Abstract
Modern biomedical imaging has revolutionized life science by providing anatomical, functional and molecular information of biological species with high spatial resolution, deep penetration depth, enhanced temporal responsiveness, and improved chemical specificity. In recent years, these imaging techniques have been increasingly tailored for characterizing biomaterials and probing their interactions with biological tissues. This in turn has spurred substantial advances in engineering material properties to accommodate different imaging modalities that was previously unattainable. Here we review advancements in engineering both imaging modalities and material properties with improved capacity, providing a timely practical guide to better assess biomaterial-tissue interactions both in vitro and in vivo.
Keywords: Biomaterials, imaging, contrast agent, tissue engineering, regenerative medicine
The Need to Visualize Biomaterial-Tissue Interactions
Biomaterials have been playing increasingly pivotal roles in biomedicine by, for example, serving as scaffolds to support engineering functional tissues in regenerative medicine [1] and carriers to deliver bioactive agents and/or therapeutic molecules in precision medicine [2]. To optimize the performance of biomaterials, it is desirable to visualize biomaterials-tissue interactions with minimal invasiveness and high fidelity [3–5]. Imaging biomaterial-tissue interactions has been historically challenging [6], because it usually requires proper imaging contrast to be engineered into the biomaterials and/or the cells. It becomes even more challenging in in vivo settings due to the increased imaging depth, complex tissue environment, and interference from the functioning biological system.
To address this challenge, coherent efforts from imaging science and materials science have been spent over the past decades, resulting in tremendous achievements that have revolutionized biomaterials research. On one hand, modern biomedical imaging has been specially tailored to image biomaterials with significantly enhanced spatial resolution, penetration depth, temporal response, detection sensitivity, and chemical specificity; on the other hand, engineering approaches have been advanced to endow biomaterials/cells with novel contrast mechanisms, enabling characterization of biomaterial-tissue interactions at relative ease. In this Review, we first summarize the advantages and limitations of different imaging modalities for characterizing biomaterial-tissue interactions, followed by discussing physical backgrounds that enable clear visualization of such interactions by these various imaging modalities. We then briefly outline design principles of next-generation contrasts with enhanced properties as well as application-specific choice over the most suitable imaging techniques. We finally conclude with perspectives on expanding our imaging capacity with unconventional toolkits and potential improvements through the use of multi-modality imaging. We aim to provide a practical guide for materials scientists to choose the optimal imaging methods and contrast mechanisms to characterized biomaterial-tissue interactions.
Modern Biomedical Imaging Modalities for Characterizing Biomaterials/Tissues
All types of biomedical imaging modalities rely on the interactions between the applied probing energy format (e.g., light, sound, magnetic field, or x-ray photon) and the biomaterials/tissues (Fig. 1a). These interactions generally include absorption, scattering, and polarization of the probing energy by the objects. There are many different criteria to group imaging technologies, such as resolution, imaging depth, and contrast mechanism. Here, we specifically choose the contrast mechanism as it is most relevant to the physical, chemical, and biological properties of a given material or tissue type, and thus can provide the most practical guide for selecting the best-matched technologies for probing biomaterials and biological entities. As such, the common biomedical imaging modalities can be classified as acoustic imaging, magnetic imaging, optical imaging, electron imaging, x-ray imaging, and nuclear imaging by their contrast mechanisms. We discuss each type of the imaging modalities for potential use in characterizing biomaterial-tissue interactions, sorted by their probing energies from low to high, summarized in Table 1. Notably, the probing energy level of an imaging modality is a good indicator of its damage potential: a higher probing energy holds a higher damage potential and thus should be used with more cautions, particularly for dedicated biomaterials and studies involving vulnerable cell types.
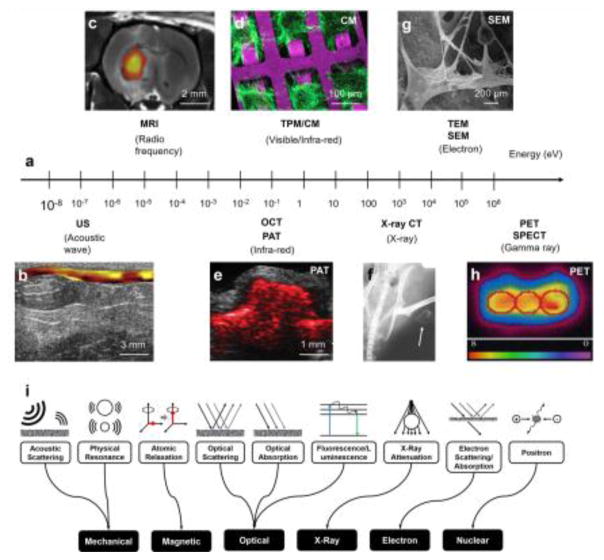
Figure 1. Biomedical imaging of biomaterials.
(a) Representative biomedical imaging modalities used in biomaterial characterization and their corresponding probing energies. US, ultrasound; MRI, magnetic resonance imaging; PAT, photoacoustic tomography; OCT, optical coherence tomography; CM, confocal microscopy; MPM, multi-photon microscopy; TEM, transmission electron microscopy; SEM, scanning electron microscopy; x-ray CT, x-ray computed tomography; PET, positron-emission tomography; SPECT, single-photon position emission tomography. (b) US elasticity imaging of a polyurethane-based tissue construct in a rat abdominal repair model at week 12. The tissue stiffness is colored from yellow (soft) to red (hard). Adapted with permission from [8]. (c) MRI image of transplanted 18F-labeled human neural stem cells (shown in color) and ECM scaffold remodeling in the stroke-damaged rat brain. Adapted with permission from [15]. (d) Confocal microscopy image of cardiomyocytes grown on a poly(glycerol sebacate) (PGS) scaffold. Adapted with permission from [20]. (e) PAT image of SWCNT-PLGA scaffold acquired at 680 nm embedded in a chicken breast tissue at the depth of 0.5 mm. SWCNT, single-walled carbon nanotube; PLGA, poly(lactic-co-glycolic acid). Adapted with permission from [7]. (f) X-ray CT image of thigh muscle pouches of a nude mouse at week 12 after implantation of zein scaffolds with rabbit mesenchymal stem cells. The white arrows indicate ectopic bone formation. Adapted with permission from [25]. (g) SEM image of a porous scaffold prepared from a composite of PLGA and hydroxyapatite (Hap), and the deposition of minerals from preosteoblasts cultured on the scaffold for 28 days. Adapted with permission from [48]. (h) PET image of transverse sections of a bioreactor chamber containing a cardiomyocyte-encapsulated fibrin scaffold under perfusion culture showing metabolic activity of the cells an hour after 18FDG labeling. 18FDG, 18F-fluordeoxyglucose. Adapted with permission from [29]. (i) Schematics showing contrast mechanisms to enhance imaging capacity.
