Abstract
The casein kinase 1 (CK1) family of serine (Ser)/threonine (Thr) protein kinases participates in a myriad of cellular processes including developmental signaling. Hedgehog (Hh) and Wnt pathways are two major and evolutionarily conserved signaling pathways that control embryonic development and adult tissue homeostasis. Deregulation of these pathways leads to many human disorders including birth defects and cancer. Here I review the role of CK1 in the regulation of Hh and Wnt signal transduction cascades from the membrane reception systems to the transcriptional effectors. In both Hh and Wnt pathways, multiple CK1 family members regulate signal transduction at several levels of the pathways and play either positive or negative roles depending on the signaling status, individual CK1 isoforms involved, and the specific substrates they phosphorylate. A common mechanism underlying the control of CK1-mediated phosphorylation of Hh and Wnt pathway components is the regulation of CK1/substrate interaction within large protein complexes. I will highlight this feature in the context of Hh signaling and draw interesting parallels between the Hh and Wnt pathways.
Keywords: CK1, kinase, GSK3, PKA, phosphorylation, Hh, Smo, Wnt, β-catenin, signaling, development, cancer
I. Introduction
Overview of the CK1 family kinases
CK1 protein kinases belong to a large family of evolutionarily conserved monomeric Ser/Thr kinases in eukaryotes (Cheong and Virshup, 2011). The CK1 family has been implicated in the regulation of diverse cellular processes, including membrane receptor trafficking, cytoskeleton maintenance, cell division, DNA damage response and repair, and nuclear translocation (Gross and Anderson, 1998; Knippschild et al., 2005). CK1 also participates in complex developmental and physiological processes such as Hh and Wnt signaling and circadian rhythms by phosphorylating key regulatory components in these processes (Chen and Jiang, 2013; MacDonald et al., 2009).
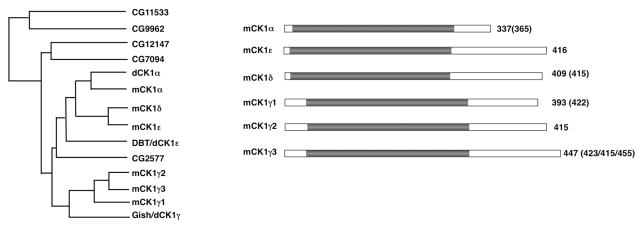
CK1 family members are found in eukaryotic organisms ranging from yeast to human with mammals possessing seven family members: α, β, γ1, γ 2, γ 3, δ, and ε. Drosophila has eight CK1 family members whereas C elegans has up to 87 members (Plowman et al., 1999; Zhang et al., 2006a). The CK1 family members contain highly conserved kinase domains but differ in their N- and C-terminal domains in terms of the length and amino acid sequence. Unlike other CK1 family members that are found in the cytosol, CK1γ is membrane associated due to palmitoylation of its C-terminus (Davidson et al., 2005). Due to their sharing of a highly conserved kinase domain, the CK1 family kinases phosphorylate similar target sites that conform the following consensus: D/E/(p)S/T(X)1-3S/T, with underlined S/T as the phospho-acceptor site, (p)S/T as phosphorylated residue, and X representing any amino acid (Knippschild et al., 2005). Although the spacing between the phospho-acceptor site and the upstream acid residue or phosphorylated S/T can be one to three amino acids, an optimal space is usually two amino acids and the phosphorylation efficiency can also be influenced by the surrounding sequence. For example, a cluster of acidic residues at the N terminus of the consensus site confer increased phosphorylation efficiency by CK1. Many CK1 target sites contain S/T instead of an acid residue at -2, -3, or -4 position, thus require a prior phosphorylation by a priming kinase. In general, CK1 family kinases are constitutively active; however, the C-terminal extensions of CK1δ and CK1ε are auto-phosphorylated, which inhibits the activity of their kinase domains, although in vivo phosphatases keep them constitutively active in many cases (Rivers et al., 1998).
Overview of canonical Hh and Wnt pathways
Hh and Wnt pathways are two major signaling pathways that control embryonic development and adult tissue homeostasis (Jiang and Hui, 2008; MacDonald et al., 2009). Malfunction of these signaling pathways have been attributed to numerous human diseases including birth defects and cancer. The canonical Hh and Wnt pathways share many pathway components, employ the same logic for pathway regulation, and thus are often considered as “sister” pathways.
Hh acts through a largely conserved pathway to regulate the balance between activator and repressor forms of the Gli family of zinc finger transcription factors (GliA and GliR; Fig. 2). While Drosophila only has one Hh and one Gli protein, Cubitus interruptus (Ci), mammals have three Hh family members Shh, Ihh and Dhh and three Gli proteins: Gli1, Gli2, and Gli3 (Jiang and Hui, 2008). In mice, GliR function is mainly derived from Gli3 through proteolytic processing whereas GliA activity is primarily contributed by Gli2. Gli1 is a transcriptional target of Hh signaling and acts in a positive feedback to reinforce GliA activity.
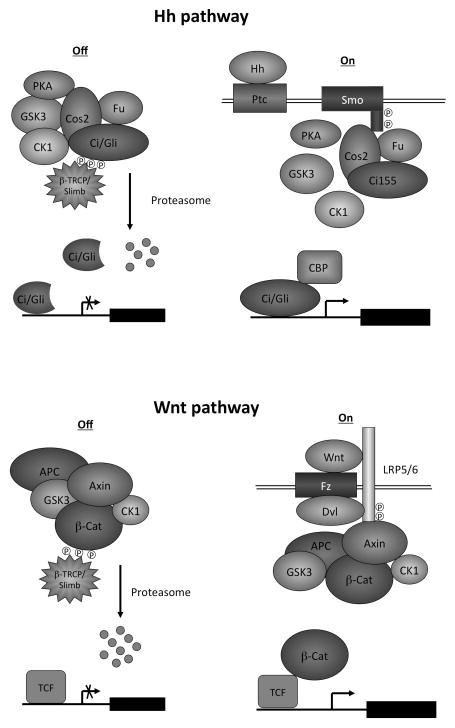
Fig. 2. Hh and Wnt pathways.
(Top) Drosophila Hh pathway. In the “signaling off “ state, Ci and its kinases PKA, GSK3 and CK1 form a large protein complex scaffolded by Cos2 and Fu, leading to its phosphorylation and proteolysis to generate a truncated repressor form that inhibits the expression of Hh target genes. In the “signaling on “ state, Hh binds Ptc and releases its inhibition of Smo, leading to Smo phosphorylation and activation. Smo interacts with Cos2/Fu and promotes Fu phosphorylation and activation but inhibits PKA/CK1-mediated Ci phosphorylation and processing. Activated Fu converts full-length Ci into an active form that stimulates the expression of Hh target genes.
(Bottom) Wnt/β-catenin pathway. In the “signaling off “ state, β-catenin and its kinases GSK3 and CK1α form a destruction complex scaffolded by Axin and APC, leading to its phosphorylation and degradation. In the “signaling on “ state, Wnt binds Fz and LRP5/6, leading to phosphorylation of LRP5/6 by GSK3 and CK1. LRP5/6 phosphorylation on the PPPSP motifs recruits the destruction complex through Axin, leading to a blockage of β-catenin phosphorylation. As a consequence, β-catenin accumulates, translocate to the nucleus, and binds TCF to turn on Wnt responsive genes.
The reception system for Hh signals consists of a twelve transmembrane protein Patched (Ptc) that binds directly to Hh and a seven transmembrane GPCR family protein Smoothened (Smo) that transduces the signal into the cytoplasm (Jiang and Hui, 2008). In the absence of Hh, the constitutive activity of Ptc blocks Smo activation, allowing the proteolytic processing of full-length Gli/Ci to generate C-terminally truncated GliR/CiR that represses a subset of Hh target genes. Binding of Hh to Ptc alleviates inhibition of Smo, and activated Smo signals intracellularly to block GliR/CiR production and promotes GliA/CiA activation. The fundamentals of Drosophila and mammalian Hh signal transduction pathways are very similar, although major differences can be found in several regulatory steps including the utilization of primary cilia for Hh signal transduction in mammals but not in Drosophila (Huangfu and Anderson, 2006).
The primary cilium is a microtubule-based membrane protrusion and antenna-like cellular structure (Goetz and Anderson, 2010). Genetic screens in mice identified multiple intraflagellar transport proteins (IFTs) critical for appropriate Hh signaling (Garcia-Garcia et al., 2005; Huangfu and Anderson, 2005). The major pathway components including Ptc, Smo, and Gli proteins are present in the primary cilium in a manner regulated by Hh signaling. Hh promotes ciliary exit of Ptc but ciliary accumulation of Smo (Corbit et al., 2005; Rohatgi et al., 2007). Hh also stimulates the accumulation of Gli proteins at the cilia tip, a step likely reflecting GliA formation. Disruption of primary cilia impedes the formation of both GliR and GliA (Haycraft et al., 2005; Huangfu and Anderson, 2005; Liu et al., 2005). Thus, the primary cilium may function as a signaling center to orchestrate the molecular events leading to Gli processing in the absence of Hh as well as Gli activation in response to Hh although the exact biochemical mechanisms remain poorly understood.
In the canonical Wnt pathway, Wnt acts through a receptor complex consisting of Frizzle (FZ) family of seven transmembrane proteins related to Smo and lipoprotein receptor-related protein LRP5/6, which signal intracellularly to stabilize the transcription effector β-catenin (Fig. 2) (MacDonald et al., 2009). In the absence of Wnt, β-catenin is degraded by the action of a destruction complex consisting of Axin, APC and several kinases including CK1 and GSK3. Binding of Wnt to FZ and LRP5/6 leads to phosphorylation and activation of LRP5/6, which binds to and inhibits the Axin/APC destruction complex, leading to stabilization and nuclear translocation of β-catenin. In the nucleus, β-catenin binds the TCF family of transcription factor to regulate Wnt pathway target genes.
Hence, in both Hh and Wnt pathways, the absence of pathway ligands allows proteolytic degradation of the pathway transcription factor/effector by large “destruction complexes” whereas ligand stimulation stabilize the pathway transcription factor/effector by inhibiting the destruction complexes”. In addition, Hh and Wnt pathways share many common pathway components including GSK3, CK1, and E3 ubiquitin ligase SCF-Slimb/βTRCP.
