Brenner et al. show that mutations in a C-terminal hotspot of kinesin-5A (KIF5A) can cause a classical ALS phenotype. Experiments using patient-derived cell lines suggest haploinsufficiency as the molecular genetic mechanism. This underlines the relevance of intracellular transport processes for ALS, and is important for clinico-genetic diagnosis and counselling.
Keywords: ALS, KIF5A mutations, axonal transport, whole exome sequencing
Abstract
Heterozygous missense mutations in the N-terminal motor or coiled-coil domains of the kinesin family member 5A (KIF5A) gene cause monogenic spastic paraplegia (HSP10) and Charcot-Marie-Tooth disease type 2 (CMT2). Moreover, heterozygous de novo frame-shift mutations in the C-terminal domain of KIF5A are associated with neonatal intractable myoclonus, a neurodevelopmental syndrome. These findings, together with the observation that many of the disease genes associated with amyotrophic lateral sclerosis disrupt cytoskeletal function and intracellular transport, led us to hypothesize that mutations in KIF5A are also a cause of amyotrophic lateral sclerosis. Using whole exome sequencing followed by rare variant analysis of 426 patients with familial amyotrophic lateral sclerosis and 6137 control subjects, we detected an enrichment of KIF5A splice-site mutations in amyotrophic lateral sclerosis (2/426 compared to 0/6137 in controls; P = 4.2 × 10−3), both located in a hot-spot in the C-terminus of the protein and predicted to affect splicing exon 27. We additionally show co-segregation with amyotrophic lateral sclerosis of two canonical splice-site mutations in two families. Investigation of lymphoblast cell lines from patients with KIF5A splice-site mutations revealed the loss of mutant RNA expression and suggested haploinsufficiency as the most probable underlying molecular mechanism. Furthermore, mRNA sequencing of a rare non-synonymous missense mutation (predicting p.Arg1007Gly) located in the C-terminus of the protein shortly upstream of the splice donor of exon 27 revealed defective KIF5A pre-mRNA splicing in respective patient-derived cell lines owing to abrogation of the donor site. Finally, the non-synonymous single nucleotide variant rs113247976 (minor allele frequency = 1.00% in controls, n = 6137), also located in the C-terminal region [p.(Pro986Leu) in exon 26], was significantly enriched in familial amyotrophic lateral sclerosis patients (minor allele frequency = 3.40%; P = 1.28 × 10−7). Our study demonstrates that mutations located specifically in a C-terminal hotspot of KIF5A can cause a classical amyotrophic lateral sclerosis phenotype, and underline the involvement of intracellular transport processes in amyotrophic lateral sclerosis pathogenesis.
Introduction
KIF5A is a member of the kinesin family of proteins that is mainly expressed in neurons (Niclas et al., 1994; Fagerberg et al., 2014). As part of a multi-subunit complex, it acts as a microtubule motor in intracellular protein and organelle transport, including mitochondria (Hirokawa et al. 2009). Missense mutations in the kinesin family member 5A (KIF5A) gene at 12q13.3 are the third most frequent cause of autosomal dominant hereditary spastic paraplegia (HSP10, SPG10, OMIM#604187) in European populations, affecting primarily the long pyramidal tracts and sometimes also the peripheral nervous system (Reid et al., 2002; Goizet et al., 2009; Morais et al., 2017). Additionally, the known phenotypic spectrum of KIF5A mutations comprises also an autosomal dominant axonal sensorimotor peripheral neuropathy (Charcot-Marie-Tooth disease type 2; CMT2) (Liu et al., 2014) and a complex infantile neurological syndrome with leukencephalopathy, myoclonus, hypotonia, optic nerve abnormalities, dysphagia, apnoea, hearing loss, and early developmental arrest [neonatal intractable myoclonus (NEIMY; OMIM#617235); Duis et al., 2016; Rydzanicz et al., 2017].
Some 5% of patients with the motor neuron disease amyotrophic lateral sclerosis (ALS) self-report a positive family history (familial ALS), most frequently as a Mendelian autosomal dominant trait. Since 1993, mutations in over 36 genes have been associated with ALS pathogenesis, and mutations in several of these have been predicted to disrupt cytoskeletal function and intracellular transport (PFN1, NEFH, PRPH, ALSIN, DCTN1; TUBA4AFiglewicz et al., 1994; Yang et al., 2001; Puls et al., 2003; Gros-Louis et al., 2004; Wu et al., 2012; Smith et al., 2014). Datasets of two large studies based on whole exome sequencing or genome-wide association testing suggested also an association between variants in KIF5A and ALS (Kenna et al., 2016; McLaughlin et al., 2017). In both studies, the association did not achieve genome-wide statistical significance and the studies also lacked data on a possible co-segregation of KIF5A variants with ALS. Furthermore, detailed clinical information beyond the phenotype ‘ALS’ was not available with regard to the possibly KIF5A-linked patients. Altogether, we hypothesized that mutations affecting KIF5A can also be a cause of ALS. Consequently, we here assessed a possible association between KIF5A and ALS by first comparing the mutation burden of KIF5A in a cohort of 426 familial ALS patients with 144 769 control individuals (comprising 6137 in-house controls and the gnomAD dataset) followed by co-segregation analysis, detailed clinical description of the patients, as well as RNA expression and splicing analysis of KIF5A mutations in patient-derived cell lines.
Material and methods
Patients and ethics statement
All ALS patients were diagnosed according to the EFNS Consensus criteria (Andersen et al., 2005, 2012). With informed written consent and approval by the national medical ethical review boards in accordance with the Declaration of Helsinki, EDTA blood samples were drawn from controls, ALS patients, and their unaffected relatives. DNA was extracted from EDTA blood samples according to standard procedures.
