouping of samples in the >95% I-Hg range we performed a Spearman’s rank analysis, which yielded ρ = 0.591 at a significance of p = .0003. This is consistent with the notion that a faster elimination rate results from higher rates of MeHg biotransformation. We also investigated whether or not gender, age, habitual fish consumption or body mass index correlates with kel; however, no significant associations were found (Supplementary Figure 2).
Figure 7.
Correlation of MeHg elimination rate with biotransformation. Elimination rate (kel) determined by longitudinal hair analysis is plotted against I-Hg (expressed as % of T-Hg) measured in fecal samples, the latter an indicator of MeHg biotransformation. Spearman coefficient ρ is shown with the p value for the correlation.
DISCUSSION
This study represents the largest sampling of people that we are aware of for a prospective controlled intervention study of MeHg elimination kinetics. For the 37 subjects evaluated, we found an average elimination rate of kel = 0.0157 days−1 (t1/2 = 44 days) amongst a more than 2-fold variation in elimination rate across individuals in this cohort (kel = 0.0248–0.0112 days−1; t1/2 = 28–62 days). Consistent with the results of our prior pilot study, we continue to find examples of variation in MeHg elimination rate over time within some individuals, while other individuals display a relatively consistent MeHg elimination rate over a period spanning 2 years. In 2 fortuitous cases we observed an acute slowing in MeHg elimination subsequent to antibiotic administration, highlighting a potential role for the gut microbiome in biotransformation and excretion of MeHg as has been observed in rodents (Rowland et al., 1978, 1980). Consistent with this mechanism, a positive correlation was seen between elimination rate and MeHg biotransformation, the latter detected as I-Hg in fecal samples. Together, these findings bring a new perspective to elaborating potential sources for inter- and intraindividual variation in MeHg kinetics. With the methods and approach used here it is now possible to attain an individual’s MerMES in a short window of time (eg, approximately 2.5 months), which could potentially reduce uncertainty with respect to MeHg kinetics, and facilitate more personalized and informed approaches to defining an individual’s tolerable weekly intake of fish. We discuss the implications of our findings and the availability of this “tool” to characterize human variation in MeHg toxicokinetics below.
A survey of prior kinetic studies reinforces the observation that MeHg elimination rates vary widely. However, discerning methodological variation from biological variation is critical for advancing a true understanding of MeHg toxicokinetics. Potential sources of experimental variation, and instances of biological variation, can be seen by comparing and contrasting our results with prior studies. The average elimination rate and t1/2 we observed is in very close agreement with that of Smith et al. (1994)t1/2 = 44 days (range 35–53 days, n = 7). Notably, the Smith et al. study is one of the more highly controlled kinetic studies, whereby 203Hg-labeled MeHg was used as a tracer with intravenous administration, followed by separation and independent measurement of MeHg and I-Hg in blood, feces and urine. It is noteworthy that t1/2 determinations by Smith et al. were measured in blood, compared with hair analyses used in the present study, reinforcing the concept that hair is a reliable proxy for blood MeHg levels for kinetic analyses. Our average t1/2 value is also close to that of 2 studies, by Kershaw et al. (1980) and Sherlock et al. (1984), which found an average t1/2 of 52 and 50 days, respectively (range of 39–70 days across both studies, n = 5 and 20, respectively). Like our study, these 2 studies relied on consumption of fish, albeit with elevated MeHg levels in the fish (1.3–9.7 ppm), however, measurements of T-Hg and I-Hg were in blood samples. Our average t1/2 value differs significantly from that found in several other studies, which likely reflects a difference in methods and study controls. For example, Birke et al. (1972), with segmental hair analysis, reported an average t1/2 = 80 days (range 33–120 days, n = 5). A more recent and comprehensive study by Yaginuma-Sakurai et al. (2012) was performed with parallel hair and blood analysis in 27 subjects who had consumed bigeye tuna and swordfish for 14 weeks followed by a 15-week elimination period. These investigators reported average t1/2 = 102 days with hair (range 60–192 days) and 94 days with blood (range 58–155,). The high background Hg levels in these subjects (eg, 2.3 ppm Hg in hair prior to the study Yaginuma-Sakurai et al., 2012) presented important considerations for how to interpret a baseline Hg level in such subjects (note: t1/2 = 64 days [hair] and 57 days [blood] were seen with baseline subtraction [Yaginuma-Sakurai et al., 2012]). We have adopted the recommendation from the Yaginuma-Sakurai et al. study to forgo baseline subtraction in elimination rate calculations. An important consideration in both the Birke et al. (1972) and Yaginuma-Sakurai et al. (2012) studies is the incomplete abstinence from fish eating by the subjects during the elimination period which likely influenced a longer observed half-life. In