Abstract
Background: Membranous nephropathy (MN) is a common cause of nephrotic syndrome in adults. Previous genome-wide association studies (GWAS) of 300 000 genotyped variants identified MN-associated loci at HLA-DQA1 and PLA2R1.
Methods: We used a combined approach of genotype imputation, GWAS, human leucocyte antigen (HLA) imputation and extension to other aetiologies of chronic kidney disease (CKD) to investigate genetic MN risk variants more comprehensively. GWAS using 9 million high-quality imputed genotypes and classical HLA alleles were conducted for 323 MN European-ancestry cases and 345 controls. Additionally, 4960 patients with different CKD aetiologies in the German Chronic Kidney Disease (GCKD) study were genotyped for risk variants at HLA-DQA1 and PLA2R1.
Results: In GWAS, lead variants in known loci [rs9272729, HLA-DQA1, odds ratio (OR) = 7.3 per risk allele, P = 5.9 × 10−27 and rs17830558, PLA2R1, OR = 2.2, P = 1.9 × 10−8] were significantly associated with MN. No novel signals emerged in GWAS of X-chromosomal variants or in sex-specific analyses. Classical HLA alleles (DRB1*0301-DQA1*0501-DQB1*0201 haplotype) were associated with MN but provided little additional information beyond rs9272729. Associations were replicated in 137 GCKD patients with MN (HLA-DQA1: P = 6.4 × 10−24; PLA2R1: P = 5.0 × 10−4). MN risk increased steeply for patients with high-risk genotype combinations (OR > 79). While genetic variation in PLA2R1 exclusively associated with MN across 19 CKD aetiologies, the HLA-DQA1 risk allele was also associated with lupus nephritis (P = 2.8 × 10−6), type 1 diabetic nephropathy (P = 6.9 × 10−5) and focal segmental glomerulosclerosis (P = 5.1 × 10−5), but not with immunoglobulin A nephropathy.
Conclusions: PLA2R1 and HLA-DQA1 are the predominant risk loci for MN detected by GWAS. While HLA-DQA1 risk variants show an association with other CKD aetiologies, PLA2R1 variants are specific to MN.
Keywords: chronic kidney disease, genome-wide association study, membranous nephropathy
INTRODUCTION
Membranous nephropathy (MN) is one of the most common causes of nephrotic syndrome in adults [1, 2]. It is considered an immune complex–mediated disease, with subepithelial deposits of immune complexes detectable using immunofluorescence or electron microscopy. In some patients, MN is secondary and consequential to tumours, infections or exposure to environmental factors. The majority of patients, however, suffer from so-called primary MN, termed ‘idiopathic’ MN, until its recent recognition as an autoimmune disease.
A genetic component to MN has long been suspected based on familial clustering [3]. The involvement of a risk allele at the human leucocyte antigen (HLA) locus, and more specifically of DQA1, had already been proposed in MN pathogenesis in the late 1980s [4]. Additional insights into MN pathophysiology from animal experiments showed that immune complex formation can result from interactions of antibodies with a local antigen on podocytes [1]. In humans, the M-type phospholipase A2 receptor protein at the podocyte surface was identified as the major autoantigen in patients with MN [5]. More recently, thrombospondin type-1 domain-containing 7A (THSD7A) was identified as another autoantigen in phospholipase A2 receptor antibody–negative patients with MN [6].
Genome-wide association studies (GWAS) can be used to evaluate the association between genome-wide genetic markers and a disease in a comprehensive and unbiased way. The first meta-analysis of a GWAS of MN identified strong associations between MN and single nucleotide polymorphisms (SNPs) at the HLA-DQA1 locus and in PLA2R1, encoding the M-type phospholipase A2 receptor [7]. This study not only provided evidence for the influence of an individual's genetic make-up in shaping the interaction between the adaptive immune system and autoantigens, but also provided striking evidence that the combination of risk alleles at only two genetic loci was associated with large increases in disease risk of a magnitude that is typically only observed for single gene disorders.
However, previous GWAS of MN only evaluated a limited set of ∼300 000 genotyped genetic variants and did not use imputation methods [8, 9] to increase marker coverage or to obtain information on classical HLA alleles [10]. Genotype imputation can increase the statistical power to detect genetic associations [9]. Imputation to the 1000 Genomes reference panel in previous studies has led to the identification of additional risk loci that were missed in previous association studies of at least equal size, e.g. for body mass index, fasting glucose and coronary artery disease [11, 12]. In addition, GWAS conditioning on the associated index variants can lead to the identification of secondary independent signals in associated regions [13]. Moreover, although MN shows a male predilection [1], previous studies have combined both sexes to evaluate genetic associations and only studied autosomes. Finally, whether the previously identified genetic risk variants in the HLA locus and PLA2R1 confer risk that is specific to MN or if they also increase the risk for other aetiologies of chronic kidney disease (CKD) has not been studied systematically. Therefore, the present study aimed to address these important gaps.
MATERIALS AND METHODS
For the GWAS of MN, DNA samples from 336 British MN cases who provided written informed consent were collected by the MRC/Kidney Research UK National DNA Bank for Glomerulonephritis and racially matched to samples from 349 control subjects of the 1958 UK birth control study (http://www.b58cgene.sgul.ac.uk). Study protocol and sample characteristics have been described previously [7]. On average, MN patients were 52.5 (±13.3) years old at the time of diagnosis and 69% of them were men.
