Summary
Advances in the classification of acute leukaemias have led to improved outcomes for a substantial fraction of patients. However, chemotherapy resistance remains a major problem for specific subsets of acute leukaemias. Here, we propose that a molecularly distinct subtype of acute leukaemia with shared myeloid and T cell lymphoblastic features, which we term acute myeloid/T-lymphoblastic leukaemia (AMTL), is divided across 3 diagnostic categories owing to variable expression of markers deemed to be defining of myeloid and T-lymphoid lineages, such as myeloperoxidase and CD3. This proposed diagnostic group is supported by i) retained myeloid differentiation potential during early T cell lymphoid development, ii) recognition that some cases of acute myeloid leukaemia (AML) harbour hallmarks of T cell development, such as T-cell receptor gene rearrangements and iii) common gene mutations in subsets of AML and T cell acute lymphoblastic leukaemia (T-ALL), including WT1, PHF6, RUNX1 and BCL11B. This proposed diagnostic entity overlaps with early T cell precursor (ETP) T-ALL and T cell/myeloid mixed phenotype acute leukaemias (MPALs), and also includes a subset of leukaemias currently classified as AML with features of T-lymphoblastic development. The proposed classification of AMTL as a distinct entity would enable more precise prospective diagnosis and permit the development of improved therapies for patients whose treatment is inadequate with current approaches.
Keywords: acute leukaemia, classifications, AML, ALL
Introduction
Acute leukaemias are aggressive neoplasms characterized by the pathological accumulation of immature haematopoietic progenitors. Improvements in clinical outcomes for patients with acute leukaemias treated with modern chemotherapy regimens have been variable. The introduction of molecularly targeted therapies has had a major impact on a small subset of genetically-defined acute leukaemias. For example, arsenic trioxide and retinoic acid, which target the PML-RARA fusion oncoprotein via distinct mechanisms (de The, 2015), can now cure the majority of patients with acute promyelocytic leukaemia without exposure to cytotoxic chemotherapies (Lo-Coco et al, 2013). However, clinical outcomes for a substantial fraction of acute leukaemias have seen little improvement despite the application of treatment regimens that are among the most intensive and toxic used for any disease. Consequently, improving clinical outcomes for these patients will require linking an improved molecular understanding to the application of effective targeted therapies.
Classification of acute leukaemias
Acute leukaemias have traditionally been classified based on the normal cell types most closely resembling the leukaemic cell population. Lymphoblastic leukaemias are those with evidence of differentiation arrest at immature stages of B- or T-cell lymphoid development, whereas acute myeloid leukaemias encompass malignancies with immunophenotypic features that lie along a broad spectrum of haematopoietic progenitors, ranging from minimally differentiated leukaemias to those with evidence of granulocytic, monocytic, erythroid or megakaryocytic differentiation. Although this classification scheme is intuitive, lineage assignment is complicated by the high frequency of aberrant differentiation states in acute leukaemias. Indeed, the simultaneous expression of markers that are not known to be co-expressed in any normal haematopoietic progenitor is common. To address the requirement for consistent diagnostic criteria for the successful performance and interpretation of clinical trials, the World Health Organization (WHO) has devised a classification scheme that allows most acute leukaemias to be unambiguously classified as specific diagnostic entities, which has been regularly revised (Swerdlow et al 2008; Arber et al, 2016).
The genetics of leukaemia are beginning to be incorporated into the WHO classification, with increasing weight being given to specific chromosomal translocations and, in some cases, somatic gene mutations, as disease-defining genetic lesions (Arber et al, 2016). Thus, the presence of a characteristic genetic feature can define a disease, independent of the phenotypic lineage which can be difficult to assign in practice. For example, leukaemias with FGFR1 translocations that can present with T cell lymphoblastic or myeloid markers are defined as a single diagnostic entity, regardless of the immunophenotype of the presenting leukaemic cells (Arber et al, 2016; Macdonald et al, 2002). Similarly, acute myeloid leukaemia (AML) cases with t(8;21) translocations exhibit aberrant expression of CD19, a marker typically associated with B cell lymphoid malignancies, due to lineage-inappropriate PAX5 expression induced by the RUNX1-RUNX1T1 (previously termed AML1-ETO) oncogene (Walter et al, 2010). While we anticipate that genomic classification will have an increasingly important role in defining leukaemia subsets in the future, the current diagnostic classification for most acute leukaemias does not rely on genetic mutations. Thus, phenotypic lineage assignment remains a central component of diagnostic classification and treatment assignment for most patients with acute leukaemia.