Genotyping for SOD1 and C9orf72 mutations
Mutations in SOD1 and C9orf72 were excluded prior to exome sequencing of familial ALS cases as described before (Freischmidt et al., 2015).
Whole-exome sequencing
We sequenced exomes of 426 European familial ALS index patients and 6137 control subjects. Controls comprised healthy parents of children with various diseases, healthy tissues of individuals with tumour diseases, and 200 individuals from the KORA studies (Kooperative Gesundheitsforschung in der Region Augsburg) (Herder et al., 2013). Sequencing, read mapping and variant calling was performed on HiSeq2000/2500 systems (Illumina) as described previously (Freischmidt et al., 2015).
RNA expression and splicing analysis
RNA was isolated from the immortalized lymphoblast cell lines derived from the mutation carriers and their unaffected mutation-negative relatives. To test the effect of the variants on mRNA level, fragments were amplified using the cDNA template with primers binding to exon 25 and 28/29. Primer sequences are available on request. PCR products were sequenced on ABI 3130xl Genetic Analyzer using BigDye v3.1 cycle sequencing kit (Life Technologies), according to the manufacturer’s protocol. Mutation nomenclature is according to the transcript NM_004984.
To calculate the relative expression of KIF5A mRNA in the subjects, quantitative real-time PCR was performed with SYBR® Green chemistry. Primers spanning exons 2/3 and exon 3/4 were used. TBP (TATA-binding protein) was used as an internal control for normalization. The ΔΔct method was used for quantification (Livak and Schmittgen, 2001).
Statistics
Fisher’s exact test was used to compare sequence variant frequencies between ALS and control groups. A significance level α < 0.05 was applied in all tests statistical tests (two-tailed).
For linkage analysis of Families A–C, we assumed an autosomal dominant model. Penetrance was set at 0.8. The frequency of the deleterious allele was set at 0.0001, the phenocopy rate at 0.003, and the marker allele frequency to 0.0001. Linkage analysis was performed using Merlin software (version 1.1.2). We set the phenotype to unknown if an unaffected individual was either younger than 60 years or had died before the age of 60 years.
Results
Exome sequencing and association analysis
To assess a possible association between KIF5A variants and familial ALS we analysed whole exome sequence data of 426 familial ALS index patients. The frequency of KIF5A variants was compared to 6137 in-house whole exome datasets from control individuals with non-neurological diseases and the 138 632 exomes and genomes of the gnomAD dataset (http://gnomad.broadinstitute.org/, Lek et al., 2016). The familial ALS index patients were selected for whole exome sequencing from families with at least two individuals affected by ALS or frontotemporal dementia from the six European countries: Germany, Denmark, Finland, Sweden, Switzerland, and Portugal, subsequent to a negative screen for pathogenic mutations in SOD1 and C9orf72.
We analysed missense variants of KIF5A at a minor allele frequency (MAF) below the thresholds of 1% and 0.1%, respectively. We did not observe a significant overall enrichment of KIF5A rare missense variants in the familial ALS group when compared to our in-house control group (n = 6137) or the gnomAD dataset (n = 138 632; Tables 1 and 2).
Table 1.
KIF5A splice site and rare missense variants (MAF <1%) found in this study (426 index patients) and basic clinical characteristics of index patients
| Varianta | Predicted consequence at protein level | MAF, % | Onset | Age at onset, | Disease duration, | Phenotype |
|---|---|---|---|---|---|---|
| c: NM_004984.2: | Allele count | years | months | |||
| g: NC_000012.11 | (gnomAD) | |||||
| Missense variants | ||||||
| c.1238A>G | p.Glu413Gly | 4.062 × 10−6 | Spinal, right UL both MN | 35 | 28 | Classical ALS |
| g.57968879A>G | (1/246 210) | LMN>UMN | ||||
| c.1422A>T | p.Gln474His | 4,065 × 10−6 | Spinal, left LL both MN | 68 | 41 | Classical ALS plus FTD |
| g.57965903A>T | (1/246 010) | |||||
| c.1729A>G | p.Ser577Gly | 4.062 × 10−6 | Bulbar | 35 | 60 | ALS |
| g.57968879A>G | (1/246 210) | |||||
| c.3019A>G | p.(Arg1007Gly)b | 0 | Spinal, LMN right UL | 53 | 45 | Classical ALS |
| g.57976411A>G | LMN > UMN | |||||
| Splice site | ||||||
| c.2993-1G> Ag.57976384G>A | c | 4.061 × 10−6 | Spinal, UMN left LL | 56 | >36 (alive) | Classical ALS |
| (1/246 246) | UMN = LMN | |||||
| c.3020+2T>C | p.(Asn999Valfs*39) | 0 | Spinal, LMN left UL | 29 | 34 | Classical ALS |
| g.57976414T>C | UMN > LMN | |||||
| c.3020+1G>Ad | p.(Asn999Valfs*39) | 0 | n.a. | n.a. | n.a. | n.a. |
| g.57976413G>A | ||||||
aGenomic positions according to the GRCh37/hg19.
bPredicted missense mutation, experimentally shown to abrogate function of splice donor site in intron 27 resulting in the predicted change p.Asn999Valfs*39 (see ‘Results’ section).
cSplice acceptor site predicted to be abrogated, resulting protein change unpredictable.
dSplice site variant found in the familial ALS exome data of the ALS Variant Server (AVS) (http://als.umassmed.edu/), no clinical information available.
LL = lower limb; LMN = lower motor neuron; n.a. = not available; UMN = upper motor neuron; UL = upper limb.
Table 2.
