Why was the cohort consortium set up?
Worldwide globalization and Westernization in social and economic aspects have led to drastic changes in South Korea during the past several decades. These changes include individual health behaviours, which were reflected as increased prevalence of non-communicable chronic diseases (NCDs), such as type 2 diabetes mellitus (T2DM), hypertension, obesity and cardiovascular disease (CVD).1 These NCDs are known to be caused by both environmental risk factors and predisposing genetic factors. Population decline is another issue in South Korea; the recorded fertility rate was ≤ 1.3 births per woman, and ≥ 10% of the population were elderly individuals aged ≥ 65 years according to the Population and Housing Census results of 2005-2010.2,3 We have also been observing an increased influx and efflux of the population due to globalization. In particular, there has been a rising tendency in the marriage-based inflow of South Asian women during the last decade.
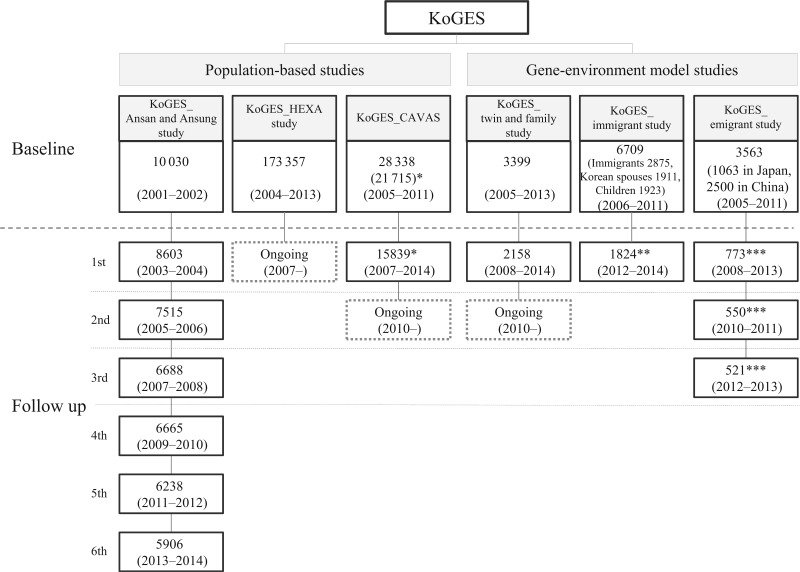
To attempt to solve public health issues resulting from these population trends and prepare for personalized and preventive health care in the future, the Korean government (National Research Institute of Health (NIH), Centers for Disease Control and Prevention and the Ministry of Health and Welfare, Korea) initiated a large prospective cohort study with government funding, named the Korean genome and epidemiology study (KoGES). The study is a consortium project consisting of six prospective cohort studies that would be categorized into population-based and gene-environment model studies (Figure 1). The aim of the KoGES was to establish a genome epidemiological study platform for the research community with a health database and biobank, to investigate the genetic and environmental aetiology of common complex diseases in Koreans (i.e. T2DM, hypertension, obesity, metabolic syndrome, osteoporosis, CVD,and cancer) and causes of death with long-term follow-up. The ultimate goal of the KoGES was to develop comprehensive and applicable health care guidelines for common complex diseases in Koreans, reduce the burden of chronic diseases and improve the quality of life.
Figure 1.
Flow diagram of baseline recruitment and follow-up for the Korean genome and epidemiology study (KoGES). *The follow-up survey was conducted in 6 out of 11 counties (baseline number from the six counties = 21 715). **The follow-up recruitment included only immigrant mothers and children. ***The follow-up survey was continued only in Japan due to difficulties in the follow-up of Chinese participants (see the ‘Who is in the cohort consortium and how often have they been followed up?’ section).
Who is in the cohort consortium and how often have they been followed up?
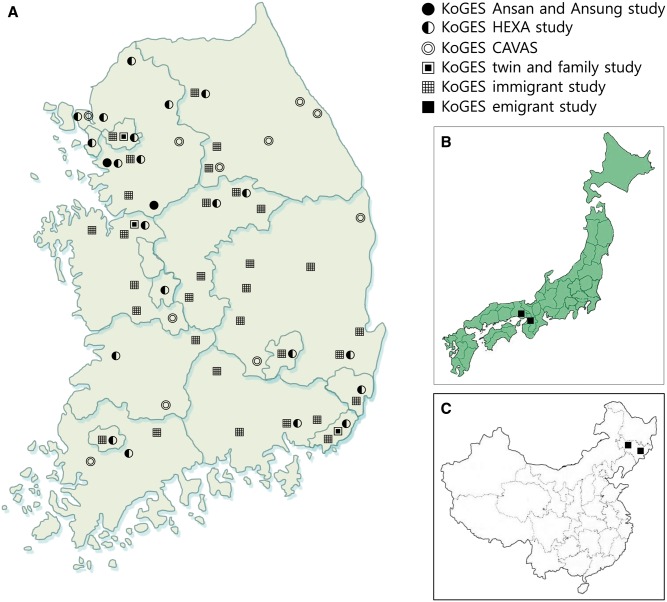
The population-based cohorts in the KoGES, including the KoGES_Ansan and Ansung study, the KoGES_health examinee (HEXA) study and the KoGES_cardiovascular disease association study (CAVAS), consist of community-dwellers and participants recruited from the national health examinee registry, men and women, aged ≥ 40 years at baseline (Figure 1). The KoGES gene-environment model studies include the KoGES_twin and family study, the KoGES_immigrant study and KoGES_emigrant study (Japan and China). For baseline recruitment, eligible participants were asked to volunteer through on-site invitation, mailed letters, telephone calls, media campaign or community leader-mediated conferences. The responders were invited to visit the survey sites, including ≥ 50 national and international medical schools, hospitals and health institutions (Figure 2), for an interview, a questionnaire administered by trained staff and physical examination. The followed-up participants were invited to complete the surveys by mail and telephone calls periodically. The inclusion criteria for baseline and follow-up recruitment for each cohort are described in Supplementary material (available as Supplementary data at IJE online).