Table 1.
Property summary of the biomedical imaging modalities.
| Imaging modality |
Contrast mechanism |
Typical spatial resolution |
Typical penetration depth |
Advantages | Disadvantages | Representative applications |
|---|---|---|---|---|---|---|
| US | Acoustic reflection (back scattering) | 0.3 mm | 10 cm | Non-invasive, high speed, deep penetration | Low resolution, low chemical sensitivity, coupling medium needed | Mechanics, flow dynamics, scaffold cavitation |
| MRI | Proton magnetization and relaxation (spinning frequency) | 1 mm | 50 cm | Non-invasive, deep penetration | Expensive, low imaging speed | Water content and transport |
|
MPM CM |
Fluorescent emission, optical scattering | 1 μm | 1 mm | Non-invasive, cellular-level resolution, high chemical sensitivity | Superficial penetration | Cell attachment of scaffolds, gene expression |
| OCT | Optical back scattering | 10 μm | 2 mm | Non- invasive, cellular-level resolution, high imaging speed | Superficial penetration, low chemical sensitivity | Vascularization, cell tracking, scaffold degradation |
| PAT | Optical absorption | 0.1 mm | 10 cm | Non-invasive, high functional and chemical sensitivity deep penetration | Coupling medium needed | Vascularization, oxygenation, cell tracking |
| X-ray imaging | X-ray absorption | 0.1 mm | 40 cm | Non-invasive, deep penetration, high resolution | Ionizing radiation, low chemical sensitivity | Engineered bone, pore structures |
| TEM/SEM | Electron scattering | 1 nm | 0.1 μm | Nano-scale resolution | Invasiveness (needing sample fixation), complex sample preparation, superficial penetration | Cell-material interaction, mineralization |
| PET/SPECT | Gamma emission | 5 mm | 50 cm | Non-invasive, deep penetration, high molecular sensitivity | Low resolution, radiative labelling | Cell metabolism, cell tracking |
US, ultrasound imaging; MRI, magnetic resonance imaging; CM, confocal microscopy; MPM, multi-photon microscopy; OCT, optical coherence tomography; PAT, photoacoustic tomography; TEM, transmission electron microscopy; SEM, scanning electron microscopy; PET, positron-emission tomography; SPECT, single-photon emission computed tomography
Acoustic imaging of mechanics
Ultrasound (US) imaging is a commonly used imaging modality that can provide three-dimensional (3D) anatomical information of the engineered tissue constructs and newly formed tissues at depths up to tens of centimetres [7, 8]. Because of the low acoustic wave energy, US imaging is considered the safest imaging method with high imaging speeds and penetration depths. So far, different US imaging methods have been applied to studying the mechanical properties of the biomaterials and their interactions with surrounding tissues, especially their mechanical properties, including the mass density, modulus, and cavitation. For example, conventional B-mode US imaging has been used to assess the mineralization (i.e., the change in mass density) in collagen hydrogels [9]; Doppler US imaging has been used to track the flow dynamics in engineered vascular grafts and the vascularization of hydrogels in vivo [10]; US elastography has been used to monitor the degradation of scaffolds (i.e., changes in modulus) in vivo (Fig. 1b) [8]. US imaging has two major drawbacks: i) it lacks sensitivity to the functional and molecular status of the biomaterials and tissues (e.g., chemical composition and functional status) due to its mechanical contrast mechanism; and ii) its spatial resolution can hardly reach the single-cell levels due to the long acoustic wavelengths used for forming the images.
Magnetic imaging of water content
Magnetic resonance imaging (MRI) can detect the water content or the proton density of an object [11] and has long been applied to monitoring the development of engineered tissues, including adipose [12], cartilage [13], and vasculature [14], with endogenous or exogenous contrasts (Fig. 1c) [15]. MRI has also been applied to track the stem cell differentiation and degradation of engineered scaffolds and hydrogels, with the specific macromolecules bonded by functionalized super paramagnetic iron oxide nanoparticles [16–18]. MRI is non-invasive and non-ionizing, and hence it has been widely applied to in vivo small-animal studies of tissue-biomaterials interactions, operating at a high magnetic field (>7T) with much improved spatial resolutions [19]. Nevertheless, MRI is an expensive imaging modality with limited accessibility for the general research community. Moreover, the influence of the strong magnetic field on normal cell functions and small animal behaviour remains to be demonstrated conclusively.
Optical imaging of chemical composition
With its flexible imaging contrasts, high spatial resolutions, and ease of use, optical imaging has become an indispensable tool in essentially any biomedical laboratory. Optical imaging is most sensitive to the chemical information of an object. Among these techniques, confocal microscopy (CM) and multi-photon microscopy (MPM) have been extensively used to image biomaterials and tissues in vitro and in vivo, typically with fluorescence markers (Fig. 1d) [20]. The cell viability in 3D scaffolds can be assessed based on the intracellular auto-fluorescence of nicotinamide adenine dinucleotide (NADH) [21]. Photoacoustic tomography (PAT) has been proven useful in evaluating the tissue angiogenesis and oxygenation saturation of hemoglobin in vivo in hydrogel scaffolds (Fig. 1e) [22, 23]. An exception lies in optical coherence tomography (OCT), which is based on scattering contrast and insensitive to the chemical composition, is a powerful tool in evaluating the 3D structural change of the engineered scaffolds caused by biomaterial degradation and tissue matrix deposition [24]. Overall, optical imaging is the dominant method for charactering the biomaterial-tissue interactions, but they collectively suffer from the relatively shallow penetration depth due to the strong optical attenuation by both biological tissues and biomaterials.
X-ray, electron, and nuclear imaging of material structures at scales
Using high-energy electromagnetic waves, electron, X-ray, and nuclear imaging have also been widely used for characterizing material’s structures at dramatically different scales. It is worth noting that we group the three imaging modalities together because they possess the most damage potential to the imaged materials, due to their high probing energy.
