Abstract
Background
Cross-sectional genome-wide association studies have identified hundreds of loci associated with blood lipids and related cardiovascular traits, but few genetic association studies have focused on long-term changes in blood lipids.
Methods
Participants from the GLACIER Study (Nmax = 3492) were genotyped with the MetaboChip array, from which 29 387 SNPs (single nucleotide polymorphisms; replication, fine-mapping regions and wildcard SNPs for lipid traits) were extracted for association tests with 10-year change in total cholesterol (ΔTC) and triglycerides (ΔTG). Four additional prospective cohort studies (MDC, PIVUS, ULSAM, MRC Ely; Nmax = 8263 participants) were used for replication. We conducted an in silico look-up for association with coronary artery disease (CAD) in the Coronary ARtery DIsease Genome-wide Replication and Meta-analysis (CARDIoGRAMplusC4D) Consortium (N ∼ 190 000) and functional annotation for the top ranking variants.
Results
In total, 956 variants were associated (P < 0.01) with either ΔTC or ΔTG in GLACIER. In GLACIER, chr19:50121999 at APOE was associated with ΔTG and multiple SNPs in the APOA1/A4/C3/A5 region at genome-wide significance (P < 5 × 10-8), whereas variants in four loci, DOCK7, BRE, SYNE1 and KCNIP1, reached study-wide significance (P < 1.7 × 10-6). The rs7412 variant at APOE was associated with ΔTC in GLACIER (P < 1.7 × 10-6). In pooled analyses of all cohorts, 139 SNPs at six and five loci were associated with ΔTC and for ΔTG, respectively (P < 10-3). Of these, a variant at CAPN3 (P = 1.2 × 10-4), multiple variants at HPR (Pmin = 1.5 × 10-6) and a variant at SIX5 (P = 1.9 × 10-4) showed evidence for association with CAD.
Conclusions
We identified seven novel genomic regions associated with long-term changes in blood lipids, of which three also raise CAD risk.
Keywords: Prospective cohort study, longitudinal analysis, total cholesterol, morbidity, triglycerides, single nucleotide polymorphism, genetic epidemiology
Key Messages
In the past few years, large studies have identified hundreds of genetic variants that are associated with blood lipid levels, but these studies were almost exclusively carried out in cross-sectional settings.
We identified and replicated associations between common genetic variants and long-term deteriorations in triglyceride and total cholesterol concentrations.
We identified seven novel genomic regions associated with either triglyceride or total cholesterol changes.
Future studies might shed light on whether interactions with environmental factors that change with age result in time-dependent genetic associations and whether these associations can be used in disease prediction.
Introduction
The estimated heritability of lipid traits is ∼ 40-70%,1 with genome-wide significant variants (P < 5 × 10-8) accounting for ∼ 20% of the heritable fraction.2 Recent large-scale meta-analyses have identified hundreds of loci for cardiovascular traits including blood lipid levels.2–5 The findings of these well-powered studies elucidate the molecular pathways underlying cardiovascular disease (CVD), but their predictive accuracy is often low. Multiple lipid variants also associate with coronary artery disease (CAD), blood pressure traits, fasting glucose, type 2 diabetes (T2D) and body mass index (BMI).2
Genetic variants associated with long-term changes in blood lipid concentrations are plausibly more clinically relevant than those only associated with cross-sectional values; however, very few studies have comprehensively examined this topic.6–9 In earlier analyses in the GLACIER Study, we showed that whereas some of the 157 loci robustly associated with cross-sectional lipid concentrations2,3 also convey robust effects on long-term changes in lipids, most loci do not.8 Thus, discovering loci related to long-term changes in blood lipids, and not merely focusing on variants that are known to bear cross-sectional associations,10 might be informative for the early identification of persons at high risk of atherosclerotic heart disease; such studies might also yield leads for targeted prevention, as loci that predict changes with age might do so owing to interactions with ageing-related risk factors or cumulative environmental exposures.7
Large-scale cross-sectional studies that are adequately powered for detecting cross-sectional genetic associations may not be sufficiently powered to identify loci with time-varying effects, particularly in meta-analyses of cohorts that differ by age.11 To address this, we leveraged the extensive lipid-related content of the MetaboChip array, which contains a large number of variants that did not reach a genome-wide level of significance (P > 5 × 10-8) in early genome-wide associations studies (GWAS) for cross-sectional lipid levels. We hypothesized that some of these loci had weaker effects in cross-sectional studies because their effects might be time-varying.
The purpose of this study was to discover loci that hitherto were not known to bear relationships with long-term deteriorations in blood lipid levels and CAD.
Materials and Methods
Ethics statement
Ethical approval for the GLACIER Study was obtained from the Regional Ethical Review Board in Umeå, Sweden. The Ethics Committee at Lund University approved the MDC Study. All participants of the PIVUS and ULSAM studies gave written informed consent and the Ethics Committee of Uppsala University approved the study protocols. All participants of the MRC Ely Study gave informed consent and the study has received ethical clearance from the local ethics committee.
Study participants
The GLACIER Study12 is a population-based cohort study nested within the Northern Sweden Health and Disease Study13 and the Västerbotten Health Survey (VHU) in Northern Sweden (N ∼ 110 000); participants were initially free of CAD and received lifestyle counseling as part of their health examination at their primary care center. Recruitment and clinical measures are described in detail elsewhere.8,13 A total of 3492 (for total cholesterol: TC) and 2209 (for triglycerides: TG) participants had relevant prospective genetic and phenotypic data available. As high-density lipoprotein cholesterol (HDL-C) was measured with a different method compared with the other lipid traits, HDL-C and low-density lipoprotein cholesterol (LDL-C) were only available in a smaller subset of samples. Therefore, in this work we only analysed TC and TG levels.
Study characteristics, clinical measures, genotyping and statistical methods for the replication cohorts (MDC, PIVUS, ULSAM and MRC Ely) are reported in Method S1, available as Supplementary data at IJE online.
Clinical measures
Clinical measures relevant to the GLACIER Study are described in detail elsewhere.8 Before blood draw, 80% of the cohort had fasted for > 8 h, 5% for 4-8 h and fasting status was unknown in 15% of the cohort; 1% of participants reported using lipid-lowering medications, which we controlled for in analyses using constants for statins reported by Wu et al. (TC: + 1.336 mmol/l, TG: + 0.207 mmol/l),14 as described previously.8 Although the type of lipid-lowering medication was not available in the GLACIER Study, statins were the most commonly prescribed lipid-lowering agents within this population at the time of recruitment.15 Using constants to correct for medication effects has been shown to yield less biased estimates and to be more powerful than adjusting with a binary variable or excluding participants on medication.16
Genotyping and SNP selection
DNA was extracted from peripheral white blood cells and genomic DNA samples were diluted to 4 ng/µl, as previously described.12,17 Samples were genotyped using the Illumina CardioMetaboChip (Illumina iSelect) array. We selected all replication SNPs for TC, TG, LDL-C and HDL-C as well as those that fine-map previously established loci and wildcard SNPs, which were included on the chip owing to previous evidence indicating a role in, for example, lipid-related pathways (N = 43 690 SNPs).18 Rare variants (minor allele frequency < 1%) (N = 14 280 SNPs) and SNPs deviating from Hardy-Weinberg equilibrium (P < 0.0001) (N = 23 SNPs) were excluded. Accordingly, 29 387 SNPs were analysed in GLACIER, for which the average genotyping success rate was 99.9%.
