Abstract
Background
Plasmodium knowlesi is recognised as the main cause of human malaria in Southeast Asia. The disease is often misdiagnosed as P. falciparum or P. malariae infections by microscopy, and the disease is difficult to eliminate due to its presence in both humans and monkeys. P. knowlesi infections can rapidly cause severe disease and require prompt diagnosis and treatment. No protein biomarker exists for the rapid diagnostic test (RDT) detection of P. knowlesi infections. Plasmodium knowlesi infections can be diagnosed by PCR.
Methods and principal findings
Phosphoethanolamine-N-methyltransferase (PMT) is involved in malaria lipid biosynthesis and is not found in the human host. The P. falciparum, P. vivax and P. knowlesi PMT proteins were recombinantly expressed in BL21(DE3) Escherichia coli host cells, affinity purified and used to raise antibodies in chickens. Antibodies against each recombinant PMT protein all detected all three recombinant proteins and the native 29 kDa P. falciparum PMT protein on western blots and in ELISA. Antibodies against a PMT epitope (PLENNQYTDEGVKC) common to all three PMT orthologues detected all three proteins. Antibodies against unique peptides from each orthologue of PMT, PfCEVEHKYLHENKE, PvVYSIKEYNSLKDC, PkLYPTDEYNSLKDC detected only the parent protein in western blots and P. falciparum infected red blood cell lysates or blood lysates spiked with the respective proteins. Similar concentrations of PfPMT and the control, PfLDH, were detected in the same parasite lysate. The recombinant PfPMT protein was detected by a human anti-malaria antibody pool.
Conclusion
PMT, like the pan-specific LDH biomarker used in RDT tests, is both soluble, present at comparable concentrations in the parasite and constitutes a promising antimalarial drug target. PMT is absent from the human proteome. PMT has the potential as a biomarker for human malaria and in particular as the first P. knowlesi specific protein with diagnostic potential for the identification of a P. knowlesi infection.
Introduction
The Plasmodium genus includes over a hundred species that infect vertebrate hosts including birds, rodents, reptiles, amphibians and simians, by dipteran vectors [1]. Four Plasmodium species, P. falciparum, P. vivax, P. ovale and P. malariae are infective and transmissible to humans by natural mosquito bites. Thanks to control efforts, human malaria has been restricted to tropical and subtropical regions where it remains endemic primarily due to Anopheline mosquito habitats. An estimated 148–304 million malaria cases occurred in 2015, resulting in 429 thousand deaths [2]. A fifth species, P. knowlesi, known to be experimentally infective to humans [3], was not regarded as naturally transmissible to humans despite a case noted in 1965 [4]. In 2004, however, Singh et al. [5] detected P. knowlesi infections in a human population in Malaysia and subsequently P. knowlesi has been included as the fifth human infecting species [6]. It remains unclear if natural transmission between humans is common and the species is still considered a zoonosis [7–9].
Since the Singh et al. study [5], P. knowlesi has been identified as the main cause of malaria in Malaysia [10], with positive diagnoses reported in Cambodia, Indonesia, Myanmar, Philippines, Singapore, Thailand, Brunei, Vietnam and the Nicobar and Andaman islands of India [11–18]. Only Laos and East Timor remain unaffected in the Southeast Asia region [19]. Since Fong et al. demonstrated experimental human-to-human transmission in 1971 [20] the asymptomatic human infections recorded by Fornace et al. [21] may present an additional reservoir to the natural monkey infections. This makes malaria elimination in Southeast Asia difficult as it would entail eliminating P. knowlesi from both hosts. The A. leucosphyrus group of mosquitoes that transmit P. knowlesi infections, actively feeds outdoors, making conventional vector control measures less effective [9, 22]. Fortunately P. knowlesi remains in Southeast Asia for the time being as there are no known P. knowlesi carrying Anopheles vectors beyond the region. P. knowlesi cases in travellers returning to Europe, USA and Australasia have been reported [22]. A changing global climate may also affect the vector distribution as new suitable habitats may arise [23]. Accurate diagnosis of P. knowlesi infections is a critical tool for treatment and to understand the dynamics of this species and its impact on human populations within Southeast Asia and in visitors returning home from this region.
Genomic evidence suggests the P. knowlesi infections in Southeast Asia were present in wild macaques prior to human settlement [7]. The morphological similarity between P. knowlesi and the late blood stages of P. malariae and the early trophozoite stages of P. falciparum leads to misdiagnosis and allowed P. knowlesi infections to escape detection [3, 24, 25]. Minor morphological differences between early trophozoite and late schizonts of P. knowlesi and P. malariae, can be identified in well stained thin blood film slides under careful examination by an expert microscopist [25, 26]. Busy routine diagnostic laboratories often only examine thick blood films and large numbers of slides, increasing the chance of misdiagnosis [27]. It has therefore been recommended that in P. knowlesi endemic regions, microscopic identification of P. malariae be diagnosed as P. knowlesi/P. malariae [26]. Importantly, prompt treatment of P. knowlesi infections is essential to prevent the onset of severe disease due to its short (24 hour) red blood cell cycle [28]. P. knowlesi, fortunately remains sensitive to chloroquine [29] and is highly sensitive to artemisinins [9] making treatment of the disease simple if diagnosed quickly and accurately. Misdiagnosis as a mild P. malariae infection (72 hour red blood cell cycle) may delay such treatment [5] resulting in the onset of severe disease and potential fatality [28]. P. knowlesi was shown to be three times as likely as P. falciparum to cause severe infections [30], emphasising the need for a rapid test capable of detecting P. knowlesi infections at point-of-care.
Diagnosis is essential for appropriate treatment, conserving resources and prevention of fatal malaria infections. Today the WHO recommends confirmative point-of-care malaria diagnosis prior to drug treatment for malaria, to improve treatment efficacy and limit the selective pressure for antimalarial drug resistance [31]. Malaria diagnosis has evolved to encompass microscopic to molecular biology based methods [32, 33]. One of these methods, immunochromatographic separation and detection of proteins with antibodies on rapid diagnostic test (RDT) devices allows for point-of-care diagnosis in a field setting. RDTs are cheap, rapid and easy to perform and interpret [32]. The first malaria RDTs targeting specific malaria proteins were introduced in 1995 [33]. From 2008 to 2015 RDT sales have increased by 182 million units [2], attesting to the popularity of RDTs for a disease where testing in settings with limited infrastructure is common [32].
Three malaria protein biomarkers are commonly targeted by RDTs: P. falciparum histidine rich protein 2 (PfHRP2), lactate dehydrogenase (LDH) and aldolase [33]. The amino-acid sequence of LDH is conserved across malaria species and “pan-malaria” RDTs detecting the LDH protein can diagnose the presence of P. falciparum, P. vivax, P. malariae, P. ovale and P. knowlesi parasites as malaria but do not differentiate between species [33, 34]. LDH was first introduced for malaria diagnostics in 1999 [35] and over 20 monoclonal antibodies with specificity for various Plasmodium LDH orthologues have since been developed [34]. Attempts to raise P. knowlesi specific monoclonal antibodies against LDH have been unsuccessful [36, 37]. Using a combination of the current monoclonal antibodies against LDH, specific diagnosis of P. knowlesi is possible [34] with good specificity (96%), but unacceptably low sensitivity (32–42, 0–45, 24–73%) [19, 22, 38]. Species-specific epitopes have been identified in the structure of the LDH protein allowing for the detection and differentiation of P. falciparum and P. vivax infections [39]. A recently identified new P. falciparum biomarker, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) has two unique P. falciparum GAPDH epitopes and an epitope common to GAPDH from all malaria species [40]. Antibodies against each of the GAPDH peptides detected the native protein. A unique epitope in P. knowlesi GAPDH was identified, but antibody based evidence to support targeting the epitope is not yet available [40].
To date the only definitive diagnosis of P. knowlesi is by PCR [21], which is more expensive than microscopy or RDTs and requires specialised training and equipment [25, 32]. PCR is considered a reference laboratory conformational tool/test. The first set of nested PCR primers specific for P. knowlesi detection was developed against the small subunit ribosomal RNA gene [5]. Multiple primer sets and several target genes for nested and real time PCR have since been identified for the detection of P. knowlesi as reviewed by [25] including a single step PCR target, Pkr140, unique to P. knowlesi. Loop mediated isothermal amplification methods (LAMP) offer a simpler alternative to PCR but are not as affordable or rapid as RDTs. P. knowlesi specific LAMP assays target mitochondrial DNA [41], small subunit ribosomal RNA [42], beta tubulin [43, 44], or apical membrane antigen-1 [45] genes.
Currently there are no RDTs or protein biomarkers for the detection of P. knowlesi infections [8, 21, 22, 44]. This study provides evidence for phosphoethanolamine-N-methyltransferase (PMT) as a malaria biomarker. The PMT gene is absent from the human genome, but present in all 5 human infecting Plasmodium species with PMT protein expression confirmed in P. falciparum, P. vivax and P. knowlesi malaria parasites [46–48]. This is a similar characteristic to the popular P. falciparum PfHRP2 diagnostic target, but unlike PfHRP2, PMT appears to be essential for parasite development [48–50]. PMT is involved in Plasmodial lipid metabolism and has potential as a drug target [46, 47, 51–55]. Here it is shown that antibodies against the protein or a peptide region of the protein detected Plasmodial PMT and thus could detect a malaria infection. P. knowlesi PMT and P. falciparum PMT-specific epitopes were identified and antibodies against these particular epitopes enable the possible identification of P. knowlesi parasites and differentiation among P. knowlesi, P. vivax and P. falciparum parasites.
Methods
Ethics
University of KwaZulu-Natal Animal Ethics Committee approval for the study was obtained. Reference:004/15//Animal. This approval complies with the South African National Standards:SANS 10386:2008, The care and use of animals for scientific purposes, ISBN 978-0-626-22296-3.
In silico and bioinformatics analysis to identify Plasmodial protein and peptide targets
Identification of phosphoethanolamine-N-methyltransferase (PMT) as a potential diagnostic protein target was done in silico as described previously [40]. Proteins were ranked based on abundance from transcription and proteomic studies [56, 57]. Peptides that were conserved in all three orthologues (common epitope) or unique to P. falciparum (PfPMT), P. vivax (PvPMT) and P. knowlesi (PkPMT) PMT proteins were selected based on sequence alignment and predicted immunogenicity (Predict7™ analyses [58]). The crystal structure of PfPMT was used to assess antibody accessibility of the peptides [55, 59]. The four selected peptide epitopes were synthesized with either N- or C-terminal cysteines for coupling to rabbit albumin and affinity resins (GL Biochem Ltd. Shanghai, China).