KIF5A variants with the respective allele frequencies in case and control whole exome datasets
| Familial ALS | In-house exomes (AF) | P-value (Fisher’s exact test) | gnomAD dataset (AF) | P-value (Fisher’s exact test) | |
|---|---|---|---|---|---|
| n | 426 | 6137 | - | 138 632 | - |
| Loss-of-function | 2 | 0 | 4.2 × 10−3 | 13 | 9.6 × 10−4 |
| (0.23%) | (0%) | (4.7 × 10−3%) | |||
| Missense | 5 | 88 | n.s. | 1596 | n.s. |
| (MAF ≤ 1%) | (0.59%) | (0.72%) | (0.58%) | ||
| Missense | 4 | n.a | n.a. | 714 | n.s. |
| (MAF ≤ 0.01%) | (0.47%) | (0.26%) | |||
| SNV rs113247976 | 29 | 123 | 1.28 × 10−7 | 3132 | 3.11 × 10−7 |
| [p.(Pro986Leu)] | (3.40%) | (1.00%) | (1.13%) |
n.a. = not applicable; n.s. = not significant.
Although we did not detect a significant enrichment of rare missense variants in the patient group, the non-synonymous single nucleotide variant (SNV) rs113247976 showed a trend towards enrichment in ALS patients in a previous GWAS (McLaughlin et al., 2017). We found this SNV highly enriched in our familial ALS cohort [29/426 patients; allele frequency (AF) = 3.4%] compared to in-house controls (123/6137 individuals; AF = 1.00%; P = 1.28 × 10−7) or the gnomAD database (3132/138 632 individuals; AF = 1.13%; P = 3.11 × 10−7). This SNV predicts an amino acid exchange in KIF5A [hg19:g.57,975,700C>T; c.2957C>T; p.(Pro986Leu)] (Table 2). Rs113247976 represents the only non-synonymous variant with a frequency >0.1% in the normal population (gnomAD dataset and in-house controls); thus the rest of KIF5A displays high evolutionary conservation. Remarkably, 11 of 29 patients carrying rs113247976 also had a heterozygous genetic variant in one of the following ALS genes (Supplementary Table 2): HNRNPA1 (p.M137V), VCP (p.R95C), ERBB4 (p.T271I), TARDBP (p.A315T and p.N352S each in one patient), FUS (p.G405R and p.R514T each in one case), FIG4 (p.R699H), SPG11 [c.7152-1G>C (acceptor splice site) and p.V2426M, each in one case; only bi-allelic mutations regarded to be pathogenic], as well as NEK1 (p.Ser1036Ter) and UBQLN2 (p.P509S) in the same individual.
Furthermore, we separately analysed loss-of-function variants, defined as nonsense, canonical splice sites (within two nucleotides of exon boundary), read-through, and frameshift variants. We identified a significant enrichment of KIF5A loss-of-function variants in the patient group (2/426 patients; AF = 0.23%) compared to the in-house control group (0/6137 individuals; AF = 0%; P = 4.2 × 10−3) or the gnomAD dataset (13/138 632 individuals; AF = 4.7 × 10−3; P = 9.6 × 10−4) (Tables 1 and 2). Consistent with the low abundance of loss-of-function variants in control datasets, the pLI score of KIF5A is 1 (http://exac.broadinstitute.org/gene/ENSG00000155980), which indicates a high probability of KIF5A loss-of-function mutation intolerance. Mutations in established ALS disease genes were not detected in the patients with KIF5A loss-of-function mutation.
Segregation analysis
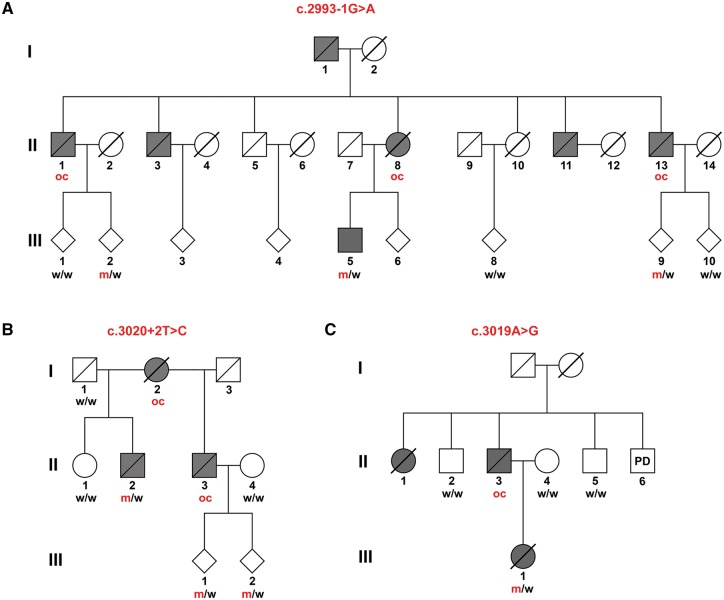
Next, we aimed to further corroborate the observed association of deleterious mutations with ALS by segregation analysis. We extended the two families with loss-of-function mutations and reanalysed them using Sanger sequencing. As shown in Fig. 1A and B, both splice site variants (c.2993-1G>A and c.3020+2T>C) co-segregate with the disease. Overall, in two families seven patients were either DNA-proven loss-of-function mutation carriers or could be deduced to have been obligate loss-of-function mutation carriers (Fig. 1A and B).
Figure 1.
Genetic analysis shows co-segregation of the KIF5A splice site variants and the apparent missense variant c.3019A>G with ALS. (A) c.2993-1G>A (B) c.3020+2T>C. (C) c.3019A>G. Obligate carriers of the respective variant are abbreviated as ‘oc’. All currently asymptomatic mutation carriers are 45 years old or younger. m = mutant allele; PD = Parkinson’s disease; w = wild-type allele.