Statistical methods
Statistical analyses were undertaken using STATA (version 13.1, StataCorp LP, TX, USA) and PLINK (version 1.07).19,20 In the discovery (GLACIER) and the replication (MDC, PIVUS, ULSAM and MRC Ely) cohorts, mean differences for continuous variables between baseline and follow-up were assessed by paired-samples t tests, and genetic effects were estimated with generalized linear models (GLMs) using genotypes (additive genetic model) as the independent variable with the lipid level change between baseline and follow-up divided by follow-up years (i.e. change rate or slope) as the dependent variable; models were adjusted for the baseline lipid measure, age, age2, follow-up age, sex and population structure (first four principal components: PC) in all models. By adjusting for both baseline and follow-up age, we account for follow-up period, which might otherwise confound results. In GLACIER, variables for fasting status were included in all models, as described previously.21 Formally, the model is:
where γ represents a trait (TC or TG), γB and γF are baseline and follow-up lipid values, ageBand ageB are baseline and follow-up ages, respectively, α is the intercept, βi represents effect size parameters for the corresponding variables and ε is an error term. Both TC and TG change rates were normally distributed; therefore, no transformations were applied to these variables. Although linear mixed models (LMMs) are commonly used in prospective association studies with more than two time points, utilizing LMMs in studies with only two time points is computationally burdensome and the covariance structure between observations derived from the same individuals cannot be properly ascertained.22,23 Comparisons between various statistical approaches to detect genetic associations with trait changes were not the main aim of this paper; however, we conducted comparisons between GLMs with change rate as the outcome (the model used in this paper) and LMMs on a subset of ∼ 1000 SNPs, which showed strong correlations of beta coefficients, standard errors and P-values (Spearman and Pearson r > 0.7 for all comparisons). Methodology and results from the comparison of GLMs and LMMs are presented in Method S2 (available as Supplementary data at IJE online).
To assess whether the variants associated with lipid changes (P < 0.01) are also associated with cross-sectional lipid measures, we conducted an in silico look-up in the Global Lipids Genetics Consortium (GLGC) (Nmax = 188 577).2
Replication and meta-analysis
We anticipated heterogeneity between cohorts due to differences in the participants’ age, follow-up periods and medication use; therefore we implemented a two-stage approach using GLACIER as the discovery cohort from which a set of SNPs associated with TG and TC at a significance threshold of P < 0.01 were identified. This significance threshold was selected to balance type I error and type II error rates, as we hypothesized that the discovery cohort may be underpowered to detect the loci of interest, and for practical reasons we chose not to carry forward a very large number of loci for replication owing to the high penalty this would incur for multiple-test correction. This multi-stage approach has been shown to be successful elsewhere where the discovery cohort was likely underpowered.24
Fixed-effects and random-effects inverse variance weighted meta-analyses were undertaken using genome-wide association meta-analysis (GWAMA)25 and heterogeneity between studies was assessed using the Cochran’s Q test (P-value from this test and I2 statistics are reported) from the random-effects meta-analysis. As the estimates from the random-effects meta-analysis demonstrated low heterogeneity for most loci (see Table 1), we present results from the fixed-effect meta-analysis. In two separate meta-analyses we pooled: (i) only the replication cohorts; and (ii) the replication cohorts and the discovery cohort (GLACIER). This was done to determine which signals replicated independently of the discovery cohort (analysis i) and thereafter to maximize power (analysis ii).26
Table 1.
Lead SNPs from the fixed-effect inverse variance weighted meta-analysis of GLACIER, MDC, PIVUS, ULSAM and MRC Ely. (Nmax=8,263)
| Trait | Chr | BP* | SNP | EA | OA | Nearest locus | β (mmol/l/yr) | SE (mmol/l/yr) | β L95 (mmol/l/yr) | β U95 (mmol/l/yr) | P | Pheterogeneity | I2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ΔTC | 1 | 62912318 | chr1:62912318 | C | T | DOCK7/ANGPTL3 | −0.004 | 0.001 | −0.006 | −0.002 | 2.5 × 10-4 | 0.13 | 0.43 |
| ΔTC | 8 | 19669744 | rs13282247 | T | C | LPL/CSGALNACT1 | −0.004 | 0.001 | −0.006 | −0.002 | 1.7 × 10-4 | 0.12 | 0.45 |
| ΔTC | 15 | 40469997 | chr15:40469997 | A | T | CAPN3 | 0.015 | 0.004 | 0.007 | 0.023 | 1.5 × 10-4 | 0.36 | 0.08 |
| ΔTC | 16 | 70636491 | chr16:70636491 | T | G | HP/HPR | 0.008 | 0.002 | 0.004 | 0.013 | 2.3 × 10-4 | 0.08 | 0.56 |
| ΔTC | 19 | 11001703 | rs17304534 | C | T | LDLR | −0.004 | 0.001 | −0.006 | −0.001 | 1.0 × 10-3 | 0.07 | 0.53 |
| ΔTC | 19 | 50103919 | rs7412 | T | C | APOE/C1 | −0.014 | 0.002 | −0.018 | −0.01 | 2.0 × 10-12 | 0.17 | 0.38 |
| ΔTG | 1 | 62954723 | chr1:62954723 | C | T | DOCK7/ANGPTL3 | −0.002 | 0.001 | −0.004 | −0.001 | 5.2 × 10-4 | 0.31 | 0.17 |
| ΔTG | 8 | 126560154 | rs2954029 | T | A | TRIB1 | −0.003 | 0.001 | −0.004 | −0.002 | 8.1 × 10-6 | 0.07 | 0.53 |
| ΔTG | 11 | 116167789 | rs651821 | C | T | ZNF159/APOA5/A1 | 0.009 | 0.001 | 0.007 | 0.012 | 1.4 × 10-10 | 7.9 × 10-5 | 0.83 |
| ΔTG | 19 | 50848859 | rs10406431 | G | A | GIPR | −0.003 | 0.001 | −0.004 | −0.001 | 1.7 × 10-4 | 0.24 | 0.27 |
| ΔTG | 19 | 50902205 | rs11668847 | G | T | QPCTL | 0.003 | 0.001 | 0.002 | 0.004 | 2.7 × 10-5 | 0.07 | 0.54 |
β - beta coefficient; β L95 - beta coefficient 95 % confidence interval lower border; β U95 - beta coefficient 95 % confidence interval upper border; BP - basepair position (Build36); Chr - chromosome; N - samples size; EA - effect allele; OA - other allele; SNP - single nucleotide polymorphism, TC - total cholesterol; TG - triglyceride.
Build36.
P values are obtained by fixed-effect inverse variance weighted meta-analysis.
Heterogeneity was measured by a Cochran's Q test. P value of this test and I2 statistics are reported.