Recombinant expression and affinity purification of three PMT protein orthologues
Plasmids encoding the genes for P. falciparum, P. vivax, and P. knowlesi orthologues of PMT were kindly provided by B. Mamoun (Yale University) [46, 52, 54]. All three PMT orthologues were recombinantly expressed in BL21(DE3) E. coli (Novagen, Darmstadt, Germany) cells and the sequences of the cloned gene for each protein confirmed by DNA sequencing. The P. falciparum protein was expressed from a pET 15(b) vector in lysogeny broth (LB) (1% (w/v) tryptone; 0.5% (w/v) yeast extract; 85 mM NaCl; 11 mM glucose) supplemented with 100 μg/ml ampicillin and induced with 1 mM isopropyl thioglucopyranoside (IPTG) for 4 h at 37°C. The other two orthologues were expressed overnight at 37°C from a pET 28(a) vector using auto-inducing terrific broth (1.2% (w/v) tryptone, 2.4% (w/v) yeast extract, 0.4% (w/v) glycerol, 0.231% (w/v) KH2PO4, 1.254% (w/v) K2HPO4) [60] supplemented with 25 μg/ml kanamycin. The histidine tagged recombinant proteins were purified on TALON® cobalt affinity resins, according to manufacturer’s instructions as described previously [40]. A 50 mM NaH2PO4, 300 mM NaCl, 0.02% (w/v) NaN3 at pH 8.0 buffer was used throughout with the addition of 10 mM imidazole in the sample loading buffer to reduce the binding of E. coli proteins to the TALON® affinity resin and 250 mM imidazole in the elution buffer.
Molecular exclusion chromatography
Four milligrams of the purified recombinant proteins were passed over a molecular exclusion chromatography column (HiPrep 16/60 Sephacryl S-200 column, 120 ml column volume) in 50 mM NaH2PO4, 150 mM NaCl at pH 8.0 to verify their respective molecular mass. Buffer flow was 0.5 ml/min and the absorbance of eluents monitored at 280 nm and 4 ml fractions collected (ÄKTA Prime Plus, GE Healthcare Life Sciences). The column was calibrated and samples prepared as described previously [40].
Raising antibodies in chickens against proteins and peptides
Ethical clearance for this study was granted by the animal research ethics committee of the University of KwaZulu-Natal (004/15//Animal) and all institutional guidelines for animal husbandry were adhered to. The chickens used were Hyline Brown, sourced at the University of KwaZulu-Natal, Ukulinga research facility. Animals (layers, 10 weeks old) were fed layers Mash (Meadow feeds, South Africa) ad libitum and had constant access to water by means of nipple drip-feed. Animals were euthanased at the end of the experiment by decapitation (AVMA guidelines for the Euthanasia of Animals 2013). Each animal was housed individually and monitored twice daily when eggs were collected and marked. Chickens were used in the experiment because antibodies can be harvested from egg yolks, thus avoiding invasive procedures required for taking blood, fewer animals are required and local inflammatory responses associated with Freund’s complete adjuvant are not seen in chickens [61, 62]. Antigen was emulsified in Freund’s complete adjuvant for the initial immunization only (week 0) and Freund’s incomplete adjuvant for booster immunizations (weeks 2, 4, 6). Animals were immunized in the breast muscle, with 50 μg of the recombinant PMT proteins or 500 μg of the respective peptides conjugated to rabbit albumin used per immunization. Principles recommended by the Hyline Brown Management Guide were adhered to (www.hyline.com/UserDocs/Pages/BRN_COM_ENG.pdf). Antibodies against the whole recombinant proteins or the selected peptides coupled to rabbit albumin carrier were raised and affinity purified as described [39, 63]. Briefly, the isolated IgY was passed over recombinant PMT or peptide affinity columns. Non-specific antibodies were removed by extensive washing and the bound antibodies were then eluted with a change in pH. The resulting polyclonal affinity purified antibodies were used in western blotting and ELISA assays.
SDS-PAGE and western blotting
Reducing SDS-PAGE gels comprising 4% stacking and 12.5% resolving gels were used throughout [64]. All reference gels were stained with Coomassie Brilliant Blue R-250 [65]. Proteins were transferred electrophoretically to nitrocellulose and the nitrocellulose was blocked with 5% (w/v) low fat milk powder in TBS (20 mM tris; 200 mM NaCl at pH 7.4) for 1 h [66]. Primary chicken IgY or a mouse anti-His6 antibody (1:6000) (Merck, Darmstadt, Germany, cat # 05–949) and secondary goat anti-mouse-HRPO (1:10000, cat # 115-035-003), rabbit anti-chicken-HRPO (1:15000, cat # 303-035-003) and rabbit anti-human-HRPO (1:6000, cat # 309-035-003) (Jackson IR laboratories Inc., Baltimore, PA, USA) antibodies were prepared in 0.5% (w/v) BSA-TBS and incubated for 1 or 2 h respectively. All other antibody concentrations are stated in the text. Protein was visualised by developing nitrocellulose bound proteins for 30 min with 3.4 mM 4-chloro-1-naphthol and 0.04% (v/v) H2O2 as substrate. Enhanced chemiluminescence (ECL) was used to detect native PfPMT in a Pf(D10) lysate using a previously described protocol [40]. All images were captured using the G:Box Chemi XR5 system (Syngene).
Measuring protein concentration
Protein concentrations were determined using the Bradford assay [67]. IgY and human IgG concentrations were calculated using A 280 nm values and the respective extinction coefficients: IgY (ε = 1.25) [68] or human IgG (ε = 1.35) [69].
Coupling IgY to HRPO
Eight milligrams of horse radish peroxidase (HRPO) (1360 Units, Boehringer Mannheim) was conjugated using the periodate coupling method (omitting the fluorodinitrobenzine reaction) to an equivalent concentration of affinity purified IgY raised against the whole recombinant PvPMT protein.
ELISA
ELISA plates were coated with sample in PBS overnight at 4°C. Wash steps comprising three PBS-Tween 20 (0.1% (v/v)) and three PBS washes were performed between each incubation step. Wells were blocked with 0.5% (w/v) BSA-PBS (1 h at 37°C) and antibody incubations were performed (1 h at 37°C) in this buffer with Tween 20 (0.1% (v/v)). Either 2,2’-azino-bis(ethylbenzothiazoline-6-sulphonic acid (ABTS) or 3,3’,5,5’-tetramethylbenzidine (TMB) substrate prepared in a 150 mM citrate-phosphate buffer at pH 5.0, was added to the wells and incubated in the dark at room temperature for 1 h. Antibody production in chickens was monitored using an indirect ELISA as described [63]. A double antibody sandwich (DAS) ELISA was optimized using the anti-peptide antibodies as the capture antibodies (between 0.5 to 1 μg) and the anti-rPvPMT-HRPO coupled antibodies (0.5 μg) for detection. For the uninfected whole blood lysate spiked ELISAs, a fresh 1 ml human A-positive blood sample was lysed in 14 ml of 150 mM NH4Cl, 10 mM NaHCO3, 1 mM Na2EDTA, pH 7.4 buffer [70]. Tween 20 was added (0.1% (v/v)) and the lysed blood samples were aliquoted and spiked with the respective recombinant PMT protein orthologues alone or in combinations, or spiked with recombinant P. falciparum lactate dehydrogenase (rPfLDH control), at 100 ng per ELISA well. Statistical analysis was done using the Student’s t-test, with p ≤ 0.05 and ≤ 0.001 indicated with “*” or “**” respectively, where applicable.
Additivity index
To assess if the antibodies bound the PMT proteins additively or competitively, their respective additivity indices were determined from average absorbance readings in duplicate ELISAs [71]. The recombinant PMT orthologues were coated at 250 ng/well with the following saturating primary antibody concentrations: anti-rPfPMT, anti-PvVYSIKEYNSLKDC (PvPMT peptide), anti-PLENNQYTDEGVKC (common PMT peptide) and anti-rPkPMT at 125 ng; anti PfCEVEHKYLHENKE (PfPMT peptide) and anti-rPvPMT at 250 ng; anti-PkLYPTDEYNSLKDC (PkPMT peptide) at 1 μg.
Human IgG anti-malarial antibodies
Three hundred milligrams of a human anti-malaria antibody pool [72, 73] was used to affinity purify human IgG antibodies against rPfPMT as described for other proteins [74].
Results
Recombinant expression, affinity purification and molecular exclusion chromatography of P. falciparum, P. vivax and P. knowlesi phosphoethanolamine-N-methyltransferase (PMT)
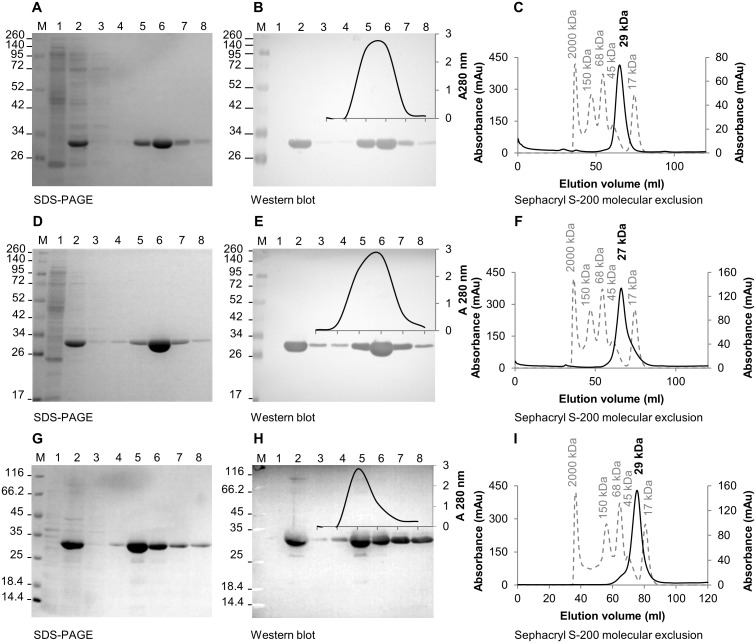
The orthologues of PMT, from P. falciparum, P. vivax and P. knowlesi were recombinantly expressed in BL21(DE3) E. coli host cells as histidine tagged proteins and the proteins were affinity purified, using a TALON® resin (Fig 1). Induced E. coli lysates contained a prominent 29 kDa rPf- and rPkPMT (Fig 1A and 1G, lane 2) and 27 kDa rPvPMT protein band (Fig 1D, lane 2). The same protein bands were detected by an anti-His6 tag antibody (Fig 1B, 1E and 1H, lane 2). The intensity of each protein band on the gel corresponded with the A 280 nm elution profile from an affinity matrix (insert in Fig 1B, 1E and 1H, lanes 3–8). No proteins were detected in the untransformed E. coli lysates (Fig 1B, 1E and 1H, lane 1). The purified recombinant proteins resolved as monomers of 29 kDa for the P. falciparum and P. knowlesi proteins and 27 kDa for the P. vivax protein on a Sephacryl S-200 molecular exclusion chromatography column (Fig 1C, 1F and 1I respectively). The purified recombinant PMT proteins were the source material for raising antibodies in chickens.
Fig 1. Purification and molecular exclusion chromatography of recombinant P. falciparum, P. vivax and P. knowlesi PMT.