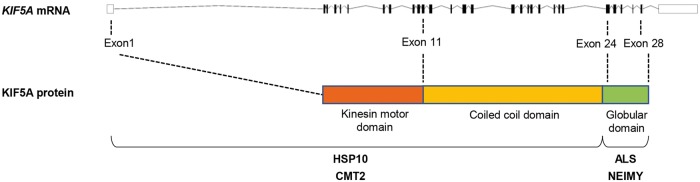
HSP- and CMT-linked KIF5A mutations reside in the N-terminal KIF5A domains (Fig. 3). In contrast, both the ALS-linked loss-of-function mutations described above, the ALS-associated non-synonymous SNV rs113247976 as well as another splice site mutation (c.3020+1G>A) found in familial ALS exome data of the ALS Variant Server (AVS) (http://als.umassmed.edu/), reside in close proximity and are predicted to affect the C-terminal exons 26 or 27 (Figs 2E and 3). The same holds true for a rare apparent missense variant detected in our familial ALS exome dataset [c.3019A>G; p.(Arg1007Gly)], directly adjacent to the above-mentioned splice site mutations c.3020+1G>A and c.3020+2T>C (Fig. 1C and Table 1). The variant was detected in the index patient and was absent in the patient’s healthy mother. This suggests that the affected father (deceased; no DNA available) of the patient also carried the variant, although a de novo mutation cannot be completely ruled out. Moreover, two unaffected uncles of the patient, one paternal, one maternal, of the patient did not carry the variant. Linkage analysis of Families A–C (Fig. 1) resulted in LOD scores of 0.68, 0.52 and 0.44, respectively, leading to an aggregate logarithm of odds (LOD) score of 1.64.
Figure 3.
Localizations of mutations in the KIF5A gene in the respective diseases caused by KIF5A mutations. HSP10 = hereditary spastic paraplegia type 10; NEIMY = neonatal intractable myoclonus.
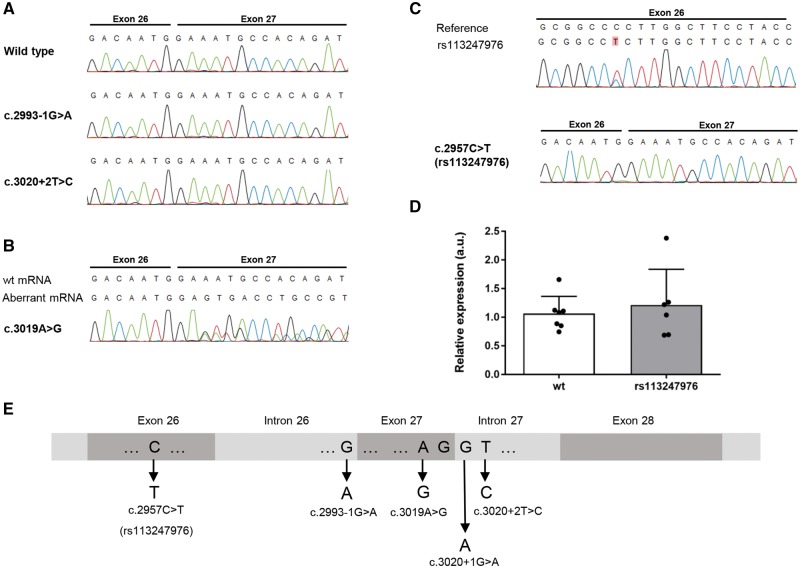
Figure 2.
Sequencing and qPCR analysis of mRNA of lymphoblast cell lines from KIF5A SNV and mutation carriers. (A) mRNA sequencing of the splice site mutations c.2993-1G>A and c.3020+2T>C shows absence of the mutated allele. (B) mRNA sequencing of the apparent missense variant c.3019A>G reveals a disruption of the splice donor site of intron 27. (C) mRNA sequencing of the SNV rs113247976 [c.2957C>T; p.(Pro986Leu)] suggests unaltered pre-mRNA splicing. (D) As analysed by quantitative PCR, heterozygous carriers of the SNV rs113247976 show equal KIF5A mRNA levels as wild-type carriers. (E) Overview of the mutational hotspot around exons 26 and 27 linked to ALS.
RNA analysis in patient-derived lymphoblast cell lines
Availability of immortalized lymphoblast cell lines from patients and controls allowed us to further delineate the functional effects of the above-described mutations at the mRNA level. KIF5A protein expression is tightly restricted to neurons preventing KIF5A protein analysis by western blotting; however, KIF5A mRNA was detectable in lymphoblast cell lines by PCR. Sequencing of the PCR products confirmed the presence of wild-type but failed to detect mis-spliced mRNA in lymphoblast cell lines from patients with the splice site variants c.2993-1G>A and c.3020+2T>C (Fig. 2A). This is most likely attributable to nonsense-mediated decay (NMD) of the RNA transcribed from the mutant alleles. To confirm a loss of expression of the splice site mutant alleles, we tried to quantify the total KIF5A mRNA levels in c.2993-1G>A and c.3020+2T>C splice site mutant and wild-type lymphoblast cell lines. However, possibly as a result of generally low KIF5A expression in non-neuronal cells, the results were too variable for quantification. Interestingly, the above-mentioned C-terminal rare variant, which theoretically leads to a single amino acid exchange in exon 27 [c.3019A>G; p.(Arg1007Gly)], deviates from the expected effect at the mRNA level: Bioinformatic splice site analysis (Berkeley Drosophila Genome Project; Celniker et al., 2002; http://www.fruitfly.org/seq_tools/splice.html) already predicts a decrease of the splice site score from 0.79 for the wild-type splice donor site in intron 27 to 0.16 upon the c.3019A>G change. In agreement with this prediction, sequencing of the PCR product from the respective lymphoblast cell line shows that this variant, separated only by one nucleotide from the 5′ end of intron 27, disrupts the splice donor site of intron 27 (Fig. 2B). This results in a skipping of exon 27 and the predicted truncating change p.Asn999Valfs*39 at the protein level. Thus, this variant, one of the ALS-linked canonical splice site mutations from this study (c.3020+2T>C) and the splice site mutation (c.3020+1G>A) found in familial ALS exome data of the AVS, are all expected or experimentally proven to disrupt the splice donor site of intron 27. However, contrary to the canonical splice site mutations, mis-spliced mRNA resulting from the c.3019A>G [p.(Arg1007Gly)] missense mutation is detectable in the lymphoblast cell lines from the respective patient (data not shown). We speculate that this result may be explained by a differential stability of the respective pre-mRNAs before or during the splicing event.