In silico look-up
We performed an in silico look-up for all SNPs (± 200 kb) that were associated with lipid change at P < 10-3 in the fixed-effect meta-analysis described above in the CARDIoGRAMplusC4D consortium database5 (Nmax = 191 650). For the pragmatic reasons outlined above for the replication analyses, a probability threshold of P < 1 × 10-3 was used for SNP look-ups. The purpose of the look-up was to detect suggestive novel associations with CAD at or near (± 200 kb) the genomic locations where associations with lipid level changes have been detected; because all variants reaching genome-wide significance in this database have been reported elsewhere,5 this search was restricted to the identification of variants yielding association signals in the P = 5 × 10-8 to P = 1 × 10-3 range. We also performed an in silico look-up of our top hits in the DIAbetes Genetics Replication And Meta-analysis (DIAGRAM)27 (Nmax = 86 531) and the International Consortium for Blood Pressure (ICBP)28 (Nmax = 203 056) consortia in order to investigate potential associations of our index variants and T2D and blood pressure, respectively.
Functional annotation
In order to characterize the regulatory potential of candidate loci, we characterized SNP overlaps with evolutionarily conserved elements determined using the GERP algorithm.29 We conducted functional annotation for the seven novel lipid change variants identified through the fixed-effect meta-analysis. These variants include those that were also associated with CAD in CARDIoGRAMplusC4D. Using data on hypersensitive sites (DHSs) in 125 cell lines generated by The ENCODE Project,30 we determined the frequencies with which DHS sites overlapped with candidate loci. Ernst et al. generated genome-wide chromatin state maps for nine cell lines using a hidden Markov model trained on histone modification patterns as well as binding of CCCTC-binding factor;31 to gauge the diversity of chromatin states associated with each SNP, we mapped these chromatin states for each of the nine cell lines onto our candidate loci. We used ChIP-seq data generated by ENCODE30 in order to assign binding capacity of specific regulatory factors to our candidate SNPs. Genomic coordinates (hg19) as well as sequence variants were obtained from the MetaboChip consortium web page [sph.umich.edu/csg/kang/MetaboChip]. Data on conserved elements were obtained from the Sidow-lab web page [mendel.stanford.edu/SidowLab/downloads/gerp]. DHS tracks (narrowPeak-pipeline), the Ernst et al. chromatin tracks and regulatory factor ChIP-seq binding tracks were obtained through the UCSC genome browser [genome.ucsc.edu]. Overlaps between genomic annotation tracks and SNPs were calculated using the GenomicRanges package in R.32
Results
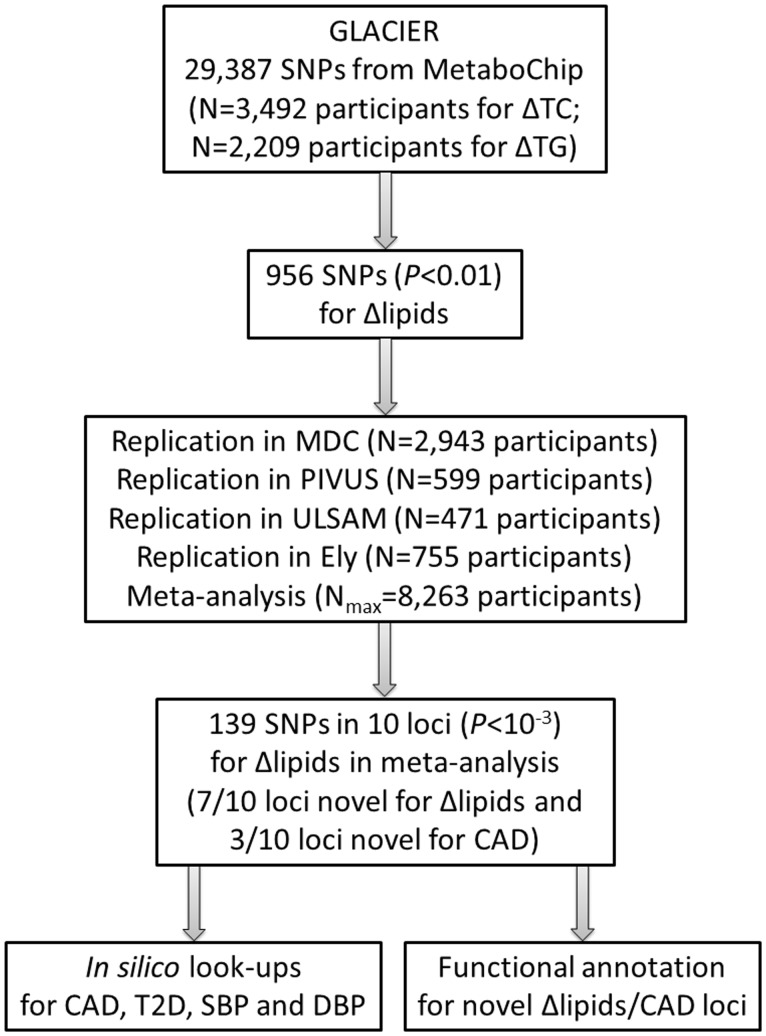
The project flowchart is shown in Figure 1. Participant baseline and follow-up characteristics are shown for each cohort in Tables S1-S5, available as Supplementary data at IJE online. The average follow-up period was 9.9 yrs (years) [standard deviation (SD) = 0.3 yrs] in the GLACIER Study, 15.8 yrs (SD = 1.4 yrs) in the MDC Study, 5.1 yrs (SD = 0.1 yrs) in PIVUS, 6.6 yrs (SD = 0.5 yrs) in ULSAM and 9.4 yrs (SD = 1.9 yrs) in the MRC Ely Study.
Figure 1.
Project Flowchart. CAD, coronary artery disease; DBP, diastolic blood pressure; N, sample size; SBP, systolic blood pressure; SNP, single nucleotide polymorphism; T2D, type 2 diabetes; TC, total cholesterol; TG, triglycerides.
Discovery analyses
An overall decrease in plasma TC concentrations from baseline to follow-up (mean change = -0.18 ± 1.12 mmol/l; P < 0.0001), but no change in TG concentrations (mean change = 0.02 ± 1 mmol/l; P = 0.32) was observed, as previously reported.8
In individual SNP analyses, a single low-frequency (MAF = 1.4%) SNP in APOE [chr19:50121999; β = 0.064 mmol/l/yr; standard error (SE) = 0.011; 95% confidence interval (CI) = 0.042 to 0.086; P = 1.7 × 10-8] and multiple SNPs in the APOA1/A4/C3/A5 gene cluster (top SNP rs9326246, β = 0.032 mmol/l/yr; SE = 0.005; 95% CI = 0.021 to 0.043; P = 4.4 × 10-9) reached genome-wide significance (P < 5 × 10-8) in relation to ΔTG. SNPs in DOCK7 (chr1:62714800; β = 0.059 mmol/l/yr; SE = 0.011; 95% CI = 0.038 to 0.081; P = 9.6 × 10-8), BRE (chr2:28165690; β = 0.059 mmol/l/yr; SE = 0.012; 95% CI = 0.036 to 0.082; P = 6.7 × 10-7), KCNIP1 (rs10041010; β = 0.054 mmol/l/yr; SE = 0.011; 95% CI = 0.032 to 0.075; P = 8.7 × 10-7) and SYNE1 (rs594522; β = 0.055 mmol/l/yr; SE = 0.011; 95% CI = 0.033 to 0.078; P = 1.4 × 10-6) reached study-wide significance for ΔTG. One SNP in APOE (rs7412; β = -0.023 mmol/l/yr; SE = 0.004; 95% CI = -0.032 to -0.014; P = 1.8 × 10-7) reached study-wide significance (α = 0.05/29,387 ∼ 1.7 × 10-6) for ΔTC; 478 and 490 SNPs associated (P < 0.01) with ΔTC and ΔTG, respectively. All associations reaching P < 0.01 for ΔTC and ΔTG are shown in Table S6 and Table S7, respectively (available as Supplementary data at IJE online).