Figure 2.
Geographical location of the survey sites in the Korean genome and epidemiology study. In Korea (A); Japan (B); China (C).
The selected baseline characteristics of participants and disease prevalence are summarized by cohort in Tables 1, 2 and 3, with additional data in Supplementary Table 2 (available as Supplementary data at IJE online). The age-standardized prevalence rates for T2DM, hypertension and obesity among the population-based cohort studies are compared in Table 2. When compared with the age-standardized prevalence rates (≥ 30 years) reported in the Korea National Health and Nutrition Examination Survey (KNHANES III, 2005: hypertension, 28.0%; diabetes, 9.1; obesity, 34.8),1 the prevalence of hypertension appeared to be higher in the KoGES population-based studies. A total of 7224 incident cancer cases and 4351 all-cause deaths were identified in the population-based studies between 2001 and 2013 (Table 3). The distribution patterns of leading primary cancer sites by sex were observed to be similar in the constituent studies, which were also comparable with the national cancer statistics, Korea Central Cancer Registry (KCCR, age-standardized incidence rates of KCCR between 1999 and 2012: stomach > colon and rectum > lung > liver > prostate > thyroid in men; thyroid > breast > colon and rectum > stomach > lung > liver in women).4 Although the details of non-responders at baseline not available from all the studies in the consortium to examine the representativeness of the baseline responder population, the comparisons of health outcomes between KNHANES and the national cancer statistics present supportive evidence that the KoGES data are generalizable to the Korean population.
Table 1.
Baseline characteristics of participants in the cohort studies of the Korean genome and epidemiology study
| Characteristics | Population-based studies |
Gene-environment model studies |
||||
|---|---|---|---|---|---|---|
| KoGES_Ansan and Ansung study | KoGES_HEXA | KoGES_CAVAS | KoGES_twin and family study | KoGES_ immigrant studye | KoGES_ emigrant study | |
| n | 10030 | 173357 | 28338 | 3399 | 4786 | 3563 |
| Age (years)a | 52.29 ± 8.93 | 53.10 ± 8.37 | 58.58 ± 9.38 | 43.96 ± 13.70 | 30.68 ± 9.61 | 52.31 ± 8.89 |
| Gender, n (%) | ||||||
| Men | 4758 (47.44) | 59294 (34.20) | 10821 (38.19) | 1397 (41.10) | 1911 (39.93) | 1516 (42.55) |
| Women | 5272 (52.56) | 114063 (65.80) | 17517 (61.81) | 2002 (58.90) | 2875 (60.07) | 2047 (57.45) |
| Questionnaires | ||||||
| Smoking status, n (%) | ||||||
| Non-smokers | 5808 (58.69) | 125162 (72.88) | 19893 (70.32) | 2201 (64.89) | 3394 (72.31) | 2125 (60.25) |
| Ex-smokers | 1539 (15.55) | 25218 (14.68) | 4254 (15.04) | 453 (13.35) | 272 (5.79) | 342 (9.70) |
| Current smokers | 2549 (25.76) | 21352 (12.43) | 4143 (14.64) | 738 (21.76) | 1028 (21.90) | 1060 (30.05) |
| Drinking status, n (%) | ||||||
| Non-drinkers | 4596 (46.24) | 87241 (50.75) | 14476 (51.23) | 938 (27.82) | 3058 (65.50) | 1683 (47.73) |
| Ex-drinkers | 652 (6.56) | 6911 (4.02) | 2026 (7.17) | 310 (9.19) | 194 (4.16) | 161 (4.57) |
| Current drinkers | 4691 (47.20) | 77740 (45.23) | 11753 (41.60) | 2124 (62.99) | 1417 (30.35) | 1682 (47.70) |
| Regular exercise, n (%)b | N/A | 90381 (52.55) | 8860 (31.34) | 1161 (34.93) | 1434 (30.46) | 875 (24.63) |
| Total energy intake (kcal)cc | 1957.02 ± 714.10 | 1759.23 ± 592.01 | 1658.98 ± 575.45 | 1916.83 ± 802.52 | 1865.54 ± 743.76 | 2010.93 ± 894.81 |
| Anthropometric measurements | ||||||
| Height (cm)a | 159.98 ± 8.66 | 160.49 ± 8.03 | 157.83 ± 8.50 | 162.30 ± 8.70 | 159.42 ± 8.25 | 160.46 ± 8.59 |
| Weight (kg)a | 63.01 ± 10.10 | 61.83 ± 9.92 | 60.92 ± 10.07 | 62.74 ± 11.65 | 56.99 ± 12.21 | 60.85 ± 10.91 |
| Body mass index (kg/m2)a | 24.57 ± 3.15 | 23.94 ± 2.91 | 24.40 ± 3.15 | 23.71 ± 3.29 | 22.24 ± 3.36 | 23.56 ± 3.37 |
| Systolic blood pressure (mmHg)a | 121.69 ± 18.48 | 122.71 ± 15.47 | 126.28 ± 17.93 | 114.81 ± 16.29 | 115.56 ± 16.65 | 126.36 ± 20.09 |
| Diastolic blood pressure (mmHg)a | 80.34 ± 11.44 | 76.24 ± 10.03 | 79.41 ± 11.12 | 72.35 ± 10.50 | 68.65 ± 12.31 | 81.23 ± 13.18 |
| Clinical examination | ||||||
| Fasting blood glucose (mg/dl)a | 87.31 ± 21.41 | 95.18 ± 21.59 | 98.67 ± 24.16 | 93.31 ± 18.67 | 91.15 ± 19.19 | 94.67 ± 32.39 |
