Abstract
Plasticizers with estrogenic activity, such as bisphenol A (BPA), have potential adverse health effects in humans. Due to mounting evidence of these health effects, BPA is being phased out and replaced by other bisphenol variants in “BPA-free” products. We have compared estrogenic activity of BPA with 6 bisphenol analogues [bisphenol S (BPS); bisphenol F (BPF); bisphenol AP (BPAP); bisphenol AF (BPAF); bisphenol Z (BPZ); bisphenol B (BPB)] in 3 human breast cancer cell lines. Estrogenicity was assessed (10−11–10−4 M) by cell growth in an estrogen receptor (ER)-mediated cell proliferation assay, and by the induction of estrogen response element-mediated transcription in a luciferase assay. BPAF was the most potent bisphenol, followed by BPB > BPZ ∼ BPA > BPF ∼ BPAP > BPS. The addition of ICI 182,780 antagonized the activation of ERs. Data mining of ToxCast high-throughput screening assays confirm our results but also show divergence in the sensitivities of the assays. Gene expression profiles were determined in MCF-7 cells by microarray analysis. The comparison of transcriptome profile alterations resulting from BPA alternatives with an ERα gene expression biomarker further indicates that all BPA alternatives act as ERα agonists in MCF-7 cells. These results were confirmed by Illumina-based RNA sequencing. In conclusion, BPA alternatives are not necessarily less estrogenic than BPA in human breast cancer cells. BPAF, BPB, and BPZ were more estrogenic than BPA. These findings point to the importance of better understanding the risk of adverse effects from exposure to BPA alternatives, including hormone-dependent breast cancer.
Keywords: bisphenol, toxicogenomics, endocrine, estrogens, cell culture
Plasticizers such as bisphenol A (BPA) have been reported to have potential adverse health effects in humans, including reproductive endocrine disorders and neurobehavioral problems (Rochester, 2013). BPA is one of the best-studied endocrine disrupting chemicals (EDCs), with more than 75 out of 91 published studies showing associations between BPA exposure and adverse human health effects as of May 2013 (Rochester, 2013). The primary BPA mode of action is the activation of estrogen receptor (ER)-mediated transcription (Shioda et al., 2013). Biomonitoring studies suggest that the urine from a majority of individuals in industrialized countries contain measurable BPA and metabolites (Gerona et al., 2016). BPA has been detected in breast milk (Nakao et al., 2015). The handling of a thermal receipt paper before eating French fries is sufficient to cause a rapid increase of serum BPA levels within 30 min (Hormann et al., 2014). A causal link between BPA and breast cancer remains equivocal because epidemiological studies have reported conflicting results (Rochester, 2013). Nonetheless, there is ample evidence from laboratory rodent and non-human primate studies that prenatal exposure to BPA may increase the propensity to develop breast cancer during adulthood (Mandrup et al., 2016; Tharp et al., 2012). In these studies, low-doses of BPA have been shown to affect mammary gland development by causing hyperplastic lesions in rats, which are parallel to early signs of breast neoplasia in women (Mandrup et al., 2016). These findings suggest that mammary tumors provoked by BPA may be initiated in the womb. Due to mounting evidence of harm and public pressure, BPA is being phased out by plastics manufacturers and is being replaced by other BP variants in “BPA-free” products (Rochester and Bolden, 2015).
Bisphenol F (BPF), Bisphenol S (BPS), and Bisphenol AF (BPAF) are among the main substitutes of BPA in polycarbonate plastics and epoxy resins (Chen et al., 2016). They are commonly found in baby feeding bottles, on thermal receipt papers, glues, dental sealants, food packaging or personal care products. BPA alternatives are detected in different categories of food items (Liao and Kannan, 2013). There is a strong negative correlation between the levels of BPA and BPS on thermal paper, whereby the papers containing high quantities of BPS have low quantities of BPA, suggesting that BPS has in part replaced BPA in this context (Liao et al., 2012). Epidemiological data on the human body burden of different BPA alternatives are very limited, but evidence suggests that they are generally widespread. In an investigation of the presence of BPS and BPF in human urine, these compounds were found to be present in 78% and 55% of respective samples in the United States (Zhou et al., 2014). Another investigation revealed that although the average urinary concentration of BPA in 130 individuals from Saudi Arabia was 4.92 ng/ml, 7 other bisphenols were found with the total bisphenol concentration reaching 19 ng/ml (Asimakopoulos et al., 2015), suggesting that BPA is only one of many BP alternatives that are found in humans.
Some of these BPA replacements are structurally related to BPA and have endocrine disrupting effects. Numerous studies have suggested that BPS and BPF have potencies similar to that of BPA resulting in endocrine disrupting effects such as uterine growth in rodents (Rochester and Bolden, 2015). A number of in vitro studies have looked at endocrine mode of action of some BPA alternatives in nuclear hormone receptor interactions (Chen et al., 2016). The range of EC50 values for estrogen activity (0.02 to >1000 µM) and androgen activity (0.29 to >1000 µM) of 20 BP congeners (5 of which are being tested here) indicates remarkable differences (Kitamura et al., 2005). Another study showed that 5 BPA alternatives (3 studied here) exhibited potencies within the same range as BPA on steroidogenesis, androgen receptor, aryl hydrocarbon receptor, and retinoic acid receptor activity (Rosenmai et al., 2014). Some BPA alternatives have also been tested in ER high-throughput screening assays in the context of the US Environmental Protection Agency (EPA) ToxCast program and shown in some cases to have estrogen-like effects (Judson et al., 2015).
Perturbation of a cell’s gene expression profile by chemicals results in specific signatures, allowing comparisons between transcriptome alterations caused by drugs and diseases to predict therapeutic and off-target effects (Iorio et al., 2010). The clinical utility of gene expression profiles is exemplified by studies showing that transcriptome signatures may affect clinical decision-making in breast cancer (Rhodes and Chinnaiyan, 2010). Gene expression profiling has been shown to be a more powerful predictor of breast cancer survival than standard systems based on clinical and histologic criteria (van de Vijver et al., 2002). Exposure of breast cancer cells to BPA presents a gene expression profile of tumor aggressiveness associated with poor clinical outcomes for breast cancer patients (Dairkee et al., 2008). While ER activation drives two-thirds of breast cancer (Green and Carroll, 2007), there have been no studies aimed at elucidating transcriptome alterations underlying estrogenic effects of a broad range of BPA alternatives to which human populations are exposed. In order to address this deficiency, we studied alterations in gene expression profiles of estrogen-dependent MCF-7 human breast cancer cells caused by BPA and 6 alternatives (Figure 1) using Affymetrix microarray and Illumina RNA sequencing platforms. The transcriptome signatures obtained were compared with a gene expression biomarker, which has been demonstrated to accurately predict ER modulation, including by “very weak” agonists (Ryan et al., 2016). We also used an E-screen assay (cell proliferation in estrogen-responding cells) as well as an estrogen response element (ERE)-luciferase reporter gene assay system. Our results clearly demonstrate that all 6 BPA alternative compounds are estrogenic. Alterations of MCF-7 transcriptome profiles caused by BPA alternatives bear the signature of ER activation with three bisphenols (BPAF, BPB, BPZ) having more estrogenic potency than BPA.
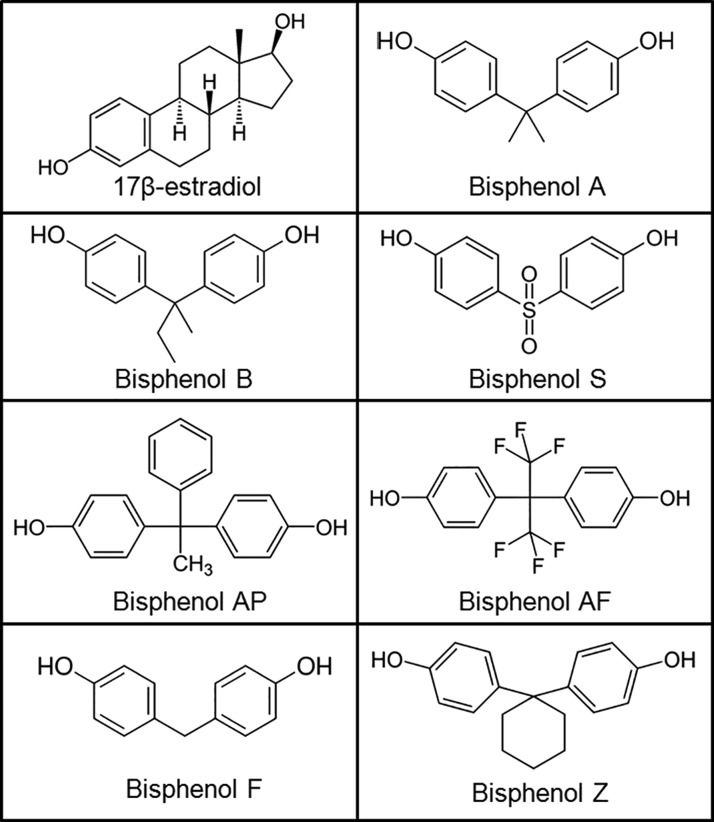
Figure 1.