the Yaginuma-Sakurai et al. study fish consumption was within the tolerable weekly intake for Japanese adults (3.4 µg/kg body weight/week), however, results in almost 5-times the U.S. EPA RfD (0.1 µg/kg body weight/d). Thus, the findings of longer half-life in this study, and that of Birke et al., as compared with ours pose the interesting possibility of a non-linear dose dependent behavior of the elimination rate. Early studies by Aberg et al. (1969) and Miettinen et al. (1971), which used 203Hg-labeled MeHg administered orally, resolved average t1/2 = 70 and 76 days, respectively (range 52–93 days, n = 3 and 15, respectively). Methodologically, these early studies, which used whole body radioactive counting methods, were complicated by the fact that loss of MeHg due to biotransformation to I-Hg could not be distinguished. Thus, the slower observed MeHg elimination likely reflects radioactive counts from the I-Hg retained in the kidney. Al-Shahristani et al. (1974), in a retrospective study, used segmental hair analysis of subjects from the high-level MeHg poisoning incident in Iraq to determine an average t1/2 = 72 days (range 35–189 days, n = 48). This longer half-life may also reflect altered kinetics that could occur at such elevated MeHg doses. In addition, as noted by Smith et al. (1994), the longer half-life seen by Al-Shahristani et al. may be due to an inability to account for hair growth rate as well as the possibility of misalignment of strands within the bundles of hair that were used for the analysis; the multiple hair strands being necessary for Hg signal detection with the CVAA method that was used. In the present study, we overcome both of these technical hurdles of longitudinal hair analysis by using a single strand of hair and by calibrating its growth rate according to the timing of fish meals. By comparison to these prior studies we conclude that our method faithfully reports the kinetics of MeHg elimination in the human body.
In this study, we find that elimination rate varied more than 2-fold among the 37 subjects. This range of t1/2 is somewhat narrower, and lower in average value, than we observed previously in our pilot study (Rand et al., 2016). In our pilot, over both trials there was a wide spread in the MeHg elimination rates between and within individuals over time (kel =0.0163–0.0054 days−1; t1/2 = 43–128 days). For the 2 trials the average t1/2 was 70 and 63 days, respectively. Given that subjects were recruited from the same geographical area as this study, this large variation in average t1/2 between our pilot and this study raised concern for methodological variation. It was noted that the Hg:S data in the pilot study appeared “noisier” giving linear regression fits ranging from R2 = 0.842–0.444 in contrast to this study where we obtained much “cleaner” data with values of R2 = 0.975–0.767. We attribute this to using an upgraded and more sensitive and stable LA-ICP-MS system in the present study. Nonetheless, analysis of 4 subjects who participated in both the pilot and this study indicates that the variation in t1/2 between the studies most likely stems from a large intraindividual biological variation in MeHg metabolism and elimination seen in a few subjects. This is illustrated by contrasting determinations for subjects S3 and S22 with subjects S34 and S38 at 3 time points across the 2 studies (Table 1). These findings show that MeHg elimination status can be dynamic, emphasizing the importance of being able to determine a person’s MerMES relative to a window where Hg exposure with fish consumption is of concern, eg, prior to and during pregnancy and during child development.
Determinations with the 33 subjects lacking dental amalgam surfaces showed a correlation whereby faster elimination rates measured with hair correspond with greater MeHg biotransformation, seen as a higher I-Hg (% of T-Hg) in feces. This corroborates a trend we observed in 4 amalgam-free subjects in our prior pilot study (Rand et al., 2016). This trend is best illustrated by the 2 slowest eliminators, S3 (kel = 0.0117 days−1, t1/2 = 59 days) and S14 (kel = 0.0112 days−1, t1/2 = 62 days), who also showed the lowest MeHg biotransformation in their feces (29% and 35% I-Hg, respectively). Although other sources of I-Hg exposure cannot be ruled out for confounding these measurements, this data is consistent with the model whereby MeHg biotransformation to I-Hg by gut microbiota contributes to the MeHg elimination process. Accordingly, disruption of microbial biotransformation would predict not only an increased proportion of residual MeHg trapped in the feces but a slower elimination rate since more MeHg would be available to return to the circulation by intestinal reabsorption. Data from subjects S2 and S34 support this model showing that with administration of antibiotics the MeHg elimination rate decreases significantly. This observation is consistent with what was previously seen in rats and mice treated with antibiotics (Rowland et al., 1977). Nonetheless, the influence of the antibiotics on other sites of xenobiotic transport and metabolism cannot be ruled out. More rigorous testing of human subjects with defined treatments that alter gut microbiota, eg, with pro- and prebiotics, are needed to further characterize this phenomenon.
