Abstract
Purpose
To determine whether deep features extracted from digital mammograms using a pre-trained deep convolutional neural network (CNN) are prognostic of occult invasive disease for patients with ductal carcinoma in situ (DCIS) on core needle biopsy.
Materials and Methods
In this retrospective study, we collected digital mammography magnification views for 99 subjects with DCIS at biopsy, 25 of which were subsequently upstaged to invasive cancer. We utilized a deep CNN model that was pre-trained on non-medical images (e.g., animals, plants, instruments) as the feature extractor. Through a statistical pooling strategy, we extracted deep features at different levels of convolutional layers from the lesion areas, without sacrificing the original resolution or distorting the underlying topology. A multivariate classifier was then trained to predict which tumors contain occult invasive disease. This was compared to the performance of traditional “handcrafted” computer vision (CV) features previously developed specifically to assess mammographic calcifications. The generalization performance was assessed using Monte Carlo cross validation and receiver operating characteristic (ROC) curve analysis.
Results
Deep features were able to distinguish DCIS with occult invasion from pure DCIS, with an area under the ROC curve (AUC-ROC) equal to 0.70 (95% CI: 0.68–0.73). This performance was comparable to the handcrafted CV features (AUC-ROC = 0.68, 95% CI: 0.66–0.71) that were designed with prior domain knowledge.
Conclusion
In spite of being pre-trained on only non-medical images, the deep features extracted from digital mammograms demonstrated comparable performance to handcrafted CV features for the challenging task of predicting DCIS upstaging.
Keywords: Ductal carcinoma in situ, digital mammography, convolution neural network, deep learning, computer vision
Introduction
Over 60,000 women in the United States are diagnosed with ductal carcinoma in situ (DCIS) every year, representing approximately 20% of all new breast cancer cases [1]. However, the risk of progression from DCIS to invasive cancer is still unclear, with estimates ranging from 14% to 53% [2]. In addition, up to 50% of lesions diagnosed as pure DCIS by coreneedle biopsy will be upstaged to contain invasive disease at definitive surgery [3]. Methods that could predict an occult invasive component associated with this upstaging may affect treatment planning and avoid delays in definitive diagnosis.
Many studies have sought to identify preoperative predictors of DCIS upstaging. Various immunohistochemical biomarkers, histological features, and medical image findings have shown limited predictive power [3–13]. In contrast, computer extracted features, such as those designed for breast cancer screening [14–18], may be a promising alternative due to the quantitative and reproducible methodology. Previously, our group (reference withheld) demonstrated that computer vision (CV) mammographic features can be used to predict DCIS upstaging with performance comparable to a radiologist. Nevertheless, this process of feature engineering is time consuming and might not capture all the image information.
Deep learning, especially deep convolutional neural network (CNN), has emerged as a promising approach for many image recognition or classification tasks [19], demonstrating human or even superhuman performance [20]. Typically deep learning requires training on very large image datasets with appropriate labeling, and has been applied to several areas in medical imaging [21, 22]. Deep CNN models learn a multiple-level, latent feature representation during the training procedure, without any input of prior expert knowledge. Used as feature extractor, some pre-trained CNN models can match or surpass the performance of domain-specific, handcrafted features [23–26]. Several recent studies applied this to medical tasks including classification of images from chest radiography, chest CT, otoscopy, and endoscopy [27–31].
The purpose of this investigation is to determine whether DCIS upstaging can be predicted using deep features extracted from digital mammograms by a pre-trained deep CNN. Additionally, we provide a head-to-head performance comparison between these non-medical deep features and handcrafted CV features developed with breast cancer domain knowledge.
Materials and Methods
Digital mammogram dataset
The study was approved by the Institutional Review Board with a waiver of informed consent. We identified women aged 40 years or greater with stereotactic biopsy-proven DCIS presenting with only calcifications. Exclusion criteria included the presence of any masses, asymmetries, or architectural distortion; history of breast cancer or prior surgery; and presence of microinvasion at the time of initial biopsy. 99 subjects met these criteria, 25 of whom were upstaged. We collected the diagnostic digital magnification views, all produced by GE Senographe Essential systems.