Conventional X-ray imaging based on x-ray attenuation has very high sensitivity to hard structures such as engineered bones (Fig. 1f) [25], but low sensitivity to soft tissue and biomaterials in vivo [26]. Phase-contrast X-ray imaging can provide greatly improved sensitivity to image polymer scaffolds in culture and implanted in tissues [27]. Nevertheless, X-ray imaging requires ionizing radiation and thus can potentially damage samples. Electron microscopy (EM), including scanning electron microscopy (SEM) and transmission electron microscopy (TEM), has been instrumental in the nanoscale characterization of biomaterials, especially cell-biomaterial interactions (Fig. 1g) [28]. The nanoparticle dimension, the scaffold pore size, and the fiber orientation are all commonly imaged by electron microscopy. For biomaterials, perhaps no other imaging methods can provide a higher-resolution image of the intimate cell-biomaterial interfaces. The downsides of EM include its expensive maintenance, thin penetration depth, challenging sample preparation, and limited ability to image specimens containing live cells. Nuclear imaging, including both positron-emission tomography (PET) and single-photon emission computed tomography (SPECT), can monitor engineered tissues and cells labelled by radioactive tracers (Fig. 1h) [29]. The advantage of nuclear imaging includes its deep penetration (whole human body) and the picomolar-level detection sensitivity. One common application of PET is to monitor the metabolism of cells by using 18F-fluordeoxyglucose (FDG), a radiative glucose analogue. However, nuclear imaging has poor spatial resolution due to the tissue scattering of the gamma rays and the random annihilation events. Therefore, nuclear imaging is normally combined with MRI or X-ray CT to provide more accurate structural and molecular information.
Engineering Properties of Materials and Tissues to Enable Visual Characterization
As mentioned above, while the various types of imaging modalities can all directly visualize certain properties of specimens of interest, the sensitivity achieved may not be satisfactory to characterize the desired biomaterial-tissue interactions. Consequently, it is often necessary to introduce exogenous contrast mechanisms to either the non-biological materials or the biological entities, or both, to make them clearly visible under the selected imaging modalities.
Physicochemical contrast mechanisms
Because a given imaging modality is usually capable of imaging only a certain property of the sample, rarely can both biomaterials and biological entities be imaged at the same time when a single modality is applied. However, if this property sufficiently differs between the material and the cell, the two components may still be distinguished, which is achieved usually through exogenously introduced contrast mechanisms. The contrast mechanism strongly depends on the imaging modality, spanning across acoustic scattering and physical resonance for mechanical imaging, atomic relaxation for magnetic resonance imaging, optical scattering, absorption, and fluorescence/luminescence for optical imaging, X-ray attenuation for CT imaging, electron scattering for electron imaging, and positron for nuclear imaging (Fig. 1i).
For example, most biomaterials, due to the relatively high refractive indices for sound and mechanical properties, possess contrasts for US imaging and US elasticity imaging themselves to enable mechanical imaging of scaffold degradation [30] or regeneration of engineered cartilage tissues [31]. An effective means to distinguish biomaterial scaffolds from the surrounding cells or tissues would then be increasing the acoustic scattering of the biomaterials. This typically requires the use of high-density biomaterials such as polymer scaffolds, leaving water-rich, less-scattering hydrogels unsuitable for US imaging. Alternatively, contrast agents, mostly gas-filled synthetic microbubbles or biogenic nanostructures that work via resonating to achieve rapid contractions and expansion upon pressure changes of the sound wave when US is applied, may be used [32]. Contrarily, MRI that relies on characterizing atomic relaxation, could easily image water-rich hydrogels that are similar in relaxation properties with biological tissues [33, 34]. Differentiating the scaffolds and the cells grown inside, nevertheless, still necessitates external labels, such as magnetic nanoparticles for the cells [35, 36].
Several contrast mechanisms exist for different optical imaging modalities. Optical scattering enabled by polymeric biomaterials and the cell membrane allows for convenient OCT imaging both in vitro and in vivo [37–39]. The contrast between the cells and the scaffolds may be improved through labels that feature higher refractive indices such as magnetic beads [37] and gold nanocages [40]. Besides scattering contrasts, absorption and fluorescence have also found broad applications in optical imaging, which usually require the introduction of external contrast mechanisms. While numerous fluorescent molecules have been developed and optimized to enable confocal and multi-photon microscopy, or whole-body luminescence imaging including the use for studying scaffold degradation [41], PAT is a more recent imaging modality based on absorption contrast that is gaining increasing interest in observing cell-scaffold interactions. Contrasts including those based on chemical agents such as chromogenic dyes [42], formazans metabolized by living cells [43] or doped in scaffolds [44], or gold nanoparticles and carbon nanomaterials [45, 46], are among the common mechanisms to improve the signals of biomaterials and/or cells.
Due to the unique contrast mechanism relying on attenuation of X-rays, CT imaging has been almost exclusively used for studying hard tissues (i.e., bones) that possess intrinsic contrasts, including engineered bone tissues [47, 48]. It is anticipated that adopting existing CT contrast agents, such as those based on metal [49] and iodine-containing [50] nanoparticles, may expand the scope of CT imaging to other tissue types beyond bone. EM, on the other hand, is extensively applied to imaging various cells, tissues, and biomaterials at extraordinarily high resolution but thin depth. While SEM achieves surface imaging of the specimens often without the need for external contrasts, TEM imaging of polymers and cells in ultrathin sections usually requires additional staining with heavy metal-containing dyes to improve the contrast [51].
Compared to the other imaging modalities discussed above, PET has been scarcely utilized in characterizing scaffold-tissue interactions, possibly owing to its limited availability and relatively low spatial resolution. In one early example, 18F-FDG, a common positron-emitting contrast agent employed clinically, was used to label cardiomyocytes embedded in a fibrin matrix for PET imaging [29]. The use of 18F-FDG and other radioactive species can be, in theory, expanded to many other cell types of interest and even the labelling of biomaterial scaffolds.