| Total cholesterol (mg/dl)a | 191.17 ± 35.83 | 197.45 ± 35.63 | 199.75 ± 37.33 | 189.15 ± 35.92 | 184.46 ± 38.74 | 189.17 ± 43.52 |
| HDL cholesterol (mg/dl)a | 44.64 ± 10.07 | 54.02 ± 12.94 | 45.36 ± 11.12 | 50.47 ± 12.65 | 45.65 ± 11.06 | 49.75 ± 17.92 |
| Triglyceride (mg/dl)a | 162.47 ± 104.96 | 126.88 ± 89.90 | 149.87 ± 100.31 | 110.13 ± 64.64 | 124.71 ± 97.95 | 143.90 ± 121.74 |
| Biospecimend | ||||||
| DNA | 10011 | 170106 | 28134 | 3232 | 4251 | 2146 |
| Plasma | 8277 | 171110 | 28312 | 3381 | 4440 | 2092 |
| Serum | 9863 | 171074 | 28310 | 3340 | 4442 | 2105 |
| Spot urine | 9380 | 169514 | 28270 | 3371 | 4398 | 3315 |
| Number of participants with genotype dataf | 8840 | 3696 | 12233 | 1803 | 348 | 934 |
N/A, not applicable.
aMeans ± standard deviation (SD).
bDefined from the questionnaire regarding whether or not a subject routinely performs 30 min-exercise/day.
cTotal energy intake was calculated using food frequency questionnaires. (The Japanese public health centre-based prospective study (JPHC) FFQ was employed in the KoGES_emigrant study).
dNumber of participants with quality-controlled biospecimens stored at the National biobank of Korea.
eBaseline characteristics of the KoGES_immigrant study included only immigrant women and their spouses.
fNumber of quality-controlled genotype data collected as of 2015. Those who participated in the study in Japan were included in the KoGES_emigrant study.
Table 2.
Prevalence of major disease targets at baseline in the Korean genome and epidemiology study
| Characteristics | Population-based studiesd |
Gene-environment model studiese |
||||
|---|---|---|---|---|---|---|
| KoGES_Ansan and Ansung study | KoGES_HEXA | KoGES_CAVAS | KoGES_twin and family study | KoGES_ immigrant study | KoGES_ emigrant study | |
| n | 10030 | 173357 | 28338 | 3399 | 4786 | 3563 |
| Prevalence | ||||||
| Hypertension n (%)a | 3249 (33.88) | 50984 (30.98) | 12614 (40.10) | 645 (18.98) | 516 (10.78) | 1315 (36.91) |
| Type 2 diabetes n (%)b | 849 (8.76) | 15157 (9.35) | 3376 (10.29) | 228 (6.71) | 148 (3.09) | 264 (7.41) |
| Obesity n (%)c | 4290 (43.03) | 56857 (34.80) | 11295 (40.16) | 1070 (31.48) | 943 (19.70) | 1088 (30.54) |
aHypertension was defined as either a systolic or a diastolic blood pressure of ≥ 140 mmHg or ≥ 90 mmHg, respectively, or when participants self-reported the diagnosed diseases.
bType 2 diabetes was defined as either a fasting blood glucose level of ≥ 126 mg/dl or when participants self-reported the diagnosed diseases.
cObesity was defined as a body mass index ≥ 25 kg/m2.
dAge-standardized prevalence rate in population-based studies was based on the 2014 mid-year resident population data.
eCrude prevalence rates were shown for the gene-environment model studies due to lack of standard population data.
Table 3.
Secondary outcomes of the Korean genome and epidemiology study
| Characteristics | Population-based studies |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Total |
KoGES_Ansan and Ansung study (n = 10030) |
KoGES_HEXA (n = 173357) |
KoGES_CAVAS (n = 28338) |
||||||
| Men | Women | Total | Men | Women | Men | Women | Men | Women | |
| N (%) | 74,873 (35.36) | 136,852 (64.64) | 211,725 (100.00) | 4,758 (47.44) | 5,272 (52.56) | 59,294 (34.20) | 114,063 (65.80) | 10,821 (38.19) | 17,517 (61.81) |
| Number of death, na | 2,722 | 1,629 | 4,351 | 454 | 239 | 1,381 | 909 | 887 | 481 |
| All causes of cancer, nb | 3,125 | 4,099 | 7,224 | 426 | 344 | 1,968 | 3,156 | 731 | 599 |
| Stomach | 683 | 375 | 1,058 | 114 | 47 | 389 | 252 | 180 | 76 |
| Colorectal | 544 | 460 | 1,004 | 70 | 37 | 368 | 339 | 106 | 84 |
| Liver | 330 | 131 | 461 | 52 | 12 | 204 | 94 | 74 | 25 |
| Lung | 377 | 211 | 588 | 54 | 31 | 208 | 136 | 115 | 44 |
| Breast | 3 | 702 | 705 | 1 | 43 | 1 | 594 | 1 | 65 |
| Thyroid | 144 | 1,317 | 1,461 | 15 | 95 | 113 | 1,090 | 16 | 132 |
| Prostate | 352 | - | 352 | 33 | - | 243 | - | 76 | - |
aMortality outcomes were ascertained by the death records from 2001 to 2013 provided by the Korea National Statistical Office.
bCancer incidence was identified by the cancer registry data from 2001 to 2012 (Korea Central Cancer Registry).
