Abstract
MicroRNAs (miRNAs) are small non-coding RNA molecules, which function in RNA silencing and post-transcriptional regulation of gene expression. Psoriasis is an inflammatory skin disease characterized by the dysfunction of keratinocytes, with the immune dysregulation. We reviewed the recent studies on the roles of miRNAs in psoriasis and showed that miRNAs play key roles in psoriasis, including the regulation of hyperproliferation, cytokine and chemokine production in keratinocyte, as well as mediating immune dysfunction in psoriasis. Furthermore, miRNAs, particularly, circulating miRNAs may serve as novel biomarkers for diagnosis, monitoring therapy response and reflecting the disease severity. Thus, targeting specific miRNAs may be used to develop new therapeutic methods for psoriasis.
Keywords: biomarkers, immunological mechanisms, miRNA, psoriasis
1 INTRODUCTION
MicroRNAs (miRNAs) are a class of small non-coding RNAs as critical post-transcriptional regulators, and they are endogenously short, 21–23 nucleotides on average, single-stranded and high sequence conservation RNAs. More than 2500 have been identified in humans from the microRNA database 2014, and miRNA comprise the most abundant class of regulators in the human genome. Base-pairing at position 2–8 nucleotides relative to the 5′ end of small RNA termed the “seed” region, which is important for target recognition.[1] miRNA can bind to the complementary sequences in the 3′ untranslated regions (UTR) of targeting genes, resulting in inhibiting their translation into protein, suppressing the expression of protein-coding genes and causing accelerated turnover or transcript degradation of the mRNA transcript (Figure 1).[2,3] Currently, the most widely used methods to profile miRNAs are microarray and 384 wells RT-PCR with high throughput advantage, while Northern blotting and single qRT-PCR are used to verify and confirm miRNA with high sensitivity (Table 1). The miRNA-mediated regulation is often detectable on the protein level, in some cases even undetectable at the mRNA level.[2,4] However, the changes in protein level can be due to miRNA even though there is no detectable change in mRNA. Interestingly, a unique miRNA can regulate hundreds of proteins, and the expression of a specific protein may be controlled by several miRNA.[5] Furthermore, miRNA have been implicated in immune response and cell proliferation, differentiation and apoptosis.[6–8] Accordingly, miRNA regulate a broad range of biological processes, and they are potentially important in the pathogenesis of the immunological disorders like psoriasis.
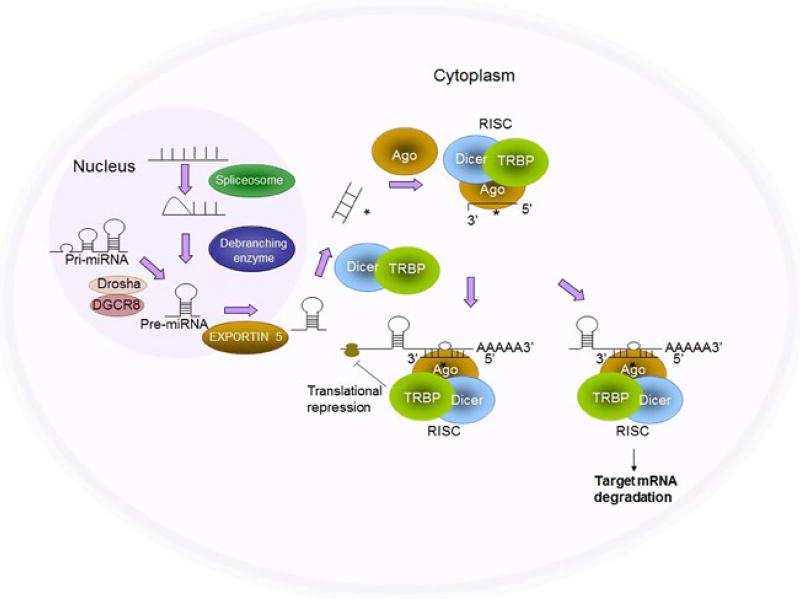
FIGURE 1.
Biogenesis of miRNAs. RNA polymerase II transcribes miRNA genes, generating long primary transcripts (pri-miRNAs). In the nucleus, the RNase III-type enzyme Drosha processes the long primary transcripts (pri-miRNAs), yielding hairpin precursors (pre-miRNA). Exportin 5 transports the pre-miRNAs from the nucleus into the cytoplasm. The pre-miRNA hairpins are further processed by Dicer into mature miRNAs, which are incorporated into the Argonaute (Ago) protein and form the RNA-induced silencing complex (RISC) together with Dicer. Once incorporated into the RISC, the miRNAs then guide the RISC to the target genes and repress target gene expression by destabilizing the target mRNAs or suppressing protein translation (Refer to our paper [50])
TABLE 1.
Current methods used to profile miRNA
| Methods | Pros | Cons |
|---|---|---|
| Northern blotting | Verify and confirm miRNA | Unable to perform high throughput detection |
| High specificity | ||
| Low sensitivity | ||
|
| ||
| qRT-PCR | Quantitative analysis of low miRNA expression | Unable to perform high throughput detection |
| Verify and confirm miRNA | Unable to identify new miRNA | |
| High sensitivity and dynamic range | ||
|
| ||
| Microarray | High throughput | Low quality, stability and repeatability |
| Non-quantitative | ||
| Low specificity | ||
| Unable to identify new miRNA | ||
|
| ||
| Small RNA sequencing | High specificity Identify new miRNA | Complex computation analysis |
| Inefficient and high expense | ||
As the most prevalent skin disease in adults, psoriasis affects 1%–3% of the population worldwide. The most common type of psoriasis, psoriasis vulgaris, appears as erythematous plaques clinically. Immunologically, psoriasis is characterized by intense proliferation and abnormal differentiation of keratinocytes as well as infiltration of lymphocytes and neutrophils, in which T cells, dendritic cells and inflammatory cytokines, including TNF-α and IL-17 act as key players.[8–11] However, antibodies, infliximab and etanercept, have been in clinical use and have dramatic effects on psoriasis.[8,12,13] However, psoriasis is recognized as a disease also occurred with a genetic susceptibility and an environmental trigger.[2,8,14] Recently, epigenetics has attracted attention for its involvement in the pathogenesis of psoriasis, and the role of miRNA in psoriasis is gradually highlighted.[8,15] miRNAs might play a major role in the regulation of hyperproliferation, abnormal differentiation of keratinocytes and abnormal immune activation in psoriasis. Herein, we collected the immunological functions of miRNA played in psoriasis, and their potential to act as biomarkers in the diagnosis and treatment of psoriasis, in order to reveal and summarize the roles of miRNA in psoriasis.
2 IMMUNOLOGICAL FUNCTION OF MIRNA IN PSORIASIS
From the miRNA profiling studies using full-depth psoriatic skin biopsies,[2,16–18] about a hundred of miRNAs were differentially expressed in psoriatic skin compared with healthy skin, and most highly changed miRNAs were consistent with the elevated inflammation and hyperproliferation. Recently, more disease-related microRNAs in psoriatic lesions were reported.[19] and miRNA expression patterns can distinguish psoriasis from healthy skin and atopic eczema, in particular miR-21, miR-125b, miR-146a and miR-203.[2,20–24] However, the regulatory networks and relevant mechanisms are far from being understood. The following results reported currently may reveal a new layer of regulatory mechanisms in psoriasis.
