Abstract
Background
Increasing cannabis consumption among adolescents, studies that link its early use with mental illnesses, and the political debate on cannabis legalization together call for an urgent need to study molecular underpinnings of adolescent brain vulnerability. The emerging role of epigenetic mechanisms in psychiatric diseases led us to hypothesize that epigenetic alterations could play a role in causes and subsequent development of the depressive/psychotic-like phenotype induced by adolescent, but not adult, Δ9-tetrahydrocannabinol (THC) exposure in female rats.
Methods
We performed a time-course analysis of histone modifications, chromatin remodelling enzymes and gene expression in the prefrontal cortex of female rats after adolescent and adult THC exposure. We also administered a specific epigenetic drug (chaetocin) with THC to investigate its impact on THC-induced behavioural alterations.
Results
Adolescent THC exposure induced alterations of selective histone modifications (mainly H3K9me3), impacting the expression of genes closely associated with synaptic plasticity. Changes in both histone modifications and gene expression were more widespread and intense after adolescent treatment, suggesting specific adolescent susceptibility. Adolescent THC exposure significantly increased Suv39H1 levels, which could account for the enhanced H3K9me3. Pharmacological blockade of H3K9me3 during adolescent THC treatment prevented THC-induced cognitive deficits, suggesting the relevant role played by H3K9me3 in THC-induced effects.
Limitations
Only female rats were investigated, and the expression studies were limited to a specific subset of genes.
Conclusion
Through a mechanism involving SUV39H1, THC modifies histone modifications and, thereby, expression of plasticity genes. This pathway appears to be relevant for the development of cognitive deficits.
Introduction
Despite epidemiological studies suggesting that heavy cannabis use during adolescence increases the risk for neuropsychiatric disorders later in life,1–3 cannabis remains the world’s most widely used illicit substance,4 especially among adolescents. It is therefore necessary to comprehend thoroughly the molecular underpinnings enhancing adolescent brain vulnerability to adverse effects from cannabis.
A growing body of literature on animal models has uncovered a clear association between adolescent cannabinoid exposure and the development of specific traits of psychiatric disorders.5–7 We have shown previously that chronic administration of Δ9-tetrahydrocannabinol (THC), the psychoactive ingredient of cannabis, in adolescent but not adult female rats induces behavioural and biochemical signs relevant for depression and psychosis8–13 later in life. Interestingly, long-term effects of THC seem to be sex-specific. In fact, adolescent male rats exposed to the same THC treatment as females experienced only cognitive deficits.8,14 In view of the more complex phenotype in adolescent female rats exposed to THC, we decided to focus the present study on female rats.
In recent years, many studies have focused on the involvement of epigenetic mechanisms in the causes and subsequent development of psychiatric illnesses, such as drug addiction and depression.15,16 However, hints on epigenetic effects of THC exposure are generally scarce, particularly regarding adolescent THC administration.17 Despite the limited evidence, the few studies addressing this topic, although based on very different experimental models, suggest that THC may principally act on the epigenome through the modulation of the repressive histone marker H3K9me3.18–23
In the present study we investigated whether specific epigenetic mechanisms may contribute to the molecular events triggered by adolescent THC exposure and whether these modifications are different after adult THC exposure. We show that different histone H3 modifications — mainly increase of H3K9me3 — occur in the prefrontal cortex (PFC) of female rats exposed to THC during adolescence. These epigenetic alterations are translated into disease-relevant transcriptional changes involving a subset of genes closely associated with the endocannabinoid system (ECS) and neuroplasticity processes. Adulthood THC administration showed milder and short-lasting effects on the epigenome, reflecting minor changes on the transcriptional point of view. Administration of a specific epigenetic drug — inhibiting the main enzymes responsible for THC-induced H3K9me3 upregulation — restores cognitive THC-related issues. We propose that an epigenetic-driven molecular mechanism contributes to adolescence-specific THC vulnerability, suggesting a causal relationship between epigenome modification and cognitive deficits, a core sign of the complex depressive/psychotic-like phenotype representing the adult consequence of THC exposure during adolescence.
Methods
Drugs and treatments
The THC was given to us by GW Pharmaceuticals; it was dissolved in ethanol, kolliphor and saline (1:1:18). We purchased the chaetocin from Cayman Chemical; it was dissolved in dimethyl sulfoxide, polysorbate 80 and saline (1:1:8).
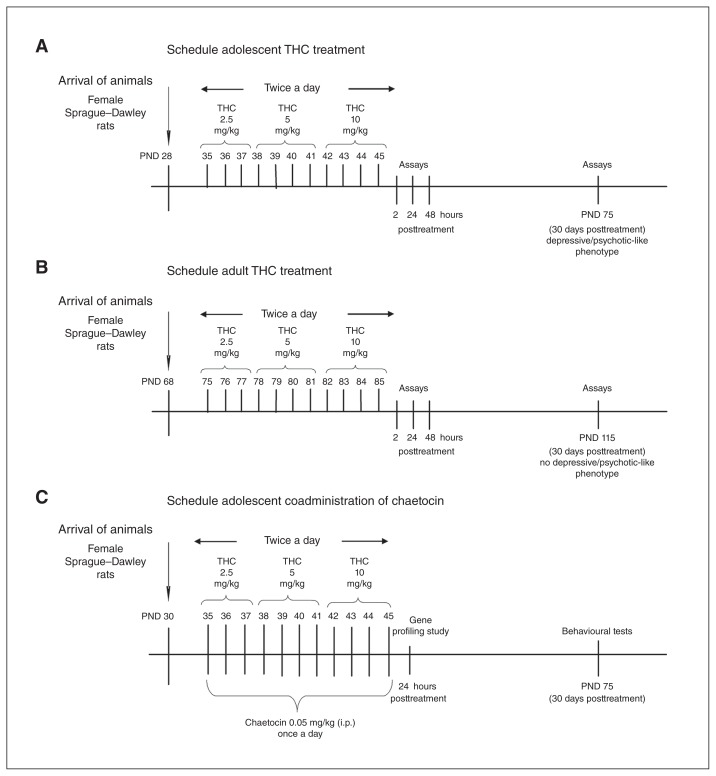
Female Sprague-Dawley rats received increasing intraperitoneal (IP) doses of THC twice a day (2.5, 5 and 10 mg/kg) or its vehicle, as previously described.8 In the adolescent setting, THC injections began at 35 postnatal days (PND) and lasted until 45 PND (Fig. 1A), whereas in the adult setting they started at 75 PND and lasted until 85 PND (Fig. 1B). This protocol mimics heavy Cannabis use according to the human-equivalent dose proposed by the United States Food and Drug Administration and the average content of THC in a marijuana cigarette; our first dose roughly corresponded to 1 marijuana cigarette containing 7% THC, the second dose corresponded to 2, and the higher dose corresponded to 4 marijuana cigarettes.24 However, according to data coming from the United States and Europe, we now know that strains of cannabis with a THC content of up to 14% are also used. In this case, our treatment would mimic the consumption of half (first dose), 1 (second dose) and 2 marijuana cigarettes (third dose). This protocol was based on the observation that the adverse consequences of cannabis use are most evident among heavy users in early adolescence.25
Fig. 1.
Δ9-Tetrahydrocannabinol (THC) treatment schedule in (A) adolescent and (B) adult female rats. (C) Protocol of chaetocin administration during adolescent THC treatment. i.p. = intraperitoneal; PND = postnatal day.
For cotreatment, we administered chaetocin intraperitoneally at the dose of 0.05 mg/kg once a day during THC treatment (Fig. 1C). No overt signs of toxicity were detected after chaetocin treatment. All experiments were performed according to the guidelines for care and use of experimental animals in the European Communities Council directive (2010/63/UE L 276 20/10/2010) and approved by the Ethical Committee for Animal Research at the University of Insubria and by the Italian Ministry of Health (Aut. N.302/2015-PR). All efforts were made to reduce the suffering and the number of animals used.
Behavioural test
The rats were subjected to the novel object recognition (NOR), social interaction (SI) and forced swim tests (FST) as previously described.13 For detailed information, see Appendix 1, available at jpn.ca/170082-a1. Behavioural data were expressed as means ± standard errors of the mean (SEM) and analyzed using 2-way analysis of variance (ANOVA) followed by a Tukey multiple comparison post hoc test.
Molecular assays
We obtained PFC samples from the rats via regional dissection on ice. The tissue was immediately frozen in liquid nitrogen and stored at −80°C until processing. Nuclear protein and histone extraction was performed as described in Appendix 1.
Western blot was performed as reported previously.12 For detailed information, see Appendix 1.
