Abstract
PRDM1 /BLIMP-1, a master regulator of plasma-cell differentiation, is frequently inactivated in activated B-cell-like (ABC) diffuse large B-cell lymphoma (DLBCL) patients. Little is known about its genetic aberrations and relevant clinical implications. We assessed a large cohort of patients with de novo DLBCL for PRDM1/BLIMP-1 deletion, mutation, and protein expression. BLIMP-1 expression was frequently associated with the ABC phenotype and plasmablastic morphologic subtype of DLBCL, yet 63% of the ABC-DLBCL patients were negative for BLIMP-1 protein expression. In these patients, loss of BLIMP-1 was associated with Myc overexpression and decreased expression of p53 pathway tumor-suppressor molecules. Also, we found that homozygous PRDM1 deletions and PRDM1 mutations within exons 1 and 2 which encode for domains crucial for transcriptional repression had a poor prognostic impact in patients with ABC-DLBCL but not those with germinal center B-cell-like DLBCL. Gene expression profiling revealed that loss of PRDM1/BLIMP-1 expression correlated with a decreased plasma-cell differentiation signature and upregulation of genes involved in B-cell receptor signaling and tumor-cell proliferation. In conclusion, these results provide clinical and biological insight into the tumor-suppressive role of PRDM1/BLIMP-1 in ABC-DLBCL patients and suggest that loss of PRDM1/BLIMP-1 function contributes to the overall poor prognosis of ABC-DLBCL patients.
Keywords: BLIMP-1, PRDM1, Deletion, Mutation, DLBCL, GEP
INTRODUCTION
Diffuse large B-cell lymphoma (DLBCL) is the most common type of lymphoid malignancy, accounting for 30–40% of all non-Hodgkin lymphoma.1 Anthracycline-based combination chemotherapy, first introduced in the 1970s, is the backbone of therapy for patients with DLBCL, and currently the standard therapy regimen includes the anti-CD20 antibody rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP).2–4 Despite encouraging complete remission and overall survival (OS) rates using this regimen, up to one third of DLBCL patients suffer from relapse or refractory primary disease.5
The heterogeneous clinical outcome of DLBCL results in part from variable genetic profiles of this tumor. Gene expression profiling (GEP) has identified two distinct types of DLBCL: germinal center B-cell–like and activated B-cell–like (GCB and ABC). Patients with ABC-DLBCL have markedly poorer survival than do patients with GCB-DLBCL.6, 7 However, the molecular mechanism responsible for this difference is not completely understood.
ABC-DLBCL is characterized by constitutive nuclear factor (NF)-κB activation and genetic alterations that interfere with terminal B-cell differentiation.8 PRDM1, located on chromosome 6q21, encodes for BLIMP-1, a zinc-finger-containing DNA-binding transcriptional repressor. BLIMP-1 expression is required for the development of immunoglobulin-secreting cells and maintenance of long-lived plasma cells.9 Conditional PRDM1-knockout mice do not have production of plasma cells or serum immunoglobulins.10 PRDM1 is frequently inactivated in ABC-DLBCL cases as a result of genetic deletions or mutations or transcriptional repression of it.11, 12 Studies have demonstrated that an inactivation mutation of PRDM1 is recurrent in approximately 25% of ABC-DLBCL cases.11–14 Moreover, conditional deletion of PRDM1 in murine B cells facilitates the development of lymphoproliferative disease resembling human ABC-DLBCL.8, 11
Limited research has been done on the clinical implications of PRDM1/BLIMP-1 abnormality in DLBCL patients. In the present study, we investigated PRDM1 deletion and mutation and BLIMP-1 protein expression in a large cohort of de novo DLBCL patients treated with R-CHOP. Our results demonstrated that disruption of PRDM1/BLIMP-1 is associated with poor prognosis for ABC-DLBCL. We further characterized the potential molecular mechanisms underlying the tumor-suppressive function of PRDM1/BLIMP-1 using GEP.
MATERIALS AND METHODS
Patients
We studied 520 biopsy specimens obtained from R-CHOP-treated patients with de novo DLBCL. The diagnostic criteria for DLBCL, patient-selection process, therapy, and treatment responses were described previously.15 The study was approved by the institutional review boards of the participating institutions.
Immunohistochemistry
Tissue microarrays prepared using formalin-fixed, paraffin-embedded (FFPE) tissue blocks of the diagnostic biopsy specimens obtained from the studied patients were stained with an anti-BLIMP-1 antibody (EPR16655; Epitomics, Burlingame, CA, USA). BLIMP-1 expression levels were determined by estimating the percentage of BLIMP-1+ tumor cells in the tissue blocks. Evaluation of other biomarkers was performed using immunohistochemistry with corresponding antibodies. Details of the immunohistochemical procedures and scoring processes were described previously.15–19
Fluorescence in situ hybridization, and PRDM1 and TP53 sequencing
Fluorescence in situ hybridization (FISH) probes spanning the PRDM1 gene and chromosome 6 centromere were generated as described previously.13 Dual-color FISH was performed using standard procedures. Briefly, 4-μ m-thick tissue sections were backed overnight at 56°C, deparaffinized in CitriSolv, treated with 0.2 N hydrochloric acid for 20 min at room temperature, pretreated with 1 N sodium thiocyanate at 80°C for 30 min, protease-digested at 37°C for 50 min, refixed in 10% buffered formalin for 10 min, and derehydrated in a 70%, 85%, and 100% ethanol series before hybridization. The tissue sections were co-denatured with the addition of a probe for 8 min at 74°C and incubated for 24 h at 37°C in a hybridization oven (ThermoBrite). Posthybridization washes were performed in 0.4× SSC/0.3% NP-40 for 2 min and 2× SSC/0.1% NP-40 for 1 min at 72°C. Slides were then mounted with 4,6-diamidino-2-phenylindole (0.5 g/ml) containing an antifade solution. For one slide containing 25 cases, 200 nuclei of cells were evaluated independently by two observers (A.T. and V.M.). The overall concordance of their evaluations was nearly perfect (κ = 0.95), so all other slides (200 nuclei/case) were scored by A.T. only. The ratio of PRDM1 signals (green) to CEP6 signals (red) was calculated. If this ratio was lower than 0.72, heterozygous PRDM1 deletion was considered to be present. Ratios lower than 0.38 were considered to be suggestive of homozygous PRDM1 deletions. These ratios were calculated as ratios below the mean plus three standard deviations of green to orange signal ratios in reference cases (tonsils; n = 6) and subtraction of tumor-infiltrating T cells, which accounted for approximately 30% of undeleted alleles. MYC, BCL2, and BCL6 gene arrangements and copy-number aberrations were detected using FISH, as well.16, 20
Sequencing of PRDM1 coding regions was performed by Polymorphic DNA Technologies (Alameda, CA, USA) using Sanger sequencing. Sequencing results were compared to the NCBI NM_001198 reference sequence, followed by exclusion of all the single nucleotide polymorphisms (SNPs) documented by NCBI dbSNP database (build 147). The remaining variates detected by sequencing were considered as PRDM1 somatic mutations. TP53 was sequenced using a p53 AmpliChip (Roche Molecular Diagnostics, Pleasanton, CA, USA) as described previously.21
GEP and GCB/ABC classification
GEP was performed using a GeneChip Human Genome U133 Plus 2.0 Array (Affymetrix, Santa Clara, CA, USA) with total RNAs as described previously.15, 21 The CEL files were deposited in the National Center for Biotechnology Information Gene Expression Omnibus repository (GSE #31312). GEP classified 407 cases as GCB/ABC subtypes, and the cell-of-origin of another 111 cases was determined and compared by immunohistochemical algorithms (Hans, Visco-Young and Choi).15 Cell-of-origin classification based on B-cell-associated gene signatures has been described previously.22
MicroRNA profiling
An HTG EdgeSeq Whole Transcriptome Assay (HTG Molecular Diagnostics, Tucson, AZ, USA) coupled with a HiSeq system (Illumina, San Diego, CA, USA) was used for measuring the expression of microRNAs (miRNAs) in FFPE tissues. Selection of regulatory miRNAs for BLIMP-1 expression was based on TargetScan and published data.23–27
Statistical analysis
Clinical and molecular features of patient tumors were compared using the Fisher exact or chi-square test. OS and progression-free survival (PFS) in the study population were analyzed using the Kaplan-Meier method, and differences between DLBCL subgroups were compared using the log-rank test with the Prism 6 software program (GraphPad Software, San Diego, CA, USA). Multivariate analysis was performed using the Cox proportional hazards regression model with the SPSS software program (version 19.0; IBM Corporation, Armonk, NY, USA). P values of up to 0.05 were considered statistically significant.
RESULTS
Patients
The clinical characteristics of the study patients with DLBCL (n = 520) are summarized in Table 1. Their median age was 65 years (range, 16–82 years), the median follow-up duration was 46.7 months (range, 20.0–186.7 months), and the male : female ratio was 1.2:1. Two hundred sixty-eight patients (52%) had GCB-DLBCL, and 250 patients (48%) had ABC-DLBCL.
Table 1.