Among 3 subjects that have consumed various portion sizes of fish, and thus various amounts of MeHg relative to their body weight, we find that a corresponding level of Hg is observed in their hairs. Such a finding supports the overall assertion that hair is an effective biomarker for assessing MeHg exposures (Cernichiari et al., 2007). Nonetheless, the hair to blood ratio has been highlighted in the recent study by Yaginuma-Sakurai et al. (2012) as an additional source of inter-individual variation in MeHg kinetics and warrants further investigation. For example, differences between females and males for distribution of MeHg to hair relative to blood have yet to be investigated. Such a study will require a larger sample size and more comprehensive and quantitative Hg measurements of both hair Hg and the external dose (fish Hg concentration and amounts consumed), in conjunction with blood Hg determinations, to unambiguously establish the variation in this relationship.
Our finding of t1/2 = 44 days is close to, yet somewhat shorter, than the value of t1/2 = 49.5 (kel = 0.014 days−1) used to derive the EPA RfD (EPA, 2001). However, the variability in t1/2 we document here, together with that seen in prior studies, highlights a critical aspect of toxicokinetics in formulating MeHg advisories; namely, that MeHg kinetics in people cannot be described by a single number. The current EPA RfD formulation takes into consideration that toxicokinetic variability and toxicodynamic variability can each contribute as much as 3-fold uncertainty, which has lead to an overall “uncertainty factor” of 10 being applied to a Benchmark Dose to derive the RfD (EPA, 2001). The ability to determine an individual’s MerMES presents the possibility of removing toxicokinetic uncertainty, and thus could lead to more accurate and personalized approach to formulating a RfD, and a corresponding fish consumption advisory, for the individual.
One limitation to this study is that little ethnic diversity was achieved in recruiting the cohort. All but 2 individuals were Caucasian. Furthermore, kinetic values were determined for relatively low levels of MeHg exposure in this instance. Thus, the general validity of the observed half-life warrants confirmation in other populations. This shortcoming presents an important future endeavor, ie, to investigate the possibility for variation in MeHg kinetics at the population level. Indeed, the variation in MeHg half-life reported for studies spanning Japanese, Middle Eastern and European subjects may reflect genetic and/or environmental (ie, dietary) factors, as well as gut microbiome specific traits, that are intrinsic to these populations. This is particularly relevant for evaluating existing epidemiological cohorts from various geographical regions. We were also limited in our attempt to quantify Hg concentration in the hairs. Hg quantitation would afford a more concise correlation of MeHg dose amounts with body burden within the window of the study. Such determinations may also facilitate analyses of absorption kinetics and efficiency that have recently been highlighted as factors affecting body burden in fish consumers (Li et al., 2016). Another limitation is in the lack of replicate determinations of subject’s hair samples in this study. Nonetheless, the density of the laser ablation spots (>90 Hg determinations along each hair) provides a robust data set for the regression analyses that yield very good coefficients of determination (R2) and confidence in the kinetic rate determinations. Also, minimal variation in replicate determinations of an individual was reported previously in our pilot study (Rand et al., 2016).
In summary, we demonstrate the efficacy of determining a MerMES for 37 human subjects, which reveals significant variation in MeHg toxicokinetics between individuals and within certain individuals over time. Nonetheless, we resolve an average elimination rate equivalent to a half-life of t1/2 = 44 days. The short time needed to obtain these metrics, the enhanced sensitivity of LA-ICP-MS instrumentation, and the non-invasiveness of the protocol make it a powerful tool for MeHg toxicokinetic studies at the individual and population level. We anticipate a wider application of a MerMES determination approach will aid in reducing uncertainty in toxicokinetic parameters used to formulate fish consumption guidelines. A MerMES approach will also support investigations into genetic and dietary factors that moderate the risks for MeHg toxicity associated with eating fish.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
FUNDING
This work was supported by the National Institute of Environmental Health Sciences (NIEHS) at the NIH (R21-ES024859) (M.D.R., PI) and the University of Rochester Environmental Health Science Center (P30 ES001247). The Trace Element Analysis Lab at Dartmouth College is funded by NIH/NIEHS (P42 ES007373, 1UG3OD023275-01 and P01ES022832), NIH/NCI (P30CA023108), and EPA (RD83544201).