Engineered biological contrast mechanisms
Contrast agents externally introduced into cells may be inefficient because the uptake efficacy could be low and this process may adversely affect cell behaviors. Alternatively, efforts have also been put into developing contrast mechanisms including fluorescence, luminescence, and chromogenic, mainly suited for optical imaging methods, through genetically engineering target cells. Although the purification of green fluorescent protein was initially reported in the 1960s [52], its utility in providing labels in biological species did not begin to be realized until the 1990s when they were expressed in bacterial and eukaryotic cells [53]. Since then, numerous variants of fluorescent proteins have been derived to effectively label cells to enable wide-spectrum fluorescence imaging [54], for applications spanning from molecular biology to tissue engineering. Similarly, bioluminescence, in this case referring to light emitted spontaneously without the need for excitation, has also been widely used in biomedicine for visualizing engineered cells. It can be generated either by activation of a protein by another molecule, such as in the case of aequorin activation by calcium [52, 55], or by chemoreactions of a substrate with an enzyme, such as in the case of luciferase oxidation of luciferin [56].
Besides luminescence/fluorescence, chromogens have also been adopted for various imaging applications of biological species where absorption is the primary contrast mechanism. Unfortunately, this has been conventionally limited to thin layers of samples since most pure optical imaging modalities are hindered by the inability to penetrate deep and extract proper volumetric information. Until recently, the advent of PAT successfully rescued this insufficiency by providing 3D imaging capacity of chromogenic contrast mechanisms genetically engineered into the cells, including, for example, LacZ reporter that encodes β-galactosidase to convert X-gal molecules into blue precipitates in the cells [57], low-extinction coefficient fluorescent proteins for enhanced photothermal conversion [58–60], tyrosinase-melanin [61, 62], and chromoproteins that enable direct chromogen production [63].
The majority of these engineered biological contrast mechanisms have been limited to the use in optical imaging, primarily due to the much greater compatibility of these mechanisms with improvement of optical properties. However, other possible ways to generate contrasts, such as in situ synthesis of nanomaterials directly within biological cells using green chemistry [64], similar to cellular metabolism of tetrazolium salts to formazan crystals in the case of PAT imaging [43], may provide additional opportunities for other imaging modalities as well, such as US, OCT, MRI, and EM. Significant investigations may need to be conducted to enhance the biocompatibility of these methods.
Roadmap for Selecting Imaging Systems to Probe Biomaterial-Tissue Interactions
The modern biomedical imaging modalities have been proven indispensable for characterizing a variety of biomaterials and their interactions with biological tissues in vitro and in vivo. Each imaging modality has its pros and cons, as any technology does, and thus should be applied to the best-matched need for the optimal outcomes. To provide a roadmap for selection of the optimal imaging modalities for probing biomaterial-tissue interactions, we have classified these interactions into several major categories, including morphology, vascularization, molecular expression, and ECM deposition, which are all important for understanding the performance of the biomaterials as substrates to enhance tissue regeneration. For example, cell attachment and proliferation on/in the biomaterial scaffolds and scaffold degradation are main morphological characteristics of biomaterial-tissue interactions; during the processes of further interactions, the cells may continue to deposit ECM components, and change their phenotypes through maturation and/or differentiation, marked by expression of particular molecular imprints. Furthermore, vascularization is a critical component in engineering functional tissues, and the invasion of the blood vessels from the surrounding tissue, how they interact with the biomaterial scaffolds, and their functional maturation in supporting blood flows can all determine the performance of the biomaterials [65].
Once an intended interaction of interest is defined, the intrinsic contrasts of the biomaterial and the biological entity (cell or tissue) should be immediately identified, whether they are scattering, absorption, fluorescence, or others. These intrinsic contrasts should be registered to the various imaging modalities to screen for these techniques on their ability in characterizing these existing contrasts, with respect to parameters such as spatial resolution and penetration depth (see the “Modern Biomedical Imaging Modalities for Characterizing Biomaterials/Tissues” section). In general, biomaterials present better intrinsic contrasts than biological components since it is usually their physical properties that are to be characterized, but this strongly depends on the imaging modality. Only very few cell types, such as erythrocytes and melanoma cells, possess intrinsic contrasts that potentially enable label-free imaging. However, the physical properties of the tissues, such as blood flow, may be readily characterized using US-based imaging without need for any labelling. In case the intrinsic contrasts cannot meet the requirements for proper characterization, extrinsic contrast mechanisms may be analyzed to enhance the capacity of the imaging modalities to visualize the desired properties of the biomaterial and the biological component (see the “Engineering Properties of Materials to Enable Visual Characterization” section). Besides the contrast mechanism of the specimens, resolution, and penetration depth, other considerations may include the invasiveness – whether live imaging is required or if end-point assessment is sufficient, the speed of imaging to fit the time frame needed for completing the characterization, and the availability of the existing imaging modalities.
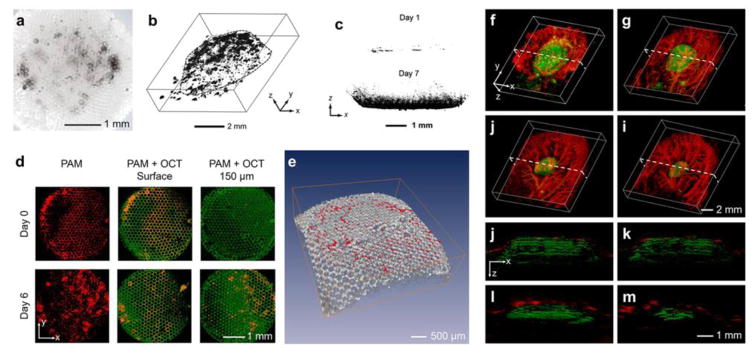
Fig. 2 shows a few concrete examples that illustrate how different imaging modalities can be selected based on the specific properties of the biomaterials and biological components to be characterized for desired interactions. As aforementioned, melanoma cells containing the intrinsic contrast melanin can be directly imaged with absorption contrast-reliant imaging modalities. When these cells are seeded in a polymer scaffold made from poly(lactic-co-glycolic acid) (PLGA), they are easily seen by bright-field optical imaging, which provides however, only surface imaging (Fig. 2a) [38]. Relying on the same absorption contrast mechanism, when switched to PAT for imaging the melanoma cells in the scaffolds, depth information can be further revealed (Fig. 2b) [66]. If other cell types other than melanoma cells are used, then exogeneous contrasts, such as formazan crystals, need to be introduced for them to be properly visualized using the same PAT platform (Fig. 2c) [43]. Similarly, the scaffolds may be imaged with either intrinsic or external contrasts by choosing the suitable imaging modality. These PLGA scaffolds possess strong intrinsic scattering contrast, and therefore, if an imaging technique based on such a contrast is used, the scaffolds should be able to be characterized. As shown in Fig. 2d–e, a dual-modality platform combining OCT and PAT could nicely characterize the proliferation of melanoma cells in a PLGA scaffold by imaging both the cells through the absorption contrast with PAT and the scaffold through the scattering contrast with OCT [38]. But if the dual-modality system is not available, the biomaterial component may still be imaged with only a single PAT platform by doping the material with an absorption contrast agent such as formazan to differentiate it from the biological component, such as the invading vasculature, using a multiple-wavelength scanning strategy (Fig. 2f–m) [44]. These examples illustrate the decision-making process involved in determining how contrast mechanisms and imaging modalities are selected to best match the needs for the specific biomaterial-tissue interactions to be characterized.