Clinical features of DLBCL patients with BLIMP-1 expression, PRDM1 mutation and homozygous deletion
| Variables | BLIMP-1 protein expression | PRDM1 mutation | Homozygous PRDM1 deletion | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | P | Positive | Negative | P | Positive | Negative | P | |
| No. of patients | 132 | 388 | 94 | 274 | 19 | 273 | |||
| Cell-of-origin | |||||||||
| GCB | 39 (30%) | 229 (59%) | <0.0001 | 48 (51%) | 141 (52%) | 0.91 | 7 (37%) | 138 (51%) | 0.34 |
| ABC | 92 (70%) | 158 (41%) | 46 (49%) | 131 (48%) | 12 (63%) | 134 (49%) | |||
| Plasmablastic subtype | |||||||||
| No | 55 (47%) | 259 (94%) | <0.0001 | 25 (80%) | 48 (87%) | 0.41 | 9 (100%) | 185 (87%) | 0.27 |
| Yes | 29 (53%) | 15 (6%) | 6 (20%) | 7 (13%) | 0 | 25 (13%) | |||
| Age, years | |||||||||
| < 60y | 53 (40%) | 166 (43%) | 0.61 | 39 (41%) | 117 (43%) | 0.90 | 10 (53%) | 114 (42%) | 0.47 |
| ≥ 60y | 79 (60%) | 222 (57%) | 55 (59%) | 157 (57%) | 9 (47%) | 159 (58%) | |||
| Gender | |||||||||
| Male | 81 (61%) | 224 (58%) | 0.46 | 51 (54%) | 162 (59%) | 0.47 | 11 (58%) | 143 (52%) | 0.81 |
| Female | 51 (39%) | 164 (42%) | 43 (46%) | 112 (41%) | 8 (42%) | 130 (48%) | |||
| Stage | |||||||||
| I – II | 60 (47%) | 179 (47%) | 0.96 | 36 (41%) | 120 (45%) | 0.54 | 11 (58%) | 126 (48%) | 0.48 |
| III – IV | 67 (53%) | 198 (53%) | 52 (59%) | 147 (55%) | 8 (42%) | 135 (52%) | |||
| B-symptoms | |||||||||
| No | 78 (61%) | 244 (67%) | 0.24 | 48 (55%) | 176 (68%) | 0.039 | 15 (79%) | 165 (63%) | 0.22 |
| Yes | 50 (39%) | 122 (33%) | 39 (45%) | 84 (32%) | 4 (21%) | 98 (37%) | |||
| LDH level | |||||||||
| Normal | 49 (41%) | 134 (38 %) | 0.58 | 30 (34%) | 107 (43%) | 0.17 | 8 (44%) | 102 (40%) | 0.80 |
| Elevated | 72 (59%) | 222 (62%) | 57 (66%) | 140 (57%) | 10 (56%) | 156 (60%) | |||
| No. of extranodal sites | |||||||||
| 0 – 1 | 93 (74%) | 287 (77%) | 0.51 | 65 (72%) | 200 (77%) | 0.39 | 17 (94%) | 197 (78%) | 0.13 |
| ≥ 2 | 33 (26%) | 87 (23%) | 25 (28%) | 59 (23%) | 1 (6%) | 57 (22%) | |||
| ECOG Performance status | |||||||||
| 0 – 1 | 99 (84%) | 289 (83%) | 0.83 | 61 (72 %) | 208 (86%) | 0.005 | 16 (89%) | 204 (83%) | 0.75 |
| ≥ 2 | 19 (16%) | 59 (17%) | 24 (28%) | 34 (14%) | 2 (11%) | 43 (17%) | |||
| Size of largest tumor | |||||||||
| < 5cm | 61 (64%) | 162 (56%) | 0.19 | 38 (61%) | 129 (59%) | 0.88 | 10 (67%) | 129 (54%) | 0.43 |
| ≥ 5cm | 35 (36%) | 128 (44%) | 24 (39%) | 88 (41%) | 5 (33%) | 108 (46%) | |||
| IPI score | |||||||||
| 0 – 2 | 77 (59%) | 231 (62%) | 0.59 | 50 (54%) | 166 (63%) | 0.14 | 13 (68%) | 160 (61%) | 0.51 |
| 3 – 5 | 53 (41%) | 142 (38%) | 42 (46%) | 96 (37%) | 6 (32%) | 103 (39%) | |||
| Therapy response | |||||||||
| CR | 103 (78%) | 286 (74%) | 0.32 | 65 (69%) | 206 (75%) | 0.28 | 14 (74%) | 214 (78%) | 0.58 |
| PR | 17 (13%) | 52 (13%) | 18 (19%) | 35 (13%) | 4 (21%) | 29 (11%) | |||
| SD | 6 (4.5%) | 18 (5%) | 4 (4%) | 12 (4%) | 1 (5%) | 11 (4%) | |||
| PD | 6 (4.5%) | 32 (8%) | 7 (7%) | 21 (8%) | 0 | 19 (7%) | |||
Abbreviations: ECOG, Eastern Cooperative Oncology Group; LDH, lactate dehydrogenase; IPI, International Prognostic Index; CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease.
Homozygous PRDM1 deletion predicts poor prognosis for ABC-DLBCL
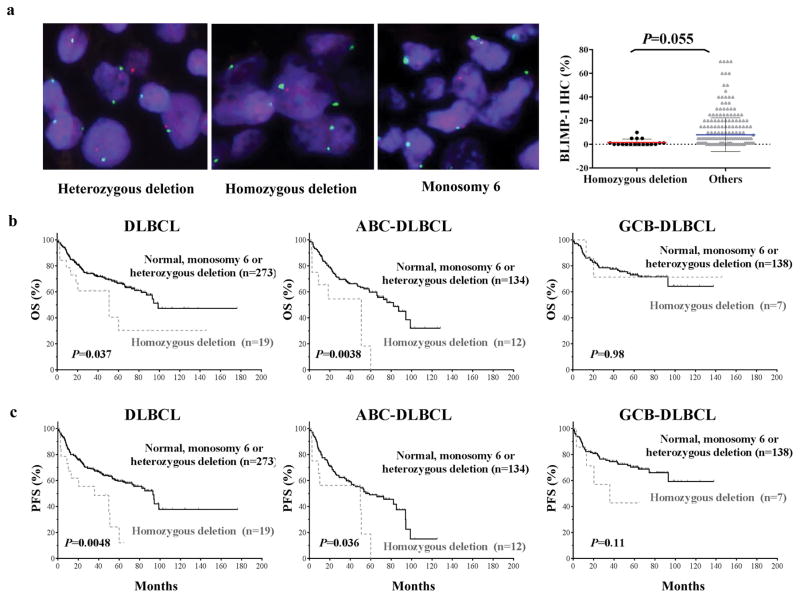
We detected homozygous deletion of PRDM1 in 19 (7%) of 292 patients with available FISH results (Figure 1a and Supplementary Table 1). Patients with homozygous PRDM1 deletions had lower BLIMP-1 protein expression than did those without these deletions (P = 0.055, Figure 1a). Homozygous PRDM1 deletion was not associated with the ABC phenotype (P = 0.34) or any other clinical parameters. OS was significantly shorter in patients with homozygous PRDM1 deletions than in those with normal or heterozygous deletions of PRDM1 or monosomy 6 (P = 0.037) (Figure 1b). This difference was remarkable in patients with ABC-DLBCL (P = 0.004) but not in those with GCB-DLBCL (P = 0.98) (Figure 1b). Homozygous PRDM1 deletion was also associated with significantly poorer PFS in the entire DLBCL cohort (P = 0.0048) and in ABC-DLBCL (P = 0.036) (Figure 1c). Multivariate analysis adjusting clinical features confirmed homozygous PRDM1 deletion as an independent prognostic factor for both OS (P = 0.032) and PFS (P = 0.037). In the ABC-DLBCL patients, homozygous PRDM1 deletion was an independent prognostic factor for OS (P = 0.032) but not for PFS (P = 0.13) (Table 2).
Figure 1.
Homozygous PRDM1 deletion in DLBCL cases. (a) Representative examples of FISH results with heterozygous PRDM1 deletion (of note are a mixture of cells with two green signals, corresponding to centromere 6, but lacking red signals, corresponding to PRDM1, and cells with two green signals and one red signal), homozygous PRDM1 deletion (of note are two green signals but a lack of red signals in the majority of the cells; one cell in the center of the microphotograph has both red and green signals and serves as an internal positive control), and monosomy 6 (all cells have only one green and one red signal). DLBCL patients with homozygous PRDM1 deletions had lower levels of BLIMP-1 protein expression than did the rest of the studied patients. (b–c) The impact of homozygous PRDM1 deletion on OS and PFS in all patients with DLBCL, patients with ABC-DLBCL, and patients with GCB-DLBCL. Patients with this deletion had shorter OS and PFS durations than did patients with normal FISH signals, heterozygous PRDM1 deletions, or monosomy 6. This trend was greater in patients with ABC-DLBCL than in GCB-DLBCL.
Table 2.