Samples (A, D and G) from the respective purification steps of three recombinantly expressed PMT orthologues, purified with an anti-His6 affinity resin, were resolved on 12.5% reducing SDS-PAGE gels. Western blots (B, E and H) of the gels were probed with the appropriate antibodies. (M) Molecular weight marker; (lane 1) untransformed E. coli lysate; (lane 2) induced E. coli lysate; (lane 3) final wash and (lanes 4 to 8) eluents 1 to 5 from the Talon affinity matrix. The elution profiles (280 nm) were overlaid on the respective blots (B, E and H). Elution profiles (C, F and I) in milli absorbance units (mAu) of each PMT orthologue from a Sephacryl S-200 chromatography matrix. The calibration standards were plotted on the primary axis (dashed line), with the respective sizes indicated above each peak. Each PMT profile (black line) was plotted on the secondary axis and the estimated protein size indicated above the respective peaks in bold.
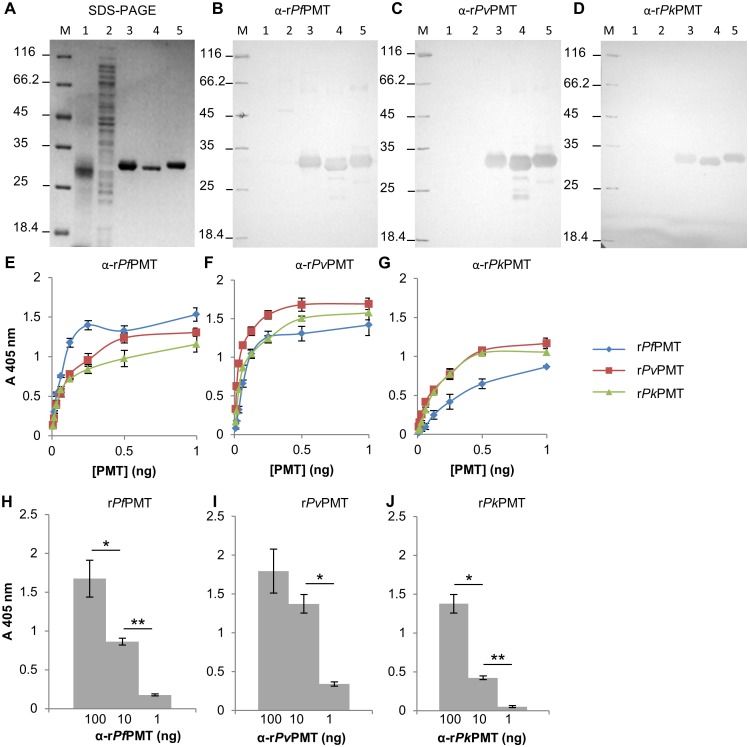
Detection of the recombinantly expressed PMT orthologues with chicken immunoglobulin Y (IgY) raised against PMT from three malaria species
Antibodies (IgY) were isolated from the eggs of chickens injected with each of the three PMT proteins and affinity purified using the parent protein coupled to an AminoLink® resin. All the affinity purified antibodies were evaluated in western blots and ELISA and none of the antibodies detected proteins in an uninfected red blood cell lysate or an untransformed E. coli lysate (Fig 2B to 2D, lanes 1 and 2). Each of the antibodies against each of the PMT proteins from a different species detected the parent protein and both of the other two PMT orthologues in western blots (Fig 2B to 2D, lanes 3 to 5). The size of the protein detected corresponded to that obtained in the SDS-PAGE gels described above. In an ELISA format (Fig 2E to 2G), antibodies against PfPMT and PvPMT had slightly higher reactivity for the parent protein than that for the other orthologues. The anti-rPkPMT antibodies detected the rPvPMT and its rPkPMT parent protein equally. Interestingly the anti-rPvPMT and anti-rPkPMT antibodies detected both rPvPMT and rPkPMT better than the rPfPMT orthologue (Fig 2F and 2G). The lower limits of detection of the antibodies were assessed by titration of the antibodies against a set PMT concentration. The anti-rPfPMT and anti-rPvPMT antibodies detected 1 ng of PMT (Fig 2, 2H and 2I respectively), and the anti-rPkPMT antibodies detected 10 ng of PMT (Fig 2J).
Fig 2. Detection of the recombinant PMT orthologues with the respective anti-recombinant PMT IgY.
(A) Reducing 12.5% SDS-PAGE reference gel for (B to D) western blots. The reference gel was the same as shown in Fig 4(A) as all experiments were performed at the same time on the same batch of samples. (M) Molecular weight marker, (lane 1) uninfected red blood cell lysate (25 μg), (lane 2) untransformed E. coli lysate (25 μg), (lanes 3 to 5) rPfPMT, rPvPMT and rPkPMT respectively. (B) Western blots probed with anti-rPfPMT or (C) anti-rPvPMT or (D) anti-rPkPMT IgY (10 μg) and detected with a rabbit anti-chicken-HRPO secondary antibody. (E to G) IgY against each of the whole recombinant PMT proteins (100 ng) was used to detect a range (0.8 to 100 ng) of concentrations of (E) rPfPMT, (F) rPvPMT and (G) rPkPMT in an ELISA. (H to J) IgY was diluted (100 to 1 ng) and used to detect a single concentration (100 ng) of rPfPMT, rPvPMT and rPkPMT respectively. Antibodies against a protein were denoted as “α” and the protein name. ELISA results present averages of triplicate values with standard deviations. Student’s t-test with p ≤ 0.05 and ≤ 0.001 are indicated with “*” or “**” respectively.
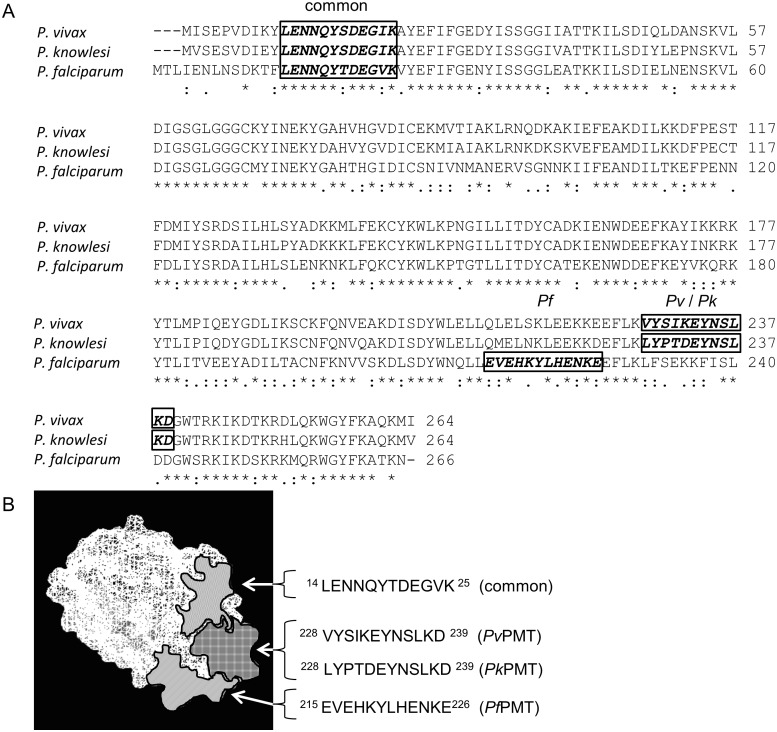
Selection of a shared or a unique peptide epitope for PMT from each species
Peptides in each PMT protein from a single species were selected as antibody targets. Peptide epitopes were identified by sequence alignment, epitope prediction software (Predict7™), 3D crystal structure and BLASTp analyses as described previously (Fig 3; [39, 40]). The four selected peptides are illustrated on an image showing the alignment of the three amino acid sequences (Fig 3A). The image depicts the “common” PLENNQYTDEGVKC, the P. falciparum specific PfCEVEHKYLHENKE, the P. vivax specific PvVYSIKEYNSLKDC and P. knowlesi specific PkLYPTDEYNSLKDC peptide sequences. The predicted surface location of each of the peptides was highlighted on a picture of the PfPMT crystal structure (Fig 3B; [55, 59]) showing that the peptide sequences are surface accessible for antibody interaction, are separate and do not overlap.
Fig 3. Sequence alignment of PMT orthologues and location of peptide epitopes on the PfPMT crystal structure.
(A) Alignment of P. vivax (XP_001614208.1), P. knowlesi (XP_002259925.1) and P. falciparum (Pf3D7_1343000.1) PMT protein orthologues. Potential epitopes selected by Predict7™ analysis are indicated on the sequences as the common (boxed “common”) and species specific epitopes (boxed “Pf” or “Pv” or “Pk”). (B) The surface location of each selected peptide epitope is indicated on the 3D crystal structure of PfPMT (3uj6).
BLASTp analysis (Table 1) was used to compare the amino acid sequences of the chosen PMT peptides with the corresponding amino-acid sequences found in PMT from other Plasmodium species. The BLASTp analysis of the peptides identified Plasmodial PMT sequences with 100% identity and no epitopes with significant sequence identity were found for either human or human pathogen proteins. The peptide epitopes were synthesized, coupled to a rabbit albumin carrier protein and injected into chickens to raise antibodies.
Table 1. BLASTp analysis of selected PMT peptide epitopes.
| Species | PfPMT1 | PvPMT | PkPMT | common PMT |
|---|---|---|---|---|
| P. falciparum | EVEHKYLHENKE (100) | LFSEKKFISLDD (42) | LFSEKKFISLDD (33) | LENNQYTDEGVK (100) |
| P. vivax | QLELSKLEEKKE (42) | VYSIKEYNSLKD (100) | VYSIKEYNSLKD (67) | LENNQYSDEGIK (83) |
| P. knowlesi | QMELNKLEEKKD (33) | LYPTDEYNSLKD (67) | LYPTDEYNSLKD (100) | LENNQYSDEGIK (83) |
| P. ovale | EMELHRLNEKKE (50) | EYSLKDYNTLKD (67) | EYSLKDYNTLKD (50) | LESYQYSDESIK (58) |
| P. malariae | EMEVNRLEQKKE (42) | KYSTKEYESLIN (58) | KYSTKEYESLIN (50) | LENNQYSDEGIK (83) |
| Overall identity | ::* *.::*: | :..:: *: | :..:: *: | **. **:**.:* |
| BLASTp | P. falciparum | P. vivax | P. knowlesi | P. falciparum, P. reichenowi |
1. The candidate epitopes were used as BLASTp query sequences to assess their specificity to the Plasmodium PMT orthologues and any matches with 100% identity were listed. The amino acid sequences from putative P. ovale and P. malariae PMT were included.