2.1 miRNA and potential targets interactions in psoriasis
Initially, after miRNA genes are transcribed into long primary transcripts, one strand of the pair miRNA duplex associates with an Argonaute (Ago) protein to form a functional silencing complex miRISC, which is guided by mature miRNA to target mRNA. miRNA can trigger Ago-catalysed endonucleolytic cleavage of target mRNA, leading to the translational repression or mRNA decay.[25] Moreover, Prolyl 4-hydroxylation appears to play an important role in this “miRNA-targets interaction” by regulating the Ago2 stability.[26] To date, many miRNA and their targeting mRNA or protein interactions have been revealed. The transcription of miRNA miR-31 can be induces NF-κB activation triggered by inflammatory cytokines. Furthermore, a negative regulator of G1 to S phase progression protein phosphatase 6 (ppp6c) is targeted by miR-31.[27] miR-26b-5p was upregulated in subcutaneous adipose tissue underneath lesional psoriasis skin, which targets and downregulates NCEH1 (neutral cholesterol ester hydrolase 1).[28] Keratinocyte-expressed miR-203 targets the Stat3 inhibitor SOCS-3 and induces cell cycle exit by targeting p63,[2,27], whereas JunB is a positive regulator of miR-203.[2] Although not in psoriasis, miR-21 was positively regulated by Stat3,[28,29] and SHIP2 was targeted by miR-205 in apoptosis.[2,30] miR-143 was predicted to target SLC26A4 and miR-223 with GLUL, SMAD3.[31,32] Based on the miRNA–mRNA or miRNA–protein target interactions (as indicated in Table S1 and Table S2 with *), miRNAs are potential regulators in the cell signalling pathways. However, more experimental detections and computer deduction methods may be involved to confirm these interactions.[33]
2.2 miRNA regulate keratinocytes proliferation and inflammation in psoriasis
Psoriasis is characterized by intense hyperproliferation and aberrant differentiation of keratinocytes, accompanied by their production of inflammatory cytokines and chemokines,[34,35] in which a specific miRNA expression profile was shown to be involved. On the one side, miRNA modulate keratinocytes proliferation via MAPK1, FGFR2 and SOCS-3.[8,36] miR-424 functioned on inhibiting MAPK1 or cyclin E1,[8] while miR-125b mediated keratinocyte proliferation by targeting FGFR2.[21] The keratinocyte-expressed miR-203 targeted Stat inhibitor SOCS-3, resulting in hyperproliferation.[37] Besides, miR-21, miR-205, miR-221 and miR-222 are involved in modulating cell growth and apoptosis.[2] miR-136 was stated as a regulatory element during TGF-β1-induced proliferation arrest by targeting PPP2R2A in keratinocytes.[40] The increased ib-miR135b-transfected cells may induce EGF, and the inhibition of miR135b may increase the proliferative potential and improve the microenvironment of basal cells.[41] miR-4516 targeted STAT3 protein by binding to its 3′UTR and down-regulating it in HaCaT keratinocytes.[42] As some of the targeting molecules of miRNA, just like MAPK1 and cyclin E1, may belong to the same signalling pathway for cell cycle regulation,[8,38] these miRNA–mRNA interactions may regulate cell proliferation more effectively than inhibiting these molecules individually. While some other miRNA, like miR-125, express in all layers of epidermis, indicating that the roles of these miRNA in skin may be mediated by different target gene sets in basal and suprabasal cell layers.[21] Investigation of these regulatory mechanisms may lead to new disease activity markers and therapeutic methods. For limitation, except the validated miRNA–targets interactions (indicated in Table S1 and S2 with *), most miRNAs did not determine their direct interaction yet between miRNA and their targeting mRNA by luciferase reporter assay, and further studies are needed to clarify this point.[8]
On the other side, miRNAs also modulate the cytokine and chemokine productions from keratinocytes. The epidermis functions as not only a physical barrier but also a chemical and immunologic barrier, because keratinocytes produce many inflammatory mediators, including cytokines, chemokines and antimicrobial peptides.[39,40] As miR-31 was one of the most highly overexpressed miRNA in psoriasis, it is found miR-31 in psoriasis keratinocytes could modulate the production of IL-1β, CXCL1/5/8 and IL-8 via targeting STK40, a negative regulator of the NF-kB, while TGF-β1 was a regulator of miR-31 in vitro and in vivo.[41] Hence, inflammatory cytokine/chemokines are induced by miRNA, which contributes to endothelial cell activation, leukocyte attraction and clinically, skin inflammation.
2.3 miRNAs modulate T-cell-mediated immune dysfunction
Psoriasis is a chronic inflammatory skin disease and mediated by T cell,[42,43] and miRNAs are revealed to be involved in regulating immune homeostasis and inflammation.[24,42] On the one side, miRNAs modulate the activation and proliferation of T cells. Evidences suggest a T-cell pathogenesis for psoriasis, and murine psoriasis-like disorder was successfully induced by naive CD4+ T cells.[44] Fu et al. found that the decreased expression of miR-138 in CD4+T cells from patients with psoriasis induced increased expression of runt-related transcription factor 3 (RUNX3). And the inhibition of miR-138 increases RUNX3 expression, as well as increased the ratio of Th1/Th2.[50] miR-223 were expressed in Th17 cells implied that miR-223 may play a role in the increased IL-17 production in the Th17 cells.[19] miR-125b was decreased during activation of naive CD4+ T cells.[51] From our studies, we found that lack of miR-223 significantly increased Langerhans cell (LC)-mediated antigen-specific CD8+ T-cell proliferation, while LC from KO and WT mice showed comparable stimulation for antigen-specific CD4+ T cells,[45], and miR-17-92 cluster was highly expressed in epidermal LC.[46] Treg cells are essential for limiting chronic inflammatory in psoriasis,[42,47] while FOXP3 is required for Treg cell development and function.[48] Overexpression of miR-210 could inhibit its target FOXP3 and impaired Treg in psoriasis, while inhibiting miR-210 reversed this immune dysfunction.[42] Our review showed that ablation of miRNAs in Treg cells completely phenocopied the loss of FOXP3 cells, which clearly indicated that multiple immunosuppressive mechanisms used by Treg cells were ultimately controlled by miRNAs.[49,50] Not only the T-cell function, but also the T-cell growth and apoptosis can be modulated by miRNA. miR-142-3p/-5p were upregulated during antigen-induced T-cell proliferation in mice,[2] whereas miR-21 suppressed T-cell apoptosis in psoriasis.[20,51] Refers to the iNKT cells, our data indicated that miRNAs were potent regulators of iNKT cell development, function and homeostasis.[52] Although we found that miR-223 deletion neither significantly interrupt iNKT cell development nor its cytokine production,[53,54] it cannot obliterate the potential roles of other miRNAs in the immunopathogenesis of psoriasis.
miRNAs also modulate the cytokine and chemokine productions from T cells. While TNF and its receptors were upregulated in psoriasis,[55–57] circulating miR-19a and miR-130a were associated with TNF-α signalling pathway in psoriasis,[58] and overexpressing miR-210 in normal CD4+ T cells could induce IFN-γ, IL-17 and inhibit IL-10, TGF-β.[42] These studies suggest miRNA may exert their functions on both proinflammatory and anti-inflammatory sides of immunological dysfunction. Furthermore, circulating miRNA were investigated in the post-transcriptional regulation of genes associated with lipid metabolism (miR-33) and vascular function and angiogenesis (miR-126),[59] which were recently proved to be associated with the pathogenesis of psoriasis.[60,61] Plasma levels of miR-33 were shown to be positively correlated with plasma insulin levels.[62,63] Although not all of these miRNAs discussed here have been directly investigated in the pathogenesis of psoriasis, it is conceivable that they may play important roles. Interestingly, the study of circulating miR-33 may provide new insights into the associated systemic inflammatory abnormalities with psoriasis.