For replication and characterization, we examined patients taking part in the German Chronic Kidney Disease (GCKD) Study, a prospective observational study of patients with CKD with either an estimated glomerular filtration rate (eGFR) between 30 and 60 mL/min/1.73 m² or overt proteinuria (albumin:creatinine ratio >300 mg/g or protein:creatinine ratio >500 mg/g) upon study inclusion [14]. The study was approved by local review boards at each participating academic institution and is described in detail elsewhere [15]. Briefly, 5217 patients who gave their written informed consent were enrolled into the study between 2010 and 2012. The patients' long-term treating nephrologists were asked to select the leading, i.e. primary, cause of CKD for each patient. For disease categories in which biopsy was not performed for the majority of patients, such as diabetic or hypertensive kidney disease, diagnosis was established by the nephrologists based on clinical grounds. The assignment of these causes was consistent with comorbidities reported by the patients as well as with the presence of elevated biomarkers at the study visit (elevated haemoglobin A1c and blood pressure) and the intake of antidiabetic and anti-hypertensive medication, respectively. All patients were of Caucasian ancestry and 60% were men. At the time of entry, patients were on average 60.0 (±12.0) years old [16]. MN as the leading cause of CKD was identified in 151 of 5217 patients, 144 of whom had a biopsy to confirm the diagnosis (80% men). In addition, other specific leading causes of CKD that comprised at least 40 patients were evaluated, as well as one smaller group of patients with rapidly progressive glomerulonephritis (GN) (pauci-immune; n = 28). Analyses were restricted to patients with a biopsy-proven diagnosis when the proportion of patients with biopsies in a given group was >70%. Information on steroid sensitivity of focal segmental glomerulosclerosis (FSGS) was not available. As a control group, genotypes from 379 individuals (47% men) comprising the European subgroup of the 1000 Genomes project were used (http://www.1000genomes.org/home; Phase 1, release v3, panel EUR). Data were downloaded from http://www.sph.umich.edu/csg/abecasis/MaCH/download/1000G.2012-03-14.html and genotypes for selected SNPs were subsequently extracted.
Because of the complexity, detailed information on genotyping, quality control and imputation as well as on statistical analyses can be found in the Supplementary data, Methods). In brief, GWAS samples were genotyped using the HumanCNV370-Quad SNP chip or HumanHap300 SNP chip. After stringent quality control using Plink and Eigensoft, the cleaned data set comprised 282 462 SNPs of 323 cases and 345 controls [17, 18]. Subsequently, genotype imputation was carried out using Shapeit and Impute2 [19, 20]. Data from the 1000 Genomes project (Phase 1, release v3, panel ALL) served as a reference panel. The final data set contained 8.9 million SNPs of high quality. In the GCKD study, genotyping of index variants was carried out using an Agena (formerly Sequenom) iPLEX assay (PLA2R1) and using a Life Technologies TaqMan assay (HLA-DQA1) with call rates of 99% and genotype concordance of 100% among duplicate samples.
Association tests relating MN case status to autosomal genetic variants were carried out using genotype probabilities (dosages) and an additive genetic model with the software SNPtest version 2.4.1 [21]. The first five principle components were included as covariates. Statistical significance was set to the standard threshold of 5.0 × 10−8. Independent replication was defined as a one sided P-value <0.05. Inverse-variance weighted, fixed-effect meta-analysis was conducted to pool results.
In GCKD, genotype information was also evaluated for association with CKD of 17 additional specific aetiologies as well as using all GCKD participants as a CKD case group, all in comparison with the 1000 Genomes control group. The statistical significance cut-off for these analyses was set to 2.6 × 10−3.
RESULTS
GWAS of MN
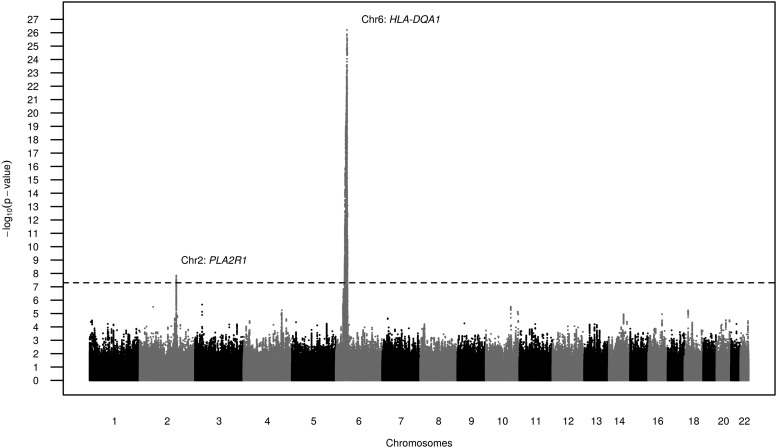
We carried out a GWAS among 323 MN cases and 345 controls of European ancestry using imputed genotypes as illustrated in the detailed analysis plan in Supplementary data, Figure S1. GWAS results did not show any evidence for inflation of the test statistics, indicating no systematic biases (quantile–quantile plot, Supplementary data, Figure S2). The two previously reported genetic regions, PLA2R1 and HLA-DQA1 [7], contained SNPs associated with MN at genome-wide significance (Figure 1). The index SNPs at the two loci used in subsequent analyses were rs9272729 in HLA-DQA1 (P = 5.9 × 10−27) and rs17830558 in PLA2R1 (P = 1.9 × 10−8, Table 1). In both instances, the index SNPs were imputed with high imputation quality and were correlated with the previously published SNPs (Table 1). Supplementary data, Figure S3A and B display the two associated regions in more detail. The previously described extensive linkage disequilibrium in the HLA region is evident, extending several hundred kilobases around HLA-DQA1 and including other HLA genes such as HLA-DQB1, HLA-DRB1, HLA-DRB6 and HLA-DRB5.
FIGURE 1.
Results from the GWAS of MN using imputed genotype data (GCKD-corrected P-values, filtered by MAF > 10%).
Table 1.