Supplementary Material
ACKNOWLEDGMENTS
We thank all the participants who volunteered for this study. We thank Lisa Prince and Daria Vorojeikina for valuable feedback on the study findings.
REFERENCES
- Aberg B., Ekman L., Falk R., Greitz U., Persson G., Snihs J. O. (1969). Metabolism of methyl mercury (203Hg) compounds in man. Archives of Environmental Health 19, 478–484. [DOI] [PubMed] [Google Scholar]
- Al-Shahristani H., Shihab K. M. (1974). Variation of biological half-life f methylmercury in man. Arch. Environ. Health 28, 342–344.http://dx.doi.org/10.1080/00039896.1974.10666505 [DOI] [PubMed] [Google Scholar]
- Ali Aldroobi K. S., Shukri A., Bauk S., Abdel Munem E., Abuarra A. M. A. (2013). Determination of arsenic and mercury level in scalp hair from a selected population in Penang, Malaysia using XRF technique. Radiat. Phys. Chem. 91, 9–14. [Google Scholar]
- Amin-Zaki L., Majeed M. A., Elhassani S. B., Clarkson T. W., Greenwood M. R., Doherty R. A. (1979). Prenatal methylmercury poisoning. Clinical observations over five years. Am. J. Dis. Child. 133, 172–177. [PubMed] [Google Scholar]
- Birke G., Johnels A. G., Plantin L. O., Sjostrand B., Skerfving S., Westermark T. (1972). Studies on humans exposed to methyl mercury through fish consumption. Arch. Environ. Health 25, 77–91. [DOI] [PubMed] [Google Scholar]
- Bradley M. A., Barst B. D., Basu N. (2017). A review of mercury bioavailability in humans and fish. Int. J. Environ. Res. Public Health 14, 169.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernichiari E., Myers G. J., Ballatori N., Zareba G., Vyas J., Clarkson T. (2007). The biological monitoring of prenatal exposure to methylmercury. Neurotoxicology 28, 1015–1022. [DOI] [PubMed] [Google Scholar]
- Engstrom K., Love T. M., Watson G. E., Zareba G., Yeates A., Wahlberg K., Alhamdow A., Thurston S. W., Mulhern M., McSorley E. M., et al. (2016). Polymorphisms in ATP-binding cassette transporters associated with maternal methylmercury disposition and infant neurodevelopment in mother-infant pairs in the Seychelles Child Development Study. Environ. Int. 94, 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EPA. (2001). Methylmercury (MeHg) (CASRN 22967-92-6): Chronic Oral Exposure (RfD). Available at: https://www.epa.gov/. Accessed May 2017. Keyword CASRN 22967-92-6 accessed 2017.
- EPA. (2017). 2017 EPA-FDA Advice about Eating Fish and Shellfish. http://www.fda.gov/Food/FoodborneIllnessContaminants/Metals/ucm115644.htm. Accessed May 2017.
- FDA. (1991–2010). Mercury Levels in Commercial Fish and Shellfish. Available at: http://www.fda.gov/Food/FoodborneIllnessContaminants/Metals/ucm115644.htm. Accessed May 2017.
- Harada M. (1995). Minamata disease: Methylmercury poisoning in Japan caused by environmental pollution. Crit. Rev. Toxicol. 25, 1–24.http://dx.doi.org/10.3109/10408449509089885 [DOI] [PubMed] [Google Scholar]
- Harkey M. R. (1993). Anatomy and physiology of hair. Forens. Sci. Int. 63, 9–18.http://dx.doi.org/10.1016/0379-0738(93)90255-9 [DOI] [PubMed] [Google Scholar]
- IAEA (2017). IAEA-086, Human Hair (Methyl Mercury) Available at: https://nucleus.iaea.org/rpst/referenceproducts/ReferenceMaterials/Trace_Elements_Methylmercury/IAEA-086.htm. Accessed July 2017.