Extracting handcrafted CV features as reference
As the reference baseline, we previously presented a model (reference withheld) based on handcrafted CV features. Three types of mammographic features were extracted from segmented individual microcalcifications (MCs) and the whole cluster for each DCIS lesion, including: (i) shape features to describe morphology and size of MCs and clusters; (ii) topological features from weighted graphs associated with the clusters; (iii) texture features such as from gray level co-occurrence matrices. Within a cluster, 25 CV features were computed for individual MCs. To describe the whole cluster, four global statistical measures were computed across all MCs: mean, standard deviation, minimum, and maximum. Overall, we obtained a set of 113 handcrafted CV features for each subject: 25×4 based on individual MCs and 13 cluster features.
Extracting deep features using a pre-trained CNN model
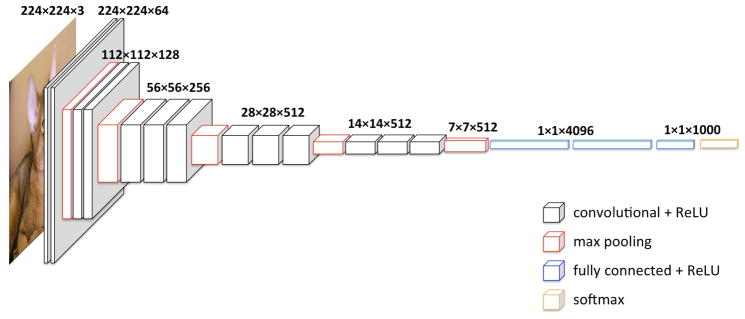
In this study, we selected the widely used deep CNN model VGG [32] (specifically the configuration with 16 layers, VGG-16) because it has been very successful in many different localization and classification tasks [25, 33–35]. Moreover, the VGG model adopts the most straightforward CNN architecture, which facilitates the extraction of deep features from multiple levels. As shown in Figure 1, the used VGG-16 model consists of 16 weight layers, including 13 convolutional layers with filter size of 3×3, and three fully connected layers at the end. The convolutional layers are divided into five groups and each group is followed by a max-pooling layer. The number of filters of convolutional layer group starts from 64 in the first group and then increases by a factor of 2 after each max-pooling layer, until it reaches 512. The overall trainable parameters are over 130 million.
Fig. 1.
Architecture of VGG16.
We utilized the VGG-16 model with parameters pre-trained on the ImageNet dataset [36], which is a large database with more than 20,000,000 images arranged into more than 15,000 non-medical concepts/categories (e.g., animals, plants, instruments). During training, ImageNet images are usually resized into square, low resolution (224×224×3 RGB) versions. Following that convention resulted in consistently poor performance for this study, however, because that resizing led to sacrificing the resolution or distorting the underlying topology of each lesion. Therefore, we directly input the region of interest (ROI) from the digital magnification view at its full resolution into the pre-trained VGG-16 model. This means the input image size varies due to different lesion sizes. Accordingly, the last three fully-connected layers cannot be utilized to extract features, and we only extracted deep features from convolutional layers, which are not affected by the input image size. As representatives for different depth levels, the convolutional layers before max-pooling in each of five groups were chosen, hereafter annotated as Layer-0, 1, 2, 3, and 4.
In spite of different input sizes, the dimensionality of the extracted deep features from convolutional layers for each subject can be kept the same by ROI pooling [37], which scales feature maps to a pre-defined size. Inspired by our handcrafted CV features study, we proposed an alternative strategy of statistical pooling. For each feature map, we calculated the mean, standard deviation, maximum, and summation. Take Layer-0 for example: Considering two input ROIs with different sizes of 200×100 and 158×158 respectively, the dimensions of corresponding Layer-0 feature maps are 200×100×64 (height × width × No. of maps) and 158×158×64. Using the four statistical metrics, each input results in 64 mean, standard deviation, maximum and summation values respectively, i.e., a set of 64×4 deep features from Layer-0 for both inputs. This statistical pooling was applied on each of the chosen five convolutional layers, thereby for each subject we extracted 5 sets of deep features from each lesion ROI with sizes of 64×4, 128×4, 256×4, 512×4 and 512×4, respectively.