Figure 2. Interrogating biomaterial-tissue interactions.
(a) Bright-field optical image showing melanoma cells grown in a porous PLGA scaffold. Adapted with permission from [38]. (b) PAT reconstruction image showing 3D distribution of melanoma cells in a PLGA scaffold. Adapted with permission from [66]. (c) PAT images showing invasion of NIH/3T3 fibroblasts into PLGA scaffolds at days 1 and 7; the cells were stained with formazan to generate exogenous contrast. Adapted with permission from [43]. (d) Top view of the PAT/OCT images showing the ingrowth of melanoma cells from the surface into the center of the PLGA scaffolds. The melanoma cells were imaged by the PAT subsystem whereas the scaffold by the OCT subsystem, both in a label-free manner. (e) Volumetric rendering of the OCT-imaged scaffold (gray) with PAT-imaged melanoma cells (red). Adapted with permission from [38]. (f–i) Co-registered 3D reconstruction PAT images showing both the degradation of an individual PLGA scaffold and the remodeling of vasculature simultaneously in vivo. (j–m) Co-registered cross-sectional PAT images at the dotted planes as indicated in (f–i), respectively. Adapted with permission from [44].
Conclusions and Perspectives
We have reviewed advances in both imaging modalities that enable better characterization of biomaterial-tissue interactions as well as contrast mechanisms that allow for improved visualization by these different imaging modalities. We further proposed some design principles in improving such capacities, which is of critical importance to fields ranging from tissue engineering and regenerative medicine to drug delivery and cancer theranostics.
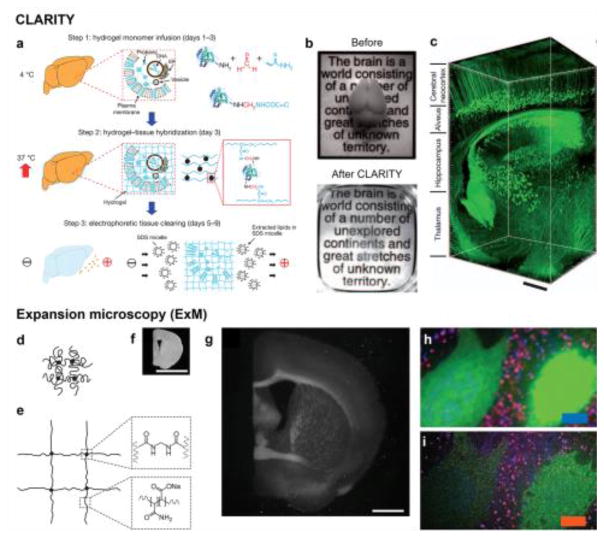
In addition to hardware refinements for imaging modalities and advances in contrast mechanisms that have advanced our ability to study tissue-biomaterial interactions, recent years have witnessed a shift towards developing unconventional approaches for improving our ability to interrogate biological tissues (and possibly biomaterials). Two major breakthroughs include CLARITY and expansion microscopy (ExM). CLARITY adopts a tissue clearing approach, where a biological specimen is first embedded in a hydrogel matrix, followed by electrophoretic removal of highly scattering molecules, in particular the lipids in the membranes, from the cells (Fig. 3a) [67]. As such, the biological tissue becomes immediately transparent (Fig. 3b), allowing a much deeper penetration depth with an optical imaging modality compared with that before clearance (Fig. 3c). ExM entirely overcomes the conventional needs for hardware or software improvements in optical microscopy, such as high-end optics or stimulated illumination and associated algorithms [68, 69]. On the contrary, in ExM, the biological specimen is also embedded in a hydrogel matrix, after which it is physically expanded through swelling, to 4–5 times its original size (Fig. 3d–g), thus achieving magnification and super-resolution without the need to alter any existing configurations of the optical imaging modality (Fig. 3h–i) [70]. Interestingly, since ExM also removes all undesired molecules from the fixed samples, they automatically become transparent, similar to the effect of CLARITY, in addition to the physical expansion. Although these unconventional toolkits are still entirely limited to optical imaging, they may find future opportunities in expanding into use for various imaging modalities with further optimization.
Figure 3. Advanced technologies in tissue processing for enhanced optical imaging capability.
(a–c) CLARITY. (a) Procedures for CLARITY: tissue is crosslinked with formaldehyde (red) in the presence of hydrogel monomers (blue), covalently linking tissue elements to monomers that are then polymerized into a hydrogel mesh (followed by a day-4 wash step); electric fields are subsequently applied across the sample in ionic detergent actively transport micelles into, and lipids out of, the tissue, leaving fine structure and crosslinked biomolecules in place. (b) Images of an adult mouse brains (3-month old) before and after CLARITY. (c) Confocal reconstructed image of a non-sectioned mouse brain tissue showing cortex, hippocampus, and thalamus (310 objective; stack size, 3,400 mm; step size, 2 mm). Scale bar: 400 μm. Adapted with permission from [77]. (d–i) ExM. (d, e) Schematics of (d) collapsed polyelectrolyte network, showing crosslinker (dot) and polymer chain (line), and (e) expanded network after H2O dialysis. (f, g) Photographs of (f) a fixed mouse brain slice and (g) the sample post-ExM. Scale bars: 5 mm. Note that the size of (g) is 4.5 times that of (f) to give direct comparison of the same specimen pre-/post-expansion. (h, i) Confocal fluorescence images of a mouse brain slice stained with presynaptic (anti-Bassoon, blue) and postsynaptic (anti-Homer1, red) markers, in addition to antibody to GFP (green), (h) pre- versus (i) post-expansion. Scale bar: 5 μm. Adapted with permission from [70]. SDS, sodium dodecyl sulfate.