II. CK1 in the regulation of Hh pathway
CK1 was initially identified as a negative regulator of Hh signaling at the level of transcription factor Ci by genetic studies in Drosophila and genome wide RNAi screen in Drosophila cultured cells (Jia et al., 2005; Lum et al., 2003; Price and Kalderon, 2002). Overexpression of a dominant negative form of CK1ε resulted in an ectopic expression of an Hh target gene decapentaplegic (dpp) in wing imaginal discs and duplication of adult wings (Jia et al., 2005), phenocopying ectopic Hh expression (Basler and Struhl, 1994). However, further genetic study revealed that CK1 also plays a positive role in the Hh pathway at the level of Smo (Jia et al., 2004). A kinome –wide RNAi screen identified CK1α as a positive regulator of Hh signaling in mammalian cultured cells (Evangelista et al., 2008), and a subsequent study demonstrated that CK1α regulates Hh signaling by phosphorylating and activating Smo (Chen et al., 2011). In Drosophila, CK1 also exerts its positive role by phosphorylating Ci to stabilize the CiA and by phosphorylating Fused (Fu) to promote its activation (Shi et al., 2014; Zhou and Kalderon, 2011). Hence CK1 play a dual role in Hh signaling and acts at multiple levels
Regulation of Ci/Gli processing by CK1
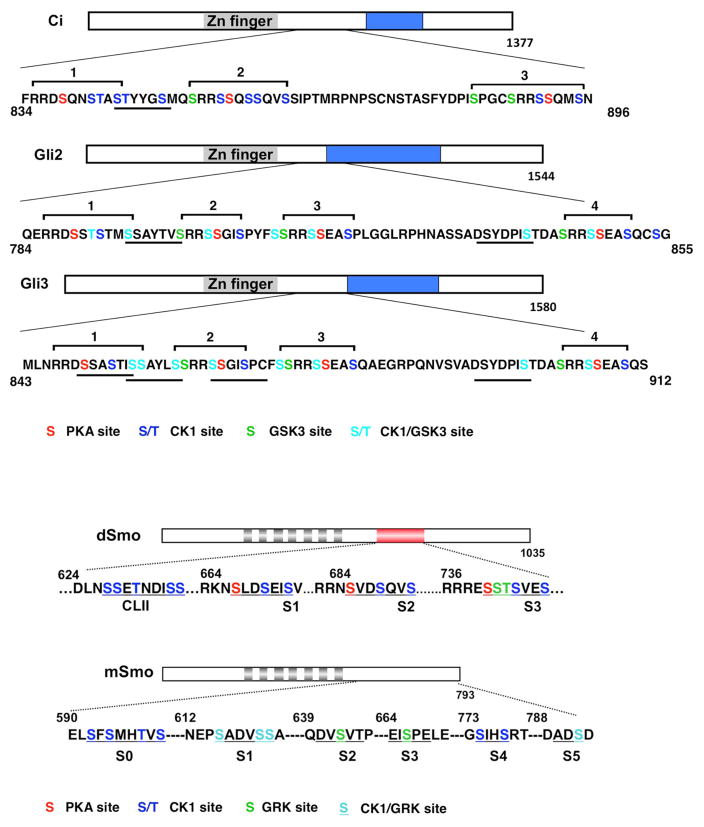
An initial hint that CK1 plays a role in Hh signaling came from the study on Ci regulation in Drosophila. Ci existed in a full-length form (CiF) and a C-terminally truncated repressor form (Ci76 or CiR) that is derived from CiF by proteolytic processing in the absence of Hh signal (Aza-Blanc et al., 1997; Methot and Basler, 1999). Ci processing requires protein kinase A (PKA) and the F-box protein Slimb, the Drosophila Orthologue of mammalian β-TRCP that functions as a substrate recognition component of the CUL1-based SCF E3 ubiquitin ligase complex (Jiang and Struhl, 1995; Jiang and Struhl, 1998; Li et al., 1995; Price and Kalderon, 1999; Wang et al., 1999). Genetic inactivation of either PKA or Slimb in wing imaginal discs resulted in accumulation of CiF at the expense of CiR and ectopic activation of dpp, leading to wing duplication, a phenotype similar to that caused by ectopic Hh expression (Jiang and Struhl, 1995; Jiang and Struhl, 1998; Li et al., 1995; Pan and Rubin, 1995). An important feature of the SCF family of E3s is that they only recognize substrates after the substrates are phosphorylated, thus providing a link between kinase-mediated phosphorylation and ubiquitin/proteasome-mediated proteolysis (Spencer et al., 1999; Winston et al., 1999). Indeed, PKA can directly phosphorylate five Ser/Thr residues in the C-terminal half of Ci, and mutating any one of the three PKA sites (sites 1–3) abolishes Ci processing (Fig. 3)(Price and Kalderon, 1999; Wang et al., 1999). However, phosphorylation of Ci by PKA alone does not confer recognition by Slimb, implying that additional phosphorylation events are required for Ci processing. Further genetic studies identified CK1 and Shaggy (Sgg), the Drosophila GSK3, as two kinases that act in conjunction with PKA to promote Ci processing (Jia et al., 2002; Jia et al., 2005; Price and Kalderon, 2002). Inactivation of either CK1α or CK1ε/DBT alone did not significantly affect Ci processing but their combined inactivation resulted in a blockage of Ci processing in wing discs (Jia et al., 2005). PKA, GSK3 and CK1 phosphorylate Ci sequentially at three S/T clusters, with PKA serving as the priming kinase for GSK3 and CK1 (Fig. 3), and these phosphorylation events create docking sites for SCFSlimb (Jia et al., 2005; Smelkinson and Kalderon, 2006). Mutating individual phosphorylation clusters diminished the production of CiR in vivo, suggesting that these phosphorylation events act in concert to promote Ci processing (Jia et al., 2002; Jia et al., 2005). A more careful analysis revealed that an extended phosphorylation cluster primed by PKA sites 1 and 2 (SpTpYYGSp) closely resembles the Slimb/β-TRCP binding site consensus DSpGX2-4Sp and provides the primary contact site for Slimb (Fig. 3) (Smelkinson et al., 2007). In addition, multiple phosphorylation events may recruit two copies of SCFSlimb complex that bind Ci simultaneously (Smelkinson et al., 2007). Thus, efficient Slimb binding and Ci processing require coordinated phosphorylation at multiple sites by these three kinases, which may render the regulation of Ci processing very sensitive to Hh. Indeed, low levels of Hh signaling appear to be sufficient to block Ci processing (Strigini and Cohen, 1997). Similar phosphorylation events mediated by PKA, GSK3 and CK1 regulate proteolysis of Gli2 and Gli3 by recruiting β-TRCP (Bhatia et al., 2006; Pan et al., 2006; Tempe et al., 2006; Wang et al., 2000a; Wang and Li, 2006; Wen et al., 2010). In contrast to Gli3 where β-TRCP - mediated proteolysis leads to partial degradation and therefore the production of GliR, Gli2 proteolysis often leads to complete degradation of the protein, consistent with the genetic studies suggesting that Gli3 is the major contributor of GliR (Hui and Angers, 2011).
Fig. 3. CK1 phosphorylates both Ci/Gli and Smo in the Hh pathway.
(Top) Diagrams of Ci, mouse Gli2, and human Gli3 showing the PKA/GSK3/CK1 phosphorylation clusters in Ci/Gli. Putative Slimb/β-TRCP binding sites in Ci/Gli are underlined. Grey and blue boxes denote a Zn-finger DNA binding domain and a trans-activation domain, respectively. Phosphorylation sites for the indicated kinases are color-coded. (Bottom) Diagrams showing the Smo phosphorylation sites in Drosophila (top) and mammalian Smo (bottom). Grey boxes indicate transmembrane helixes. The red box in Drosophila Smo C-tail denotes the SAID domain. The phosphorylation clusters are underlined and phosphorylation sites for the indicated kinases are color-coded.
In the absence of Hh, Ci forms a complex with several Hh pathway components including the kinesin-like protein Costal2 (Cos2) and the S/T kinase Fused (Fu) (Robbins et al., 1997; Sisson et al., 1997). In addition to restricting Ci nuclear translocation, the Cos2/Fu complex also interacts with PKA, GSK3 and CK1 to facilitate Ci phosphorylation and processing (Fig. 2) (Zhang et al., 2005). In the presence of Hh, the ability of Cos2/Fu complex to recruit PKA, GSK3 and CK1 is compromised (Shi et al., 2011; Zhang et al., 2005), leading to diminished Ci phosphorylation by these kinases; as a consequence, Ci processing and the production of CiR are blocked. Exactly how Hh inhibits Cos2-Fu-Ci-kinase complex formation is still not understood. Ci also forms a complex with Suppressor of Fused (Sufu), another inhibitory component of the Hh pathway, which impedes Ci nuclear translocation and inhibits CiA activity in the nucleus (Methot and Basler, 2000; Ohlmeyer and Kalderon, 1998; Wang et al., 2000b).
In mammals, Gli proteins also interact with Sufu and Kif7, the mammalian homolog of Cos2. In contrast to Drosophila where Cos2 plays a major role in inhibiting Ci in the absence of Hh, Sufu plays a major role whereas Kif7 plays a minor role in restricting Hh pathway activity in mammals (Cheung et al., 2009; Cooper et al., 2005; Endoh-Yamagami et al., 2009; Kise et al., 2009; Liem et al., 2009; Svard et al., 2006). Both Sufu and Kif7 are required for Gli3 processing (Chen et al., 2009; Cheung et al., 2009; Endoh-Yamagami et al., 2009; Humke et al., 2010; Kise et al., 2009; Liem et al., 2009; Wang et al., 2010), raising a possibility that they may also regulate Gli3 phosphorylation. Indeed, Sufu can simultaneously bind GSK3 and Gli3 and thus recruit GSK3 to phosphorylate Gli3 (Kise et al., 2009). Shh signaling may inhibit Gli phosphorylation by dissociating Sufu-Gli-kinase complex (Humke et al., 2010; Kise et al., 2009; Tukachinsky et al., 2010). It remains to be determined whether Sufu/Kif7 recruit PKA and CK1 to promote Gli3 phosphorylation and processing.