In our analysis of lymphoblast cell line RNA, the above-described SNV rs113247976 [p.(Pro986Leu)] did not have an impact on splicing of introns 26 or 27, and the respective mRNA level was unaltered compared to wild-type lymphoblast cell lines (Fig. 2C and D). Together with our data and the splice site mutation derived from the AVS, we identified four different mutations predicted or experimentally shown to affect splicing of introns 26 or 27 of KIF5A in familial ALS (Fig. 2E).
Clinical phenotypes of KIF5A mutation carriers
The phenotypes of the patients with a canonical KIF5A splice site mutation or with the c.3019A>G predicted missense variant that led to abrogation of the intron 27 splice donor site were compatible with a classical ALS syndrome: Specifically, adult-onset of initial focal asymmetric onset of affection of both the upper and lower motor neuron systems with later generalization, bulbar motor involvement, emotional lability, absence of vegetative symptoms and sensory symptoms in most patients, as well as rapid disease progression and early death—all of which were fully compatible with a typical ALS phenotype rather than HSP or CMT (Table 1 and Supplementary Table 1)
Patients carrying the SNV rs113247976 [c.2957C>T; p.(Pro986Leu)] found to be enriched in familial ALS tended to have a (non-significantly) shorter disease duration and a significantly decreased proportion of FTD co-morbidity compared to the background cohort of German patients with familial ALS (Supplementary Table 3). However, the number of and clinical information on SNV carriers rs113247976 needs to be extended before it can be definitely ascertained whether rs113247976 is modifying the disease phenotype.
Finally, we screened nine patients with sporadic ALS, from whom post-mortem brain tissue was available, for the SNV rs113247976. One of nine patients carried this SNV. Neuropathological examination revealed typical stage 2 TDP-43 neuropathology (Braak et al., 2013, 2017; Brettschneider et al., 2014; Tan et al., 2013) (Supplementary Fig. 1).
Discussion
The present data, supported by previous suggestive findings (Kenna et al., 2016; McLaughlin et al., 2017), indicate that KIF5A is a novel ALS gene. In our study, the association of rare KIF5A variants with ALS was predominantly carried by loss-of-function mutations, in particular the two splice site mutations found among the studied 426 ALS families. Notably, in the familial ALS group of the AVS, another KIF5A splice variant is reported in a familial ALS patient (c.3020+1G>A), affecting the same splice donor site as the c.3020+2T>C mutation found in our cohort.
Corroborating the link to ALS, both of the splice site mutations (c.2993-1G>A and c.3020+2T>C) we identified co-segregated with the disease. The splice site mutations were present in all seven patients from whom DNA or indirect information about their mutation status (obligate carriers) was available. Furthermore, no healthy individual older than the expected age of disease onset and carrying the mutation was identified in the two families studied. This information, although in its present form limited, is suggestive of a high penetrance of KIF5A loss-of-function mutations.
In contrast to loss-of-function mutations, we found no significant association between familial ALS and rare missense variants (MAF < 1%). Nevertheless, it is well possible that at least a subset of missense variants is pathogenic. KIF5A has a z-score of 4.38 (http://exac.broadinstitute.org/gene/ENSG00000155980), i.e. fewer missense variants than expected, which indicates a general intolerance towards variation in this gene. Remarkably, RNA analysis of patient-derived cell lines revealed that one of the rare (predicted) missense variants (c.3019A>G; predicting p.Arg1007Gly) turned out to abrogate the splice donor site in intron 27, the same donor site that is affected by two of the above mentioned canonical splice site mutations (c.3020+1G>A and c.3020+2T>C).
The KIF5A mutations previously described in HSP or CMT2 patients are restricted to missense mutations in the kinesin motor domain (amino acid positions 9–327) and in the alpha-helical coiled-coil domain (amino acid positions 331–906) (summarized in Kaji et al., 2016; Guinto et al., 2017). By contrast, the ALS-associated mutations described here (Fig. 2E) are predicted to affect the C-terminal part of KIF5A or to represent loss-of-function mutations, as experimentally shown for two of them (overview in Fig. 3). Moreover, deletion mutations in the same C-terminal domain that are predicted to cause a frame-shift with stop loss and an elongated protein (c.2854delC, c.2921delC, c.2934delC), have been associated with the severe developmental syndrome neonatal intractable myoclonus (NEIMY) (Duis et al., 2016; Rydzanicz et al., 2017). Given our and previously reported data, it is thus possible to link the N-terminal and the C-terminal mutational hotspots in KIF5A to HSP and to CMT or to ALS and NEIMY (Fig. 3). The C-terminal globular tail has been demonstrated to be necessary for the binding of cargo-adaptor proteins (e.g. TRAK1/2, GABARAP) (Nakajima et al., 2012; Randall et al., 2013). Mutations predicted to affect the C-terminal tail, if translated into protein, might therefore lead to altered binding of cargo to KIF5A, although this awaits experimental confirmation. By contrast, mutations in the N-terminal kinesin motor domain linked to HSP and CMT decrease the velocity and flux of cargo transport (Wang et al., 2010). This could explain why missense mutations in the N-terminal kinesin motor or the central coiled-coil domain cause the milder HSP/CMT2 phenotype, whereas mutations in the C-terminal small globular domain induce the more severe ALS and NEIMY syndromes, possibly owing to altered cargo binding, haploinsufficiency or a dominant-negative effect of truncated KIF5A.