Multivariate analysis for homozygous PRDM1 deletions in DLBCL and ABC-DLBCL
| DLBCL | ABC-DLBCL | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Overall survival | Progression-free survival | Overall survival | Progression-free survival | |||||||||
| P | HR | 95% CI | P | HR | 95% CI | P | HR | 95% CI | P | HR | 95% CI | |
| Female Sex | 0.60 | 0.89 | 0.58–1.37 | 0.69 | 0.92 | 0.62–1.37 | 0.63 | 1.15 | 0.66–1.99 | 0.71 | 1.10 | 0.67–1.82 |
| B-symptoms | 0.031 | 1.62 | 1.04–2.50 | 0.072 | 1.45 | 0.97–2.19 | 0.26 | 1.37 | 0.79–2.37 | 0.21 | 1.39 | 0.83–2.32 |
| IPI >2 | <0.001 | 3.02 | 1.96–4.64 | <0.001 | 2.62 | 1.76–3.89 | <0.001 | 2.67 | 1.57–4.53 | 0.001 | 2.29 | 1.40–3.75 |
| Tumor Size >5cm | 0.18 | 1.33 | 0.87–2.02 | 0.56 | 1.12 | 0.64–1.66 | 0.26 | 1.35 | 0.80–2.26 | 0.60 | 1.14 | 0.70–1.84 |
| PRDM1 homozygous deletion | 0.032 | 2.23 | 1.07–4.65 | 0.037 | 2.09 | 1.05–4.18 | 0.032 | 2.58 | 1.08–6.15 | 0.13 | 1.95 | 0.83–4.59 |
Abbreviations: ABC, activated B-cell like; HR, hazard ratio; CI, confidence interval.
PRDM1 mutation predicts poor prognosis for ABC-DLBCL
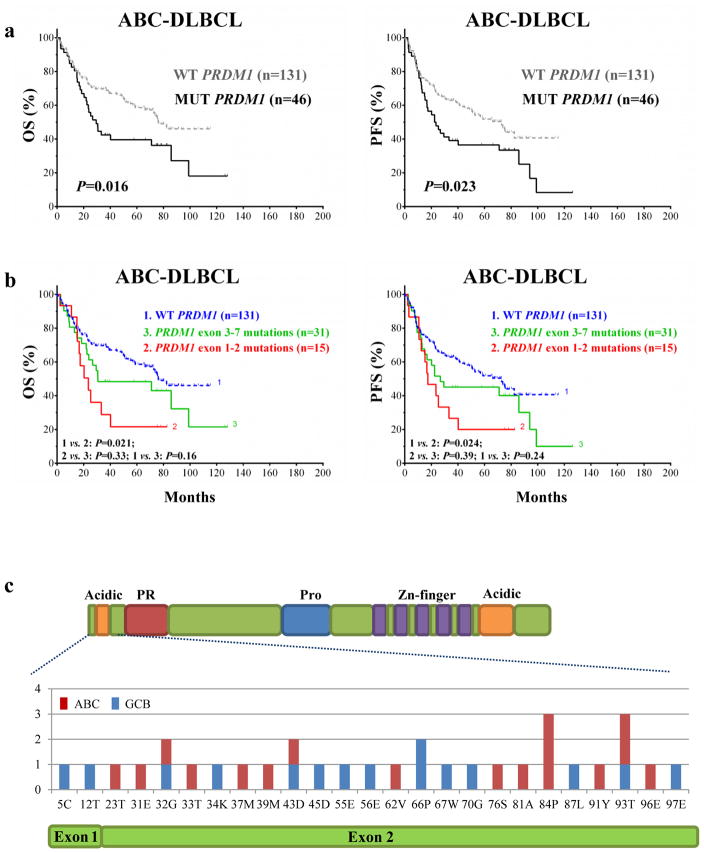
We studied PRDM1 mutations in 368 patients with available genomic DNA. Using a quality score cutoff of 16 (97% confidence), we identified 94 patients (25.5%) had PRDM1 mutations within the coding DNA sequence region of PRDM1 (Supplementary Table 2). Of these, seven patients had truncating (nonsense) mutations and two had a frameshift mutation. Seven of these nine patients had ABC-DLBCL, including four patients with short OS and PFS (Supplementary Table 3). PRDM1 mutation was not associated with the ABC phenotype (P = 0.91) and did not impact survival in the overall cohort or in GCB-DLBCL patients in particular (Supplementary Figure 1). However, PRDM1 mutation did correlate with poorer prognosis for ABC-DLBCL (Figure 2a). Moreover, ABC-DLBCL patients harboring PRDM1 mutations within exon 1 or 2 had markedly shorter survival durations than did patients with mutations in exons 3–7 or wild-type for PRDM1 (Figures 2b–c). In comparison, the prognostic impact of exons 1 and 2 PRDM1 mutations on GCB-DLBCL was minimal (Supplementary Figure 2). Consistently, we found that patients with exons 1 and 2 PRDM1 mutations were more likely to have overexpression of both Myc and Bcl-2 than were patients with exons 3–7 mutations in the overall cohort (P = 0.03) and those with wild-type PRDM1 in ABC-DLBCL (P = 0.05) but not in those with GCB-DLBCL (P = 0.44). Multivariate analysis of clinical parameters, Myc/Bcl-2 expression scores, and PRDM1 mutations indicated that Myc/Bcl-2 co-expression (hazard ratio: 2.9, P < 0.001), but not PRDM1 mutations (hazard ratio: 1.54, P = 0.12), was an independent prognostic factor.
Figure 2.
Correlation of PRDM1 mutations with poor prognosis for DLBCL. (a) In ABC-DLBCL, patients with PRDM1 mutations had shorter OS and PFS durations than did patients with wild-type PRDM1. (b) PRDM1 mutations within exons 1 and 2 had even greater poor impact on OS and PFS compared to did the overall mutation cohort. (c) The detailed distribution of PRDM1 mutations within exons 1 and 2.
We also studied the correlations between various types of PRDM1 mutations and BLIMP-1 protein expression as well as PRDM1 deletion. Patients with nonsense mutations or exon 1/2 mutations had lower BLIMP-1 expression than patients without these aberrations, but the P values for the differences were not significant (Supplementary Figure 3). No significant associations were observed between PRDM1 mutations and PRDM1 allelic deletions.
Loss of BLIMP-1 protein expression correlates with Myc overexpression and decreased p53 pathway molecule expression in ABC-DLBCLs
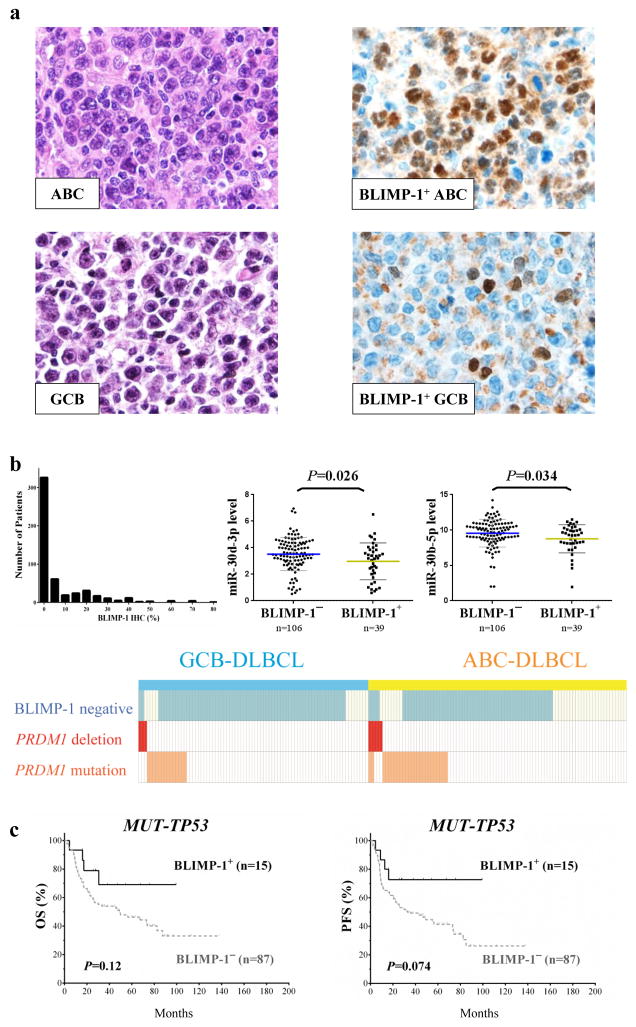
Representative immunohistochemical staining of FFPE tissue sections for BLIMP-1 protein expression is shown in Figure 3a. Using a cutoff of 10%, we found that 132 (25%) of the 520 patients were positive for BLIMP-1. BLIMP-1 protein expression was more common in patients with the ABC phenotype (37% vs. 15% in GCB-DLBCL, P < 0.0001) and plasmablast subtype22 of DLBCL than others (P < 0.0001; Table 1). The histogram of BLIMP-1 expression in the study cohort, and distribution of PRDM1 deletions and mutations and BLIMP-1 protein expression in 180 patients with all data available is shown in Figure 3b. Ninety-six of these patients were in the ABC subgroup, 59 of whom (61%) had BLIMP-1− DLBCL. Thirty-seven (63%) of the BLIMP-1− ABC-DLBCL cases had no detectable PRDM1 deletions or mutations. MiRNA profiling identified that miR-30d-3p, miR-30d-5p and miR-30b-5p were significantly upregulated in BLIMP-1− patients without apparent PRDM1 genetic aberrations, suggesting epigenetic regulations may also play a role in loss of PRDM1/BLIMP-1 expression.