Regulation of Smo activation by CK1 in Drosophila
Smo belongs to the GPCR family of transmembrane proteins and is an obligatory signal transducer of the canonical Hh signaling pathway in both Drosophila and vertebrates (Jiang and Hui, 2008). Activation mutations in Smo have been found in basal cell carcinoma (BCC) and medulloblastoma (Jiang and Hui, 2008; Xie et al., 1998). Hence, Smo has emerged as a prominent target for cancer therapeutics (Rubin and de Sauvage, 2006). Indeed, the FDA has proved the use of Vismodegib, a potent synthetic oral Smo inhibitor, for the treatment of advanced BCC.
In Drosophila, Hh induces cell surface accumulation and phosphorylation of Smo (Denef et al., 2000). Identification of Smo kinases came from unexpected findings that both PKA and CK1 play dual roles in Hh signaling in Drosophila embryos and imaginal discs (Jia et al., 2004; Ohlmeyer and Kalderon, 1997). Gain of PKA function promotes Smo accumulation and Hh pathway activation whereas loss of PKA function blocks Hh-induced Smo accumulation as well as high levels of Hh signaling (Jia et al., 2004). Similarly, inactivation of CK1α/ε by RNAi blocks Hh-induced Smo accumulation and high levels of Hh signaling activity (Jia et al., 2004). Biochemical studies demonstrated that PKA and CK1 sequentially phosphorylate three clusters of S/T residues in the Smo carboxyl-terminal cytoplasmic tail (C-tail), with PKA serving as the priming kinase for CK1 phosphorylation (Fig. 3)(Apionishev et al., 2005; Jia et al., 2004; Zhang et al., 2004). Phospho-deficient Smo variants with either PKA or CK1 sites mutated to nonphosphorable Ala exhibit reduced cell surface expression and diminished Hh signaling activity whereas phospho-mimetic Smo variants with both PKA or CK1 sites converted to acidic residues exhibit increased cell surface level and constitutive activity (Apionishev et al., 2005; Jia et al., 2004; Zhang et al., 2004). Interestingly, increasing the number of phospho-mimetic clusters in Smo results in a progressive elevation of Smo activity, suggesting that graded Hh signals may induce different levels of Smo activity through differential phosphorylation (Jia et al., 2004). Indeed, using a phospho-specific antibody to monitor Smo phosphorylation, a later study showed that Hh induces Smo phosphorylation in a dose-dependent manner (Fan et al., 2012).
Mechanistically, PKA/CK1-meidated phosphorylation promotes Smo activity via at least two paralleled mechanisms: regulating its subcellular localization and conformation. Two recent studies revealed that Hh-induced phosphorylation promotes Smo cell surface expression by inhibiting ubiquitination-mediated endocytosis and degradation of Smo (Li et al., 2012; Xia et al., 2012). In the absence of Hh, Smo is both mono- and poly-ubiquitinated at multiple Lys residues in the carboxyl-terminal cytoplasmic tail (C-tail), leading to Smo endocytosis and degradation by both lysosome and proteasome-dependent mechanisms. In addition, Smo endocytosis is also promoted by the binding of Kurtz (Krz), the Drosophila non-visual arrestin (Li et al., 2012; Molnar et al., 2011). Hh inhibits Smo ubiquitination and attenuates Smo/Krz interaction, thereby stabilizing Smo on the cell surface (Li et al., 2012; Xia et al., 2012). How Hh-induced phosphorylation of Smo inhibits its ubiquitination has not be resolved. One possibility is that PKA/CK1-mediated phosphorylation of Smo C-tail may inhibits the binding of one or more Smo E3 ubiquitin ligases. The identification of Smo E3s may allow a direct test of this model.
In addition to regulating Smo trafficking, PKA/CK1-mediated phosphorylation also controls the conformation of Smo C-tail (Zhao et al., 2007). In the absence of Hh, Smo adopts a closed conformation in which the Smo C-terminus folds back to form salt bridge with multiple basic clusters (mostly Arg) in the SAID domain (Smo auto-inhibition domain), which is located in the middle region of the Smo C-tail (Zhao et al., 2007). Hh-induced phosphorylation by PKA/CK1 brings negative charges to neutralize the positive charges of the nearby Arg motifs in the SAID domain, which disrupts the intramolecular electrostatic interaction that maintains the closed conformation and promotes an open conformation and clustering of Smo C-tails. The pairing of positive and negative regulatory elements offers a more precise regulation of Smo activity in response to graded Hh signals as increasing phosphorylation may gradually neutralize the negative effect of multi-Arg clusters, leading to a progressive change in Smo cell surface accumulation, conformation, and activity (Zhao et al., 2007). Indeed, decreasing the number of functional Arg motifs has the same effect of increasing the number of phospho-mimetic mutations, both leading to a progressive increase of Smo activity (Jia et al., 2004; Zhao et al., 2007).
Phospho-mimetic mutations in the three PKA/CK1 phosphorylation clusters render Smo constitutively active but fail to confer full pathway activity (Jia et al., 2004), suggesting that Smo activation may involve additional mechanisms. A mass spec analysis identified many phosphorylation sites in Smo C-tail other than the three PKA/CK1 phosphorylation clusters (Zhang et al., 2004). Subsequent studies identified both casein kinase 2 (CK2) and G protein coupled receptor kinase 2 (Gprk2 or GRK2) as additional Smo kinases (Chen et al., 2010; Jia et al., 2010). Interestingly, GRK2 promotes high-level Hh signaling through both kinase-dependent and kinase-independent mechanisms (Chen et al., 2010; Cheng et al., 2010). GRK2 phosphorylates Smo C-terminal tail at Ser741/Thr742, which is facilitated by PKA/CK1-mediated phosphorylation at adjacent Ser residues (Chen et al., 2010). In addition, GRK2 forms a dimer and binds Smo to stabilize its open conformation and promote the dimerization of Smo C-tail (Chen et al., 2010). A recent study identified Gish/CK1γ as another Smo kinase that fine-tunes Hh pathway activity as loss of Gish affects the expression of high threshold Hh target genes (Li et al., 2016). Gish is a membrane-associated kinase due to its C-terminal lipid modification and membrane association appears to be critical for its function the Hh pathway. Mechanistically, Gish interacts with Smo and phosphorylates a membrane proximal S/T cluster (CLII) on the Smo C-tail. Interestingly, interaction between Gish/Smo and subsequent phosphorylation of CLII are stimulated by Hh and promoted by PKA-mediated phosphorylation of the distal region on the Smo C-tail (Li et al., 2016). Hence the Smo kinases fall into two categories: PKA and CK1α/ε are the primary kinases whose phosphorylation of Smo at multiple sites is critical for Smo activation whereas CK2, Gprk2 are Gish/CK1γ are the secondary kinases whose phosphorylation of Smo is promoted by the primary phosphorylation to further boost Hh pathway activity.
A conserved role of CK1 in mammalian Smo activation
Mammalian Smo (mSmo) and Drosophila Smo (dSmo) diverge significantly in their primary sequences. For example, mSmo does not contain three PKA/CK1 phosphorylation clusters found in dSmo (Fig. 2). In addition, mSmo traffics into primary cilia to transduce the Hh signal whereas the primary cilium is dispensable for Drosophila Hh signal transduction (Corbit et al., 2005; Rohatgi et al., 2007). These observations have led to the proposal that mSmo and dSmo are regulated by fundamentally distinct mechanisms (Huangfu and Anderson, 2006; Varjosalo et al., 2006). However, subsequent recent revealed striking similarities in the activation mechanism between dSmo and mSmo: both dSmo and mSmo are regulated by multi-site phosphorylation in a dose dependent manner and phosphorylation regulates both their subcellular localization and conformation (Chen et al., 2011; Zhao et al., 2007). Furthermore, both CK1α and CK1γ are involved in the phosphorylation and activation of mSmo (Chen et al., 2011; Li et al., 2016).
A kinome RNAi screen identified CK1α as a positive regulator of mammalian Hh pathway in cultured cells (Evangelista et al., 2008). Several studies also suggested that GRK2 (orthologue of Drosophila Gprk2) positively regulate mammalian Hh pathways and GKR2 promotes mSmo phosphorylation (Chen et al., 2004; Evangelista et al., 2008; Meloni et al., 2006). Further study demonstrated that CK1α and GRK2 bind mSmo in response to Hh stimulation and phosphorylate mSmo C-tail at six Ser/Thr clusters (S0–S5; Fig. 3) (Chen et al., 2011). Studies using both cultured mammalian cells and chick neural tubes suggested that multiple CK1α/GRK2 phosphorylation sites regulate mSmo activity in a dose dependent manner, with the two membrane-proximal clusters (S0 and S1) playing a major role (Chen et al., 2011). As is the case for dSmo (Fan et al., 2012; Jia et al., 2004), mSmo phosphorylation is regulated in a dose dependent manner with increasing amounts of Hh inducing a progressive increase of mSmo phosphorylation, suggesting that Hh gradient is translated into mSmo phosphorylation and activity gradient (Chen et al., 2011).
CK1α/GRK2-mediated phosphorylation promotes mSmo ciliary localization as phosphorylation deficient form of mSmo failed to accumulate to the primary cilium in response to Hh stimulation or oncogenic mutations whereas phospho-mimetic mutations resulted in constitutive mSmo ciliary accumulation (Chen et al., 2011). How phosphorylation promotes mSmo ciliary localization remains an unresolved issue but it correlates with enhanced binding of β-arrestin, which is thought to link mSmo to the kinesin-II motor responsible for anterograde ciliary transport (Chen et al., 2011; Kovacs et al., 2008). Besides regulating mSmo ciliary localization, Hh induces a conformational switch that results in dimerization/oligomerization of mSmo C-tail (Zhao et al., 2007). Hh-induced conformational switch is also governed by CK1α/GRK2-mediated multi-site phosphorylation of mSmo C-tail in a similar manner to dSmo (Chen et al., 2011). Interestingly, cyclopamine traps ciliary localized mSmo in an unphosphorylated form that adopts an inactive conformation whereas ciliary localized mSmo in response to Hh or Smo agonists is phosphorylated and thus adopts an active conformation (Chen et al., 2011). Hence, Smo phosphorylation is a more faithful readout for pathway activation than Smo ciliary localization and can serve as a biomarker for cancers caused by deregulated Hh pathway activation.