The current WHO classification considers strong expression of a small number of immunophenotypic markers to be lineage-assigning: myeloperoxidase (MPO) or at least two monocytic markers (CD11c, CD14, CD64 or lysozyme) for myeloid lineage; surface or cytoplasmic CD3 expression for T cell lineage; and CD19 with at least one additional B cell marker (CD79a, cytoplasmic CD22, or CD10) for B cell lineage (Arber et al, 2016). While the specificity of these markers is widely agreed upon, some prominent clinical trials groups use subtly different criteria for lineage assignment (Dworzak et al, 2017). Nevertheless, it is recognized that these classification schemes are imperfect, and the strict application of the WHO criteria for lineage assignment is not required to make a diagnosis of AML, T cell acute lymphoblastic leukaemia (T-ALL) or B cell acute lymphoblastic leukaemia (B-ALL) unless a diagnosis of mixed phenotype acute leukaemia (MPAL) is also being considered (Arber et al, 2016). Thus, while MPALs that meet formal criteria for assignment to two distinct lineages are very rare, leukaemias classified as AML, T-ALL or B-ALL based on their predominant morphology and immunophenotype commonly co-express markers that indicate aberrant differentiation towards another lineage.
Acute myeloid/T-lymphoblastic leukaemia (AMTL): Acute leukaemias with shared T cell lymphoid and myeloid features
The classical model of haematopoiesis postulates an early binary split between common myeloid and lymphoid progenitors that subsequently give rise to both B- and T-cell lymphocytes. However, more recent work using clonal tracking assays has shown the existence of progenitors that retain T cell/myeloid or B cell/myeloid bi-lineage potential. By contrast, individual progenitors whose potential is restricted to T cell and B cell (but not myeloid) lineages have been difficult to identify (Kawamoto et al, 2010). These findings fit the clinical observation that MPALs most commonly present with B cell/myeloid or T cell/myeloid marker co-expression, whereas B/T cell MPALs are very rare (Matutes et al, 2011).
Here, we propose AMTL as a molecularly distinct subtype of acute leukaemias associated with shared T cell lymphoid and myeloid features. Expression of markers deemed to be defining of the myeloid and T cell lymphoid lineages, including myeloperoxidase and CD3, is variable in this disease, resulting in its separation across three diagnostic categories in the current WHO classification scheme: early T cell precursor (ETP) T-ALL, T/myeloid MPAL, and a specific subset of AML harbouring hallmarks of T-lymphoblastic differentiation.
This proposed diagnostic entity overlaps with subsets of ETP T-ALL, which is characterized by differentiation arrest at early stages of T cell development. While a number of biomarkers have been described to identify these cases (Coustan-Smith et al, 2009; Gutierrez et al, 2010; Homminga et al, 2011; Zuurbier et al, 2014), these are most commonly defined clinically using an immunophenotypic classifier that includes absent expression of CD1a and CD8, CD5 expression that is substantially lower than that of normal peripheral blood T cells, and the presence of one or more markers of myeloid or haematopoietic progenitors (CD117, CD34, HLA-DR, CD13, CD33, CD11b or CD65) (Conter et al, 2016; Coustan-Smith et al, 2009; Patrick et al, 2014). Currently, ETP is classified as a subtype of T-ALL due to the expression of CD3, but has a genetic mutational profile that is similar to those observed in myeloid malignancies, such as AML, whereas mutations characteristic of other subtypes of T-ALL, such as CDKN2A deletions, are less common (Gutierrez et al, 2010; Van Vlierberghe et al, 2011a; Zhang et al, 2012).
While similarities between ETP T-ALL and AML have been previously recognized (Van Vlierberghe et al, 2011a; Zhang et al, 2012; Zuurbier et al, 2014), we propose that the biological overlap of AMTL is not with AML per se, but with a specific subset of AML cases that also exhibit T cell lymphoid features. Namely, a subset of AMLs has long been recognized to harbour clonal T-cell receptor (TCR) or immunoglobulin (Ig) gene rearrangements, indicating activity of the RAG recombinase that is responsible for generating somatic V(D)J recombination at these loci at specific stages of lymphoid development (Norton et al, 1987; Parreira et al, 1992; Schmidt et al, 1992). These AML cases can also express the lymphoid marker terminal deoxynucleotidyltransferase (TdT, also known as DNTT), which generates diversity at the TCR and Ig genes by mutating the junctions of rearrangements during V(D)J recombination (Drexler et al, 1993; Patel et al, 2013). Non-megakaryoblastic AMLs with TCR rearrangements typically co-express phenotypic markers of T-lymphoblastic differentiation, such as CD7, CD2 and CD4 (Schmidt et al, 1992). We postulate that this is the subset of AMLs that also harbour mutations that are commonly observed in a distinct subset of T-ALL, including WT1, PHF6, RUNX1 and BCL11B (The Cancer Genome Atlas Research Network, 2013; Della Gatta et al, 2012; Ding et al, 2012; Klco et al, 2015; Tosello et al, 2009; Van Vlierberghe et al, 2010; Van Vlierberghe et al, 2011b; Zhang et al, 2012). Thus, a similar group of acute leukaemias exhibit shared features of myeloid and T cell lymphoid differentiation, and shared genetic mutations, which will need to be formally assessed in future studies.