Molecular structures of BPA and bisphenol alternatives in comparison to the natural hormone 17β-estradiol.
MATERIALS AND METHODS
Cell culture and treatments
All reagents and chemicals, unless otherwise specified, were of analytical grade and purchased from Sigma-Aldrich (UK). MCF-7, MDA-MB-231, and T47D human breast cancer cell lines were a gift from Prof. Joy Burchell (Research Oncology Department, King’s College London, United Kingdom). The T47D-KBluc cells were purchased from the American Type Culture Collection (ATCC, Teddington, United Kingdom) and harbor a stably integrated copy of a luciferase reporter gene under control of a promoter containing EREs. All cells were grown at 37°C (5% CO2) in 75 cm2 flasks (Corning, Tewksbury, USA) in a maintenance medium composed of phenol red-free DMEM (Life Technologies, Warrington, United Kingdom), 10% fetal bovine serum (FBS; GE Healthcare Life Sciences, Buckinghamshire, United Kingdom), 2-mM glutamine (GE Healthcare Life Sciences), and 10 µg/ml penicillin/streptomycin (Life Technologies). Test substances were diluted in 100% ethanol to prepare stock solutions. All treatments were prepared in a test medium containing phenol red-free DMEM, 5% charcoal stripped FBS (Life Technologies), 2-mM glutamine (GE Healthcare Life Sciences), and 10 µg/ml penicillin/streptomycin (Life Technologies) and contained no more than 0.1% ethanol. The solvent control also contained 0.1% (v/v) absolute ethanol diluted in test medium. Cells were detached from the flask substrate using 0.05% trypsin EDTA (Life Technologies) and counted using a haemocytometer prior to seeding. After a 24-h recovery period in DMEM maintenance medium, test substances were applied.
E-screen assay
The E-screen assay allows the determination of estrogenic effects by determining ER-mediated cell proliferation in hormone-responsive cells. This assay was performed as originally described (Soto et al., 1995), except that the bioassay was terminated using an MTT assay, indirectly measuring cell number by testing the activity of mitochondrial succinate dehydrogenase, in order to assess cell proliferation. The MTT assay has also been proven to be a reliable measure of toxicity (Mosmann, 1983). In brief, MCF-7, MDA-MB-231, and T47D cells were seeded into 48-well plates (Dutscher Scientific, Brentwood, United Kingdom) at a density of 20 000 cells per well in 250 µl maintenance medium. Following a 24-h incubation to allow cell attachment, medium was changed to include various concentrations of treatment substances. A 6-day long incubation with the test compounds allowed detection of variations in proliferative rates, which were reflected by differences in cell number measured at the end of the incubation period. The test medium was refreshed after 3 days. After another 3-day incubation, an MTT assay was performed as follows. Cells were incubated with 250 µl of MTT solution (1 mg/ml) for 2 h and the test terminated by lysing the cells with dimethyl sulfoxide (DMSO). As a measure of cell proliferation, the optical density of the cell lysate was determined at 570 nm using the SPECTROstar Nano plate reader (BMG Labtech, Ortenberg, Germany). The number of cells was directly proportional to the intensity of the signal. The cell proliferative effect was expressed as a percentage of the control, untreated samples.
ERE-mediated luciferase reporter gene assay
The ERE-mediated transcription of a luciferase reporter gene was determined in the T47D-KBluc cells using the Steady-Glo luciferase assay system following the manufacturer’s instructions (Promega, Southampton, United Kingdom). The T47D-KBluc cells were seeded in white 96 well plates (Greiner Bio-One, Germany) at a density of 20 000 cells per well in 50 µl of maintenance medium and allowed to attach overnight. An initial 24-h incubation was performed in the absence of test substances to purge cells of hormone residues in order to improve estrogen deprivation. Test substances were added and plates were incubated for another 24 h before the addition of 50 µl Steady-Glo luciferase reagent. The plates were left to stand for 10 min in the dark at room temperature to allow cell lysis and establishment of the luciferase reaction. Luminescence was measured using the Orion II microplate luminometer (Berthold Detection Systems, Bad Wildbad, Germany). ER modulation was confirmed by measuring the inhibitory effect of ICI 182,780 on the concentration of BPA resulting in a 20-fold increase in luciferase activity.
Microarray gene expression profiling
MCF-7 cells were seeded into 96-well plates with maintenance medium at a density of 20 000 cells per well. After 24 h of steroid hormone deprivation in hormone-free medium, the cells were stimulated with test substances at a concentration corresponding to the E-Screen AC50 for 48 h in triplicate in 3 independent experiments. RNA extraction was performed using the Agencourt RNAdvance Cell V2 kit according to the manufacturer’s instructions (Beckman Coulter Ltd, High Wycombe, United Kingdom). The samples were checked for RNA quality using the Agilent 2100 Bioanalyzer (Agilent Technologies LDA UK Limited, Stockport, United Kingdom) and quantified using the Nanodrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Wilmington, USA). The RNA integrity number ranged from 7.9 to 10 (mean of 9.7 ± 0.3) (Schroeder et al., 2006). Subsequently, triplicate samples which passed quality control (QC) criteria were pooled appropriately such that the final input amount of each sample was 3 ng.
The gene expression profiles were determined using the Affymetrix Human Transcriptome 2.0 Array as follows. Single Primer Isothermal Amplified (SPIA) cDNA was generated using the Ovation Pico WTA System V2 kit (Nugen, AC Leek, The Netherlands) following the manufacturer’s instructions. In addition, the SPIA cDNA was subjected to a QC check to assess quality (Agilent 2100 Bioanalyzer) and quantity (Nanodrop ND-1000 Spectrophotometer) in preparation for the next stage. The SPIA cDNA was fragmented and biotin-labeled using the Encore Biotin Module (Nugen) according to the manufacturer’s instructions. The fragmented and biotin-labeled cDNA was subjected to a further round of QC checks to assess fragmentation size (Agilent 2100 Bioanalyzer). Hybridization cocktails were prepared from the fragmented labeled-cDNA according to Nugen’s recommendations and hybridized to the microarrays at 45°C overnight. The arrays were washed and stained using the wash protocol FS450_0001 recommended for Affymetrix Human Transcriptome 2.0 Arrays on the GeneChip Fluidics 450 station. Ultimately, the arrays were scanned using the Affymetrix GeneChip Scanner. CEL files were QC assessed in the Expression Console software package (Affymetrix) by using standard metrics and guidelines for the Affymetrix microarray system. Data were imported and normalized together in Omics Explorer 3.0 (Qlucore, New York, NY, USA), using the Robust Multi-array Average (RMA) sketch algorithm. These microarray data have been submitted to Gene Omnibus and are accessible through accession number GSE85350.
Gene expression profiling by RNA-Sequencing
RNA-Sequencing was performed by applying Illumina sequencing by synthesis technology as follows. The amount of RNA for each library (100 ng) was a pool made up of 33 ng of RNA from each of the replicate wells for each sample. The preparation of the library was done by NEBNext Ultra Directional RNA (New England Biolabs, Hitchin, United Kingdom) following the manufacturer’s protocol. The amplified library was assessed using the Agilent 2100 Bioanalyzer for size and presence of adapter/primers/dimers—sized at ∼400 bp (including ∼130 bp adapter). The rRNAs were removed using the rRNA depletion module (New England Biolabs) following the manufacturer’s protocol. Libraries were pooled together and sequenced on a HiSeq2500 using a Rapid Run v2 flowcell with on-board clustering in a 2 × 100 paired-end (PE) configuration. BCL files were processed and deconvoluted using standard techniques. The sequencing output FASTQ files contained the sequences for each read and also a quality score. We analyzed the quality scores and other metrics using FASTQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/; last accessed May 23, 2017). Contamination from rRNA was measured using an alignment script (http://genomespot.blogspot.co.uk/2015/08/screen-for-rrna-contamination-in-rna.html; last accessed May 23, 2017). Adapter sequences (standard TruSeq LT adapter seq) were removed/trimmed using cutadapt (Martin, 2011). Sequences were then aligned to the genome (hg38 database) using the hierarchical indexing for spliced alignment of transcripts program HISAT2 (Kim et al., 2015). Binary Alignment/Map (BAM) files were imported to Qlucore omics explorer, along with the Gene Transfer Format (GTF) file for known genes in hg38 (downloaded from UCSC). Qlucore normalizes data using a method similar to the trimmed mean of M-values normalization method (TMM), which corrects for transcript length, applies a log transformation (Robinson and Oshlack, 2010) and gives values similar to the quantification of transcript levels in reads per kilobase of exon model per million mapped reads (RPKM) (Robinson and Oshlack, 2010), but also incorporates the TMM normalization factor for an improved between-sample normalization.
