Abstract
Many drugs designed to inhibit kinases have their clinical utility limited by cardiotoxicity-related label warnings or prescribing restrictions. While this liability is widely recognized, designing safer kinase inhibitors (KI) requires knowledge of the causative kinase(s). Efforts to unravel the kinases have encountered pharmacology with nearly prohibitive complexity. At therapeutically relevant concentrations, KIs show promiscuity distributed across the kinome. Here, to overcome this complexity, 65 KIs with known kinome-scale polypharmacology profiles were assessed for effects on cardiomyocyte (CM) beating. Changes in human iPSC-CM beat rate and amplitude were measured using label-free cellular impedance. Correlations between beat effects and kinase inhibition profiles were mined by computation analysis (Matthews Correlation Coefficient) to identify associated kinases. Thirty kinases met criteria of having (1) pharmacological inhibition correlated with CM beat changes, (2) expression in both human-induced pluripotent stem cell-derived cardiomyocytes and adult heart tissue, and (3) effects on CM beating following single gene knockdown. A subset of these 30 kinases were selected for mechanistic follow up. Examples of kinases regulating processes spanning the excitation–contraction cascade were identified, including calcium flux (RPS6KA3, IKBKE) and action potential duration (MAP4K2). Finally, a simple model was created to predict functional cardiotoxicity whereby inactivity at three sentinel kinases (RPS6KB1, FAK, STK35) showed exceptional accuracy in vitro and translated to clinical KI safety data. For drug discovery, identifying causative kinases and introducing a predictive model should transform the ability to design safer KI medicines. For cardiovascular biology, discovering kinases previously unrecognized as influencing cardiovascular biology should stimulate investigation of underappreciated signaling pathways.
Keywords: cardiotoxicity, kinase, kinase inhibitor, cellular impedance
Because kinases play a critical role in regulating diverse human biology, their modulation has become a mainstay for pharmaceutical discovery particularly for treating cancer (Wu et al., 2015). More than 25 small molecule kinase inhibitor (SMKI) drugs are now approved (Wu et al., 2015), yet many have cardiac liabilities requiring label warnings (Bellinger et al., 2015; Fabbro et al., 2012; Orphanos et al., 2009; and Supplementary References). Cardiotoxicity is also a cause for SMKI failure in clinical development (Ho et al., 2011) and preclinical discovery (Lim et al., 2011). Thus, the current preclinical testing cascades need supplemental tools to help mitigate cardiotoxicity. In principle, understanding the particular kinase(s) that cause cardiotoxicity could allow rational design of safer drugs. Unfortunately, SMKI are notoriously promiscuous and this polypharmacology has been too complex to unravel within a single drug discovery program.
A dedicated study to deconvolute kinases contributing to functional cardiotoxicity has been prohibited by at least one of three hurdles. First, the human genome encodes 518 protein kinases, 478 of which belong to the “typical” (or “eukaryotic”) superfamily with a conserved catalytic domain (Manning et al., 2002). The highly conserved ATP binding is the site of interaction for the majority of SMKI. This creates polypharmacology with kinome-scale complexity thereby requiring higher throughput assays and large data sets to deconvolute. Second, kinases modulate cardiac function by coordinated regulation of proteins involved in the action potential, calcium flux, and mechanical contraction (Bers, 2002). This necessitates functional assays that are sensitive to endpoints downstream in the excitation–contraction signaling cascade to capture diverse kinase effects on excitation, calcium flux and contraction. Third, interspecies variation in kinases raises the possibility that kinase inhibitor polypharmacology profiles will also vary between species. It follows that both human kinome pharmacology profiles and human functional assays are needed to align in vitro data with clinical cardiotoxicity. Thus, while existing cardiotoxicity assays perform well for most drug classes, they lack the throughput, downstream functional readout, and/or human basis that is required for unraveling mechanisms of SMKI cardiotoxicity.
Recent advances have addressed each of these three hurdles. Kinase selectivity profiles across the majority of human kinome have been publicly disclosed for diverse sets of compounds (Anastassiadis et al., 2011; Davis et al., 2011). These data sets are now larger than the proprietary sets used to successfully generate predictive models of SMKI-induced chromosomal damage (Olaharski et al., 2009) and bone marrow toxicity (Olaharski et al., 2010). Advances in stem cell biology and cell manufacturing have enabled abundant quantities of human induced pluripotent stem cell-derived cardiomyocytes (hiPSC CM), making human cell-based cardiotoxicity screening a practical option. Finally, impedance-based assays with hiPSC CM have proven highly predictive for each of the leading functional cardiotoxicity hazards including arrhythmia (hERG and non-hERG related), chronotropy, inotropy, and KI induced functional cardiotoxicity (Guo et al., 2013; Lamore et al., 2013; Peters et al., 2015; Scott et al., 2014).
The present study tests a large panel of SMKI for effects on human CM beating and mines their kinome-scale pharmacology profiles for correlations with the aim of achieving two independent but complementary goals. First, can kinase(s) whose inhibition alters CM function be discovered, thereby providing a mechanistic basis for future studies? Second, can a predictive model be developed that flags compounds based on kinase inhibition profiles for earlier testing and refinement?
MATERIALS AND METHODS
Cell culture
iCell Cardiomyocytes (cat #CMC-100-010, Cellular Dynamics International, Madison, Wisconsin) were thawed according to the manufacturer’s suggestion and plated in 96-well xCELLigence Cardio E-plates (ACEA Biosciences, San Diego, California) coated with fibronectin (10 µg/ml in DPBS; cat # F1114, Sigma, St Louis, Missouri) at 30 000 plateable cells per well. Cells were cultured at 7% CO2 and 37 °C for twelve days prior to experimentation. The maintenance medium was changed every 48 h and also 4 h prior to drug addition.
Drug source preparation and addition
Compounds were obtained from EMD Millipore (Billerica, Massachusetts), Tocris Biosciences (Bristol, UK), Selleck Chemicals (Houston, Texas), or were synthesized at AstraZeneca Pharmaceuticals. Compound stock solutions (1000 × g) were prepared in DMSO. hiPSC-CM were treated with compound in a final concentration of 0.1% DMSO or 0.1% DMSO as a vehicle control. Because temperature decreases reduce CM beat rates, liquid handling steps were automated on a BioMek FX in order to minimize the time that cell cultures were out of the incubator to fewer than 90 s.
Measurement of beat rate and amplitude as determined by cellular impedance recordings
Cellular impedance was selected as the detection method because it provides high throughput and exquisite sensitivity to changes in cell morphology although it does not quantitate contractile force. Impedance data were collected with the xCELLigence Cardio System instrument (ACEA Biosciences, 92121 San Diego, California) with three consecutive 58 s reads separated by 2 s intervals. SMKI responses were collected at single time, 2 h postdrug addition. In Excel, data were transformed first to percent of baseline for each well and subsequently to percent of time-matched vehicle control. Data were then exported to Prism (GraphPad Software, La Jolla, California) for graphing and statistical analysis. Data are representative of at least three independent experiments. To ensure robust assessment of cytotoxicity, cellular ATP levels were measured 24 h postdrug addition. Cell index (measured as the relative change in impedance), which is dependent on cell attachment and viability, was also measured 24 h postdrug addition or 72 h post siRNA transfection.
Quantification of cellular ATP levels
ATP content was determined using the CellTiter-Glo luminescent assay (Promega, Madison, Wisconsin) according to the manufacturer’s protocol. All but 75 µl of cell culture supernatant was removed from each well and an equal volume of CellTiter-Glo reagent was added. Plates were shaken for 1 min, incubated at ambient temperature for 10 min and 125 µl was transferred to a white-walled 96 well plate (Costar, Corning, New York). Luminescence was recorded on a Victor Light Luminescence Counter (PerkinElmer, Santa Clara, California). Data are representative of at least three independent experiments.
Method to assess the co-variability of two variables
Because the underlying distributions of the variables in these data sets are unknown the statistical significance was quantified by sampling. For each pair of variables (v1, v2) the Matthews Correlation Coefficient (MCC; Baldi et al., 2000) was calculated with respect to v1 using the n examples tested. MCC is widely used in machine learning approaches to compare models with binary data where the two classes differ in size. It is a balanced measure that takes into account both true and false positives and negatives, which makes the measure less biased toward the majority class. MCC values range from -1 to +1, where +1 indicates perfect positive prediction, 0 indicates no better than random prediction, and -1 indicates perfect inverse prediction. To evaluate if the obtained correlation was significantly different from what could be expected from a random correlation, m samples with length n where drawn from v2 and MCC was calculated with respect to v1 for each sample. This resulted in an estimate of the distribution describing the random co-variation between v1 and a random variable with the same outcome as v2. By sorting the sampled MCC values, a two sided 90% confidence interval (CI) was calculated by retrieving the MCC from the 0.05*m:th position in the ordered list as well as the value of the .95*m:th position. A P-value was also calculated by counting the number of MCCs more extreme than the observed MCC for v2 and dividing by m. To easily rank and compare the co-variation of the variables an enrichment factor, which incorporates information about both magnitude (MCC) and statistical significance (CI) of the correlation, was calculated. The enrichment factor was defined as the obtained MCC for v2 divided by the largest limit of the CI with the same sign, thus for positive v2 MCCs the upper 90% CI limit was used. This ordering was used to rank the co-variation and thus the importance of the individual variables. This method was applied to the analysis of correlation between functional beat changes and individual kinases as well as the importance of kinase groups.