Meanwhile, as imaging technologies evolve with new capabilities, several considerations may be taken to improve their biomaterial-specific performance besides the adoption of proper contrast mechanisms. For US imaging, the spatial resolution can be substantially improved by the newly developed super-resolution methods [71]. For optical imaging including PAT, the imaging depth may be potentially improved with the state-of-the-art wavefront engineering technologies that can compensate for the tissue scattering [72]. For MRI and X-ray CT, small-animal implementations with compressed sensing technologies need be developed to accelerate the imaging speed while maintaining the spatial resolutions [73]. For EM, sample preparation methods that do not need thin cutting or conductive coating will great improve its accessibility for biomaterial research [74]; alternatively, the combination of e-beam layer-by-layer etching followed by in situ imaging has been shown to generate volumetric SEM imaging data [75]. For nuclear imaging, combination with more high-resolution imaging technologies such as PAT and US imaging can provide a more comprehensive understanding of structural and functional changes during the tissue engineering process [76].
Ultimately, the development of specialized multi-modality imaging systems and multiplexed contrast mechanisms, possibly in combination with new toolkits beyond these two conventional considerations, will likely eventually enable multi-scale characterization of biomaterial-tissue interactions in high capacity (see Outstanding Questions). These developments will in turn facilitate our understanding of their respective pros and cons, allowing us to choose the optimal imaging modalities and contrasts for probing specific biomaterial-tissue interactions.
Outstanding Questions Box.
Can the imaging modalities be combined in a way that they are customizable to individual imaging requirements for interrogating biomaterial-tissue interactions?
Can the contrast mechanisms be engineered to enable multi-modality imaging of both biomaterials and biological entities without interfering with their intrinsic properties?
Can the unconventional toolkits, or their variations, be expanded to a significantly wider variety of imaging modalities beyond optical imaging?
Trends Box.
Interrogating biomaterials-tissue interactions with minimal invasiveness and high fidelity through imaging analyses is desired.
Various imaging modalities that are based on individually unique contrast mechanisms can be used to characterize biomaterial-tissue interactions in different manners.
Contrast mechanisms are constantly undergoing revolution and enable improved visualization of biomaterial-tissue interactions with relevant imaging modalities.
Unconventional toolkits have also been developed to promote the ability to image biological tissues and possibly biomaterials.
The development of multi-modality imaging platforms and multiplexed contrast mechanisms in combination with new toolkits will allow for multi-scale characterization of biomaterial-tissue interactions in high capacity.
Acknowledgments
Y.S.Z. acknowledges funding from the National Cancer Institute of the National Institutes of Health (K99CA201603), the Lush Prize, the Science and Technology Commission of Shanghai Municipality (STCSM) 17JC 1400200, and the National Natural Science Foundation of China (U1605225).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhang YS, et al. Advances in engineering hydrogels. Science. 2017;356:eaaf3627. doi: 10.1126/science.aaf3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilhelm S, et al. Analysis of nanoparticle delivery to tumours. Nat Rev Mater. 2016;1:16014. [Google Scholar]
- 3.Nam SY, et al. Imaging strategies for tissue engineering applications. Tissue Eng B. 2014;21:88–102. doi: 10.1089/ten.teb.2014.0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmadi A, et al. Biomaterials for cardiac regeneration. Springer; 2015. Imaging of the biomaterial structure and function; pp. 275–293. [Google Scholar]
- 5.Trachtenberg J, et al. Pre-clinical characterization of tissue engineering constructs for bone and cartilage regeneration. Ann Biomed Eng. 2015;43:681–696. doi: 10.1007/s10439-014-1151-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Appel AA, et al. Imaging challenges in biomaterials and tissue engineering. Biomaterials. 2013;34:6615–6630. doi: 10.1016/j.biomaterials.2013.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Talukdar Y, et al. Multimodal ultrasound-photoacoustic imaging of tissue engineering scaffolds and blood oxygen saturation in and around the scaffolds. Tissue Eng C. 2014;20:440–449. doi: 10.1089/ten.tec.2013.0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu J, et al. Non-invasive characterization of polyurethane-based tissue constructs in a rat abdominal repair model using high frequency ultrasound elasticity imaging. Biomaterials. 2013;34:2701–2709. doi: 10.1016/j.biomaterials.2013.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gudur M, et al. Noninvasive, quantitative, spatiotemporal characterization of mineralization in three-dimensional collagen hydrogels using high-resolution spectral ultrasound imaging. Tissue Eng C. 2012;18:935–946. doi: 10.1089/ten.tec.2012.0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang KT, et al. Bioengineered human vascular networks transplanted into secondary mice reconnect with the host vasculature and re-establish perfusion. Blood. 2011;118:6718–6721. doi: 10.1182/blood-2011-08-375188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu HH, et al. Monitoring tissue engineering using magnetic resonance imaging. J Biosci Bioeng. 2008;106:515–527. doi: 10.1263/jbb.106.515. [DOI] [PubMed] [Google Scholar]
- 12.Xu JL, et al. Reconstruction of epidural fat with engineered adipose tissue from adipose derived stem cells and plga in the rabbit dorsal laminectomy model. Biomaterials. 2012;33:6965–6973. doi: 10.1016/j.biomaterials.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 13.Kotecha M, et al. Monitoring cartilage tissue engineering using magnetic resonance spectroscopy, imaging, and elastography. Tissue Eng B. 2013;19:470–484. doi: 10.1089/ten.teb.2012.0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beaumont M, et al. Monitoring angiogenesis in soft-tissue engineered constructs for calvarium bone regeneration: An in vivo longitudinal dce-mri study. NMR Biomed. 2010;23:48–55. doi: 10.1002/nbm.1425. [DOI] [PubMed] [Google Scholar]