Potential mechanisms for combined T-lymphoblastic and myeloid differentiation in acute leukaemia
At least two non-mutually exclusive mechanisms can explain the development of acute leukaemias with differentiation potential shared with myeloid and T-lymphoid lineages:
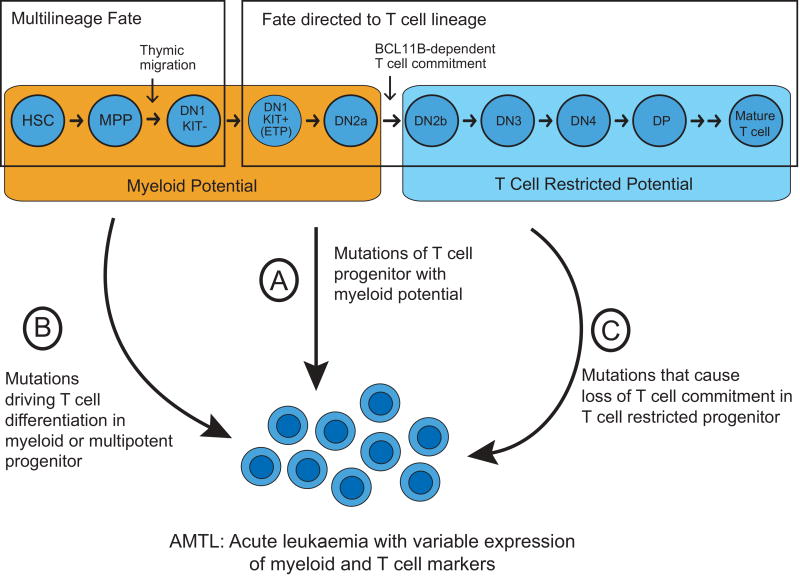
Transformation of normal haematopoietic progenitors with T-lymphoblastic and myeloid bi-lineage potential. While studies of endogenous haematopoiesis have revealed limited evidence of a putative normal progenitor whose fate is restricted to T-lymphoid and myeloid lineages, the identification of such a progenitor may have been hindered by differences in the lifespan of mature myeloid versus T-lymphoid cells. Nevertheless, even if a normal progenitor whose differentiation fate is restricted to T-lymphocytes and myeloid cells is lacking, such leukaemias can arise from cells that retain multi-lineage potential, even though their fate is normally restricted to a single lineage by microenvironmental signals. Immature intrathymic T cell progenitors at early double-negative stages of differentiation represent one potential cell of origin for such leukaemias. Indeed, the most immature intrathymic T cell progenitors retain myeloid (but not B-lymphoid) potential until they progress through a BCL11B-dependent commitment to the T cell lineage (Balciunaite et al, 2005; Ikawa et al, 2010; Li et al, 2010) (Figure 1). Further, lineage tracing studies have demonstrated that immature T cell progenitors give rise to some macrophages and neutrophils harbouring TCR gene rearrangements in vivo (Bell & Bhandoola, 2008; Wada et al, 2008). Ectopic expression of Myc and Bcl2 in mouse early double-negative T cell progenitors can drive development of acute leukaemias with variable expression of markers of myeloid and T-lymphoid lineages (Riemke et al, 2016), demonstrating that early T cell progenitors can function as the potential cell of origin of AMTL.
Induction of aberrant trans-differentiation by leukemogenic mutations. It is now established that specific gene mutations can induce T cell lymphoid or myeloid differentiation. For example, activation of NOTCH1 signalling in mouse bone marrow cells causes aberrant T cell differentiation independent of thymic microenvironmental signals (Pui et al, 1999). Likewise, mutations inactivating Runx1 in haematopoietic progenitors cause, aberrant myeloid differentiation (Goyama et al, 2013). Thus, aberrant trans-differentiation of AMTL cells can in principle, occur as a result of combinations of mutations that cooperatively result in combined myeloid and T-lymphoid lineage commitment with differentiation arrest (Figure 1). These mutations include genes encoding regulators of chromatin and DNA remodelling, and transcription factors with key functions in controlling lymphoid and myeloid cell fate specification. Future studies using emerging approaches for combinatorial gene editing and conditional gene manipulation in vivo may reveal how the specific combinations of mutations in specific cell progenitor populations cooperate to induce AMTL and other non-canonical leukaemia subtypes.
Figure 1. Potential mechanisms leading to acute leukaemia with shared myeloid and T-lymphoblastic features.