Assessment of correlation between functional beat changes and individual kinases
The categorical data set described in “Assessment of correlation between functional beat changes and kinase groups” (see below) was used to assess the correlation between specific kinases and beat response. The correlation between effects on CM beating and kinase inhibition was assessed as described in “Method to assess the co-variability of two variables”. Because the underlying distribution of the kinase data is unknown it was important to use a methodology that does not assume any underlying distribution of the variables. The co-variability of beat response and kinase inhibition was evaluated by MCC, P-value, and enrichment (see “Method to assess the co-variability of two variables”).
Kinase gene expression obtained from GEO
Kinase gene expression data were obtained from the NCBI Gene Expression Omnibus (GEO dataset record number GSE35671: sample numbers GSM873327-9 and GSM873336-8 for adult heart samples; GSM873330-2 for left ventricular sample; and GSM873300-2 for hiPSC 45 days after initiation of differentiation; Babiarz et al., 2012).
siRNA-mediated gene knockdown
iCell Cardiomyocytes were thawed into 96-well xCELLigence Cardio E-plates and cultured as described in “Cell culture”. Ten days postthaw, cells were transiently transfected with a pool of four small interfering RNA (siRNA) oligonucleotides (oligos) targeting the specific gene of interest (see Supplementary Table 2) or 20–100 nM of non-targeting siRNA oligos (siControl; D-001210-01) using DharmaFECT 1 transfection reagent (0.4 µl/well; Thermo Fisher Scientific). For each transfection, a siRNA oligo stock and a Dharmafect 1 stock were prepared in Optimem (Life Technologies) and incubated separately for 5 min. The two stocks were then mixed and incubated for approximately 30 min. iCell Cardiomyocyte maintenance medium (without antibiotics) was added to the mixture and 100 µl was added to each well. Following a 6 h incubation, transfection medium was removed and replaced with fresh maintenance medium. Data are representative of at least three independent experiments.
Quantitative RT-PCR
Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed using the Ambion Gene Expression Cells-to-CT kit from Life Technologies (Carlsbad, California) according to the manufacturer’s instructions. Cells were lysed and cDNA was synthesized using the provided reverse transcriptase. Quantitative PCR was performed using an Applied Biosystems 7900HT SDS instrument (Life Technologies) with the following cycling conditions: 50 °C for 2 min, 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s alternating with 60 °C for 1 min. PCR reactions contained TaqMan Universal PCR Master Mix, nuclease free water, VIC-labeled glyceraldehyde 3-phosphate dehydrogenase (GAPDH) TaqMan Gene Expression Assay (assay ID Hs02758991_g1) as endogenous control, and FAM-labeled TaqMan Gene Expression Assays specific to the gene of interest (see Supplementary Table 2). For each sample, gene-specific product was normalized to GAPDH and quantified using the comparative Ct (ΔΔCt) method. Data are representative of at least three independent experiments.
Assessment of correlation between functional beat changes and kinase groups
A categorical data set was devised where both beat response and kinase inhibition were categorized into binary variables. Significant effects on CM beating were defined by changes in beat rate or amplitude >3 * standard deviation (SD) of the vehicle-treated group. For each of the 65 inhibitors, the beat response separates alteration in beat rate or amplitude from no alteration (ie, “effect” vs “no effect”). For kinase inhibition, the variable separates > 50% binding at 3 µM (active) from <50% (inactive). This results in a binary matrix with inhibitor and associated beat response versus kinase assays. In addition, all kinases were labeled with their respective kinase group (Manning et al., 2002). The Wilcoxon Rank Sum test (Toutenburg 1975), a non-parametric test that thus can be used without assuming an underlying distribution of the variables, was used to assess if kinase groups could be used to identify inhibitors that affect beat response. Within each kinase group the sum of active kinases was calculated for each compound in the data set. The data were separated into two sets, active and inactive with respect to the beat response. The null hypothesis was that there was no difference in the number of active kinases within a group between the active and inactive compounds. For each kinase group the low limit of the CI for the estimated shift from the Wilcoxon Rank Sum test was used to separate predicted active and inactive compounds. Comparing the predicted classification to the beat response for each compound resulted in a contingency table for each kinase group. The MCC, P-value, and enrichment were calculated from these contingency tables (see “Method to assess the covariability of two variables”). Wilcoxon Rank Sum calculations were made using R (The R Project for Statistical Computing, version 2.5.0, 2007).
Recursive partitioning analysis
Categorical data as described above were analyzed for kinase activity correlations with alterations in beat activity using the Recursive Partitioning function in JMP (SAS Institute, Inc). The y variable was set to the beat activity and the kinase categorical activities were assessed for their contribution to that activity. Because of the low number of actives in the dataset, the minimum split size was set at 3.
In vitro kinase assays
Competition binding assays were performed with human enzymes using the KinomeScan profiling service (KinomeScan, a division of DiscoveRx, San Diego, California). The selectivity profile of the internal set of 102 AstraZeneca compounds was evaluated at a concentration of 1 µM. Clinically tested SMKI (Table 5) were evaluated against RPS6KB1, FAK, and STK35 at four different concentrations, centered around human Cmaxss values, and IC50s were estimated from a four point concentration–response curve.
Table 5.
Retrospective Analysis of Sentinel Kinase Model Using Clinically Tested SMKI
| SMKI | Primary Target | Cardiotoxicity | Cmaxss (µM) | IC50 (µM) |
||
|---|---|---|---|---|---|---|
| FAK | RPS6KB1 | STK35 | ||||
| Ibrutinib1,2 | BTK | FDA-approved: no warnings | 0.3 | >3.2 | >3.2 | 1.1 |
| Dabrafenib3,4 | B-Raf | FDA-approved: no warnings | 4.7 | 78.8 | 0.5 | 14.5 |
| Palbociclib5,6 | CDK4/6 | FDA-approved: no warnings | 0.1 | >3.2 | >3.2 | >3.2 |
| Tofacitinib7,8 | JAK1/3 | FDA-approved: no warnings | 0.1 | >3.2 | >3.2 | >3.2 |
| Telatinib9 | VEGFR2/3, c-Kit | DLT (phase I): hypertension | 2.0–3.6 | >32 | >32 | >32 |
| Tivozanib10 | VEGFR1/2/3 | DLT (phase I): hypertension | 0.2 | 3.1 | >3.2 | >3.2 |
| Ponatinib11 | BCR-ABL | FDA approved with black box warning: Arterial Thrombosis. Other hazards: thrombosis, occlusions, fatal myocardial infarction, stroke, stenosis, heart failure, left ventricular dysfunction | 0.1 | 3.1 | 0.2 | 0.1 |
| Regorafenib12 | VEGFR2, TIE2 | FDA approved with warnings and precautions: hypertension and cardiac ischemia and infarction | 8.1 | >100 | 10.9 | 7.5 |
| Vemurafenib13 | B-Raf | FDA approved with warnings and precautions: prolonged QT. Atrial fibrillation also listed as a clinically relevant adverse event. | 126.6 | >100 | >100 | 39.7 |
| Cabozantinib 14,15 | c-Met VEGFR2 | FDA approved with warnings and precautions: hypertension and thrombotic events | 2.8 | 2.8 | 13.2 | 5.7 |
| Ceritinib16,17 | ALK | FDA approved with warnings and precautions: bradycardia and QT interval prolongation | 1.4 | <0.1 | >32 | 5.9 |
| Lenvatinib18,19 | VEGFR1/2/3 FGFR1/2/3/4 PDGFRα c-Kit, RET | FDA approved with warnings and precautions: hypertension, cardiac dysfunction, thrombolytic events, QT interval prolongation | 1.4 | 65.2 | 25.4 | 1.3 |
| Sotrastaurin20,21 | PKC | Significant finding (phase II trial): tachycardia | 1.4* | 7.3 | 0.3 | >32 |
| PF-381473522 | Aurora A/B | DLTs (Phase I): left ventricular dysfunction | 4.4 | <0.3 | <0.3 | 0.4 |
| AT-928323 | Aurora A/B, Jak2/3, c-ABL | DLTs (phase I): myocardial infarction, hypertension, and cardiomyopathy | 0.05–3.3 | 0.8 | 0.03 | 0.6 |
| ENMD-207624 | Aurora A, Flt3 | DLTs (phase I): hypertension and congestive heart failure | 0.6–1.5 | 10.9 | 0.6 | >10 |
| AZD776225 | Chk1/2 | DLTs (phase I): elevated troponin, myocardial ischemia, and myocardial infarction with ventricular dysfunction | 0.1–1.1 | 0.1 | 3.0 | >3.2 |
| SCH90077626 | Chk1 | DLTs (phase I): QT interval prolongation (alone), and supraventrical tachycardia and atrial fibrillation (in combination with gemcitabine) | 3.2–15.2 | >30 | 1.0 | 30.0 |
| Trametinib27,28 | FDA approved with warnings and precautions: cardiomyopathy. Hypertension also listed as a clinically relevant adverse event. | 0.04 | >1 | >1 | >1 | |
Comparison of clinical cardiac liabilities, human plasma exposure [steady state Cmax (Cmaxss)], and binding profile to FAK, RPS6KB1, and STK35 for clinically tested SMKI. Cardiac liability data were derived from FDA inserts and Pharmaprojects, January 2015 (Pharma Intelligence) and Trialtrove, January 2015 (Pharma Intelligence). “Cardiac safe” SMKI are highlighted in white, VEGF inhibitors with hypertension DLT are highlighted in light grey, and “cardiotoxic” SMKI and their associated cardiotoxicities are highlighted in dark grey. Cmax at steady state (Cmaxss) or Cmax after a single dose (indicated by *) are displayed for each SMKI. The kinase binding IC50 is highlighted in dark grey if the IC50 ≤ Cmaxss. IC50 values are listed as “>X” for compounds showing <50% inhibition at the highest concentration tested (specified as X µM). References denoted by superscript numbers are included in Supplementary Text.