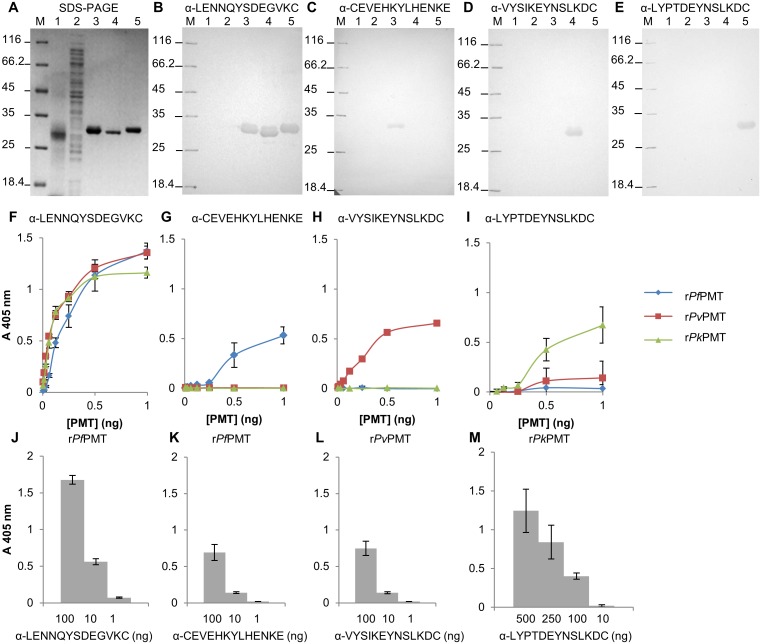
Detection of the recombinantly expressed PMT orthologues with the respective anti-peptide IgY
Antibodies raised against each peptide in chickens were affinity purified using the respective parent peptides coupled to a SulfoLink® affinity resin and evaluated in a western blot and a direct ELISA (Fig 4). None of the anti-peptide antibodies detected any proteins in either an uninfected red blood cell lysate, or an untransformed E. coli lysate (Fig 4B to 4E, lanes 1 and 2). The antibodies against the common peptide (PLENNQYTDEGVKC) detected all three PMT orthologues at the appropriate sizes in a western blot (Fig 4B, lanes 3 to 5) and with similar intensity of signal in an ELISA format (Fig 4F). The antibodies against the P. falciparum PMT unique peptide (PfCEVEHKYLHENKE), the P. vivax (PvVYSIKEYNSLKDC) or P. knowlesi (PkLYPTDEYNSLKDC) peptides only detected their respective PMT proteins from the parent species in a western blot (Fig 4C, lane 3, D and E, lanes 4 and 5 respectively) and an ELISA (Fig 4G, 4H and 4I respectively). The strongest signals were obtained for antibodies against the common peptide (Fig 4J to 4M). The limit of detection of the anti-peptide antibodies was assessed by titration of the antibodies against a set series of PMT concentrations. The anti- P. falciparum (PfCEVEHKYLHENKE) and P. vivax (PvVYSIKEYNSLKDC) specific epitope antibodies detected their specific PMT protein orthologues at similar concentrations (10 ng; Fig 4K and 4L respectively), but the anti- P. knowlesi peptide (PkLYPTDEYNSLKDC) antibodies detected rPkPMT at a higher concentration (100 ng; Fig 4M).
Fig 4. Detection of the recombinant PMT orthologues with the respective anti-PMT peptide IgY.
(A) Reducing 12.5% SDS-PAGE reference gel for (B to E) western blots. The reference gel was the same as shown in Fig 2(A). All experiments were performed at the same time on the same samples. (M) Molecular weight marker, (lane 1) uninfected red blood cell lysate (25 μg), (lane 2) untransformed E. coli lysate (25 μg) and (lanes 3 to 5) rPfPMT, rPvPMT and rPkPMT respectively. (B) Western blots probed with anti-PLENNQYTDEGVKC, (C) anti-PfCEVEHKYLHENKE, (D) anti-PvVYSIKEYNSLKDC, or (E) anti-PkLYPTDEYNSLKDC IgY (10 μg) and detected with a rabbit anti-chicken-HRPO secondary antibody using 4-chloro-1-naphthol and H2O2. (F to I) Detection of rPfPMT, rPvPMT and rPkPMT respectively (0.8 to 100 ng) in an ELISA with anti-peptide IgY (100 ng). (J to M) ELISA plates were coated with the PMT orthologues at 100 ng and detected with different dilutions of anti-PMT peptide IgY, with 100 to 1 ng (J to L) and 500 to 10 ng (M). All ELISAs were done in triplicate and the standard deviations included. Antibodies against a peptide were denoted as “α” and the peptide sequence. Student’s t-test with p ≤ 0.05 and ≤ 0.001 are indicated with “*” or “**” respectively.
Evaluating epitopes as antibody targets for the capture and detection of PMT in an ELISA
To develop an ELISA or RDT where both a capture and a detection antibody against the same protein are required, it is necessary to evaluate potential competition in binding to the protein between the two antibodies. This was done by evaluating the additivity of the different antibody combinations (anti-peptide and anti-whole protein antibodies) in ELISA formats. If the signal increases with the addition of the second antibody there is little competition. All the whole protein and anti-peptide antibodies generated a signal that increased by 50% or more indicative of a lack of competition (Table 2). Additivity values above 70% were obtained for 5 of the combinations and the highest (100%) were obtained with the anti-PvVYSIKEYNSLKDC anti-PLENNQYTDEGVKC combination; the anti-rPkPMT whole protein anti-PkLYPTDEYNSLKDC combination and the anti-PkLYPTDEYNSLKDC and anti-PLENNQYTDEGVKC combination (Table 2).
Table 2. Additivity indices of antibodies raised against the recombinant PMT orthologues and the selected peptide epitopes.
| Antibody | α-PfCEVEHKYLHENKE | α-PLENNQYTDEGVKC |
| α-rPfPMT | 55.9* | 59.5 |
| α-PfCEVEHKYLHENKE | NA | 83 |
| Antibody | α-PvVYSIKEYNSLKDC | α-PLENNQYTDEGVKC |
| α-rPvPMT | 83.3 | 61.8 |
| α-PvVYSIKEYNSLKDC | NA | 100 |
| Antibody | α- PkLYPTDEYNSLKDC | α-PLENNQYTDEGVKC |
| α-rPkPMT | 100 | 71.7 |
| α-PkLYPTDEYNSLKDC | NA | 100 |
*All additivity index values represent increased signal percentages calculated from average absorbance values.
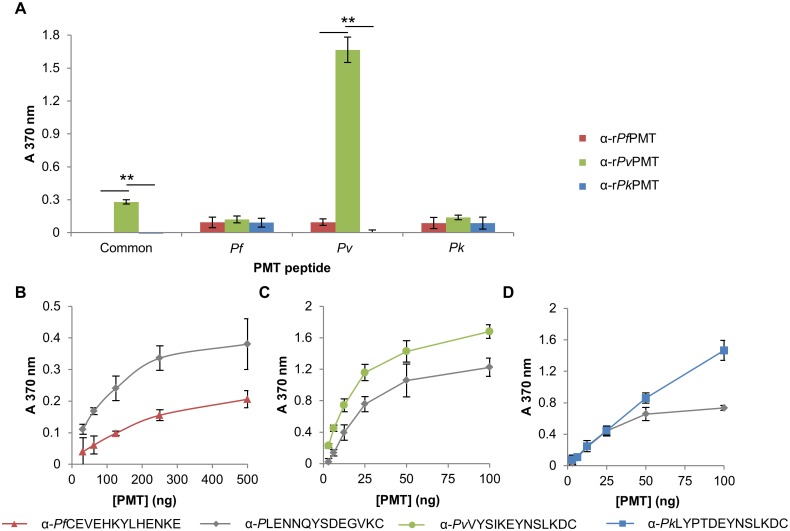
It was important to evaluate the peptide epitopes as possible antibody targets for the capture and detection of the PMT protein (Fig 5). The IgY antibodies raised against the whole recombinant PMT orthologues were tested for their reactivity with the selected peptides (Fig 5A). Only the anti-rPvPMT IgY reacted strongly with the P. vivax specific (PvVYSIKEYNSLKDC) and all other interactions were negligible (Fig 5A). When combining the antibodies in a capture and detection ELISA (Fig 5B, 5C and 5D), using the antibodies against the common peptide as the capture antibody, resulted in lower detection signals compared to using the species specific anti-peptide antibodies, with rPfPMT being the only exception. This was in agreement with the additivity results (Table 2). Antibodies or anti-peptide antibodies against rPfPMT produced the lowest signals in the ELISA tests in comparison to the ELISAs detecting the two other PMT orthologues.
Fig 5. PMT peptide detection with the anti-rPMT antibodies and DAS-ELISA based capture of rPMT orthologues.
(A) PMT peptides coated at 100 ng were detected with the anti-recombinant PMT IgY (100 ng). (B to D) The recombinant PMT proteins were captured with the anti-peptide antibodies and detected with the anti-PvPMT-HRPO conjugate. All results represent triplicate values with standard deviations. Antibodies against a peptide were denoted as “α” and the peptide sequence. Student’s t-test with p ≤ 0.05 and ≤ 0.001 are indicated with “*” or “**” respectively. Note the different scale in (B) compared to (C and D).
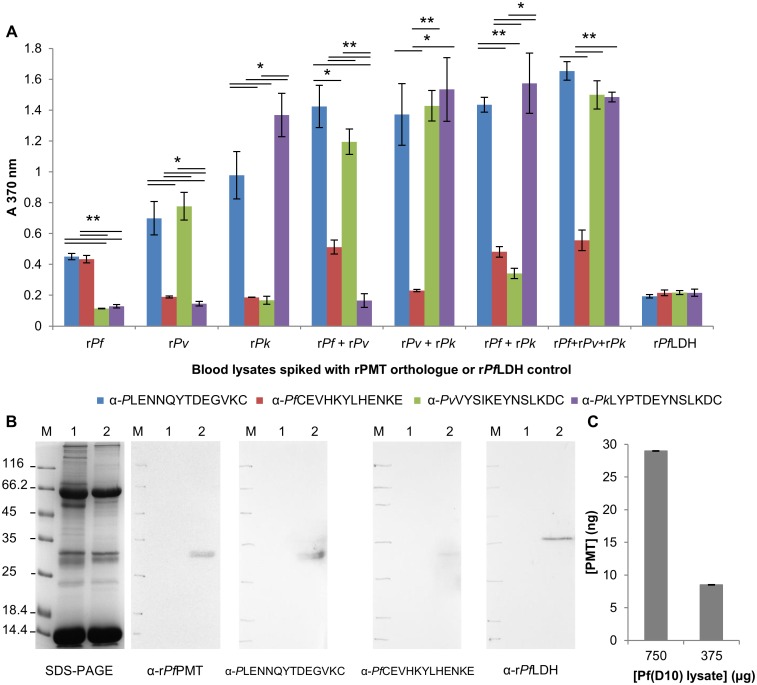
Detecting PMT in spiked uninfected human blood lysates and native PMT in a Pf(D10) culture lysate by DAS ELISA
During the red blood cell cycle, as the parasites rupture and invade new host red cells, parasite proteins, like LDH and PfHRP2 are released into peripheral circulation and can be detected to diagnose infection. Therefore any potential interference by blood cell proteins when detecting the PMT protein was evaluated. In an ELISA, PMT proteins were detected by all the antibodies and blood cell lysate components did not influence the assay (Fig 6). Antibodies raised against the common epitope captured all three PMT orthologues from the spiked blood samples. The anti-peptide antibodies against PMT from each species detected only the parent proteins in the spiked blood cell lysates (Fig 6A). When blood was spiked with recombinant PfPMT and PvPMT there was a high signal for the anti-PvPMT peptide antibody. This signal was lower than that for the anti-PfPMT antibody (P<0.05). The anti-PfPMT peptide antibody had the lowest signal of all the anti-peptide antibody combinations in the ELISA. Native PfPMT was detected as a 29 kDa protein in a Pf(D10) infected culture lysate by western blot (Fig 6B) with the anti-rPfPMT IgY as well as the antibodies raised against the common (PLENNQYTDEGVKC) and P. falciparum specific (PfCEVEHKYLHENKE) peptides. In the ELISA format PfPMT was captured using the anti-PfCEVEHKYLHENKE antibodies (Fig 6C) and detected approximately 28 ng in the 750 μg of Pf(D10) lysate protein.