3 TARGETING MIRNAS TO TREAT PSORIASIS
Ten years ago, biological agents targeting chief molecules in psoriasis have become successful in clinical.[20,64,65] Based on the immunological functions in psoriasis discussed above, recent report showed the feasibility of targeting key genes in psoriasis employing RNA interference-mediated down-regulation by viral vectors or liposome-encapsulated siRNA, and miRNA had been tested with promising results.[20,66] These disease-specific miRNA identified may be helpful to represent potential therapeutic targets in treating psoriasis (Figure 2).
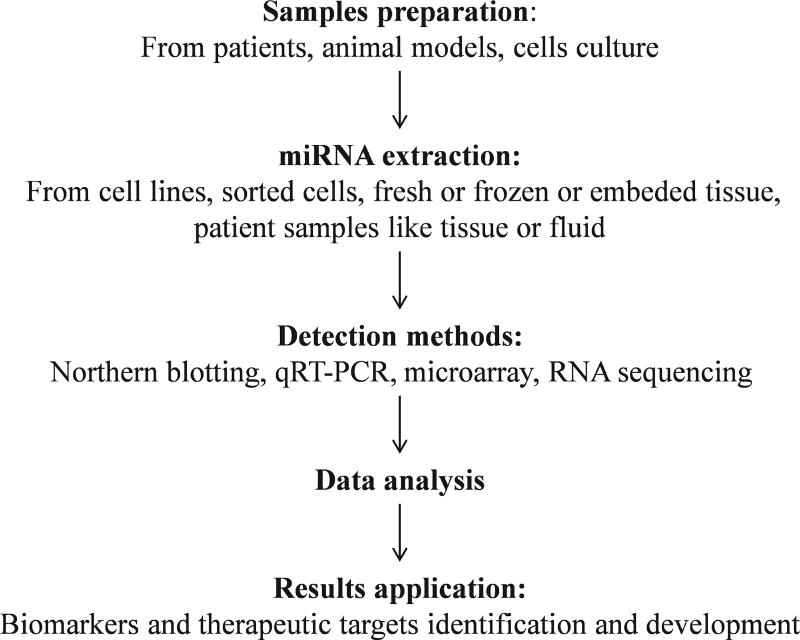
FIGURE 2.
Materials for miRNA extraction and process of miRNA profiles, including the sample preparation, miRNA extraction, detection, data analysis and results application
A number of miRNAs have been described to be upregulated in psoriasis, thereby inhibiting these miRNA may be effective. Increased miR-21 leads to activation of IL-6 /Stat3,[67,68] while miR-21 was downregulated after therapeutic ultraviolet B treatment of psoriatic skin,[69] suggesting that miR-21 down-regulation in psoriasis could be beneficial. In addition, miR-21 was found increased in psoriatic T cells,[20] and inhibiting miR-21 had a functional effect on non-malignant T cells.[20,70] Overall, blocking miR-21 may have advantages over current biologics and small molecules in development for treating psoriasis such as anti-IL-12/ IL-23, anti-IL-17/ IL-17R (IL-17 receptor), anti-PDE4 (phosphodiesterase 4) and anti-S100 proteins, but more tests need to be performed.[71] Besides, psoriasis is a risk factor for skin carcinoma including squamous cell carcinoma (SSC)[72,73] and shares many pathogenic pathways, such as TIMP-3.[74] As miR-21 was overexpressed both in psoriasis and carcinomas, treatments to these two diseases may both effect by suppressing miR-21.[75]
Many studies showed that modulating specific miRNAs had therapeutic effect on keratinocytes. Xu et al.[21] reported that downregulated miR-125b was involved in mediating the pathological proliferation and differentiation of keratinocytes in psoriasis, while the overexpression of miR-125b inhibited keratinocyte proliferation and promoted differentiation via inhibiting its direct target, FGFR2, in primary human keratinocytes. Moreover, anti-miR-21 has also been shown to be effective in treating psoriasis in animal models.[68] By contrast, it was identified that TGF-β1 could upregulate miR-31, while inhibition of miR-31 resulted in suppression of IL-1β and IL-8 in human primary keratinocytes.[41] Overexpression of miR-210 led to increased proinflammatory cytokine (IFN-γ and IL-17) expression and decreased regulatory cytokine (IL-10 and TGF-β) expression in CD4+ T cells.[42] Collectively, these experimental data provide important clues to help elucidate the pathogenesis of psoriasis and implicate that promotion of miR-125b while inhibition of miR-31 or miR-210 may also be potential therapeutic options to psoriasis (Figure 3).
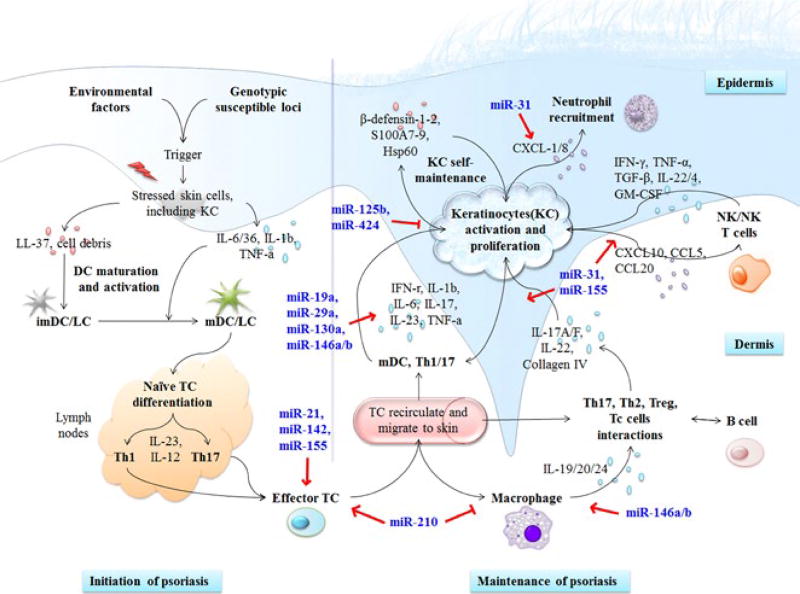
FIGURE 3.