What is attrition like?
The follow-up surveys are ongoing in most cohorts and therefore detailed information regarding attrition is limited and inconclusive. In some pilot surveys, most of the non-attenders were lost to follow-up because we were unable to contact them (i.e. change of contact information), or they were too busy to participate or not interested in continuing to participate in the study. In particular, difficulty in following up in the KoGES_twin and family and KoGES_immigrant studies was attributed to the family-based survey. General characteristics of participants and non-participants in the follow-up surveys in each study are compared in Supplementary Table 3 (available as Supplementary data at IJE online). According to these attrition analyses, there were statistically significant differences (P ≤ 0.05) in some variables between responders and non-responders in most of the cohorts; however, there was no particular directionality in the disparity (i.e. healthy cohort effect).
We have relatively detailed information regarding attrition in the KoGES_Ansan and Ansung study in which biannual repeated surveys were continued since baseline recruitment in 2001–2002 up to the 6th follow-up. Out of the 10 030 baseline participants, 633 cumulative deaths were recorded between baseline and the 5th follow-up (2001 to 2012), and the 6th follow-up was conducted in 5906 participants (out of 9397 survivor; Figure 1, and Supplementary Figure 1, available as Supplementary data at IJE online). The participation rates for the 1st to 6th follow-ups were 86.1%, 76.0%, 68.2%, 68.9%, 65.3% and 62.8%, respectively. Approximately 90% of baseline participants completed at least one follow-up survey over the course of 12 years. There were continuous attempts to contact participants by means of annual birthday cards and telephone calls. The main reasons for refusing to participate included changes in the telephone number or mailing address (25%), reported to be too busy to attend (19%) and not answering telephone calls (10%). We observed no particular differences in the baseline characteristics between responders and non-responders (i.e. those who participated in the baseline survey only), except that some distinctions in smoking, diet habits (total energy intake) and fasting glucose level were observed (Supplementary Table 3, available as Supplementary data at IJE online).
What has been measured?
All participants provided informed consent for the baseline data and biospecimens and underwent an interview and physical examination. Ethical approval was obtained from the institutional review boards of NIH and collaborators of the KoGES groups.
The six constituent cohort studies shared core questionnaire and examination items. The identical questionnaires, physical examinations and clinical investigations were mostly used during the baseline and follow-up phases (Table 4). The participants were questioned by trained interviewers regarding their socio-demographic status, lifestyle (i.e. diet, smoking, drinking and physical activity), reproductive history, psychological stress, social relationships and disease history (i.e. disease status of the participants and his/her family members). For dietary assessment, a semi-quantitative food frequency questionnaire (FFQ) involving 103 items was developed for the KoGES. Detailed information regarding the protocol and results of a validation study for the FFQ are described elsewhere.5,6 Although an FFQ is the most practical and common dietary assessment method used in prospective cohort studies, it contains a limited list of food items, and individuals are unable to accurately report their food intake retrospectively over a long period of time. To compensate for the limitations, a 24-h diet recall survey has been conducted in some of the cohorts.
Table 4.
Summary of core variables collected in the Korean genome and epidemiology study
| Questionnaires/measurements | Baseline | Follow-up |
|---|---|---|
| Questionnaires | ||
| Socio-demographic data | ||
| Education | √ | √ |
| Household income | √ | √ |
| Marital status | √ | √ |
| Occupation | √ | √ |
| Medical history and related questionnaires | ||
| Self-rated health | √ | √ |
| Past disease history | √ | √ |
| Current status of disease treatment | √ | √ |
| Medication history | √ | √ |
| Family disease history | √ | √ |
| Psychosocial battery (e.g. PWI-SF,a CES-Db) | √(subset) | √(subset) |
| Lifestyle | ||
| Self-reported smoking and alcohol habits | √ | √ |
| Physical activity | √ | √ |
| Dietary habit (e.g. FFQc 24-h recall) | √(subset) | √(subset) |
| Sleep disorder | – | √(subset) |
| Reproductive factors (for women) | ||
| Menstrual factors (age at menarche length, of menstrual cycle) | √ | √ |
| Menopausal status | √ | √ |
| Reproductive history (number of pregnancies, age at each | √ | √ |
| Pregnancy, duration and outcome of pregnancies, | ||
| Breastfeeding, infertility) | ||
| Anthropometric measures | ||
| Height and weight | √ | √ |
| Waist and hip circumference | √ | √ |
| Body composition | √ | √ |
| Blood pressure and pulse rate | √ | √ |
| Clinical examination | ||
| Blood test | ||
| Complete blood cell count | √ | √ |
| Glucose (fasting) | √ | √ |
| Glucose (1-h/2-h on oral glucose tolerance test) | √(subset) | √(subset) |
| Total protein | √(subset) | √(subset) |
| Albumin | √ | √ |
| Blood urea nitrogen | √ | √ |
| Creatinine | √ | √ |
| Total bilirubin | √(subset) | √(subset) |
| AST (SGOT) | √ | √ |
| ALT (SGPT) | √ | √ |
| γ-GTP | √ | √ |
| Total cholesterol | √ | √ |
| HDL-cholesterol | √ | √ |
| LDL-cholesterol | √(subset) | √(subset) |
| Triglyceride | √ | √ |
| hs-CRP | √ | √ |
| Haemoglobin (Hb) | √ | √ |
| HbA1C | √(subset) | √(subset) |
| Insulin (fasting) | √(subset) | √(subset) |
| Insulin (1-h/2-h) | √(subset) | √(subset) |
| Calcium (Ca) | √(subset) | √(subset) |
| Homocysteine | √(subset) | √(subset) |
| Urine test | ||
| pH | √ | √ |
| Protein | √ | √ |
| Glucose | √ | √ |
| Ketone | √ | √ |
| Bilirubin | √ | √ |
| Blood | √ | √ |
| Electrocardiography | √(subset) | √(subset) |
| Spirometry | √(subset) | √(subset) |
| Bone mineral density | ||
| Bone strength (sonometer) | √(subset) | √(subset) |
| Bone mineral density (dual energy x-ray absorptiometry) | √(subset) | √(subset) |
| DNA genome sequencing (GWAS chip-based) | √(subset) | √(subset) |
aPWI-SF, Psychosocial Well-being Index.