Hh enhances the recruitment of both CK1α and GRK2 to mSmo, which may explain the signal-stimulated phosphorylation (Chen et al., 2011). In the absence of Hh, the kinase-binding pockets appear to be masked when mSmo C-tail adopts a closed conformation. Shh stimulates CK1α binding to a juxtamembrane site, likely by inducing a conformational change in the transmembrane helixes, to initiate phosphorylation and conformational change of mSmo C-tail. This further increases the binding of CK1α/GRK2 to the mSmo Ctail, forming a feed forward mechanism to increase mSmo phosphorylation (Chen et al., 2011). Interestingly, CK1α accumulates in primary cilia in response to Hh stimulation, which may explain, at least in part, why phosphorylation of mSmo is more effective in primary cilia (Chen et al., 2011).
A recent study revealed that exogenously expressed CK1γ is localized to the primary cilia of NIH3T3 cells in a manner depending on its membrane association (Li et al., 2016). Using a phospho-specific antibody against one of the major CK1 sites (S1) on mSmo to monitor mSmo phosphorylation, Li et al found that transfection of wild type, but not a cytosolic form (CK1γ–ΔC) of CK1γ stimulated S1 phosphorylation in a manner enhanced by Shh treatment whereas transfection of dominant negative forms of CK1γ attenuated Hh-induced S1 phosphorylation (Li et al., 2016). In addition, overexpression of wild type CK1γ, but not CK1γ–ΔC, increased the expression of Gli-luc reporter gene while the dominant negative forms attenuated Hh-induced Gli-luc expression (Li et al., 2016). The ability of CK1γ to promote mSmo phosphorylation and activity appears to depend on primary cilia as blockage of ciliogenesis abolished the effect of CK1γ on mSmo phosphorylation and activation (Li et al., 2016). Taken together, these observations suggest that CK1γ plays a conserved role in the Hh pathway by directly phosphorylating Smo and boosting its activity after it accumulates on the cell surface in Drosophila or on the primary cilia in mammals.
CK1 positively regulates Hh signaling downstream of Smo
Besides phosphorylating Smo to promote Hh pathway activation, CK1 plays additional positive roles downstream of Smo because inactivation of CK1 inhibits Hh pathway activity elicited by expressing a phospho-mimetic and constitutively active form of Smo in which both the PKA and CK1 sites are converted into acidic residues (Shi et al., 2014; Zhou and Kalderon, 2011). In Drosophila, the Fu kinase acts downstream of Smo to convert CiF into an activator form (CiA) in response to high levels of Hh signal (Ohlmeyer and Kalderon, 1998). Fu forms a stoichiometric complex with Cos2 and is recruited to the dimerized intracellular tail of activated Smo, leading to its own dimerization, phosphorylation, and activation (Shi et al., 2011; Zhang et al., 2011; Zhou and Kalderon, 2011). Phosphorylation of Fu occurs at multiple Ser/Thr residues in the activation loop of its kinase domain as well as in its C-terminal regulatory domain (Shi et al., 2011; Zhou and Kalderon, 2011). Phosphorylation of the Fu activation loop and C-terminal regulatory domain is mediated by Fu trans-autophosphorylation, which may prime their further phosphorylation by CK1 although direct evidence for phosphorylation of Fu by CK1 is still lacking (Shi et al., 2011; Zhou and Kalderon, 2011). Phosphorylation of Fu activation loop appears to promote Fu activation in a dose dependent manner: increasing levels of Hh induced increasing levels of Fu phosphorylation, which in turn resulted in increasing levels of Fu activity (Shi et al., 2011). Mutating a CK1 consensus site in the activation loop severely compromised Fu activity whereas mutating a cluster of CK1 consensus sites in the regulatory domain also attenuated Fu activity (Zhou and Kalderon, 2011). Taken together, these studies suggest that CK1 may positively regulate Hh pathway by phosphorylating both the kinase and regulatory domains of Fu.
CiA is a labile form of Ci that is degraded by a Cul3-based E3 ubiquitin ligase containing the BTB family protein HIB (also called Rdx) (Kent et al., 2006; Zhang et al., 2009; Zhang et al., 2006b). }. Interestingly, hib expression is induced by Hh signaling in both Drosophila embryos and imaginal discs, thus forming a negative feedback loop to attenuate Hh pathway activity (Kent et al., 2006; Zhang et al., 2006b). SPOP, which is the vertebrate homolog of HIB, may play an analogous role in fine-tuning Hh signaling by degrading Gli2/3 proteins (Chen et al., 2009; Wang et al., 2010; Wen et al., 2010). The degradation of CiA/GliA by HIB/SPOP is likely to be under tight control as excessive degradation can lead to premature loss of Hh pathway activity. Indeed, a recent study showed that Hh stimulates Ci phosphorylation by CK1 at multiple Ser/Thr-rich degrons to inhibit its recognition by HIB (Shi et al., 2014). In Hh-receiving wing disc cells, reduction of CK1 activity accelerated HIB-mediated degradation of CiA, leading to premature loss of pathway activity (Shi et al., 2014). Furthermore, this study showed that GliA is regulated by CK1 in a similar fashion and that CK1 acts downstream of Sufu to promote Shh signaling. Hence, depending on availability of the Hh ligand, CK1 can either inhibit or activate Ci by phosphorylating distinct sites.
III. CK1 in Wnt signaling
Similar to its role in the Hh pathway, CK1 also plays both positive and negative roles in the Wnt pathway by phosphorylating different pathway components. The first indication that CK1 family kinase regulates Wnt signaling is that injection of CK1ε mRNA into the ventral side of Xenopus embryos led to dorsalization and axial duplication similar to injection of Wnts (Peters et al., 1999; Sakanaka et al., 1999). It was further shown that CK1ε forms a complex with Disheveled (Dvl) and Axin and positively regulates Wnt signaling by phosphorylating Dvl on multiple sites (McKay et al., 2001; Peters et al., 1999; Sakanaka et al., 1999). However, the physiological relevance of Dvl phosphorylation by CK1 has not been established. Other CK1 family members including CK1α and CK1γ have also been implicated in Wnt signaling (see below). Although CK1ε and CK1γ mainly play a positive role in Wnt signaling through their influence on LRP6 and Dvl whereas CK1α mainly plays a negative role through its role in the regulation of β-catenin degradation, a genetic study in Drosophila also reveals a negative role for CK1ε and a positive role for CK1α in a genetic sensitized background (Zhang et al., 2006a).
Regulation of β-catenin destruction: a negative role of CK1 in Wnt signaling
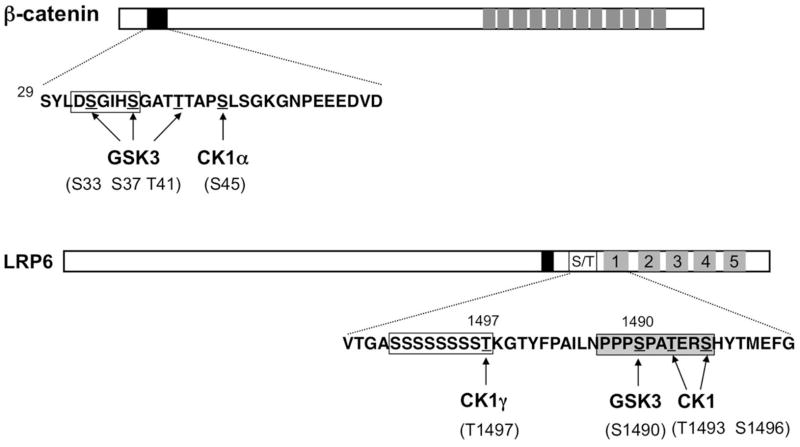
Regulation of the cytoplasmic β-catenin protein abundance is the hallmark of the canonical Wnt signal transduction pathway. In the absence of Wnt, β-catenin is phosphorylated by CK1α and GSK3, which target it for ubiquitin/proteasome-mediated degradation. β-catenin contains a degron, DS33GIHS37GAVT41QAPS45, in its N-terminal region (Fig. 4), and mutations of the S/T residues stabilized β-catenin in human cancers (Polakis, 2000). Two studies showed that phosphorylation of S45 by CK1α primes GSK3 to phosphorylation T41, S37, and S33 consecutively (Amit et al., 2002; Liu et al., 2002), which creates a docking site for the SCF family of E3 ubiquitin ligase containing the F-box protein Slimb/β-TRCP (Jiang and Struhl, 1998; Winston et al., 1999), leading to its ubiquitination and degradation of by the proteasome.
Fig. 4. Phosphorylation of β-catenin and LRP6 by CK1 and GSK3.
(A) Diagram of βcatenin with the CK1 and GSK3 phosphorylation cluster shown underneath. β-TRCP binding site is boxed. Grey boxes denote the Armadillo (Arm) repeats and the black box indicates the destruction box. (B) Diagram of LRP6 with the membrane proximal S/T rich sequence and the first PPPSP motif shown underneath. Filled, open, and grey boxes indicate the transmembrane domain, S/T rich domain, and multiple PPPSP motifs.
Both CK1α and GSK3 are present in the destruction complex containing the scaffold proteins Axin and APC (Amit et al., 2002). The Axin/APC also binds β-catenin, thus bringing the substrate and kinases in close proximity to facilitate β-catenin phosphorylation (Liu et al., 2002). Interestingly, APC is also phosphorylated by CK1 (in this case, CK1ε and CK1δ) as well as GSK3 at several sites within the 20 amino acid repeats that mediate its binding to β-catenin, and these phosphorylation events greatly enhance its binding to β-catenin (Ha et al., 2004; Xing et al., 2004), and its ability to downregulate β-catenin (Rubinfeld et al., 2001). Phosphorylation of APC by CK1ε is greatly enhanced by the presence of an Axin fragment containing the β-catenin and CK1-binding sites (Ha et al., 2004; Rubinfeld et al., 2001). Thus, Axin functions as a scaffold in the destruction complex to facilitate phosphorylation of both β-catenin and APC.