Roles of miRNAs in the modulation of immunological mechanism of psoriasis. Both the cells interaction network and the cytokines and chemokines network are regulated by miRNA, which indicates a critical role of miRNA playing in the immunopathogenesis of psoriasis. KC, keratinocyte; imDC, immature dendritic cell; mDC, mature dendritic cell; LC, Langerhans cell; TC, T cell; NK, natural killer cell; NKT, natural killer T cell; Treg, regulatory T cell; miR, microRNA
Based on the data above, miR-21 is the only miRNA whose modulation has been tested in psoriasis in vivo. Although miRNA may become the promising therapeutic targets, most of the effects of their inhibition have not been tested in vivo. Anyway, targeting miRNA may represent a potential therapeutic option for the treatment of psoriasis.[67] But miRNAs may not trigger the disease directly, they are deregulated because of some unknown environmental or immunological trigger, and therefore, therapeutics targeting this initial trigger may exert potential effects. However, the beneficial outcome of targeting miRNA needs to be compared to the anti-TNF-α biologics currently used in the clinical management of psoriasis as well as new emerging therapies, like anti-IL-17, anti-IL-17R, anti-IL-12/ IL-23 and anti-PDE4 inhibitors.[68,76]
4 MIRNA AS PO TENTIAL BIOMARKERS IN PSORIASIS
Some of the innate properties of miRNAs make them highly attractive as potential biomarkers. They are highly conserved between species,[77] allowing the use of animal models of disease for preclinical studies.[78] After the miRNA was firstly reported in the circulation, the use of miRNA in the biomarker field was then emerged given their stability and relative ease of detection. On the one side, miRNA in serum are resistant to circulating ribonucleases and severe physicochemical conditions, such as extended storage, freeze-thawing and extreme pH.[78,79] On the other side, miRNA can be readily detected in small volume blood samples using specific and sensitive quantitative real-time PCR (qRT-PCR); besides of serum and plasma, miRNA have also been isolated from most other body fluids.[78,80–82] Our laboratory used real-time PCR miRNA arrays technology and identified 5 miRNAs, miR-146a, miR-16, miR-195, miR-30e and miR-744, to be stably expressed in different mice strains, which could serve as mouse serum miRNA endogenous references for single assay experiments.[83]
Promisingly, the serum level of miRNA can be useful for diagnosis, prognosis and therapeutic evaluation in various diseases. As biomarkers that reflect the diagnosis and disease activity have not been in clinical use for psoriasis, there is currently growing interest in the study of circulating miRNA given their potential as biomarkers. We have found that the expressions of 20 serum miRNAs were changed in vitiligo mice compared to control mice. Three increased miRNAs, miR-146a, miR-191 and miR-342-3p, were further confirmed by qRT-PCR. Our findings suggest that serum miRNAs could serve as serum biomarkers for vitiligo in mice.[84] To date, miRNA have been shown to be deregulated in psoriasis and regulate relevant immunological processes in structural and infiltrating immune cells, and serum miRNA have been shown to change after anti-TNF-α treatment.[85] Thus, miRNA can potentially serve as therapeutic targets and become the biomarkers to reflect the therapy response (Figure 3).
4.1 miRNA as diagnostic markers for psoriasis
There is a specific miRNA expression profile in psoriasis. Hence, miRNAs may become novel regulators involved in the diagnosis of psoriasis. Peripheral blood was attractive for biomarker discovery.[86] miR-223 and miR-143 were found to significantly upregulate in the PBMCs from patients with psoriasis, and these miRNA significantly correlated with PASI score. ROC analysis showed that miR-223 and miR-143 have the potential to distinguish between psoriasis and healthy controls.[31] Hence, it is suggested that miR-223 and miR-143 may serve as a novel biomarker for disease activity of psoriasis.[31] Moreover, miR-19a, a regulatory miRNA of TNF-α, were found significantly upregulated only in psoriasis and showed significant difference in ROC analysis.[87] Rie Oyama et al. identified the serum miR-19a, reflecting the activation of TNF, could be biomarkers.[58,88] In a study of Ichihara et al.[8], it was reported that serum miR-424 reflected the proliferative activity of keratinocytes. In the later study, they reported serum levels of miR-1266, a putative regulator of IL-17A, in patients with psoriasis showed weak inverse correlations with PASI scores and Body Surface Areas (BSA).[89] In the similar disease scleroderma, Takemoto et al.[90] showed the result that decreased miR-29a levels had contracture of the phalanges at a significantly higher prevalence, which indicated serum miRNA could be served as independent biomarkers. Five miRNAs, miR-146a, miR-16, miR-195, miR-30e and miR-744, were identified using real-time PCR miRNA arrays technology, which were stably expressed in different mice strains, could serve as mouse serum miRNA endogenous references for single assay experiments.[83] Another study found that by real-time PCR study, levels of miR-125b, miR-146a, miR-203 and miR-205 in serum were significantly decreased in patients with psoriasis compared with normal subjects.[94] Besides of blood elements, miRNA have also been isolated from most other body fluids and tissues.[78,80–82] Laser capture microdissection (LCM) and next-generation sequencing (NGS) were used to identify that miR-193b expression and miR-223 expression were decrease in the epidermis and dermal inflammatory infiltrates of psoriatic skin compared with normal psoriatic skin.[19] Compared with normal controls and those with atopic dermatitis, miR-424 in hair shaft were significantly upregulated in patients with psoriasis.[98]
Present data suggest that certain miRNAs could potentially serve as psoriasis activity markers. Complementary to the results from Lovendorf et al., another study showed that miR-223 controlled the migration of inflammatory cells to tissue via regulating inflammatory molecules.[31,91] Hirao et al.[87] showed that relative miR-19a levels were inversely and significantly correlated with the duration between symptom onset and the first visit to the hospital in patients with psoriasis. However, unlike in psoriatic skin, Ichihara et al.[8] did not find a statistically significant difference in serum miR-424 levels between patients with psoriasis and healthy controls, which may because of the small number of patients.
Among the patients with psoriasis, approximately 10%–30% develop psoriatic arthritis (PsA). As skin manifestations precede joint symptoms in nearly all patients with PsA, identification of biomarkers for early prediction of joint damage is an important clinical need. Recent studies from our laboratory and others have demonstrated that serum contains stable miRNAs derived from immune cells and various tissues, establishing their potential value as biomarkers for changes in physiologic and pathologic conditions of psoriasis and PsA.[92] To date, not much is known regarding miRNA especially the circulating miRNA expression in patients with PsA. However, it has been suggested that serum miRNAs may provide a source of candidate biomarkers for PsA.[93, 94]
4.2 miRNA levels monitor therapeutic effects
Pathological T cells and dendritic cells can trigger the abnormal keratinocyte proliferation in psoriasis progression via many cytokines especially TNF.[58,95] Besides, antibodies against TNF (e.g., infliximab and adalimumab) or soluble TNF receptor (etanercept) and biological therapy have been in clinical use.[13,58,95] Thus, TNF is essential for the pathogenesis of psoriasis.[58] Anti-TNF-α biological drug etanercept significantly suppressed a panel of 38 miRNA and validated serum levels of miR-106b, miR-26b, miR-142-3p, miR-223 and miR-126 were significantly downregulated,[85] while adalimumab could increase miR-23b[96] indicating changes in the miRNA level may reflect a previously unknown effect of anti-TNF-α therapy and may become a biomarker monitoring therapeutic effects. Along with this study, other miRNAs were tested as biomarkers reflecting the therapy responses in cancers. For example, miR-494 and miR-1973 reflected as biomarkers for the therapy response of combination chemotherapy to early stage of Hodgkin lymphoma, miR-608 may be an important factor for modulating colorectal cancer prognosis and predicting therapy response, and miR-425-5p and miR-93-5p may be represented as biomarkers for radiochemotherapy monitoring for head and neck cancer. As a result, miR-21, miR-93, miR-219, miR-222, miR-425, miR-494, miR-549, miR-608 and miR-1973 were identified as potential biomarkers for therapy responses.[97–100] These tissue-or cell-related biomarkers could be released to blood, which could be used as potential serum biomarkers for monitoring therapeutic effects in psoriasis. However, current studies on the miRNA as biomarkers monitoring therapeutic effects in psoriasis are very limited. More studies on the serum miRNA biomarkers should be further performed to expand and confirm their values on psoriasis.