In this study, we only included ALS patients who had been diagnosed by experienced ALS clinical specialists adhering to stringent ALS criteria (Andersen et al., 2005, 2012). This is important, not the least because two previous reports suggested a possible link between ALS and KIF5A, but lacked precise clinical information (Kenna et al., 2016; McLaughlin et al., 2017). Considering the relative phenotypic similarities between ALS, HSP, and CMT, it cannot be excluded that part of the association signal in the previous large studies arose from HSP or CMT patients misdiagnosed as (slowly progressive) ALS. The patients in our study showed a classical ALS phenotype. Nevertheless, overlapping syndromes may exist and could complicate the distinction between ALS, CMT and HSP in some instances. Along this line, it remained unclear if the pronounced sensory symptoms of Patient A/II.13 were the consequence of a paraneoplastic syndrome or indicated CMT2 co-morbidity.
Interestingly, we observe a co-occurrence of rs113247976 with rare genetic missense variants in other known ALS genes in 11 of 29 patients carrying this particular SNV. Half of these variants have been reported to be pathogenic for ALS in earlier studies (Gitcho et al., 2008; Kimonis et al., 2008; Kühnlein et al., 2008; Chiò et al., 2009; Örlén et al., 2009; Deng et al., 2011; Brenner et al., 2016; Nguyen et al., 2018), while the rest are of uncertain significance (Supplementary Table 2). Supporting our observation of a bigenic effect for ALS causality, one of the patients who carried the rs113247976 also had a TARDBP p.N352S mutation, in agreement with van Blitterswijk et al. who described that 50% of patients with familial ALS carrying the TARDBP p.N352S mutation also have a mutation in another ALS gene (Van Blitterswijk et al., 2012). Moreover, only homozygous or compound heterozygous SPG11 mutations have been shown to cause juvenile ALS, HSP or CMT2. Nevertheless, considering that both SPG11 and KIF5A are involved in axonal transport and cause the same phenotypic spectrum when mutated, one is inclined to speculate about an additive effect of the observed co-occurrence of heterozygous SPG11 mutations and the SNV rs113247976 in KIF5A. Similarly, the NEK1 variant (p.Ser1036Ter) observed in combination with rs113247976 and the UBQLN2 mutation p.P509S—previously described by Deng et al. (2011)—shows incomplete penetrance (Brenner et al., 2016; Nguyen et al., 2018). Thus, penetrance of NEK1 mutations may require one or more additional genetic variants in the same patient. Taken together, we therefore hypothesize that the SNV rs113247976 lowers the threshold for phenoconversion in carriers of additional ALS gene mutations. This would be compatible with an oligogenic mode of inheritance and could possibly also explain a substantial proportion of sporadic ALS cases.
A patient with the rare KIF5A missense variant (c.1422A>T; p.Gln474His, Table 1) suffered from ALS and FTD. Co-segregation analysis of this variant was unfortunately not possible. Preliminary results suggested a reduced frequency of FTD co-morbidity among ALS patients carrying the SNV rs113247976 (Supplementary Table 3); however, this result requires replication in larger cohorts.
In conclusion, we demonstrate here that highly penetrant C-terminal KIF5A splice site mutations can cause ALS and we present detailed clinical information on the KIF5A-linked ALS phenotype. The type of mutations together with RNA analysis in patient-derived cell lines indicates that haploinsufficiency is the most likely molecular genetic mechanism for highly penetrant KIF5A mutations. In addition, we report that the SNV rs113247976 is associated with familial ALS and possibly involved in digenic/polygenic inheritance of the disease, representing thus far, to our knowledge, the most frequent genetic factor contributing to ALS pathogenesis. Our findings underline the importance of intracelluar transport molecules for ALS pathogenesis. Finally, we outline a hypothesis on how the type and location of KIF5A variants determine the manifestation of four different neurological syndromes.
Web resources
ALS Variant Server, Worcester, MA (http://als.umassmed.edu/).
Supplementary Material
Acknowledgements
We are indebted to the patients and their families for their participation in this project. We are grateful to Eva Jonsson, Ann-Charloth Nilsson, and Helena Alstermark for skilful technical assistance. The authors would like to thank the ALS Variant Server (als.umassmed.edu), which is supported by funds from NIH/NINDS (1R01NS065847), AriSLA (EXOMEFALS, NOVALS), the ALS Association, and the Motor Neuron Disease Association.