Axin is also phosphorylated by CK1, but the biological significance of this phosphorylation event is unclear (Gao et al., 2002). A recent study reveals that APC promotes efficient Axin multimerization through multivalent interactions and that phosphorylation of the R2/B region of APC by GSK3/CK1 induces a conformational change in APC to release APC’s Arm-repeats from Axin, allowing the release of phosphorylated β-catenin to the SCFβ-TRCP E3 ligase, leading to ubiquitination and degradation of β-catenin (Pronobis et al., 2015).
Regulation of LRP6 phosphorylation: a positive role of CK1 in Wnt signaling
The Wnt receptor complex contains the Fz family seven transmembrane proteins and the co-receptor LRP5/6. In the presence of Wnt, the destruction complex is recruited to activated LRP5/6 through its binding to Axin, leading to inhibition of β-catenin phosphorylation, β-catenin stabilization in the cytoplasm, and Wnt pathway activation (Mao et al., 2001; Tamai et al., 2004). LRP6 and its Drosophila homologue Arrow contain five PPPSP motifs that mediate Axin binding when phosphorylated in response to Wnt (Fig. 4) (Tamai et al., 2004; Zeng et al., 2005). Phosphorylation of PPPSP is mediated by GSK3, followed by CK1-mediated phosphorylation at one or more adjacent sites C-terminal to the PPPSP motifs. Multiple CK1 isoforms including CK1α, CK1ε/δ, and CK1γ have been implicated in the phosphorylation the sites adjacent to the PPPSP motifs (Davidson et al., 2005; Zeng et al., 2005). Phosphorylation of LRP6 by both GSK3 and CK1 is required for Axin binding and phosphorylated PPPSP motifs inhibit GSK3-mediated phosphorylation of β-catenin within the destruction complex, leading to β-catenin stabilization and Wnt pathway activation (Piao et al., 2008; Wu et al., 2009; Zeng et al., 2005). In addition, CK1γ is involved in the phosphorylation of T1497, which is adjacent to a S/T cluster (Fig. 4) (Davidson et al., 2005). Similar to its binding to Smo, CK1γ binds LRP6 depending on its membrane association through its C-terminal palmitoylation (Davidson et al., 2005).
Wnt-induce LRP6 phosphorylation requires the Fz receptor and its downstream partner Dvl that promotes the formation of a large multimeric protein complex called LRP6 signalosome (Bilic et al., 2007; Zeng et al., 2008). Further studies reveal that Wnt3a induces the formation of PI(4 5)P2 to promote LRP6 phosphorylation and that Amer1/WTX couples the Wnt3a-induced PI(4 5)P2 to LRP6 phosphorylation (Pan et al., 2008; Tanneberger et al., 2011). Several other proteins including the transmembrane protein 198 and p120-catenin have also been implicated in the regulation of LRP6 phosphorylation (Casagolda et al., 2010; Liang et al., 2011). Furthermore, Wnt-induced LRP6 phosphorylation occurs in an acidic vesicular compartment depending on the action of prorenin receptor (PRR) and vacuolar ATPase (Cruciat et al., 2010).
Like in the Hh pathway, CK1 also exerts positive influence on Wnt signaling at multiple levels. In addition to regulating the phosphorylation of LRP6 and Dvl, CK1 also phosphorylates TCF3 to promote its interaction with β-catenin (Lee et al., 2001).
IV. Regulation of CK1 in Hh and Wnt signaling
The involvement of CK1 in both Hh and Wnt signal transduction pathways and the findings that CK1 regulates each pathway at multiple levels to exert both positive and negative influence have raised import questions of how phosphorylation of CK1 substrates is regulated and how Hh and Wnt signaling achieve pathway specificity despite being regulated by a similar set of kinases. A prominent feature of Hh and Wnt signaling is that the kinases and their substrates are present in large protein complexes organized by pathway-specific scaffolding proteins (Fig. 2). In the Hh pathway, Ci and its kinases PKA, CK1, and GSK3 are present in protein complexes organized by Cos2 and Fu (Chen and Jiang, 2013; Zhang et al., 2005). Similarly in the Wnt pathway, β-catenin and its kinases CK1 and GSK3 form a complex scaffolded by Axin and APC (MacDonald et al., 2009). Therefore, to achieve pathway specificity, Hh and Wnt regulate different pools of kinases by eliciting interaction between their receptor complexes and the pathway specific scaffold proteins i.e., Smo/Cos2 interaction in the Hh pathway and LRP6/Axin interaction in the Wnt pathway (Jia et al., 2003; Jiang and Hui, 2008; MacDonald et al., 2009; Tamai et al., 2004). In other words, Hh and Wnt signaling are regulated by specific pool of kinases that are compartmentalized with their pathway effectors.
Another layer of CK1 regulation in the Hh and Wnt pathways is to employ different CK1 isoforms to phosphorylate distinct pathway components or even distinct sites on the same substrates. In this regard, it has been shown recently that the membrane-associated CK1 isoform CK1γ, but not the cytosolic isoform CK1α or CK1ε, is responsible for phosphorylating a membrane proximal cluster of S/T residues on the Smo C-tail to promote high-level Hh pathway activity (Li et al., 2016). Similarly, CK1γ has been shown to phosphorylate a membrane proximal site (T1497) on the LRP6 intracellular domain to modulate the activity of Wnt signaling (Davidson et al., 2005), and in Drosophila, CK1γ is the most potent CK1 isoform that phosphorylates Arrow, the Drosophila homolog of LRP6, likely due to its membrane association (Zhang et al., 2006a). On the other hand, CK1α is the major CK1 isoform responsible for β-catenin phosphorylation and degradation. However, both CK1α and CK1ε are involved in the regulation of Ci phosphorylation and degradation in the Hh way (Jia et al., 2005). Interestingly, Hh switches the CK1α/ε substrate from Ci to Smo, thus converting CK1α and CK1ε from negative to positive regulators of the pathway (Jia et al., 2004; Zhang et al., 2005). This conversion is achieved, at least in part, by regulating kinase/substrate interaction because Hh signaling inhibits the formation of Cos2-Ci-kinase complex but promotes the formation of Cos2-Smo-kinase complexes (Li et al., 2014; Zhang et al., 2005).
Although it is generally thought that CK1 activity is not regulated but rather its substrate accessibility is regulated in the Hh and Wnt pathways, a recent study reveals a novel mechanism of CK1 regulation by DDX3, which belongs to a family of ATP-dependent DEAD-box RNA helicases (Cruciat et al., 2013). DDX3 was identified by a genome-wide siRNA screen for novel Wnt regulators in cultured mammalian cells (Cruciat et al., 2013). DDX3 directly binds to CK1ε in a manner stimulated by Wnt, and DDX3 stimulates CK1ε kinase activity and promotes phosphorylation of Dvl2 although the physiological relevance of Dvl2 phosphorylation has not been directly tested (Cruciat et al., 2013). The enzymatic activities of DDX3, i.e., ATP hydrolysis and RNA unwinding, appear to be dispensable for binding to CK1ε and, suggesting that DDX3 activates CK1ε through an allosteric mechanism (Cruciat et al., 2013). Interestingly, mutations in DDX3 were found in Wnt-subgroup medulloblastoma, illustrating its physiological role in Wnt/β-catenin signaling in humans (Jones et al., 2012; Pugh et al., 2012). It remains to be determined whether DDX3 regulates other CK1 isoforms in vivo and whether other DDX family helicases are involved in CK1 regulation.
Conclusion
Multiple CK1 family members regulate both Hh and Wnt signaling, and in each pathway, CK1 exerts both positive and negative influences depending on the signaling status, the specific CK1 isoforms involved, and which pathway components CK1 phosphorylates. Although many relevant CK1 substrates have been identified and the specific phosphorylation sites on individual pathway components defined, not all the biological relevant sites have been determined. For example, it has shown that CK1 activates the Hh pathway at the level of Smo as well as downstream of Smo by phosphorylating Fu and Ci (Shi et al., 2014; Zhou and Kalderon, 2011); however, the precise role and relevant CK1 sites on Fu remain to be determined. In addition, the precise function of individual CK1 isoforms in the Hh and Wnt pathways remains to be clarified, especially in the case of LRP6 phosphorylation where multiple CK1 isoforms have been implicated (Davidson et al., 2005; Zeng et al., 2005). In the past, loss of function study mainly employed dominant negative forms of individual isoforms, which may invoke cross-regulation among different CK1 isoforms and thus may not be exclusively “isoform specific”. In addition, redundancy among different CK1 family members might have underscored the role of individual CK1 isoforms in certain signaling processes, which is particularly problematic for CK1γ because of the presence of three CK1γ family members in mammals. Recent advance in gene editing technology, especially the CRISPR/Cas9 technology that offers an efficient way to knock out multiple genes in the same cells (Doudna and Charpentier, 2014), will undoubtedly facilitate the loss-of-function study of individual CK1 family members in different cellular and developmental contexts and help elucidating their roles in human diseases.
Fig. 1.
CK1 family of kinases. (A) Family tree of CK1 isoforms from Drosophila and mammals. (B) Schematic drawings of mammalian CK1 family members with kinase domains depicted in grey. Numbers in parentheses indicate the lengths of CK1 isoforms generated from alternative splicing.
Acknowledgments
This work is supported by grants from National Institutes of Health (GM118063) and Welch foundation (I-1603). J. Jiang is a Eugene McDermott Endowed Scholar in Biomedical Science at the University of Texas Southwestern.