From the 956 variants nominally associated with lipid changes, 274 SNPs had no cross-sectional associations with TC or TG in the GLGC (148 variants associated with TG, 183 variants associated with TC and 351 variants associated with both traits; P < 5 × 10-8).
Replication and meta-analysis
The 956 variants that were associated with Δlipids in the discovery cohort were further examined in the replication cohorts (excluding the discovery cohort). The variant at rs7412 at APOE replicated for ΔTC (P = 1.5 × 10-7), as did multiple variants at APOA1/A4/C3/A5 for ΔTG (Pmin = 6.5 × 10-7); 135/478 SNPs (∼ 28%) for ΔTC and 66/490 (∼ 13%) SNPs for ΔTG reached nominal statistical significance (P < 0.05). None of the APOE association signals replicated for ΔTG.
In the meta-analysis including all cohorts (discovery and replication), 88 SNPs at six loci for ΔTC and 51 SNPs at five loci for ΔTG (total of 10 loci, as one locus associated with both traits) replicated (P < 1.0 × 10-3). The lead SNPs for the 10 replicating loci in relation to their respective traits are shown in Table 1.
All 10 of the associated loci have been linked to cross-sectional lipid levels in previous meta-analyses.2 The DOCK7/ANGPTL3 locus associated with both ΔTC and ΔTG in the meta-analysis (chr1:62912318; β = -0.004 mmol/l/yr; SE = 0.001; 95% CI = -0.006 to -0.002; P = 2.5 × 10-4 for ΔTC and chr1:62954723; β = -0.002 mmol/l/yr; SE = 0.001; 95% CI = -0.004 to -0.001; P = 5.3 × 10-4 for ΔTG). Two loci, APOE/C1 for ΔTC (β = -0.014 mmol/l/yr; SE = 0.002; 95% CI = -0.018 to -0.01; P = 2.0 × 10-12) and ZNF159/APOA5/A1 for ΔTG (β = 0.009 mmol/l/yr; SE = 0.001; 95% CI = 0.007 to 0.012; P = 1.4 × 10-10) reached genome-wide significance (P < 5 × 10-8). All associations with lipid level changes remained statistically significant at P < 0.05 after removing the discovery cohort, GLACIER, from the meta-analysis (Table 2).
Table 2.
Lead SNPs from the fixed-effect inverse variance weighted meta-analysis of the replication cohorts, MDC, PIVUS, ULSAM and MRC Ely. (Nmax=4,768)
| Trait | Chr | BP* | SNP | EA | OA | Nearest locus | β (mmol/l/yr) | SE (mmol/l/yr) | β L95 (mmol/l/yr) | β U95 (mmol/l/yr) | P | Pheterogeneity | I2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ΔTC | 1 | 62912318 | chr1:62912318 | C | T | DOCK7/ANGPTL3 | −0.003 | 0.001 | −0.005 | −0.0004 | 0.02 | 0.39 | 0 |
| ΔTC | 8 | 19669744 | rs13282247 | T | C | LPL/CSGALNACT1 | −0.003 | 0.001 | −0.006 | −0.001 | 0.02 | 0.39 | 0 |
| ΔTC | 15 | 40469997 | chr15:40469997 | A | T | CAPN3 | 0.012 | 0.004 | 0.004 | 0.021 | 0.005 | 0.42 | 0 |
| ΔTC | 16 | 70636491 | chr16:70636491 | T | G | HP/HPR | 0.011 | 0.004 | 0.002 | 0.02 | 0.01 | 0.04 | 0.69 |
| ΔTC | 19 | 11001703 | rs17304534 | C | T | LDLR | −0.002 | 0.001 | −0.004 | −0.001 | 0.01 | 0.99 | 0 |
| ΔTC | 19 | 50103919 | rs7412 | T | C | APOE/C1 | −0.012 | 0.002 | −0.016 | −0.007 | 1.5 × 10-7 | 0.81 | 0 |
| ΔTG | 1 | 62954723 | chr1:62954723 | C | T | DOCK7/ANGPTL3 | −0.002 | 0.001 | −0.004 | −0.001 | 0.004 | 0.66 | 0 |
| ΔTG | 8 | 126560154 | rs2954029 | T | A | TRIB1 | −0.003 | 0.001 | −0.004 | −0.001 | 2.4 × 10-4 | 0.47 | 0 |
| ΔTG | 11 | 116167789 | rs651821 | C | T | ZNF159/APOA5/A1 | 0.008 | 0.002 | 0.005 | 0.011 | 3.6 × 10-7 | 0.06 | 0.60 |
| ΔTG | 19 | 50848859 | rs10406431 | G | A | GIPR | −0.002 | 0.001 | −0.004 | −0.001 | 0.002 | 0.6 | 0 |
| ΔTG | 19 | 50902205 | rs11668847 | G | T | QPCTL | −0.003 | 0.001 | −0.004 | −0.001 | 0.003 | 0.13 | 0.48 |
β - beta coefficient; β L95 - beta coefficient 95 % confidence interval lower border; β U95 - beta coefficient 95 % confidence interval upper border; BP - basepair position (Build36); Chr - chromosome; N - samples size; EA - effect allele; OA - other allele; SNP - single nucleotide polymorphism, TC - total cholesterol; TG - triglyceride.
Build36.
P values are obtained by fixed-effect inverse variance weighted meta-analysis.
Heterogeneity was measured by a Cochran's Q test. P value of this test and I2 statistics are reported.
Associations with CAD, blood pressure and T2D
When conducting the CARDIoGRAMplusC4D in silico look-up for the 10 top loci (Table 3), we allocated these to three distinct categories of association signals:
Table 3.