Total RNA was extracted from the PFC using the TRIzol protocol and incubated with DNase I to digest DNA contamination for 10 minutes at 25°C (RNase-Free DNase set, Qiagen Sciences Inc.). After the digestion, the RNA was purified and concentrated according to the manufacturer’s manual using the RNeasy MinElute Cleanup kit (Qiagen). The RNA concentration was ensured spectrophotometrically. All RNA samples had A260:280 ratios of 1.8:2.0 after purification using an RNeasy MinElute Cleanup kit. We reverse-transcribed 1 μg of total RNA from each sample into complementary DNA (cDNA) using an RT2 First Strand Kit (Qiagen) according to the manufacturer’s protocol.
Gene profiling was done using the RT2 Profile PCR Array Custom Rat Synaptic Plasticity kit (Qiagen; see the complete list of genes in Appendix 1, Table S1). We designed the array using a 96-well format to simultaneously analyze the expression of 41 genes for each experimental group (control v. THC). The panel included genes associated with the ECS (Cnr1, Faah, Mgll, Ppara and Pparg), the glutamatergic and GABAergic system (Gria1, Gria2, Grin1, Grin2a, Grin2b, Grm1, Grm2, Grm3, Grm5, Gabbr1, Gabbr2, Gabra1, Gabra2, Gabrb1, Gad1 and Abat) and the serotonergic system (Htr2a); the remodelling chromatin enzyme Sirt1; genes encoding proteins associated with plasticity (Bdnf, Dlg4, Ntrk12, Camk2a, Camk2g, Pdyn, Prkaca, Nos1, Creb1, Homer1, Pick1, Synpo, Crebbp, Arc, Reln and Adcy1); and housekeeping genes (Bact and Gapdh). In addition, 1 well contains a genomic DNA control, 3 wells contain reverse-transcription controls, and 3 wells contain a positive polymerase chain reaction (PCR) control. The realtime PCR cycling program was run on a Chromo4 thermal cycler (Biorad).
Gene expression was normalized to Gapdh because it was more stable, and no significant changes were detected between the groups. The threshold and baseline were set manually according to the manufacturer’s instructions. Gene expression was calculated using the PCR Array Data Analysis website (http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php). Values were obtained for the threshold cycle (Ct) for each gene, normalized using the Gapdh housekeeping gene on the same array, and then transformed into base-2 logarithms. Faah and Pparg Ct values were too high (> 35) to be detectable with this approach. Differences between groups were assessed using a Student t test with Welch correction or 2-way ANOVA followed by a Tukey multiple comparison post hoc test. Data were analyzed using GraphPad Prism 4 software.
Chromatin immunoprecipitation was performed as reported previously.26 The PFC was crosslinked in formaldehyde and underwent chromatin ultrasound fragmentation. For each sample, 40 mg of sonicated chromatin was immunoprecipitated with H3K9me3 antibody (AbCam). Subsequently, phenol-chloroform precipitation of DNA was followed by qualitative PCR analysis. To choose the specific target region to be analyzed by chromatin immunoprecipitation we searched the ENCODE data for levels of H3K9 trimethylation in the UCSC Genome browser for every analyzed gene in the most comparable annotated tissue (i.e., mouse whole brain). According to the literature, H3K9me3-enriched regions were mainly found in the gene bodies. Regarding Grin1, we designed primers encompassing the genomic region, including the proximal promoter and the first part of the transcribed sequence. For all other genes (Abat, Mgll, Homer1 and Dlg4), we selected intronic and exonic regions mapping in the gene bodies. Primer sequences are shown in Appendix 1, Table S2. The DNA enrichments are expressed over the input samples. We assessed differences between treated and control animals using a Student t test.
Results
Adolescent THC exposure impacts global histone modifications
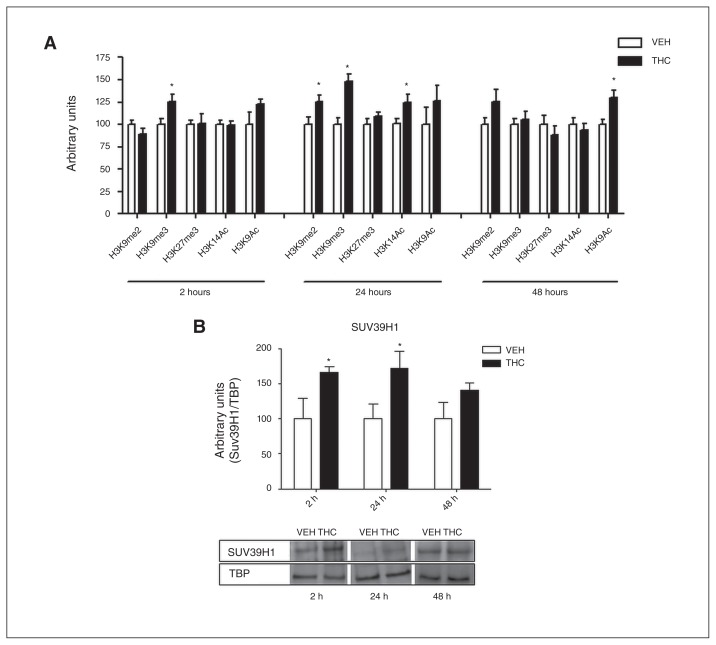
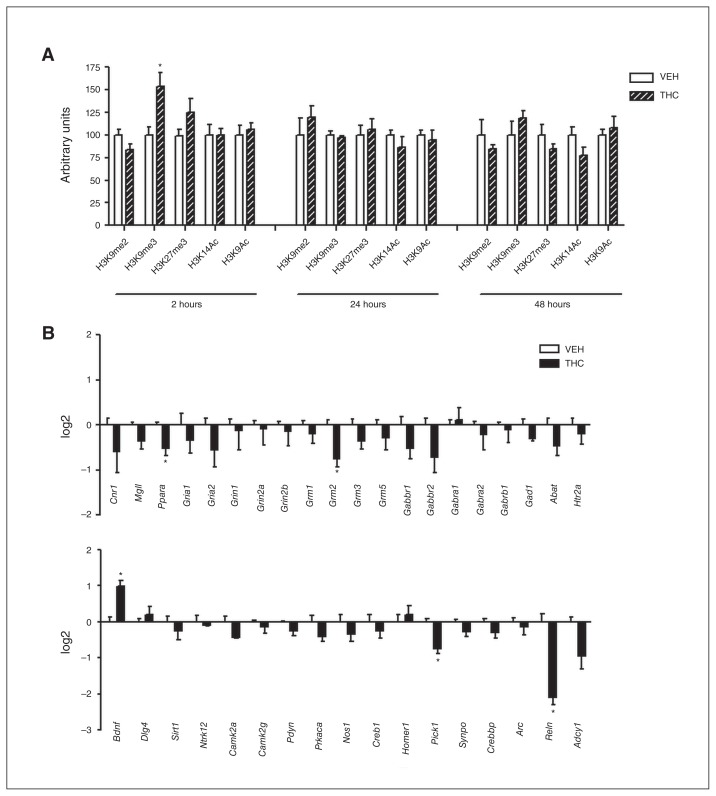
To assess the effect of adolescent THC exposure on histone modifications in the rat PFC, we performed a time-course analysis of 5 different histone H3 modifications (Fig. 2A). We investigated markers associated with both transcriptional repression (di- and trimethylation of lysine 9 [H3K9me2 and H3K9me3] as well as trimethylation of lysine 27 [H3K27me3]) and transcriptional activation (acetylation of lysine 9 [H3K9ac] and lysine 14 ]H3K14ac]) 2, 24 and 48 hours after the last THC injection. Two hours after the last THC injection, H3K9me3 was significantly increased (25%). Twenty-four hours later, the increase of H3K9me3 was evident with even enhanced intensity (48%) together with a significant increase of H3K9me2 (25%). Meanwhile, at 24 hours the positive histone mark H3K14ac started to increase (24%). Forty-eight hours after the last treatment, H3K9me3, H3K9me2 and H3K14ac returned to control values, whereas H3K9ac significantly increased (30%). This picture suggests that 2 waves of epigenetic modifications occur following chronic THC treatment, with the former (immediately after chronic THC exposure) being associated with transcriptional repression and the latter (at later time points) promoting a switch toward transcriptional activation.
Fig. 2.
(A) Effect of adolescent Δ9-tetrahydrocannabinol (THC) exposure on histone modifications 2, 24 and 48 hours after the last THC injection. Data are expressed as means ± standard errors of the mean (SEM) for at least 8 animals per experimental group. *p < 0.05 compared with controls (Student t test with Welch correction). (B) Effect of adolescent THC exposure on Suv39H1 levels 2, 24 and 48 hours after the last THC injection. Data are expressed as means ± SEM for at least 6 animals per experimental group. *p < 0.05 compared with controls (Student t test with Welch correction). TBP = tata binding protein; VEH = vehicle.