References
- Amit S, Hatzubai A, Birman Y, Andersen JS, Ben-Shushan E, Mann M, Ben-Neriah Y, Alkalay I. Axin-mediated CKI phosphorylation of beta-catenin at Ser 45: a molecular switch for the Wnt pathway. Genes Dev. 2002;16:1066–76. doi: 10.1101/gad.230302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apionishev S, Katanayeva NM, Marks SA, Kalderon D, Tomlinson A. Drosophila Smoothened phosphorylation sites essential for Hedgehog signal transduction. Nat Cell Biol. 2005;7:86–92. doi: 10.1038/ncb1210. [DOI] [PubMed] [Google Scholar]
- Aza-Blanc P, Ramirez-Weber F, Laget M, Schwartz C, Kornberg T. Proteolysis that is inhibited by Hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell. 1997;89:1043–53. doi: 10.1016/s0092-8674(00)80292-5. [DOI] [PubMed] [Google Scholar]
- Basler K, Struhl G. Compartment boundaries and the control of Drosophila limb pattern by hedgehog protein. Nature. 1994;368:208–14. doi: 10.1038/368208a0. [DOI] [PubMed] [Google Scholar]
- Bhatia N, Thiyagarajan S, Elcheva I, Saleem M, Dlugosz A, Mukhtar H, Spiegelman VS. Gli2 is targeted for ubiquitination and degradation by beta-TrCP ubiquitin ligase. The Journal of biological chemistry. 2006;281:19320–6. doi: 10.1074/jbc.M513203200. [DOI] [PubMed] [Google Scholar]
- Bilic J, Huang YL, Davidson G, Zimmermann T, Cruciat CM, Bienz M, Niehrs C. Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science. 2007;316:1619–22. doi: 10.1126/science.1137065. [DOI] [PubMed] [Google Scholar]
- Casagolda D, Del Valle-Perez B, Valls G, Lugilde E, Vinyoles M, Casado-Vela J, Solanas G, Batlle E, Reynolds AB, Casal JI, de Herreros AG, Dunach M. A p120-catenin-CK1epsilon complex regulates Wnt signaling. J Cell Sci. 2010;123:2621–31. doi: 10.1242/jcs.067512. [DOI] [PubMed] [Google Scholar]
- Chen MH, Wilson CW, Li YJ, Law KK, Lu CS, Gacayan R, Zhang X, Hui CC, Chuang PT. Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev. 2009;23:1910–28. doi: 10.1101/gad.1794109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Ren XR, Nelson CD, Barak LS, Chen JK, Beachy PA, de Sauvage F, Lefkowitz RJ. Activity-dependent internalization of smoothened mediated by beta-arrestin 2 and GRK2. Science. 2004;306:2257–60. doi: 10.1126/science.1104135. [DOI] [PubMed] [Google Scholar]
- Chen Y, Jiang J. Decoding the phosphorylation code in Hedgehog signal transduction. Cell Res. 2013;23:186–200. doi: 10.1038/cr.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Li S, Tong C, Zhao Y, Wang B, Liu Y, Jia J, Jiang J. G protein-coupled receptor kinase 2 promotes high-level Hedgehog signaling by regulating the active state of Smo through kinase-dependent and kinase-independent mechanisms in Drosophila. Genes Dev. 2010;24:2054–67. doi: 10.1101/gad.1948710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Sasai N, Ma G, Yue T, Jia J, Briscoe J, Jiang J. Sonic Hedgehog dependent phosphorylation by CK1alpha and GRK2 is required for ciliary accumulation and activation of smoothened. PLoS Biol. 2011;9:e1001083. doi: 10.1371/journal.pbio.1001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Maier D, Neubueser D, Hipfner DR. Regulation of smoothened by Drosophila G-protein-coupled receptor kinases. Dev Biol. 2010;337:99–109. doi: 10.1016/j.ydbio.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong JK, Virshup DM. Casein kinase 1: Complexity in the family. Int J Biochem Cell Biol. 2011;43:465–9. doi: 10.1016/j.biocel.2010.12.004. [DOI] [PubMed] [Google Scholar]
- Cheung HO, Zhang X, Ribeiro A, Mo R, Makino S, Puviindran V, Law KK, Briscoe J, Hui CC. The kinesin protein Kif7 is a critical regulator of Gli transcription factors in mammalian hedgehog signaling. Sci Signal. 2009;2:ra29. doi: 10.1126/scisignal.2000405. [DOI] [PubMed] [Google Scholar]
- Cooper AF, Yu KP, Brueckner M, Brailey LL, Johnson L, McGrath JM, Bale AE. Cardiac and CNS defects in a mouse with targeted disruption of suppressor of fused. Development. 2005;132:4407–17. doi: 10.1242/dev.02021. [DOI] [PubMed] [Google Scholar]
- Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–21. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- Cruciat CM, Dolde C, de Groot RE, Ohkawara B, Reinhard C, Korswagen HC, Niehrs C. RNA helicase DDX3 is a regulatory subunit of casein kinase 1 in Wnt-beta-catenin signaling. Science. 2013;339:1436–41. doi: 10.1126/science.1231499. [DOI] [PubMed] [Google Scholar]
- Cruciat CM, Ohkawara B, Acebron SP, Karaulanov E, Reinhard C, Ingelfinger D, Boutros M, Niehrs C. Requirement of prorenin receptor and vacuolar H+-ATPase-mediated acidification for Wnt signaling. Science. 2010;327:459–63. doi: 10.1126/science.1179802. [DOI] [PubMed] [Google Scholar]
- Davidson G, Wu W, Shen J, Bilic J, Fenger U, Stannek P, Glinka A, Niehrs C. Casein kinase 1 gamma couples Wnt receptor activation to cytoplasmic signal transduction. Nature. 2005;438:867–72. doi: 10.1038/nature04170. [DOI] [PubMed] [Google Scholar]
- Denef N, Neubuser D, Perez L, Cohen SM. Hedgehog induces opposite changes in turnover and subcellular localization of patched and smoothened. Cell. 2000;102:521–31. doi: 10.1016/s0092-8674(00)00056-8. [DOI] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Endoh-Yamagami S, Evangelista M, Wilson D, Wen X, Theunissen JW, Phamluong K, Davis M, Scales SJ, Solloway MJ, de Sauvage FJ, Peterson AS. The mammalian Cos2 homolog Kif7 plays an essential role in modulating Hh signal transduction during development. Curr Biol. 2009;19:1320–6. doi: 10.1016/j.cub.2009.06.046. [DOI] [PubMed] [Google Scholar]
- Evangelista M, Lim TY, Lee J, Parker L, Ashique A, Peterson AS, Ye W, Davis DP, de Sauvage FJ. Kinome siRNA screen identifies regulators of ciliogenesis and hedgehog signal transduction. Sci Signal. 2008;1:ra7. doi: 10.1126/scisignal.1162925. [DOI] [PubMed] [Google Scholar]
- Fan J, Liu Y, Jia J. Hh-induced Smoothened conformational switch is mediated by differential phosphorylation at its C-terminal tail in a dose- and position-dependent manner. Developmental biology. 2012;366:172–84. doi: 10.1016/j.ydbio.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao ZH, Seeling JM, Hill V, Yochum A, Virshup DM. Casein kinase I phosphorylates and destabilizes the beta-catenin degradation complex. Proc Natl Acad Sci U S A. 2002;99:1182–7. doi: 10.1073/pnas.032468199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Garcia MJ, Eggenschwiler JT, Caspary T, Alcorn HL, Wyler MR, Huangfu D, Rakeman AS, Lee JD, Feinberg EH, Timmer JR, Anderson KV. Analysis of mouse embryonic patterning and morphogenesis by forward genetics. Proc Natl Acad Sci U S A. 2005;102:5913–9. doi: 10.1073/pnas.0501071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–44. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross SD, Anderson RA. Casein kinase I: spatial organization and positioning of a multifunctional protein kinase family. Cell Signal. 1998;10:699–711. doi: 10.1016/s0898-6568(98)00042-4. [DOI] [PubMed] [Google Scholar]
- Ha NC, Tonozuka T, Stamos JL, Choi HJ, Weis WI. Mechanism of phosphorylation-dependent binding of APC to beta-catenin and its role in beta-catenin degradation. Mol Cell. 2004;15:511–21. doi: 10.1016/j.molcel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Haycraft CJ, Banizs B, Aydin-Son Y, Zhang Q, Michaud EJ, Yoder BK. Gli2 and gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1:e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Anderson KV. Cilia and Hedgehog responsiveness in the mouse. Proc Natl Acad Sci U S A. 2005;102:11325–30. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Anderson KV. Signaling from Smo to Ci/Gli: conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development. 2006;133:3–14. doi: 10.1242/dev.02169. [DOI] [PubMed] [Google Scholar]
- Hui CC, Angers S. Gli proteins in development and disease. Annual review of cell and developmental biology. 2011;27:513–37. doi: 10.1146/annurev-cellbio-092910-154048. [DOI] [PubMed] [Google Scholar]
- Humke EW, Dorn KV, Milenkovic L, Scott MP, Rohatgi R. The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev. 2010;24:670–82. doi: 10.1101/gad.1902910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Liu Y, Xia R, Tong C, Yue T, Jiang J, Jia J. Casein kinase 2 promotes Hedgehog signaling by regulating both smoothened and Cubitus interruptus. The Journal of biological chemistry. 2010;285:37218–26. doi: 10.1074/jbc.M110.174565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Amanai K, Wang G, Tang J, Wang B, Jiang J. Shaggy/GSK3 antagonizes Hedgehog signalling by regulating Cubitus interruptus. Nature. 2002;416:548–52. doi: 10.1038/nature733. [DOI] [PubMed] [Google Scholar]
- Jia J, Tong C, Jiang J. Smoothened transduces Hedgehog signal by physically interacting with Costal2/Fused complex through its C-terminal tail. Genes Dev. 2003;17:2709–20. doi: 10.1101/gad.1136603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Tong C, Wang B, Luo L, Jiang J. Hedgehog signalling activity of Smoothened requires phosphorylation by protein kinase A and casein kinase I. Nature. 2004;432:1045–50. doi: 10.1038/nature03179. [DOI] [PubMed] [Google Scholar]
- Jia J, Zhang L, Zhang Q, Tong C, Wang B, Hou F, Amanai K, Jiang J. Phosphorylation by double-time/CKIepsilon and CKIalpha targets cubitus interruptus for Slimb/beta-TRCP-mediated proteolytic processing. Dev Cell. 2005;9:819–30. doi: 10.1016/j.devcel.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15:801–12. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Struhl G. Protein kinase A and hedgehog signaling in Drosophila limb development. Cell. 1995;80:563–72. doi: 10.1016/0092-8674(95)90510-3. [DOI] [PubMed] [Google Scholar]