Replicated SNPs associated with MN at genome-wide significance
| Gene |
||
|---|---|---|
| PLA2R1 | HLA-DQA1 | |
| SNP ID | rs17830558a | rs9272729b |
| chr:pos (hg19) | chr2:160878364 | chr6:32609594 |
| Function | Intronic | Intronic |
| Imputed (quality) | Yes (0.938) | Yes (0.901) |
| A1/A2 | T/G | A/G |
| Discovery analysis | ||
| Allele frequency A1 | 0.54 | 0.24 |
| OR | 2.16 | 7.29 |
| 95% CIc | 1.64–2.85 | 4.82–11.04 |
| P-valuec | 1.9 × 10−8 | 5.9 × 10−27 |
| Replication analysis | ||
| Allele frequency A1 | 0.49 | 0.16 |
| OR | 1.63 | 6.86 |
| 95% CI | 1.23–2.15 | 4.55–10.34 |
| P-value | 5.0 × 10−4 | 6.4 × 10−24 |
| Meta-analysis | ||
| Allele frequency A1 | 0.52 | 0.20 |
| OR | 1.87 | 7.07 |
| 95% CI | 1.54–2.28 | 5.28–9.47 |
| P-value | 3.7 × 10−10 | 2.1 × 10−39 |
OR refers to A1.
aSNP rs17830558 was used in all subsequent analyses of replication and meta-analysis as proxy for rs17830307, which showed the lowest P-value in the association analysis (results can be found in Supplementary data, Table S1).
bIn GCKD, of the proxy rs2187668 was genotyped instead (pairwise correlations within our data) of rs9272729 and previously reported rs2187668 (r2 = 0.87) and of rs17830558 and previously reported rs4664308 (r2 = 0.40).
cGCKD-corrected values.
No additional loci on the autosomes or the X chromosome contained SNPs associated with MN at genome-wide significance. Supplementary data, Table S1 contains all SNPs on autosomes and chromosome X with minor allele frequency (MAF) >5% and corrected P < 10−5.
Secondary analyses
To further examine the genetic architecture of MN, several secondary analyses were carried out: first, additional GWAS were conducted separately for men (222 cases and 106 controls) and women (101 cases and 239 controls). These analyses were carried out because of the strong observed male predilection for MN, although the smaller sample sizes limit statistical power. No additional signals of genome-wide significance were identified in either men or women (data not shown). Second, we evaluated whether a recessive model instead of an additive model, which is typically used in discovery GWAS, better fit the data. Consistent with lower statistical power of a recessive model, P-values were larger than those obtained from the additive model for both PLA2R1 and HLA-DQA1. No other genomic regions contained significantly associated markers (data not shown).
We evaluated the identified regions containing PLA2R1 and HLA-DQA1 more closely in order to assess the presence of more than one independent signal of genome-wide significance. After conditioning on the index SNP of the corresponding region, no additional SNP across the respective chromosome was associated with MN at genome-wide significance. Thus, based on the variants assessed in this study, there was no evidence of multiple common independent variants in the known regions that contributed to the observed associations at genome-wide significance.
Finally, a very recent publication by Gbadegesin et al. [22] reported on the association of an HLA-DQA1 risk allele and steroid-sensitive nephrotic syndrome (SSNS) in children. We, therefore, assessed whether the reported index variant, rs1129740, was also associated with MN in our genome-wide data. The reported risk allele for SSNS, the major A allele, was significantly associated with MN risk in our data {odds ratio [OR] = 2.48 [95% confidence interval (CI) 1.81–3.40], P = 4.45 × 10−9}. The OR for MN was similar to the one published for SSNS. The SSNS index variant rs1129740 was correlated with the MN index SNP evaluated in this study, rs9272729 (r2 = 0.19, D′ = 1, based on the MN GWAS data). Besides MN, the SSNS risk allele at rs1129740 was also nominally associated with minimal change disease [OR = 1.64 (95% CI 1.09–2.45), P = 0.014] and FSGS [OR = 1.52 (95% CI 1.16–1.98), P = 0.002] in the GCKD study.
Replication and interactions
Replication of SNP associations and meta-analysis
We assessed evidence for replication of the association at the index variants in PLA2R1 and HLA-DQA1 in 137 participants of the GCKD study with biopsy-proven MN and 379 ethnically matched controls (see Materials and Methods). Both index SNPs were associated with MN after correction for multiple testing in this replication sample (Table 1; P = 5.0 × 10−4 for PLA2R1 and 6.4 × 10−24 for HLA-DQA1). The ORs for MN after meta-analysis of the discovery and replication samples were 1.87 (95% CI 1.54–2.28) for PLA2R1 and 7.07 (95% CI 5.28–9.47) for HLA-DQA1 for each additional copy of a risk allele.
Combined allelic effects and interactions
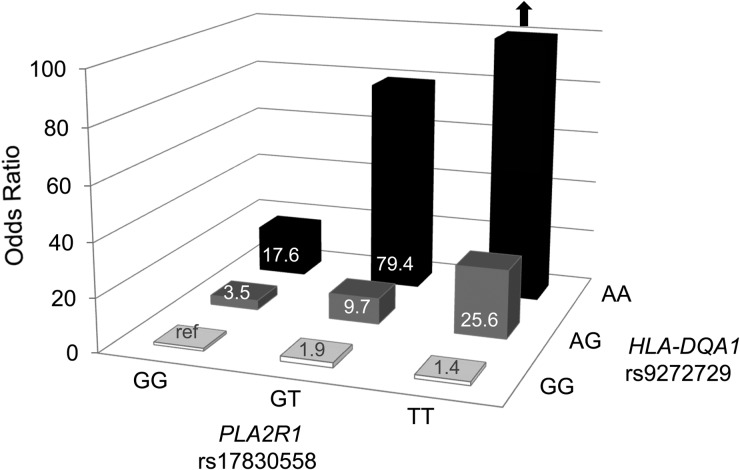
Figure 2 presents the association with MN for the combination of risk alleles at PLA2R1 and HLA-DQA1. The risk for MN increased dramatically with a higher number of risk alleles: almost all persons carrying two risk alleles at HLA-DQA1 were affected with MN [14 of 16 (88%)] and all three individuals who carried two risk alleles at both loci were affected with MN (so that no risk estimates could be provided; see arrow in Figure 2). The second-highest risk of MN was observed for individuals carrying two risk alleles at HLA-DQA1 and one risk allele at PLA2R1. Compared with individuals who did not carry any risk alleles, their MN risk was increased almost 80-fold (OR = 79.4, P = 7.1 × 10−5). Supplementary data, Table S2 displays case and control numbers for all risk categories and, to obtain more stable numerical estimates, the corresponding results when combining individuals carrying one or two risk alleles at the HLA-DQA1 locus into a single category.