Measurements of action potential, intracellular Ca2+ and contraction
Human iPS-CM were transiently loaded with 6 μM di-4-ANEPPS (Biotium Ref. 61010) in serum-free media (DMEM Gibco #11966, 10 mM Galactose, 1 mM Na-Pyruvate). The media containing the dye was replaced by fresh serum free media and kept in this condition throughout the experiment. Intracellular Ca2+ was measured after Fura4-AM incubation (Molecular Probes Ref. F14201 3 μM, 30 min incubation, 37 °C). Cell contractility was assessed using 10 s of recorded video using a Hammatsu Flash4 camera recorded at 100 Hz. The cell motion was analyzed using a previously established method (Hortigon-Vinagre et al., 2016). A multi-well plate was placed on the stage incubator (37 °C, 5% CO2, water-saturated air atmosphere) of the CellOPTIQ platform (Clyde Biosciences Ltd, Glasgow, Scotland) and the electrical activity, intracellular Ca2+ and contractility was measured from areas (0.2 × 0.2 mm) of hiPS-CM in individual wells visualized using a 40× (NA0.6) objective. Fluorescence signals were digitized at 10 kHz and the records were subsequently analyzed off-line using proprietary software (CellOPTIQ, Clyde Biosciences Ltd).
Statistical analysis
Graphing and statistical analyses were performed using Prism (GraphPad Software). Data in text and figures are represented as mean ± standard error of the mean (SEM). The relationship between a selectivity score (S-score) (Davis et al., 2011) and beat rate/amplitude inhibition was determined by Pearson correlation analysis. For experiments determining compound effects, significance was judged as any change >3 * SD of the vehicle-treated group. For experiments determining consequence of siRNA-based knockdown on beat rate and amplitude, P-values were adjusted for multiple comparisons using the false discovery rate (FDR) approach (Benjamini and Hochberg, 1995). Knockdown groups with FDR-adjusted P ≤ .01 compared with the non-targeting siRNA group were deemed as having an effect on CM beating.
RESULTS
Effects of KIs on hiPSC Cardiomyocyte Beating
Cellular impedance was used to evaluate the effect on hiPSC CM beating by 65 SMKI whose affinity for 385 wild type kinases has been published (Davis et al., 2011). Effects on CM beat rate and amplitude were measured following a 2 h exposure based on our previous experience that KI effects on CM beating are more robust at 2 h than 20 min or 24 h (Lamore et al., 2013). The total compound concentration was 3 µM to match the selectivity profiling by Davis et al., (2011). Of the 65 compounds tested, 22 resulted in a significant change in beat rate or amplitude (Figure 1 and Supplementary Table 1). The most frequent functional effect was a decrease in beat rate independent of amplitude changes (17 compounds; Figure 1). One compound (GDC-0879) increased beat rate independent of a change in amplitude whereas two compounds (CEP-701 and PKC-412) increased beat rate with a concomitant decreased amplitude. Two compounds (A-674563 and GSK-690693) increased amplitude independent of a change in beat rate. With the exception of staurosporine, these functional effects occurred in absence of cytotoxicity as judged by decreased cellular ATP levels and decreased cell index measured 24 h after compound addition (Figure 1 and Supplementary Table 1). This set of data meets criteria for further correlation analysis by the presence of ample active and inactive compounds with respect to functional effects on CM beating in the absence of cytotoxicity.
Figure 1.
Small molecule kinase inhibitors affect human induced pluripotent stem cell-derived cardiomyocytes beating function at concentrations that do not affect cell viability. Changes in beat rate (BR), amplitude (AMP), cell index (CI), and cellular ATP levels expressed as percent control of hiPSC CM treated with 3 µM of compounds for 2 h (BR and AMP) or 24 h (CI and ATP). Points are representative of the mean of at least three independent experiments. Only those compounds that had an effect on BR or AMP are shown.
Identification of Individual Kinases Critical for CM Function
We sought to identify and evaluate individual kinases contributing to functional cardiotoxicity. Inhibition of each of the 385 kinases and functional cardiotoxicity were both scored in binary form (see Methods section) and univariate analysis was used to determine correlation between kinase inhibition and effect on CM beating. Of the 385 kinases, 58 correlated with functional cardiotoxicity (MCC > 0.4, P ≤ .001, and enrichment score > 2.0; Table 1). Eliminating kinases not expressed in both hiPSC CM and adult heart tissue (Babiarz et al., 2012) reduced the candidate list to 46 kinases (Table 1).
Table 1.
Kinases Identified by a Univariate Analysis With SMKI-Induced Effects on hiPSC CM
| Kinase | Group | Enrichment | MCC | P-value | |
|---|---|---|---|---|---|
| Expressed in hiPSC CM and heart | PAK1 | STE | 3.06 | 0.50 | 1.0E-04 |
| RPS6KB1 | AGC | 2.84 | 0.56 | 0.0E+00 | |
| MST4 | STE | 2.82 | 0.49 | 1.6E-04 | |
| RPS6KA3 | AGC | 2.63 | 0.49 | 2.4E-04 | |
| FAK | TK | 2.63 | 0.49 | 8.0E-05 | |
| PAK3 | STE | 2.55 | 0.42 | 1.2E-03 | |
| MAP3K4 | STE | 2.55 | 0.42 | 1.0E-03 | |
| STK33 | CAMK | 2.54 | 0.56 | 0.0E+00 | |
| MAP4K2 | STE | 2.50 | 0.43 | 6.4E-04 | |
| FER | TK | 2.47 | 0.49 | 1.6E-04 | |
| MAP3K12 | TKL | 2.47 | 0.49 | 1.0E-04 | |
| STK35 | OTHER | 2.47 | 0.49 | 1.8E-04 | |
| MAP3K2 | STE | 2.44 | 0.44 | 4.6E-04 | |
| MAP4K3 | STE | 2.39 | 0.46 | 3.8E-04 | |
| PIP4K2B | LIPID | 2.36 | 0.41 | 1.6E-03 | |
| PRKAA2 | CAMK | 2.36 | 0.41 | 1.3E-03 | |
| PIM3 | CAMK | 2.36 | 0.41 | 1.3E-03 | |
| SGK3 | AGC | 2.36 | 0.41 | 1.5E-03 | |
| STK24 | STE | 2.36 | 0.41 | 1.3E-03 | |
| PRKCQ | AGC | 2.36 | 0.59 | 0.0E+00 | |
| PRKD3 | CAMK | 2.34 | 0.49 | 1.0E-04 | |
| TBK1 | OTHER | 2.34 | 0.49 | 1.2E-04 | |
| CAMKK2 | OTHER | 2.34 | 0.49 | 1.0E-04 | |
| MAP4K1 | STE | 2.32 | 0.45 | 2.4E-04 | |
| CDK7 | CMGC | 2.27 | 0.47 | 2.0E-04 | |
| ROCK1 | AGC | 2.22 | 0.41 | 1.3E-03 | |
| PRKD1 | CAMK | 2.22 | 0.41 | 1.5E-03 | |
| INSR | TK | 2.22 | 0.41 | 1.5E-03 | |
| MAP2K3 | STE | 2.22 | 0.41 | 1.4E-03 | |
| NUAK1 | CAMK | 2.22 | 0.41 | 1.5E-03 | |
| LATS2 | AGC | 2.22 | 0.41 | 1.2E-03 | |
| CAMK1D | CAMK | 2.22 | 0.41 | 1.4E-03 | |
| SRPK3 | CMGC | 2.22 | 0.41 | 1.4E-03 | |
| ULK2 | OTHER | 2.19 | 0.52 | 6.0E-05 | |
| PRKD2 | CAMK | 2.17 | 0.46 | 3.2E-04 | |
| CHEK2 | CAMK | 2.17 | 0.46 | 4.2E-04 | |
| PRKCE | AGC | 2.17 | 0.46 | 3.8E-04 | |
| NUAK2 | CAMK | 2.14 | 0.50 | 1.0E-04 | |
| PRKX | AGC | 2.12 | 0.42 | 1.8E-03 | |
| PRKACA | AGC | 2.12 | 0.42 | 1.9E-03 | |
| ZAP70 | TK | 2.12 | 0.42 | 1.7E-03 | |
| PRKCD | AGC | 2.12 | 0.52 | 6.0E-05 | |
| PRKG1 | AGC | 2.12 | 0.52 | 2.0E-05 | |
| STK4 | STE | 2.10 | 0.42 | 1.3E-03 | |
| IKBKE | OTHER | 2.10 | 0.42 | 9.8E-04 | |
| PKN2 | AGC | 2.02 | 0.50 | 4.0E-05 | |
| Expressed in heart, not in hiPSC CM | SYK | TK | 2.36 | 0.41 | 1.5E-03 |
| AKT2 | AGC | 2.12 | 0.42 | 1.8E-03 | |
| FES | TK | 2.10 | 0.42 | 1.2E-03 | |
| Not expressed in hiPSC CM or heart | MINK1 | STE | 2.71 | 0.51 | 4.0E-05 |
| JAK3 | TK | 2.65 | 0.52 | 4.0E-05 | |
| GRK1 | AGC | 2.47 | 0.49 | 8.0E-05 | |
| YSK4 | STE | 2.45 | 0.46 | 4.0E-05 | |
| GRK7 | AGC | 2.23 | 0.49 | 1.0E-04 | |
| CAMKK1 | OTHER | 2.10 | 0.42 | 1.3E-03 | |
| HIPK1 | CMGC | 2.53 | 0.46 | 3.6E-04 | |
| MAP3K13 | TKL | 2.47 | 0.49 | 1.4E-04 | |
| LATS1 | AGC | 2.17 | 0.46 | 4.0E-04 |
Fifty-eight kinases in total were identified as statistically significant; 46 are expressed in adult heart and hiPSC CM at time of culture for experimentation; 3 kinases are expressed in heart but not in hiPSC CM; 9 kinases are not expressed in either hiPSC CM or adult heart.