4.3 miRNA levels reflect psoriasis severity
Meaningfully, serum and skin miR-369-3p levels in patients with psoriasis were detected and confirmed their correlation with disease severity,[67] in which miR-369-3p levels in skin had a positive linear relation with PASI scores.[67,101] Similar results were obtained in the investigation of miR-223 and miR-143.[31] However, the study from Pivarcsi et al.[85] did not observed the anticipated relation of the changed miRNA to the disease severity of psoriasis. Hence, although some of the serum miRNA could be used as potential biomarkers for disease severity, the changes of these miRNA levels may not always be related to the biological or systemic treatment of psoriasis. Further studies need to find the overlapped ones between the miRNA which reflect disease severity and the other miRNA which reflect the therapy response.
Overall, these data above suggested that certain miRNA could be developed as diagnostic markers, some could monitor therapy response, while the others may be used as the markers to reflect the disease severity of psoriasis. However, not all of the miRNA with important immunological modulation effects will become the best biomarker. Besides, the miRNA as biomarkers detection procedures were still not completely established.[102] For instance, the accurate measurement of these miRNA has been associated with many challenges.[78] Another prerequisite to develop circulating miRNA-based diagnostics is the ability to quantify miRNA from plasma and serum with sufficient sensitivity and precision.[78,103] To reduce the risk of introducing technical variation, whole blood samples which require a minimal effort of sample handling can be used initially.[31] Anyway, although future studies need to be performed to test and confirm the roles of miRNA as biomarkers and to improve the detection procedures, circulating miRNAs can be used as biomarkers and will be helpful for disease management.
5 SUMMARY AND PROSPECT
Psoriasis is a common inflammatory skin disease with limited treatment options that is characterized by a complex interplay between keratinocytes, immune cells and inflammatory mediators. miRNAs are important gene regulatory molecules which target multiple mRNAs and regulate numerous cellular processes, and they may play a key role in the pathogenesis of psoriasis. Investigation of the regulatory mechanisms of keratinocyte proliferation and immune dysfunction by miRNA may lead to new markers for diagnosis, monitoring the therapy response and reflect the disease severity. New treatments are also implicated such as using miRNA by transfection into the lesional skin, and it has been suggested that deregulation at the post-transcriptional level could lead to a direct antisense targeting of miRNA.[2,104]
To date, the disease severity of psoriasis is assessed by PASI score and BSA.[58] However, serum marker which reflects disease activity, similar to eosinophils, serum IgE and TARC for atopic dermatitis, has not been in clinical use in psoriasis.[58] Thus, it will be helpful to develop such markers as circulating miRNA in this disease. Circulating miRNA in the biomarker field were then investigated given their stability and relative ease of detection from minimal volume of blood sample. The researches above qualified miRNA as potential biomarkers for psoriasis. However, the miRNA that are the most important in pathogenesis are not necessarily the best biomarkers; for example, miRNA in the circulation show limited or no correlation with miRNA expression in skin. Moreover, whether the deregulated miRNA in the blood are psoriasis-specific or related to the systemic inflammation is unclear in some studies. Accordingly, comparative larger studies including blood from patients with other skin diseases such as atopic dermatitis may be helpful to clarify the specific roles of miRNA in psoriasis.[31]
In the past few years, miRNAs are gradually recognized to play very import roles in disease development. The role of miRNAs in the pathogenesis of psoriasis has been revealed progressively. Despite miRNA biomarkers of psoriasis achieved some accomplishments, there is still no validated biomarkers being used in clinical yet. For instance, the sensitivity and specificity of miRNAs as the diagnosis, treatment and prognosis markers need to be further detected. New medicine based on miRNA modulation will be discovered soon, which will bring a new light for the psoriasis treatment. In short, with further researches, the diagnostic and therapeutic value of miRNAs in psoriasis and other immune-related diseases will probably achieve greater breakthroughs.
Supplementary Material
Table S1 Immunological mechanisms of miRNA involved in psoriasis
Table S2 miRNA in psoriasis as potential biomarkers and therapeutic targets
Data S1 Supplementary References
Acknowledgments
Funding information
Guangdong Natural Science Fund, Grant/Award Number: S2013030011515; National Natural Science Foundation of China, Grant/Award Number: 81473681; NIH/National Institute of Allergy and Infectious Diseases, Grant/Award Number: 1R56AI119041 and 1R01AI119041; NIH/National Institute of Arthritis and Musculoskeletal and Skin Diseases, Grant/Award Number: 1R01AR069681; Henry Ford Immunology Program, Grant/Award Number: T7107 and T7106.
This research was partially supported by Guangdong Natural Science Fund (S2013030011515), National Natural Science Foundation of China (81473681 to CL), NIH/National Institute of Allergy and Infectious Diseases (1R56AI119041, 1R01AI119041 to QSM), NIH/National Institute of Arthritis and Musculoskeletal and Skin Diseases (1R01AR069681 to QSM) and Henry Ford Immunology Program (T7107 to LZ and T7106 to QSM).
Abbreviations
- ADAM17
A disintegrin and metalloproteinase 17
- AP-1
Activating protein 1
- BSA
Body Surface Areas
- EGF
Epidermal growth factor
- FGFR2
Fibroblast growth factor receptor 2
- GLUL
Glutamate-ammonia ligase
- IL
Interleukin
- LCM
Laser capture microdissection
- MAPK
Mitogen activated protein kinase
- miRISC
miRNA-induced silencing complex
- NCEH1
Neutral cholesterol ester hydrolase 1
- NGS
Next-generation sequencing
- NHEK
Human epidermal keratinocytes
- PASI
Psoriasis area severity index
- PBMCs
Peripheral blood mononuclear cells
- PDCD4
Programmed cell death 4
- PDE4
phosphodiesterase 4
- ppp6c
Protein phosphatase 6
- ROC
Receiver-operating characteristic analysis
- RUNX3
Runt-related transcription factor 3
- SHIP-2
SH2 Domain-Containing 5′-Inositol Phosphatase-2
- SLC26A4
Solute carrier family 26 member 4
- SMAD3
SMAD family member 3
- SOCS-3
Suppressor of cytokine signalling 3
- SREBP-2
Sterol-regulatory element-binding protein 2
- Stat3
Signal transducer and activator of transcription 3
- STK40
Serine/Threonine kinase 40
- TACE
Tumor necrosis factor-α-converting enzyme
- TIMP-3
Tissue inhibitor of matrix metalloproteinase 3
- TNF
Tumor necrosis factor
- TSPM1
Tumor suppressor protein tropomyosin 1.
Footnotes
AUTHOR CONTRIBUTIONS
QSM, LZ and CL designed the topic of this review; QL, DW, LH, LZ, RH, CL and QSM wrote the manuscript; and all authors have read and revised the manuscript critically.
CONFLICT OF INTERESTS
The authors have declared no conflicting interests.