FIGURE 2.
Combined effect of genetic risk variants at replicated loci on risk of MN in the GCKD study. See Supplementary data, Table S2 for tabular presentation of the results.
Evidence for statistical interaction between the two index variants at PLA2R1 and HLA-DQA1 was assessed by simultaneous inclusion of the variants as well as of an interaction term into the regression model (see Materials and Methods). Compared with a model containing both variants but no interaction term, the inclusion of the interaction term lead to a decrease of Akaike's information criterion (AIC) from 491.9 to 487.2, which was statistically significant (likelihood ratio test P-value = 0.009). The OR of the interaction term in this model was 2.20 (P = 0.01), which together with the lower AIC supports the finding that the combination of risk alleles increases MN risk to a greater degree than expected based on the effect of the individual risk alleles at the two loci by themselves.
Characterization across CKD aetiologies reveals shared associations at HLA-DQA1
Subsequently we evaluated whether the association at PLA2R1 and HLA-DQA1 is specific to MN or whether it extends to additional aetiologies of CKD in the large GCKD study, which collected information on the underlying cause of CKD (see Materials and Methods). Table 2 shows the association of the two index variants with CKD defined based on eGFR and/or albuminuria, as well as with 17 specific aetiologies of CKD. PLA2R1 was only associated with MN but no other CKD aetiology after correction for multiple testing. Conversely, a significant association of the HLA-DQA1 risk variant was observed not only with MN, but also with lupus nephritis (P = 2.8 × 10−6), CKD in type 1 diabetes mellitus (P = 6.9 × 10−5) and FSGS (P = 5.1 × 10−5). The association with FSGS remained significant even after the exclusion of eight patients with HIV and hepatitis, potential causes of secondary FSGS (P = 2.3 × 10−5). The observed association with CKD (P = 1.3 × 10−3) was mainly due to the inclusion of patients with MN, lupus nephritis, type 1 diabetes mellitus and FSGS.
Table 2.
Association of replicated SNPs with different CKD aetiologies in the GCKD study
| CKD aetiology (leading cause) | n | % biopsy proven | rs17830558 (PLA2R1) |
rs9272729 (HLA-DQA1)a |
||||
|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | P-value | OR | 95% CI | P-value | |||
| MNb | 137 | 100 | 1.63 | 1.23–2.15 | 5.0 × 10−4 | 6.86 | 4.55–10.34 | 6.4 × 10−24 |
| Lupus nephritisb | 104 | 100 | 1.10 | 0.82–1.47 | 5.2 × 10−1 | 3.00 | 1.90–4.72 | 2.8 × 10−6 |
| Diabetes mellitus, type 1 | 89 | 3.4 | 0.76 | 0.55–1.05 | 9.2 × 10−2 | 2.69 | 1.67–4.34 | 6.9 × 10−5 |
| FSGSb | 139 | 100 | 0.87 | 0.66–1.14 | 3.1 × 10−1 | 2.36 | 1.56–3.57 | 5.1 × 10−5 |
| Membranoproliferative GNb | 36 | 100 | 1.36 | 0.85–2.19 | 1.9 × 10−1 | 2.15 | 1.10–4.22 | 3.5 × 10−2 |
| Minimal change GNb | 53 | 100 | 1.30 | 0.87–1.94 | 2.0 × 10−1 | 1.70 | 0.90–3.22 | 1.2 × 10−1 |
| Interstitial nephritis | 135 | 19.3 | 0.98 | 0.75–1.29 | 9.1 × 10−1 | 1.56 | 1.00–2.43 | 5.3 × 10−2 |
| Nephrosclerosis | 1087 | 7.9 | 0.97 | 0.82–1.15 | 7.4 × 10−1 | 1.47 | 1.10–1.96 | 7.8 × 10−3 |
| Diabetes mellitus, type 2 | 649 | 4.2 | 0.89 | 0.74–1.07 | 2.0 × 10−1 | 1.38 | 1.02–1.87 | 3.3 × 10−2 |
| Renal artery stenosis | 47 | 0 | 1.32 | 0.87–2.00 | 1.9 × 10−1 | 1.29 | 0.64–2.62 | 4.9 × 10−1 |
| Wegener's granulomatosisb | 77 | 100 | 0.93 | 0.67–1.31 | 6.9 × 10−1 | 1.18 | 0.66–2.13 | 5.8 × 10−1 |
| IgA nephropathyb | 308 | 100 | 1.02 | 0.83–1.25 | 8.6 × 10−1 | 1.14 | 0.79–1.63 | 4.8 × 10−1 |
| Autosomal dominant polycystic kidney disease | 170 | 1.8 | 1.07 | 0.83–1.37 | 6.2 × 10−1 | 1.13 | 0.73–1.77 | 5.8 × 10−1 |
| Tumour nephrectomy | 61 | 11.5 | 0.91 | 0.63–1.32 | 6.2 × 10−1 | 1.07 | 0.55–2.08 | 8.4 × 10−1 |
| Analgesic nephropathy | 52 | 3.8 | 1.12 | 0.76–1.67 | 5.6 × 10−1 | 1.03 | 0.49–2.16 | 9.4 × 10−1 |
| Acute kidney injury | 59 | 10.2 | 0.90 | 0.61–1.32 | 5.9 × 10−1 | 1.02 | 0.50–2.08 | 9.5 × 10−1 |
| Micropolyangiitisb | 54 | 100 | 0.82 | 0.55–1.22 | 3.3 × 10−1 | 0.52 | 0.20–1.33 | 1.4 × 10−1 |
| Rapidly progressive GN, pauci-immuneb | 25 | 100 | 0.72 | 0.41–1.29 | 2.7 × 10−1 | 0.45 | 0.11–1.88 | 2.2 × 10−1 |
| KDIGO-defined CKD (all GCKD participants) | 4960 | 25.8 | 0.99 | 0.86–1.15 | 9.1 × 10−1 | 1.50 | 1.16–1.94 | 1.3 × 10−3 |
P-values that are statistically significant after correction for multiple testing are in bold type (P-value < 2.6 × 10−3 = 0.05/19).