- Ishihara N. (2000). Excretion of methyl mercury in human feces. Archives of Environmental Health 55, 44–47.http://dx.doi.org/10.1080/00039890009603384 [DOI] [PubMed] [Google Scholar]
- Kempson I. M., Lombi E. (2011). Hair analysis as a biomonitor for toxicology, disease and health status. Chem. Soc. Rev. 40, 3915–3940.http://dx.doi.org/10.1039/c1cs15021a [DOI] [PubMed] [Google Scholar]
- Kershaw T. G., Clarkson T. W., Dhahir P. H. (1980). The relationship between blood levels and dose of methylmercury in man. Arch. Environ. Health 35, 28–36.http://dx.doi.org/10.1080/00039896.1980.10667458 [DOI] [PubMed] [Google Scholar]
- Legrand M., Lam R., Jensen-Fontaine M., Salin E. D., Chan H. M. (2004). Direct detection of mercury in single human hair strands by laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS). J. Anal. Atom. Spectrom. 19, 1287–1288. [Google Scholar]
- Legrand M., Lam R., Passos C. J., Mergler D., Salin E. D., Chan H. M. (2007). Analysis of mercury in sequential micrometer segments of single hair strands of fish-eaters. Environ. Sci. Technol. 41, 593–598. [DOI] [PubMed] [Google Scholar]
- Li M., von Stackelberg K., Rheinberger C. M., Hammitt J. K., Krabbenhoft D. P., Yin R., Sunderland E. M. (2016). Insights from mercury stable isotopes into factors affecting the internal body burden of methylmercury in frequent fish consumers. Elem. Sci. Anth. 4, 103. [Google Scholar]
- Li Y. F., Chen C., Li B., Wang J., Gao Y., Zhao Y., Chai Z. (2008). Scalp hair as a biomarker in environmental and occupational mercury exposed populations: Suitable or not? Environ. Res. 107, 39–44. [DOI] [PubMed] [Google Scholar]
- Lind B., Holmgren E., Friberg L., Vahter M. (1994). Demethylation of methylmercury during inorganic determination by teh Magos cold vapor atomic absorption technique. Fresnius J. Anal. Chem. 348, 815–819.http://dx.doi.org/10.1007/BF01780983 [Google Scholar]
- Llop S., Tran V., Ballester F., Barbone F., Sofianou-Katsoulis A., Sunyer J., Engstrom K., Alhamdow A., Love T. M., Watson G. E., et al. (2017). CYP3A genes and the association between prenatal methylmercury exposure and neurodevelopment. Environ. Int. 105, 34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magos L., Clarkson T. W. (1972). Atomic absorption determination of total, inorganic, and organic mercury in blood. Journal - Association of Official Analytical Chemists 55, 966–971. [PubMed] [Google Scholar]
- Marsh D. O., Clarkson T. W., Cox C., Myers G. J., Amin-Zaki L., Al-Tikriti S. (1987). Fetal methylmercury poisoning. Relationship between concentration in single strands of maternal hair and child effects. Arch. Neurol., 44, 1017–1022. [DOI] [PubMed] [Google Scholar]
- Mazzaron Barcelos G. R., de Marco K. C., Grotto D., Valentini J., Garcia S. C., Leite Braga G. U., Barbosa F. Jr. (2012). Evaluation of glutathione S-transferase GSTM1 and GSTT1 polymorphisms and methylmercury metabolism in an exposed Amazon population. J. Toxicol. Environ. Health 75, 960–970. [DOI] [PubMed] [Google Scholar]
- Miettinen J. K., Rahola T., Hattula T., Rissanen K., Tillander M. (1971). Elimination of 203Hg-methylmercury in man. Ann. Clin. Res. 3, 116–122. [PubMed] [Google Scholar]
- Myers R. J., Hamilton J. B. (1951). Regeneration and rate of growth of hairs in man. Ann. N. Y. Acad. Sci. 53, 562–568.http://dx.doi.org/10.1111/j.1749-6632.1951.tb31957.x [DOI] [PubMed] [Google Scholar]
- Norseth T., Clarkson T. W. (1971). Intestinal transport of 203Hg-labeled methyl mercury chloride. Role of biotransformation in rats. Arch. Environ. Health 22, 568–577.http://dx.doi.org/10.1080/00039896.1971.10665903 [DOI] [PubMed] [Google Scholar]