A total of 376.6 million raw reads were obtained (25.1 ± 5.4 million reads per sample). The average value of Q30, representing the probability of an incorrect base call 1 in 1000 times, was above 96%. The GC content (%GC) of the reads was on average 49%. A total of 90.92% ± 6.54% of the clean reads were mapped onto the human reference genome hg38. Among them, an average of 71.01% ± 5.82% and 18.25% ± 3.79% reads align concordantly exactly one time and more than one time, respectively. These RNA-Seq data have been submitted to Gene Omnibus and are accessible through accession number GSE87701.
ToxCast data mining
Publically available data from the ToxCast program was analyzed using the iCSS ToxCast Dashboard (http://actor.epa.gov/dashboard/; last accessed May 23, 2017). In the ToxCast program, out of 18 ER assays, 5 measured ERE-mediated transcription and gave results comparable to those of our T47D-Kluc assay. These assays measured ERα and ERβ activities. One assay was a single-readout fluorescent protein induction system measuring interaction of ER with ERE at 2 time points by microscopy technology (OT_ERa_EREGFP_0120 and _0048). Two other assays were reporter gene assays measuring mRNA in HepG2 cells (ATG_ERa_TRANS_up and ATG_ERE_CIS_up). The last 2 assays measured reporter protein levels in HEK293T (Tox21_ERa_BLA_Agonist_ratio) and BG-1 (Tox21_ERa_LUC_BG1_Agonist) cell lines. One assay of protein complementation measures formation of ERα dimers (OT_ER_ERaERa_0480 and _1440). Finally, NVS_NR_bER and NVS_NR_hER are radioligand receptor binding assays. Not all assays were performed on all bisphenols. For instance, BPAP was only tested in Attagene assays (ATG_ERa_TRANS_up and ATG_ERE_CIS_up). In this study, the AC50 value was used as a quantitative measure to reflect the potency of BP alternatives.
Comparison of the microarray and RNA-Seq data to the ER biomarker
The Running Fisher test (a rank-based nonparametric analysis) implemented within the NextBio database (Illumina, San Diego, California) was used to compare gene lists derived from the microarray and RNA-Seq data to the ER biomarker characterized earlier (Ryan et al., 2016). The Running Fisher test is a normalized ranking approach which enables comparability across data from different studies, platforms, and analysis methods by removing dependence on absolute values of fold-change, and minimizing some of the effects of normalization methods used, while accounting for the level of genome coverage by the different platforms. The Running Fisher algorithm computes statistical significance of similarity between ranked fold-change values of 2 gene lists using a Fisher’s exact test (Kupershmidt et al., 2010). A P-value ≤1E−4 was selected as our cutoff for significance based on prior evaluation of the cutoff as predictive of activation of ER (Ryan et al., 2016).
Statistical analysis
The concentration required to elicit a 50% response (AC50) was determined using a nonlinear regression fit using a sigmoid (5-parameters) equation calculated with GraphPad software (GraphPad Software, Inc., La Jolla, California, USA). For the transcriptome analysis, pair-wise comparisons of each tested substance to the negative control were performed using a t test controlling for batch effects in Omics Explorer 3.0 (Qlucore). Data used for the functional analysis were selected at cut-off P-values <.05 with fold-change >1.2 to study the ER activation signature as previously described (Ryan et al., 2016). Gene and disease ontology were analyzed using the Thomson Reuters MetaCore Analytical Suite version 6.28 recognizing network objects (proteins, protein complexes or groups, peptides, RNA species, compounds among others). The P-values are determined by hypergeometric calculation and adjusted using a Benjamini & Hochberg approach. All experiments were performed 3 times in triplicate (n = 3).
RESULTS
E-Screen and ERE-Luciferase Reporter Gene Assays
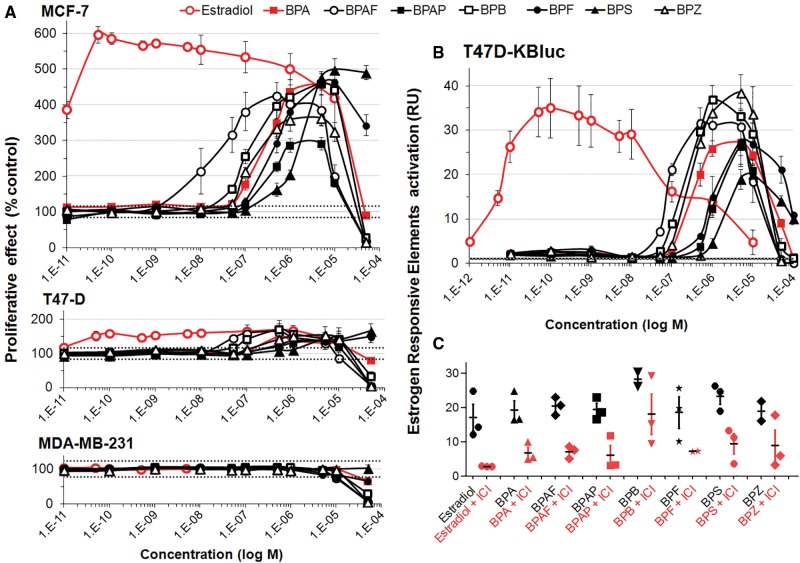
We have studied the estrogenic activities of 7 bisphenols (BP) found in foodstuffs and human biological fluids in three human breast cancer cell lines. As an initial investigative step of estrogenic potential, an E-Screen assay was performed using two hormone dependent (MCF-7, T47D) and one hormone independent (MDA-MB-231) human breast cancer cell lines (Figure 2A). The positive control 17β-estradiol was very potent in inducing the proliferation of MCF-7 cells (AC50 = 8 pM). All BP derivatives were also able to promote cell growth at concentrations 10 000–100 000 higher than 17β-estradiol (Figure 2A, top panel). BPAF was the most potent BP (AC50 = 0.03 µM) followed by BPZ (0.11 µM) > BPB (0.24 µM) > BPA (0.36 µM) > BPAP (0.39 µM) > BPF (0.55 µM) > BPS (1.33 µM). The same overall trend was observed with the T47-D cell line (Figure 2A, middle panel), albeit to a lesser extent which can be explained by the lower levels of ER expression in T47-D cells (Pais and Degani, 2016). As expected no cell proliferative effects were observed with the hormone independent, ER-negative MDA-MB-231 cell line suggesting that proliferative effects were mediated by ER (Figure 2a, bottom panel).
Figure 2.
BPA alternatives can effectively substitute for estradiol in promoting growth through ERs of human breast cancer cells. (A) Proliferative effect of BPA and bisphenol alternatives on mammary cells in an E-Screen bioassay. After 24 h of steroid hormone starvation cells were treated for 6 days with the test compounds. Numbers of cells was measured by an MTT colorimetric assay. Results are expressed as proliferative effect percentage relative to the proliferation of cells under hormone-free conditions. Data are the mean ±SE of 3 independent experiments, with each performed in triplicate. (B) BPA alternatives stimulate ERE-mediated transcription. After 24 h of steroid hormone starvation, T47D-Kluc cells were treated with the test compounds for 24 h. Cells were then lysed and subjected to a bioluminescence luciferase reporter gene assay. (C) Luciferase assays of T47D-Kluc cells treated with BPA alternatives in the absence (black) or presence (red) of ICI 182,780, an ER antagonist. Results show that ICI 182,780 represses ERE-mediated transcription induced by BPA alternatives.
Since the observed BP-derivative induced proliferation of MCF-7 and T47D cells (Figs. 2A and 2B) could be mediated by different receptors, estrogenic effects of BPA alternatives was investigated employing a luciferase reporter gene assay allowing measurement of ERE-mediated transcription. The results (Figure 2B, upper panel) were very similar to those obtained from the E-Screen assay: BPAF was the most potent bisphenol (AC50 = 0.08 µM) at stimulating ERE-luciferase reporter gene expression followed by BPB (0.3 µM) > BPZ (0.4 µM) ∼ BPA (0.4 µM) > BPF (1 µM) ∼ BPAP (1 µM) > BPS (1.5 µM) having estrogenic effects in the same range as BPA. Moreover, the addition of ICI 182,780 (100 nM) antagonized the activation of ER by estradiol and bisphenols confirming reporter gene expression via this receptor (Figure 2C). However, estrogenic effects of some BP derivatives, such as BPB and BPZ, were not completely antagonized by ICI addition, suggesting estrogenic activation mechanisms independent of ER.