Lipid change associated variants after correction for multiple testing in relation to CAD in the CARDIoGRAMplusC4D consortium. (Nmax=191,650)
| Lipid change meta-analysis |
CARDIoGRAMplusC4D |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Trait | Locus | Chr | BP* | SNP | P | BP* | SNP | C4D OR | C4D P | Category |
| ΔTC | APOE/C1 | 19 | 50103919 | rs7412 | 2.0 × 10-12 | 50087459 | rs2075650 | 1.11 | 5.9 × 10−11 | published for CAD and Δlipids |
| ΔTG | TRIB1 | 8 | 126560154 | rs2954029 | 8.1 × 10-6 | 126560154 | rs2954029 | 1.05 | 4.5 × 10−8 | published for CAD and Δlipids |
| ΔTG | ZNF159/APOA5/A1 | 11 | 116167789 | rs651821 | 1.4 × 10-10 | 116116943 | rs9326246 | 1.09 | 1.5 × 10−7 | published for CAD and Δlipids |
| ΔTC | DOCK7/ANGPTL3 | 1 | 62912318 | chr1:62912318 | 5.2 × 10-4 | 62912318 | chr1:62912318 | 1.00 | NS** | not significant for CAD, novel for Δlipids |
| ΔTC | LPL/CSGALNACT1 | 8 | 19669744 | rs13282247 | 1.7 × 10-4 | 19857460 | rs264 | 1.07 | 5.1 × 10−9 | published for CAD, novel for Δlipids |
| ΔTC | LDLR | 19 | 11001703 | rs17304534 | 1.0 × 10-3 | 11024601 | rs1122608 | 1.10 | 6.3 × 10−14 | published for CAD, novel for Δlipids |
| ΔTG | GIPR | 19 | 50848859 | rs10406431 | 1.7 × 10-4 | 50841458 | rs1029846 | 1.02 | NS** | not significant for CAD, novel for Δlipids |
| ΔTG | DOCK7/ANGPTL3 | 1 | 62954723 | chr1:62954723 | 5.2 × 10-4 | 62912318 | chr1:62912318 | 1.00 | NS** | not significant for CAD, novel for Δlipids |
| ΔTC | CAPN3 | 15 | 40469997 | chr15:40469997 | 1.5 × 10-4 | 40471079 | rs2412710 | 1.14 | 2.4 × 10-4 | novel for CAD and Δlipids |
| ΔTC | HP/HPR | 16 | 70636491 | chr16:70636491 | 2.3 × 10-4 | 70636628 | rs7197453 | 1.04 | 6.1 × 10-5 | novel for CAD and Δlipids |
| ΔTG | QPCTL | 19 | 50902205 | rs11668847 | 2.7 × 10-5 | 50960742 | rs2341097 | 1.03 | 3.5 × 10-4 | novel for CAD and Δlipids |
BP - basepair; C4D - CARDIoGRAMplusC4D; CAD - coronary artery disease; Chr - chromosome; NS - not significant; OR - odds ratio; SNP - single nucleotide polymorphism; TC - total cholesterol; TG - triglyceride.
Build36
Few variants with nominal statistical significance (P∼0.05) in the region
P values are based on linear regression models. SNP associations were tested by fitting the genetic variants (additive model) as the independent variables with follow-up lipid measures as dependent variables.
C4D P values are obtained by a fixed-effect inverse variance weighted meta-analysis in the CARDIoGRAMplusC4D consortium for the top SNPs in the loci associated with lipid changes.
three loci (APOE/C1, TRIB1, ZNF159/APOA5/A1) that have been previously reported for their associations with both lipid changes8 and CAD;5
loci not previously associated with lipid changes, but either associated with (P < 5 × 10-8 for LPL/CSGALNACT and LDLR) or not associated (P > 0.05 for DOCK7/ANGPTL3 and GIPR) with CAD;
loci for which novel associations with both lipid changes and CAD were observed in the current meta-analysis (CAPN3, HP/HPR and QPCTL).
Associations for the 10 loci in the DIAGRAM and the ICBP are shown in Tables S8 and Table S9, respectively (available as Supplementary data at IJE online). Three loci (APOE/C1, TRIB1 and CAPN3) show suggestive associations with T2D (5 × 10-8 < P < 1 × 10-3), whereas two loci (CAPN3 and DOCK7/ANGPTL3) show suggestive associations with systolic blood pressure (P ∼ 7 × 10-3).
Functional annotation
We functionally annotated the top SNPs and SNPs in close proximity to our top SNPs (n = 11 SNPs) for the seven loci (LPL/CSGALNACT1, LDLR, DOCK7/ANGPTL3, CAPN3, HP/HPR, GIPR, QPCTL) in the second and third categories described above. All results from the functional annotation are shown in Table S10 (available as Supplementary data at IJE online). Two SNPs, rs1050362 near HPR and rs2341097 in SIX5 (in close proximity to QPCTL and GIPR) overlapped with elements displaying excess constraint. Expanding the overlaps to a 501 base pair window centred on each SNP yielded an additional variant, rs13282247 in LPL/CSGALNACT1 with overlapping elements. Variants rs2412710 in CAPN3, rs1050362 near HPR, rs2341097 in SIX5 and rs13282247 in LPL/CSGALNACT1 disrupt CpG-dinucleotides, potentially altering the local balance of DNA methylation and providing a common mechanism by which both variants may influence regulatory factor binding. In total, 6/11 SNPs exhibited DNaseI hypersensitive sites (DHS) in at least one ENCyclopedia Of DNA Elements (ENCODE) cell line, including rs2412710 at CAPN3. We found a high degree of correspondence between active states, e.g. ‘transcription elongation’ as well as ‘weak transcribed’ and other indicators of regulatory potential such as DHS occurrence across a broad range of ENCODE cell lines. In the meta-analysis, some SNPs that were associated with lipid changes and CAD displayed enhancer-related chromatin marks, rs2412710 at CAPN3 was indicated by a ‘weak enhancer’ state in HepG2 cells, rs10406431 at GIPR by ‘weak enhancer’ in K562 cells and ‘repressed’ in GM12878 cells, and rs11668847 at QPCTL by ‘strong enhancer’ state in GM12878 cells. In total, 5/11 SNPs were in regions with ChIP-seq peaks. Variants rs11668847 at QPCTL, rs10406431 at GIPR, rs17304534 at LDLR, chr1:62954723 in DOCK7/ANGPTL3 and rs13282247 at LPL/CSGALNACT1 displayed evidence of binding multiple enhancer and gene regulation-related factors, including FOS, GATA1 and GATA3, all in the K562 cell line. The variant rs11668847 at QPCTL, which displayed a ‘strong enhancer’ state in GM12878 cells, displayed concomitant RNA polymerase II (POLR2A) binding.
Discussion
We sought to discover causal loci affecting CAD risk by virtue of their associations with long-term deteriorations in blood lipid concentrations. The discovery analyses identified a SNP associated with worsening TG concentrations at the APOA1/A4/C3/A5 locus, which reached genome-wide significance (P < 5 × 10-8); a further four loci (DOCK7, BRE, KCNIP1 and SYNE1) were associated with worsening TG at a study-wide level of statistical significance. One variant at APOE reached study-wide statistical significance for worsening TC in the discovery cohort. In the meta-analysis, 10 loci were associated with lipid changes, of which three were also associated with CAD. Functional annotation of these variants and those in close proximity revealed compelling evidence across a range of ENCODE cell lines that some of these loci might influence regulatory factor binding and transcription factor binding.