Increased H3K9me3 levels represent the main effect induced by adolescent THC exposure from 2 to 24 hours after the end of the treatment. The delayed increase (from 24 to 48 hours after treatment) in acetylation of H3K9/14 residues could instead be interpreted as a homeostatic mechanism aimed at counterbalancing the strong repressional state initially induced by THC. Since the main histone methyltransferases (HMTs) responsible for lysine 9 methylation are Suv39H1 and G9a,27–30 we next asked whether THC exposure during adolescence could alter the expression of these enzymes. Adolescent THC treatment induced a significant increase in Suv39H1 from 2 to 24 hours after the last injection, in the same time frame of H3K9me3 increase. Consistently, 24 hours later, when H3K9me3 returned to control levels, Suv39H1 protein expression also underwent normalization (Fig. 2B). Levels of Suv39H1 were already enhanced during the adolescent THC treatment (PND 39; Appendix 1, Fig. S1A). In contrast, THC treatment did not induce any alteration in G9a protein expression (Appendix 1, Fig. S1B).
Adolescent THC exposure alters gene expression
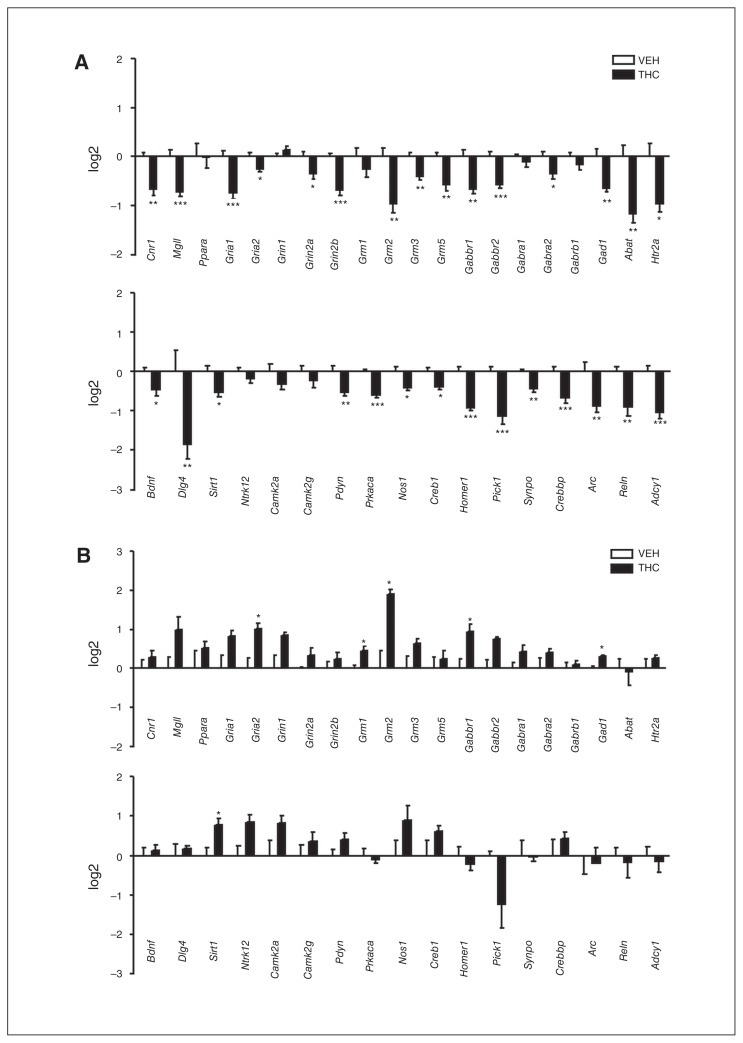
Because histone modifications impact transcriptional activity, we investigated the effect of adolescent THC exposure on gene expression. It is known that adolescence is characterized by intense processes of neuronal refinement,31 in which the ECS seems to play a crucial role.10 On this basis, we focussed our analyses on the expression of genes closely associated with the ECS and genes involved in plasticity processes. Two hours after the last THC injection statistical analysis revealed a significant decrease of Crebbp (−1.42 ± 0.13, n = 4, p < 0.01) and the immediate early gene Arc (−2.10 ± 0.44, n = 4, p < 0.01) messenger RNA (mRNA) levels. Twenty-four hours after the treatment (Fig. 3A), downregulation of THC-induced gene expression became more evident. Of the 37 plasticity-related genes analyzed, 29 underwent significant downregulation of mRNA levels. Alterations included genes belonging to the ECS (Cnr1 and Mgll), the glutamatergic system (Gria1, Gria2, Grin2a, Grin2b, Grm2, Grm3 and Grm5), the GABAergic system (Gabbr1, Gabbr2, Gabra2, Gad1 and Abat) and the serotonergic system (Htr2a) and genes encoding proteins involved in the modulation of neuronal plasticity (Bdnf, Dlg4, Sirt1, Pdyn, Prkaca, Nos1, Creb1, Homer1, Pick1, Synpo, Crebbp, Arc, Reln and Adcy1). This picture suggests that adolescent THC exposure induces significant transcriptional repression in this subset of plasticity genes. In contrast, 48 hours after the last THC injection (Fig. 3B) all the previously downregulated mRNAs returned to control levels, and Gria2, Grm2, Gabbr1, Gad1 and Sirt1 expression significantly increased over control values.
Fig. 3.
Effect of adolescent Δ9-tetrahydrocannabinol (THC) exposure on the expression of genes closely associated with the endocannabinoid system or synaptic plasticity (A) 24 hours and (B) 48 hours after the last THC injection. Data are expressed as log2 ± standard errors of the mean for at least 4 animals per experimental group. *p < 0.05, **p < 0.01, ***p < 0.001 (Student t test with Welch correction). VEH = vehicle.
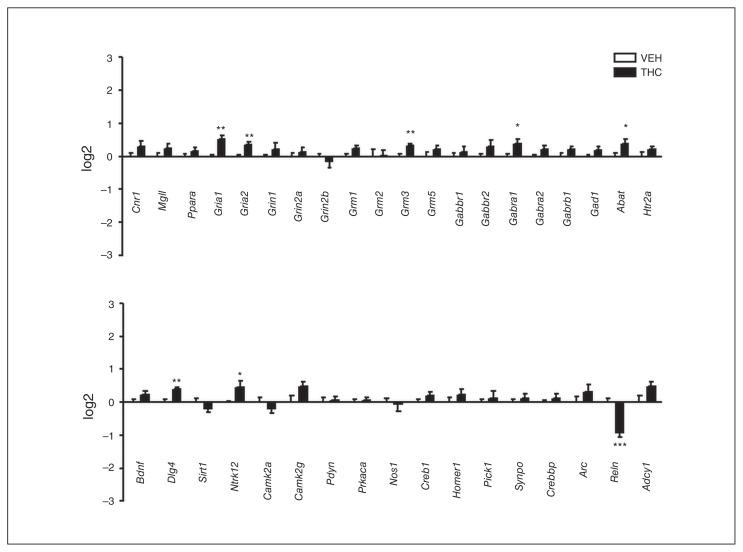
To evaluate long-term alterations in gene expression induced by adolescent THC exposure, we performed gene profiling at 75 PND (Fig. 4), when the depressive/psychotic-like phenotype becomes evident. Seven genes, Gria1, Gria2, Grm3, Gabra1, Abat, Dlg4 and Ntrk12, showed upregulated mRNA levels, whereas only 1 gene, Reln, showed significantly downregulated mRNA. Reelin level variations are highly relevant in psychotic disorders,32 hence we also investigated Reelin protein expression upon THC exposure. We found decreased levels of Reelin isoforms 410 KDa and 180 KDa, whereas Reelin 330 KDa was unchanged (Appendix 1, Fig. S2).
Fig. 4.
Effect of adolescent Δ9-tetrahydrocannabinol (THC) exposure on the expression of genes closely associated with the endocannabinoid system or synaptic plasticity at 75 postnatal days. Data are expressed as log2 ± standard errors of the mean for 8 animals per experimental group. *p < 0.05, **p < 0.01, ***p < 0.001 compared with controls (Student t test with Welch correction). VEH = vehicle.