Fig 6. Spiked blood DAS-ELISAs and detection of native PfPMT in a Pf(D10) parasite culture lysate.
(A) Uninfected, A-positive human whole blood lysates were spiked with recombinant PMT orthologues (100 ng) and captured with anti-peptide IgY (all coated at 500 ng, except anti-PfCEVEHKYLHENKE at 1 μg). Captured PMT proteins were detected with anti-rPvPMT-HRPO coupled IgY (500 ng). rPfLDH (100 ng) spiked blood lysates served as a negative control. (B) Native P. falciparum PMT was detected on western blots, with the Coomassie stained reference gel on the left (SDS-PAGE). (M) Molecular weight marker, (lane 1) uninfected A-positive human whole blood lysate (100 μg) and (lane 2) Pf(D10) culture lysate (100 μg). Western blots were probed with: anti-rPfPMT (10 μg); anti-PLENNQYTDEGVKC or anti-PfCEVEHKYLHENKE or anti-rPfLDH IgY (100 μg). (C) Detection of native P. falciparum PMT in a Pf(D10) culture lysate with the anti-PfCEVEHKYLHENKE as capture and anti-rPvPMT-HRPO as the detection antibody. TMB + H2O2 was used as the substrate in all ELISAs, each performed in triplicate with standard deviations shown. Antibodies were denoted as “α” and the peptide sequence or protein name. Student’s t-test with p ≤ 0.05 and ≤ 0.001 are indicated with “*” or “**” respectively.
Presence of anti-PfPMT antibodies in a human anti-malaria antibody pool
A human anti-malaria antibody pool [72, 73] was passed consecutively over four affinity resins with recombinant P. falciparum LDH, GAPDH, Cox17 and PMT coupled to the resins. Approximately 0.41 mg of affinity purified human antibodies detecting rPfPMT was eluted from the rPfPMT affinity resin (Table 3). The yield of antibodies against rPfPMT was lower than those that bound to recombinant P. falciparum LDH and similar (0.42 mg) to those binding recombinant P. falciparum GAPDH ([40]) on an affinity matrix. The lowest yield was from antibodies binding to recombinant P. falciparum Cox17, a copper chaperone [74].
Table 3. Yields of human IgG against four recombinant P. falciparum proteins.
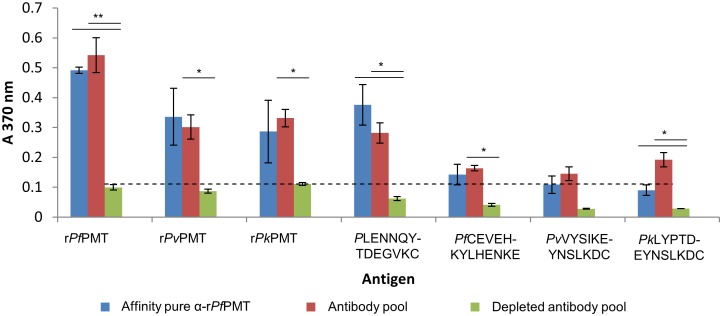
The human antibodies affinity purified on the rPfPMT affinity matrix detected the PMT protein in an ELISA format (Fig 7), and the limit of detection was 100-fold higher than the crude human antibody pool before isolation of the antibody. The unbound human antibodies from the rPfPMT affinity resin did not react with any of the proteins tested at the same concentration. Both the crude and rPfPMT specific human antibodies reacted with both the P. vivax and the P. knowlesi PMT orthologues as predicted from the similarity in amino acid sequence between the proteins and from data with IgY raised against the whole recombinant PMT orthologues (Fig 2B to 2D and 2E to 2G). Interestingly the human antibodies detected the common peptide (PLENNQYTDEGVKC), (Fig 7), unlike the IgY (Fig 5A). Reactivity with the other PMT peptides was negligible compared to the background control.
Fig 7. Detection of anti-rPfPMT antibodies in a human anti-malaria hyperimmune antibody pool.
A human anti-malaria hyperimmune antibody pool was passed over a rPfPMT affinity column. The affinity purified anti-rPfPMT human antibodies (100 ng) were used to detect each of the recombinant PMT orthologues or each of the PMT peptide epitopes coated directly onto ELISA plates at 100 ng per well. The pool before (antibody pool) and the pool after (depleted antibody pool) passing the human antibodies over the rPfPMT affinity resin were used at 10 μg. All readings were done in triplicate with standard deviations shown. A cut-off value for positive reactions of three times the standard deviation of the background control was included as a horizontal dashed line. Student’s t-test with p ≤ 0.05 and ≤ 0.001 are indicated with “*” or “**” respectively.
Discussion
A P. knowlesi infection is difficult to diagnose with microscopy [25, 26] and can be diagnosed with PCR [25] or LAMP assays [41–45]. A specific point-of-care rapid diagnostic test to detect a P. knowlesi infection is not available at present [8, 21, 22, 44]. Such a test would improve diagnosis and treatment of the infection and aid in tracking the spread of this species in the Southeast Asia region. We identified phosphoethanolamine-N-methyltransferase (PMT) as a P. knowlesi specific protein biomarker with the potential to identify a P. knowlesi infection and to differentiate P. knowlesi from a P. falciparum or P. vivax infection.
PMT selection and background to the protein
The Plasmodial database [75] was screened to identify proteins that are highly expressed, are expressed by the parasite and not the host or have unique peptide sequences, and can be recombinantly expressed as soluble proteins [40]. Based on these parameters PMT was identified as an attractive target. The enzyme is expressed by P. falciparum, P. vivax and P. knowlesi species and is not expressed by the host [46–48]. Based on in silico data, the protein is expressed at similar or higher levels than the current diagnostic target, LDH [40, 56, 57]. An additional advantage is that the protein, unlike PfHRP2, the most popular protein diagnostic target, has been shown to be essential for the parasite in different stages of development [48–50]. The implication is that the gene for the protein is unlikely to be deleted from the parasite’s genome as has been recorded for the PfHRP2 gene in a number of geographic locations [76–79]. PMT has been characterised and is being explored as an antimalarial drug target [46, 47, 51–55].
Recombinant PMT orthologue model
The PMT gene is present and the protein is expressed by P. falciparum, P. vivax and P. knowlesi parasites [46, 54]. The gene is also present in P. ovale and P. malariae genomes ([75], NCBI accessed Aug. 2017). PMT appears to be expressed in P. reichenowi and P. gallinaceum, but is absent from all rodent malaria species [53]. The affinity purified recombinant PMT proteins resolved on a SDS-PAGE gel and in gel chromatography at 29 kDa for the P. falciparum and P. knowlesi proteins and a 27 kDa P. vivax protein as predicted from the gene sequences (Fig 1; [46, 52, 54, 75]). The closest human homolog to PMT is a histamine methyltransferase, which is also a S-adenocyl-L-methionine dependent methyl transferase with 7–16% sequence identity and 31% sequence similarity around the substrate binding site [80]. This human protein has insufficient amino-acid identity to be detected by antibodies raised against the malarial proteins. A human pathogen, Cyclospora cayetanensis protein shared 39% identity overall. Affinity purified chicken antibodies against each of the three PMTs detected all three orthologues at the appropriate molecular masses in western blots but did not detect uninfected human red blood cell proteins (Figs 2 and 6). This result confirms the purity of the immunisation material and suggests that the antibodies have potential in a diagnostic test.
Peptide selection
Antibodies against peptide epitopes within the amino acid sequences of the P. falciparum HRP2 and LDH have been used to identify both the parent protein and used to detect malaria infections [81, 82]. Anti-peptide antibodies against unique peptides in the amino acid sequence of malaria LDH or GAPDH were shown to differentiate between the proteins [39, 40]. An antibody against a common peptide present in the amino acid sequence of orthologues of the malaria specific protein would enable the detection of the protein in all species expressing the protein and by implication detect an infection by any of those species. Similarly antibodies against a peptide unique to a P. knowlesi protein sequence would enable identification of a P. knowlesi infection. Anti-peptide antibodies raised against both common and species specific peptide motifs within the amino acid sequence of the PMT protein detected the parental proteins (Fig 3 and Table 1). The species-specific antibodies detected only their parental PMT protein and differentiated between the PMT protein from each species, as predicted by sequence alignment and found for LDH and GAPDH peptides (Fig 4 and Table 1; [39, 40]). PMT malaria orthologues share 61 to 88% sequence identity, which is lower than that described for malarial LDH [54]. Since nonsynonymous sequence mutations on PfHRP2 have had detrimental effects on PfHRP2 based RDTs the potential presence of mutations in PMT sequences was evaluated [76–79]. Alignment of the PMT amino acid sequences from all available isolates of the same species showed a single I192T mutation which lies outside of any of the chosen peptide sequences. The P. vivax and P. knowlesi isolates sequenced to date appear not to have any mutations in the sequences of the PMT genes (accessed August 2017). The essentiality of the gene for the parasite, coupled with the lack of mutations found in gene sequences available to date, support the potential of PMT as a diagnostic reagent.
Antibody characterisation and compatibility
Antibodies raised against peptides and the whole protein were evaluated for use in an antigen capture and detection ELISA (Table 2). Two antibodies detecting the same protein target molecule may either bind competitively (with minimal additivity) or additively i.e. combine to increase the signal [71]. The antibodies raised against the whole proteins combined with the anti-peptide antibodies had additivity indices above 50%, suggesting all the combinations of antibodies could be used in an ELISA or RDT format [83]. Combining the species specific antibodies with anti-common peptide antibodies gave the highest additivity suggesting these to be the best combinations for capture and detection of the PMT protein (Figs 5 and 6). Antibodies against each whole protein detected all the orthologues. The anti-rPvPMT and anti-rPkPMT antibodies detected both rPvPMT and rPkPMT better than the rPfPMT orthologue. This is thought to be due to 88% sequence identity shared between PvPMT and PkPMT in comparison to 64 and 62% shared with PfPMT respectively [54]. Interestingly only the anti-rPvPMT antibodies detected the corresponding specific species epitope in a direct ELISA (Fig 6), which suggests that the other epitopes are not immunogenic in chickens when displayed on the protein in the context of the whole amino-acid sequence.