ToxCast data mining (Table 1) confirms our results but also reveals some discrepancies. BPF, BPZ, and BPB were classified as inactive in the ToxCast gene reporter ERa_BLA_Agonist_ratio assay, because they did not reach the response threshold, possibly due to the lower sensitivity of this assay. For the 4 BPs that were examined in the ER computational network model, BPA, BPB, and BPAF were active and BPF was inactive (Judson et al., 2015).
Table 1.
Comparison to ToxCast Data
| CASRN | Name | ATG_ERa_ TRANS_up | ATG_ERE_ CIS_up | NVS_NR_ bER | NVS_NR_ hER | OT_ER_ ERaERa_0480 | OT_ER_ ERaERa_1440 | OT_ERa_ EREGFP_0120 | OT_ERa_ EREGFP_0480 | TOX21_ERa_BLA _Agonist_ratio | TOX21_ERa_ LUC_BG1_Agonist | AC50 MCF7 this study | AC50 LUC this study |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1478-61-1 | Bisphenol AF | 0.36 | 0.09 | 0.07 | 0.03 | 1.05 | 0.712 | 0.1 | 0.11 | 0.36 | 0.05 | 0.03 | 0.08 |
| 843-55-0 | Bisphenol Z | #### | 0.31 | 0.11 | 0.4 | ||||||||
| 77-40-7 | Bisphenol B | 0.13 | 0.17 | 0.21 | 0.27 | 1.82 | 1.41 | 0.18 | 0.22 | #### | 0.11 | 0.24 | 0.3 |
| 80-05-7 | BPA | 0.12 | 0.1 | 0.42 | 0.23 | 5.73 | 4.33 | 0.42 | 0.63 | 1.01 | 0.36 | 0.36 | 0.4 |
| 1571-75-1 | Bisphenol AP | 0.12 | 0.29 | 0.39 | 1 | ||||||||
| 2467-02-9 | BPF | 34.9 | 30.3 | #### | #### | #### | #### | #### | #### | #### | #### | 0.55 | 1 |
| 80-09-1 | Bisphenol S | 0.8 | 1.42 | 17.7 | 5.45 | 43.8 | 56.1 | 59.2 | 26.6 | 9.29 | 1.87 | 1.33 | 1.5 |
Publically available data was analyzed from the ToxCast program using the iCSS ToxCast Dashboard. AC50 values (µM) for ToxCast assays relative to ESR1 activation are compared with the results presented here. Negative results are displayed by bolded values. Non-bold values show assays in which bisphenols have not been tested.
Transcriptome Profiling
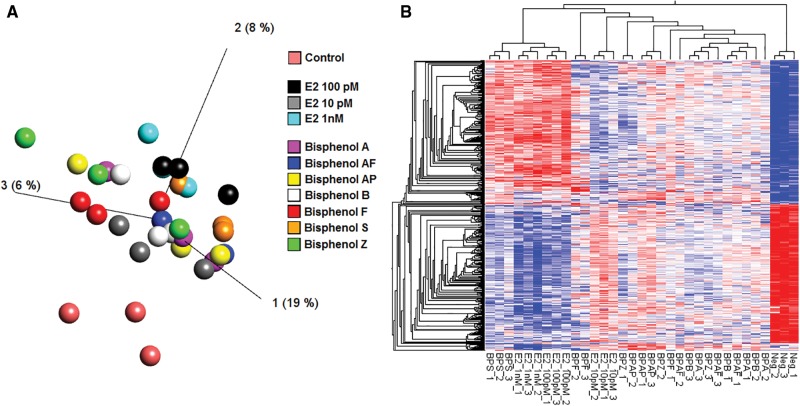
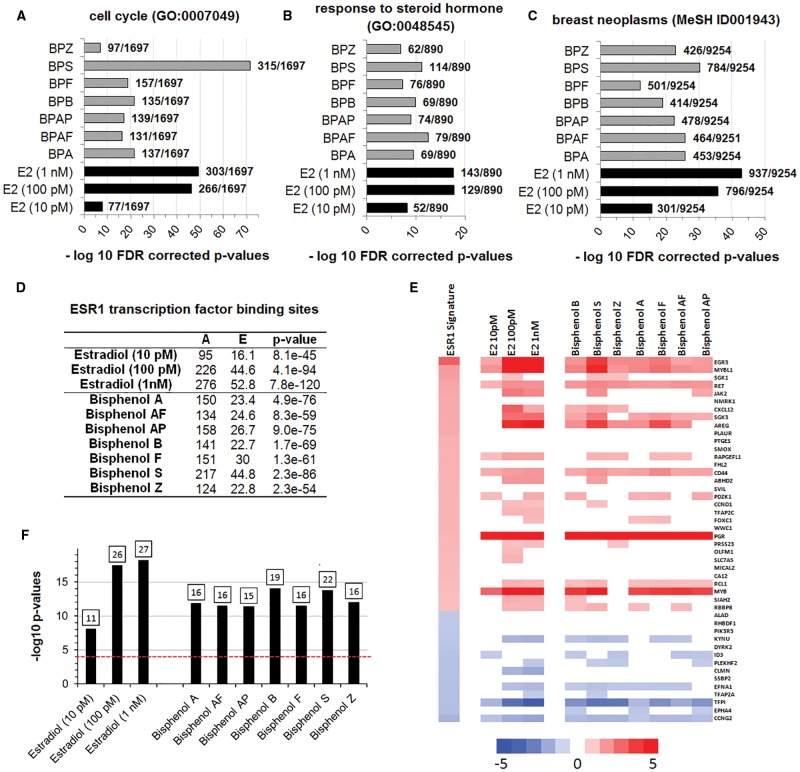
MCF-7 cells treated for a period of 48 h with BPA, 6 BP alternatives, or 17β-estradiol, were subjected to full transcriptome profiling using the Affymetrix microarray platform. The length of exposure (48 h) and the concentration of test agent were selected based on data from previous studies showing that these conditions produce robust gene expression changes (Shioda et al., 2006). Supplementary File 1 shows the statistical significance of differential transcript cluster expression with respective fold-changes by volcano plots. The visual representation of the variance structure obtained by a principal component analysis indicates that samples treated with BPA and BP alternatives separates from the control samples (Figure 3A) and cluster along samples treated with 17β-estradiol. This result is corroborated by the heatmap representation of the 1000 most differentially expressed genes according to a one-way ANOVA (Figure 3B). Surprisingly, samples treated with BPS and the 2 highest concentrations of 17β-estradiol clustered together, distantly of the negative control (Figure 3B). They were also segregated from other samples presenting a mixed pattern (Figure 3B). The set of genes altered by treatment with BPA and BP alternatives (Supplementary File 2) were highly enriched in genes involved in the regulation of the cell cycle (Figure 4A), as well as in genes regulated by steroid hormones (Figure 4B). Samples treated with BPS and the 2 highest concentrations of 17β-estradiol were the most enriched for cell cycle annotation, confirming their discordant pattern (Figure 3B). In addition, an enrichment analysis of MeSH terms reveals that exposure to both BPA or the 6 BP alternatives result in changes in the expression of genes involved in the etiology of breast cancer (Figure 4C). Genes having the highest fold-changes were similar and indicative of a hormone-induced proliferative effect (Table 2). The gene encoding the progesterone receptor (PGR) consistently exhibited the highest fold-change after stimulation with BPA or all the BP alternatives. ER binding motifs were overrepresented in the promoter of the differentially expressed genes (DEGs). There were 4.8–6.4 more ER binding sequences (EREs) in the promoters of the DEGs than what would be expected by chance (Figure 4D).
Figure 3.
Variation in the transcriptome profiles of MCF-7 cells exposed to BPA alternatives and 17β-estradiol. (A) PCA analysis of gene expression profiles showed a clear separation between the negative control and the BPA alternatives. The different colors indicate different treatments. (B) Expression of the 1000 genes presenting the most pronounced variations between the different samples was evaluated according to a one-way ANOVA. Hierarchical clustering revealed different patterns in gene expression between the BPA alternatives and 17β-estradiol-treated cells.
Figure 4.