Extraction of deep features is not a computationally expensive process, since it only requires feeding the images forward through the model, without any backpropagation. For the 99 cases in this study, it took only 26 seconds (MXNet package on a Linux/Intel Core i7-6700k and Nvidia GeForce GTX 1017) compared to 14 seconds for the typical process of using resized 224*224 images.
Evaluation of performance
For both deep features and hand-crated CV features, we developed logistic regression classifiers with L2 norm regularization. Considering the high dimensionality of deep features as well as the relative small size of our dataset, we then explored feature selection for improved feature representations. For this purpose, we performed “stability feature selection” [38] that has been widely used in high-dimensional settings with logistic regression.
The generalization performance of the predictive model was assessed using receiver operating characteristic (ROC) curve analysis with Monte Carlo cross validation [39]. Specifically, 20% cases (5 positives, 15 negatives) were randomly selected for validation, while the remaining 80% (20 positives, 59 negatives) were used for training. Average ROC curves were generated using the ‘cvROC’ package within R (V.3.3.1) [40] and area under ROC curve (AUC-ROC) were compared semi-parametrically [41].
Results
General performance for raw features
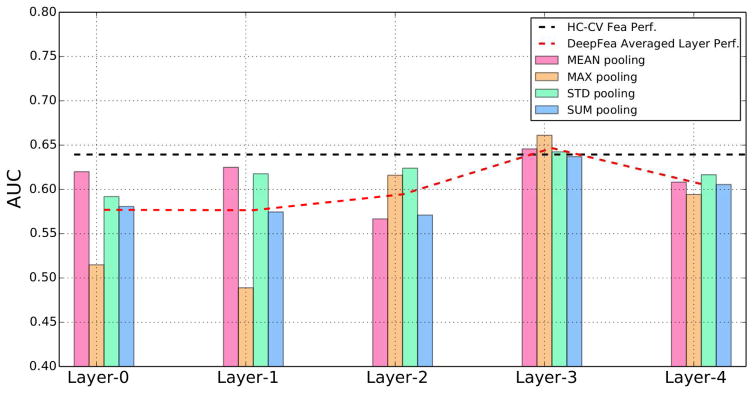
The AUC-ROC performances of 113 handcrafted CV features and deep features from 5 different levels are shown in Table 1. For a direct comparison between performances of deep features from different layers, we further provide the corresponding plots in Figure 2.
Table 1.
Classification performance - AUC-ROC for handcrafted CV features and deep features from different layers using a logistic regression classifier without feature selection.
| Handcrafted CV features | Number of features | Classification performance: AUC-ROC | ||||
|---|---|---|---|---|---|---|
| 113 | 0.64 | |||||
| Deep Features | Number of feature maps | Classification performance: AUC-ROC | ||||
| Mean-pooling | MAX-pooling | STD-pooling | SUM-pooling | Averaged | ||
| Layer-0 | 64 | 0.62 | 0.51 | 0.59 | 0.58 | 0.58 |
| Layer-1 | 128 | 0.62 | 0.49 | 0.62 | 0.57 | 0.58 |
| Layer-2 | 256 | 0.57 | 0.62 | 0.62 | 0.57 | 0.59 |
| Layer-3 | 512 | 0.65 | 0.66 | 0.64 | 0.64 | 0.65 |
| Layer-4 | 512 | 0.61 | 0.59 | 0.62 | 0.61 | 0.61 |
Fig. 2.
Classification performances (AUC-ROC) for deep features at different layers without feature selection. The reference performance of handcrafted CV features is shown as the black dashed line. The averaged performance of deep features at each level is shown as the red dashed line.