References
- 1.Jones-Rhoades MW, Bartel DP. Mol. Cell. 2004;14:787. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 2.Zibert JR, Lovendorf MB, Litman T, Olsen J, Kaczkowski B, Skov L. J. Dermatol. Sci. 2010;58:177. doi: 10.1016/j.jdermsci.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Pasquinelli AE. Nat. Rev. Genet. 2012;13:271. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 4.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Nature. 2008;455:58. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 5.Coronnello C, Benos PV. Nucleic Acids Res. 2013;41:W159. doi: 10.1093/nar/gkt379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindsay MA. Trends Immunol. 2008;29:343. doi: 10.1016/j.it.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Baumjohann D, Ansel KM. Nat. Rev. Immunol. 2013;13:666. doi: 10.1038/nri3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ichihara A, Jinnin M, Yamane K, Fujisawa A, Sakai K, Masuguchi S, Fukushima S, Maruo K, Ihn H. Br. J. Dermatol. 1003;2011:165. doi: 10.1111/j.1365-2133.2011.10497.x. [DOI] [PubMed] [Google Scholar]
- 9.Ortega C, Fernandez AS, Carrillo JM, Romero P, Molina IJ, Moreno JC, Santamaria M. J. Leukoc. Biol. 2009;86:435. doi: 10.1189/JLB.0109046. [DOI] [PubMed] [Google Scholar]
- 10.Van Belle AB, de Heusch M, Lemaire MM, Hendrickx E, Warnier G, Dunussi-Joannopoulos K, Fouser LA, Renauld JC, Dumoutier L. J. Immunol. 2012;188:462. doi: 10.4049/jimmunol.1102224. [DOI] [PubMed] [Google Scholar]
- 11.Xu YP, Qi RQ, Chen W, Shi Y, Cui ZZ, Gao XH, Chen HD, Zhou L, Mi QS. Aging. 2012;4:742. doi: 10.18632/aging.100501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Micali G, Wilsmann-Theis D, Mallbris L, Gallo G, Marino V, Brault Y, Germain JM. Acta Derm. Venereol. 2014;95:57. doi: 10.2340/00015555-1845. [DOI] [PubMed] [Google Scholar]
- 13.Shear NH, Hartmann M, Toledo-Bahena M, Katsambas A, Connors L, Chang Q, Yao R, Nograles K, Popmihajlov Z on behalf of All REALITY investigators. Br. J. Dermatol. 2014;171:631. doi: 10.1111/bjd.13004. [DOI] [PubMed] [Google Scholar]
- 14.Luszczek W, Kubicka W, Cislo M, Nockowski P, Manczak M, Woszczek G, Baran E, Kusnierczyk P. Immunol. Lett. 2003;85:59. doi: 10.1016/s0165-2478(02)00212-2. [DOI] [PubMed] [Google Scholar]
- 15.Trowbridge RM, Pittelkow MR. J. Drugs Dermatol. 2014;13:111. [PubMed] [Google Scholar]
- 16.Sonkoly E, Wei T, Janson PC, Saaf A, Lundeberg L, Tengvall-Linder M, Norstedt G, Alenius H, Homey B, Scheynius A, Stahle M, Pivarcsi A. PLoS ONE. 2007;2:e610. doi: 10.1371/journal.pone.0000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lerman G, Avivi C, Mardoukh C, Barzilai A, Tessone A, Gradus B, Pavlotsky F, Barshack I, Polak-Charcon S, Orenstein A, Hornstein E, Sidi Y, Avni D. PLoS ONE. 2011;6:e20916. doi: 10.1371/journal.pone.0020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joyce CE, Zhou X, Xia J, Barzilai A, Tessone A, Gradus B, Pavlotsky F, Barshack I, Polak-Charcon S, Orenstein A, Hornstein E, Sidi Y, Avni S. Hum. Mol. Genet. 2011;20:4025. [Google Scholar]
- 19.Lovendorf MB, Mitsui H, Zibert JR, Ropke MA, Hafner M, Dyring-Andersen B, Bonefeld CM, Krueger JG, Skov L. Exp. Dermatol. 2015;24:187. doi: 10.1111/exd.12604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meisgen F, Xu N, Wei T, Janson PC, Obad S, Broom O, Nagy N, Kauppinen S, Kemeny L, Stahle M, Pivarcsi A, Sonkoly E. Exp. Dermatol. 2012;21:312. doi: 10.1111/j.1600-0625.2012.01462.x. [DOI] [PubMed] [Google Scholar]
- 21.Xu N, Brodin P, Wei T, Meisgen F, Eidsmo L, Nagy N, Kemeny L, Stahle M, Sonkoly E, Pivarcsi A. J. Invest. Dermatol. 2011;131:1521. doi: 10.1038/jid.2011.55. [DOI] [PubMed] [Google Scholar]
- 22.Meisgen F, Xu Landen N, Wang A, Rethi B, Bouez C, Zuccolo M, Gueniche A, Stahle M, Sonkoly E, Breton L, Pivarcsi A. J. Invest. Dermatol. 1931;2014:134. doi: 10.1038/jid.2014.89. [DOI] [PubMed] [Google Scholar]
- 23.Wei T, Xu N, Meisgen F, Stahle M, Sonkoly E, Pivarcsi A. Eur. J. Dermatol. 2013;23:66. doi: 10.1684/ejd.2013.1997. [DOI] [PubMed] [Google Scholar]
- 24.Ha TY. Immune Netw. 2011;11:227. doi: 10.4110/in.2011.11.5.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee D, Shin C. Ann. N. Y. Acad. Sci. 2012;1271:118. doi: 10.1111/j.1749-6632.2012.06745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi HH, Ongusaha PP, Myllyharju J, Cheng D, Pakkanen O, Shi Y, Lee SW, Peng J, Shi Y. Nature. 2008;455:421. doi: 10.1038/nature07186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yi R, Poy MN, Stoffel M, Fuchs E. Nature. 2008;452:225. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Y, Chaerkady R, Beer MA, Mendell JT, Pandey A. Proteomics. 2009;9:1374. doi: 10.1002/pmic.200800551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujita S, Ito T, Mizutani T, Minoguchi S, Yamamichi N, Sakurai K, Iba H. J. Mol. Biol. 2008;378:492. doi: 10.1016/j.jmb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 30.Yu J, Peng H, Ruan Q, Fatima A, Getsios S, Lavker RM. FASEB J. 2010;24:3950. doi: 10.1096/fj.10-157404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lovendorf MB, Zibert JR, Gyldenlove M, Gyldenlove M, Ropke MA, Skov L. J. Dermatol. Sci. 2014;75:133. doi: 10.1016/j.jdermsci.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 32.Iizuka H, Takahashi H, Ishida-Yamamoto A. J. Dermatol. Sci. 2004;35:93. doi: 10.1016/j.jdermsci.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 33.Zhu W, Chen YP, Syst BMC. Biol. 2014;8:14. doi: 10.1186/1752-0509-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong T, Hsu L, Liao W. J. Cutan. Med. Surg. 2013;17:6. doi: 10.2310/7750.2012.11124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tonel G, Conrad C. Int. J. Biochem. Cell B. 2009;41:963. doi: 10.1016/j.biocel.2008.10.022. [DOI] [PubMed] [Google Scholar]
- 36.Haase I, Hobbs RM, Romero MR, Broad S, Watt FM. J. Clin. Investig. 2001;108:527. doi: 10.1172/JCI12153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lena AM, Shalom-Feuerstein R, Rivetti di Val Cervo P, Aberdam D, Knight RA, Melino G, Candi E. Cell Death Differ. 2008;15:1187. doi: 10.1038/cdd.2008.69. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J, Hu S, Schofield DE, Sorensen PH, Triche TJ. Cancer Res. 2004;64:6026. doi: 10.1158/0008-5472.CAN-03-2594. [DOI] [PubMed] [Google Scholar]