Capture and detection of PMT in solution
Antibodies against the PMT protein from each of the three species, detected their partner protein in an ELISA and there was no interference when a blood lysate was added to the assay (Fig 6). Each of the anti-peptide antibodies bound to the respective PMT protein harbouring the appropriate peptide, including in a blood lysate spiked with combinations of the PMT orthologues. Antibodies against the common PMT peptide (PLENNQYTDEGVKC), detected all three P. falciparum, P. vivax and P. knowlesi PMT proteins in a spiked blood lysate. Antibodies against the P. falciparum PMT protein or the common epitope or the P. falciparum epitope detected PMT in a P. falciparum infected lysate (Fig 6). The antibody against the PfPMT peptide produced the lowest signal in the ELISA (Fig 6A) where it was marginally better than the anti-PvPMT peptide antibody and gave a poor signal in the western blot (Fig 6B). We are looking for a better Pf-specific PMT peptide antibody combination. The antibodies used here were raised in chickens. Chicken IgY does not cross-react with human rheumatoid factor and chicken antibodies are stable at 4°C for long periods indicating their diagnostic potential [84]. A combination of RDTs detecting LDH could be used to detect P. knowlesi infections [34] albeit with unacceptably low sensitivity [19, 22, 38]. As P. knowlesi parasitemia during infection increases so rapidly compared to P. falciparum, it is important to diagnose the correct species as soon as possible. At present this cannot be done with microscopy or RDTs, but can be done with PCR based methods [21]. The PMT protein is suggested as a candidate protein for evaluation in rapid diagnostic tests.
PMT was detected at 28 ng per 1 ml of a 1% Pf(D10) culture lysate, which is a similar concentration to PfLDH in the same lysate sample (Fig 6; [40, 85]) and corresponds to a ranking of protein abundance based on mRNA and expression data [40, 86]. This concentration is about five to six times lower than GAPDH (139 ng/ml; [40]) or PfHRP2 (164.5 ng/ml; [85]). In culture PfPMT expression increased by three-fold as the parasite progressed from ring to trophozoite stages [46, 57, 87]. The protein is also expressed in gametocyte [48] and sporozoite stages of the life cycle [75]. The presence of the PMT protein in all stages of parasite development, like LDH, supports its potential for diagnosis. The antibodies raised here are predicted to detect the protein in gametocytes and sporozoites, though this has not yet been evaluated.
PMT is involved in the synthesis of the major membrane phospholipid, phosphatidylcholine and is a soluble protein that localises to the Golgi apparatus [46, 87, 88]. Interestingly PMT mRNA transcript and protein levels are carefully regulated by the parasite, dependent on the choline concentration and membrane biogenesis [88]. The predicted half-lives of PfLDH, PfPMT and PfHRP2 proteins based on amino acid sequences are similar, however the half-life determined in vivo of PfLDH and PfHRP2 are very different [89, 90]. Proteins with short half-lives, like PfLDH are more accurate indicators of current infections and useful for tracking disease progress and treatment success and it would be interesting to evaluate the half-life of PMT in vivo for this reason.
Human anti-malaria antibody pool
Circulating host antibodies against malarial proteins have been suggested to interfere with PfHRP2 based RDTs, while LDH based tests appear to be unaffected [91]. A pool of human anti-malaria antibodies from 800 donors was screened for the presence of antibodies against several P. falciparum proteins (Table 3; [40, 74]). The human antibodies, like those raised in chickens against the whole PMT proteins, detected PMT from all three malaria species (Figs 2 and 7). Interestingly the common epitope was the only peptide of the three detected by this pool of human antibodies and differs from the epitope detected by IgY suggesting that either the recombinant protein assumes a different conformation to that of the native protein, or the response to native protein in humans differs to a response generated in the presence of adjuvant in chickens (Figs 7 and 6 respectively). Since the recombinant protein has been shown to have enzyme activity, it is thought that the latter explanation is the more likely [45 – 48]. The detection of the recombinant protein by the human antibody pool indicates that the recombinant protein and the native protein share structural features. The low concentration of anti-PMT antibodies in the pool, which are lower than anti-PfLDH antibodies, suggests that circulating antibodies in the host are unlikely to reduce the efficacy of a RDT using PMT as the target antigen. Several LDH based diagnostic tests detect parasite LDH despite the presence of antibodies against PLDH in human serum [39, 88].
Peptide epitope comparisons
A P. vivax specific LDH epitope was identified and antibodies against the peptide differentiated between P. vivax and P. falciparum LDH protein [38]. The P. vivax LDH specific peptide, has been found with further analysis, to share 92% sequence identity with P. knowlesi LDH. The antibodies against the P. vivax peptide are likely to detect a P. knowlesi infection and enable differentiation between a P. knowlesi and a P. falciparum infection, but not between P. vivax and P. knowlesi infections. In a similar manner a GAPDH peptide was identified that could be a target for anti-peptide antibodies to differentiate between P. falciparum and P. knowlesi GAPDH [39]. The P. knowlesi GAPDH peptide shares 79% sequence identity with the P. vivax GAPDH sequence which may be sufficient to generate a P. knowlesi specific anti-peptide antibody. Given that P. vivax and P. knowlesi share a common ancestry and are predicted to be evolutionarily closer to each other than to P. falciparum, it is likely that identifying proteins as targets to differentiate between P. vivax and P. knowlesi infections may prove difficult as shown above for the LDH and GAPDH sequences. In this context, the description of a unique P. knowlesi epitope in the PMT protein sequence strengthens the importance of the PMT protein as a diagnostic candidate. The recommendation in P. knowlesi endemic regions is that microscopic identification of P. malariae be diagnosed as P. knowlesi/P. malariae [26]. Based on the amino acid sequences of the P. knowlesi and P. malariae PMT proteins, antibodies against the P. knowlesi specific peptide will not detect the P. malariae PMT protein. Conversely there is a unique region in the P. malariae amino acid sequence (within the same region on the protein as the P. knowlesi specific peptide) that could be used to raise antibodies to detect P. malariae PMT and hence a P. malariae infection. At present we do not have P. malariae genomic DNA or the recombinant P. malariae PMT protein to evaluate this possibility. Given the presence of species-specific epitopes able to generate polyclonal antibodies in chickens in this study, it is predicted that either mouse monoclonal antibodies or phage expressed single chain variable fragment (scFv) antibodies can be raised/selected to detect P. knowlesi PMT and diagnose a P. knowlesi infection.
Conclusion
Phosphoethanolamine-N-methyltransferase has several favourable characteristics supporting its use as a diagnostic target: PMT is absent from the human proteome; low concentration of circulating human antibodies against the P. falciparum PMT protein were detected; PMT is present throughout the human infecting life-cycle stages; the PMT protein is present at similar or slightly higher concentrations than the current RDT target LDH; the protein is essential for parasite survival and propagation and is considered an antimalarial drug target; and PMT has lower shared identity between Plasmodium species orthologues compared to LDH allowing species-specific epitope selection. We describe a common epitope as well as species-specific epitopes for differentiation between P. vivax, P. falciparum, P. knowlesi, P. malariae and to our knowledge describe the first P. knowlesi specific protein target with potential for development in malaria diagnostic tests, including RDTs. It is important to assess these antibodies and epitopes with clinical samples and in a RDT format. Though we have evaluated anti-peptide antibodies produced in chickens, it is likely that mouse monoclonal antibodies raised against the same peptides would have the same specificity as that described here. A phage display library expressing single chain variable fragment antibodies is currently being screened to isolate additional antibodies against the PMT protein as has been reported for the PfHRP2 protein [92] to include in our repertoire of potential diagnostic reagents for malaria. PMT appears to be a promising target for evaluation in RDTs and has potential as a P. knowlesi specific target for consideration as a point-of-care diagnostic test to differentiate and detect P. knowlesi infection in the Southeast Asia region [8, 21, 22]).
Acknowledgments
The authors would like to thank Ben Mamoun (Yale University School of Medicine) for vectors expressing the three PMT orthologues, all members of the Goldring laboratory, Theresa Coetzer for reading the manuscript and Annemarie Krause for blood collections. We thank the South African National Research Foundation, Medical Research Council and the University of KwaZulu-Natal for funding. RK thanks the South African NRF for PostDoctoral funding.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by the South African National Research Foundation. Grant No: 92740 (http://www.nrf.ac.za/) to DG; South African Medical Research Council to DG. Self Initiated Grant Goldring 2011 (http://www.mrc.ac.za/) to DG; University of KwaZulu-Natal to DG; South African National Research Foundation. Post Doctoral funding to RGEK and DG. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Martinsen ES, Perkins SL, Schall JJ, (2008) A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): evolution of life-history traits and host switches. Mol. Phylogenet. 2008; Evol. 47: 261–73. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization (2016) World malaria report 2016. Geneva: World Health Organization.