In the scenario when no feature selection is performed, deep features for one of the layers showed comparable performance to that for the handcrafted features. Specifically, using 113 handcrafted CV features, the proposed multivariate predictive model using logistic regression without feature selection achieved AUC-ROC of 0.64 (95% CI: 0.61–0.67). The same predictive model using deep features from different layers resulted in AUC-ROC ranging from 0.49 to 0.66. The best averaged performance is from Layer-3 with AUC-ROC of 0.65. Among different ways to perform statistical pooling for different layers, the deep features using max-pooling at Layer-3 achieved the best overall classification performance, with AUC-ROC of 0.66 (95% CI: 0.63–0.69). The multivariate model using this set of deep features resulted in a slightly better performance than using handcrafted CV features (as shown in Fig. 3a), but with no statistically significant difference (p<0.061).
Fig. 3.
Comparison of averaged ROC curves from 113 handcrafted CV features (black) and the best deep features from Layer-3 with Max pooling (red), for the scenarios: (a) without feature selection and (b) with feature selection.
Improved performance using feature selection
We next present the performance of the logistic regression classifier with stability feature selection. Specifically, we applied the model on the 113 handcrafted CV features and the best set of deep features (Layer-3 with max-pooling) respectively. Using 113 handcrafted CV features and feature selection, the model achieved AUC-ROC of 0.68 (95% CI: 0.66–0.71); using the deep features extracted from Layer-3 with max-pooling, the model achieved AUC-ROC of 0.70 (95% CI: 0.68–0.73). Both of them are statistically significantly better than a random guess. Furthermore, using feature selection improved performance for both models significantly (p <0.0001), and the model using deep features still achieved a slightly better performance than handcrafted CV features (as shown in Fig. 3b), but with no statistically significant difference (p <0.074).
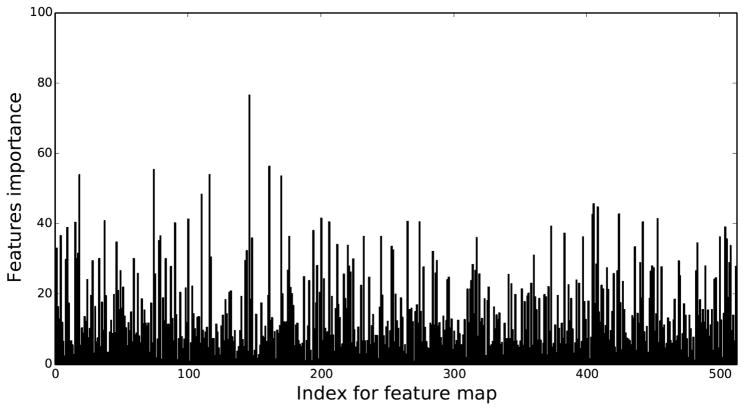
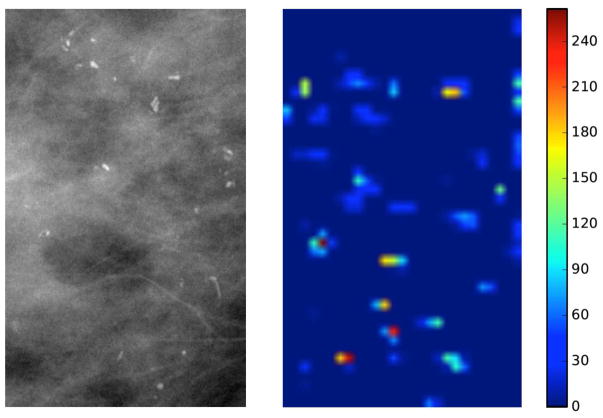
To further investigate the potential of deep features, we calculated the feature importance for each deep feature at Layer-3 (as shown in Fig. 4). Since the logistic regression classifier learns a linear combination of selected features, the normalized absolute value of the learned weight for each feature can suggest this feature’s importance within the multivariate model. The overall feature importance was summed over the 100 repeats. We can see only relatively few deep features in Layer-3 contribute significantly, while the majority have very low importance. The most important feature map is at index 145. To visualize the impact of that feature map, we input ROIs from one positive subject and one negative subject into the VGG16 model respectively, and showed their respective activations for the index 145 feature map at Layer-3 in Figure 5. However, since feature maps at Layer-3 have a much smaller spatial dimension than the original inputs and also extract more complicated feature representations, there was no discernible difference in the visual patterns.