THC-induced modifications of gene expression are accompanied by inherent changes in chromatin structure
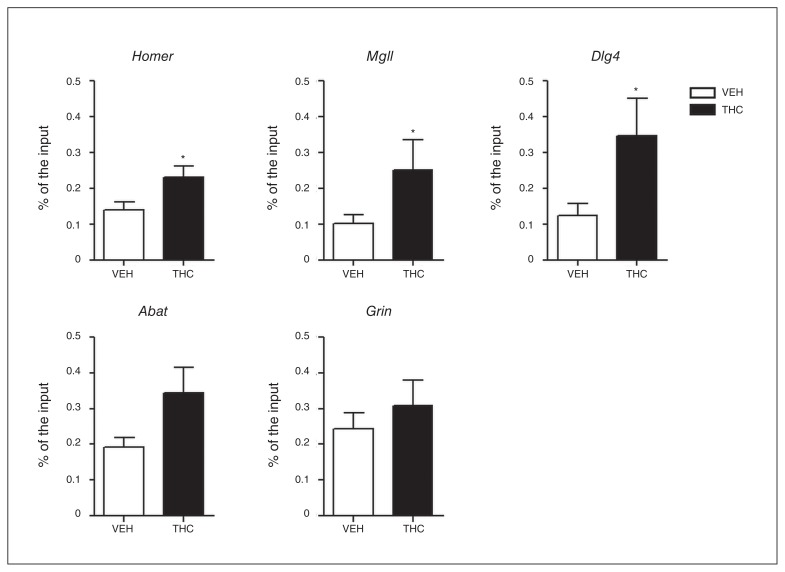
Because the major THC-dependent gene repression occurs 24 hours after the end of chronic treatment, we investigated whether the observed global increase in H3K9me3 specifically entailed chromatin changes in genes whose expression was modified by THC. We performed quantitative chromatin immunoprecipitation, selecting those genes displaying either stronger or highly significant modulation among all the THC-downregulated genes (Homer1, Mgll, Abat and Dlg4). In parallel we analyzed a gene, Grin1, whose expression was not modified after THC treatment. Analyzed genomic regions were selected within the gene bodies. H3K9me3 is generally distributed at the level of coding sequences rather than peaking at proximal promoter regions.33 Interestingly, we detected a THC-dependent increase of H3K9me3 levels in all the THC-downregulated genes, except for Abat. With regard to Grin1, whose expression was unaffected by the treatment, no differences were observed in terms of H3K9me3 between THC- and vehicle-treated rats (Fig. 5).
Fig. 5.
H3K9me3 levels at specific gene promotor bodies measured by chromatin immunoprecipitation–qualitative polymerase chain reaction 24 hours after the end of Δ9-tetrahydrocannabinol (THC) treatment. Values are displayed as fold enrichment over the input samples. Data are expressed as means ± standard errors of the mean for 10 animals per experimental group. *p < 0.05 compared with controls (Student t test with Welch correction). VEH = vehicle.
Adult v. adolescent THC exposure differentially affects histone modifications and gene expression
Because the onset of the depressive/psychotic-like phenotype is restricted to adolescent THC exposure,8–14 we investigated whether described alterations in histone modifications and gene expression were restricted to the adolescent treatment. We performed the same protocol of injections (Fig. 1B), and the same histone modifications and gene expression analysis in adult female rats. Two hours after the last THC injection, we observed a significant increase in H3K9me3 levels (54%) reminiscent to that of adolescent treatment. However, in contrast to the effects observed in adolescent rats, adult THC exposure did not lead to any significant alteration 24 and 48 hours after the end of THC treatment (Fig. 6A). The most divergent profile of THC-induced histone modifications between rats treated in adolescence and rats treated in adulthood was observed 24 hours after the end of THC administration, which is why we chose this time point to perform gene expression analysis in adult animals (Fig. 6B). Statistical analysis showed that only a few genes (Ppara, Grm2, Pick1, and Reln) were significantly downregulated in the PFC of adult THC–treated animals. Interestingly, Bdnf gene expression was significantly increased only in THC-treated adult rats. These data suggest that transcriptional repression induced by adult THC exposure was less intense and widespread than that induced by adolescent THC exposure.
Fig. 6.
(A) Effect of adult Δ9-tetrahydrocannabinol (THC) exposure on histone modification 2, 24 and 48 hours after the last THC injection. Data are expressed as means ± standard errors of the mean (SEM) for at least 4 animals per experimental group. *p < 0.05 compared with controls (Student t test with Welch correction). (B) Effect of adult THC exposure on the expression of genes closely associated with the endocannabinoid system or synaptic plasticity 24 hours after the last THC injection. Data are expressed as log2 ± SEM for 4 animals per experimental group. *p < 0.05 compared with controls (Student t test with Welch correction). VEH = vehicle.
Chaetocin-induced H3K9me3 inhibition during adolescent THC exposure prevents downregulation of gene expression and adult cognitive deficits
To investigate the possible role of H3K9me3 increase induced by adolescent THC exposure,8–13 we administered chaetocin, an inhibitor of the HMTs SUV39H1 and G9a,34,35 simultaneously with THC treatment. To establish the active dose of chaetocin in vivo, we first tested 2 different intraperitoneal doses (0.05 mg/kg and 0.1 mg/kg), and 24 hours after the injection, we investigated H3K9me3 levels. Only the 0.05 mg/kg dose of chaetocin significantly decreased H3K9me3 (Appendix 1, Fig. S3A). Further studies are needed to understand the reason why the higher chaetocin dosage was not effective in modulating SUV39H1 and, therefore, its specific target histone marker. Thus, we administered the 0.05 mg/kg dose once a day during the adolescent THC treatment. Twenty-four hours after the end of the treatment, we checked its effect on H3K9me3. As expected, THC induced a significant increase in H3K9me3 levels (p < 0.05), and chaetocin coadministration prevented such an increase (Appendix 1, Fig. S3B). Regarding gene expression, chaetocin coadministration prevented most of the THC-induced downregulation observed 24 hours after the end of the treatment. In fact, downregulation of 19 of 29 genes was prevented by chaetocin coadministration (Table 1), whereas the decrease of 10 mRNAs was not recovered by this cotreatment (Table 2).
Table 1.
Genes downregulated by Δ9-tetrahydrocannabinol and prevented by chaetocin coadministration
| Condition; mean ± SEM (n) | THC × chaetocin interaction | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Gene | Vehicle | Chaetocin | THC | THC + chaetocin | F | p value* |
| Cnr1 | 0.000 ± 0.080 (11) | 0.515 ± 0.179 (5) | −0.652 ± 0.148 (10) | 0.954 ± 0.174 (7) | F1–29 = 14.1 | < 0.001 |
| Mgll | 0.000 ± 0.138 (11) | 0.059 ± 0.227 (5) | −0.720 ± 0.083 (11) | 0.469 ± 0.192 (7) | F1–30 = 13.43 | < 0.001 |
| Gria1 | 0.000 ± 0.110 (11) | −0.056 ± 0.248 (5) | −0.734 ± 0.124 (10) | 0.161 ± 0.122 (7) | F1–29 = 10.72 | 0.003 |
| Grin2a | 0.000 ± 0.101 (12) | −0.304 ± 0.093 (5) | −0.346 ± 0.116 (14) | 0.393 ± 0.187 (7) | F1–33 = 11.23 | 0.002 |
| Grin2b | 0.000 ± 0.074 (11) | −0.474 ± 0.123 (5) | −0.682 ± 0.115 (13) | −0.086 ± 0.172 (7) | F1–32 = 17.04 | < 0001 |
| Grm2 | 0.000 ± 0.175 (9) | −0.337 ± 0.157 (5) | −0.951 ± 0.196 (11) | 0.211 ± 0.267 (7) | F1–28 = 11.63 | 0.002 |
| Grm3 | 0.000 ± 0.076 (9) | −0.050 ± 0.102 (5) | −0.395 ± 0.084 (12) | 0.401 ± 0.272 (7) | F1–28 = 7.21 | 0.012 |
| Grm5 | 0.000 ± 0.085 (10) | 0.386 ± 0.158 (5) | −0.573 ± 0.129 (12) | 0.999 ± 0.212 (7) | F1–30 = 15.32 | < 0.001 |
| Gabbr1 | 0.000 ± 0.141 (11) | 0.050 ± 0.255 (5) | −0.657 ± 0.095 (11) | 0.336 ± 0.280 (6) | F1–29 = 6.88 | 0.010 |
| Gabra2 | 0.000 ± 0.102 (11) | 0.144 ± 0.173 (5) | −0.349 ± 0.109 (13) | 0.217 ± 0.254 (7) | F1–31 = 5.56 | 0.025 |
| Gad1 | 0.000 ± 0.151 (11) | −0.403 ± 0.111 (4) | −0.640 ± 0.082 (14) | −0.360 ± 0.425 (7) | F1–32 = 12.09 | 0.001 |
| Bdnf | 0.000 ± 0.104 (11) | 0.438 ± 0.213 (5) | −0.465 ± 0.148 (10) | 1.349 ± 0.234 (6) | F1–28 = 7.84 | < 0.001 |
| Dlg4 | 0.000 ± 0.528 (12) | 0.018 ± 0.209 (5) | −1.855 ± 0.376 (14) | 0.755 ± 0.195 (7) | F1–34 = 7.07 | 0.010 |
| Prkaca | 0.000 ± 0.052 (12) | 0.171 ± 0.177 (5) | −0.593 ± 0.085 (12) | 0.477 ± 0.348 (7) | F1–32 = 7.21 | 0.010 |
| Nos1 | 0.000 ± 0.113 (10) | −0.114 ± 0.103 (5) | −0.332 ± 0.101 (13) | 0.886 ± 0.757 (7) | F1–29 = 5.56 | 0.030 |
| Creb1 | 0.000 ± 0.099 (11) | −0.356 ± 0.181 (5) | −0.310 ± 0.103 (14) | 0.147 ± 0.191 (7) | F1–31 = 17.35 | < 0.001 |
| Homer1 | 0.000 ± 0.123 (11) | −0.267 ± 0.183 (5) | −0.928 ± 0.073 (11) | 0.194 ± 0.149 (7) | F1–30 = 28.56 | < 0.001 |
| Crebbp | 0.000 ± 0.119 (10) | −0.450 ± 0.189 (5) | −0.676 ± 0.131 (14) | 0.135 ± 0.195 (7) | F1–32 = 14.78 | < 0.001 |
| Reln | 0.000 ± 0.119 (10) | 0.129 ± 0.108 (4) | −0.894 ± 0.226 (12) | −0.210 ± 0.251 (7) | F1–29 = 23.33 | < 0.001 |
SEM = standard error of the mean; THC = Δ9-tetrahydrocannabinol.