Funding
This work was supported in whole or in part by grants from the German society for patients with muscular diseases (DGM), the German Federal Ministry of Education and Research [JPND ‘STRENGTH’ consortium (01ED1408); JPND ‘PreFrontAls’ (01ED1512), German Network for ALS Research MND-NET (01GM1103A), German FTLDc network (O1GI1007A)], the DFG-funded Swabian ALS Registry, SFB1279, the ALS association, EU: FAIR-PARK II 633190, the foundation of the state Baden-Württemberg (D.3830), Boehringer Ingelheim Ulm University BioCenter (D.5009), Thierry Latran Foundation, the Swedish Brain Foundation, the Swedish Science Council, the Knut and Alice Wallenberg Foundation, the Bertil Hållsten Foundation, the Ulla-Carin Lindquist Foundation, the Neuroförbundet Association, the Torsten and Ragnar Söderberg Foundation, the Stratneuro Initiative, and the Västerbotten County Council. The work of A.E.V was funded by the Deutsche Forschungsgemeinschaft (DFG, VO 2028/1-1).
Supplementary material
Supplementary material is available at Brain online.
Appendix 1
List of participants of The German ALS network MND-NET (see also Supplementary material):
Glossary
Abbreviations
- ALS
amyotrophic lateral sclerosis
- CMT
Charcot-Marie-Tooth disease
- HSP
hereditary spastic paraplegia
- MAF
minor allele frequency
- SNV
single nucleotide variant
Contributor Information
The German ALS network MND-NET:
Ute Weyen, Andreas Hermann, Tim Hagenacker, Jan Christoph Koch, Paul Lingor, Bettina Göricke, Stephan Zierz, Petra Baum, Joachim Wolf, Andrea Winkler, Peter Young, Ulrich Bogdahn, Johannes Prudlo, and Jan Kassubek
References
- Andersen PM, Borasio GD, Dengler R, Hardiman O, Kollewe K, Leigh PN, et al. EFNS task force on management of amyotrophic lateral sclerosis: guidelines for diagnosing and clinical care of patients and relatives. An evidence-based review with good practice points. Eur J Neurol 2005; 12: 921–38. [DOI] [PubMed] [Google Scholar]
- Andersen PM, Abrahams S, Borasio GD, de Carvalho M, Chio A, Van Damme P, et al. EFNS guidelines on the Clinical Management of Amyotrophic Lateral Sclerosis (MALS)—revised report of an EFNS task force. Eur J Neurol 2012; 19: 360–75. [DOI] [PubMed] [Google Scholar]
- Braak H, Brettschneider J, Ludolph AC, Lee VM, Trojanowski JQ, Del Tredici K. Amyotrophic lateral sclerosis–a model of corticofugal axonal spread. Nat Rev Neurol 2013; 9: 708–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H, Ludolph AC, Neumann M, Ravits J, Del Tredici K. Pathological TDP-43 changes in Betz cells differ from those in bulbar and spinal α-motoneurons in sporadic amyotrophic lateral sclerosis. Acta Neuropathol 2017; 133: 79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner D, Müller K, Wieland T, Weydt P, Böhm S, Lulé D, et al. NEK1 mutations in familial amyotrophic lateral sclerosis. Brain 2016; 139: e28. [DOI] [PubMed] [Google Scholar]
- Brettschneider J, Arai K, Del Tredici K, Toledo JB, Robinson JL, Lee EB, et al. TDP-43 pathology and neuronal loss in amyotrophic lateral sclerosis spinal cord. Acta Neuropathol 2014; 128: 423–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker SE, Wheeler DA, Kronmiller B, Carlson JW, Halpern A, Patel S, et al. Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol 2002; 3: RESEARCH0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiò A, Restagno G, Brunetti M, Ossola I, Calvo A, Mora G, et al. Two Italian kindreds with familial amyotrophic lateral sclerosis due to FUS mutation. Neurobiol Aging 2009; 30: 1272–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H-X, Chen W, Hong S-T, Boycott KM, Gorrie GH, Siddique N, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature 2011; 477: 211–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duis J, Dean S, Applegate C, Harper A, Xiao R, He W, et al. KIF5A mutations cause an infantile onset phenotype including severe myoclonus with evidence of mitochondrial dysfunction. Ann Neurol 2016; 80: 633–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics 2014; 13: 397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figlewicz DA, Krizus A, Martinoli MG, Meininger V, Dib M, Rouleau GA, et al. Variants of the heavy neurofilament subunit are associated with the development of amyotrophic lateral sclerosis. Hum Mol Genet 1994; 3: 1757–61. [DOI] [PubMed] [Google Scholar]
- Freischmidt A, Wieland T, Richter B, Ruf W, Schaeffer V, Müller K, et al. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat Neurosci 2015; 18: 631–6. [DOI] [PubMed] [Google Scholar]
- Gitcho MA, Baloh RH, Chakraverty S, Mayo K, Norton JB, Levitch D, et al. TDP-43 A315T mutation in familial motor neuron disease. Ann Neurol 2008; 63: 535–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goizet C, Boukhris A, Mundwiller E, Tallaksen C, Forlani S, Toutain A, et al. Complicated forms of autosomal dominant hereditary spastic paraplegia are frequent in SPG10. Hum Mutat 2009; 30: E376–85. [DOI] [PubMed] [Google Scholar]
- Gros-Louis F, Larivière R, Gowing G, Laurent S, Camu W, Bouchard J-P, et al. A frameshift deletion in peripherin gene associated with amyotrophic lateral sclerosis. J Biol Chem 2004; 279: 45951–6. [DOI] [PubMed] [Google Scholar]