Of the 10 loci robustly associated with lipid changes, seven (DOCK7/ANGPTL3, LPL/CSGALNACT, HP/HPR, LDLR, APOE/C1, DOCK7/ANGPTL3, TRIB1, ZNF159/APOA5/A1, CAPN3) are known genetic determinants of blood lipid levels from large-scale cross-sectional analyses,2,3 and three (APOE/C1, TRIB1, ZNF159/APOA5/A1) have also been previously associated with prospective changes in blood lipid concentrations.8 In a previous analysis investigating known lipid-associated loci, the rs4420638 variant at APOE was robustly associated with TC changes.8 In the current meta-analysis, the rs7412 variant was the lead SNP in the APOE/C1 locus. This SNP defines the ε2, ε3 and ε4 status in humans, which plays a crucial role in CVD.33 However, the lead variant at APOE for TG changes in the discovery analysis was another variant, chr19:50121999, but this variant was not associated with TG changes in the meta-analysis. We discovered SNP associations with worsening lipids at seven additional loci, with three (CAPN3, HP/HPR, QPCTL) of these also being associated with CAD.
The CAPN3 variant was associated with worsening TC (P = 1.5 × 10-4) and CAD (P = 1.2 × 10-4), as well as with T2D (P = 1.6 × 10-3) and cross-sectional systolic blood pressure (SBP) (P = 6.9 × 10-3). In addition, functional annotation showed that this variant disrupts CpG-dinucleotides and displays DNaseI hypersensitivity sites in two cell lines and ‘weak enhancer’ status in HepG2 cells. CAPN3 encodes for calpain3, which is strongly expressed in muscle and weakly expressed in heart with calpainopathy (symmetrical and progressive weakness of proximal muscles).34 Although calpain3’s function has not been fully elucidated, it is known to have proteolytic activity in skeletal muscle.34 In murine models, targeted Capn3 knockout reverses myopathy,35 and restriction of calpain3 transgene expression in heart prevents cardiac toxicity and reverts the pathological signs of calpain3 deficiency.36 In addition, targeted knock-in of calpain3 leads to increased circulating creatine kinase levels,37 a marker for myocardial onfarction (MI). A Japanese autopsy study recently reported that two unrelated patients with calpainopathy (LGMD2A), who had died of ischaemic cardiomyopathy and systemic circulatory failure respectively, carried mutations in CAPN3.38 We note that the paralog of CAPN3 (CAPN10) was once considered a major candidate locus for T2D based on linkage scans, but GWAS has failed to validate this hypothesis.39
Associations were observed for rs10406431 at GIPR (P = 1.7 × 10-4) and rs11668847 at QPCTL (P = 2.7 × 10-5) (variants 53 kB apart, r2 = 0.09, D’ = 0.33) for worsening TG concentrations. These variants display DNaseI hypersensitivity and regulatory factor binding potential in multiple cell lines. The rs11668847 variant at QPCTL displayed a ‘strong enhancer’ status, whereas rs10406431 at GIPR was characterized by a ‘polycomb-repressed’ status in GM12878 cells. The rs2341097 SIX5 variant is in close proximity to these loci (111 kB, r2 = 0.09, D’ = 0.52 to GIPR variant and 59 kB, r2 = 0.17, D’ = 0.66 to QPCTL variant) and was associated with CAD (P = 1.9 × 10-4). The GIPR gene encodes a G-protein coupled receptor for gastric inhibitory polypeptide (GIP). Saxena et al. reported that the A allele of the rs10423928 variant is associated with increased 2-h glucose concentrations and decreased insulin secretion.40 In addition, interactions between GIPR and carbohydrate and fat intake have been reported for incident T2D. In that study, rs10423928 A allele homozygotes were at a lower risk of T2D when consuming high-fat, low-carbohydrate diets, and two-thirds of the population homozygous for the T-allele were at lower risk of T2D when consuming high-carbohydrate, low-fat diets.41
QPCTL is responsible for the biosynthesis of pyroglutamyl peptides. The rs2287019 variant’s C allele at the QPCTL locus has been previously associated with increased insulinogenic index and disposition index.42 Furthermore, genome-wide significant associations with BMI near the QPCTL locus have been reported.43,44 We found that the rs2341097 SIX5 variant disrupts a CpG site and resides in an evolutionarily conserved region. SIX5 encodes for the homeodomain-containing transcription factor that appears to regulate organogenesis. SIX5 is associated with myotonic dystrophy, a highly variable multisystem disease in which the classic adult-onset form displays progressive muscle wasting with myotonia, cataracts, heart block, gonadal atrophy, insulin resistance and neuropsychiatric impairment.45 The SIX5 rs16980013 variant has been nominally associated with myocardial infarction in Japanese patients.46 In Six5+/- (heterozygote) adult mice, prolonged QRS duration was observed, which is one of the initial phenotypes of adult-onset cardiac conduction abnormalities in humans with myotonic dystrophy.47
The variant at the HP/HPR locus was associated with worsening TC (P = 2.3 × 10-4) and with CAD (P = 3.5 × 10-5) in our analyses. HPR is located 2.2 kb downstream of the haptoglobin (HP) gene and shares 91% of common DNA sequence variation with HP gene. HPR encodes the haptoglobin-like protein, which binds to haemoglobin with high affinity.48,49 It is known to be associated with TC, LDL-C, haptoglobin, liver enzymes, haemoglobin levels and red cell count in adults.2,3,50,51 Although high blood haptoglobin level is a known risk factor for acute MI, carotid atherosclerosis, stroke and heart failure,52,53 the HP/HPR locus has not previously been associated with CAD to our knowledge.
Large-scale cross-sectional meta-analyses have detected hundreds of genetic associations.2 However, some time-dependent genetic association signals are likely to be hard or impossible to detect using cross-sectional data.11 In the discovery phase of this study, we used the MetaboChip array and selected genetic variants genome-wide or sub-genome-wide significant cross-sectional lipid association signals for follow-up. We hypothesized that loci that convey time-dependent genetic effects might be hard to detect (i.e. the association P-value may be weak) in cross-sectional data, owing to the heterogeneous nature of these signals. By consequence, such loci may not have reached genome-wide significance in GWAS meta-analyses, but may still have been included on the MetaboChip as the array is enriched with sub-genome-wide significant loci. Despite these advantages, this array is clearly not genome-wide in coverage and we may have overlooked other signals for lipid change; hence, larger-scale GWAS meta-analyses focused on long-term changes in lipid levels may yield additional signals. Future studies might also benefit from focusing on other lipids (e.g. LDL-C and HDL-C) and subfractions, which we did not have access to in the present study. It is also the case that most of the top-ranking loci in this analysis convey small effects, consistent with previous findings on the genetics of dyslipidaemia.2,3,8,54 It is important to keep in mind, however, that variants with small effect sizes may still be physiologically relevant. For example, variants at HMGCR (encoding the HMG-CoA reductase enzyme), which plays a crucial role in cholesterol synthesis, have relatively small effects on lipids,3 but inhibiting the HMG-CoA reductase enzyme by statins is one of the most powerful and widely-used medical interventions with large effects.55 A further limitation to our study is that the meta-analysis only involves Northern European participants, so the results might not be generalizable to other ethnicities.