Alteration of MCF-7 transcriptome profiles caused by BPA alternatives bear the signature of ER activation. Transcriptome analysis of MCF-7 cells treated with BPA and alternative bisphenols reflect cell cycle changes (A), response to hormone (B), as well as an association to breast cancer (C). The P-values are determined by hypergeometric calculation and adjusted using a Benjamini and Hochberg approach. Ratios show the total number of network objects belonging to each term in comparison to those disturbed by the treatments. The total of network objects (ie, all DEG) recognized by Metacore are indicated in parentheses (D). Overrepresentation of ER binding motifs in the promoters of the differentially expressed genes. The analysis conducted with the transcription factor analysis tool of Metacore. A total of 1262 ER binding sites is found in the 42 909 protein-based objects in the Metacore background list. A, number of targets in the activated dataset regulated by ESR1; E, expected mean of hypergeometric distribution; P-value calculated using hypergeometric distribution. (E) A gene expression biomarker confirms that BPA and bisphenol alternatives are ER agonists. Lists of statistically significant genes from MCF-7 cells treated with BPA and bisphenol alternatives or the natural hormone 17β-estradiol were examined against an ER gene expression biomarker signature consisting of 46 genes. The heat map shows the expression of genes in the biomarker after exposure to the indicated compound. Fold-change values for the ER biomarker are the average across 7 agonist treatments. (F) Bar plots showing the significance of the correlation by their −log10 P-values. Classification of activation or suppression required P < .0001. The number of genes overlapping the ER biomarker is indicated at the top of each bar.
Table 2.
List of the 5 Most Up- or Down-Regulated Genes After Treatment With BPA, BPAF or Estradiol (E2)
| E2 (10 pM) | E2 (100 pM) | E2 (1 nM) | BPA | BPAF | |||||
|---|---|---|---|---|---|---|---|---|---|
| ALDH1A3 | 0.4 | ALDH1A3 | 0.2 | ALDH1A3 | 0.2 | SEMA5A | 0.3 | ALDH1A3 | 0.3 |
| SEMA5A | 0.4 | PRICKLE2-AS3 | 0.2 | PSCA | 0.2 | ALDH1A3 | 0.3 | PSCA | 0.4 |
| PPARG | 0.5 | SEMA5A | 0.3 | TFPI | 0.2 | PSCA | 0.3 | RP11-706C16.7 | 0.4 |
| PSCA | 0.5 | PSCA | 0.3 | PRICKLE2-AS3 | 0.2 | PRICKLE2-AS3 | 0.3 | SEMA5A | 0.4 |
| IGFBPL1 | 0.5 | EPAS1 | 0.3 | EPAS1 | 0.3 | GABBR2 | 0.3 | EPAS1 | 0.4 |
| AC005534.8 | 2.6 | MYBL1 | 5.5 | AC106875.1 | 6.4 | AC106875.1 | 3.3 | AGR3 | 3.2 |
| MYB | 3.0 | AC106875.1 | 6.3 | MYBL1 | 6.5 | MYB | 3.7 | MYB-AS1 | 3.4 |
| AC106875.1 | 3.2 | GREB1 | 7.1 | GREB1 | 7.1 | GREB1 | 3.8 | MGP | 3.5 |
| GREB1 | 3.7 | PGR | 10.0 | PGR | 9.2 | MGP | 4.2 | MYB | 4.1 |
| PGR | 4.6 | MGP | 12.4 | MGP | 15.0 | PGR | 6.2 | PGR | 5.7 |
| BPB | BPF | BPS | BPZ | BPAP | |||||
| ALDH1A3 | 0.3 | ALDH1A3 | 0.3 | ALDH1A3 | 0.2 | ALDH1A3 | 0.3 | ALDH1A3 | 0.3 |
| GABBR2 | 0.3 | SEMA5A | 0.3 | PSCA | 0.3 | TFPI | 0.3 | SEMA5A | 0.4 |
| PSCA | 0.3 | EPAS1 | 0.4 | SEMA5A | 0.3 | GABBR2 | 0.3 | GABBR2 | 0.4 |
| PRICKLE2-AS3 | 0.4 | PSCA | 0.4 | TFPI | 0.3 | PSCA | 0.4 | TFPI | 0.4 |
| EPAS1 | 0.4 | HCAR3 | 0.4 | PRICKLE2-AS3 | 0.3 | EPAS1 | 0.4 | EPAS1 | 0.4 |
| AGR3 | 3.3 | MYB | 3.9 | AC106875.1 | 5.4 | STC1 | 2.8 | STC1 | 3.1 |
| GREB1 | 3.8 | MYB-AS1 | 4.4 | MYB | 5.7 | ASCL1 | 2.9 | AL121578.2 | 3.1 |
| MYB | 4.1 | GREB1 | 4.5 | GREB1 | 6.0 | AGR3 | 3.3 | MYB | 3.8 |
| MGP | 4.4 | MGP | 5.0 | MGP | 7.8 | MYB-AS1 | 3.4 | AGR3 | 3.8 |
| PGR | 6.8 | PGR | 6.5 | PGR | 9.5 | PGR | 5.8 | PGR | 5.8 |
The transcriptome profiling was performed using the Affymetrix microarray platform.
Statistical values derived from gene ontology analysis can have limited reliability due to the background set including only the genes that are likely to be expressed in the experiment (Tipney and Hunter 2010). Genes disturbed by chance have a higher probability to be associated with endocrine ontologies in an endocrine sensitive tissue such as the breast. This could result in an increased false positive rate. In order to circumvent this problem, we applied a biomarker approach to predict ER modulation. The gene expression biomarker consists of 46 genes which exhibit consistent expression patterns after exposure to 7 ER agonists and 3 ER antagonists determined using MCF-7 microarray data derived from the CMAP 2.0 dataset (Ryan et al., 2016). This gene expression biomarker has a balanced accuracy for prediction of ER activation of 94%. Gene expression profiles from the cells treated with BPA and BP alternatives were compared to the ER biomarker using the Running Fisher algorithm as previously described (Ryan et al., 2016). The cut-off value for statistical significance was P-value ≤.0001 after a Benjamini-Hochberg correction of α = 0.001. All transcriptome profile alterations resulting from exposure to BPA and all 6 BP alternatives (Figure 4E) achieved statistical significance and exhibited a pattern highly similar to that of the biomarker (Figure 4F).
We finally investigated differences in gene expression patterns provoked by BPA alternatives in MCF-7 cells. This was done by identifying pathways, which were disturbed by BPA alternatives but not by BPA itself, using the Metacore pathway maps tool. The list of all pathways altered by the different treatments is shown in Supplementary File 3. BPS had the most discordant pattern with 33 pathways statistically significantly altered, which were not disturbed by BPA. The high concentration necessary to activate ER could also possibly induce toxic effects as indicated by the disturbance of apoptotic pathways (FDR corrected P-value = 2.5e−2). This Metacore analysis also suggested that BPAF is a modulator of the glucocorticoid receptor (GR) (FDR corrected P-value = 1.9e−4). A synergistic activation of GR and ER could possibly explain the strong cell proliferation stimulatory effect of BPAF starting from 10 nM (Figure 2A), a concentration at which this compound does not appear to be estrogenic (Figure 2B).