- 15.Bible E, et al. Non-invasive imaging of transplanted human neural stem cells and ecm scaffold remodeling in the stroke-damaged rat brain by f-19- and diffusion-mri. Biomaterials. 2012;33:2858–2871. doi: 10.1016/j.biomaterials.2011.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roeder E, et al. Dose-response of superparamagnetic iron oxide labeling on mesenchymal stem cells chondrogenic differentiation: A multi-scale in vitro study. Plos One. 2014;9:e98451. doi: 10.1371/journal.pone.0098451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mertens ME, et al. Fmn-coated fluorescent uspio for cell labeling and non-invasive mr imaging in tissue engineering. Theranostics. 2014;4:1002–1013. doi: 10.7150/thno.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leferink AM, et al. Methods of monitoring cell fate and tissue growth in three-dimensional scaffold-based strategies for in vitro tissue engineering. Tissue Eng B. 2016;22:265–283. doi: 10.1089/ten.TEB.2015.0340. [DOI] [PubMed] [Google Scholar]
- 19.Hsueh YS, et al. Laminin-alginate beads as preadipocyte carriers to enhance adipogenesis in vitro and in vivo. Tissue Eng A. 2017;23:185–194. doi: 10.1089/ten.TEA.2016.0290. [DOI] [PubMed] [Google Scholar]
- 20.Kolewe ME, et al. 3d structural patterns in scalable, elastomeric scaffolds guide engineered tissue architecture. Adv Mater. 2013;25:4459–4465. doi: 10.1002/adma.201301016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dittmar R, et al. Assessment of cell viability in three-dimensional scaffolds using cellular autofluorescence. Tissue Eng C. 2012;18:198–204. doi: 10.1089/ten.tec.2011.0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan D, et al. Molecular photoacoustic imaging of angiogenesis with integrin-targeted gold nanobeacons. FASEB J. 2011;25:875–882. doi: 10.1096/fj.10-171728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cai X, et al. Photoacoustic microscopy in tissue engineering. Mater Today (Kidlington) 2013;16:67–77. doi: 10.1016/j.mattod.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang X, et al. Imaging engineered tissues using structural and functional optical coherence tomography. J Biophoton. 2009;2:643–655. doi: 10.1002/jbio.200910048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tu JW, et al. The in vivo bone formation by mesenchymal stem cells in zein scaffolds. Biomaterials. 2009;30:4369–4376. doi: 10.1016/j.biomaterials.2009.04.054. [DOI] [PubMed] [Google Scholar]
- 26.Zou D, et al. Blood vessel formation in the tissue-engineered bone with the constitutively active form of hif-1alpha mediated bmscs. Biomaterials. 2012;33:2097–2108. doi: 10.1016/j.biomaterials.2011.11.053. [DOI] [PubMed] [Google Scholar]
- 27.Zhu N, et al. X-ray diffraction enhanced imaging as a novel method to visualize low-density scaffolds in soft tissue engineering. Tissue Eng C. 2011;17:1071–1080. doi: 10.1089/ten.tec.2011.0102. [DOI] [PubMed] [Google Scholar]
- 28.Kim UJ, et al. Three-dimensional aqueous-derived biomaterial scaffolds from silk fibroin. Biomaterials. 2005;26:2775–2785. doi: 10.1016/j.biomaterials.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 29.Kofidis T, et al. Pulsatile perfusion and cardiomyocyte viability in a solid three-dimensional matrix. Biomaterials. 2003;24:5009–5014. doi: 10.1016/s0142-9612(03)00429-0. [DOI] [PubMed] [Google Scholar]
- 30.Kim K, et al. Non-invasive monitoring of tissue scaffold degradation using ultrasound elasticity imaging. Acta Biomater. 2008;4:783–790. doi: 10.1016/j.actbio.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hattori K, et al. Measurement of the mechanical condition of articular cartilage with an ultrasonic probe: Quantitative evaluation using wavelet transformation. Clin Biomech. 2003;18:553–557. doi: 10.1016/s0268-0033(03)00048-2. [DOI] [PubMed] [Google Scholar]
- 32.Shapiro MG, et al. Biogenic gas nanostructures as ultrasonic molecular reporters. Nat Nanotechnol. 2014;9:311–316. doi: 10.1038/nnano.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ravindran S, et al. Biological and mri characterization of biomimetic ecm scaffolds for cartilage tissue regeneration. Biomaterials. 2015;71:58–70. doi: 10.1016/j.biomaterials.2015.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kotecha M, et al. Monitoring tissue engineering and regeneration by magnetic resonance imaging and spectroscopy. J Tissue Sci Eng. 2013;11:007. [Google Scholar]
- 35.Mahmoudi M, et al. Infection-resistant mri-visible scaffolds for tissue engineering applications. BioImpacts: BI. 2016;6:111–115. doi: 10.15171/bi.2016.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poirier-Quinot M, et al. High-resolution 1.5-tesla magnetic resonance imaging for tissue-engineered constructs: A noninvasive tool to assess three-dimensional scaffold architecture and cell seeding. Tissue Eng C. 2009;16:185–200. doi: 10.1089/ten.TEC.2009.0015. [DOI] [PubMed] [Google Scholar]
- 37.Ying Y, et al. Investigation of optical coherence tomography as an imaging modality in tissue engineering. Phys Med Biol. 2006;51:1649. doi: 10.1088/0031-9155/51/7/001. [DOI] [PubMed] [Google Scholar]
- 38.Zhang YS, et al. Seeing through the surface: Non-invasive characterization of biomaterial–tissue interactions using photoacoustic microscopy. Ann Biomed Eng. 2016;44:649–666. doi: 10.1007/s10439-015-1485-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cai X, et al. Investigation of neovascularization in 3d porous scaffolds in vivo by photoacoustic microscopy and optical coherence tomography. Tissue Eng C. 2013;19:196–204. doi: 10.1089/ten.tec.2012.0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cang H, et al. Gold nanocages as contrast agents for spectroscopic optical coherence tomography. Opt Lett. 2005;30:3048–3050. doi: 10.1364/ol.30.003048. [DOI] [PubMed] [Google Scholar]