bCES-D Center for Epidemiologic Studies Depression Scale.
cFFQ Food Frequency Questionnaire.
(√: collected on all the participants in all 6 cohorts; √ (subset): collected in subsets of the 6 cohorts).
Anthropometric and clinical measurements were also obtained (i.e. height, weight, waist circumference, blood pressure, blood count, biochemical analysis, including blood sugar, lipid profiles and other biomarkers to evaluate current health status; Table 4). In addition to the core variables, the KoGES has also been promoting various ancillary and collaborative measurements and studies, including oral glucose tolerance test, carotid intima-media thickness test, pulse wave velocity (PWV) test, electrocardiography, pulmonary function test, bone mineral density, brain magnetic resonance imaging, osteoporosis, periodontal diseases and oriental medicine study,7,8 that lead to numerous in-depth research outcomes. A list of these special clinical tests is shown in Supplementary Table 4 (available as Supplementary data at IJE online).
Bio-specimens included fasting blood samples that were collected in a serum separator tube and two ethylenediaminetetraacetic acid (EDTA) tubes, and a 10-ml midstream urine sample. For long-term storage, both serum and plasma were prepared and aliquoted in 6–10 vials (300–500 μL per vial), and 80–100 μg samples of blood DNA were also prepared. All samples were then transported to the National Biobank of Korea9 and stored for future research purposes.
Genome-wide single nucleotide polymorphism (SNP) data are available for a subset of the KoGES participants using Affymetrix or Illumina platforms (Table 1), and the imputed data based on the 1000-genome sequence or the Korean HapMap data.10,11 The KoGES data have been linked to national data sources, including death records (Korea National Statistical Office) and cancer registry (KCCR4) to evaluate the mortality and cancer incidence rates, respectively (Table 3).
What has it found? Key findings and publications
The KoGES has been contributing to the research community by publishing ≥ 400 articles since the initiation of the KoGES_Ansan and Ansung study in 2001. Given that the baseline recruitment was completed in 2013, specific analyses of the combined consortium data have recently been initiated. Therefore, the current findings have been focused on individual cohort studies and are selectively summarized in the following paragraphs.
Genome-wide association studies (GWAS)
The focus of some of the key findings in the GWAS includes the identification of genetic variants associated with various disease-related phenotypes, which was facilitated by the availability of genome-wide genotype data and repeatedly measured disease-related variables in the KoGES_Ansan and Ansung study. In particular, genetic variants associated with T2DM, blood pressure, waist-hip-ratio, bone mineral density and serum lipid level in Asians and Koreans have been found by GWAS for the KoGES data (known as ‘KARE’ for ‘Korean Association Resource’) in collaboration with several international GWAS consortia (such as the Asian Genetic Epidemiology Network (AGEN) and Meta-Analyses of Glucose and Insulin-related Traits Consortium (MAGIC)).12–17 For example, our electrocardiography analysis data allowed us to identify the genetic variants in SLC8A1 (sodium/calcium exchanger 1 precursor) and PRDM16 (PR domain-containing 16) loci associated with electrocardiographic traits including QT and PR intervals and with QRS duration in the Asian population, respectively, which implicated the genes in cardiac function.18,19
Metabolic syndrome and T2DM
Extensive findings of the KoGES have been concentrated on metabolic syndrome and T2DM because of their relatively high prevalence in the population and clear disease ascertainment. It has been found that an increased baseline serum adiponectin level was a protective factor for incident metabolic syndrome.20 In one of our nested case-control studies, high plasma concentration of isoflavones was associated with a decreased risk of T2DM in women, suggesting a beneficial effect of soy-based food intake which is rich in isoflavones.21 We have estimated the relative risk for metabolic syndrome in heavy drinkers (> 30 g/day), and T2DM in ex- and current smokers.22,23 It was also demonstrated that the haemoglobin A1c (HbA1c) cut-off values of 5.9% and ≥ 5.6% can be effectively used to identify undiagnosed T2DM and an increased risk for disease incidence, respectively.24
Hypertension and CVD
According to our prospective study, healthy obesity (without the metabolic syndrome component) confers a 2-fold increased risk of hypertension.25 Habitual snoring (≥ 4 days per week) was found to be independently associated with an increased incidence rate of hypertension.26 We also demonstrated that serum uric acid level was positively correlated with brachial-ankle pulse wave velocity (PWV), a marker for arterial stiffness and carotid atherosclerosis.27 It is well known that the risks of CVD morbidity and mortality increase with occurrence of T2DM, which is attributed to the effects of hyperglycaemia on vasculature and the coexistence of other metabolic risk factors. Based on the 10-year follow-up of the KoGES_Ansan and Ansung study, we reported that CVD mortality was much higher in individuals with diabetes alone, compared twith those with metabolic syndrome alone. The CVD risk and mortality in individuals with diabetes were not additively influenced by the presence of metabolic syndrome.28
Ageing studies