Of note, LMMs are often used in prospective association studies with more than two time points. However, in this analysis only two time points were available for analysis. Thus, we assessed genetic associations using slope (lipid level change) as outcome with generalized linear models, as described in Methods. When slope is estimated from data with multiple time points, P-values for the SNPs’ main effects agree perfectly with P-values for an SNP × time interaction term from LMMs.56 When data at only two time points are available, however, independently of method, one cannot accurately distinguish between measurement error and real change. Hence, the magnitude of the effect and its statistical significance may be under- or overestimated. To help address these limitations, we undertook replication analyses and functional annotation in independent materials.
In conclusion, we discovered and replicated associations between numerous genetic variants and long-term deteriorations in blood lipid concentrations. Through meta-analysis of data from multiple Northern European prospective cohort studies, we identified seven additional loci robustly associated with long-term lipid changes, from which three regions also demonstrated suggestive evidence for association with CAD. We also demonstrate that several of these loci are likely to interfere with gene function across different tissues. Future studies will be required to determine whether interactions with environmental factors that change with age underlie these time-dependent associations.
Supplementary Data
Supplementary data are available at IJE online.
Supplementary Material
Acknowledgements
The authors thank Dr. Robert Hanson for his advise on the rebuttal regarding mixed linear models. GLACIER: We thank the participants, health professionals and data managers involved in the Västerbotten Intervention Project. We are also grateful to the staff of the Northern Sweden Biobank for preparing materials and to K. Enqvist and T. Johansson (Västerbottens County Council, Umeå, Sweden) for DNA preparation. MDC: We thank the participants, health professionals, data managers and biobank staff involved in the Malmö Diet and Cancer Study. We also thank M. Svensson and W. Tas for excellent technical assistance. PIVUS/ULSAM: Parts of the analyses were performed with resources provided by SNIC through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under Project b2011036. Genotyping was performed by the SNP&SEQ Technology Platform in Uppsala [www.genotyping.se]. MRC Ely: We are most grateful to all study participants and to the staff of the St. Mary's Street Surgery, Ely. We thank all the staff who worked on the study.
Funding
GLACIER: This research was undertaken as part of a research program supported by the European Commission (CoG-2015_681742_NASCENT), Swedish Research Council (Distinguished Young Researchers Award in Medicine), Swedish Heart-Lung Foundation, and the Novo Nordisk Foundation, all grants to PWF. MDC: This study was supported by the Swedish Research Council, the Swedish Heart and Lung Foundation, the Novo Nordic Foundation, the Swedish Diabetes Foundation, and the Påhlsson Foundation, and by equipment grants from the Knut and Alice Wallenberg Foundation, the Region Skåne, Skåne University Hospital, the Linneus Foundation for the Lund University Diabetes Center and the European Research Council (Consolidator Grant nr. 649021, MO-M). PIVUS, ULSAM: These studies were supported by grants from the Knut och Alice Wallenberg Foundation (Wallenberg Academy Fellow), European Research Council (ERC-2013-StG; Grant no. 335395), Swedish Diabetes Foundation, Swedish Heart-Lung Foundation (Grant no. 20120197), Swedish Research Council (Grant no. 2012-1397), and Wellcome Trust (Grant no. WT098017, WT090532, and WT064890). MRC Ely: The MRC Ely Study was funded by the MRC (MC_U106179471) and Diabetes UK. Genotyping in the Ely study was supported in part by an MRC-GlaxoSmithKline pilot programme Grant (G0701863).
Conflict of interest: PWF has been a paid consultant for Eli Lilly and Sanofi Aventis and has received research support from several pharmaceutical companies as part of European Union Innovative Medicines Initiative (IMI) projects.
References
- 1. Shah S, Casas JP, Gaunt TR. et al. Influence of common genetic variation on blood lipid levels, cardiovascular risk, and coronary events in two British prospective cohort studies. Eur Heart J 2013;34:972–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Global Lipids Genetics Consortium, Willer CJ, Schmidt EM, Sengupta S. et al. Discovery and refinement of loci associated with lipid levels. Nat Genet 2013;45:1274–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Teslovich TM, Musunuru K, Smith AV. et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 2010;466:707–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Do R, Willer CJ, Schmidt EM. et al. Common variants associated with plasma triglycerides and risk for coronary artery disease. Nat Genet 2013;45:1345–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. CARDIoGRAMplusC4D Consortium Deloukas P, Kanoni S. et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet 2013;45:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lu Y, Feskens EJ, Boer JM. et al. Exploring genetic determinants of plasma total cholesterol levels and their predictive value in a longitudinal study. Atherosclerosis 2010;213:200–05. [DOI] [PubMed] [Google Scholar]
- 7. Lutsey PL, Rasmussen-Torvik LJ, Pankow JS. et al. Relation of lipid gene scores to longitudinal trends in lipid levels and incidence of abnormal lipid levels among individuals of European ancestry: the Atherosclerosis Risk in Communities (ARIC) study. Circ Cardiovasc Genet 2012;5:73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Varga TV, Sonestedt E, Shungin D. et al. Genetic determinants of long-term changes in blood lipid concentrations: 10-year follow-up of the GLACIER study. PLoS Genet 2014;10:e1004388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brouwers MC, Kono N, van Greevenbroek MM. et al. Longitudinal differences in familial combined hyperlipidemia quantitative trait loci. Arterioscler Thromb Vasc Biol 2006;26:e118–19. [DOI] [PubMed] [Google Scholar]
- 10. Dumitrescu L, Carty CL, Franceschini N. et al. Post-genome-wide association study challenges for lipid traits: describing age as a modifier of gene-lipid associations in the Population Architecture using Genomics and Epidemiology (PAGE) study. Ann Hum Genet 2013;77:416–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kurbasic A, Poveda A, Chen Y. et al. Gene-Lifestyle Interactions in Complex Diseases: design and description of the GLACIER and VIKING Studies. Curr Nutr Rep 2014;3:400–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Franks PW, Rolandsson O, Debenham SL. et al. Replication of the association between variants in WFS1 and risk of type 2 diabetes in European populations. Diabetologia 2008;51:458–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hallmans G, Agren A, Johansson G. et al. Cardiovascular disease and diabetes in the Northern Sweden Health and Disease Study Cohort - evaluation of risk factors and their interactions. Scand J Public Health Suppl 2003;61:18–24. [DOI] [PubMed] [Google Scholar]
- 14. Wu J, Province MA, Coon H. et al. An investigation of the effects of lipid-lowering medications: genome-wide linkage analysis of lipids in the HyperGEN study. BMC Genet 2007;8:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eliasson M, Janlert U, Jansson JH, Stegmayr B.. Time trends in population cholesterol levels 1986-2004: influence of lipid-lowering drugs, obesity, smoking and educational level. The northern Sweden MONICA study. J Intern Med 2006;260:551–59. [DOI] [PubMed] [Google Scholar]
- 16. Tobin MD, Sheehan NA, Scurrah KJ, Burton PR.. Adjusting for treatment effects in studies of quantitative traits: antihypertensive therapy and systolic blood pressure. Stat Med 2005;24:2911–35. [DOI] [PubMed] [Google Scholar]
- 17. Renstrom F, Payne F, Nordstrom A. et al. Replication and extension of genome-wide association study results for obesity in 4923 adults from northern Sweden. Hum Mol Genet 2009;18:1489–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Voight BF, Kang HM, Ding J. et al. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet 2012;8:e1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Purcell S, Neale B, Todd-Brown K. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Statacorp. STATA 12.1 SL. http://www.stata.com/.