- 3.Knowles R, Das Gupta BM, (1932) A study of monkey-malaria and its experimental transmission to man. Ind Med Gaz. 67: 301–21. [PMC free article] [PubMed] [Google Scholar]
- 4.Chin W, Contacos PG, Coatney GR, Kimball HR, (1965) A Naturally Acquired Quotidian-Type Malaria in Man Transferable to Monkeys. Science. 149: 865 [DOI] [PubMed] [Google Scholar]
- 5.Singh B, Kim Sung L, Matusop A, Radhakrishnan A, Shamsul SS, Cox-Singh J, et al. , (2004) A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 363: 1017–24. doi: 10.1016/S0140-6736(04)15836-4 [DOI] [PubMed] [Google Scholar]
- 6.White NJ, (2008) Plasmodium knowlesi: the fifth human malaria parasite. Clin. Infect. Dis. 46: 172–3. doi: 10.1086/524889 [DOI] [PubMed] [Google Scholar]
- 7.Lee KS, Divis PC, Zakaria SK, Matusop A, Julin RA, Conway DJ, et al. , (2011) Plasmodium knowlesi: reservoir hosts and tracking the emergence in humans and macaques. PLoS pathog.7: e1002015 doi: 10.1371/journal.ppat.1002015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richards J, Mueller I, (2017) Identifying the risks for human transmission of Plasmodium knowlesi. Lancet Planet. Health. 1: e83–e5. [DOI] [PubMed] [Google Scholar]
- 9.Grigg MJ, Cox J, William T, Jelip J, Fornace KM, Brock PM, et al. , (2017) Individual-level factors associated with the risk of acquiring human Plasmodium knowlesi malaria in Malaysia: a case-control study. Lancet Planet. Health. 1: e97–e104. doi: 10.1016/S2542-5196(17)30031-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yusof R, Lau YL, Mahmud R, Fong MY, Jelip J, Ngian HU, et al. , (2014) High proportion of knowlesi malaria in recent malaria cases in Malaysia. Malar J. 13: 168 doi: 10.1186/1475-2875-13-168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khim N, Siv S, Kim S, Mueller T, Fleischmann E, Singh B, et al. , (2011) Plasmodium knowlesi infection in humans, Cambodia, 2007–2010. Emerg. Infect. Dis. 17: 1900–2. doi: 10.3201/eid1710.110355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Figtree M, Lee R, Bain L, Kennedy T, Mackertich S, Urban M, et al. , (2010) Plasmodium knowlesi in human, Indonesian Borneo. Emerg. Infect. Dis. 16: 672–4. doi: 10.3201/eid1604.091624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang N, Chang Q, Sun X, Lu H, Yin J, Zhang Z, et al. , (2010) Co-infections with Plasmodium knowlesi and other malaria parasites, Myanmar. Emerg. Infect. Dis. 16: 1476–8. doi: 10.3201/eid1609.100339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luchavez J, Espino F, Curameng P, Espina R, Bell D, Chiodini P, et al. , (2008) Human Infections with Plasmodium knowlesi, the Philippines. Emerg. Infect. Dis. 14: 811–3. doi: 10.3201/eid1405.071407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeslyn WP, Huat TC, Vernon L, Irene LM, Sung LK, Jarrod LP, et al. , (2011) Molecular epidemiological investigation of Plasmodium knowlesi in humans and macaques in Singapore. Vector Borne Zoonotic Dis. 11: 131–5. doi: 10.1089/vbz.2010.0024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jongwutiwes S, Buppan P, Kosuvin R, Seethamchai S, Pattanawong U, Sirichaisinthop J, et al. , (2011) Plasmodium knowlesi Malaria in humans and macaques, Thailand. Emerg. Infect. Dis. 17: 1799–806. doi: 10.3201/eid1710.110349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sermwittayawong N, Singh B, Nishibuchi M, Sawangjaroen N, Vuddhakul V, (2012) Human Plasmodium knowlesi infection in Ranong province, southwestern border of Thailand. Malar J. 11: 36 doi: 10.1186/1475-2875-11-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marchand RP, Culleton R, Maeno Y, Quang NT, Nakazawa S, (2011) Co-infections of Plasmodium knowlesi, P. falciparum, and P. vivax among Humans and Anopheles dirus Mosquitoes, Southern Vietnam. Emerg. Infect. Dis. 17: 1232–9. doi: 10.3201/eid1707.101551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grigg MJ, William T, Barber BE, Parameswaran U, Bird E, Piera K, et al. , (2014) Combining parasite lactate dehydrogenase-based and histidine-rich protein 2-based rapid tests to improve specificity for diagnosis of malaria Due to Plasmodium knowlesi and other Plasmodium species in Sabah, Malaysia. J Clin Microbiol. 52: 2053–60. doi: 10.1128/JCM.00181-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fong YL, Cadigan FC, Coatney GR, (1971) A presumptive case of naturally occurring Plasmodium knowlesi malaria in man in Malaysia. Trans R Soc Trop Med Hyg. 65: 839–40. [DOI] [PubMed] [Google Scholar]
- 21.Fornace KM, Nuin NA, Betson M, Grigg MJ, William T, Anstey NM, et al. , (2016) Asymptomatic and Submicroscopic Carriage of Plasmodium knowlesi Malaria in Household and Community Members of Clinical Cases in Sabah, Malaysia. J Infect Dis. 213: 784–7. doi: 10.1093/infdis/jiv475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh B, (2016) Plasmodium knowlesi: an update. Microbiology Australia. 39–42. [Google Scholar]
- 23.Burke A, Dandalo L, Munhenga G, Dahan-Moss Y, Mbokazi F, Ngxongo S, et al. , (2017) A new malaria vector mosquito in South Africa. Sci. Rep. 7: 43779 doi: 10.1038/srep43779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee KS, Cox-Singh J, Singh B, (2009) Morphological features and differential counts of Plasmodium knowlesi parasites in naturally acquired human infections. Malar J. 8: 73 doi: 10.1186/1475-2875-8-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh B, Daneshvar C, (2013) Human infections and detection of Plasmodium knowlesi. Clin. Microbiol. Rev. 26: 165–84. doi: 10.1128/CMR.00079-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiodini P, Bain BJ, (2017) Plasmodium knowlesi. American journal of hematology. 92: 716 doi: 10.1002/ajh.24697 [DOI] [PubMed] [Google Scholar]
- 27.Cox-Singh J, Davis TM, Lee KS, Shamsul SS, Matusop A, Ratnam S, et al. , (2008) Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening. Clin. Infect. Dis. 46: 165–71. doi: 10.1086/524888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daneshvar C, Davis TM, Cox-Singh J, Rafa’ee MZ, Zakaria SK, Divis PC, et al. , (2009) Clinical and laboratory features of human Plasmodium knowlesi infection. Clin. Infect. Dis. 49: 852–60. doi: 10.1086/605439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fatih FA, Staines HM, Siner A, Ahmed MA, Woon LC, Pasini EM, et al. , (2013) Susceptibility of human Plasmodium knowlesi infections to anti-malarials. Malar J. 12: 425 doi: 10.1186/1475-2875-12-425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barber BE, William T, Grigg MJ, Menon J, Auburn S, Marfurt J, et al. , (2013) A prospective comparative study of knowlesi, falciparum, and vivax malaria in Sabah, Malaysia: high proportion with severe disease from Plasmodium knowlesi and Plasmodium vivax but no mortality with early referral and artesunate therapy. Clinical infectious diseases. 56: 383–97. doi: 10.1093/cid/cis902 [DOI] [PubMed] [Google Scholar]
- 31.White NJ, Pongtavornpinyo W, Maude RJ, Saralamba S, Aguas R, Stepniewska K, et al. , (2009) Hyperparasitaemia and low dosing are an important source of anti-malarial drug resistance. Malar J. 8: 253 doi: 10.1186/1475-2875-8-253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moody A, (2002) Rapid diagnostic tests for malaria parasites. Clin. Microbiol. Rev. 15: 66–78. doi: 10.1128/CMR.15.1.66-78.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mouatcho JC, Goldring JP, (2013) Malaria rapid diagnostic tests: challenges and prospects. J. Med. Microbiol. 62: 1491–505. doi: 10.1099/jmm.0.052506-0 [DOI] [PubMed] [Google Scholar]
- 34.McCutchan TF, Piper RC, Makler MT, (2008) Use of malaria rapid diagnostic test to identify Plasmodium knowlesi infection. Emerg. Infect. Dis. 14: 1750–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piper R, Lebras J, Wentworth L, Hunt-Cooke A, Houze S, Chiodini P, et al. , (1999) Immunocapture diagnostic assays for malaria using Plasmodium lactate dehydrogenase (pLDH). Am J Trop Med Hyg. 60: 109–18. [DOI] [PubMed] [Google Scholar]
- 36.Kaushal DC, Kaushal NA, Chandra D, (1995) Monoclonal antibodies against lactate dehydrogenase of Plasmodium knowlesi. Indian J Exp Biol. 33: 6–11. [PubMed] [Google Scholar]
- 37.Singh V, Kaushal DC, Rathaur S, Kumar N, Kaushal NA, (2012) Cloning, overexpression, purification and characterization of Plasmodium knowlesi lactate dehydrogenase. Protein Expr Purif. 84: 195–203. doi: 10.1016/j.pep.2012.05.008 [DOI] [PubMed] [Google Scholar]
- 38.Foster D, Cox-Singh J, Mohamad DS, Krishna S, Chin PP, Singh B, (2014) Evaluation of three rapid diagnostic tests for the detection of human infections with Plasmodium knowlesi. Malar J. 13: 60 doi: 10.1186/1475-2875-13-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hurdayal R, Achilonu I, Choveaux D, Coetzer TH, Dean Goldring JP, (2010) Anti-peptide antibodies differentiate between plasmodial lactate dehydrogenases. Peptides. 31: 525–32. doi: 10.1016/j.peptides.2010.01.002 [DOI] [PubMed] [Google Scholar]
- 40.Krause RGE, Hurdayal R, Choveaux D, Przyborski JM, Coetzer THT, Goldring JPD, (2017) Plasmodium glyceraldehyde-3-phosphate dehydrogenase: A potential malaria diagnostic target. Exp Parasitol. 179: 7–19. doi: 10.1016/j.exppara.2017.05.007 [DOI] [PubMed] [Google Scholar]
- 41.Britton S, Cheng Q, Grigg MJ, William T, Anstey NM, McCarthy JS, (2016) A Sensitive, Colorimetric, High-Throughput Loop-Mediated Isothermal Amplification Assay for the Detection of Plasmodium knowlesi. Am J Trop Med Hyg. 95: 120–2. doi: 10.4269/ajtmh.15-0670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lau YL, Lai MY, Fong MY, Jelip J, Mahmud R, (2016) Loop-Mediated Isothermal Amplification Assay for Identification of Five Human Plasmodium Species in Malaysia. Am J Trop Med Hyg. 94: 336–9. doi: 10.4269/ajtmh.15-0569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iseki H, Kawai S, Takahashi N, Hirai M, Tanabe K, Yokoyama N, et al. , (2010) Evaluation of a loop-mediated isothermal amplification method as a tool for diagnosis of infection by the zoonotic simian malaria parasite Plasmodium knowlesi. J Clin Microbiol. 48: 2509–14. doi: 10.1128/JCM.00331-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piera KA, Aziz A, William T, Bell D, Gonzalez IJ, Barber BE, et al. , (2017) Detection of Plasmodium knowlesi, Plasmodium falciparum and Plasmodium vivax using loop-mediated isothermal amplification (LAMP) in a co-endemic area in Malaysia. Malar J. 16: 29 doi: 10.1186/s12936-016-1676-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lau YL, Fong MY, Mahmud R, Chang PY, Palaeya V, Cheong FW, et al. , (2011) Specific, sensitive and rapid detection of human Plasmodium knowlesi infection by loop-mediated isothermal amplification (LAMP) in blood samples. Malar J. 10: 197 doi: 10.1186/1475-2875-10-197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pessi G, Kociubinski G, Mamoun CB, (2004) A pathway for phosphatidylcholine biosynthesis in Plasmodium falciparum involving phosphoethanolamine methylation. Proc. Natl. Acad. Sci. U.S.A. 101: 6206–11. doi: 10.1073/pnas.0307742101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bobenchik AM, Augagneur Y, Hao B, Hoch JC, Ben Mamoun C, (2011) Phosphoethanolamine methyltransferases in phosphocholine biosynthesis: functions and potential for antiparasite therapy. FEMS Microbiol Rev. 35: 609–19. doi: 10.1111/j.1574-6976.2011.00267.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bobenchik AM, Witola WH, Augagneur Y, Nic Lochlainn L, Garg A, Pachikara N, et al. , (2013) Plasmodium falciparum phosphoethanolamine methyltransferase is essential for malaria transmission. Proc. Natl. Acad. Sci. U.S.A. 110: 18262–7. doi: 10.1073/pnas.1313965110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bushell E, Gomes AR, Sanderson T, Anar B, Girling G, Herd C, et al. , (2017) Functional Profiling of a Plasmodium Genome Reveals an Abundance of Essential Genes. Cell.170: 260–72 doi: 10.1016/j.cell.2017.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Witola WH, El Bissati K, Pessi G, Xie C, Roepe PD, Mamoun CB, (2008) Disruption of the Plasmodium falciparum PfPMT gene results in a complete loss of phosphatidylcholine biosynthesis via the serine-decarboxylase-phosphoethanolamine-methyltransferase pathway and severe growth and survival defects. J. Biol. Chem. 283: 27636–43. doi: 10.1074/jbc.M804360200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pessi G, Choi JY, Reynolds JM, Voelker DR, Mamoun CB, (2005) In vivo evidence for the specificity of Plasmodium falciparum phosphoethanolamine methyltransferase and its coupling to the Kennedy pathway. J. Biol. Chem. 280: 12461–6. doi: 10.1074/jbc.M414626200 [DOI] [PubMed] [Google Scholar]