aIn GCKD, the proxy SNP rs2187668 was genotyped.
bAnalysis based on patients with biopsy-proven (leading) cause only because the proportion of available biopsy results was >70%.
Associations with classical HLA alleles
Lastly, we attempted to assess whether MN showed stronger associations with carrier status of classical HLA alleles than with the index variant identified in the GWAS, and thus imputed classical HLA alleles from the genotyped SNPs in the discovery sample (see Materials and Methods). MN was significantly associated with seven of the classical HLA alleles at genome-wide significance (Table 3). Whereas the lowest P-value was observed for HLA-DRB1*0301 carrier status (OR = 6.15, P = 1.6 × 10−28), HLA-DQA1*0501 conferred the largest risk (OR = 6.23, P = 1.4 × 10−27). Carrier status at these two HLA alleles was almost perfectly correlated (Spearman r = 0.995), indicating a long shared risk haplotype, to which HLA-DQB1*0201 also belongs (Spearman correlation with HLA-DQA1*0501 r = 1). Supplementary data, Table S3 presents the associations of all imputed common HLA alleles (MAF >5%) with MN.
Table 3.
Association of imputed classical HLA alleles with MN
| HLA allele | MAF | OR | 95% CI | P-value |
|---|---|---|---|---|
| DRB1-0301 | 0.27 | 6.15 | 4.27–8.85 | 1.6 × 10−28 |
| DQA1-0501 | 0.30 | 6.23 | 4.29–9.04 | 1.4 × 10−27 |
| DQB1-0201 | 0.27 | 5.89 | 4.11–8.45 | 1.5 × 10−27 |
| B-0801 | 0.24 | 4.47 | 3.16–6.33 | 7.5 × 10−20 |
| DRB3-0100 | 0.26 | 3.98 | 2.83–5.59 | 1.3 × 10−17 |
| DRB3-9901 | 0.47 | 0.29 | 0.21–0.39 | 1.6 × 10−17 |
| C-0701 | 0.28 | 2.94 | 2.14–4.03 | 2.4 × 10−12 |
Using the classical HLA allele HLA-DQA1*0501 as a proxy for this risk haplotype, we assessed whether the index SNP at HLA-DQA1 in the discovery GWAS, rs9272729, provided independent information. Individually, the association of rs9272729 with MN was of similar magnitude as the association of HLA-DQA1*0501 with MN: OR = 6.51 (P = 7.16 × 10−19) for the SNP versus OR = 5.87 (P = 1.25 × 10−18) for the HLA allele among individuals with both types of information available. Upon simultaneous inclusion of rs9272729 and HLA-DQA1*0501 into the model, the effects of both variables were attenuated but still significant. There was a significant improvement of model fit (likelihood ratio test P = 0.02), suggesting that the SNP captures most of the information but other variants in the genomic region and/or classical HLA alleles may still add some additional information.
DISCUSSION
In this first GWAS of MN using imputed genotypes, we confirm the predominant importance of risk alleles at PLA2R1 and HLA-DQA1 in modulating MN risk. With the available number of cases, we did not find evidence for additional or independent common risk alleles or of sex-specific risk loci. While we found that genetic variation in PLA2R1 exclusively associated with MN across a wide range of CKD aetiologies studied, the risk allele at HLA-DQA1 was also associated with higher odds of a number of other autoimmunity-associated kidney diseases—lupus nephritis, type 1 diabetic nephropathy and FSGS—but not with immunoglobulin A (IgA) nephropathy.
In accordance with other studies [23], our results from the GCKD study confirmed the association between MN and previously reported risk variants or their proxies in PLA2R1 and HLA-DQA1. In addition, in the GWAS of high coverage, we did not identify additional common risk variants of similar importance. While imputation to the 1000 Genomes reference panel has led to the identification of additional risk loci missed in earlier studies of similar size for a variety of traits and diseases [11, 12], this was not the case for MN. Our study, therefore, highlights the oligogenic architecture of MN, informs future studies to detect genes encoding for additional antigens and confirms PLA2R1 as a risk locus of major and specific importance. This is consistent with the presence of anti-PLA2R1 antibodies in ∼70–80% of MN patients [1]. In the independent GCKD study, we also confirmed the large increase in MN risk conferred by the combination of risk variants at PLA2R1 and HLA-DQA1, as well as a statistical interaction between the risk variants at both loci [24]. In fact, all GCKD patients who were homozygous for the risk alleles at both loci were affected with MN, as were the great majority of persons homozygous for the risk allele at HLA-DQA1 and heterozygous at PLA2R1. There are several potential explanations for the higher proportion of MN patients among carriers of high-risk genotype combinations in the GCKD study compared with the initial study [7], including random fluctuations due to small numbers, differences in the prevalence of contributing environmental triggers and recruitment of patients under nephrological care with reduced eGFR into the GCKD study, which may increased the number of MN patients with advanced disease and high-risk genotypes. Only long-term prospective studies, if not lifetime studies, will be able to answer how many individuals carrying high-risk genotype combinations will not develop MN over their life course. Likewise, conclusive answers relating to long-term outcomes require standardized observational studies that follow patients for at least 10 years or until death while collecting information on immunosuppressive treatments and other comorbidities.