THC + chaetocin v. THC; 2-way analysis of variance followed by Tukey multiple comparison test.
Table 2.
Genes downregulated by Δ9-tetrahydrocannabinol and not prevented by chaetocin coadministration
| Condition; mean ± SEM (n) | THC × chaetocin interaction | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Gene | Vehicle | Chaetocin | THC | THC + chaetocin | F | p value* |
| Gria2 | 0.000 ± 0.074 (10) | 0.164 ± 0.133 (5) | −0.254 ± 0.050 (10) | 0.322 ± 0.160 (7) | F1–27 = 2.68 | 0.11 |
| Gabbr2 | 0.000 ± 0.099 (11) | 0.032 ± 0.157 (5) | −0.567 ± 0.079 (11) | 0.070 ± 0.373 (7) | F1–30 = 2.60 | 0.12 |
| Abat | 0.000 ± 0.227 (8) | −1.173 ± 0.398 (4) | −1.158 ± 0.183 (11) | −1.253 ± 0.402 (7) | F1–25 = 3.36 | 0.07 |
| Htr2a | 0.000 ± 0.273 (10) | −0.567 ± 0.128 (4) | −0.949 ± 0.175 (12) | −1.023 ± 0.314 (7) | F1–29 = 0.81 | 0.37 |
| Sirt1 | 0.000 ± 0.135 (10) | −0.503 ± 0.179 (5) | −0.523 ± 0.130 (13) | −0.356 ± 0.232 (6) | F1–30 = 3.80 | 0.06 |
| Pick1 | 0.000 ± 0.119 (9) | −0.370 ± 0.232 (5) | −1.121 ± 0.212 (12) | −0.614 ± 0.227 (6) | F1–28 = 4.00 | 0.06 |
| Pdyn | 0.000 ± 0.145 (11) | −0.191 ± 0.126 (4) | −0.541 ± 0.086 (13) | −0.211 ± 0.258 (6) | F1–30 = 2.46 | 0.13 |
| Synpo | 0.000 ± 0.052 (11) | 0.133 ± 0.196 (5) | −0.429 ± 0.107 (13) | −0.267 ± 0.522 (7) | F1–31 = 3.92 | 0.06 |
| Arc | 0.000 ± 0.229 (12) | −0.529 ± 0.632 (5) | −0.870 ± 0.181 (11) | 0.318 ± 0.336 (7) | F1–30 = 1.08 | 0.31 |
| Adcy1 | 0.000 ± 0.135 (11) | 0.154 ± 0.300 (4) | −1.040 ± 0.163 (14) | −0.114 ± 0.239 (7) | F1–32 = 3.34 | 0.08 |
SEM = standard error of the mean; THC = Δ9-tetrahydrocannabinol.
THC + chaetocin v. THC; 2-way analysis of variance.
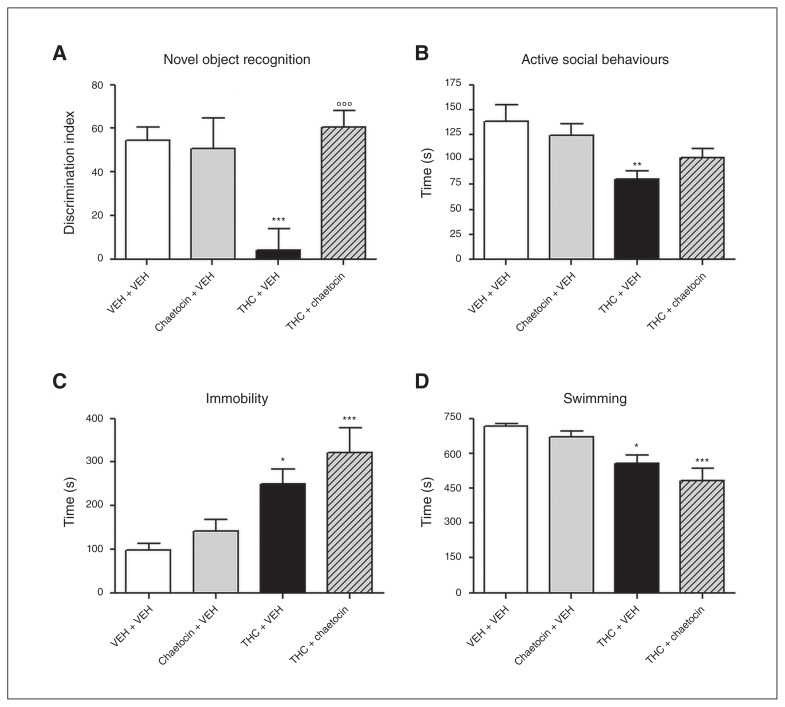
Finally, to investigate the effect of H3K9me3 inhibition on the development of the depressive/psychotic-like phenotype, adult animals (PND 75) that had been pre-exposed to THC and chaetocin during adolescence were submitted to a battery of behavioural tests. In the NOR test, 2-way ANOVA showed a significant effect of THC treatment (F1–32 = 38.52, p < 0.001) as well as a significant effect of chaetocin (F1–32 = 63.63, p < 0.001) and a significant THC × chaetocin interaction (F1–32 = 84.68, p < 0.001). In fact, as expected, adult rats pre-exposed to THC showed a lower discrimination index (DI) than controls (p < 0.001); the DI did not differ between controls and rats cotreated with THC and chaetocin (Fig. 7A). There were no differences in total exploration time between experimental groups (data not shown), suggesting the absence of an anxiety state or alterations in motor activity. In the SI test, 2-way ANOVA showed a significant effect of THC treatment only (F1–30 = 12.43, p < 0.01). In fact, the time spent in active social behaviours was significantly reduced (42%) in animals pretreated with THC. With chaetocin coadministration, animals showed a milder reduction in social behaviour (27%), which did not reach the significance threshold when compared with control animals (Fig. 7B). In the FST, 2-way ANOVA showed a significant effect of THC treatment only (Fig. 5E and F). Adult animals pre-exposed to THC spent more time in immobility (F1–32 = 19.63, p < 0.001) and less time swimming (F1–32 = 23.96, p < 0.001), suggesting the presence of a passive coping strategy. However, these parameters were not prevented by chaetocin coadministration (Fig. 7C and 7D).
Fig. 7.
Effects of chaetocin administration during adolescent Δ9-tetrahydrocannabinol (THC) exposure on adult behaviour (postnatal day 75): (A) discrimination index in the novel object recognition test, (B) active social behaviours in the social interaction test, (C) immobility and (D) swimming parameters in the forced swim test. Data are expressed as means ± standard errors of the mean for at least 5 animals per group. *p < 0.05, **p < 0.01, ***p < 0.001 compared with controls; °°°p < 0.001 compared with THC-treated animals (2-way analysis of variance followed by Tukey post hoc test). VEH = vehicle.
Discussion
The present study shows that adolescent THC exposure in female rats induces alterations in specific histone modifications in the PFC, with complex kinetics entailing a first round of global increased trimethylation of H3K9, followed by a wave of histone H3 acetylation with an opposite outcome at the transcriptional level. Interestingly, alterations in both histone modifications and gene expression were more intense and long-lasting following adolescent THC treatment compared with adult treatment (a further graphical representation of our results is summarized in Fig. 8 and Fig. 9). This differential effect clearly indicates age-dependency in THC-induced molecular alterations, at least in the PFC, shedding new light on molecular mechanisms underlying adolescent susceptibility to substance-induced psychopathologies. The PFC is mostly involved in the modulation of cognitive and emotional behaviour through top–down inhibition of subcortical affective circuitry,36,37 playing an important role in the onset of cannabis-induced neuropsychiatric disorders.38 Notably, THC-induced epigenetic changes impact the expression of a set of genes associated with plasticity, likely contributing to circuitry maladaptation instrumental to disease pathogenesis. These changes include genes encoding some elements of the cAMP transduction pathways together with Homer1, Pick1, Pdyn and Reln. However, we cannot exclude that gene repression mediated by THC could also involve other genes that are not associated with neuronal plasticity. Moreover, H3K9me3 increases 24 hours after THC treatment, and this modification is likely mediated by the upregulation of Suv39H1, a histone methyltransferase specifically involved in the trimethylation of H3K9. Meanwhile, transcription of selected genes associated with plasticity is silenced, which is in line with the development of cognitive impairments later in life. Indeed, the pharmacological inhibition of specific HMTs and consequent H3K9me3 normalization during THC treatment is able to prevent the onset of THC-mediated cognitive deficits in adulthood.