- Oken E., Radesky J. S., Wright R. O., Bellinger D. C., Amarasiriwardena C. J., Kleinman K. P., Hu H., Gillman M. W. (2008). Maternal fish intake during pregnancy, blood mercury levels, and child cognition at age 3 years in a US cohort. Am. J. Epidemiol. 167, 1171–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps R. W., Clarkson T. W., Kershaw T. G., Wheatley B. (1980). Interrelationships of blood and hair mercury concentrations in a North American population exposed to methylmercury. Arch. Environ. Health 35, 161–168. [DOI] [PubMed] [Google Scholar]
- Rand M. D., Vorojeikina D., van Wijngaarden E., Jackson B. P., Scrimale T., Zareba G., Love T. M., Myers G. J., Watson G. E. (2016). Methods for individualized determination of methylmercury elimination rate and de-methylation status in humans following fish consumption. Toxicol. Sci. 149, 385–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodushkin I., Axelsson M. D. (2003). Application of double focusing sector field ICP-MS for multielemental characterization of human hair and nails. Part III. Direct analysis by laser ablation. Sci. Total Environ. 305, 23–39.http://dx.doi.org/10.1016/S0048-9697(02)00463-1 [DOI] [PubMed] [Google Scholar]
- Rowland I. R., Davies M. J., Evans J. G. (1980). The effect of the gastrointestinal flora on tissue content of mercury and organomercurial neurotoxicity in rats given methylmercuric chloride. Dev. Toxicol. Environ. Sci. 8, 79–82. [PubMed] [Google Scholar]
- Rowland I. R., Davies M. J., Grasso P. (1977). The effect of elimination of the gastrointestinal flora on the accumulation of methylmercuric chloride by the rat. Biochem. Soc. Trans. 5, 423–425.http://dx.doi.org/10.1042/bst0050423 [DOI] [PubMed] [Google Scholar]
- Rowland I. R., Davies M. J., Grasso P. (1978). Metabolism of methylmercuric chloride by the gastro-intestinal flora of the rat. Xenobiotica 8, 37–43.http://dx.doi.org/10.3109/00498257809060381 [DOI] [PubMed] [Google Scholar]
- Rowland I. R., Robinson R. D., Doherty R. A. (1984). Effects of diet on mercury metabolism and excretion in mice given methylmercury: Role of gut flora. Arch. Environ. Health 39, 401–408.http://dx.doi.org/10.1080/00039896.1984.10545872 [DOI] [PubMed] [Google Scholar]
- Sherlock J., Hislop J., Newton D., Topping G., Whittle K. (1984). Elevation of mercury in human blood from controlled chronic ingestion of methylmercury in fish. Hum. Toxicol. 3, 117–131. [DOI] [PubMed] [Google Scholar]
- Smith J. C., Allen P. V., Turner M. D., Most B., Fisher H. L., Hall L. L. (1994). The kinetics of intravenously administered methyl mercury in man. Toxicol. Appl. Pharmacol. 128, 251–256. [DOI] [PubMed] [Google Scholar]
- Stadlbauer C., Prohaska T., Reiter C., Knaus A., Stingeder G. (2005). Time-resolved monitoring of heavy-metal intoxication in single hair by laser ablation ICP-DRCMS. Anal. Bioanal. Chem. 383, 500–508. [DOI] [PubMed] [Google Scholar]
- Starling P., Charlton K., McMahon A. T., Lucas C. (2015). Fish intake during pregnancy and foetal neurodevelopment–a systematic review of the evidence. Nutrients 7, 2001–2014.http://dx.doi.org/10.3390/nu7032001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorojeikina D., Broberg K., Love T. M., Davidson P. W., van Wijngaarden E., Rand M. D. (2017). Editor’s highlight: Glutathione S-transferase activity moderates methylmercury toxicity during development in Drosophila. Toxicol. Sci. 157, 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaginuma-Sakurai K., Murata K., Iwai-Shimada M., Nakai K., Kurokawa N., Tatsuta N., Satoh H. (2012). Hair-to-blood ratio and biological half-life of mercury: Experimental study of methylmercury exposure through fish consumption in humans. J. Toxicol. Sci. 37, 123–130. [DOI] [PubMed] [Google Scholar]
- Zareba G., Cernichiari E., Goldsmith L. A., Clarkson T. W. (2008). Validity of methyl mercury hair analysis: Mercury monitoring in human scalp/nude mouse model. J. Appl. Toxicol. 28, 535–542. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.