Imputation of classical HLA alleles revealed the strongest associations with HLA-DQA1*0501, HLA-DRB1*0301 and HLA-DQB1*0201. The almost perfect correlation between these alleles implicates one long shared MN risk haplotype on which these alleles reside, DRB1*0301-DQA1*0501-DQB1*0201. This is supported by the reported finding that circulating anti-PLA2R1-antibody levels in MN cases are linked to the number of alleles of DQA1*0501 and DQB1*0201 [25]. Therefore, although we refer to the HLA risk allele as the HLA-DQA1 locus, the underlying causal variant could also be situated in HLA-DQB1 or HLA-DRB1. Given the large inter-individual genetic variability of the HLA region, it is noteworthy that our analyses suggest that most of the strong association with MN at this locus can be captured by knowledge of the genotype at a single common SNP in the region, which may have clinical implications for risk prediction. Our observations are consistent with a relatively common genetic predisposition that controls antigen processing and/or presentation, that only a combination of additional factors, such as the presence of specific auto antigen-processed peptides from, for example, specific forms of PLA2R1 for MN—and/or certain environmental triggers can give rise to a specific autoantibody to cause disease [7]. In support of this, the DRB1*0301-DQA1*0501-DQB1*0201 haplotype has also been described to increase the risk for other common autoimmune diseases, including type 1 diabetes [26], systemic lupus erythematosus (SLE) [27], coeliac disease [28] and thyroid disease [29].
Our data indicate that this observation may extend to SSNS. As SSNS is commonly observed in children, it was not among the aetiologies that could be evaluated in the GCKD study. However, the association of the reported SSNS risk allele with MN risk in our GWAS and the high correlation of the risk allele of the reported SSNS variant and the risk allele of the MN index variant in our data suggest that the genetic predisposition at the HLA-DQA1 locus shares at least some degree of overlap between SSNS and MN. The results from the GCKD study further suggest that the shared susceptibility between the SSNS variant extends to an association with minimal change disease and FSGS in adulthood. In contrast, there was no statistically significant association between the HLA-DQA1 risk allele for MN and IgA nephropathy, another autoimmune kidney disease. This is consistent with the fact that previous GWAS of IgA nephropathy have highlighted other associated alleles in the HLA-DQA1 region [30].
While genetic variants in the HLA-DQA1 locus were associated with a variety of underlying causes of CKD, those in PLA2R1 uniquely associated with MN across almost 20 different aetiologies of CKD. The genetic associations between HLA risk alleles and nephropathy resulting from SLE or type 1 diabetes are biologically plausible, given the shared genetic susceptibility to these autoimmune diseases. Because of the case-only design of the GCKD study, we could not specifically assess whether the HLA risk allele increases the risk for nephropathy in the setting of type 1 diabetes and SLE, or whether the observed associations are due to an association with the primary cause of the respective disease. The mechanisms underlying the observed association with FSGS need further study to delineate whether they are indicative of a common mechanism resulting in FSGS-defining features in the biopsies or whether some of the cases in whom the primary CKD aetiology was assigned as FSGS may in fact have additional specific causes contributing to their nephropathy, such as SLE. Although not significant after correction for multiple testing, the risk allele at HLA-DQA1 also more than doubled the odds of membranoproliferative GN. This could be indicative of an autoimmune component specific to membranoproliferative GN or represent cases of secondary membranoproliferative GN such as those observed in SLE. Our approach highlights the power of collecting detailed information on kidney disease phenotypes, as the specific evaluation of associations across disease subtypes can reveal significant associations that would be missed in GWAS of these CKD aetiologies. A similar observation has been made for the APOL1 risk alleles, where associations with FSGS and nondiabetic end-stage renal disease—originally identified using genome-wide approaches—were shown to extend to HIV-associated nephropathy without conducting a separate genome-wide genetic screen [31, 32].
Strengths of our study include the availability of high-coverage genome-wide SNP data as well as the variety of CKD aetiologies in the GCKD study that allowed us to evaluate the presence of novel associations, carefully characterize known associations and identify a shared genetic predisposition to several autoimmune kidney diseases. Our study also has potential limitations: genotypes were generated on different platforms, which we addressed through careful data cleaning and combined imputation based on a common SNP set. Although—for a rare disease such as MN—we studied a sizeable number of cases, our study is still underpowered to identify associations with rare alleles or alleles conferring moderate or small increases in risk. For example, we did not find evidence for an association between the THSD7A variant and MN, which may not be surprising given the fact that this autoantigen has only been described in a minority of patients. In addition, because we evaluated chip-based and imputed genotypes, there may be rare risk variants that can only be discovered through sequencing-based approaches. At least for PLA2R1, however, targeted re-sequencing of the exons and canonical splice sites did not yield rare variants that could explain the observed association [33]. In our study of prevalent MN cases, we did not have information on PLA2R1 antibody concentrations and could, therefore, not investigate antibody-negative patients separately. In any case, the statistical power to detect novel associated loci among antibody-negative patients in the current study, assuming 20% antibody-negative patients among the cases, would have been <30% even for very common alleles. The assembly of larger collectives of MN patients with antibody-status and genome-wide genotypes is, therefore, an important future endeavour. Likewise, larger collectives of patients with other primary forms of specific CKD aetiologies such as FSGS are needed to confirm our findings or to establish statistical significance such as with membranoproliferative GN. Although in the GCKD study the patients' treating nephrologists were asked to identify the leading cause of CKD, misclassification can occur, especially for the leading causes of CKD that are typically not confirmed by kidney biopsy or for which no specific diagnostic criteria are available. Finally, as in all association studies, we cannot directly pinpoint causal variants, but associated variants may still be valuable for risk stratification.