Our consortium population of > 40-year-old participants and their longitudinal follow-up data are a suitable test-bed for ageing studies. We have presented the sex-specific reference range for fraction of exhaled nitric oxide, which is a useful non-invasive biomarker for asthma diagnosis, and its diagnostic optimal cut-off value for asthma prediction in elderly participants.29 We have also evaluated the prevalence of neck and low back pain in the elderly populations, which is associated with quality of life and substantial medical costs.30,31 It has been recently shown that obstructive sleep apnoea in the middle-aged and older population is a risk factor for cerebral white matter changes which are associated with incident stroke, dementia and mortality.32
Nutrition studies
We have been collecting dietary assessment data from the FFQ as the consortium’s core variables, providing a valuable resource for nutrition studies. In our preliminary study involving data from ≥ 160 000 participants, it was shown that a high diet quality, estimated by the healthy eating index, was positively correlated with the prevalence of hypertension and T2DM.33,34 Dietary intake of zinc has been proposed to be associated with atherosclerosis risk. Phytate is known to be a dietary inhibitor of zinc. We have demonstrated that lower zinc bioavailability, based on the phytate-zinc molar ratio estimated from the FFQ data, is linked to a higher risk of atherosclerosis.35 We observed that the inclusion of dietary predictors such as consumption of poultry, legumes, carbonated soft drinks or green tea, into the CVD risk prediction model has improved model performance and prediction ability.36 The association between the effects of environmental changes (i.e. diet habit) experienced by immigrants or emigrants and CVD37 or metabolic syndrome38 has also been shown.
We are currently preparing for combined consortium data analysis and meta-analyses of individual cohorts, which will allow replication and validation of previous cross-sectional or individual cohort study results and will ultimately provide valuable findings in the future when the long-term follow-up phases are completed. Researchers who are interested in the comprehensive list of publications as of the end of 2014 can refer to [http://www.nih.go.kr/NIH/eng/main.jsp > Research infrastructure > KoGES > Scientific accomplishment].
What are the main strengths and weaknesses of the study?
A key strength of the KoGES is the richness of health- and disease-related phenotype information and the comprehensive list of biospecimens collected from all the participants who have provided the informed consent (i.e. genomic DNA, serum, plasma and spot urine; Table 1). These resources are used to capture information regarding the epidemiological characteristics of the Korean population, elucidate genetic and environmental risk factors for common diseases and develop preventive and therapeutic measures. Other strengths include, first, the availability of GWAS data for identifying genetic variants associated with traits and diseases and environmental factors interacting with genes. These data will allow us to conduct causal model analyses (i.e. Mendelian randomization studies) to estimate direct and indirect effects of exposures on outcomes.39,40 Second, all biospecimens were prepared and stored using uniformed standard protocols to be used for validation of already measured markers and the development of new biomarkers. Finally, a wide range of disease outcomes can be followed by data linkage with national data sources based on the unique personal identification key code system. The secondary data include national health insurance and medical care records, nationwide cancer registry data (KCCR4) and death records provided by the Korea national statistical office (Table 3).
The study has some limitations to be considered. The first stage of recruitment was completed during an extended time period for the KoGES_HEXA and the KoGES_CAVAS studies (2004–2013 and 2004–2011, respectively), which probably led to variations in exposures between participants recruited at early and late stages. Furthermore, the retention rate in some of the cohorts significantly decreased during the second phase of follow-up (∼ 40%). Despite efforts to engage participants, our follow-up strategy of recruiting volunteers proved to be inefficient for some cohorts. Although disease outcomes and other health endpoints can be ascertained and followed by secondary data linkage, these may result in potential selection bias in studies involving repeated measurement of exposures or intermediate outcomes, which should be considered when interpreting the study results. Secondly, the study population is not a statistically random sample that is representative of the entire population, which occurs in many of prospective cohort studies.41 This might not be critical for identifying exposure-outcome associations, but needs to be considered when applying the results for the entire population.
Finally, the composite entity of the KoGES, including the six cohorts, could be considered as a weakness. Nonetheless, the cohorts commonly share core items and biospecimens collected using standardized procedures, and each study involves a specific data collection designed to study a unique hypothesis making it possible to conduct integrative analyses and in-depth studies using data from the subgroups (Supplementary Table 4, available as Supplementary data at IJE online). Moreover, the gene-environment model studies will provide unique data resources to elucidate attributable risk factors and modifiable effects for the target NCDs and other health outcomes experienced in future.