- Jiang J, Struhl G. Regulation of the Hedgehog and Wingless signalling pathways by the F- box/WD40-repeat protein Slimb. Nature. 1998;391:493–6. doi: 10.1038/35154. [DOI] [PubMed] [Google Scholar]
- Jones DT, Jager N, Kool M, Zichner T, Hutter B, Sultan M, Cho YJ, Pugh TJ, Hovestadt V, Stutz AM, Rausch T, Warnatz HJ, Ryzhova M, Bender S, Sturm D, Pleier S, Cin H, Pfaff E, Sieber L, Wittmann A, Remke M, Witt H, Hutter S, Tzaridis T, Weischenfeldt J, Raeder B, Avci M, Amstislavskiy V, Zapatka M, Weber UD, Wang Q, Lasitschka B, Bartholomae CC, Schmidt M, von Kalle C, Ast V, Lawerenz C, Eils J, Kabbe R, Benes V, van Sluis P, Koster J, Volckmann R, Shih D, Betts MJ, Russell RB, Coco S, Tonini GP, Schuller U, Hans V, Graf N, Kim YJ, Monoranu C, Roggendorf W, Unterberg A, Herold-Mende C, Milde T, Kulozik AE, von Deimling A, Witt O, Maass E, Rossler J, Ebinger M, Schuhmann MU, Fruhwald MC, Hasselblatt M, Jabado N, Rutkowski S, von Bueren AO, Williamson D, Clifford SC, McCabe MG, Collins VP, Wolf S, Wiemann S, Lehrach H, Brors B, Scheurlen W, Felsberg J, Reifenberger G, Northcott PA, Taylor MD, Meyerson M, Pomeroy SL, Yaspo ML, Korbel JO, Korshunov A, Eils R, Pfister SM, Lichter P. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–5. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent D, Bush EW, Hooper JE. Roadkill attenuates Hedgehog responses through degradation of Cubitus interruptus. Development. 2006;133:2001–10. doi: 10.1242/dev.02370. [DOI] [PubMed] [Google Scholar]
- Kise Y, Morinaka A, Teglund S, Miki H. Sufu recruits GSK3beta for efficient processing of Gli3. Biochemical and biophysical research communications. 2009;387:569–74. doi: 10.1016/j.bbrc.2009.07.087. [DOI] [PubMed] [Google Scholar]
- Knippschild U, Gocht A, Wolff S, Huber N, Lohler J, Stoter M. The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cell Signal. 2005;17:675–689. doi: 10.1016/j.cellsig.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Kovacs JJ, Whalen EJ, Liu R, Xiao K, Kim J, Chen M, Wang J, Chen W, Lefkowitz RJ. Beta-arrestin-mediated localization of smoothened to the primary cilium. Science. 2008;320:1777–81. doi: 10.1126/science.1157983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Salic A, Kirschner MW. Physiological regulation of [beta]-catenin stability by Tcf3 and CK1epsilon. J Cell Biol. 2001;154:983–93. doi: 10.1083/jcb.200102074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Chen Y, Shi Q, Yue T, Wang B, Jiang J. Hedgehog-regulated ubiquitination controls smoothened trafficking and cell surface expression in Drosophila. PLoS biology. 2012;10:e1001239. doi: 10.1371/journal.pbio.1001239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Li S, Han Y, Tong C, Wang B, Chen Y, Jiang J. Regulation of Smoothened Phosphorylation and High-Level Hedgehog Signaling Activity by a Plasma Membrane Associated Kinase. PLoS Biol. 2016;14:e1002481. doi: 10.1371/journal.pbio.1002481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Ma G, Wang B, Jiang J. Hedgehog induces formation of PKA-Smoothened complexes to promote Smoothened phosphorylation and pathway activation. Sci Signal. 2014;7:ra62. doi: 10.1126/scisignal.2005414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Ohlmeyer JT, Lane ME, Kalderon D. Function of protein kinase A in hedghehog signal transduction and Drosophila imaginal disc development. Cell. 1995;80:553–562. doi: 10.1016/0092-8674(95)90509-x. [DOI] [PubMed] [Google Scholar]
- Liang J, Fu Y, Cruciat CM, Jia S, Wang Y, Tong Z, Tao Q, Ingelfinger D, Boutros M, Meng A, Niehrs C, Wu W. Transmembrane protein 198 promotes LRP6 phosphorylation and Wnt signaling activation. Mol Cell Biol. 2011;31:2577–90. doi: 10.1128/MCB.05103-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liem KF, Jr, He M, Ocbina PJ, Anderson KV. Mouse Kif7/Costal2 is a cilia-associated protein that regulates Sonic hedgehog signaling. Proc Natl Acad Sci U S A. 2009;106:13377–82. doi: 10.1073/pnas.0906944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Wang B, Niswander LA. Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development. 2005;132:3103–11. doi: 10.1242/dev.01894. [DOI] [PubMed] [Google Scholar]
- Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, Zhang Z, Lin X, He X. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–47. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- Lum L, Yao S, Mozer B, Rovescalli A, Von Kessler D, Nirenberg M, Beachy PA. Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science. 2003;299:2039–45. doi: 10.1126/science.1081403. [DOI] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Developmental cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, Wang J, Liu B, Pan W, Farr GH, 3rd, Flynn C, Yuan H, Takada S, Kimelman D, Li L, Wu D. Low-density lipoprotein receptor-related protein-5 binds to Axin and regulates the canonical Wnt signaling pathway. Mol Cell. 2001;7:801–9. doi: 10.1016/s1097-2765(01)00224-6. [DOI] [PubMed] [Google Scholar]
- McKay RM, Peters JM, Graff JM. The casein kinase I family in Wnt signaling. Dev Biol. 2001;235:388–96. doi: 10.1006/dbio.2001.0308. [DOI] [PubMed] [Google Scholar]
- Meloni AR, Fralish GB, Kelly P, Salahpour A, Chen JK, Wechsler-Reya RJ, Lefkowitz RJ, Caron MG. Smoothened signal transduction is promoted by G protein-coupled receptor kinase 2. Mol Cell Biol. 2006;26:7550–60. doi: 10.1128/MCB.00546-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methot N, Basler K. Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and repressor forms of Cubitus interruptus. Cell. 1999;96:819–31. doi: 10.1016/s0092-8674(00)80592-9. [DOI] [PubMed] [Google Scholar]
- Methot N, Basler K. Suppressor of Fused opposes Hedgehog signal transduction by impeding nuclear accumulation of the activator form of Cubitus interruptus. Development. 2000;127:4001–4010. doi: 10.1242/dev.127.18.4001. [DOI] [PubMed] [Google Scholar]
- Molnar C, Ruiz-Gomez A, Martin M, Rojo-Berciano S, Mayor F, de Celis JF. Role of the Drosophila non-visual ss-arrestin kurtz in hedgehog signalling. PLoS Genet. 2011;7:e1001335. doi: 10.1371/journal.pgen.1001335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlmeyer JT, Kalderon D. Dual pathways for induction of wingless expression by protein kinase A and Hedgehog in Drosophila embryos. Genes Dev. 1997;11:2250–8. doi: 10.1101/gad.11.17.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlmeyer JT, Kalderon D. Hedgehog stimulates maturation of Cubitus interruptus into a labile transcriptional activator. Nature. 1998;396:749–53. doi: 10.1038/25533. [DOI] [PubMed] [Google Scholar]
- Pan D, Rubin GM. cAMP-dependent protein kinase and hedgehog act antagonistically in regulating decapentaplegic transcription in Drosophila imaginal discs. Cell. 1995;80:543–552. doi: 10.1016/0092-8674(95)90508-1. [DOI] [PubMed] [Google Scholar]
- Pan W, Choi SC, Wang H, Qin Y, Volpicelli-Daley L, Swan L, Lucast L, Khoo C, Zhang X, Li L, Abrams CS, Sokol SY, Wu D. Wnt3a-mediated formation of phosphatidylinositol 4,5-bisphosphate regulates LRP6 phosphorylation. Science. 2008;321:1350–3. doi: 10.1126/science.1160741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Bai CB, Joyner AL, Wang B. Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Molecular and cellular biology. 2006;26:3365–77. doi: 10.1128/MCB.26.9.3365-3377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, McKay RM, McKay JP, Graff JM. Casein kinase I transduces Wnt signals. Nature. 1999;401:345–50. doi: 10.1038/43830. [DOI] [PubMed] [Google Scholar]
- Piao S, Lee SH, Kim H, Yum S, Stamos JL, Xu Y, Lee SJ, Lee J, Oh S, Han JK, Park BJ, Weis WI, Ha NC. Direct inhibition of GSK3beta by the phosphorylated cytoplasmic domain of LRP6 in Wnt/beta-catenin signaling. PLoS One. 2008;3:e4046. doi: 10.1371/journal.pone.0004046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowman GD, Sudarsanam S, Bingham J, Whyte D, Hunter T. The protein kinases of Caenorhabditis elegans: a model for signal transduction in multicellular organisms. Proc Natl Acad Sci U S A. 1999;96:13603–10. doi: 10.1073/pnas.96.24.13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–51. [PubMed] [Google Scholar]
- Price MA, Kalderon D. Proteolysis of cubitus interruptus in Drosophila requires phosphorylation by protein kinase A. Development. 1999;126:4331–9. doi: 10.1242/dev.126.19.4331. [DOI] [PubMed] [Google Scholar]
- Price MA, Kalderon D. Proteolysis of the Hedgehog signaling effector Cubitus interruptus requires phosphorylation by Glycogen Synthase Kinase 3 and Casein Kinase 1. Cell. 2002;108:823–35. doi: 10.1016/s0092-8674(02)00664-5. [DOI] [PubMed] [Google Scholar]
- Pronobis MI, Rusan NM, Peifer M. A novel GSK3-regulated APC:Axin interaction regulates Wnt signaling by driving a catalytic cycle of efficient betacatenin destruction. Elife. 2015;4:e08022. doi: 10.7554/eLife.08022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J, Carneiro MO, Carter SL, Cibulskis K, Erlich RL, Greulich H, Lawrence MS, Lennon NJ, McKenna A, Meldrim J, Ramos AH, Ross MG, Russ C, Shefler E, Sivachenko A, Sogoloff B, Stojanov P, Tamayo P, Mesirov JP, Amani V, Teider N, Sengupta S, Francois JP, Northcott PA, Taylor MD, Yu F, Crabtree GR, Kautzman AG, Gabriel SB, Getz G, Jager N, Jones DT, Lichter P, Pfister SM, Roberts TM, Meyerson M, Pomeroy SL, Cho YJ. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–10. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivers A, Gietzen KF, Vielhaber E, Virshup DM. Regulation of casein kinase I epsilon and casein kinase I delta by an in vivo futile phosphorylation cycle. J Biol Chem. 1998;273:15980–4. doi: 10.1074/jbc.273.26.15980. [DOI] [PubMed] [Google Scholar]