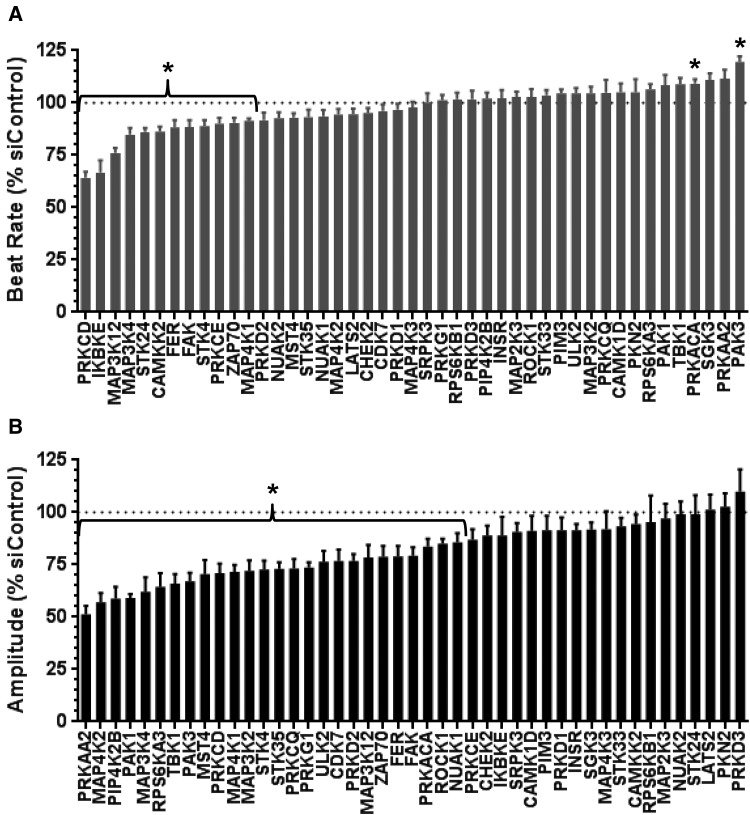
Experimentally testing the univariate results with additional pharmacological probes is complicated by the lack of highly selective compounds. Therefore, as an independent test, expression of each of the 46 candidate kinases was knocked down and effects on CM beating were evaluated. Robust knockdown (>75% at the mRNA level) was achieved for 45 of 46 the genes (Supplementary Table 2). For those 45 kinases, the effects of gene knockdown on CM beat rate or amplitude were assessed for significance in accordance with the FDR (Benjamini and Hochberg, 1995; FDR-adjusted P < .01). 30 out of the 45 kinases resulted in significant changes in CM beat function upon knockdown; these effects occurred in the absence of cytotoxicity as judged by cellular ATP and cell index (Table 2, Figure 2, and Supplementary Table 3). Knockdown of four kinases (CAMKK2, IKBKE, PRKCE, and STK24) decreased beat rate independent of changes in amplitude, whereas sixteen kinases (CDK7, MAP3K2, MAP4K2, MST4, NUAK1, PAK1, PIP4K2B, PRKAA2, PRKCQ, PRKD2, PRKG1, ROCK1, RPS6KA3, STK35, TBK1, and ULK2) decreased beat amplitude without affecting beat rate. Knockdown of either PAK3 or PRKACA decreased beat amplitude with a concomitant increased beat rate, whereas knockdown of eight genes (FAK, STK4, PRKCD, MAP3K12, MAP3K4, FER, ZAP70, and MAP4K1) simultaneously decreased beat rate and amplitude. This functional specificity may be a valuable first step for follow-up studies on either understanding their biological role or providing context for toxic liability.
Table 2.
Individual Kinases Discovered as Critical for CM Function
| Amplitude |
Beat Rate |
|||
|---|---|---|---|---|
| Kinase | % siControl | FDR-Adjusted P-value | % siControl | FDR-Adjusted P-value |
| CAMKK2 | — | — | 86.3 | 2.3E–05 |
| CDK7 | 76.8 | 2.1E–04 | — | — |
| FAK | 79.2 | 4.0E–05 | 88.5 | 1.4E–03 |
| FER | 79.1 | 2.2E–04 | 88.4 | 1.2E–03 |
| IKBKE | — | — | 66.8 | 1.7E–06 |
| MAP3K12 | 78.5 | 4.1E–04 | 76.0 | 7.6E–09 |
| MAP3K2 | 72.1 | 4.5E–07 | — | — |
| MAP3K4 | 62.1 | 2.3E–07 | 84.9 | 6.9E–05 |
| MAP4K1 | 71.6 | 4.0E–05 | 91.4 | 2.9E–03 |
| MAP4K2 | 57.1 | 2.0E–10 | — | — |
| MST4 | 70.5 | 2.2E–06 | — | — |
| NUAK1 | 85.7 | 3.5E–03 | — | — |
| PAK1 | 59.1 | 1.7E–10 | — | — |
| PAK3 | 67.0 | 4.9E–09 | 119.5 | 1.1E–06 |
| PIP4K2B | 58.8 | 1.2E–10 | — | — |
| PRKAA2 | 51.3 | 8.7E–13 | — | — |
| PRKACA | 83.7 | 6.6E–04 | 109.2 | 2.7E–03 |
| PRKCD | 71.0 | 1.1E–06 | 64.3 | 1.1E–11 |
| PRKCE | — | — | 90.1 | 2.1E–03 |
| PRKCQ | 73.1 | 6.6E–07 | — | — |
| PRKD2 | 76.9 | 2.5E–05 | — | — |
| PRKG1 | 73.7 | 7.7E–08 | — | — |
| ROCK1 | 85.1 | 5.2E–03 | — | — |
| RPS6KA3 | 64.4 | 3.1E–08 | — | — |
| STK4 | 72.6 | 3.3E–07 | 89.0 | 9.4E–04 |
| STK24 | — | — | 86.0 | 3.9E–06 |
| STK35 | 73.0 | 1.7E–06 | — | — |
| TBK1 | 66.0 | 2.7E–09 | — | — |
| ULK2 | 76.4 | 1.6E–05 | — | — |
| ZAP70 | 78.8 | 9.6E–04 | 90.5 | 2.1E–03 |
The effect of siRNA-based knockdown on beat rate or amplitude expressed as % siControl is listed for each kinase. “—” indicates no significant change compared with siControl. Only those kinases for which there was a significant change in beat rate or amplitude upon knockdown (FDR-adjusted P < .01) are listed in this table. A complete description of knockdown effects on beat rate, beat amplitude, cell index, and cellular ATP level is listed for all 45 genes knocked down in Supplementary Table 3.
Figure 2.
siRNA-based knockdown of Matthews Correlation Coefficient (MCC)-correlated kinases validates kinases critical for hiPSC CM function. hiPSC CM were transfected with either non-silencing control siRNA (siControl) or a pool of siRNA targeted against the indicated gene. Cardiomyocyte beat rate (A) or amplitude (B) expressed as percent siControl was determined by impedance measurements 48 h post siRNA transfection. All data are expressed as mean + SEM of three independent experiments (≥ 12 wells/experiment). Asterisks indicate significant differences relative to siControl-transfected cells (*False discovery rate [FDR]-adjusted P < .01).