Can I get hold of the data? Where can I find out more?
The KoGES provides valuable resources for the research community, including a wide range and depth of phenotype information and biospecimen archives (i.e. serum, plasma, urine and DNA). The genotype (genome-wide SNP data) and epidemiological dataset are made available to researchers after completing the quality control process. Researchers can access the dataset after receiving approval from a designated research proposal review committee of the NIH. Sample sharing is restricted to genomic DNA among the archived biological materials for future use. Further information is available at the KoGES website [http://www.nih.go.kr/NIH/eng/main.jsp > Research infrastructure > KoGES > Data]. International researchers are welcome to send us an e-mail [kimye@korea.kr] for additional information regarding collaboration and data access.
Cohort profile in a nutshell Korean genome and epidemiology study (KoGES) profile in a nutshell
KoGES consortium was designed to investigate and assess genetic and environmental factors as correlates or determinants of the incidence of chronic diseases in Koreans, such as type 2 diabetes, hypertension, cardiovascular diseases and cancer.
This study has been managed as an umbrella project that includes six ongoing cohort studies with approximately 245 000 participants at baseline, recruited in ≥ 50 national and international survey sites between 2001 and 2013.
The repeated follow-up surveys are being conducted during intervals of 2–4 years.
The dataset comprises a wide range of phenotypic and environmental measures, biological samples (i.e. DNA, serum, plasma and urine), genome-wide genotype information and linkage to health and administrative records.
KoGES is an open access resource for domestic research community. We also encourage international collaborations with researchers [http://www.nih.go.kr/NIH/eng/main.jsp > Research infrastructure > KoGES > Data].
Supplementary Data
Supplementary data are available here.
Funding
The work was funded by the Ministry for Health and Welfare, Republic of Korea [4845‐301 and 4851‐302].
Supplementary Material
Acknowledgments
The authors appreciate the efforts made by the clinical laboratories (Seoul Clinical Laboratories, Seegene Medical Foundation (previously Neodin) and Green Cross Laboratories), which have been in contract for transportation, preparation and biochemical analyses of the biospecimens. The authors are grateful to all the researchers, including Drs Hyun Kyung Moon, Jung Han Song and Yeo-min Yun for their valuable expert advice on KoGES, and Drs Sun Ha Jee, Seung Ku Lee and Ae Sun Shin for their insightful comments regarding the manuscript. Finally, we acknowledge all participants of the KoGES, and the coordinators, staff and interviewers of each cohort study.
Conflict of interest: None declared.
References
- 1. Korea Ministry of Health and Welfare. Korea Health Statistics 2010. Seoul: National Health and Nutrition Examination Survey (KNHANES III-V), Korea Centers for Disease Control and Prevention, 2011. [Google Scholar]
- 2. United Nations. World Population Prospects: 2006 Revision. New York, NY: United Nations Department of Economic and Social Affairs, Population Division, 2007. [Google Scholar]
- 3. Jung H-W. Statistics highlight scale of the aging population. http://koreajoongangdaily.joins.com/news/article/article.aspx?aid=2912868. Published November 21, 2009. [Google Scholar]
- 4. Jung KW, Won YJ, Kong HJ et al. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2012. Cancer Res Treat 2015;47:127–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ahn Y, Kwon E, Shim JE et al. Validation and reproducibility of food frequency questionnaire for Korean genome epidemiologic study. Eur J Clin Nutr 2007;61:1435–41. [DOI] [PubMed] [Google Scholar]
- 6. Kim J, Kim Y, Ahn YO et al. Development of a food frequency questionnaire in Koreans. Asia Pac J Clin Nutr 2003;12:243–50. [PubMed] [Google Scholar]
- 7. Cho NH, Kim JY, Kim SS, Lee SK, Shin C. Predicting type 2 diabetes using Sasang constitutional medicine. J Diabetes Investig 2014;5):525–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yoon DW, Lee SK, Yi H et al. Total nasal resistance among Sasang constitutional types: a population-based study in Korea. BMC Complement Altern Med 2013;13:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cho SY, Hong EJ, Nam JM, Han B, Chu C, Park O. Opening of the national biobank of Korea as the infrastructure of future biomedical science in Korea. Osong Public Health Res Perspect 2013;3):177–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. The 1000 Genomes Project Consortium; McVean GA. An integrated map of genetic variation from 1092 human genomes. Nature 2012;491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim YU, Kim S, Jin HS, Park Y, Ji M, Kim YJ. The Korean HapMap project website. Genomics Informatics 2008;6:91–94. [Google Scholar]
- 12. The HUGO Pan-Asian SNP Consortium; Abdulla MA, Ahmed I et al. Mapping human genetic diversity in Asia. Science 2009;326:1541–45. [DOI] [PubMed] [Google Scholar]
- 13. Cho YS, Chen CH, Hu C et al. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians. Nat Genet 2012;44:67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cho YS, Go MJ, Kim YJ et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet 2009;41: 527–34. [DOI] [PubMed] [Google Scholar]