Fig. 4.
Regression coefficients used to infer feature importance for each deep feature map from Layer-3.
Fig. 5.
The digital magnification view input ROI and its activation of index 145 feature map from Layer-3 for a (a) pure DCIS case and (b) upstaged case.
Discussion
In this work, we showed the diagnostic potential for deep features extracted from a deep convolutional neural network, even though it was originally optimized for the seemingly unrelated task of classifying non-medical images. We demonstrated this potential for a challenging medical task of using a small set of mammograms to predict DCIS upstaging. With no further training, the deep features showed significantly better-than-chance predictive power, and in fact achieved a comparable performance as carefully handcrafted CV features with guidance of prior domain knowledge. To the best of our knowledge, our study is the first of its kind to examine the feasibility of deep feature approach for a radiomics task in breast imaging.
We can draw several notable conclusions from these findings. First, deep features extracted using pre-trained deep convolutional neural network can match handcrafted features with guidance of prior domain knowledge. This finding is consistent with previous studies [27–31], and demonstrates the extraordinary feature engineering ability of deep CNN models. It is truly remarkable that such a general image recognition framework, albeit one trained on over a million images, can provide useful data representations for a variety of very specific medical imaging tasks. Second, we observed that our best classification performance is from deep features extracted at the middle level layer, similar to another study for a different medical task [27]. Deep features from low level layers (near the input images) are too general and simple for a complicated task, while deep features from the high level layers (near the output) are too specific for its original task domain and lack transferability [42]. In addition, deep features at middle-level layers are also regarded to be associated with different textural patterns [43]. This agrees with the findings from our previous study (reference withheld) that texture-related CV features were among the most frequently selected for this task. Third, in order to preserve the high resolution of magnification views of digital mammograms, as well as the underlying topology for the DCIS lesion, we proposed to extract deep features directly from the complete ROI for the lesion area at its full resolution, instead of utilizing the commonly adopted resizing or decomposing solutions. Even though all spatial information was lost during the subsequent statistical pooling procedure, the deep features extracted via this method still contain significantly better-than-chance predictive power for our task. Last but not least, using feature selection significantly boosted the performance of deep features. Based on the feature importance maps, most deep features contain very little information, and the feature selection is an important step to mitigate the curse of dimensionality and improve performance.
The current study has several limitations. First, the dataset size is relatively small because of the single-institution design and the strict exclusion criteria. This affects the performance of our predictive model, especially considering the high-dimensionality of the original, dense deep features. Applying statistical pooling and utilizing feature selection alleviates the problem to a certain degree, but is still not optimal. Second, recent studies showed other alternative ways of utilizing deep features, such as exploring cross-layer representation [44], aggregating densely pooled deep features [45], and ensemble learning between the deep features and traditional handcrafted features [46]. The goal of this paper was to validate the feasibility of applying deep feature idea on our specific task, but these advanced strategies may be fruitful in future work. Third, we attempted to understand the latent deep feature representation related to our specific task by visualizing the feature activation map, but did not discern any useful information. Nevertheless, understanding CNN features and models is still a difficulty task even in the deep learning research community. Despite the generic feature engineering ability of deep CNN models, little is known about how these models represent invariances at each depth level. Recently, several attempts have been made from different perspectives [43, 47–49], and we will explore these methodologies in subsequent work. Fourth, this preliminary study was an unblinded, retrospective study. We were intentionally conservative with feature selection and modeling algorithms, as well as limiting the number of trials. However, our reported results were not adjusted by a multiple-hypothesis testing correction procedure, so there is still potential for overfitting bias. Finally, as indicated in some previous studies [42], the performance and transferability of deep features are significantly affected by the similarity between the original task and the target task. Thus, the VGG-16 model pre-trained on ImageNet might not be the optimal one to extract the off-the-shelf deep features from digital mammograms, although it is well recognized as one of the most generalizable models for many different tasks. For the breast imaging tasks, we believe that better representation of deep features can be learned if deep learning models can be trained on more similar domains, such as the texture datasets, or medical image datasets on other human body parts.