- 41.Artzi N, et al. In vivo and in vitro tracking of erosion in biodegradable materials using non-invasive fluorescence imaging. Nat Mater. 2011;10:704–709. doi: 10.1038/nmat3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang YS, et al. Optical-resolution photoacoustic microscopy for volumetric and spectral analysis of histological and immunochemical samples. Angew Chemie Int Ed. 2014;53:8099–8103. doi: 10.1002/anie.201403812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, et al. Noninvasive photoacoustic microscopy of living cells in two and three dimensions through enhancement by a metabolite dye. Angew Chem Int Ed. 2011;50:7359–7363. doi: 10.1002/anie.201101659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, et al. Non-invasive and in situ characterization of the degradation of biomaterial scaffolds by photoacoustic microscopy. Angew Chem Int Ed. 2014;53:184–188. doi: 10.1002/anie.201306282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cai X, et al. Multiscale photoacoustic microscopy of single-walled carbon nanotube-incorporated tissue engineering scaffolds. Tissue Eng C. 2012;18:310–317. doi: 10.1089/ten.tec.2011.0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Talukdar Y, et al. Multimodal ultrasound-photoacoustic imaging of tissue engineering scaffolds and blood oxygen saturation in and around the scaffolds. Tissue Eng C. 2014;20:440–449. doi: 10.1089/ten.tec.2013.0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Lenthe GH, et al. Nondestructive micro-computed tomography for biological imaging and quantification of scaffold–bone interaction in vivo. Biomaterials. 2007;28:2479–2490. doi: 10.1016/j.biomaterials.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 48.Choi S-W, et al. In vitro mineralization by preosteoblasts in poly(dl-lactide-co-glycolide) inverse opal scaffolds reinforced with hydroxyapatite nanoparticles. Langmuir. 2010;26:12126–12131. doi: 10.1021/la101519b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hainfeld JF, et al. Gold nanoparticles: A new x-ray contrast agent. Br J Radiol. 2006;79:248–253. doi: 10.1259/bjr/13169882. [DOI] [PubMed] [Google Scholar]
- 50.Bai M-Y, et al. A facile and general method for the encapsulation of different types of imaging contrast agents within micrometer-sized polymer beads. Adv Funct Mater. 2012;22:764–770. doi: 10.1002/adfm.201102582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arakawa H, et al. Protein images obtained by stm, afm and tem. Nature. 1992;358:171–173. doi: 10.1038/358171a0. [DOI] [PubMed] [Google Scholar]
- 52.Shimomura O, et al. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, aequorea. J Cell Comp Physiol. 1962;59:223–239. doi: 10.1002/jcp.1030590302. [DOI] [PubMed] [Google Scholar]
- 53.Chalfie M, et al. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 54.Heim R, et al. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 55.Shimomura O. A short story of aequorin. Biol Bulletin. 1995;189:1–5. doi: 10.2307/1542194. [DOI] [PubMed] [Google Scholar]
- 56.Saito K, et al. Recent progress in luminescent proteins development. Curr Opin Chem Biol. 2015;27:46–51. doi: 10.1016/j.cbpa.2015.05.029. [DOI] [PubMed] [Google Scholar]
- 57.Cai X, et al. Multi-scale molecular photoacoustic tomography of gene expression. PLoS One. 2012;7:e43999. doi: 10.1371/journal.pone.0043999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Filonov GS, et al. Deep-tissue photoacoustic tomography of a genetically encoded near-infrared fluorescent probe. Angew Chem Int Ed. 2012;51:1448–1451. doi: 10.1002/anie.201107026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krumholz A, et al. Multicontrast photoacoustic in vivo imaging using near-infrared fluorescent proteins. Sci Rep. 2014;4:3939. doi: 10.1038/srep03939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Daniel R, et al. Multispectral opto-acoustic tomography of deep-seated fluorescent proteins in vivo. Nat Photonics. 2009;3:412–417. [Google Scholar]
- 61.Paproski RJ, et al. Tyrosinase as a dual reporter gene for both photoacoustic and magnetic resonance imaging. Biomed Opt Express. 2011;2:771–780. doi: 10.1364/BOE.2.000771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jathoul AP, et al. Deep in vivo photoacoustic imaging of mammalian tissues using a tyrosinase-based genetic reporter. Nat Photon. 2015;9:239–246. [Google Scholar]
- 63.Yao J, et al. Multiscale photoacoustic tomography using reversibly switchable bacterial phytochrome as a near-infrared photochromic probe. Nat Methods. 2016;13:67–73. doi: 10.1038/nmeth.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Duan H, et al. Green chemistry for nanoparticle synthesis. Chem Soc Rev. 2015;44:5778–5792. doi: 10.1039/c4cs00363b. [DOI] [PubMed] [Google Scholar]
- 65.Zhang YS, et al. Multiple facets for extracellular matrix mimicking in regenerative medicine. Nanomedicine. 2015;10:689–692. doi: 10.2217/nnm.15.10. [DOI] [PubMed] [Google Scholar]
- 66.Zhang Y, et al. Chronic label-free volumetric photoacoustic microscopy of melanoma cells in three-dimensional porous scaffolds. Biomaterials. 2010;31:8651–8658. doi: 10.1016/j.biomaterials.2010.07.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chung K, et al. Clarity for mapping the nervous system. Nat Methods. 2013;10:508–513. doi: 10.1038/nmeth.2481. [DOI] [PubMed] [Google Scholar]
- 68.Moneron G, et al. Two-photon excitation sted microscopy. Opt Express. 2009;17:14567–14573. doi: 10.1364/oe.17.014567. [DOI] [PubMed] [Google Scholar]
- 69.Rust MJ, et al. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (storm) Nat Methods. 2006;3:793–796. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen F, et al. Expansion microscopy. Science. 2015;347:1260088. doi: 10.1126/science.1260088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Errico C, et al. Ultrafast ultrasound localization microscopy for deep super-resolution vascular imaging. Nature. 2015;527:499–502. doi: 10.1038/nature16066. [DOI] [PubMed] [Google Scholar]
- 72.Wang YM, et al. Deep-tissue focal fluorescence imaging with digitally time-reversed ultrasound-encoded light. Nat Commun. 2012;3:928. doi: 10.1038/ncomms1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arai K, et al. Fourier magnetic imaging with nanoscale resolution and compressed sensing speedup using electronic spins in diamond. Nat Nanotechnol. 2015;10:859–864. doi: 10.1038/nnano.2015.171. [DOI] [PubMed] [Google Scholar]
- 74.Mikula S, et al. High-resolution whole-brain staining for electron microscopic circuit reconstruction. Nat Methods. 2015;12:541. doi: 10.1038/nmeth.3361. [DOI] [PubMed] [Google Scholar]
- 75.Gopinath A, et al. Engineering and mapping nanocavity emission via precision placement of DNA origami. Nature. 2016;535:401. doi: 10.1038/nature18287. [DOI] [PubMed] [Google Scholar]
- 76.Qin CX, et al. Tyrosinase as a multifunctional reporter gene for photoacoustic/mri/pet triple modality molecular imaging. Sci Rep. 2013;3:1490. doi: 10.1038/srep01490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chung K, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]