- 52.Bobenchik AM, Choi JY, Mishra A, Rujan IN, Hao B, Voelker DR, et al. , (2010) Identification of inhibitors of Plasmodium falciparum phosphoethanolamine methyltransferase using an enzyme-coupled transmethylation assay. BMC Biochem. 11: 4 doi: 10.1186/1471-2091-11-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dechamps S, Maynadier M, Wein S, Gannoun-Zaki L, Marechal E, Vial HJ, (2010) Rodent and nonrodent malaria parasites differ in their phospholipid metabolic pathways. J. Lipid Res. 51: 81–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garg A, Lukk T, Kumar V, Choi JY, Augagneur Y, Voelker DR, et al. , (2015) Structure, function and inhibition of the phosphoethanolamine methyltransferases of the human malaria parasites Plasmodium vivax and Plasmodium knowlesi. Sci. Rep. 5: 9064 doi: 10.1038/srep09064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee SG, Alpert TD, Jez JM, (2012) Crystal structure of phosphoethanolamine methyltransferase from Plasmodium falciparum in complex with amodiaquine. Bioorganic Med. Chem. Lett. 22: 4990–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Foth BJ, Zhang N, Chaal BK, Sze SK, Preiser PR, Bozdech Z, (2011) Quantitative time-course profiling of parasite and host cell proteins in the human malaria parasite Plasmodium falciparum. Mol. Cell. Proteomics. 10: M110 006411. doi: 10.1074/mcp.M110.006411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, Haynes JD, et al. , (2003) Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 301: 1503–8. doi: 10.1126/science.1087025 [DOI] [PubMed] [Google Scholar]
- 58.Carmenes RS, Freije JP, Molina MM, Martin JM, (1989) Predict7, a program for protein structure prediction. Biochem Biophys Res Commun. 159: 687–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee SG, Kim Y, Alpert TD, Nagata A, Jez JM, (2012) Structure and reaction mechanism of phosphoethanolamine methyltransferase from the malaria parasite Plasmodium falciparum: an antiparasitic drug target. J. Biol. Chem. 287: 1426–34. doi: 10.1074/jbc.M111.315267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Studier FW, (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 41: 207–34. [DOI] [PubMed] [Google Scholar]
- 61.Gassmann M, Thommes P, Weiser T, Hubscher U, (1990) Efficient production of chicken egg yolk antibodies against a conserved mammalian protein. FASEB J. 4: 2528–32. [DOI] [PubMed] [Google Scholar]
- 62.Schade R, Behn R, Erhard M, Hlinak A, Staak C. Chicken Egg Yolk Antibodies, Production and Application. Berlin Heidelberg: Springer; 2001. doi: 10.1007/978-3-662-04488-9 [Google Scholar]
- 63.Krause RG, Grobler AF, Goldring JP, (2015) Comparing Antibody Responses in Chickens Against Plasmodium falciparum Lactate Dehydrogenase and Glyceraldehyde-3-phosphate Dehydrogenase with Freund’s and Pheroid(R) Adjuvants. Immunol. Invest. 44: 627–42. doi: 10.3109/08820139.2015.1070268 [DOI] [PubMed] [Google Scholar]
- 64.Laemmli UK, (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227: 680–5. [DOI] [PubMed] [Google Scholar]
- 65.Choveaux D, Krause RG, Goldring JP, (2012) Rapid detection of proteins in polyacrylamide electrophoresis gels with Direct Red 81 and Amido Black. Methods Mol Biol. 869: 585–9. doi: 10.1007/978-1-61779-821-4_53 [DOI] [PubMed] [Google Scholar]
- 66.Towbin H, Staehelin T, Gordon J, (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. U.S.A. 76: 4350–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bradford MM, (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 72: 248–54. [DOI] [PubMed] [Google Scholar]
- 68.Goldring JP, Coetzer THT, (2003) Isolation of Chicken Immunoglobulins (IgY) from Egg Yolk. Biochem Mol Biol Educ. 31: 185–7. [Google Scholar]
- 69.Semenova VA, Steward-Clark E, Stamey KL, Taylor TH Jr., Schmidt DS, Martin SK, et al. , (2004) Mass value assignment of total and subclass immunoglobulin G in a human standard anthrax reference serum. Clin Diagn Lab Immunol. 11: 919–23. doi: 10.1128/CDLI.11.5.919-923.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McCoy JP Jr., (2001) Handling, storage, and preparation of human blood cells. Current protocols in cytometry. Chapter 5: Unit 5 1 doi: 10.1002/0471142956.cy0501s00 [DOI] [PubMed] [Google Scholar]
- 71.Friguet B, Djavadi-Ohaniance L, Pages J, Bussard A, Goldberg M, (1983) A convenient enzyme-linked immunosorbent assay for testing whether monoclonal antibodies recognize the same antigenic site. Application to hybridomas specific for the beta 2-subunit of Escherichia coli tryptophan synthase. J Immunol Methods. 60: 351–8. [DOI] [PubMed] [Google Scholar]
- 72.Goldring JD, Molyneux ME, Taylor T, Wirima J, Hommel M, (1992) Plasmodium falciparum: diversity of isolates from Malawi in their cytoadherence to melanoma cells and monocytes in vitro. Br. J. Haematol. 81: 413–8. [DOI] [PubMed] [Google Scholar]
- 73.Taylor TE, Molyneux ME, Wirima JJ, Borgstein A, Goldring JD, Hommel M, (1992) Intravenous immunoglobulin in the treatment of paediatric cerebral malaria. Clin Exp Immunol. 90: 357–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Choveaux DL, Krause RG, Przyborski JM, Goldring JP, (2015) Identification and initial characterisation of a Plasmodium falciparum Cox17 copper metallochaperone. Exp Parasitol. 148: 30–9. doi: 10.1016/j.exppara.2014.11.001 [DOI] [PubMed] [Google Scholar]
- 75.Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gajria B, et al. , (2009) PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 37: D539–43. doi: 10.1093/nar/gkn814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baker J, McCarthy J, Gatton M, Kyle DE, Belizario V, Luchavez J, et al. , (2005) Genetic diversity of Plasmodium falciparum histidine-rich protein 2 (PfHRP2) and its effect on the performance of PfHRP2-based rapid diagnostic tests. J Infect Dis. 192: 870–7. doi: 10.1086/432010 [DOI] [PubMed] [Google Scholar]
- 77.Gamboa D, Ho MF, Bendezu J, Torres K, Chiodini PL, Barnwell JW, et al. , (2010) A large proportion of P. falciparum isolates in the Amazon region of Peru lack pfhrp2 and pfhrp3: implications for malaria rapid diagnostic tests. PloS one. 5: e8091 doi: 10.1371/journal.pone.0008091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kumar N, Pande V, Bhatt RM, Shah NK, Mishra N, Srivastava B, et al. , (2013) Genetic deletion of HRP2 and HRP3 in Indian Plasmodium falciparum population and false negative malaria rapid diagnostic test. Acta Trop. 125: 119–21. doi: 10.1016/j.actatropica.2012.09.015 [DOI] [PubMed] [Google Scholar]
- 79.Deme AB, Park DJ, Bei AK, Sarr O, Badiane AS, Gueye Pel H, et al. , (2014) Analysis of pfhrp2 genetic diversity in Senegal and implications for use of rapid diagnostic tests. Malar J. 13: 34 doi: 10.1186/1475-2875-13-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Horton JR, Sawada K, Nishibori M, Zhang X, Cheng X, (2001) Two polymorphic forms of human histamine methyltransferase: structural, thermal, and kinetic comparisons. Structure. 9: 837–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Verma P, Biswas S, Mohan T, Ali S, Rao DN, (2013) Detection of histidine rich protein & lactate dehydrogenase of Plasmodium falciparum in malaria patients by sandwich ELISA using in-house reagents. Indian J. Med. Res. 138: 977–87. [PMC free article] [PubMed] [Google Scholar]
- 82.Leow CH, Jones M, Cheng Q, Mahler S, McCarthy J, (2014) Production and characterization of specific monoclonal antibodies binding the Plasmodium falciparum diagnostic biomarker, histidine-rich protein 2. Malar J. 13: 277 doi: 10.1186/1475-2875-13-277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ravyn MD, Lamb LJ, Jemmerson R, Goodman JL, Johnson RC, (1999) Characterization of monoclonal antibodies to an immunodominant protein of the etiologic agent of human granulocytic ehrlichiosis. Am J Trop Med Hyg. 61: 171–6. [DOI] [PubMed] [Google Scholar]
- 84.Carlander D, Stalberg J, Larsson A, (1999) Chicken antibodies: a clinical chemistry perspective. Ups J Med Sci. 104: 179–89. [DOI] [PubMed] [Google Scholar]
- 85.Martin SK, Rajasekariah GH, Awinda G, Waitumbi J, Kifude C, (2009) Unified parasite lactate dehydrogenase and histidine-rich protein ELISA for quantification of Plasmodium falciparum. Am J Trop Med Hyg. 80: 516–22. [PubMed] [Google Scholar]
- 86.Ginsburg H. Malaria parasite metabolic pathways Israel: The Hebrew University of Jerusalem; 2017. [cited 2017 21.8.2017]. [Google Scholar]
- 87.Witola WH, Pessi G, El Bissati K, Reynolds JM, Mamoun CB, (2006) Localization of the phosphoethanolamine methyltransferase of the human malaria parasite Plasmodium falciparum to the Golgi apparatus. J. Biol. Chem. 281: 21305–11. doi: 10.1074/jbc.M603260200 [DOI] [PubMed] [Google Scholar]
- 88.Witola WH, Ben Mamoun C, (2007) Choline induces transcriptional repression and proteasomal degradation of the malarial phosphoethanolamine methyltransferase. Eukaryot Cell. 6: 1618–24. doi: 10.1128/EC.00229-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ashley EA, Touabi M, Ahrer M, Hutagalung R, Htun K, Luchavez J, et al. , (2009) Evaluation of three parasite lactate dehydrogenase-based rapid diagnostic tests for the diagnosis of falciparum and vivax malaria. Malar J. 8: 241 doi: 10.1186/1475-2875-8-241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aydin-Schmidt B, Mubi M, Morris U, Petzold M, Ngasala BE, Premji Z, et al. , (2013) Usefulness of Plasmodium falciparum-specific rapid diagnostic tests for assessment of parasite clearance and detection of recurrent infections after artemisinin-based combination therapy. Malar J. 12: 349 doi: 10.1186/1475-2875-12-349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ho MF, Baker J, Lee N, Luchavez J, Ariey F, Nhem S, et al. , (2014) Circulating antibodies against Plasmodium falciparum histidine-rich proteins 2 interfere with antigen detection by rapid diagnostic tests. Malar J. 13: 480 doi: 10.1186/1475-2875-13-480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ravaoarisoa E, Zamanka H, Fusai T, Bellalou J, Bedouelle H, Mercereau-Puijalon O, et al. , (2010) Recombinant antibodies specific for the Plasmodium falciparum histidine-rich protein 2. mAbs.2: 416–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.