Conclusions
In conclusion, our study demonstrates the feasibility of using deep features for the radiomics prediction task of predicating DCIS upstaging. Specifically, we extracted deep features from the DCIS lesion area on magnification mammography views using a deep CNN model that was pre-trained on non-medical images, and those deep features performed comparably to handcrafted CV features that were designed with prior domain knowledge. From our work, we hope to show the medical imaging community an alternative way of utilizing the powerful deep learning technique when a large-scale dataset is not available. Future work will explore other strategies to better understand and utilize the deep features to improve the predictive power of our model for DCIS upstaging. In addition, we are also planning to include more subjects into this study via collaboration with other institutions for a better-justified validation of our model.
Take Home Points.
The extraordinary feature engineering ability of deep convolutional neural network provides an alternative way of utilizing powerful deep learning techniques for medical imaging, where large-scale datasets are often not available.
Using a CNN model pre-trained on only non-medical images, the deep features extracted from digital mammograms demonstrated comparable performance to handcrafted computer vision features in the prediction of DCIS upstaging.
Like most imaging markers, deep features contained substantial amounts of irrelevant or redundant information, requiring feature selection to reduce dimensionality and provide significant performance improvement.
Acknowledgments
Sources of support:
This work was supported in part by NIH/NCI R01-CQA185138 and DOD Breast Cancer Research Program W81XWH-14-1-0473.
Footnotes
Conflict of interest information:
The authors declare that there is no conflict of interests regarding the publication of this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Society, A.C. Cancer Facts & Figures 2015. American Atlanta; 2015. [Google Scholar]
- 2.Erbas B, Provenzano E, Armes J, et al. The natural history of ductal carcinoma in situ of the breast: a review. Breast Cancer Research and Treatment. 2006;97(2):135–144. doi: 10.1007/s10549-005-9101-z. [DOI] [PubMed] [Google Scholar]
- 3.Brennan ME, Turner RM, Ciatto S, et al. Ductal Carcinoma in Situ at Core-Needle Biopsy: Meta-Analysis of Underestimation and Predictors of Invasive Breast Cancer. Radiology. 2011;260(1):119–128. doi: 10.1148/radiol.11102368. [DOI] [PubMed] [Google Scholar]
- 4.Bagnall MJ, Evans AJ, Wilson ARM, et al. Predicting invasion in mammographically detected microcalcification. Clinical radiology. 2001;56(10):828–832. doi: 10.1053/crad.2001.0779. [DOI] [PubMed] [Google Scholar]
- 5.Dillon MF, McDermott EW, Quinn CM, et al. Predictors of invasive disease in breast cancer when core biopsy demonstrates DCIS only. Journal of surgical oncology. 2006;93(7):559–563. doi: 10.1002/jso.20445. [DOI] [PubMed] [Google Scholar]
- 6.Kurniawan ED, Rose A, Mou A, et al. RIsk factors for invasive breast cancer when core needle biopsy shows ductal carcinoma in situ. Archives of Surgery. 2010;145(11):1098–1104. doi: 10.1001/archsurg.2010.243. [DOI] [PubMed] [Google Scholar]
- 7.Lee CW, Wu HK, Lai HW, et al. Preoperative clinicopathologic factors and breast magnetic resonance imaging features can predict ductal carcinoma in situ with invasive components. European Journal of Radiology. 2016;85(4):780–789. doi: 10.1016/j.ejrad.2015.12.027. [DOI] [PubMed] [Google Scholar]
- 8.Lee CH, Carter D, Philpotts LE, et al. Ductal Carcinoma in Situ Diagnosed with Stereotactic Core Needle Biopsy: Can Invasion Be Predicted? 1. Radiology. 2000;217(2):466–470. doi: 10.1148/radiology.217.2.r00nv08466. [DOI] [PubMed] [Google Scholar]