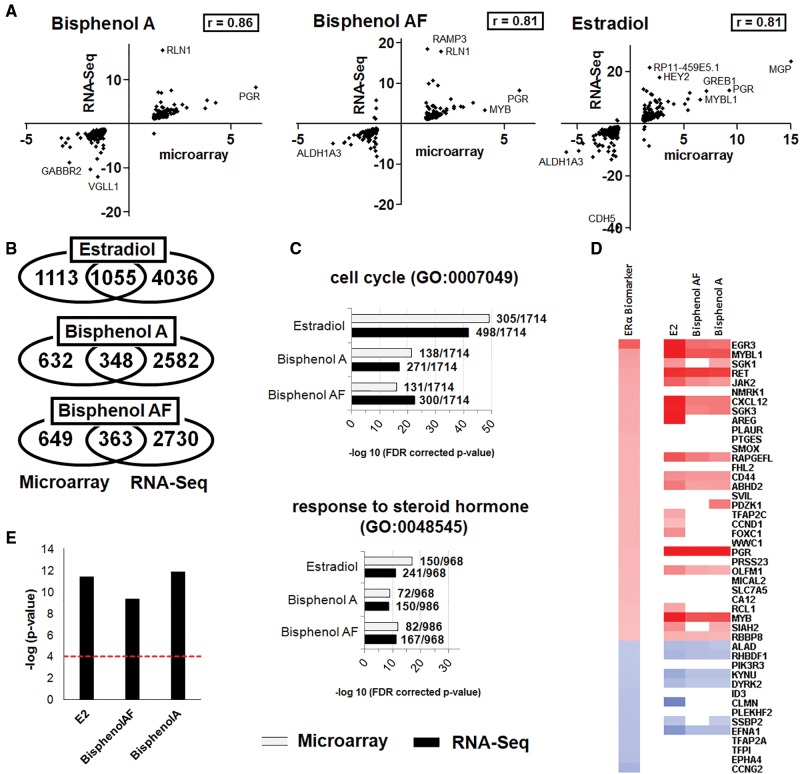
In order to confirm estrogenic effects provoked by BPA and BP alternatives, RNA extracted from MCF-7 cells treated with BPAF (0.08 µM), BPA (0.36 µM), and estradiol (1 nM) were subjected to total transcriptome RNA sequencing (RNA-Seq) analysis using the Illumina HiSeq 2500 system. This also allowed us to compare the sensitivity of RNA-Seq and microarrays to determine estrogenic effects. We have performed pairwise comparisons to determine the list of DEGs following the same criteria used for the microarray analysis. Only 12%-21% of the DEGs identified by RNA-Seq were also found to be altered on the microarrays (Figure 5B). The gene expression fold changes common to both the microarray and the RNA-Seq analysis were well correlated (Figure 5A) for 17β-estradiol (Pearson r = 0.81), BPA (r = 0.86), and BPAF (r = 0.81). Overall, the RNA-Seq method was more sensitive and identified 2-3 times more significantly altered genes compared with the microarray method (Figure 5B). A total of 5091, 2930, and 3093 genes were significantly altered by estradiol, BPA, and BPAF, respectively. Genes commonly found altered after the application of both transcriptome profiling methods generally had a higher fold change by the RNA-Seq platform (Supplementary File 4). The ontology analysis based on RNA-Seq data gave similar results to our previous calculations based on the microarray data and confirms estrogenic effects of BPA and BPAF (Figure 5C). Although more DEGs related to cell cycle function were found by RNA seq than by microarray, the −log10 P-value for BPA was lower because the total number of DEGs found by RNA seq was higher than by microarray. We similarly applied the ER gene expression biomarker to detect ER agonists after analysis with the Illumina sequencing platform (Figs. 5D and 5E). The RNA-Seq platform gave results similar to that of Affymetrix microarrays confirming estrogenic effects of the BPA alternatives.
Figure 5.
Comparison of RNA-Seq and microarray platforms in determining endocrine disrupting effects of BPA and bisphenol alternatives. RNA extracted from MCF-7 cells was subjected to a full transcriptome profiling using the Illumina RNA sequencing or the microarray technology under similar conditions. (A) Pearson correlation coefficients between the RNA-Seq and microarray data. The fold changes in gene function having an altered expression by the two methods are presented. (B) Venn diagrams showing the number of genes uniquely or commonly disturbed. (C) Gene ontology analysis of terms associated with MCF-7 hormone induced proliferation in the transcriptome profiles obtained by RNA-Seq or microarray analysis. Ratios are showing the total number of network objects belonging to each term in comparison to those disturbed by the treatments. The total of network objects (ie, all DEG) recognized by Metacore are indicated in parentheses. (D) Heat map of genes whose expression was statistically significantly altered examined against an ER gene expression biomarker. (E) Bar plots showing the significance of the correlation by their −log10 P-values.
DISCUSSION
Human populations are exposed to a wide range of BPA alternatives. Presented here is the first comprehensive, side-by-side comparison of the estrogenic effects of BPA and 6 BP variants (BPZ, BPAF, BPAP, BPB, BPF, BPS) found in foodstuffs and human fluids using both cell proliferation and gene expression profile assays. Our study revealed that the 6 bisphenols introduced in “BPA-free” plastics displayed estrogenic effects with BPAF, BPB, and BPZ being more potent than BPA in our experimental system. The comparison of transcriptome profiles revealed a signature demonstrating that these effects were mediated via the ER (Figs. 4A–F). Although data on plasma concentrations of BPA alternatives are scarce (Chen et al., 2016), the potencies reported in this study are likely to be much higher than concentrations found in most cases of human environmental exposures. Our results suggest the need to conduct studies to examine possible pathological effects in laboratory animals at concentrations relevant for human exposures. In addition, combined effects of these bisphenols should be explored in further studies to determine the relevance of these estrogenic effects in terms of human health-risk assessment. In particular, our results provide a basis for investigating the relevance of human exposure to BPA alternatives in hormone-dependent breast cancer progression. The structure of BPA alternatives (Figure 1) provides insights into the possible mechanisms of action toward ER. BPA binding to ER is mainly driven by van der Waals’ forces and hydrogen bond interactions (Li et al., 2015). BPA and 17β-estradiol bind in a similar manner, with 2 phenol rings pointing to the 2 ends of the ER hydrophobic pocket. Differences in estrogenic activities between the different BPA alternatives may be due to the different groups present at the bridge between the 2 phenol rings. Their hydrophobicity is driving their affinity, since the matching of the group carried by the methylene bridge is known to determine binding affinity toward the hydrophobic surface of the ER binding site (Endo et al., 2005). Such a mechanism of ER interaction is supported by the hydrophobicity ranking of bisphenols (logP values BPZ > BPAF > BPAP > BPB > BPA > BPF > BPS), which closely corresponds to their AC50 toxicity score.
It is important to note that in some cases EDCs can elicit a non-monotonic response that significantly deviates from the usual sigmoidal curve (Vandenberg et al., 2012). In another transcriptomics study in which MCF-7 cells were exposed to varying concentrations of BPA, a weak gene activation peak at a very low concentration range (∼0.1 nM) was observed in addition to the main peak of gene activation (Shioda et al., 2013). In addition, an investigation of the combinatorial effects of bisphenols would reflect real-world exposure conditions even if estrogenic effects of chemical mixtures in vitro are predicted by the use of concentration addition models (Evans et al., 2012).
Our data allow a direct comparison of the sensitivity of RNA-Seq and microarray hybridization in the determination of the transcriptome signature of ERα activation. Overall, the application of RNA-Seq resulted in the detection of more statistically significant alterations in gene expression than the microarray method. These genes generally had a higher fold change leading to P-values associated with gene expression biomarkers being lower. Overall, RNA sequencing appears to be more sensitive than microarray analysis. This was also the conclusion of studies performed in other cell lines (Perkins et al., 2014), mesenchymal stem cells (Li et al., 2016), and rat tissues (Perkins et al., 2014). In a study of 498 primary neuroblastomas, RNA-Seq outperformed microarrays in determining the transcriptomic characteristics of these cancers, while RNA-Seq and microarray performed similarly in clinical endpoint prediction (Zhang et al., 2015). Both technologies are suitable to detect estrogenic effects of BPA alternatives using the gene expression biomarker of ERα activation. Although the genes weakly expressed were differentially detected by the two platforms, the biomarker signatures were comparable. However, it is perhaps noteworthy that the microarray processing and analysis pipelines are better standardized and could thus be more reliable in comparing experiments performed by different laboratories. Further intra- and inter-experimental comparisons would be necessary to conclude which method is the most sensitive and reliable to determine transcriptome signatures.
The mechanisms of action resulting in toxic effects from BPA at low levels of exposure have long remained elusive due to its relatively low affinity for ER. However, BPA is very potent at inducing rapid non-genomic responses from membrane surface receptors (Wozniak et al., 2005). For example, BPA administration inhibited meiotic maturation of Zebrafish oocytes through a G protein-coupled ER-dependent epidermal growth factor receptor pathway (Fitzgerald et al., 2015). BPA can also induce adipogenesis through other receptors such as peroxisome proliferator-activated receptor gamma (PPARγ) (Ahmed and Atlas 2016). Surprisingly, this latest study provided evidence that BPS is a more potent adipogen than BPA in inducing 3T3-L1 adipocyte differentiation and lipid accumulation. Overall, effects of BPA alternatives on G protein-coupled ERs have poorly been investigated. Recent studies have revealed that less studied endocrine-related systems such as glucocorticoid, PPAR, monoamine, noradrenaline, and serotonin pathways could also be targets of endocrine disruption (Filer et al., 2014). Our transcriptome profiling of MCF-7 cells exposed to BPAF revealed that it could be a modulator of the GR. This could explain the potent cell proliferation stimulatory effects of BPAF starting from 10 nM (Figure 2A). However, targeted studies of GR modulation are needed to allow a firm conclusion on such a BPAF-ER interaction.