- 15. Kato N, Takeuchi F, Tabara Y et al. Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in East Asians. Nat Genet 2011;43:53–5–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Okada Y, Sim X, Go MJ et al. Meta-analysis identifies multiple loci associated with kidney function-related traits in East Asian populations. Nat Genet 2012;44:90–0‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang L, Choi HJ, Estrada K et al. Multistage genome-wide association meta-analyses identified two new loci for bone mineral density. Hum Mol Genet 2014;23:1923–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hong KW, Lim JE, Kim JW et al. Identification of three novel genetic variations associated with electrocardiographic traits (QRS duration and PR interval) in East Asians. Hum Mol Genet 2014;23:6659–67. [DOI] [PubMed] [Google Scholar]
- 19. Kim JW, Hong KW, Go MJ et al. A common variant in SLC8A1 is associated with the duration of the electrocardiographic QT interval. Am J Hum Genet 2012;91:180–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim JY, Ahn SV, Yoon JH et al. Prospective study of serum adiponectin and incident metabolic syndrome: the ARIRANG study. Diabetes Care 2013;36:1547–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ko KP, Kim CS, Ahn Y et al. Plasma isoflavone concentration is associated with decreased risk of type 2 diabetes in Korean women but not men: results from the Korean Genome and Epidemiology Study. Diabetologia 2015;58:726–35. [DOI] [PubMed] [Google Scholar]
- 22. Baik I, Shin C. Prospective study of alcohol consumption and metabolic syndrome. Am J Clin Nutr 2008;87:1455–63. [DOI] [PubMed] [Google Scholar]
- 23. Cho NH, Chan JC, Jang HC, Lim S, Kim HL, Choi SH. Cigarette smoking is an independent risk factor for type 2 diabetes: a four-year community-based prospective study. Clin Endocrinol (Oxf) 2009;71:679–85. [DOI] [PubMed] [Google Scholar]
- 24. Choi SH, Kim TH, Lim S, Park KS, Jang HC, Cho NH. Hemoglobin A1c as a diagnostic tool for diabetes screening and new-onset diabetes prediction: a 6-year community-based prospective study. Diabetes Care 2011;34:944–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee SK, Kim SH, Cho GY et al. Obesity phenotype and incident hypertension: a prospective community-based cohort study. J Hypertens 2013;31:145–51. [DOI] [PubMed] [Google Scholar]
- 26. Kim J, Yi H, Shin KR, Kim JH, Jung KH, Shin C. Snoring as an independent risk factor for hypertension in the nonobese population: the Korean Health and Genome Study. Am J Hypertens 2007;20:819–24. [DOI] [PubMed] [Google Scholar]
- 27. Bae JS, Shin DH, Park PS et al. The impact of serum uric acid level on arterial stiffness and carotid atherosclerosis: the Korean Multi-Rural Communities Cohort study. Atherosclerosis 2013;231:145–51. [DOI] [PubMed] [Google Scholar]
- 28. Bae JC, Cho NH, Suh S et al. Cardiovascular disease incidence, mortality and case fatality related to diabetes and metabolic syndrome: A community-based prospective study (Ansung-Ansan cohort 2001–12). J Diabetes 2015;7:791–99. [DOI] [PubMed] [Google Scholar]
- 29. Jo EJ, Song WJ, Kim TW et al. Reference ranges and determinant factors for exhaled nitric oxide in a healthy Korean elderly population. Allergy Asthma Immunol Res 2014;6:504–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cho NH, Jung YO, Lim SH, Chung CK, Kim HA. The prevalence and risk factors of low back pain in rural community residents of Korea. Spine 2012;37:2001–10. [DOI] [PubMed] [Google Scholar]
- 31. Son KM, Cho NH, Lim SH, Kim HA. Prevalence and risk factor of neck pain in elderly Korean community residents. J Korean Med Sci 2013;28:680–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim H, Yun CH, Thomas RJ et al. Obstructive sleep apnea as a risk factor for cerebral white matter change in a middle-aged and older general population. Sleep 2013;36:709–15B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hurley KM, Oberlander SE, Merry BC, Wrobleski MM, Klassen AC, Black MM. The healthy eating index and youth healthy eating index are unique, nonredundant measures of diet quality among low-income, African American adolescents. J Nutr 2009;139:359–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lyu J, Kim Y. [Healthy eating index (HEI) indicates that high diet quality is associated with low prevalence of hypertension and type 2 diabetes in Koreans: The Korean genome and epidemiology study (KoGES).]. Public Health Wkly Rep 2015;8(3):51–8. [Google Scholar]
- 35. Jung SK, Kim MK, Lee YH et al. Lower zinc bioavailability may be related to higher risk of subclinical atherosclerosis in Korean adults. PLoS One 2013;8:e80115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Baik I, Cho NH, Kim SH, Shin C. Dietary information improves cardiovascular disease risk prediction models. Eur J Clin Nutr 2013;67:25–30. [DOI] [PubMed] [Google Scholar]
- 37. Ko A, Kim H, Han CJ, Kim JM, Chung HW, Chang N. Association between high sensitivity C-reactive protein and dietary intake in Vietnamese young women. Nutr Res Pract 2014;8:445–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shin M, Kim MK, Li ZM et al. Comparison of prevalence of metabolic syndrome between Korean emigrants and host country residents in Japan and China – The Korean Emigrant Study. Epidemiol Health 2010;32:e2010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet 2014;23:R89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. The EPIC-InterAct Consortium, Burgess S, Daniel RM et al. Network Mendelian randomization: using genetic variants as instrumental variables to investigate mediation in causal pathways. Int J Epidemiol 2015;44:484–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. The E3N Study Group; Clavel-Chapelon F. Cohort Profile: The French E3N Cohort Study. Int J Epidemiol 2015;44:801–09. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.