- 9.O’Flynn E, Morel J, Gonzalez J, et al. Prediction of the presence of invasive disease from the measurement of extent of malignant microcalcification on mammography and ductal carcinoma in situ grade at core biopsy. Clinical radiology. 2009;64(2):178–183. doi: 10.1016/j.crad.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Park HS, Kim HY, Park S, et al. A nomogram for predicting underestimation of invasiveness in ductal carcinoma in situ diagnosed by preoperative needle biopsy. The Breast. 2013;22(5):869–873. doi: 10.1016/j.breast.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 11.Park HS, Park S, Cho J, et al. Risk predictors of underestimation and the need for sentinel node biopsy in patients diagnosed with ductal carcinoma in situ by preoperative needle biopsy. Journal of Surgical Oncology. 2013;107(4):388–392. doi: 10.1002/jso.23273. [DOI] [PubMed] [Google Scholar]
- 12.Renshaw AA. Predicting invasion in the excision specimen from breast core needle biopsy specimens with only ductal carcinoma in situ. Archives of pathology & laboratory medicine. 2002;126(1):39–41. doi: 10.5858/2002-126-0039-PIITES. [DOI] [PubMed] [Google Scholar]
- 13.Sim Y, Litherland J, Lindsay E, et al. Upgrade of ductal carcinoma in situ on core biopsies to invasive disease at final surgery: a retrospective review across the Scottish Breast Screening Programme. Clinical radiology. 2015;70(5):502–506. doi: 10.1016/j.crad.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 14.Mina LM, Isa NAM. A Review of Computer-Aided Detection and Diagnosis of Breast Cancer in Digital Mammography. Journal of Medical Sciences. 2015;15(3):110. [Google Scholar]
- 15.Giger ML, Karssemeijer N, Schnabel JA. Breast image analysis for risk assessment, detection, diagnosis, and treatment of cancer. Annual review of biomedical engineering. 2013;15:327–357. doi: 10.1146/annurev-bioeng-071812-152416. [DOI] [PubMed] [Google Scholar]
- 16.Jalalian A, Mashohor SB, Mahmud HR, et al. Computer-aided detection/diagnosis of breast cancer in mammography and ultrasound: a review. Clinical imaging. 2013;37(3):420–426. doi: 10.1016/j.clinimag.2012.09.024. [DOI] [PubMed] [Google Scholar]
- 17.Dromain C, Boyer B, Ferre R, et al. Computed-aided diagnosis (CAD) in the detection of breast cancer. European journal of radiology. 2013;82(3):417–423. doi: 10.1016/j.ejrad.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 18.Petrick N, Sahiner B, Armato SG, et al. Evaluation of computer-aided detection and diagnosis systems. Medical Physics. 2013;40(8):087001. doi: 10.1118/1.4816310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 20.He K, Zhang X, Ren S, et al. Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. Proceedings of the IEEE international conference on computer vision; 2015. [Google Scholar]
- 21.Kooi T, Litjens G, van Ginneken B, et al. Large scale deep learning for computer aided detection of mammographic lesions. Medical image analysis. 2017;35:303–312. doi: 10.1016/j.media.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 22.Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316(22):2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 23.Gong Y, Wang L, Guo R, et al. Multi-scale orderless pooling of deep convolutional activation features. European conference on computer vision; Springer; 2014. [Google Scholar]
- 24.Sharif Razavian A, Azizpour H, Sullivan J, et al. CNN features off-the-shelf: an astounding baseline for recognition. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops; 2014. [Google Scholar]
- 25.Cimpoi M, Maji S, Vedaldi A. Deep filter banks for texture recognition and segmentation. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sermanet P, Eigen D, Zhang X, et al. Overfeat: Integrated recognition, localization and detection using convolutional networks. 2013 arXiv preprint arXiv:1312.6229. [Google Scholar]
- 27.Tamaki T, Sonoyama S, Hirakawa T, et al. Computer-Aided Colorectal Tumor Classification in NBI Endoscopy Using CNN Features. 2016 doi: 10.1016/j.media.2012.08.003. arXiv preprint arXiv:1608.06709. [DOI] [PubMed] [Google Scholar]