Kinome-Wide Promiscuity and Effects on Cardiomyocyte Beating
Given the promiscuous nature of ATP-competitive KIs at clinically relevant concentrations, a simple hypothesis is that SMKI-induced cardiotoxicity correlates with lack of selectivity across the kinome. To quantify SMKI promiscuity, Davis et al. determined dissociation constants (Kd) and then calculated a selectivity (S) score for each compound by calculating the fraction of kinases bound at a specified concentration. This approach was used to calculate S scores at a 3 µM test concentration for the 65 SMKI that had been screened against 385 wild-type kinases. The resulting S scores varied widely (0.0078–0.8782), signifying a broad range of selectivity. To examine the correlation between kinome selectivity and functional effect on CM beating, S (3 µM) was plotted against changes in beat rate (Figure 3a) and beat amplitude (Figure 3b) following a 2 h exposure to 3 µM for each inhibitor. Some highly selective compounds including GDC-0879 [S (3 µM) = 0.042] and BMS-540215 [S (3 µM) = 0.086] induced CM beating effects whereas the relatively promiscuous inhibitors TG-101348 and SU14813 with S (3 µM) = 0.539 and 0.542, respectively, had no effect on CM beating. To minimize effects on CM beating influenced by cytotoxicity, staurosporine, the only compound to induce cell death at 3 µM, was removed and Pearson’s correlation coefficients (r) and their P-values were calculated. Overall, S score did not significantly correlate with changes in beat rate (r= −0.08, P = .48) or amplitude (r = −0.12, P = .34). Thus, the present measurement of promiscuity at the kinome level is not a predictive tool of functional cardiotoxicity.
Figure 3.
Lack of correlation between selectivity (S) score and hiPSC CM beating function. Changes in beat rate (A) or amplitude (B) expressed as percent control of iPSC CM treated with 3 µM of compounds for 2 h are plotted against S score [3 µM]. All points have error bars that represent SEM from three independent experiments. The correlation of S score and effect on CM beating function was not judged to be significant for beat rate (r2 = 0.008 and P = .48) or beat amplitude (r2 = 0.014 and P = .34).
Intragroup Promiscuity and Effects on Cardiomyocyte Beating
The weak relationship between kinome-wide selectivity and effect on CM beating warranted a more detailed analysis to identify specific kinase groups more suitable for predictive purposes. In addition to the eight groups within the typical protein kinase superfamily (CAMK, STE, AGC, CK1, CMGC, TK, TKL, and OTHER), we looked for correlation within the atypical protein kinase group, which lacks sequence similarity to the typical protein kinase superfamily, as well as the lipid kinase group. To this end, the binary classification of kinase activity and beat activity was also used to identify any potential correlations between activity within particular kinase group(s) and functional beat effects. The CAMK, AGC, STE, and OTHER kinase groups were identified as strongly correlated with functional cardiotoxicity (MCC > 0.4, P ≤ .001, and enrichment score > 2.0; Table 3). The atypical, CMGC, and CK1 groups were identified as having a weaker correlative relationship (MCC < 0.4, P < .05, and enrichment scores < 2.0). The lipid, TKL, and TK groups did not significantly correlate with functional cardiotoxicity (MCC < 0.4, P > .05, and enrichment scores < 2.0). Thus, rather than identifying concentration within a particular group, these data indicate culprit promiscuity is spread across at least four groups.
Table 3.
Correlation of Intragroup Promiscuity and Effect on hiPSC CM Beating Function
| Kinase Group | Enrichment Score | MCC | P-value |
|---|---|---|---|
| CAMK | 2.98 | 0.58 | <0.0001 |
| OTHER | 2.42 | 0.43 | 0.0006 |
| STE | 2.26 | 0.46 | <0.0001 |
| AGC | 2.23 | 0.49 | 0.0002 |
| Atypical | 1.62 | 0.34 | 0.004 |
| CK1 | 1.36 | 0.25 | 0.04 |
| CMGC | 1.35 | 0.26 | 0.04 |
| LIPID | 1.00 | 0.18 | 0.11 |
| TKL | 0.41 | 0.10 | 0.31 |
| TK | 0.18 | 0.04 | 0.53 |
Correlation between CM beating effect and promiscuity within the eight typical kinase groups, the atypical kinase group, and lipid kinase group as described by MCC and ranked by enrichment score.
Identification of Sentinel Kinases With Predictive Value
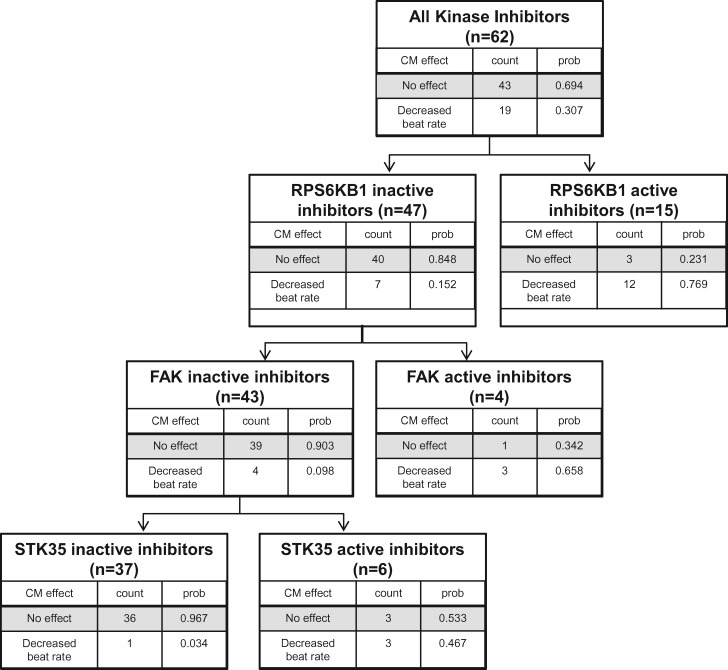
To identify a minimum set of individual kinases that could be used as “predictor” or “sentinel” kinases, the data set was probed using recursive partitioning. Using the binary classification framework to describe kinase activity and effects on CM beating, compounds were categorized into one of two endpoints: “decreased beat rate” (n = 19) or “no change in beat rate” (n = 43). To increase statistical power, the three compounds that increased beat rate were not included in construction of the sentinel kinase model because the number was limited. The classification tree was constructed by sequentially selecting kinases that best separated out compounds that decrease beat rate (Figure 4). The individual kinase whose pharmacological activity best separated “decreased beat rate” from “no change in beat rate” was RPS6KB1 (LogWorth = 5.5; P = 2.8 × 10−6). Of the compounds with RPS6KB1 inhibitor activity, 80% (12/15) decreased beat rate. Removing RPS6KB1-active inhibitors reduced the percentage of compounds with beat activity by half [from 30.7% (19/62) in the full set to 15.2% (7/47) in the RPS6KB1-inactive set]. Successively repeating this approach among these remaining compounds identified FAK (aka PTK2; LogWorth = 2.4; P = .004) as the best predictor of beat activity among the remaining RPS6KB1-inactive compounds. A third iteration identified STK35 (LogWorth = 2.6; P = .003) as the best predictor among RPS6KB1 and FAK-inactive compounds. Eighteen of the nineteen compounds that were categorized as inducing a decreased beat rate were active against at least one of the sentinel kinases. Thus, recursive partitioning analysis of the present data set generated a simple predictive model such that compounds inactive at all three sentinel kinases (RPS6KB1, FAK, and STK35) had a 3.4% probability of affecting CM beat activity compared with 30.7% in the full training set (Figure 4 and Table 4).
Figure 4.
RPS6KB1, FAK, and STK35 are key sentinel kinases of effects on cardiomyocyte beating. The decision tree constructed by recursive partitioning of kinase inhibitor-induced changes in hiPSC CM beating function predicted that RPS6KB1 was the most significant predictor of functional changes followed by FAK and then STK35.
Table 4.
Effects on iPSC CM Beating Versus Compounds Inactive at All Three Sentinel Kinases for the Training and Testing Sets
| Full Compound Set |
Negative at All Three Sentinel Kinases |
|||
|---|---|---|---|---|
| Functional CM Effect: | % No Effect | % ↓ BR | % No Effect | % ↓ BR |
| Predicted (training set) | 69.4 | 30.7 | 96.7 | 3.4 |
| Observed (testing set) | 86.6 | 13.4 | 96.4 | 3.6 |
Using inactivity at RPS6KB1, FAK, and STK35 as a classifier, 3.6% of compounds observed versus the 3.4% predicted by recursive partitioning analysis of the training set decreased CM beat rate.