Figure 3.
BLIMP-1 protein expression in DLBCL. (a) Representative immunohistochemical stains of DLBCL sections for BLIMP-1 protein expression. (b) The histograms of BLIMP-1 protein expression in all DLBCL patients, and the distribution of PRDM1 deletions and mutations and BLIMP-1 protein expression in 180 patients with all these three data available. Expression of miR-30d-3p and miR-30b-5p was elevated in BLIMP-1− DLBCL patients without PRDM1 genetic alterations. (c) The impact of BLIMP-1 protein expression on OS and PFS in DLBCL patients with mutated TP53.
In the ABC subgroup, BLIMP-1 negativity was associated with reduced protein expression for the NF-κB pathway component p65 (P = 0.028), p53 (P = 0.024; only in subjects wild-type for TP53), p53 downstream targets MDM2 (P = 0.002) and p21 (P = 0.074). BLIMP-1 negativity also was more common in patients without expression of CD30 (P = 0.0082) or phosphorylated AKT (P = 0.0095) (Table 3). Of note, Myc expression was increased in BLIMP-1− cases, with a trend of significance (P = 0.085), and this negative correlation between BLIMP-1 and Myc overexpression in ABC-DLBCL was statistically significant when using a higher cutoff (≥20%) for BLIMP-1+ (P = 0.0062).
Table 3.
BLIMP-1 protein expression and its correlation with pathologic and biological biomarkers in DLBCL
| Total | ABC-DLBCL | GCB-DLBCL | Overall DLBCL | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Variables | BLIMP-1+ | BLIMP-1− | P | BLIMP-1+ | BLIMP-1− | P | BLIMP-1+ | BLIMP-1− | P | |
| Patient No. | 520 | n = 92 | n = 158 | n = 39 | n = 229 | n = 132 | n = 388 | |||
| ABC subtype | 518 | - | - | - | - | - | - | 70% (92/131) | 41% (158/387) | <0.0001 |
| Ki-67 ≥ 70% | 516 | 76% (70/92) | 65% (102/158) | 0.066 | 56% (22/39) | 60% (136/225) | 0.72 | 71% (93/132) | 62% (236/384) | 0.092 |
| BCL2 translocation | 417 | 3.8% (3/80) | 4% (5/127) | 1.0 | 22% (8/36) | 35% (60/173) | 0.17 | 9% (11/116) | 22% (65/301) | 0.0043 |
| Bcl-2 ≥ 70% | 514 | 55% (50/91) | 59% (93/157) | 0.60 | 28% (11/39) | 47% (96/225) | 0.11 | 47% (61/131) | 49% (189/383) | 0.61 |
| MYC translocation | 332 | 9% (6/70) | 7% (7/101) | 0.77 | 3% (1/30) | 18% (24/130) | 0.049 | 7% (7/100) | 13% (31/232) | 0.13 |
| Myc ≥ 70% | 513 | 31% (28/91) | 42% (66/158) | 0.085 | 18% (7/39) | 29% (65/223) | 0.15 | 28% (36/131) | 34% (131/382) | 0.15 |
| BCL6 translocation | 362 | 44% (30/69) | 39% (39/101) | 0.53 | 40% (12/31) | 23% (36/159) | 0.06 | 42% (42/101) | 29% (75/261) | 0.019 |
| Bcl-6 ≥ 50% | 510 | 52% (47/90) | 49% (77/157) | 0.69 | 66% (26/39) | 76% (171/224) | 0.23 | 57% (73/129) | 65% (248/381) | 0.092 |
| TP53 mutation | 459 | 13% (10/80) | 21% (30/142) | 0.15 | 16% (6/38) | 30% (59/198) | 0.077 | 14% (16/118) | 26% (89/341) | 0.005 |
| p53 ≥ 20% | 450 | 39% (31/80) | 33% (46/139) | 0.46 | 26% (10/38) | 38% (75/193) | 0.20 | 35% (41/118) | 36% (121/332) | 0.82 |
| Wild type-p53 ≥ 20% | 348 | 36% (25/70) | 20% (22/110) | 0.024 | 28% (9/32) | 26% (35/136) | 0.82 | 33% (34/102) | 23% (57/246) | 0.049 |
| Mutated p53 ≥ 20% | 102 | 60% (6/10) | 83% (24/29) | 0.20 | 17% (1/6) | 70% (40/57) | 0.017 | 44% (7/16) | 74% (64/86) | 0.02 |
| p63 ≥ 10% | 499 | 51% (45/89) | 41% (63/155) | 0.15 | 53% (20/38) | 38% (81/215) | 0.10 | 51% (65/128) | 39% (145/371) | 0.023 |
| MDM2 > 10% | 513 | 55% (50/91) | 35% (24/156) | 0.0018 | 46% (18/39) | 34% (77/226) | 0.15 | 52% (68/131) | 34% (131/382) | 0.0004 |
| p21 > 5% | 450 | 40% (32/80) | 28% (39/139) | 0.074 | 42% (16/38) | 19% (26/193) | 0.0028 | 41% (48/118) | 23% (75/332) | 0.0003 |
| IRF4/MUM1 > 30% | 515 | 84% (76/91) | 73% (115/158) | 0.054 | 35% (14/39) | 22% (51/227) | 0.062 | 69% (90/130) | 43% (166/385) | <0.0001 |
| GCET1 ≥ 50% | 511 | 19% (17/90) | 8% (13/155) | 0.025 | 38% (15/39) | 54% (122/226) | 0.084 | 25% (32/129) | 35% (133/382) | 0.039 |
| CD10 > 30% | 517 | 10% (9/92) | 7% (11/158) | 0.47 | 64% (25/39) | 70% (159/228) | 0.57 | 26% (34/131) | 44% (170/386) | 0.0003 |
| Nuclear p65+ | 467 | 66% (55/84) | 50% (75/149) | 0.028 | 62% (21/34) | 61% (121/198) | 1.0 | 65% (77/119) | 56% (196/348) | 0.13 |
| Nuclear p50 ≥ 20% | 452 | 46% (36/79) | 39% (55/141) | 0.39 | 37% (13/35) | 26% (50/196) | 0.16 | 44% (50/115) | 31% (105/337) | 0.023 |
| pAKT ≥ 30% | 493 | 51% (45/88) | 34% (51/151) | 0.0095 | 51% (19/37) | 37% (79/215) | 0.1 | 51% (64/126) | 35% (130/367) | 0.0003 |
| CD30+ | 516 | 23% (21/92) | 10% (15/156) | 0.0082 | 18% (7/39) | 16% (37/227) | 0.82 | 21% (28/132) | 14% (52/384) | 0.05 |
In the GCB subgroup, BLIMP-1 negativity was associated with MYC translocation (P = 0.049), mutated p53 overexpression, and decreased p21 expression (P = 0.0028) (Table 3). We found several other alterations associated with BLIMP-1 negativity in the overall DBLCL cohort, including BCL2 translocation (P = 0.004), TP53 mutation (P = 0.005), decreased expression of p63 (P = 0.023), nuclear p50 (P = 0.023) and IRF4/MUM1 (P < 0.0001), increased expression of GCET1 (P = 0.039) and CD10 (P = 0.019), and a lack of BCL6 translocation. Bcl-6 expression tended to be upregulated in BLIMP-1− DLBCLs (P = 0.092), especially in patients with mutated TP53 (P = 0.075).
We did not observe correlations between BLIMP-1 expression and survival in the study cohort (Supplementary Figure 4). However, BLIMP-1 negativity had an unfavorable impact on OS and PFS in DLBCL patients with TP53 mutations, although the impact was not significant (OS, P = 0.12; PFS, P = 0.074) (Figure 3c).
Homozygous PRDM1 deletion contributes to gene expression signatures of transcriptional activation
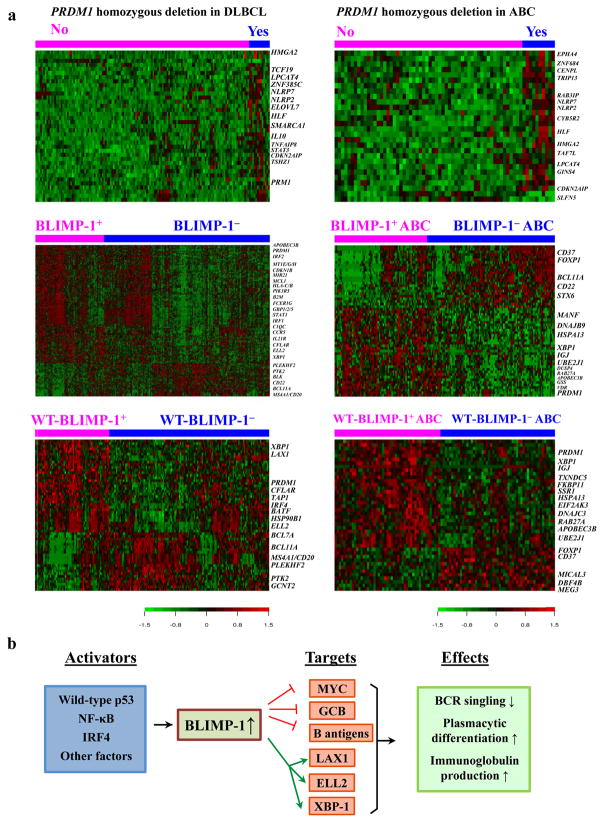
We identified 25 differentially expressed genes (DEGs) between homozygous PRDM1 deletion-positive and -negative DLBCL cases (Figure 4a). Expression of all of the DEGs was upregulated. Subjects with homozygous PRDM1 deletions had robust transcriptional activity with upregulation of transcription factors STAT3, HLF, TCF19, TSHZ1, and HMGA2. Accordingly, analysis of the ABC subgroup also identified increased transcriptional signatures (Table 4). In contrast, we found no DEGs for homozygous PRDM1 deletion in the GCB subgroup.