- 28.Bar Y, Diamant I, Wolf L, et al. SPIE Medical Imaging. International Society for Optics and Photonics; 2015. Deep learning with non-medical training used for chest pathology identification. [Google Scholar]
- 29.Wang X, Lu L, Shin H-c, et al. Unsupervised Joint Mining of Deep Features and Image Labels for Large-scale Radiology Image Categorization and Scene Recognition. 2017 arXiv preprint arXiv:1701.06599. [Google Scholar]
- 30.Shie C-K, Chuang C-H, Chou C-N, et al. Transfer representation learning for medical image analysis. Engineering in Medicine and Biology Society (EMBC), 2015 37th Annual International Conference of the IEEE; IEEE; 2015. [DOI] [PubMed] [Google Scholar]
- 31.Paul R, Hawkins SH, Balagurunathan Y, et al. Deep Feature Transfer Learning in Combination with Traditional Features Predicts Survival Among Patients with Lung Adenocarcinoma. Tomography: a journal for imaging research. 2016;2(4):388. doi: 10.18383/j.tom.2016.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. 2014 arXiv preprint arXiv:1409.1556. [Google Scholar]
- 33.Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2015. [DOI] [PubMed] [Google Scholar]
- 34.Xu Z, Yang Y, Hauptmann AG. A discriminative CNN video representation for event detection. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2015. [Google Scholar]
- 35.Xie S, Tu Z. Holistically-nested edge detection. Proceedings of the IEEE International Conference on Computer Vision; 2015. [Google Scholar]
- 36.Deng J, Dong W, Socher R, et al. Imagenet: A large-scale hierarchical image database. Computer Vision and Pattern Recognition, 2009. CVPR 2009. IEEE Conference on; IEEE; 2009. [Google Scholar]
- 37.Girshick R. Fast r-cnn. Proceedings of the IEEE International Conference on Computer Vision; 2015. [Google Scholar]
- 38.Meinshausen N, Bühlmann P. Stability selection. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2010;72(4):417–473. [Google Scholar]
- 39.Dubitzky W, Granzow M, Berrar DP. Fundamentals of data mining in genomics and proteomics. Springer Science & Business Media; 2007. [Google Scholar]
- 40.LeDell E, Petersen M, van der Laan M. Computationally efficient confidence intervals for cross-validated area under the ROC curve estimates. Electronic journal of statistics. 2015;9(1):1583. doi: 10.1214/15-EJS1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanley JA, McNeil BJ. A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology. 1983;148(3):839–843. doi: 10.1148/radiology.148.3.6878708. [DOI] [PubMed] [Google Scholar]
- 42.Yosinski J, Clune J, Bengio Y, et al. How transferable are features in deep neural networks? Advances in neural information processing systems. 2014 [Google Scholar]
- 43.Zeiler MD, Fergus R. Visualizing and understanding convolutional networks. European conference on computer vision; Springer; 2014. [Google Scholar]
- 44.Liu L, Shen C, van den Hengel A. The treasure beneath convolutional layers: Cross-convolutional-layer pooling for image classification. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2015. [Google Scholar]
- 45.Babenko A, Lempitsky V. Aggregating local deep features for image retrieval. Proceedings of the IEEE international conference on computer vision; 2015. [Google Scholar]
- 46.Wu S, Chen Y-C, Li X, et al. An enhanced deep feature representation for person reidentification. Applications of Computer Vision (WACV), 2016 IEEE Winter Conference on; IEEE; 2016. [Google Scholar]
- 47.Dosovitskiy A, Brox T. Inverting visual representations with convolutional networks. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2016. [Google Scholar]
- 48.Mahendran A, Vedaldi A. Understanding deep image representations by inverting them. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2015. [Google Scholar]
- 49.Simonyan K, Vedaldi A, Zisserman A. Deep inside convolutional networks: Visualising image classification models and saliency maps. 2013 arXiv preprint arXiv:1312.6034. [Google Scholar]