- 21. Renstrom F, Shungin D, Johansson I. et al. Genetic predisposition to long-term nondiabetic deteriorations in glucose homeostasis: Ten-year follow-up of the GLACIER study. Diabetes 2011;60:345–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gurka MJ, Edwards LJ, Muller KE.. Avoiding bias in mixed model inference for fixed effects. Stat Med 2011;30:2696–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Littell RC, Pendergast J, Natarajan R.. Modelling covariance structure in the analysis of repeated measures data. Stat Med 2000;19:1793–819. [DOI] [PubMed] [Google Scholar]
- 24. Thomas G, Jacobs KB, Kraft P. et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1). Nat Genet 2009;41:579–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Magi R, Morris AP.. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics 2010;11:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Skol AD, Scott LJ, Abecasis GR, Boehnke M.. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet 2006;38:209–13. [DOI] [PubMed] [Google Scholar]
- 27. Morris AP, Voight BF, Teslovich TM. et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet 2012;44:981–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ehret GB, Munroe PB, Rice KM. et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 2011;478:103–09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cooper GM, Stone EA, Asimenos G. et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res 2005;15:901–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bernstein BE, Birney E, Dunham I. et al. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ernst J, Kheradpour P, Mikkelsen TS. et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 2011;473:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. R Development Core Team: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing, 2013. [Google Scholar]
- 33. Eichner JE, Dunn ST, Perveen G, Thompson DM, Stewart KE, Stroehla BC.. Apolipoprotein E polymorphism and cardiovascular disease: a HuGE review. Am J Epidemiol 2002;155:487–95. [DOI] [PubMed] [Google Scholar]
- 34. Hauerslev S, Sveen ML, Duno M, Angelini C, Vissing J, Krag TO.. Calpain 3 is important for muscle regeneration: evidence from patients with limb girdle muscular dystrophies. BMC Musculoskelet Disord 2012;13:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Charton K, Daniele N, Vihola A. et al. Removal of the calpain 3 protease reverses the myopathology in a mouse model for titinopathies. Hum Mol Genet 2010;19:4608–24. [DOI] [PubMed] [Google Scholar]
- 36. Roudaut C, Le Roy F, Suel L. et al. Restriction of calpain3 expression to the skeletal muscle prevents cardiac toxicity and corrects pathology in a murine model of limb-girdle muscular dystrophy. Circulation 2013;128:1094–104. [DOI] [PubMed] [Google Scholar]
- 37. Ojima K, Kawabata Y, Nakao H. et al. Dynamic distribution of muscle-specific calpain in mice has a key role in physical-stress adaptation and is impaired in muscular dystrophy. J Clin Invest 2010;120:2672–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hashiguchi S, Adachi K, Inui T. et al. [A clinicopathological investigation of two autopsy cases of calpainopathy (LGMD2A)]. Shinkei Kenkyu No Shinpo 2014;66:1097–102. [PubMed] [Google Scholar]
- 39. Hanis CL, Boerwinkle E, Chakraborty R. et al. A genome-wide search for human non-insulin-dependent (type 2) diabetes genes reveals a major susceptibility locus on chromosome 2. Nat Genet 1996;13:161–6. [DOI] [PubMed] [Google Scholar]
- 40. Saxena R, Hivert MF, Langenberg C. et al. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat Genet. 2010;42:142–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sonestedt E, Lyssenko V, Ericson U. et al. Genetic variation in the glucose-dependent insulinotropic polypeptide receptor modifies the association between carbohydrate and fat intake and risk of type 2 diabetes in the Malmo Diet and Cancer cohort. J Clin Endocrinol Metab 2012;97:E810–18. [DOI] [PubMed] [Google Scholar]
- 42. Burgdorf KS, Gjesing AP, Grarup N. et al. Association studies of novel obesity-related gene variants with quantitative metabolic phenotypes in a population-based sample of 6,039 Danish individuals. Diabetologia 2012;55:105–13. [DOI] [PubMed] [Google Scholar]
- 43. Speliotes EK, Willer CJ, Berndt SI. et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet 2010;42:937–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wen W, Cho YS, Zheng W. et al. Meta-analysis identifies common variants associated with body mass index in east Asians. Nat Genet 2012;44:307–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sarkar PS, Appukuttan B, Han J. et al. Heterozygous loss of Six5 in mice is sufficient to cause ocular cataracts. Nat Genet 2000;25:110–14. [DOI] [PubMed] [Google Scholar]
- 46. Fujimaki T, Kato K, Yokoi K. et al. Association of genetic variants in SEMA3F, CLEC16A, LAMA3, and PCSK2 with myocardial infarction in Japanese individuals. Atherosclerosis 2010;210:468–73. [DOI] [PubMed] [Google Scholar]
- 47. Wakimoto H, Maguire CT, Sherwood MC. et al. Characterization of cardiac conduction system abnormalities in mice with targeted disruption of Six5 gene. J Interv Card Electrophysiol 2002;7:127–35. [DOI] [PubMed] [Google Scholar]
- 48. Maeda N. Nucleotide sequence of the haptoglobin and haptoglobin-related gene pair. The haptoglobin-related gene contains a retrovirus-like element. J Biol Chem 1985;260:6698–709. [PubMed] [Google Scholar]
- 49. Nielsen MJ, Petersen SV, Jacobsen C. et al. Haptoglobin-related protein is a high-affinity hemoglobin-binding plasma protein. Blood 2006;108:2846–49. [DOI] [PubMed] [Google Scholar]
- 50. Guthrie PA, Rodriguez S, Gaunt TR, Lawlor DA, Davey Smith G, Day IN.. Complexity of a complex trait locus: HP, HPR, haemoglobin and cholesterol. Gene 2012;499:8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chambers JC, Zhang W, Sehmi J. et al. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet 2011;43:1131–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Froguel P, Ndiaye NC, Bonnefond A. et al. A genome-wide association study identifies rs2000999 as a strong genetic determinant of circulating haptoglobin levels. PloS One 2012;7(3):e32327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Holme I, Aastveit AH, Hammar N, Jungner I, Walldius G.. Haptoglobin and risk of myocardial infarction, stroke, and congestive heart failure in 342,125 men and women in the Apolipoprotein MOrtality RISk study (AMORIS). Ann Med 2009;41:522–32. [DOI] [PubMed] [Google Scholar]
- 54. Chasman DI, Pare G, Mora S. et al. Forty-three loci associated with plasma lipoprotein size, concentration, and cholesterol content in genome-wide analysis. PLoS Genet 2009;5:e1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Baigent C, Keech A, Kearney PM. et al. Efficacy and safety of cholesterol-lowering treatment: prospective meta-analysis of data from 90,056 participants in 14 randomised trials of statins. Lancet 2005;366:1267–78. [DOI] [PubMed] [Google Scholar]
- 56. Sikorska K, Rivadeneira F, Groenen PJ. et al. Fast linear mixed model computations for genome-wide association studies with longitudinal data. Stat Med 2013;32:165–80. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.