Figure 4.
Gene expression profiles for the overall and ABC-DLBCL patients and the BLIMP-1 network in DLBCL. (a) Differential expression of genes between patients with and without homozygous PRDM1 deletions, and between patients with and without BLIMP-1 protein expression. Differential expression of genes were only found between wild-type BLIMP-1+ and BLIMP-1− DLBCLs but not between mutated BLIMP-1+ and BLIMP-1− DLBCLs. (b) A brief network of BLIMP-1’s functions and regulations summarizing our results. BLIMP-1 can be activated by p53, IRF4, and NF-κB signaling. BLIMP-1 represses the transcription of MYC, B-cell antigen/surface receptors, and germinal center programs, whereas activates LAX1 which inhibits B-cell receptor (BCR) signaling, therefore leading to attenuated BCR signaling and decreased tumor cell proliferation. BLIMP-1 also transactivates XBP1 and ELL2, which results in activation of plasmacytic differentiation and immunoglobulin production.
Table 4.
Differentially expressed genes between homozygous PRDM1 deletion-positive and -negative DLBCL and between BLIMP-1 protein-negative and -positive DLBCL
| Homozygous PRDM1 deletion-positive vs. - negative DLBCL |
Overall BLIMP-1 protein-negative vs. -positive DLBCL |
Wild-type BLIMP-1 protein-negative vs. -positive DLBCL |
|||||
|---|---|---|---|---|---|---|---|
| Overall DLBCL | ABC-DLBCL | Overall DLBCL | ABC-DLBCL | Overall DLBCL | ABC-DLBCL | ||
| Function category | FDR < 0.1 | FDR < 0.3 | FDR < 0.01 | FDR < 0.05 | FDR < 0.01 | FDR < 0.3 | |
| Cytokine, immune/inflammation related | Up | NLRP7, IL10 | NLRP7 | CNR2 | - | - | - |
| Down | - | - | GZMB, SLAMF7, PRF1, IL2RB, GBP1, GZMA, APOBEC3B, GBP5, GBP2, FAM46C, STAT1, FCER1G, C1QC, IRF1, SRGN, C1orf38, RAB27A, IDO1, CST7, CCR5, IL21R, HLA-C, HLA-B, IRF2, NLRC5, B2M, ZBP1, MGC29506, ZC3HAV1, TAP1 | IGJ, APOBEC3B, RAB27A | PRF1, FAM46C, LANCL2, TAP1 | IGJ, APOBEC3B, RAB27A | |
| Differentiation | Up | DPPA4 | - | BCL11A | BCL11A, FOXP1 | BCL11A | FOXP1 |
| Down | - | SLFN5 | BATF | - | BATF | - | |
| B/T antigen/surface receptor and regulation | Up | - | - | MS4A1, CD22, GCNT2, BLK | CD22, CD37 | MS4A1, GCNT2 | CD37 |
| Down | - | - | TARP /// TRGC2 | - | LAX1 | - | |
| Transcription factor; RNA processing; protein synthesis, folding, and transportation; endoplasmic reticulum stress | Up | STAT3, HLF, TCF19, TSHZ1, HMGA2 | HLF, ZNF684, TAF7L, HMGA2 | ELL3 /// SERINC4, PLEKHF2, TEAD2, DNAJC10 | - | ELL3 /// SERINC4, DNAJC10, PLEKHF2, SLC6A16 | GOLGA8B |
| Down | - | - | PRDM1, ELL2, XBP1, HSP90B1, FKBP11, CRTC3, WARS, RUNX3, SUB1, XRN1, SSR4, DNAJC3, CTSB, VAMP5, NEAT1, SAR1B, DNAJB9, LAP3, ARID3A, LITAF, SEC11C, SSR1, SSR3, ARL4C, RALB, RNF149, MANF, HSPA13, C15orf24, SEC24A, RNF213, EDEM2, UFM1, LRPAP1, SELK | PRDM1, UBE2J1, FKBP11, ZBTB38, XBP1, HERPUD1, DNAJC3, SEC24A, HSPA13, ERLEC1, MANF, EIF2AK3, DNAJB9 | PRDM1, ELL2, IRF4, FKBP11, HSP90B1, XBP1, SSR4, DNAJB9, TRAM2, MANF, DNAJC3 | PRDM1, FKBP11, UBE2J1, UBE2J1, XBP1, MANF, ERLEC1, HSPA13, SSR1, HSPA5, DNAJB9, EIF2AK3, CD2BP2 | |
| Cell cycle, DNA replication, chromatin structure, DNA damage response, proliferation, and signaling | Up | NLRP2, CDKN2AIP, EPHA4, SMARCA1, C10orf2, PRM1 | NLRP2, TRIP13, CDKN2AIP, CENPL, EPHA4, RAB3IP, GINS4 | CLLU1, SLC6A16 | CLLU1, GPR39, STX6 | BCL7A | DBF4B |
| Down | - | - | DUSP4, MIR21, VDR, CDKN1B, SERPINA1, SPRY4, PIM2, GIMAP6, TMEM184B, PIK3R5, GRIN3A, CALM1 | DUSP4, VDR, TRIB1 | - | VDR | |
| Cell death, tumor suppression | Up | TNFAIP8 | - | PAWR | - | - | MEG3 |
| Down | - | - | TXNDC5, CFLAR, TMEM49, MLKL, MCL1, GADD45A, GHITM, ARMCX3 | TXNDC5, GHITM | MLKL, TXNDC5, CFLAR | TXNDC5, TMEM59 | |
| Metabolism, autophagy, oxidative stress | Up | CYB5R2, SLC2A13, ELOVL7, MOCOS, LPCAT4 | CYB5R2, LPCAT4, SLC2A13, TGDS | RIMKLB, GYLTL1B, CCBL1, CALCA | ATP8B1, SLC39A9 | - | ACSM1, SLC39A9 |
| Down | - | - | CHST2, WIPI1, TMEM59, SOD2, TXN, GLUL, ACP2, P4HB, SLC39A8, GLA, SAT1, GALM, GSS, APOL6, GLRX, ACSL1 | GSS, TMEM59 | - | - | |
| Adhesion, motility, cytoskeleton, extracellular matrix | Up | NRCAM, C14orf45 | - | PCDHGA4, KANK1, DAAM1, PTK2, MICAL3 | - | PTK2 | MICAL3 |
| Down | - | - | CADM1, MT2A, MT1H, MT1X, MT1G, MT1E, MT1E/H/M, IQGAP2, ITGAL, TMSB10, FNDC3A, C12orf55 | ITGAL, C12orf55 | - | - | |
| Unknown | Up | ZNF385C, PSORS1C2, LOC4orf45, FAM108C1, hCG_17324 | ZNF385C, BTF3L4, MGC87042, FAM108C1, LOC401312 | SYPL1, C8orf37, LOC151162 /// MGAT5 | FAM129C, SNORA74A, TRIM34 /// TRIM6-TRIM34 | SYPL1 | - |
| Down | - | - | LOC541471 /// NCRNA00152, KIAA1618, NKG7, LOC283922, LOC100289053, KLHDC7B | - | C12orf55, C20orf196 | LOC100129637, FAM122C, C16orf70 | |
BLIMP-1 expression signatures in the overall cohort resemble those in ABC-DLBCL
We compared GEP of BLIMP-1− and BLIMP-1+ DLBCL cases in the overall DLBCL cohort, in wild-type and mutated PRDM1 subsets (Figure 4a). DEGs were identified in the overall DLBCL and the subset with wild-type PRDM1 (147 and 32 DEGs, respectively, with a false discovery rate [FDR] threshold of 0.01), but not in the subset with mutated PRDM1, suggesting loss of wild-type BLIMP-1 function (Table 4). Loss of BLIMP-1 expression in DLBCL was associated with downregulation of PRDM1, X-box binding protein 1 (XBP1), which encodes for a critical regulator of plasma differentiation,28 and its downstream target genes involved in endoplasmic reticulum, protein synthesis and transportation.29, 30 Moreover, expression of genes related to immunoglobulin production (ELL2, MGC29506/MZB1, and ARID3A), cell differentiation (BATF, and IRF4 which transactivates PRDM1 and represses BCL6 expression31), and B/T cell receptor signaling inhibition (LAX1) was also downregulated in BLIMP-1− DLBCLs. In contrast, expression of CD22, MS4A1 which encodes for CD20, BCL11A (upregulated during hematopoietic cell differentiation; a B-cell proto-oncogene and co-factor with Bcl-6), and BLK which is involved in B cell receptor (BCR) signaling was markedly upregulated (1.42-fold, 1.54-fold, 1.67-fold, and 1.2-fold, respectively).