Fig. 8.
Comparison of effects induced by adolescent and adult Δ9-tetrahydrocannabinol (THC) exposure. PND = postnatal day.
Fig. 9.
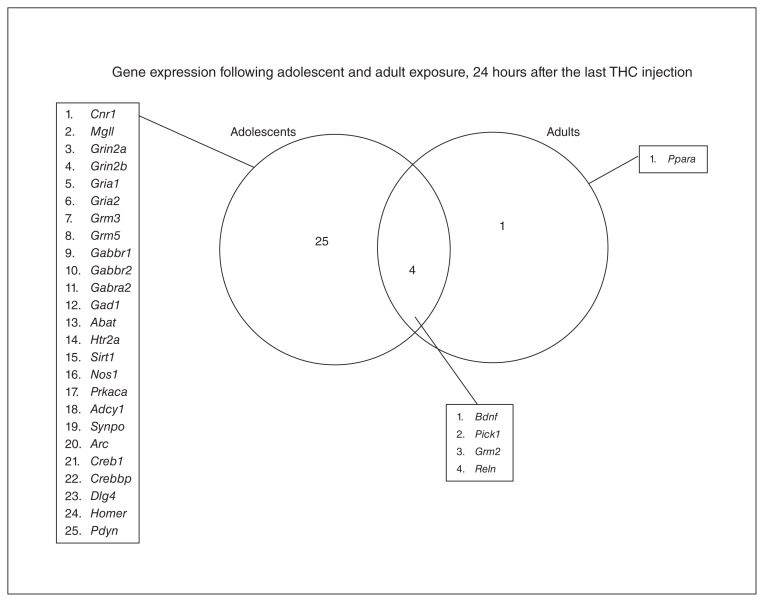
Gene expression following adolescent and adult Δ9-tetrahydrocannabinol (THC) exposure, 24 hours after the last THC injection. Twenty-five genes are specifically downregulated after adolescent THC exposure, whereas only 1 gene (Ppara) is specifically downregulated after adult THC exposure. The overlap section shows genes downregulated by both treatments, except for Bdnf, which is downregulated after adolescent THC exposure but upregulated after adult THC exposure.
The enhanced histone H3 acetylation observed from 24 to 48 hours after THC treatment may represent either a direct effect of THC, displaying a delayed onset compared with THC-mediated H3K9me3 increase, or the result of a homeostatic response directly induced by the aberrant increase of SUV39H1 and consequent H3K9me3 accumulation. In both cases, active histone H3 acetylation should represent an epigenetic mechanism that counteracts the strong transcriptional repression mediated by SUV39H1 overexpression. Indeed, 48 hours after the end of THC treatment, when H3K9 acetylation is promoted, a strong transcriptional reactivation of those same genes that were repressed at 24 hours occurs. Strikingly, 8 of these genes have altered expression at 75 PND and appear to be particularly important for the depressive/psychotic-like phenotype described in THC-exposed animals. Among these genes, Grm3, Gabra, Abat and Dlg4 are components of the glutamatergic and GABAergic systems and have been shown to play a relevant role in the pathophysiology of schizophrenia and mood disorders.39–43 Moreover, 2 other genes whose THC-mediated deregulation during adolescence persisted at 75 PND, Gria1 and Reln, have also been found to be modified in postmortem studies performed in the cortex of patients with schizophrenia.44–47 Thus, the 2 main THC-induced epigenetic events, the early (0–24 h) H3K9me3 upregulation followed by a delayed (24–48 h) increase of H3 acetylation, dramatically change the physiologic expression of the analyzed genes. These genes may play a crucial role in the correct developmental trajectory of the adolescent brain, thus their aberrant expression could affect the shaping of adult brain organization. Consistently, these alterations in gene expression fluctuations may drive changes in the PFC maturational events occurring, for example, at glutamatergic and GABAergic synapses after heavy adolescent THC exposure, as previously described.10,12 This leads to an adult brain characterized by altered functionality likely underpinning cognitive impairments present in the depressive/psychotic-like phenotype observed in female rats after adolescent THC exposure.10,11 Thus, hindering the first event (the increase in Suv39H1/H3K9me3) through chaetocin administration, we prevented, at least in part, the flow of events that lead to impairments in brain and behaviour (Fig. 10).
Fig. 10.
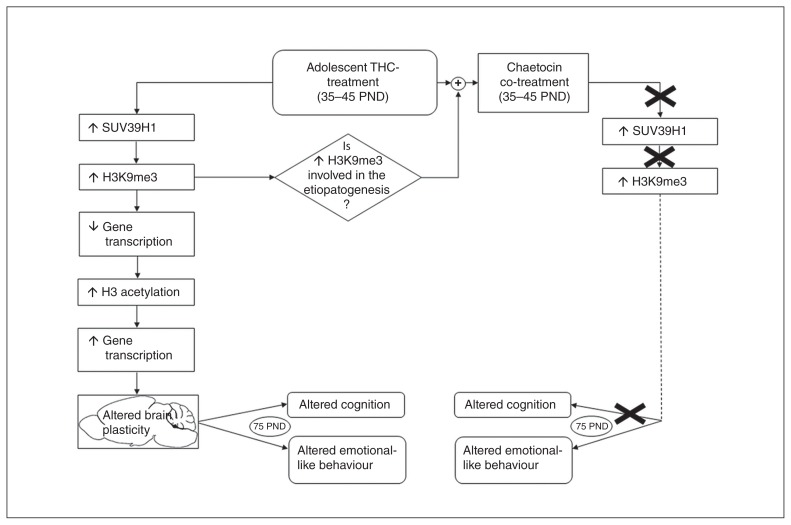
Flow of events induced by adolescent Δ9-tetrahydrocannabinol (THC) exposure and prevented by chaetocin coadministration. PND = postnatal day.
In support of evidence for major vulnerability of the adolescent brain to the long-lasting adverse effects of cannabinoids, it is worth commenting on the different results we obtained after adult and adolescent THC treatments. Exposure to THC at both ages induced a significant increase in H3K9me3 2 hours after the last THC injection. However, at the later time points, adult THC exposure did not induce any significant alteration. Thus, we can speculate that the increase in H3K9me3 is closely associated with THC treatment, regardless of the time of administration, whereas the later histone alterations (H3K9me2, H3K14ac and H3K9ac) represent a feature of adolescent exposure.
The different epigenetic response between the adolescent and adult brain might play a role in the mechanisms involved in the long-term behavioural impairments induced by adolescent, but not adult, THC exposure.11 We cannot assess whether the different epigenetic/transcriptional response induced by THC administration in adolescent compared with adult animals was due to an age-specific THC effect on the epigenome or to a differential homeostatic response promoted by H3K9me3 upregulation occurring only in young rats.
A picture emerges only in adolescent rats in which transcriptional increase of THC target genes is promoted to counteract their early THC-mediated transcriptional repression. However, this seemingly compensatory mechanism aberrantly triggers a pathologic transcriptional facilitation of a subset of genes associated with disease-inherent long-lasting effects, ultimately modifying PFC circuitry with a maladaptive mean.
Furthermore, in the adult rat brain we found upregulation of the gene coding for Bdnf, a neurotrophin that regulates synaptic function and plasticity in the mature nervous system.48 Recent studies suggest an important protective role of Bdnf in alcohol use disorders49 or implicate Bdnf in the medial PFC as a negative regulator of cocaine-seeking.50,51 Based on these observations, we might speculate that the increase in Bdnf mRNA levels restricted to adult THC exposure could represent a protective response. If this is the case, the lack of such adaptation after adolescent THC exposure is consistent with the observation that the adolescent brain is more vulnerable to THC effects.