To test the model’s predictivity for the ability to translate to a distinct compound set, we turned to our internal library of SMKI from which we drew 102 compounds with binding data for RPS6KB1, FAK, and STK35. Ten of these compounds were identical to compounds in the training set but were included because they were tested at different concentrations and therefore the kinase binding profiles differed. The remaining 92 compounds in the test set were diverse relative to the training set, as indicated by low Tanimoto similarity to the nearest neighbor in the training set (ranging from 0.22 to 0.71 and computed using ECFP4 fingerprints; Rogers and Hahn, 2010). These compounds were tested in the CM impedance assay at 1 µM to match the binding data. Five compounds that resulted in an increased beat rate were not included in analysis as the model was constructed to predict “decreased beat rate” versus “no change in beat rate”. The internal AstraZeneca set revealed fewer compounds that resulted in a decreased beat rate [13.4% (13/97) vs 30.7% (19/62) in the training set], perhaps due to the lower concentration tested or more highly selective drug-like nature of the inhibitors. None of the compounds that caused a decrease in CM beat rate affected cell viability as determined by cellular ATP and cell index (data not shown). The 97 compounds were scored with the recursive partitioning model. Of the 56 that were inactive at all three sentinel kinases, only two (3.6%) had an effect on beat rate (Table 4).
We tested how well the sentinels translate to the clinic by screening an independent set of 19 SMKI with sufficient published clinical cardiac safety profiles and human exposure data (Cmaxss). On the basis of label warnings, we categorized the four without any warnings as “cardiac safe” and the fifteen SMKI with cardiac liabilities as “cardiotoxic” [seven FDA-approved SMKI with CV warnings and eight with CV dose limiting toxicities (DLTs) in phase I or II]. We screened all 19 compounds against each of the three sentinel kinases, RPS6KB1, FAK, and STK35 at four concentrations centered around the published human Cmax values. Kinase binding IC50 ≤ Cmaxss was used to delineate inhibition at clinically relevant concentrations (Table 5, frequency of clinical CV toxicity listed in Supplementary Table 5). The model correctly predicted three of the four “cardiac safe” SMKI, in that their kinase IC50 > Cmaxss for all three kinases. Twelve of the fifteen “cardiotoxic” SMKI were correctly predicted: their activity against sentinel kinases was detected at clinically relevant concentrations (kinase IC50 < Cmaxss) for at least one of the sentinel kinases. Out of the three “cardiotoxic” SMKI that were missed, two (telatinib and tivozanib) are VEGF inhibitors with hypertension as the only cardiac-related DLT event in clinical trials. Hypertension with VEGFR inhibitors is well established (Force et al., 2007) and mediated by effects on vascular endothelial cells (Lankhorst et al., 2015) rather than on CMs. Therefore predictivity metrics were calculated with and without the two VEGF inhibitors (Table 6).
Table 6.
Performance Parameters for Sentinel Kinase Model Using Clinically Tested SMKI
| Entire Data Set | Excluding Vascular-Mediated Hypertension | |
|---|---|---|
| Sensitivity | 0.80 | 0.92 |
| Specificity | 0.75 | 0.75 |
| PPV | 0.92 | 0.92 |
| NPV | 0.50 | 0.75 |
| Accuracy | 0.79 | 0.88 |
The sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and accuracy are listed for the full set of nineteen clinically tested drugs and for the set of seventeen drugs with VEGF-targeted therapies causing hypertension excluded.
Contribution of Non-Kinase Targets to Cardiotoxicity Seen With Test and Validation Sets
The sentinel kinase discovery and validation experiments would be complicated if the sets of SMKI evaluated had direct pharmacological effects on hERG or other non-kinase targets that affect excitation–contraction coupling (ECC). To examine the contributions of non-kinase targets, we selected all of the SMKI that altered CM beating in the discovery (Ambit) and validation (AstraZeneca) sets plus a random subset that did not alter beating. Compounds were tested for activity against six ion channels known to control the cardiac action potential and eight non-channel targets considered as cardiotoxicity risks. The data were interrogated using binary and regression analysis. For binary assessment, off-target activity was classified by potency relative to concentration tested in the CM assay. Among compounds that did not alter CM beating, 50% were active against at least one target in the panel. For compounds that did alter CM beating, 47% were active against at least one target. Reviewing individual targets in the panel, there was a lack of enrichment among beat altering compounds (Supplementary Table 4a and b). Finally, regression analysis comparing percent change in beat activity with potencies at individual cardiotoxicity targets did not reveal significant correlations for any of the targets in the panel (R2 values in Supplementary Figure 1). The lack of correlation between beat effects and inhibition of ion channel or non-kinase cardiotoxicity targets suggest direct pharmacological inhibition of these targets was limited in the SMKIs used in this study.
The Contribution of Selected Kinases to the ECC Mechanism
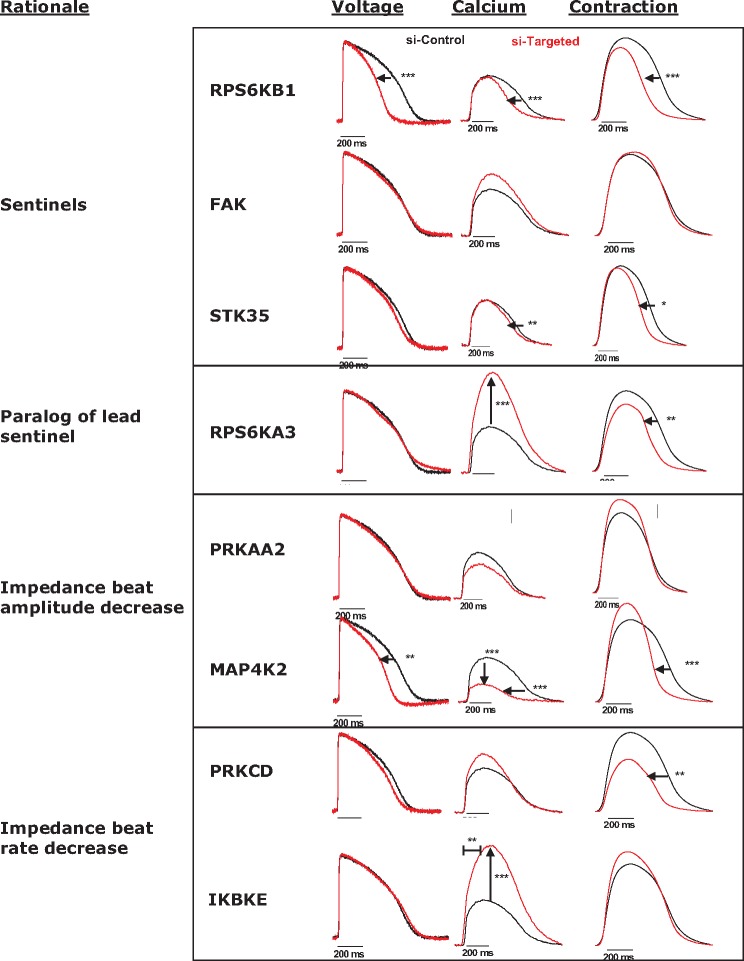
In order to achieve the broadest coverage of kinases linked to CM function, an assay sensitive to a downstream endpoint, contraction measured by impedance, was selected as the discovery screen. However, this assay does not provide mechanistic insight as to where within the ECC process particular kinases may be acting. In an effort to gain additional granularity on mechanistic site(s) of action, individual kinases were knocked down and effects on action potential duration (APD), Ca2+ transients and contractility were assessed simultaneously using an optical approach. For spontaneously beating hiPSC CM, a change in beat rate can affect the beat amplitude. Therefore, in order to specifically assess the effect on contractility, the CMs were paced to maintain a constant beat rate (1 Hz). The kinases chosen for investigation include the three sentinels (RPS6KB1, FAK, STK35), a paralog of the best sentinel (RPS6KA3), and two each that altered spontaneous beat rate (PRKCD, IKBKE) and amplitude (PRKAA2, MAP4K2).
Comparing across the kinase knockdown experiments revealed a remarkable diversity and specificity for each component of the ECC process. Reduced expression of RPS6KB1 and MAP4K2 resulted in a shortened APD, which in turn shortened the duration of the Ca2+ and contractility transients. Knockdown of STK35 had no effect on APD, but shortened both the Ca2+ and contractility transients (Figure 5, and Supplementary Figure 2). Loss of RPS6KA3 and IKBKE increased the amplitude of the Ca2+ transient, with the former also decreasing the duration of the contractility transient. Knockdown of PRKCD had no effect on APD or the Ca2+ transient, but did reduce the duration of contraction. Knockdown of FAK and PRKAA2 had no noteworthy effects detectable by optical methods.
Figure 5.
Effects of siRNA-based kinase knockdown on hiPSC CM action potentials and Ca2+and contractility transients. Each trace shows representative recordings of membrane potential, cytosolic Ca2+ and contraction made 72 h posttransfection with either non-silencing control siRNA (siControl, black traces) or a pool of siRNA targeted against the indicated kinase gene (red traces). The level of RNA knockdown achieved in each experiment were 83%, 49%, 83%, 76%, 77%, 74%, 32%, 57% (percent of control, top to bottom respectively). *P ≤ .05, **P ≤ .005, ***P ≤ .0005—when compared with siControl using ANOVA and Bonferroni post hoc.