DEGs were also identified in the ABC and GCB subgroups (38 DEGs in ABC with a FDR threshold of 0.05, Table 4; 22 DEGs in GCB with a FDR threshold of 0.15, Supplementary Table 4). When analyzed in the wild-type PRDM1 and mutated PRDM1 subsets separately, DEGs were only identified in ABC-DLBCL (but not GCB-DLBCL) with wild-type PRDM1 (30 DEGs with a FDR threshold of 0.30, Figure 4a and Table 4). The spectrum of DEGs in the ABC subgroup was similar to that in the overall cohort, including downregulation of PRDM1, XBP1, GHITM, DUSP4, TMEM59, APOBEC3B, FKBP11, DNAJC3, DNAJB9, MANF, SEC24A, VDR, TXNDC5, ITGAL, and C12orf55, as well as upregulation of BCL11A, CD22, and CLLU1 in BLIMP-1− ABC-DLBCL cases. IGJ was downregulated in in BLIMP-1− ABC-DLBCL whereas CD37 antigen was upregulated. In comparison, the DEGs identified in the overall GCB-DLBCL (regardless PRDM1 mutation status) also included PRDM1, BCL11A, and DUSP4, but were more likely to be involved in immune responses (CTLA4, GBP5, C1orf38, HLA-F, and SERPINB1) and metabolism (ACSL1, LPCAT2, GTPBP8, CTSA, and METRNL) (Supplementary Figure 5a ).
Considering the potential prognostic impact of BLIMP-1 expression in DLBCL patients with mutated TP53, we additionally compared the BLIMP-1 expression signatures in patients with mutated TP53 and those wild-type for TP53. Forty-three and 33 DEGs were identified respectively in these two subsets with a FDR threshold of 0.20 (Supplementary Figures 5b–c and Supplementary Table 4). Comparing these two BLIMP-1 GEP signatures, we found PRDM1, SSR1, and HSP90B1 were common signatures. Unique BLIMP-1 expression signatures in cases wild-type for TP53 included MS4A1/CD20, LAX1, IGJ, PRF1, DNAJB9, MOV10, LANCL2, CD2BP2, and IRF1. In the TP53-mutated subgroup, STAT3 which inhibits BCL6 was downregulated in BLIMP-1− cases.32 Other unique signatures in DLBCL with mutated TP53 included CFLAR, RBPJ, CMAH, DNAJC13, DLEU2, VAMP5, and many others.
PRDM1 mutations within exon 1/2 but not mutations in exons 3–7 showed gene expression signatures
We further compared GEP between mutated PRDM1 and wild-type PRDM1 DLBCLs regardless of BLIMP-1 expression. In the overall DLBCL and GCB subset, 103 and 193 DEGs were identified with FDR thresholds of 0.10 and 0.05 respectively. No DEGs were identified in the ABC subset, however, probably due to heterogeneity. Similarly, GEP comparisons between cases with mutated PRDM1 exon 1/2 and those with wild-type PRDM1 only identified DEGs in the overall DLBCL (Supplementary Figure 5d and Supplementary Table 5) and GCB (156 DEGs with a FDR threshold of 0.01) but not ABC subset. Interestingly, tumor suppressors PTEN and IKZF1 (a transcriptional regulator of lymphocyte differentiation) were significantly downregulated in cases with mutated PRDM1 exon 1/2. In contrast, no significant genes were found differentially expressed between cases with mutated PRDM1 exons 3–7 and those with wild-type PRDM1.
DISCUSSION
In this study, we analyzed the clinical and experimental data on 520 de novo DLBCL cases to determine the tumor-suppressive function of PRDM1/BLIMP-1. Previous studies demonstrated that loss of PRDM1/BLIMP1 function is critical for the pathogenesis of ABC-DLBCL.8, 11 Herein we provide evidence that loss of PRDM1/BLIMP1 function is a factor for poor prognosis for ABC-DLBCL. We found homozygous PRDM1 deletions and PRDM1 mutations within PRDM1 exons 1 and 2 were poor prognostic factors in patients with ABC-DLBCL. Loss of BLIMP-1 protein expression was common in ABC-DLBCL and associated with a decreased plasma-cell differentiation signature and upregulation of B-cell antigens.
In univariate and multivariate analyses, we found that the prognosis for DLBCL with homozygous PRDM1 deletion was worse than that for DLBCL without this deletion. However, this homozygous deletion only had prognostic significance in the ABC subgroup, as its impact on prognosis for GCB-DLBCLs was minimal. Although the number of patients harboring homozygous PRDM1 deletions was small, our GEP still supported reduced transcriptional repression in patients with these deletions as suggested by upregulation of transcription factors, including ZNF385C, HLF, and HMGA2. Notably, we only observed these gene expression signatures in the ABC group.
Similarly, PRDM1 mutations only influenced survival of DLBCL in the ABC subgroup. Our data further support this by demonstrating that PRDM1 mutations within exons 1 and 2 still significantly affect survival of ABC- but not GCB-DLBCLs. Therefore, we suggest that intact BLIMP-1 function is critical for the repression of ABC-DLBCLs. Exons 1 and 2 encode for the first 97 amino acids of BLIMP-1, including the N-terminal acidic domain. An intact N-terminal acidic domain is critical for the normal transcriptional repression function of BLIMP-1.33 Researchers showed that BLIMP-1 lacking this domain do not repress the MYC promoter.34 Interestingly, all but two of the PRDM1 mutations identified in our cohort were missense mutations. Mandelbaum et al proved that a subset of missense mutations of PRDM1 can directly impair BLIMP-1’s protein stability as well as its transcriptional repression function, including 2 missense mutations at P48 (corresponding to 84P in this article due to different transcripts), the most frequently mutated site within exons 1 and 2 in our ABC-DLBCLs.11 Although the decreased BLIMP-1 protein expression in patients with PRDM1 mutations within exons 1 and 2 was not significant, we found that mutations in exons 1 and 2 were closely correlated with Myc and Bcl-2 co-expression, a previously identified marker for poor DLBCL prognosis that was overrepresented in the ABC subgroup.18 GEP analysis results reinforced the notion that BLIMP-1 mutations especially those in exons 1–2 had lost functions of wild-type BLIMP-1.
At the protein level, although BLIMP-1 expression was frequently associated with the ABC phenotype of DLBCL, 63% of the ABC-DLBCL patients lacked BLIMP-1 protein expression. Over half of these BLIMP-1− ABC-DLBCL patients had no apparent genetic changes in PRDM1. In vitro studies have demonstrated that BLIMP-1 can be inactivated by constitutively active Bcl-6.9, 11 We found a negative correlation between Bcl-6 and BLIMP-1 expression with borderline significance. Also, expression of BCL11A, a co-factor with Bcl-6, was upregulated in our GEP analysis of both ABC and GCB subtypes of BLIMP-1− DLBCLs.35 Additionally, PRDM1 can be epigenetically regulated by diverse miRNAs including miR-23, miR-9, and let-7a. 23–27 Using miRNA profiling, we found upregulation of miR-30d and miR-30b-5p in BLIMP-1− cases without genetic aberrations compared to BLIMP1+ cases. Further functional evidence are needed to elucidate the regulation between these miRNAs and BLIMP-1 expression.
ABC-DLBCL patients without expression of BLIMP-1 had decreased expression of the tumor suppressor protein p53 and its downstream targets MDM2 and p21 as well as increased expression of Myc which promotes cell proliferation.36 Therefore, in BLIMP-1− DLBCL cases, decreased p53 signaling and Myc overexpression may synergistically promote tumor progression. In addition, loss of BLIMP-1 expression was associated with dysregulation of NF-κB pathway molecules. Induction of PRDM1 mRNA expression in B-cell lymphoma cell lines can be blocked by NF-κB inhibitors.37 Recently, Heise et al. proved that NF-κB subunit p65 (RELA) is required for the induction of BLIMP-1 expression, which is consistent with the positive association of p65 expression and BLIMP-1+ in our ABC DLBCL cohort.38
Surprisingly, BLIMP-1 protein expression was not a survival predictor in the present study. Gyory et al. found that the alternative splicing protein BLIMP-1β, which lacks the first three exons in the normal BLIMP-1 protein, does not have transcriptional repression activity.33 Consistently, detection of BLIMP-1β mRNA expression in tumor cells has been associated with poor prognosis for ABC-DLBCL.39 Using our immunohistochemical assay, we could not discriminate between the normal BLIMP-1 protein and BLIMP-1β. The presence of BLIMP-1β in our cohort may reduce the prognostic impact of normal BLIMP-1 by diminishing its ability to repress the activity of downstream targets such as Myc and Bcl-6.