Impairments in chromatin remodelling enzymes are increasingly being recognized as playing a crucial role in the development and maintenance of addictive disorders. The crucial role of the HMT G9a was shown in the structural and behavioural plasticity induced by cocaine.21 More interestingly, chronic amphetamine exposure induced histone H3 methylation (K4 and K9) on the c-fos promoter in parallel with the increase in protein amounts of the HMT Suv39H1.52 Consistently, our direct analysis of the histone marker H3K9me3 at selected genes revealed a correlation between THC-mediated transcriptional downregulation and increase of this repressive histone marker, strengthening the hypothesis that transcriptional repression is operated at least in part by chromatin structure modification at specific genes. However, given the global THC-mediated increase of H3K9me3, we should also take into consideration the possibility that this modification might occur at specific intergenic regions generating heterochromatic domains that might play an indirect effect on gene expression. It is therefore possible that the increase of Suv39H1 we observed in our study could be a potential mechanism through which THC acts on chromatin to induce long-lasting effects. Indeed, daily coadministration of THC and chaetocin, an inhibitor of the HMTs acting on H3K9,34,35 prevented the increase in H3K9me3 as well as the large gene downregulation induced by THC in the PFC. Specifically, chaetocin prevented the repression of 19 of 29 genes, suggesting that their expression could be mediated by H3K9me3 levels. Remarkably, these genes mainly belong to the ECS and the glutamatergic synapse. It has been suggested that the composition of the ECS may be a general feature of most glutamatergic synapses in the brain,53 including the PFC.54 Impairments of PFC glutamatergic transmission are detrimental to PFC-dependent cognitive processes, such as the ones evaluated with the NOR test.55 Thus, we can speculate that preventing glutamatergic and endocannabinoid downregulation of genes by chaetocin could account for the lack of cognitive deficits observed after adolescent THC exposure.
In contrast, the lack of effect of chaetocin on Abat downregulation and on downregulation of 9 other genes (Gria2, Gabbr2, Sirt1, Pick1, Pdyn, Synpo, Arc, Adcy1 and Htr2a) implies that these genes might be regulated by mechanisms different from H3K9me3. Accordingly, chromatin immunoprecipitation results did not detect a significant increase in H3K9me3 levels in the Abat gene. At the behavioural level, chaetocin administration during adolescent THC exposure prevented the development of cognitive deficits in adulthood, with only a partial effect, if any, on emotional behaviour. Lack of complete behavioural recovery, in particular for the emotional phenotype, could reflect the existence of 2 distinct THC downstream pathways: a former, where H3K9me3 upregulation represents the first step of a chain of events, principally impacting cognition (Fig. 10), and a latter, independent of increased H3K9me3, more related to affective behaviour. This suggests that THC alterations in an emotional behaviour pathway could be triggered by other histone modifications, such as H3 acetylation, occurring in other brain areas. Further studies are warranted to explore whether drugs inhibiting these other modifications might hold promise in reverting the adverse effects of adolescent cannabinoid exposure.
Limitations
Our study should be considered together with some limitations. We focused our study only on female rats, and because a recent study56 has suggested that the sexes differ in cannabinoid signalling and thus in response to THC, we cannot extend these results to male rats. Moreover, our gene expression analyses were limited to a specific subset of genes closely associated with the endocannabinoid system and brain plasticity, thus we cannot exclude that other genes may be affected by adolescent THC exposure.
Conclusion
Through a mechanism involving SUV39H1, THC modifies histone modifications and, thereby, expression of plasticity genes. This pathway appears to be relevant for the development of cognitive deficits. To our knowledge, chaetocin has been used for the first time as a possible modulator of behavioural traits relevant to psychiatric disorders, thus suggesting that H3K9me3 could be considered as a possible target for therapeutic interventions.
Acknowledgements
This work was supported by Fondazione Zardi Gori (bando 2014) and Dipartimento delle Politiche Antidroga (EpiCa) to T. Rubino, F. Rusconi (Cariplo 2014-0972) and E. Battaglioli (Telethon GGP14074). The authors thank Giorgio Binelli for his help with statistical analysis.
References
- 1.Di Forti M, Sallis H, Allegri F, et al. Daily use, especially of high-potency cannabis, drives the earlier onset of psychosis in cannabis users. Schizophr Bull. 2014;40:1509–17. doi: 10.1093/schbul/sbt181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lev-Ran S, Roerecke M, Le Foll B, et al. The association between cannabis use and depression: a systematic review and meta-analysis of longitudinal studies. Psychol Med. 2014;44:797–810. doi: 10.1017/S0033291713001438. [DOI] [PubMed] [Google Scholar]
- 3.Wilkinson ST, Radhakrishnan R, D’Souza DC. Impact of cannabis use on the development of psychotic disorders. Curr Addict Rep. 2014;1:115–28. doi: 10.1007/s40429-014-0018-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.United Nations Office on Drugs. World Drug Report 2016. [accessed 2017 Sept 29]. Available: www.unodc.org/wdr2016/
- 5.Chadwick B, Miller ML, Hurd YL. Cannabis use during adolescent development: susceptibility to psychiatric illness. Front Psychiatry. 2013;4:129. doi: 10.3389/fpsyt.2013.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Renard J, Krebs MO, Le Pen G, et al. Long-term consequences of adolescent cannabinoid exposure in adult psychopathology. Front Neurosci. 2014;8:361. doi: 10.3389/fnins.2014.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rubino T, Parolaro D. Cannabis abuse in adolescence and the risk of psychosis: a brief review of the preclinical evidence. Prog Neuropsychopharmacol Biol Psychiatry. 2014;52:41–4. doi: 10.1016/j.pnpbp.2013.07.020. [DOI] [PubMed] [Google Scholar]
- 8.Rubino T, Vigano D, Realini N, et al. Chronic delta 9-tetrahydrocannabinol during adolescence provokes sex-dependent changes in the emotional profile in adult rats: behavioral and biochemical correlates. Neuropsychopharmacology. 2008;33:2760–71. doi: 10.1038/sj.npp.1301664. [DOI] [PubMed] [Google Scholar]
- 9.Rubino T, Realini N, Braida D, et al. The depressive phenotype induced in adult female rats by adolescent exposure to THC is associated with cognitive impairment and altered neuroplasticity in the prefrontal cortex. Neurotox Res. 2009;15:291–302. doi: 10.1007/s12640-009-9031-3. [DOI] [PubMed] [Google Scholar]
- 10.Rubino T, Prini P, Piscitelli F, et al. Adolescent exposure to THC in female rats disrupts developmental changes in the prefrontal cortex. Neurobiol Dis. 2015;73:60–9. doi: 10.1016/j.nbd.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 11.Realini N, Vigano D, Guidali C, et al. Chronic URB597 treatment at adulthood reverted most depressive-like symptoms induced by adolescent exposure to THC in female rats. Neuropharmacology. 2011;60:235–43. doi: 10.1016/j.neuropharm.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 12.Zamberletti E, Beggiato S, Steardo L, Jr, et al. Alterations of prefrontal cortex GABAergic transmission in the complex psychotic-like phenotype induced by adolescent delta-9-tetrahydrocannabinol exposure in rats. Neurobiol Dis. 2014;63:35–47. doi: 10.1016/j.nbd.2013.10.028. [DOI] [PubMed] [Google Scholar]
- 13.Zamberletti E, Gabaglio M, Prini P, et al. Cortical neuroinflammation contributes to long-term cognitive dysfunctions following adolescent delta-9-tetrahydrocannabinol treatment in female rats. Eur Neuropsychopharmacol. 2015;25:2404–15. doi: 10.1016/j.euroneuro.2015.09.021. [DOI] [PubMed] [Google Scholar]
- 14.Rubino T, Realini N, Braida D, et al. Changes in hippocampal morphology and neuroplasticity induced by adolescent THC treatment are associated with cognitive impairment in adulthood. Hippocampus. 2009;19:763–72. doi: 10.1002/hipo.20554. [DOI] [PubMed] [Google Scholar]
- 15.Tsankova N, Renthal W, Kumar A, et al. Epigenetic regulation in psychiatric disorders. Nat Rev Neurosci. 2007;8:355–67. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- 16.Akbarian S, Nestler EJ. Epigenetic mechanisms in psychiatry. Neuropsychopharmacology. 2013;38:1–2. doi: 10.1038/npp.2012.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szutorisz H, Hurd YL. Epigenetic effects of cannabis exposure. Biol Psychiatry. 2016;79:586–94. doi: 10.1016/j.biopsych.2015.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ryu H, Lee J, Hagerty SW, et al. ESET/SETDB1 gene expression and histone H3 (K9) trimethylation in Huntington’s disease. Proc Natl Acad Sci U S A. 2006;103:19176–81. doi: 10.1073/pnas.0606373103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aguado T, Carracedo A, Julien B, et al. Cannabinoids induce glioma stem-like cell differentiation and inhibit gliomagenesis. J Biol Chem. 2007;282:6854–62. doi: 10.1074/jbc.M608900200. [DOI] [PubMed] [Google Scholar]