- 39.Guttman-Yassky E, Nograles KE, Krueger JG. J. Allergy Clin. Immunol. 2011;127:1420. doi: 10.1016/j.jaci.2011.01.054. [DOI] [PubMed] [Google Scholar]
- 40.Sa SM, Valdez PA, Wu J, Jung K, Zhong F, Hall L, Kasman I, Winer J, Modrusan Z, Danilenko DM, Ouyang W. J. Immunol. 2007;178:2229. doi: 10.4049/jimmunol.178.4.2229. [DOI] [PubMed] [Google Scholar]
- 41.Xu N, Meisgen F, Butler LM, Han G, Wang XJ, Soderberg-Naucler C, Stahle M, Pivarcsi A, Sonkoly E. J. Immunol. 2013;190:678. doi: 10.4049/jimmunol.1202695. [DOI] [PubMed] [Google Scholar]
- 42.Zhao M, Wang LT, Liang GP, Deng XJ, Tang Q, Zhai HY, Chang CC, Su YW, Lu QJ. Clin. Immunol. 2014;150:22. doi: 10.1016/j.clim.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 43.Valdimarsson H, Baker BS, Jonsdottir I, Powles A, Fry L. Immunol. Today. 1995;16:145. doi: 10.1016/0167-5699(95)80132-4. [DOI] [PubMed] [Google Scholar]
- 44.Schon MP, Detmar M, Parker CM. Nat. Med. 1997;3:183. doi: 10.1038/nm0297-183. [DOI] [PubMed] [Google Scholar]
- 45.Mi QS, Xu YP, Wang H, Qi RQ, Dong Z, Zhou L. Int. J. Biochem. Cell Biol. 2013;45:395. doi: 10.1016/j.biocel.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou L, Qi RQ, Liu M, Xu YP, Li G, Weiland M, Kaplan DH, Mi QS. Genes Immun. 2014;15:57. doi: 10.1038/gene.2013.61. [DOI] [PubMed] [Google Scholar]
- 47.Miyara M, Gorochov G, Ehrenstein M, Musset L, Sakaguchi S, Amoura Z. Autoimmun. Rev. 2011;10:744. doi: 10.1016/j.autrev.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 48.Wan YY, Flavell RA. Nature. 2007;445:766. doi: 10.1038/nature05479. [DOI] [PubMed] [Google Scholar]
- 49.Zhou L, Seo KH, Wong HK, Mi QS. Int. Immunopharmacol. 2009;9:524. doi: 10.1016/j.intimp.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 50.Zhou L, Park JJ, Zheng Q, Dong Z, Mi Q. Cell. Mol. Immunol. 2011;8:380. doi: 10.1038/cmi.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi C, Liang Y, Yang J, Xia Y, Chen H, Han H, Yang Y, Wu W, Gao R, Qin H. PLoS ONE. 2013;8:e66814. doi: 10.1371/journal.pone.0066814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou L, Seo KH, He HZ, Pacholczyk R, Meng DM, Li CG, Xu J, She JX, Dong Z, Mi QS. Proc. Natl Acad. Sci. USA. 2009;106:10266. doi: 10.1073/pnas.0811119106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li K, Seo KH, Gao T, Zheng Q, Qi RQ, Wang H, Weiland M, Dong Z, Mi QS, Zhou L. Int. Immunopharmacol. 2011;11:561. doi: 10.1016/j.intimp.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 54.Seo KH, Zhou L, Meng D, Xu J, Dong Z, Mi QS. Cell. Mol. Immunol. 2010;7:447. doi: 10.1038/cmi.2010.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Caldarola G, De Simone C, Carbone A, Tulli A, Amerio P, Feliciani C. Int. J. Immunopathol. Pharmacol. 2009;22:961. doi: 10.1177/039463200902200411. [DOI] [PubMed] [Google Scholar]
- 56.Grine L, Dejager L, Libert C, Vandenbroucke RE. Cytokine Growth Factor Rev. 2014;26:25. doi: 10.1016/j.cytogfr.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 57.Gudjonsson JE, Ding J, Johnston A, Tejasvi T, Guzman AM, Nair RP, Voorhees JJ, Abecasis GR, Elder JT. J. Invest. Dermatol. 1829;2010:130. doi: 10.1038/jid.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oyama R, Jinnin M, Kakimoto A, Kanemaru H, Ichihara A, Fujisawa A, Honda N, Masuguchi S, Fukushima S, Maruo K, Ihn H. J. Dermatol. Sci. 2011;61:209. doi: 10.1016/j.jdermsci.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 59.Davalos A, Fernandez-Hernando C. Pharmacol. Res. 2013;75:60. doi: 10.1016/j.phrs.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.El Asmi MA, Zidi W, Mebazaa A, Zayani Y, Ayadi I, Feki M, Ben Osman A, Kaabachi N. Clin. Lab. 1043;2014:60. doi: 10.7754/clin.lab.2013.130535. [DOI] [PubMed] [Google Scholar]
- 61.Feingold KR, Grunfeld C. J. Lipid Res. 2012;53:1427. doi: 10.1194/jlr.E029330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garcia-Rodriguez S, Arias-Santiago S, Orgaz-Molina J, Magro-Checa C, Valenzuela I, Navarro P, Naranjo-Sintes R, Sancho J, Zubiaur M. Actas Dermosifiliogr. 2014;105:497. doi: 10.1016/j.ad.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 63.Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernandez-Hernando C. Science. 2010;328:1570. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lowes MA, Bowcock AM, Krueger JG. Nature. 2007;445:866. doi: 10.1038/nature05663. [DOI] [PubMed] [Google Scholar]
- 65.Chamian F, Lowes MA, Lin SL, Lee E, Kikuchi T, Gilleaudeau P, Sullivan-Whalen M, Cardinale I, Khatcherian A, Novitskaya I, Wittkowski KM, Krueger JG. Proc. Natl Acad. Sci. USA. 2005;102:2075. doi: 10.1073/pnas.0409569102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Borel F, Konstantinova P, Jansen PL. J. Hepatol. 2012;56:1371. doi: 10.1016/j.jhep.2011.11.026. [DOI] [PubMed] [Google Scholar]
- 67.Deng X, Su Y, Wu H, Wu R, Zhang P, Dai Y, Chan TM, Zhao M, Lu Q. Scand. J. Immunol. 2014;81:153. doi: 10.1111/sji.12261. [DOI] [PubMed] [Google Scholar]
- 68.Guinea-Viniegra J, Jimenez M, Schonthaler HB, Navarro R, Delgado Y, Concha-Garzon MJ, Tschachler E, Obad S, Dauden E, Wagner EF. Sci. Transl. Med. 2014;6:225re221. doi: 10.1126/scitranslmed.3008089. [DOI] [PubMed] [Google Scholar]
- 69.Gu X, Nylander E, Coates PJ, Nylander K. Acta Derm. Venereol. 2011;91:392. doi: 10.2340/00015555-1086. [DOI] [PubMed] [Google Scholar]
- 70.Leone E, Morelli E, Di Martino MT, Amodio N, Foresta U, Gulla A, Rossi M, Neri A, Giordano A, Munshi NC, Anderson KC, Tagliaferri P, Tassone P. Clin. Cancer Res. 2013;19:2096. doi: 10.1158/1078-0432.CCR-12-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Agrawal U, Gupta M, Dube D, Vyas SP. Crit. Rev. Ther. Drug Carrier Syst. 2013;30:51. doi: 10.1615/critrevtherdrugcarriersyst.2013005589. [DOI] [PubMed] [Google Scholar]