Kinase knockdowns were assessed for modulation of the coupling between APD and Ca2+ as well as between Ca2+ and contraction by analyzing hysteresis loops (Supplementary Figure 3). Compared with assessment of each of these parameters in isolation, hysteresis loops allow the interdependency of each parameter to be highlighted and provide fingerprints of different mechanisms of action. RPS6KB1 hysteresis data indicated the effects on action potential were associated with minimal changes in coupling between ECC nodes. In contrast, MAP4K2 knockdown hysteresis loops revealed decreased coupling from APD to Ca2+ followed by increased coupling from Ca2+ to contraction. Knockdown of both RPS6KA3 and IKBKE revealed increased APD to Ca2+ coupling with comparably decreased coupling of Ca2+ changes to contraction (Supplementary Figure 3). See Supplementary Material for further analysis and discussion of these data.
DISCUSSION
The central aim of the present study was to discover individual kinases whose inhibition alters human CM beating. We identified 30 kinases that met criteria of having (1) pharmacological inhibition highly correlated with beat changes, (2) expression in both hiPSC CM and adult heart tissue and (3) single gene knockdown affecting CM beating. From the perspective of cardiovascular biology, this work identifies novel kinase signaling pathways regulating CM beating. From a drug discovery point-of-view, the impact may be more immediate. The identification of kinases linked to CM beating expands the ability to predict and assess mechanisms of drug-induced cardiotoxicity.
Many of the 30 kinases identified have a limited known role in cardiac function. No published findings exist directly linking MST4, STK24 (aka MST3), MAP4K1, or MAP3K12 to CM function. In contrast, calcium handling and contractility signaling pathways were well represented. In addition to CAMK and PKC isoforms, both PAK1 and PAK3 regulate CM contractility by enhancing sensitivity to calcium (reviewed in (Sheehan et al., 2007)). STK4 (aka MST1) phosphorylates cardiac troponin I in vitro (You et al., 2009). Similarly, PRKACA, the catalytic subunit of Protein Kinase A (PKA), acts downstream of the β-adrenergic receptors (Kamp and Hell, 2000; Layland et al., 2005). PIP4K2B catalyzes formation of phosphatidylinositol-5,4-bisphosphate (PIP2), known to directly regulate several potassium channels (Kruse et al., 2012; Stansfeld et al., 2009).
An additional 15 kinases met the criteria of having pharmacological inhibition that was highly correlated with effects on beat activity but were not confirmed by single gene knockdown. The criteria of knocking down mRNA by 75% (Supplementary Table 2) may be insufficient to deplete functional reserve in some kinase pathways. These kinases merit additional follow up. More complete kinase inhibition of individual kinase is a straightforward starting point. Alternatively, some of these 15 kinases may regulate CM beating in combination with another kinase such as a closely related homolog or as part of convergent signaling pathways. Any necessity for dual inhibition of pairs of kinases in the primary pharmacological screen would not be distinguished by the univariate analysis and would fail single gene knockdown confirmation. Thus, follow-up studies could explore combination effects by dual knock down of pairs or by repeating the analysis using larger panels of SMKIs sufficiently powered for multivariate analysis.
One measure to evaluate the success or limitations of the present search for kinases is to consider the kinases missed relative to literature precedent. In a review of KI cardiotoxicity, Force and Kolaja highlight four kinase pathways linked to functional effects: PKC, CAMK, PKA, and PI3Kγ (Force and Kolaja, 2011). Our analysis identified members of three of these four families; kinases identified included the PKA catalytic subunit (PRKACA and the closely related PRKX), one CAMK isoform (CAMK1D and a kinase in the pathway CAMKK2), and three PKC isoforms (PRKCD, PRKCE, PRKCQ along with the related PKN2). Interestingly, PI3Kγ was not identified. Inspection of the published PI3Kγ data reveals that gene deletion selectively increased the beat rate of sinoatrial nodal CM but not atrial CM (Rose et al., 2007). Because we used CM with a very low proportion of nodal cells, it is perhaps expected that we would miss PI3Kγ activity. When the capability emerges to direct cellular maturation and differentiation of hiPSC into adult atrial, ventricular, or sinus nodal CM, future studies could retest kinase profiles in these different CM types.
A second aim was to evaluate the kinome for predictive pharmacology tools. It is intuitive to hypothesize that compounds inhibiting more kinases will have a greater chance of inhibiting kinases that control CM beating, thereby raising the possibility that kinome promiscuity may be a metric that predicts cardiotoxicity (Chen et al., 2008; Lal et al., 2013). Another widely recognized observation is that the overwhelming majority of SMKI with clinical cardiotoxicity are tyrosine kinases inhibitors, thereby raising the possibility that promiscuity within the tyrosine kinase group may be predictive (Kruse et al., 2012; Sharma et al., 2017). The present analysis revealed a weak correlation between kinase selectivity and effect on beat rate or amplitude (Figure 3) and no particular concentration of promiscuity within the tyrosine kinase group (MCC = 0.04, P = .53). Promiscuity within four of the ten kinase groups (CAMK, Other, STE, and AGC; MCC > 0.4, P < .05, and enrichment > 2.0) were correlated with effects on CM beating (Table 3). However, these four groups comprise ∼ 51% of the human kinome. Activity against hERG, or other cardiotoxicity-related targets did not correlate with beat effects in the present sets of SMKI (Supplementary Table 4 and Supplementary Figure 1). Thus, for the present set of SMKI, neither promiscuity across the kinome, promiscuity within a particular group, or known cardiotoxicity targets proved to be a practical tool for predicting SMKI functional cardiotoxicity.
Rather than kinome or tyrosine kinase promiscuity, the predictive tools identified were three sentinel kinases, RPS6KB1, FAK, and STK35 (Table 6). The predictivity was validated by retrospective analysis of clinical SMKI. While this retrospective test is necessary, its interpretation comes with the caveats of comparing biochemical potencies with clinical concentrations. For kinases, these include differences not limited to: (1) ATP concentration, (2) protein constructs, (3) kinase binding partners, and (4) cell penetrance/pharmacokinetic profiles. Thus, rather than lending too much credence to the precise values of predictivity metrics, we submit that the presently available data indicate sentinel predictivity is highly likely to translate to the clinic. It is also important to emphasize that SMKI-associated functional cardiotoxicity was assessed in CMs which does not address vascular, neuronal, hormonal or structural cardiotoxicity/cytotoxicity. Future studies aiming to refine the choice of sentinel kinases may include compounds that selectively affect amplitude, rhythmicity or increased beat rate and thereby advance the current predictive model or focus it on functional endpoints such as contractility, arrhythmia, and tachycardia.
Most of the 30 kinases identified in this study were previously unknown or underappreciated as regulators of CM beating, hence resolving the signaling networks through which they modulate ECC will require extensive follow-up. At the molecular level, the scope of this challenge can be gauged from phosphoproteome analysis which suggests 20 phosphorylation sites per kinase (Huang et al., 2014). From a cellular perspective, it will be valuable to elucidate the modulation point(s) within the ECC cascade affected by each kinase. A suite of technologies have emerged to enable measuring the influence of KI and kinase siRNA on APD, calcium flux, and contraction either individually or simultaneously (eg, MEA, FLIPR, impedance, and CellOPTIQ; Hortigon-Vinagre et al., 2016; Peters et al., 2015). Application to the present kinases illustrates both the diversity and complexity of their ECC modulation. For example, RPS6KB1 and RPS6KA3 are close paralogs yet were found to differentially regulate APD and calcium flux, respectively (Figure 5). Both are novel findings—neither RPS6KB1’s involvement in action potential regulation nor RPS6KA3’s regulation of calcium flux were previously recognized. The two kinases that affected beat amplitude did so via different mechanisms as judged by their profiles in APD and calcium transient studies; the same is true for the two kinases that reduced beat rate. Thus, cellular function assays may provide the guidance necessary to focus future molecular studies aimed at identifying phosphorylation substrates and delineate signaling networks regulated by particular kinases.
The present study reveals (1) a novel platform to deconvolute SMKI cardiotoxicity by combining kinome-wide human kinase profiles and human stem cell-derived CMs with label-free CM beat detection, (2) a simple predictive model of three sentinel kinases, and (3) a list of kinases mechanistically linked to effects on CM beating. We envision future safety-testing cascades for SMKI drug discovery will use the sentinels as a high throughput screen for early assessment at low cost. Any compounds flagged would be tested directly for CM beating activity in vitro. If activity is confirmed, follow-up mechanistic studies would be enabled by the list of 30 candidate kinases. Once the culprit kinases are identified, testing with traditional kinase assays could be used to screen for activity, develop SAR, and design-out CV toxicity risk. Taken together, the present findings offer the opportunity for a paradigm shift from the present era where the majority of SMKI drugs exhibit cardiotoxicity in the clinic, to rational design of SMKI CV safety.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
FUNDING
Work was funded by AstraZeneca Pharmceuticals.
AUTHOR CONTRIBUTIONS
S.L., M.P., C.S., M.L, M.H.-V., V.R., S.B., and G.S. designed the overall study and experiments, conducted the research, analyzed the data and wrote the manuscript. E.A., S.B., M.L., J.S., L.C., and J.S. designed, generated, and analyzed the computational data. A.C. conducted search for SMKI clinical data. All authors offered critical comments and approved the final manuscript.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Alex Harmer, Amy Pointon, and Claudio Chuaqui for useful discussions and Alex Harmer and Chris Pollard for critical comments on the manuscript. The authors declare no competing financial interests.