However, lack of BLIMP-1 expression seemed to be an unfavorable prognostic factor for DLBCL in patients with aberrant p53 statuses. Kusam et al. reported that Bcl-6 can immortalize primary B cells and greatly increase B-cell function only in the absence of normal p53 function.40 BLIMP-1 is a known transcriptional repressor of BCL6.41 We found that BLIMP-1 positivity was closely associated with absence of Bcl-6 protein expression in our group of DLBCL patients with mutated TP53. Therefore, BLIMP-1 may inhibit the proliferation of tumor cells harboring mutated TP53 by repressing Bcl-6 activity. Consistently, GEP analysis in DLBCL patients with mutated TP53 found that expression of IRF4 and STAT3, activators of BLIMP-1 as well as inhibitors of BCL6, 31, 32 was downregulated in BLIMP-1− subgroup.
The BLIMP-1 negativity signature in the overall cohort of DLBCL patients was similar to that in the ABC subgroup: both were characterized by downregulation of plasmacytic differentiation and upregulation of B cell antigens. BLIMP-1’s role in promoting plasmacytic differentiation27 of DLBCL was indicated by downregulation of XBP1 along with several of its downstream targets involved in protein synthesis in endoplasmic reticulum and transportation in BLIMP-1− cases.42 XBP1 is a transcriptional activator that acts downstream of BLIMP-1. BLIMP-1 and XBP1 are jointly required for the establishment of terminally differentiated plasma cells.28 Moreover, we found that expression of genes involved in immunoglobulin production (ELL2, MZB1, and ARID3A) and cell differentiation (BATF) was downregulated in the BLIMP-1− group. On the other hand, loss of BLIMP-1 expression in DLBCL correlated with upregulation of CD22 and BLK and downregulation of LAX1 that have important roles in regulating BCR signaling. Importantly, expression of MS4A1 (encoding CD20) was also upregulated in BLIMP-1− cases, possibly indicating an increased therapeutic effect of rituximab in patients with BLIMP-1− DLBCL as suggested previously.43, 44 In line with the role of BLIMP-1 in plasmacytic differentiation suggested by GEP analysis, the cell-of-origin analysis based on B-cell-associated gene signatures22 also showed positive correlation between BLIMP-1 expression and the plasmablastic subtype of DLBCL (Table 1).
The anti-proliferative effect of BLIMP-1 on DLBCL was also suggested by the GEP results as cell-cycle suppressors (e.g., CDKN1B, GADD45A, GHITM, DUSP4) were upregulated in BLIMP-1+ DLBCLs. Also, in vitro experiments have demonstrated that B-cell proliferation in DLBCL cell lines was promoted upon acute ablation of BLIMP-1 and that BLIMP-1 inactivation led to B-cell lymphomagenesis and shortening of life spans in mouse models of DLBCL.8, 11 Reintroduction of BLIMP-1 into a DLBCL cell line likely represses the cells’ proliferation by turning off MYC and other genes involved in cell-cycle progression and DNA repair.45 A brief network of BLIMP-1’s functions and regulation generated from our results is presented in Figure 4b.
Collectively, the results of the present study demonstrated that loss of PRDM1/BLIMP-1 function contributed to the overall poor prognosis for ABC-DLBCL in three ways. First, genetic PRDM1 aberrations (including homozygous deletions and mutations) affected the prognosis for ABC- but not GCB-DLBCLs. Second, a lack of BLIMP-1 expression correlated with an impaired p53 signaling pathway and Myc overexpression in ABC-DLBCLs. Third, the GEP results for the overall DLBCL patient cohort resembled those for the ABC subgroup, both indicating downregulation of plasmacytic differentiation of DLBCLs and upregulation of B antigen and BCR signaling in BLIMP-1− tumors. These data may suggest that inactivated BLIMP-1 facilitates DLBCL progression through Myc and BCR signaling which are essential for survival of ABC-DLBCL.18, 46 Therapeutic approaches that restore the normal function of BLIMP-1 may help drive terminal differentiation of tumor cells and overcome the chemoresistance of ABC-DLBCL.
Supplementary Material
Acknowledgments
Authors appreciate critical discussion with Dr. Riccardo Dalla-Favera. This study was supported by grants from the National Institutes of Health/National Cancer Institute (R01CA138688 and 1RC1CA146299 to KHY). Y.X. is a recipient of a hematology and oncology scholarship award. AT was supported by the Krebsliga Beider Basel and Stiftung zur Krebsbekaempfung Zurich. KHY was supported by The University of Texas MD Anderson Cancer Center Lymphoma Moon Shot Program, the Institutional Research and Development Fund, an MD Anderson Institutional Research Grant, an MD Anderson Lymphoma Specialized Program of Research Excellence Research Development Program Award, an MD Anderson Myeloma Specialized Program of Research Excellence Research Development Program Award, and a Gundersen Medical Foundation award. Also, the study was partially supported by grants from the National Institutes of Health/National Cancer Institute (P50CA136411 and P50CA142509) and by the National Institutes of Health/National Cancer Institute under award number P30CA016672. K.H.Y. receives research support from Roche Molecular Systems, Gilead Sciences, Seattle Genetics, Daiichi Sankyo, Adaptive Biotechnologies, Incyte, and HTG Molecular Diagnostics.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.Lenz G, Staudt LM. Aggressive lymphomas. N Engl J Med. 2010;362(15):1417–1429. doi: 10.1056/NEJMra0807082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeVita VT, Jr, Canellos GP, Chabner B, Schein P, Hubbard SP, Young RC. Advanced diffuse histiocytic lymphoma, a potentially curable disease. Lancet. 1975;1(7901):248–250. doi: 10.1016/s0140-6736(75)91142-3. [DOI] [PubMed] [Google Scholar]
- 3.Coiffier B, Lepage E, Briere J, Herbrecht R, Tilly H, Bouabdallah R, et al. CHOP chemotherapy plus rituximab compared with CHOP alone in elderly patients with diffuse large-B-cell lymphoma. N Engl J Med. 2002;346(4):235–242. doi: 10.1056/NEJMoa011795. [DOI] [PubMed] [Google Scholar]
- 4.Coiffier B, Thieblemont C, Van Den Neste E, Lepeu G, Plantier I, Castaigne S, et al. Long-term outcome of patients in the LNH-98. 5 trial, the first randomized study comparing rituximab-CHOP to standard CHOP chemotherapy in DLBCL patients: a study by the Groupe d’Etudes des Lymphomes de l’Adulte. Blood. 2010;116(12):2040–2045. doi: 10.1182/blood-2010-03-276246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedberg JW. Relapsed/refractory diffuse large B-cell lymphoma. Hematology Am Soc Hematol Educ Program. 2011;2011:498–505. doi: 10.1182/asheducation-2011.1.498. [DOI] [PubMed] [Google Scholar]
- 6.Bea S, Zettl A, Wright G, Salaverria I, Jehn P, Moreno V, et al. Diffuse large B-cell lymphoma subgroups have distinct genetic profiles that influence tumor biology and improve gene-expression-based survival prediction. Blood. 2005;106(9):3183–3190. doi: 10.1182/blood-2005-04-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 8.Calado DP, Zhang B, Srinivasan L, Sasaki Y, Seagal J, Unitt C, et al. Constitutive canonical NF-kappaB activation cooperates with disruption of BLIMP1 in the pathogenesis of activated B cell-like diffuse large cell lymphoma. Cancer cell. 2010;18(6):580–589. doi: 10.1016/j.ccr.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crotty S, Johnston RJ, Schoenberger SP. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat Immunol. 2010;11(2):114–120. doi: 10.1038/ni.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19(4):607–620. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 11.Mandelbaum J, Bhagat G, Tang H, Mo T, Brahmachary M, Shen Q, et al. BLIMP1 is a tumor suppressor gene frequently disrupted in activated B cell-like diffuse large B cell lymphoma. Cancer cell. 2010;18(6):568–579. doi: 10.1016/j.ccr.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tam W, Gomez M, Chadburn A, Lee JW, Chan WC, Knowles DM. Mutational analysis of PRDM1 indicates a tumor-suppressor role in diffuse large B-cell lymphomas. Blood. 2006;107(10):4090–4100. doi: 10.1182/blood-2005-09-3778. [DOI] [PubMed] [Google Scholar]
- 13.Pasqualucci L, Compagno M, Houldsworth J, Monti S, Grunn A, Nandula SV, et al. Inactivation of the PRDM1/BLIMP1 gene in diffuse large B cell lymphoma. J Exp Med. 2006;203(2):311–317. doi: 10.1084/jem.20052204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tate G, Hirayama-Ohashi Y, Kishimoto K, Mitsuya T. Novel BLIMP1/PRDM1 gene mutations in B-cell lymphoma. Cancer Genet Cytogenet. 2007;172(2):151–153. doi: 10.1016/j.cancergencyto.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Visco C, Li Y, Xu-Monette ZY, Miranda RN, Green TM, Li Y, et al. Comprehensive gene expression profiling and immunohistochemical studies support application of immunophenotypic algorithm for molecular subtype classification in diffuse large B-cell lymphoma: a report from the International DLBCL Rituximab-CHOP Consortium Program Study. Leukemia. 2012;26(9):2103–2113. doi: 10.1038/leu.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tzankov A, Xu-Monette ZY, Gerhard M, Visco C, Dirnhofer S, Gisin N, et al. Rearrangements of MYC gene facilitate risk stratification in diffuse large B-cell lymphoma patients treated with rituximab-CHOP. Mod Pathol. 2014;27(7):958–971. doi: 10.1038/modpathol.2013.214. [DOI] [PubMed] [Google Scholar]