In adult humans, BPA is rapidly metabolized by the liver, with elimination virtually complete within 24 h of exposure, although chronic exposures could result in accumulation if BPA distributes within tissues that slowly release BPA (Stahlhut et al., 2009). In an investigation of concentrations of BPA metabolites in the urine of 112 pregnant women (ethnically and racially diverse), total BPA consisted of 71% BPA glucuronide, 15% BPA sulfate, and 14% unconjugated BPA (Gerona et al., 2016). These metabolites are believed to be biologically inactive because they are not thought to be ER agonists. However, this does not preclude effects on other receptor systems or the creation of an equilibrium between the metabolite and the parent if the metabolite is long-lived. For instance, a recent study showed that BPA glucuronide can induce lipid accumulation and differentiation of pre-adipocytes (Boucher et al., 2015). Metabolites of the main BPA alternatives have not been routinely measured in human biomonitoring studies, which may greatly underestimate real world exposures. Moreover, BPA metabolites may be deconjugated by β-glucuronidase, which is particularly active in the placenta and fetal liver resulting in increased fetal exposure (Ginsberg and Rice 2009). Although it is known that UGT2B15, the UDP-glucuronosyltransferase most effective at conjugating BPA, is expressed at low levels in the fetal liver, it increases postnatally to maximum values between 3 and 15 weeks after birth. In contrast, SULT1A1, the sulfotransferase most effective in sulfating BPA, is expressed at levels in the fetal liver equivalent to the adult. Thus, it is also important to consider the efficiency of the fetus at conjugating BPA with sulfate (Divakaran et al., 2014; Duanmu et al., 2006). Previous studies showed that UGT2B15 is active in MCF-7 breast cancer cells (Harrington et al., 2006). Further studies comparing estrogenic effects of BPA alternatives in cell lines devoid of UDP-glucuronosyltransferases could also provide interesting insights into the pathways of possible toxicity of this compound.
Even though disruption of ER function is one of the most investigated endocrine-related targets in the ToxCast data (18 assays) (Judson et al., 2015), our analysis of the high-throughput system (HTS) employed has revealed a lack of concordance between our results and the HTS ToxCast assays, with some of these ToxCast assays failing to detect estrogenic effects (Table 1). This was especially the case for BPF which was found negative in 9 out of the 11 assays scrutinized. The 2 systems of analysis detecting the estrogenic capability of BPF were the Attagene assays. BPF was found to be less potent in these assays than in this study (Table 1). Our results are in agreement with previously published estrogenic effects of BPF reporting an EC50 of 0.82 µM in an ER luciferase assay (Rosenmai et al., 2014). BPAP on the other hand was found to be more potent in the ToxCast assays, although the difference within the 2 current assays was minimal and could be explained by different grades of chemical purity. Another reason for these differences could be the different sensitivities of HTS assays. These are frequently based on heterologous expression of reporter genes in non-hormone-responsive cell lines that possibly do not possess all the co-factors necessary to transduce the hormone signal, thus resulting in a decreased sensitivity to detect relevant signals.
The US EPA has proposed the Endocrine Disruptor Screening Program Tier 1 assays with the goal of reducing the cost and time of toxicity testing, as well as animal use (US EPA, 2015). The US EPA will consider certain in vitro high throughput assays and computational modeling data as alternatives for detecting and measuring ER agonist and antagonist bioactivity by three current EDSP Tier 1 screening battery assays (that is, the ER binding in vitro assay, the ER transcriptional activation in vitro assay, and the in vivo uterotrophic assay) (US EPA, 2015). Integrating gene expression profiling into the proposed HTS framework might add value to targeted in vitro testing, because multiple targets and pathways could be evaluated simultaneously (Ryan et al., 2016). Transcriptome profiles resulting from exposure to a given chemical could be correlated to signatures of a wide range of chemicals using the Library of Integrated Network-based Cellular Signatures (LINCS) database, which contains signatures from around 4000 chemicals screened in approximately 17 cell lines (http://www.lincsproject.org; last accessed May 23, 2017) or to the ∼1300 chemicals that were examined in multiple cell lines as part of the Connectivity Map project (Lamb et al., 2006).
CONCLUSION
We have detected estrogenic effects of BPA alternatives by the application of a gene expression biomarker of ERα activation, whose results are corroborated by functional cellular assays. Our comparison of microarray and RNA-Seq technologies showed that both platforms are suitable for the use of this ERα gene expression biomarker. Recently, the plastics manufacturing industry have turned to alternative bisphenols to produce their “BPA-free” products, often with little toxicology testing. Our data show that some of these BPA alternatives are more potent ER activators than BPA, suggesting that alternative testing strategies could provide valuable information to support decisions related to chemical substitutions.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
Supplementary Material
Acknowledgments
We thank Drs Richard Judson and Charles Wood for critical review of the manuscript and Dr John Rooney for technical assistance. The authors declare they have no competing interests.
FUNDING
This study was funded by the Sustainable Food Alliance (USA) and Breast Cancer UK, whose support is gratefully acknowledged. This study has been subjected to review by the U.S. EPA National Health and Environmental Effects Research Laboratory and approved for publication. Approval does not signify that the contents reflect the views of the Agency, nor does mention of trade names or commercial products constitute endorsement or recommendation for use.
REFERENCES
- Ahmed S., Atlas E. (2016). Bisphenol s- and bisphenol a-induced adipogenesis of murine preadipocytes occurs through direct peroxisome proliferator-activated receptor gamma activation. Int. J. Obes. (Lond) 40, 1566–1573. [DOI] [PubMed] [Google Scholar]
- Asimakopoulos A. G., Xue J., De Carvalho B. P., Iyer A., Abualnaja K. O., Yaghmoor S. S., Kumosani T.A., Kannan K. (2015). Urinary biomarkers of exposure to 57 xenobiotics and its association with oxidative stress in a population in Jeddah, Saudi Arabia. Environ. Res. 150, 573–581. [DOI] [PubMed] [Google Scholar]
- Boucher J. G., Boudreau A., Ahmed S., Atlas E. (2015). In vitro effects of bisphenol a beta-d-glucuronide (bpa-g) on adipogenesis in human and murine preadipocytes. Environ. Health Perspect. 123, 1287–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Kannan K., Tan H., Zheng Z., Feng Y. L., Wu Y., et al. (2016). Bisphenol analogues other than bpa: Environmental occurrence, human exposure, and toxicity-a review. Environ. Sci. Technol. 50, 5438–5453. [DOI] [PubMed] [Google Scholar]
- Dairkee S. H., Seok J., Champion S., Sayeed A., Mindrinos M., Xiao W., et al. (2008). Bisphenol a induces a profile of tumor aggressiveness in high-risk cells from breast cancer patients. Cancer Res. 68, 2076–2080. [DOI] [PubMed] [Google Scholar]
- Divakaran K., Hines R. N., McCarver D. G. (2014). Human hepatic UGT2B15 developmental expression. Toxicol. Sci. 141, 292–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duanmu Z., Weckle A., Koukouritaki S. B., Hines R. N., Falany J., Falany C. N., Kocarek T. A., Runge-Morris M. (2006). Developmental expression of aryl, estrogen and hydoxysteroid sulfotransferases in pre- and post-natal human liver. J. Pharmacol. Exp. Therapeutics 316, 1310–1317. [DOI] [PubMed] [Google Scholar]
- Endo Y., Yoshimi T., Ohta K., Suzuki T., Ohta S. (2005). Potent estrogen receptor ligands based on bisphenols with a globular hydrophobic core. J. Med. Chem. 48, 3941–3944. [DOI] [PubMed] [Google Scholar]
- Evans R. M., Scholze M., Kortenkamp A. (2012). Additive mixture effects of estrogenic chemicals in human cell-based assays can be influenced by inclusion of chemicals with differing effect profiles. PLoS One 7, e43606.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filer D., Patisaul H. B., Schug T., Reif D., Thayer K. (2014). Test driving toxcast: endocrine profiling for 1858 chemicals included in phase II. Curr. Opin. Pharmacol. 19, 145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald A. C., Peyton C., Dong J., Thomas P. (2015). Bisphenol a and related alkylphenols exert nongenomic estrogenic actions through a g protein-coupled estrogen receptor 1 (gper)/epidermal growth factor receptor (egfr) pathway to inhibit meiotic maturation of zebrafish oocytes. Biol. Reprod. 93, 135.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerona R. R., Pan J., Zota A. R., Schwartz J. M., Friesen M., Taylor J. A., Hunt P.A., Woodruff T.J. (2016). Direct measurement of bisphenol a (bpa), bpa glucuronide and bpa sulfate in a diverse and low-income population of pregnant women reveals high exposure, with potential implications for previous exposure estimates: a cross-sectional study. Environ. Health 15, 50.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg G., Rice D. C. (2009). Does rapid metabolism ensure negligible risk from bisphenol A? Environ. Health Perspect. 117, 1639–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green K. A., Carroll J. S. (2007). Oestrogen-receptor-mediated transcription and the influence of co-factors and chromatin state. Nat. Rev. Cancer 7, 713–722. [DOI] [PubMed] [Google Scholar]