- Robbins DJ, Nybakken KE, Kobayashi R, Sisson JC, Bishop JM, Therond PP. Hedgehog elicits signal transduction by means of a large complex containing the kinesin-related protein costal2. Cell. 1997;90:225–34. doi: 10.1016/s0092-8674(00)80331-1. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–6. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- Rubin LL, de Sauvage FJ. Targeting the Hedgehog pathway in cancer. Nature reviews Drug discovery. 2006;5:1026–33. doi: 10.1038/nrd2086. [DOI] [PubMed] [Google Scholar]
- Rubinfeld B, Tice DA, Polakis P. Axin-dependent phosphorylation of the adenomatous polyposis coli protein mediated by casein kinase 1epsilon. J Biol Chem. 2001;276:39037–45. doi: 10.1074/jbc.M105148200. [DOI] [PubMed] [Google Scholar]
- Sakanaka C, Leong P, Xu L, Harrison SD, Williams LT. Casein kinase iepsilon in the wnt pathway: regulation of beta-catenin function. Proc Natl Acad Sci U S A. 1999;96:12548–52. doi: 10.1073/pnas.96.22.12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q, Li S, Jia J, Jiang J. The Hedgehog-induced Smoothened conformational switch assembles a signaling complex that activates Fused by promoting its dimerization and phosphorylation. Development. 2011;138:4219–31. doi: 10.1242/dev.067959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q, Li S, Li S, Jiang A, Chen Y, Jiang J. Hedgehog-induced phosphorylation by CK1 sustains the activity of Ci/Gli activator. Proc Natl Acad Sci U S A. 2014;111:E5651–60. doi: 10.1073/pnas.1416652111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisson JC, Ho KS, Suyama K, Scott MP. Costal2, a novel kinesin-related protein in the Hedgehog signaling pathway. Cell. 1997;90:235–45. doi: 10.1016/s0092-8674(00)80332-3. [DOI] [PubMed] [Google Scholar]
- Smelkinson MG, Kalderon D. Processing of the Drosophila hedgehog signaling effector Ci-155 to the repressor Ci-75 is mediated by direct binding to the SCF component slimb. Curr Biol. 2006;16:110–6. doi: 10.1016/j.cub.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Smelkinson MG, Zhou Q, Kalderon D. Regulation of Ci-SCFSlimb binding, Ci proteolysis, and hedgehog pathway activity by Ci phosphorylation. Dev Cell. 2007;13:481–95. doi: 10.1016/j.devcel.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer E, Jiang J, Chen ZJ. Signal-induced ubiquitination of IkappaBalpha by the F-box protein Slimb/beta-TrCP. Genes Dev. 1999;13:284–94. doi: 10.1101/gad.13.3.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strigini M, Cohen SM. A Hedgehog activity gradient contributes to AP axial patterning of the Drosophila wing. Development. 1997;124:4697–705. doi: 10.1242/dev.124.22.4697. [DOI] [PubMed] [Google Scholar]
- Svard J, Heby-Henricson K, Persson-Lek M, Rozell B, Lauth M, Bergstrom A, Ericson J, Toftgard R, Teglund S. Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev Cell. 2006;10:187–97. doi: 10.1016/j.devcel.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Tamai K, Zeng X, Liu C, Zhang X, Harada Y, Chang Z, He X. A mechanism for Wnt coreceptor activation. Mol Cell. 2004;13:149–56. doi: 10.1016/s1097-2765(03)00484-2. [DOI] [PubMed] [Google Scholar]
- Tanneberger K, Pfister AS, Brauburger K, Schneikert J, Hadjihannas MV, Kriz V, Schulte G, Bryja V, Behrens J. Amer1/WTX couples Wnt-induced formation of PtdIns(4,5)P2 to LRP6 phosphorylation. EMBO J. 2011;30:1433–43. doi: 10.1038/emboj.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempe D, Casas M, Karaz S, Blanchet-Tournier MF, Concordet JP. Multisite protein kinase A and glycogen synthase kinase 3beta phosphorylation leads to Gli3 ubiquitination by SCFbetaTrCP. Mol Cell Biol. 2006;26:4316–26. doi: 10.1128/MCB.02183-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukachinsky H, Lopez LV, Salic A. A mechanism for vertebrate Hedgehog signaling: recruitment to cilia and dissociation of SuFu-Gli protein complexes. J Cell Biol. 2010;191:415–28. doi: 10.1083/jcb.201004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varjosalo M, Li SP, Taipale J. Divergence of hedgehog signal transduction mechanism between Drosophila and mammals. Dev Cell. 2006;10:177–86. doi: 10.1016/j.devcel.2005.12.014. [DOI] [PubMed] [Google Scholar]
- Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000a;100:423–34. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- Wang B, Li Y. Evidence for the direct involvement of {beta}TrCP in Gli3 protein processing. Proc Natl Acad Sci U S A. 2006;103:33–8. doi: 10.1073/pnas.0509927103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Pan Y, Wang B. Suppressor of fused and Spop regulate the stability, processing and function of Gli2 and Gli3 full-length activators but not their repressors. Development. 2010;137:2001–9. doi: 10.1242/dev.052126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Amanai K, Wang B, Jiang J. Interactions with Costal2 and suppressor of fused regulate nuclear translocation and activity of cubitus interruptus. Genes Dev. 2000b;14:2893–905. doi: 10.1101/gad.843900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Wang B, Jiang J. Protein kinase A antagonizes Hedgehog signaling by regulating both the activator and repressor forms of Cubitus interruptus. Genes Dev. 1999;13:2828–37. doi: 10.1101/gad.13.21.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Lai CK, Evangelista M, Hongo JA, de Sauvage FJ, Scales SJ. Kinetics of hedgehog-dependent full-length Gli3 accumulation in primary cilia and subsequent degradation. Molecular and cellular biology. 2010;30:1910–22. doi: 10.1128/MCB.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, Harper JW. The SCFbeta-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and beta-catenin and stimulates IkappaBalpha ubiquitination in vitro. Genes Dev. 1999;13:270–83. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Huang H, Garcia Abreu J, He X. Inhibition of GSK3 phosphorylation of beta-catenin via phosphorylated PPPSPXS motifs of Wnt coreceptor LRP6. PLoS One. 2009;4:e4926. doi: 10.1371/journal.pone.0004926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R, Jia H, Fan J, Liu Y, Jia J. USP8 promotes smoothened signaling by preventing its ubiquitination and changing its subcellular localization. PLoS biology. 2012;10:e1001238. doi: 10.1371/journal.pbio.1001238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Murone M, Luoh SM, Ryan A, Gu Q, Zhang C, Bonifas JM, Lam CW, Hynes M, Goddard A, Rosenthal A, Epstein EH, Jr, de Sauvage FJ. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature. 1998;391:90–92. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- Xing Y, Clements WK, Le Trong I, Hinds TR, Stenkamp R, Kimelman D, Xu W. Crystal structure of a beta-catenin/APC complex reveals a critical role for APC phosphorylation in APC function. Mol Cell. 2004;15:523–33. doi: 10.1016/j.molcel.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Zeng X, Huang H, Tamai K, Zhang X, Harada Y, Yokota C, Almeida K, Wang J, Doble B, Woodgett J, Wynshaw-Boris A, Hsieh JC, He X. Initiation of Wnt signaling: control of Wnt coreceptor Lrp6 phosphorylation/activation via frizzled, dishevelled and axin functions. Development. 2008;135:367–75. doi: 10.1242/dev.013540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Tamai K, Doble B, Li S, Huang H, Habas R, Okamura H, Woodgett J, He X. A dual-kinase mechanism for Wnt co-receptor phosphorylation and activation. Nature. 2005;438:873–7. doi: 10.1038/nature04185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Williams EH, Guo Y, Lum L, Beachy PA. Extensive phosphorylation of Smoothened in Hedgehog pathway activation. Proc Natl Acad Sci U S A. 2004;101:17900–7. doi: 10.1073/pnas.0408093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Jia J, Wang B, Amanai K, Wharton KA, Jr, Jiang J. Regulation of wingless signaling by the CKI family in Drosophila limb development. Dev Biol. 2006a;299:221–37. doi: 10.1016/j.ydbio.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Shi Q, Chen Y, Yue T, Li S, Wang B, Jiang J. Multiple Ser/Thr-rich degrons mediate the degradation of Ci/Gli by the Cul3-HIB/SPOP E3 ubiquitin ligase. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:21191–6. doi: 10.1073/pnas.0912008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zhang L, Wang B, Ou CY, Chien CT, Jiang J. A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Dev Cell. 2006b;10:719–29. doi: 10.1016/j.devcel.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Zhang W, Zhao Y, Tong C, Wang G, Wang B, Jia J, Jiang J. Hedgehog-regulated costal2-kinase complexes control phosphorylation and proteolytic processing of cubitus interruptus. Dev Cell. 2005;8:267–78. doi: 10.1016/j.devcel.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Mao F, Lu Y, Wu W, Zhang L, Zhao Y. Transduction of the Hedgehog signal through the dimerization of Fused and the nuclear translocation of Cubitus interruptus. Cell research. 2011;21:1436–51. doi: 10.1038/cr.2011.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Tong C, Jiang J. Hedgehog regulates smoothened activity by inducing a conformational switch. Nature. 2007;450:252–8. doi: 10.1038/nature06225. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Kalderon D. Hedgehog Activates Fused through Phosphorylation to Elicit a Full Spectrum of Pathway Responses. Dev Cell. 2011;20:802–14. doi: 10.1016/j.devcel.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]