- 72.Richard MA, Barnetche T, Horreau C, Brenaut E, Pouplard C, Aractingi S, Aubin F, Cribier B, Joly P, Jullien D, Le Maitre M, Misery L, Ortonne JP, Paul C. J. Eur. Acad. Dermatol. Venereol. 2013;27(Suppl 3):2. doi: 10.1111/jdv.12162. [DOI] [PubMed] [Google Scholar]
- 73.Stern RS, Scotto J, Fears TR. J. Am. Acad. Dermatol. 1985;12:67. doi: 10.1016/s0190-9622(85)70011-4. [DOI] [PubMed] [Google Scholar]
- 74.Selaru FM, Olaru AV, Kan T, David S, Cheng Y, Mori Y, Yang J, Paun B, Jin Z, Agarwal R, Hamilton JP, Abraham J, Georgiades C, Alvarez H, Vivekanandan P, Yu W, Maitra A, Torbenson M, Thuluvath PJ, Gores GJ, LaRusso NF, Hruban R, Meltzer SJ. Hepatology. 2009;49:1595. doi: 10.1002/hep.22838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang X, He H, Lu Y, Ren W, Teng KY, Chiang CL, Yang Z, Yu B, Hsu S, Jacob ST, Ghoshal K, Lee LJ. Biochim. Biophys. Acta. 1853;2015:244. doi: 10.1016/j.bbamcr.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yost J, Gudjonsson JE. F1000 Med. Rep. 2009;1:30. doi: 10.3410/M1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cuperus JT, Fahlgren N, Carrington JC. Plant Cell. 2011;23:431. doi: 10.1105/tpc.110.082784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sita-Lumsden A, Dart DA, Waxman J, Bevan CL. Br. J. Cancer. 1925;2013:108. doi: 10.1038/bjc.2013.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen X, Ba Y, Ma L, et al. Cell Res. 2008;18:997. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 80.Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. Nat. Rev. Clin. Oncol. 2011;8:467. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Korzeniewski N, Tosev G, Pahernik S, Hadaschik B, Hohenfellner M, Duensing S. Urol. Oncol. 2014;33:17. doi: 10.1016/j.urolonc.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 82.Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, Wong DT, Xiao X. Clin. Chem. 2014;33:17. doi: 10.1373/clinchem.2014.230433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mi QS, Weiland M, Qi RQ, Gao XH, Poisson LM, Zhou L. PLoS ONE. 2012;7:e31278. doi: 10.1371/journal.pone.0031278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shi YL, Weiland M, Lim HW, Mi QS, Zhou L. Exp. Dermatol. 2014;23:140. doi: 10.1111/exd.12319. [DOI] [PubMed] [Google Scholar]
- 85.Pivarcsi A, Meisgen F, Xu N, Stahle M, Sonkoly E. Br. J. Dermatol. 2013;169:563. doi: 10.1111/bjd.12381. [DOI] [PubMed] [Google Scholar]
- 86.Zhong J, He Y, Chen W, Shui X, Chen C, Lei W. Int. J. Mol. Sci. 2014;15:20355. doi: 10.3390/ijms151120355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hirao H, Jinnin M, Ichihara A, Fujisawa A, Makino K, Kajihara I, Sakai K, Fukushima S, Inoue Y, Ihn H. Eur. J. Dermatol. 2013;23:807. doi: 10.1684/ejd.2013.2190. [DOI] [PubMed] [Google Scholar]
- 88.Wang Z, Jinnin M, Kudo H, Inoue K, Nakayama W, Honda N, Makino K, Kajihara I, Fukushima S, Inoue Y, Ihn H. J. Dermatol. Sci. 2013;72:134. doi: 10.1016/j.jdermsci.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 89.Ichihara A, Jinnin M, Oyama R, Yamane K, Fujisawa A, Sakai K, Masuguchi S, Fukushima S, Maruo K, Ihn H. Eur. J. Dermatol. 2012;22:68. doi: 10.1684/ejd.2011.1600. [DOI] [PubMed] [Google Scholar]
- 90.Takemoto R, Jinnin M, Wang Z, Kudo H, Inoue K, Nakayama W, Ichihara A, Igata T, Kajihara I, Fukushima S, Ihn H. Exp. Dermatol. 2013;22:832. doi: 10.1111/exd.12245. [DOI] [PubMed] [Google Scholar]
- 91.Dorhoi A, Iannaccone M, Farinacci M, Fae KC, Schreiber J, Moura-Alves P, Nouailles G, Mollenkopf HJ, Oberbeck-Muller D, Jorg S, Heinemann E, Hahnke K, Lowe D, Del Nonno F, Goletti D, Capparelli R, Kaufmann SH. J. Clin. Investig. 2013;123:4836. doi: 10.1172/JCI67604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Weiland M, Gao XH, Zhou L, Mi QS. RNA Biol. 2012;9:850. doi: 10.4161/rna.20378. [DOI] [PubMed] [Google Scholar]
- 93.Villanova F, DiMeglio P, Nestle FO. Ann. Rheum. Dis. 2013;72(Suppl 2):ii104. doi: 10.1136/annrheumdis-2012-203037. [DOI] [PubMed] [Google Scholar]
- 94.Chatzikyriakidou A, Voulgari PV, Georgiou I, Drosos AA. Scand. J Immunol. 2010;71:382. doi: 10.1111/j.1365-3083.2010.02381.x. [DOI] [PubMed] [Google Scholar]
- 95.Eltaher S, Mohammed GF, Younes S, Elakhras A. J. Dermatol. Treat. 2014;26:335. doi: 10.3109/09546634.2014.990411. [DOI] [PubMed] [Google Scholar]
- 96.Raaby L, Langkilde A, Kjellerup RB, Vinter H, Khatib SH, Hjuler KF, Johansen C, Iversen L. Br. J. Dermatol. 2015;173:436. doi: 10.1111/bjd.13721. [DOI] [PubMed] [Google Scholar]
- 97.Odenthal M, Hee J, Gockel I, Sisic L, Schmitz J, Stoecklein NH, Driemel C, Mohlendick B, Schmidt T, Knoefel WT, Lang H, Buttner R, Ott K, Vallbohmer D. Int. J. Cancer. 2014;137:230. doi: 10.1002/ijc.29363. [DOI] [PubMed] [Google Scholar]
- 98.Pardini B, Rosa F, Naccarati A, Vymetalkova V, Ye Y, Wu X, di Gaetano C, Buchler T, Novotny J, Matullo G, Vodicka P. Carcinogenesis. 2014;36:82. doi: 10.1093/carcin/bgu224. [DOI] [PubMed] [Google Scholar]
- 99.Summerer I, Niyazi M, Unger K, Pitea A, Zangen V, Hess J, Atkinson MJ, Belka C, Moertl S, Zitzelsberger H. Radiat. Oncol. 2013;8:296. doi: 10.1186/1748-717X-8-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jones K, Nourse JP, Keane C, Bhatnagar A, Gandhi MK. Clin. Cancer Res. 2014;20:253. doi: 10.1158/1078-0432.CCR-13-1024. [DOI] [PubMed] [Google Scholar]
- 101.Guo S, Zhang W, Wei C, Wang L, Zhu G, Shi Q, Li S, Ge R, Li K, Gao L, Gao T, Wang G, Li C. Eur. J. Dermatol. 2013;23:608. doi: 10.1684/ejd.2013.2148. [DOI] [PubMed] [Google Scholar]
- 102.Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, Miyaji MM, Tait JF, Tewari M. Cancer Prev. Res. 2012;5:492. doi: 10.1158/1940-6207.CAPR-11-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sita-Lumsden A, Fletcher CE, Dart DA, Brooke GN, Waxman J, Bevan CL. Biomark. Med. 2013;7:867. doi: 10.2217/bmm.13.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. Nature. 2008;452:896. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Immunological mechanisms of miRNA involved in psoriasis
Table S2 miRNA in psoriasis as potential biomarkers and therapeutic targets
Data S1 Supplementary References