In conclusion, PLA2R1 and HLA-DQA1 are the predominant risk loci for MN. Common variants in these two loci give rise to rare high-risk genotype combinations, which associate with disease risk of a magnitude otherwise observed for single-gene disorders. The risk allele at HLA-DQA1, but not PLA2R1, is also associated with nephropathy attributed to type 1 diabetes, SLE and FSGS, but not with IgA nephropathy, another autoimmune kidney disease. Future studies including those in PLA2R1 antibody–negative patients and sequence-based association studies should evaluate the presence of additional genetic risk variants for MN.
SUPPLEMENTARY DATA
Supplementary data are available online at http://ndt.oxfordjournals.org.
Supplementary Material
ACKNOWLEDGEMENTS
P.S. was supported by the BMBF (Gerontosys II NephAge Project, 031 5896 A). The German Research Foundation funded the work of P.S., Y.L., M.W. and A.K. through the Emmy Noether Programme (KO 3598/2-1 to A.K.). The work of M.W., G.W. and A.K. was additionally supported by the German Research Foundation through CRC/SFB 1140.
Work on the British study was supported in part by grants from the David and Elaine Potter Charitable Foundation (to S.H.P. and R.K.); the European Union, FP7 (EURenomics grant agreement 2013-305608; to H.C.S., D.B., S.H.P., P.B. and R.K.); St Peter's Trust for Kidney, Bladder and Prostate Research (to D.B., S.H.P. and R.K.); Kids Kidney Research UK (to D.B. and R.K.); Medical Research Council (MRC) at the Centre for Integrated Genomic Medical Research, University of Manchester; grants from the MRC (G0000934) and the Wellcome Trust (068545/Z/02); Manchester Academic Health Science Centre (MAHSC 186/200) and MRC project grant MR/J010847/1 (to P.B.). We thank the MRC and Kidney Research UK, as part of the MRC/Kidney Research UK National DNA Bank for Glomerulonephritis, for collection of the British idiopathic membranous nephropathy DNA samples.
The GCKD study is funded by grants from the German Ministry of Education and Research (BMBF, grant number 01ER0804) and the KfH Foundation for Preventive Medicine. We are grateful for the willingness of the patients to participate in the GCKD study. The enormous effort of the study personnel of the various regional centres is highly appreciated. We thank the large number of nephrologists who provide routine care for the patients and collaborate with the GCKD study. A list of nephrologists currently collaborating with the GCKD study is available at http://www.gckd.org.
GCKD Investigators are as follows:
University of Erlangen-Nürnberg, Germany: Kai-Uwe Eckardt, Stephanie Titze, Hans-Ulrich Prokosch, Barbara Bärthlein, André Reis, Arif B. Ekici, Olaf Gefeller, Karl F. Hilgers, Silvia Hübner, Susanne Avendaño, Dinah Becker-Grosspitsch, Nina Hauck, Susanne A. Seuchter, Birgit Hausknecht, Marion Rittmeier, Anke Weigel, Andreas Beck, Thomas Ganslandt, Sabine Knispel, Thomas Dressel and Martina Malzer.
Technical University of Aachen, Germany: Jürgen Floege, Frank Eitner, Georg Schlieper, Katharina Findeisen, Elfriede Arweiler, Sabine Ernst, Mario Unger and Stefan Lipski.
Charité, Humboldt-University of Berlin, Germany: Elke Schaeffner, Seema Baid-Agrawal, Kerstin Petzold and Ralf Schindler.
University of Freiburg, Germany: Anna Köttgen, Ulla Schultheiss, Simone Meder, Erna Mitsch, Ursula Reinhard and Gerd Walz.
Hannover Medical School, Germany: Hermann Haller, Johan Lorenzen, Jan T. Kielstein and Petra Otto.
University of Heidelberg, Germany: Claudia Sommerer, Claudia Föllinger and Martin Zeier.
University of Jena, Germany: Gunter Wolf, Martin Busch, Katharina Paul and Lisett Dittrich.
Ludwig-Maximilians University of München, Germany: Thomas Sitter, Robert Hilge and Claudia Blank.
University of Würzburg, Germany: Christoph Wanner, Vera Krane, Daniel Schmiedeke, Sebastian Toncar, Daniela Cavitt, Karina Schönowsky and Antje Börner-Klein.
Medical University of Innsbruck, Austria: Florian Kronenberg, Julia Raschenberger, Barbara Kollerits, Lukas Forer, Sebastian Schönherr and Hansi Weißensteiner.
University of Regensburg, Germany: Peter Oefner, Wolfram Gronwald and Helena Zacharias.
Department of Medical Biometry, Informatics and Epidemiology (IMBIE), University of Bonn: Matthias Schmid.
CONFLICT OF INTEREST STATEMENT
None declared.