- 17.Ok CY, Chen J, Xu-Monette ZY, Tzankov A, Manyam GC, Li L, et al. Clinical implications of phosphorylated STAT3 expression in De Novo diffuse large B-cell lymphoma. Clin Cancer Res. 2014;20(19):5113–5123. doi: 10.1158/1078-0432.CCR-14-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu S, Xu-Monette ZY, Tzankov A, Green T, Wu L, Balasubramanyam A, et al. MYC/BCL2 protein coexpression contributes to the inferior survival of activated B-cell subtype of diffuse large B-cell lymphoma and demonstrates high-risk gene expression signatures: a report from The International DLBCL Rituximab-CHOP Consortium Program. Blood. 2013;121(20):4021–4031. doi: 10.1182/blood-2012-10-460063. quiz 4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu-Monette ZY, Moller MB, Tzankov A, Montes-Moreno S, Hu W, Manyam GC, et al. MDM2 phenotypic and genotypic profiling, respective to TP53 genetic status, in diffuse large B-cell lymphoma patients treated with rituximab-CHOP immunochemotherapy: a report from the International DLBCL Rituximab-CHOP Consortium Program. Blood. 2013;122(15):2630–2640. doi: 10.1182/blood-2012-12-473702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Visco C, Tzankov A, Xu-Monette ZY, Miranda RN, Tai YC, Li Y, et al. Patients with diffuse large B-cell lymphoma of germinal center origin with BCL2 translocations have poor outcome, irrespective of MYC status: a report from an International DLBCL rituximab-CHOP Consortium Program Study. Haematologica. 2013;98(2):255–263. doi: 10.3324/haematol.2012.066209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu-Monette ZY, Wu L, Visco C, Tai YC, Tzankov A, Liu WM, et al. Mutational profile and prognostic significance of TP53 in diffuse large B-cell lymphoma patients treated with R-CHOP: report from an International DLBCL Rituximab-CHOP Consortium Program Study. Blood. 2012;120(19):3986–3996. doi: 10.1182/blood-2012-05-433334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dybkaer K, Bogsted M, Falgreen S, Bodker JS, Kjeldsen MK, Schmitz A, et al. Diffuse large B-cell lymphoma classification system that associates normal B-cell subset phenotypes with prognosis. J Clin Oncol. 2015;33(12):1379–1388. doi: 10.1200/JCO.2014.57.7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin R, Chen L, Chen G, Hu C, Jiang S, Sevilla J, et al. Targeting miR-23a in CD8+ cytotoxic T lymphocytes prevents tumor-dependent immunosuppression. J Clin Invest. 2014;124(12):5352–5367. doi: 10.1172/JCI76561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White CA, Pone EJ, Lam T, Tat C, Hayama KL, Li G, et al. Histone deacetylase inhibitors upregulate B cell microRNAs that silence AID and Blimp-1 expression for epigenetic modulation of antibody and autoantibody responses. J Immunol. 2014;193(12):5933–5950. doi: 10.4049/jimmunol.1401702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang L, Nong L, Zhang S, Zhao J, Ti H, Dong Y, et al. The downregulation of PRDM1/Blimp-1 is associated with aberrant expression of miR-223 in extranodal NK/T-cell lymphoma, nasal type. J Exp Clin Cancer Res. 2014;33:7. doi: 10.1186/1756-9966-33-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin J, Lwin T, Zhao JJ, Tam W, Choi YS, Moscinski LC, et al. Follicular dendritic cell-induced microRNA-mediated upregulation of PRDM1 and downregulation of BCL-6 in non-Hodgkin’s B-cell lymphomas. Leukemia. 2011;25(1):145–152. doi: 10.1038/leu.2010.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nie K, Zhang T, Allawi H, Gomez M, Liu Y, Chadburn A, et al. Epigenetic down-regulation of the tumor suppressor gene PRDM1/Blimp-1 in diffuse large B cell lymphomas: a potential role of the microRNA let-7. Am J Pathol. 2010;177(3):1470–1479. doi: 10.2353/ajpath.2010.091291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8(1):22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- 29.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415(6867):92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 30.Namba T, Chu K, Kodama R, Byun S, Yoon KW, Hiraki M, et al. Loss of p53 enhances the function of the endoplasmic reticulum through activation of the IRE1alpha/XBP1 pathway. Oncotarget. 2015;6(24):19990–20001. doi: 10.18632/oncotarget.4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saito M, Gao J, Basso K, Kitagawa Y, Smith PM, Bhagat G, et al. A signaling pathway mediating downregulation of BCL6 in germinal center B cells is blocked by BCL6 gene alterations in B cell lymphoma. Cancer cell. 2007;12(3):280–292. doi: 10.1016/j.ccr.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 32.Ding BB, Yu JJ, Yu RY, Mendez LM, Shaknovich R, Zhang Y, et al. Constitutively activated STAT3 promotes cell proliferation and survival in the activated B-cell subtype of diffuse large B-cell lymphomas. Blood. 2008;111(3):1515–1523. doi: 10.1182/blood-2007-04-087734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gyory I, Fejer G, Ghosh N, Seto E, Wright KL. Identification of a functionally impaired positive regulatory domain I binding factor 1 transcription repressor in myeloma cell lines. J Immunol. 2003;170(6):3125–3133. doi: 10.4049/jimmunol.170.6.3125. [DOI] [PubMed] [Google Scholar]
- 34.Yu J, Angelin-Duclos C, Greenwood J, Liao J, Calame K. Transcriptional repression by blimp-1 (PRDI-BF1) involves recruitment of histone deacetylase. Mol Cell Biol. 2000;20(7):2592–2603. doi: 10.1128/mcb.20.7.2592-2603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu H, Ippolito GC, Wall JK, Niu T, Probst L, Lee BS, et al. Functional studies of BCL11A: characterization of the conserved BCL11A-XL splice variant and its interaction with BCL6 in nuclear paraspeckles of germinal center B cells. Mol Cancer. 2006;5:18. doi: 10.1186/1476-4598-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gustafson WC, Weiss WA. Myc proteins as therapeutic targets. Oncogene. 2010;29(9):1249–1259. doi: 10.1038/onc.2009.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson K, Shapiro-Shelef M, Tunyaplin C, Calame K. Regulatory events in early and late B-cell differentiation. Mol Immunol. 2005;42(7):749–761. doi: 10.1016/j.molimm.2004.06.039. [DOI] [PubMed] [Google Scholar]
- 38.Heise N, De Silva NS, Silva K, Carette A, Simonetti G, Pasparakis M, et al. Germinal center B cell maintenance and differentiation are controlled by distinct NF-kappaB transcription factor subunits. J Exp Med. 2014;211(10):2103–2118. doi: 10.1084/jem.20132613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu YY, Leboeuf C, Shi JY, Li JM, Wang L, Shen Y, et al. Rituximab plus CHOP (R-CHOP) overcomes PRDM1-associated resistance to chemotherapy in patients with diffuse large B-cell lymphoma. Blood. 2007;110(1):339–344. doi: 10.1182/blood-2006-09-049189. [DOI] [PubMed] [Google Scholar]
- 40.Kusam S, Vasanwala FH, Dent AL. Transcriptional repressor BCL-6 immortalizes germinal center-like B cells in the absence of p53 function. Oncogene. 2004;23(3):839–844. doi: 10.1038/sj.onc.1207065. [DOI] [PubMed] [Google Scholar]
- 41.Martins G, Calame K. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu Rev Immunol. 2008;26:133–169. doi: 10.1146/annurev.immunol.26.021607.090241. [DOI] [PubMed] [Google Scholar]
- 42.Wright G, Tan B, Rosenwald A, Hurt EH, Wiestner A, Staudt LM. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci U S A. 2003;100(17):9991–9996. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Montes-Moreno S, Gonzalez-Medina AR, Rodriguez-Pinilla SM, Maestre L, Sanchez-Verde L, Roncador G, et al. Aggressive large B-cell lymphoma with plasma cell differentiation: immunohistochemical characterization of plasmablastic lymphoma and diffuse large B-cell lymphoma with partial plasmablastic phenotype. Haematologica. 2010;95(8):1342–1349. doi: 10.3324/haematol.2009.016113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garcia JF, Roncador G, Garcia JF, Sanz AI, Maestre L, Lucas E, et al. PRDM1/BLIMP-1 expression in multiple B and T-cell lymphoma. Haematologica. 2006;91(4):467–474. [PubMed] [Google Scholar]
- 45.Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17(1):51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- 46.Davis RE, Ngo VN, Lenz G, Tolar P, Young RM, Romesser PB, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463(7277):88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.