- Harrington W. R., Sengupta S., Katzenellenbogen B. S. (2006). Estrogen regulation of the glucuronidation enzyme UGT2B15 in estrogen receptor-positive breast cancer cells. Endocrinology 147, 3843–3850. [DOI] [PubMed] [Google Scholar]
- Hormann A. M., Vom Saal F. S., Nagel S. C., Stahlhut R. W., Moyer C. L., Ellersieck M. R., et al. (2014). Holding thermal receipt paper and eating food after using hand sanitizer results in high serum bioactive and urine total levels of bisphenol a (bpa). PLoS One 9, e110509.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio F., Bosotti R., Scacheri E., Belcastro V., Mithbaokar P., Ferriero R., et al. (2010). Discovery of drug mode of action and drug repositioning from transcriptional responses. Proc. Natl Acad. Sci. U. S. A. 107, 14621–14626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson R. S., Magpantay F. M., Chickarmane V., Haskell C., Tania N., Taylor J., et al. (2015). Integrated model of chemical perturbations of a biological pathway using 18 in vitro high-throughput screening assays for the estrogen receptor. Toxicol. Sci. 148, 137–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S. L. (2015). Hisat: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura S., Suzuki T., Sanoh S., Kohta R., Jinno N., Sugihara K., Yoshihara S., Fujimoto N., Watanabe H., Ohta S. (2005). Comparative study of the endocrine-disrupting activity of bisphenol a and 19 related compounds. Toxicol. Sci. 84, 249–259. [DOI] [PubMed] [Google Scholar]
- Kupershmidt I., Su Q. J., Grewal A., Sundaresh S., Halperin I., Flynn J., Shekar M., Wang H., Park J., Cui W, et al. (2010). Ontology-based meta-analysis of global collections of high-throughput public data. PLoS One 5, e13066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J., Crawford E. D., Peck D., Modell J. W., Blat I. C., Wrobel M. J., Lerner J., Brunet J. P., Subramanian A., Ross K. N., et al. (2006). The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935. [DOI] [PubMed] [Google Scholar]
- Li J., Hou R., Niu X., Liu R., Wang Q., Wang C., et al. (2016). Comparison of microarray and RNA-Seq analysis of mrna expression in dermal mesenchymal stem cells. Biotechnol. Lett. 38, 33–41. [DOI] [PubMed] [Google Scholar]
- Li L., Wang Q., Zhang Y., Niu Y., Yao X., Liu H. (2015). The molecular mechanism of bisphenol a (bpa) as an endocrine disruptor by interacting with nuclear receptors: Insights from molecular dynamics (md) simulations. PLoS One 10, e0120330.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C., Liu F., Kannan K. (2012). Bisphenol s, a new bisphenol analogue, in paper products and currency bills and its association with bisphenol a residues. Environ. Sci. Technol. 46, 6515–6522. [DOI] [PubMed] [Google Scholar]
- Liao C., Kannan K. (2013). Concentrations and profiles of bisphenol a and other bisphenol analogues in foodstuffs from the United States and their implications for human exposure. J. Agric. Food Chem. 61, 4655–4662. [DOI] [PubMed] [Google Scholar]
- Mandrup K., Boberg J., Isling L. K., Christiansen S., Hass U. (2016). Low-dose effects of bisphenol a on mammary gland development in rats. Andrology 4, 673–683. [DOI] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12. [Google Scholar]
- Mosmann T. (1983). Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 65, 55–63. [DOI] [PubMed] [Google Scholar]
- Nakao T., Akiyama E., Kakutani H., Mizuno A., Aozasa O., Akai Y., et al. (2015). Levels of tetrabromobisphenol a, tribromobisphenol a, dibromobisphenol a, monobromobisphenol a, and bisphenol a in Japanese breast milk. Chem. Res. Toxicol. 28, 722–728. [DOI] [PubMed] [Google Scholar]
- Pais A., Degani H. (2016). Estrogen receptor-targeted contrast agents for molecular magnetic resonance imaging of breast cancer hormonal status. Front. Oncol. 6, 100.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins J. R., Antunes-Martins A., Calvo M., Grist J., Rust W., Schmid R., et al. (2014). A comparison of RNA-Seq and exon arrays for whole genome transcription profiling of the l5 spinal nerve transection model of neuropathic pain in the rat. Mol. Pain 10, 7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D. R., Chinnaiyan A. M. (2005). Integrative analysis of the cancer transcriptome. Nat. Genet. 37, S31–S37. [DOI] [PubMed] [Google Scholar]
- Robinson M. D., Oshlack A. (2010). A scaling normalization method for differential expression analysis of RNA-Seq data. Genome Biol. 11, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochester J. R. (2013). Bisphenol a and human health: a review of the literature. Reprod. Toxicol. 42, 132–155. [DOI] [PubMed] [Google Scholar]
- Rochester J. R., Bolden A. L. (2015). Bisphenol s and f: a systematic review and comparison of the hormonal activity of bisphenol a substitutes. Environ. Health Perspect. 123, 643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenmai A. K., Dybdahl M., Pedersen M., Alice van Vugt-Lussenburg B. M., Wedebye E. B., Taxvig C., et al. (2014). Are structural analogues to bisphenol a safe alternatives?. Toxicol. Sci. 139, 35–47. [DOI] [PubMed] [Google Scholar]
- Ryan N., Chorley B., Tice R. R., Judson R., Corton J. C. (2016). Moving toward integrating gene expression profiling into high-throughput testing: a gene expression biomarker accurately predicts estrogen receptor alpha modulation in a microarray compendium. Toxicol. Sci. 151, 88–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder A., Mueller O., Stocker S., Salowsky R., Leiber M., Gassmann M., Lightfoot S., Menzel W., Granzow M., Ragg T. (2006). The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 31, 3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioda T., Chesnes J., Coser K. R., Zou L., Hur J., Dean K. L., et al. (2006). Importance of dosage standardization for interpreting transcriptomal signature profiles: evidence from studies of xenoestrogens. Proc. Natl Acad. Sci. U. S. A. 103, 12033–12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioda T., Rosenthal N. F., Coser K. R., Suto M., Phatak M., Medvedovic M., et al. (2013). Expressomal approach for comprehensive analysis and visualization of ligand sensitivities of xenoestrogen responsive genes. Proc. Natl Acad. Sci. U. S. A. 110, 16508–16513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto A. M., Sonnenschein C., Chung K. L., Fernandez M. F., Olea N., Serrano F. O. (1995). The e-screen assay as a tool to identify estrogens: an update on estrogenic environmental pollutants. Environ. Health Perspect. 103(Suppl 7), 113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahlhut R. W., Welshons W. V., Swan S. H. (2009). Bisphenol a data in nhanes suggest longer than expected half-life, substantial nonfood exposure, or both. Environ. Health Perspect. 117, 784–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharp A. P., Maffini M. V., Hunt P. A., VandeVoort C. A., Sonnenschein C., Soto A. M. (2012). Bisphenol a alters the development of the rhesus monkey mammary gland. Proc. Natl Acad. Sci. U. S. A. 109, 8190–8195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipney H., Hunter L. (2010). An introduction to effective use of enrichment analysis software. Hum. Genom. 4, 202.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. EPA. (2015). Endocrine Disruptor Screening Program: Use of High Throughput Assays and Computational Tools. Notice. Federal Register (2015-15182, Jun 19, 2015) (FRL-9928-69).
- van de Vijver M. J., He Y. D., van't Veer L. J., Dai H., Hart A. A. M., Voskuil D. W., et al. (2002). A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 347, 1999–2009. [DOI] [PubMed] [Google Scholar]
- Vandenberg L.N., Colborn T., Hayes T.B., Heindel J.J., Jacobs D.R. Jr, Lee D.H., Shioda T., Soto A.M., vom Saal F.S., Welshons W.V., et al. (2012). Hormones and endocrine-disrupting chemicals: Low-dose effects and nonmonotonic dose responses. Endocr. Rev. 33, 378–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak A. L., Bulayeva N. N., Watson C. S. (2005). Xenoestrogens at picomolar to nanomolar concentrations trigger membrane estrogen receptor-alpha-mediated ca2+ fluxes and prolactin release in gh3/b6 pituitary tumor cells. Environ. Health Perspect. 113, 431–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Yu Y., Hertwig F., Thierry-Mieg J., Zhang W., Thierry-Mieg D., et al. (2015). Comparison of RNA-Seq and microarray-based models for clinical endpoint prediction. Genome Biol. 16, 133.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Kramer J. P., Calafat A. M., Ye X. (2014). Automated on-line column-switching high performance liquid chromatography isotope dilution tandem mass spectrometry method for the quantification of bisphenol a, bisphenol f, bisphenol s, and 11 other phenols in urine. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 944, 152–156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.