REFERENCES
- 1. Beck LH Jr, Salant DJ. Membranous nephropathy: from models to man. J Clin Invest 2014; 124: 2307–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Debiec H, Ronco P. Immunopathogenesis of membranous nephropathy: an update. Semin Immunopathol 2014; 36: 381–397 [DOI] [PubMed] [Google Scholar]
- 3. Short CD, Feehally J, Gokal R et al. Familial membranous nephropathy. Br Med J 1984; 289: 1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vaughan RW, Demaine AG, Welsh KI. A DQA1 allele is strongly associated with idiopathic membranous nephropathy. Tissue Antigens 1989; 34: 261–269 [DOI] [PubMed] [Google Scholar]
- 5. Beck LH Jr, Bonegio RG, Lambeau G et al. M-type phospholipase A2 receptor as target antigen in idiopathic membranous nephropathy. N Engl J Med 2009; 361: 11–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tomas NM, Beck LH Jr, Meyer-Schwesinger C et al. Thrombospondin type-1 domain-containing 7A in idiopathic membranous nephropathy. N Engl J Med 2014; 371: 2277–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stanescu HC, Arcos-Burgos M, Medlar A et al. Risk HLA-DQA1 and PLA2R1 alleles in idiopathic membranous nephropathy. N Engl J Med 2011; 364: 616–626 [DOI] [PubMed] [Google Scholar]
- 8. Leslie S, Donnelly P, McVean G. A statistical method for predicting classical HLA alleles from SNP data. Am J Hum Genet 2008; 82: 48–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet 2010; 11: 499–511 [DOI] [PubMed] [Google Scholar]
- 10. Fernando MM, Vyse TJ. Risk alleles in idiopathic membranous nephropathy. N Engl J Med 2011; 364: 2072; author reply 2073–2074 [DOI] [PubMed] [Google Scholar]
- 11. Horikoshi M, Mgi R, van de Bunt M et al. Discovery and fine-mapping of glycaemic and obesity-related trait loci using high-density imputation. PLoS Genet 2015; 11: e1005230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Consortium CAD. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet 2015; 47: 1121–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kiryluk K. Risk alleles in idiopathic membranous nephropathy. N Engl J Med 2011; 364: 2072–2073; author reply 2073–2074 [DOI] [PubMed] [Google Scholar]
- 14. Kidney Disease: Improving Global Outcomes (KDIGO) CKD Work Group. KDIGO 2012 clinical practice guideline for the evaluation and management of chronic kidney disease. Kidney Int Suppl 2013; 3: 1–150 [Google Scholar]
- 15. Eckardt KU, Barthlein B, Baid-Agrawal S et al. The German Chronic Kidney Disease (GCKD) study: design and methods. Nephrol Dial Transplant 2012; 27: 1454–1460 [DOI] [PubMed] [Google Scholar]
- 16. Titze S, Schmid M, Kottgen A et al. Disease burden and risk profile in referred patients with moderate chronic kidney disease: composition of the German Chronic Kidney Disease (GCKD) cohort. Nephrol Dial Transplant 2015; 30: 441–451 [DOI] [PubMed] [Google Scholar]
- 17. Purcell S, Neale B, Todd-Brown K et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Price AL, Patterson NJ, Plenge RM et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006; 38: 904–909 [DOI] [PubMed] [Google Scholar]
- 19. Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods 2012; 9: 179–181 [DOI] [PubMed] [Google Scholar]
- 20. Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009; 5: e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Marchini J, Howie B, Myers S et al. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 2007; 39: 906–913 [DOI] [PubMed] [Google Scholar]
- 22. Gbadegesin RA, Adeyemo A, Webb NJ et al. HLA-DQA1 and PLCG2 are candidate risk loci for childhood-onset steroid-sensitive nephrotic syndrome. J Am Soc Nephrol 2015; 26: 1701–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bullich G, Ballarin J, Oliver A et al. HLA-DQA1 and PLA2R1 polymorphisms and risk of idiopathic membranous nephropathy. Clin J Am Soc Nephrol 2014; 9: 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lv J, Hou W, Zhou X et al. Interaction between PLA2R1 and HLA-DQA1 variants associates with anti-PLA2R antibodies and membranous nephropathy. J Am Soc Nephrol 2013; 24: 1323–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kanigicherla D, Gummadova J, McKenzie EA et al. Anti-PLA2R antibodies measured by ELISA predict long-term outcome in a prevalent population of patients with idiopathic membranous nephropathy. Kidney Int 2013; 83: 940–948 [DOI] [PubMed] [Google Scholar]
- 26. Erlich H, Valdes AM, Noble J et al. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes 2008; 57: 1084–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fernando MM, Stevens CR, Sabeti PC et al. Identification of two independent risk factors for lupus within the MHC in United Kingdom families. PLoS Genet 2007; 3: e192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Spurkland A, Sollid LM, Ronningen KS et al. Susceptibility to develop celiac disease is primarily associated with HLA-DQ alleles. Hum Immunol 1990; 29: 157–165 [DOI] [PubMed] [Google Scholar]
- 29. Yanagawa T, Mangklabruks A, Chang YB et al. Human histocompatibility leukocyte antigen-DQA1*0501 allele associated with genetic susceptibility to Graves’ disease in a Caucasian population. J Clin Endocrinol Metab 1993; 76: 1569–1574 [DOI] [PubMed] [Google Scholar]
- 30. Gharavi AG, Kiryluk K, Choi M et al. Genome-wide association study identifies susceptibility loci for IgA nephropathy. Nat Genet 2011; 43: 321–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Behar DM, Kedem E, Rosset S et al. Absence of APOL1 risk variants protects against HIV-associated nephropathy in the Ethiopian population. Am J Nephrol 2011; 34: 452–459 [DOI] [PubMed] [Google Scholar]
- 32. Kopp JB, Nelson GW, Sampath K et al. APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol 2011; 22: 2129–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Coenen MJ, Hofstra JM, Debiec H et al. Phospholipase A2 receptor (PLA2R1) sequence variants in idiopathic membranous nephropathy. J Am Soc Nephrol 2013; 24: 677–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.