REFERENCES
- Anastassiadis T., Deacon S. W., Devarajan K., Ma H., Peterson J. R. (2011). Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat. Biotechnol. 29, 1039–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz J. E., Ravon M., Sridhar S., Ravindran P., Swanson B., Bitter H., Weiser T., Chiao E., Certa U., Kolaja K. L. (2012). Determination of the human cardiomyocyte mRNA and miRNA differentiation network by fine-scale profiling. Stem Cells Dev. 21, 1956–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi P., Brunak S., Chauvin Y., Andersen C. A., Nielsen H. (2000). Assessing the accuracy of prediction algorithms for classification: An overview. Bioinformatics 16, 412–424. [DOI] [PubMed] [Google Scholar]
- Bellinger A. M., Arteaga C. L., Force T., Humphreys B. D., Demetri G. D., Druker B. J., Moslehi J. J. (2015). Cardio-oncology: How new targeted cancer therapies and precision medicine can inform cardiovascular discovery. Circulation 132, 2248–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289–300. [Google Scholar]
- Bers D. M. (2002). Cardiac excitation–contraction coupling. Nature 415, 198–205. [DOI] [PubMed] [Google Scholar]
- Chen M. H., Kerkela R., Force T. (2008). Mechanisms of cardiac dysfunction associated with tyrosine kinase inhibitor cancer therapeutics. Circulation 118, 84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. I., Hunt J. P., Herrgard S., Ciceri P., Wodicka L. M., Pallares G., Hocker M., Treiber D. K., Zarrinkar P. P. (2011). Comprehensive analysis of kinase inhibitor selectivity. Nat. Biotechnol. 29, 1046–1051. [DOI] [PubMed] [Google Scholar]
- Fabbro D., Cowan-Jacob S. W., Mobitz H., Martiny-Baron G. (2012). Targeting cancer with small-molecular-weight kinase inhibitors. Methods Mol. Biol. 795, 1–34. [DOI] [PubMed] [Google Scholar]
- Force T., Kolaja K. L. (2011). Cardiotoxicity of kinase inhibitors: The prediction and translation of preclinical models to clinical outcomes. Nat. Rev. Drug Discov. 10, 111–126. [DOI] [PubMed] [Google Scholar]
- Force T., Krause D. S., Van Etten R. A. (2007). Molecular mechanisms of cardiotoxicity of tyrosine kinase inhibition. Nat. Rev. Cancer 7, 332–344. [DOI] [PubMed] [Google Scholar]
- Guo L., Coyle L., Abrams R. M., Kemper R., Chiao E. T., Kolaja K. L. (2013). Refining the human iPSC-cardiomyocyte arrhythmic risk assessment model. Toxicol. Sci. 136, 581–594. [DOI] [PubMed] [Google Scholar]
- Ho A. L., Bendell J. C., Cleary J. M., Schwartz G. K., Burris H. A., Oakes F., Agbo F., Barker P. N., Senderowica A. M., Shapiro G. (2011). Phase I, open-label, dose-escalation study of AZD7762 in combination with irinotecan (irino) in patients (pts) with advanced solid tumors. J. Clin. Oncol. 29, 3033. [Google Scholar]
- Hortigon-Vinagre M. P., Zamora V., Burton F. L., Green J., Gintant G. A., Smith G. L. (2016). The use of ratiometric fluorescence measurements of the voltage sensitive dye di-4-ANEPPS to examine action potential characteristics and drug effects on human induced pluripotent stem cell-derived cardiomyocytes. Toxicol. Sci. 154, 320–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K. Y., Wu H. Y., Chen Y. J., Lu C. T., Su M. G., Hsieh Y. C., Tsai C. M., Lin K. I., Huang H. D., Lee T. Y., et al. (2014). RegPhos 2.0: An updated resource to explore protein kinase-substrate phosphorylation networks in mammals. Database (Oxford) 2014, bau034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamp T. J., Hell J. W. (2000). Regulation of cardiac L-type calcium channels by protein kinase A and protein kinase C. Circ. Res. 87, 1095–1102. [DOI] [PubMed] [Google Scholar]
- Kruse M., Hammond G. R., Hille B. (2012). Regulation of voltage-gated potassium channels by PI(4,5)P2. J. Gen. Physiol. 140, 189–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal H., Kolaja K. L., Force T. (2013). Cancer genetics and the cardiotoxicity of the therapeutics. J. Am. Coll. Cardiol. 61, 267–274. [DOI] [PubMed] [Google Scholar]
- Lamore S. D., Kamendi H. W., Scott C. W., Dragan Y. P., Peters M. F. (2013). Cellular impedance assays for predictive preclinical drug screening of kinase inhibitor cardiovascular toxicity. Toxicol. Sci. 135, 402–413. [DOI] [PubMed] [Google Scholar]
- Lankhorst S., Saleh L., Danser A. J., van den Meiracker A. H. (2015). Etiology of angiogenesis inhibition-related hypertension. Curr. Opin. Pharmacol. 21, 7–13. [DOI] [PubMed] [Google Scholar]
- Layland J., Solaro R. J., Shah A. M. (2005). Regulation of cardiac contractile function by troponin I phosphorylation. Cardiovasc. Res. 66, 12–21. [DOI] [PubMed] [Google Scholar]
- Lim J., Taoka B. M., Lee S., Northrup A., Altman M. D., Sloman D. L., Stanton M. G., Noucti N. (2011). Pyrazolo[1,5-A]Pyrimidines as MARK Inhibitors. Rahway, NJ: Merck Sharp & Dohme Corp.
- Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. (2002). The protein kinase complement of the human genome. Science 298, 1912–1934. [DOI] [PubMed] [Google Scholar]
- Olaharski A. J., Bitter H., Gonzaludo N., Kondru R., Goldstein D. M., Zabka T. S., Lin H., Singer T., Kolaja K. (2010). Modeling bone marrow toxicity using kinase structural motifs and the inhibition profiles of small molecular kinase inhibitors. Toxicol. Sci. 118, 266–275. [DOI] [PubMed] [Google Scholar]
- Olaharski A. J., Gonzaludo N., Bitter H., Goldstein D., Kirchner S., Uppal H., Kolaja K. (2009). Identification of a kinase profile that predicts chromosome damage induced by small molecule kinase inhibitors. PLoS Comput. Biol. 5, e1000446.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanos G. S., Ioannidis G. N., Ardavanis A. G. (2009). Cardiotoxicity induced by tyrosine kinase inhibitors. Acta Oncol. 48, 964–970. [DOI] [PubMed] [Google Scholar]
- Peters M. F., Lamore S. D., Guo L., Scott C. W., Kolaja K. L. (2015). Human stem cell-derived cardiomyocytes in cellular impedance assays: Bringing cardiotoxicity screening to the front line. Cardiovasc. Toxicol. 15, 127–139. [DOI] [PubMed] [Google Scholar]
- Rogers D., Hahn M. (2010). Extended-connectivity fingerprints. J. Chem. Inf. Model. 50, 742–754. [DOI] [PubMed] [Google Scholar]
- Rose R. A., Kabir M. G., Backx P. H. (2007). Altered heart rate and sinoatrial node function in mice lacking the cAMP regulator phosphoinositide 3-kinase-gamma. Circ Res 101, 1274–1282. [DOI] [PubMed] [Google Scholar]
- Scott C. W., Zhang X., Abi-Gerges N., Lamore S. D., Abassi Y. A., Peters M. F. (2014). An impedance-based cellular assay using human iPSC-derived cardiomyocytes to quantify modulators of cardiac contractility. Toxicol. Sci. 142, 331–338. [DOI] [PubMed] [Google Scholar]
- Sharma A., Burridge P. W., McKeithan W. L., Serrano R., Shukla P., Sayed N., Churko J. M., Kitani T., Wu H., Holmstrom A., et al. (2017). High-throughput screening of tyrosine kinase inhibitor cardiotoxicity with human induced pluripotent stem cells. Sci. Transl. Med. 9, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan K. A., Ke Y., Solaro R. J. (2007). p21-Activated kinase-1 and its role in integrated regulation of cardiac contractility. Am. J. Physiol. Regul. Integr. Comp. Physiol. 293, R963–R973. [DOI] [PubMed] [Google Scholar]
- Stansfeld P. J., Hopkinson R., Ashcroft F. M., Sansom M. S. (2009). PIP(2)-binding site in Kir channels: Definition by multiscale biomolecular simulations. Biochemistry 48, 10926–10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toutenburg H. (1975). Hollander, M., D. A. Wolfe: Nonparametric statistical methods. John Wiley & Sons, New York-Sydney-Tokyo-Mexico City 1973. 503 S., Biom. J. 17, 526. [Google Scholar]
- Wu P., Nielsen T. E., Clausen M. H. (2015). FDA-approved small-molecule kinase inhibitors. Trends Pharmacol. Sci. 36, 422–439. [DOI] [PubMed] [Google Scholar]
- You B., Yan G., Zhang Z., Yan L., Li J., Ge Q., Jin J. P., Sun J. (2009). Phosphorylation of cardiac troponin I by mammalian sterile 20-like kinase 1. Biochem. J. 418, 93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.