- Guinto CO, Diarra S, Diallo S, Cissé L, Coulibaly T, Diallo SH, et al. A novel mutation in KIF5A in a Malian family with spastic paraplegia and sensory loss. Ann Clin Transl Neurol 2017; 4: 272–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herder C, Bongaerts BWC, Rathmann W, Heier M, Kowall B, Koenig W, et al. Association of subclinical inflammation with polyneuropathy in the older population: KORA F4 study. Diabetes Care 2013; 36: 3663–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol 2009; 10: 682–96. [DOI] [PubMed] [Google Scholar]
- Kaji S, Kawarai T, Miyamoto R, Nodera H, Pedace L, Orlacchio A, et al. Late-onset spastic paraplegia type 10 (SPG10) family presenting with bulbar symptoms and fasciculations mimicking amyotrophic lateral sclerosis. J Neurol Sci 2016; 364: 45–9. [DOI] [PubMed] [Google Scholar]
- Kenna KP, van Doormaal PTC, Dekker AM, Ticozzi N, Kenna BJ, Diekstra FP, et al. NEK1 variants confer susceptibility to amyotrophic lateral sclerosis. Nat Genet 2016; 48: 1037–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimonis VE, Fulchiero E, Vesa J, Watts G. VCP disease associated with myopathy, Paget disease of bone and frontotemporal dementia: review of a unique disorder. Biochim Biophys Acta Mol Basis Dis 2008; 1782: 744–8. [DOI] [PubMed] [Google Scholar]
- Kühnlein P, Sperfeld A-D, Vanmassenhove B, Van Deerlin V, Lee VM-Y, Trojanowski JQ, et al. Two German kindreds with familial amyotrophic lateral sclerosis due to TARDBP mutations. Arch Neurol 2008; 65: 1185–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016; 536: 285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001; 25: 402–8. [DOI] [PubMed] [Google Scholar]
- Liu Y-T, Laura M, Hersheson J, Horga A, Jaunmuktane Z, Brandner S, et al. Extended phenotypic spectrum of KIF5A mutations: from spastic paraplegia to axonal neuropathy. Neurology 2014; 83: 612–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin RL, Schijven D, van Rheenen W, van Eijk KR, O’Brien M, Kahn RS, et al. Genetic correlation between amyotrophic lateral sclerosis and schizophrenia. Nat Commun 2017; 8: 14774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais S, Raymond L, Mairey M, Coutinho P, Brandão E, Ribeiro P, et al. Massive sequencing of 70 genes reveals a myriad of missing genes or mechanisms to be uncovered in hereditary spastic paraplegias. Eur J Hum Genet 2017; 25: 1217–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Yin X, Takei Y, Seog D-H, Homma N, Hirokawa N. Molecular motor KIF5A is essential for GABAA receptor transport, and KIF5A deletion causes epilepsy. Neuron 2012; 76: 945–61. [DOI] [PubMed] [Google Scholar]
- Nguyen HP, Van Mossevelde S, Dillen L, De Bleecker JL, Moisse M, Van Damme P, et al. NEK1 genetic variability in a Belgian cohort of ALS and ALS-FTD patients. Neurobiol Aging 2018; 61: 255.e1–e7. [DOI] [PubMed] [Google Scholar]
- Niclas J, Navone F, Hom-Booher N, Vale RD. Cloning and localization of a conventional kinesin motor expressed exclusively in neurons. Neuron 1994; 12: 1059–72. [DOI] [PubMed] [Google Scholar]
- Örlén H, Melberg A, Raininko R, Kumlien E, Entesarian M, Söderberg P, et al. SPG11 mutations cause Kjellin syndrome, a hereditary spastic paraplegia with thin corpus callosum and central retinal degeneration. Am J Med Genet B Neuropsychiatr Genet 2009; 150B: 984–92. [DOI] [PubMed] [Google Scholar]
- Puls I, Jonnakuty C, LaMonte BH, Holzbaur ELF, Tokito M, Mann E, et al. Mutant dynactin in motor neuron disease. Nat Genet 2003; 33: 455–6. [DOI] [PubMed] [Google Scholar]
- Randall TS, Moores C, Stephenson FA. Delineation of the TRAK binding regions of the kinesin-1 motor proteins. FEBS Lett 2013; 587: 3763–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid E, Kloos M, Ashley-Koch A, Hughes L, Bevan S, Svenson IK, et al. A kinesin heavy chain (KIF5A) mutation in hereditary spastic paraplegia (SPG10). Am J Hum Genet 2002; 71: 1189–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydzanicz M, Jagła M, Kosinska J, Tomasik T, Sobczak A, Pollak A, et al. KIF5A de novo mutation associated with myoclonic seizures and neonatal onset progressive leukoencephalopathy. Clin Genet 2017; 91: 769–73. [DOI] [PubMed] [Google Scholar]
- Smith BN, Ticozzi N, Fallini C, Gkazi AS, Topp S, Kenna KP, et al. Exome-wide rare variant analysis identifies TUBA4A mutations associated with familial ALS. Neuron 2014; 84: 324–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan RH, Shepherd CE, Kril JJ, McCann H, McGeachie A, McGinley C, et al. Classification of FTLD-TDP cases into pathological subtypes using antibodies against phosphorylated and non-phosphorylated TDP43. Acta Neuropathol Commun 2013; 1: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Blitterswijk M, van Es MA, Hennekam EAM, Dooijes D, van Rheenen W, Medic J, et al. Evidence for an oligogenic basis of amyotrophic lateral sclerosis. Hum Mol Genet 2012; 21: 3776–84. [DOI] [PubMed] [Google Scholar]
- Wang L, Brown A. A hereditary spastic paraplegia mutation in kinesin-1A/KIF5A disrupts neurofilament transport. Mol Neurodegener 2010; 5: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-H, Fallini C, Ticozzi N, Keagle PJ, Sapp PC, Piotrowska K, et al. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature 2012; 488: 499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Hentati A, Deng H-X, Dabbagh O, Sasaki T, Hirano M, et al. The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat Genet 2001; 29: 160–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.