- 20.Al-Mahdawi S, Pinto RM, Ismail O, et al. The Friedreich ataxia GAA repeat expansion mutation induces comparable epigenetic changes in human and transgenic mouse brain and heart tissues. Hum Mol Genet. 2008;17:735–46. doi: 10.1093/hmg/ddm346. [DOI] [PubMed] [Google Scholar]
- 21.Maze I, Covington HE, III, Dietz DM, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–6. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomasiewicz HC, Jacobs MM, Wilkinson MB, et al. Proenkephalin mediates the enduring effects of adolescent cannabis exposure associated with adult opiate vulnerability. Biol Psychiatry. 2012;72:803–10. doi: 10.1016/j.biopsych.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang X, Hegde VL, Rao R, et al. Histone modifications are associated with Δ9-tetrahydrocannabinol-mediated alterations in antigen-specific T cell responses. J Biol Chem. 2014;289:18707–18. doi: 10.1074/jbc.M113.545210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zamberletti E, Prini P, Speziali S, et al. Gender-dependent behavioral and biochemical effects of adolescent delta-9-tetrahydrocannabinol in adult maternally deprived rats. Neuroscience. 2012;204:245–57. doi: 10.1016/j.neuroscience.2011.11.038. [DOI] [PubMed] [Google Scholar]
- 25.Coffey C, Patton GC. Cannabis use in adolescence and young adulthood: a review of findings from the Victorian Adolescent Health Cohort Study. Can J Psychiatry. 2016;61:318–27. doi: 10.1177/0706743716645289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rusconi F, Grillo B, Ponzoni L, et al. LSD1 modulates stress-evoked transcription of immediate early genes and emotional behavior. Proc Natl Acad Sci U S A. 2016;113:3651–6. doi: 10.1073/pnas.1511974113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Carroll D, Scherthan H, Peters AH, et al. Isolation and characterization of Suv39h2, a second histone H3 methyltransferase gene that displays testis-specific expression. Mol Cell Biol. 2000;20:9423–33. doi: 10.1128/mcb.20.24.9423-9433.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rea S, Eisenhaber F, O’Carroll D, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–9. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 29.Tachibana M, Sugimoto K, Fukushima T, et al. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem. 2001;276:25309–17. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- 30.Lin Y, Dong C, Zhou BP. Epigenetic regulation of EMT: the Snail story. Curr Pharm Des. 2014;20:1698–705. doi: 10.2174/13816128113199990512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tau GZ, Peterson BS. Normal development of brain circuits. Neuropsychopharmacology. 2010;35:147–68. doi: 10.1038/npp.2009.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Folsom TD, Fatemi SH. The involvement of Reelin in neurodevelopmental disorders. Neuropharmacology. 2013;68:122–35. doi: 10.1016/j.neuropharm.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen L, Shao NY, Liu X, et al. diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS ONE. 2013;8:e65598. doi: 10.1371/journal.pone.0065598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greiner D, Bonaldi T, Eskeland R, et al. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3–9. Nat Chem Biol. 2005;1:143–5. doi: 10.1038/nchembio721. [DOI] [PubMed] [Google Scholar]
- 35.Cherblanc FL, Chapman KL, Reid J, et al. On the histone lysine methyltransferase activity of fungal metabolite chaetocin. J Med Chem. 2013;56:8616–25. doi: 10.1021/jm401063r. [DOI] [PubMed] [Google Scholar]
- 36.Arnsten AF, Rubia K. Neurobiological circuits regulating attention, cognitive control, motivation, and emotion: disruptions in neurodevelopmental psychiatric disorders. J Am Acad Child Adolesc Psychiatry. 2012;51:356–67. doi: 10.1016/j.jaac.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 37.Clarke R, Johnstone T. Prefrontal inhibition of threat processing reduces working memory interference. Front Hum Neurosci. 2013;7:228. doi: 10.3389/fnhum.2013.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bossong MG, Niesink RJ. Adolescent brain maturation, the endogenous cannabinoid system and the neurobiology of cannabis-induced schizophrenia. Prog Neurobiol. 2010;92:370–85. doi: 10.1016/j.pneurobio.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 39.Yamada K, Watanabe A, Iwayama-Shigeno Y, et al. Evidence of association between gamma-aminobutyric acid type A receptor genes located on 5q34 and female patients with mood disorders. Neurosci Lett. 2003;349:9–12. doi: 10.1016/s0304-3940(03)00611-6. [DOI] [PubMed] [Google Scholar]
- 40.Horiuchi Y, Nakayama J, Ishiguro H, et al. Possible association between a haplotype of the GABA-A receptor alpha 1 subunit gene (GABRA1) and mood disorders. Biol Psychiatry. 2004;55:40–5. doi: 10.1016/s0006-3223(03)00689-9. [DOI] [PubMed] [Google Scholar]
- 41.Kristiansen LV, Beneyto M, Haroutunian V, et al. Changes in NMDA receptor subunits and interacting PSD proteins in dorsolateral prefrontal and anterior cingulate cortex indicate abnormal regional expression in schizophrenia. Mol Psychiatry. 2006;11:737–47. doi: 10.1038/sj.mp.4001844. [DOI] [PubMed] [Google Scholar]
- 42.Harrison PJ, Lyon L, Sartorius LJ, et al. The group II metabotropic glutamate receptor 3 (mGluR3, mGlu3, GRM3): expression, function and involvement in schizophrenia. J Psychopharmacol. 2008;22:308–22. doi: 10.1177/0269881108089818. [DOI] [PubMed] [Google Scholar]
- 43.Jia Peilin, Wang Lily, Meltzer Herbert Y, et al. Common variants conferring risk of schizophrenia: a pathway analysis of GWAS data. Schizophr Res. 2010;122:38–42. doi: 10.1016/j.schres.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guidotti A, Auta J, Davis JM, et al. Decrease in reelin and glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder: a postmortem brain study. Arch Gen Psychiatry. 2000;57:1061–9. doi: 10.1001/archpsyc.57.11.1061. [DOI] [PubMed] [Google Scholar]
- 45.Eastwood SL, Harrison PJ. Cellular basis of reduced cortical reelin expression in schizophrenia. Am J Psychiatry. 2006;163:540–2. doi: 10.1176/appi.ajp.163.3.540. [DOI] [PubMed] [Google Scholar]
- 46.O’Connor JA, Hemby SE. Elevated GRIA1 mRNA expression in Layer II/III and V pyramidal cells of the DLPFC in schizophrenia. Schizophr Res. 2007;97:277–88. doi: 10.1016/j.schres.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cass DK, Flores-Barrera E, Thomases DR, et al. CB1 cannabinoid receptor stimulation during adolescence impairs the maturation of GABA function in the adult rat prefrontal cortex. Mol Psychiatry. 2014;19:536–43. doi: 10.1038/mp.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang EJ, Reichardt LF. Neurotrophins: roles in neuronal development and function. Annu Rev Neurosci. 2001;24:677–736. doi: 10.1146/annurev.neuro.24.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Logrip ML, Barak S, Warnault V, et al. Corticostriatal BDNF and alcohol addiction. Brain Res. 2015;1628:60–7. doi: 10.1016/j.brainres.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berglind WJ, See RE, Fuchs RA, et al. A BDNF infusion into the medial prefrontal cortex suppresses cocaine seeking in rats. Eur J Neurosci. 2007;26:757–66. doi: 10.1111/j.1460-9568.2007.05692.x. [DOI] [PubMed] [Google Scholar]
- 51.Whitfield TW, Jr, Shi X, Sun WL, et al. The suppressive effect of an intra-prefrontal cortical infusion of BDNF on cocaine-seeking is Trk receptor and extracellular signal-regulated protein kinase mitogen-activated protein kinase dependent. J Neurosci. 2011;31:834–42. doi: 10.1523/JNEUROSCI.4986-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Renthal W, Carle TL, Maze I, et al. Delta FosB mediates epigenetic desensitization of the c-fos gene after chronic amphetamine exposure. J Neurosci. 2008;28:7344–9. doi: 10.1523/JNEUROSCI.1043-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Katona I, Urbán GM, Wallace M, et al. Molecular composition of the endocannabinoid system at glutamatergic synapses. J Neurosci. 2006;26:5628–37. doi: 10.1523/JNEUROSCI.0309-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lafourcade M, Elezgarai I, Mato S, et al. Molecular components and functions of the endocannabinoid system in mouse prefrontal cortex. PLoS ONE. 2007;2:e709. doi: 10.1371/journal.pone.0000709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yuen EY, Wei J, Liu W, et al. Repeated stress causes cognitive impairment by suppressing glutamate receptor expression and function in prefrontal cortex. Neuron. 2012;73:962–77. doi: 10.1016/j.neuron.2011.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rubino T, Parolaro D. Sex-dependent vulnerability to cannabis abuse in adolescence. Front Psychiatry. 2015;6:56. doi: 10.3389/fpsyt.2015.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]