The fate of multipotent haematopoietic progenitors first entering the thymus is directed to the T cell lineage via thymic microenvironmental signals, but these cells retain myeloid potential until BCL11B-dependent commitment to the T cell lineage at the DN2a to DN2b transition (Hosokawa & Rothenberg, 2017). Such T cell progenitors with myeloid potential can function as a cell of origin of AMTL (Riemke et al, 2016) (A). Alternatively, AMTL can in principle arise from mutations that induce aberrant T cell differentiation in myeloid or multipotent haematopoietic progenitors (B), or from mutations in T cell restricted progenitors leading to their myeloid differentiation (C). Note that the developmental stages shown (top) are based on mouse T cell development, because human early T cell development is less well-defined.
AMTL: acute myeloid/T-lymphoblastic leukaemia; DN: CD4/CD8 double-negative T cell progenitor; DP: CD4/CD8 double-positive T cell progenitor; ETP: early T cell precursor; HSC: haematopoietic stem cell; MPP: multipotent progenitor.
Identification of Acute Myeloid/T-Lymphoblastic Leukaemia
To a first approximation, AMTL can be defined as acute leukaemias that fit the diagnostic criteria of ETP T-ALL or of T/myeloid MPALs, together with AMLs with clonal TCR gene rearrangements and evidence of T-lymphoid differentiation (CD3, CD7, CD2 or CD4 expression). However, we note that aberrant expression of CD4 and CD7 is relatively common in acute megakaryoblastic leukaemia (AMKL), a subtype of AML that is molecularly distinct from AMTL (de Rooij et al, 2017). Therefore, AMKL should be excluded from the definition of AMTL, based on absence of megakaryblastic morphology and expression of the platelet markers CD41 or CD61. Thus, our initial proposal for AMTL as a diagnostic entity includes all cases currently classified as ETP T-ALL and T/myeloid MPALs, together with the subset of AMLs with TCR rearrangements and expression of T-lymphoblastic markers.
Given the variability in cell surface marker expression in acute leukaemias, we propose that clinical genomic profiling will enhance the classification of AMTL as a specific diagnostic entity. Indeed, ETP T-ALLs harbour mutations of several genes that are commonly mutated in myeloid neoplasms, such as RAS family members, ETV6, MEF2C and EZH2, but whose mutations are rare in other T-ALL subtypes, (Homminga et al, 2011; Van Vlierberghe et al, 2011a; Zhang et al, 2012; Zuurbier et al, 2014). We also suggest that the subset of AML cases harbouring clonal TCR gene rearrangements should demonstrate substantial overlap with those AML cases harbouring mutations that are common in T-ALL, such as WT1, RUNX1, PHF6 and BCL11B (The Cancer Genome Atlas Research Network, 2013; Ding et al, 2012; Klco et al, 2015; Zhang et al, 2012). We anticipate that future investigation will lead to an improved classifier of AMTL based on both immunophenotypic and genetic diagnostic markers.
Conclusions
We propose acute myeloid/T-lymphoblastic leukaemia (AMTL) as a new diagnostic entity for acute leukaemias with shared myeloid and T cell lymphoblastic features. Defining the detailed molecular mechanisms of aberrant cell differentiation may lead to the development of improved therapies by targeting specific molecular dependencies in these cells. For example, mutant transcription factors function in the context of corepressor and coactivator complexes, which have specific enzymatic functions, such as histone deacetylase (HDAC) inhibitors (Haberland et al, 2009). Isoform-specific HDAC inhibitors are beginning to be developed (West & Johnstone, 2014), and specific inhibitors may warrant therapeutic investigation in AMTL with dysregulation of MEF2 family members, for example. Likewise, specific mutations may engender synthetic lethal dependencies, such as, for example, inhibition of the EZH2 methyltransferase in cases of cancers with mutations of genes encoding components of the SWItch/Sucrose Non-Fermentable/BRG1-associated factors (SWI/SNF/BAF) chromatin remodelling complex (Kim et al, 2015). Recently, acetyltranferase inhibitors have been found to have therapeutic efficacy in CBP-deficient lymphomas (Ogiwara et al, 2016). Finally, many leukaemias exhibit aberrant resistance to mitochondrial apoptosis, conferring resistance to intensive combination chemotherapies, such as those used for treatment of refractory AML and T-ALL. Recent data suggest that AMTLs are particularly susceptible to inhibitors of BCL2 (Chonghaile et al, 2014; Pan et al, 2014), and emerging inhibitors of other regulators of intrinsic apoptosis are expected to offer useful therapeutic strategies for AMTL patients as well.
Acknowledgments
We thank Misha Roshal, Mark Fleming, and Peter Steinherz for helpful discussions. This work was supported by NIH R01 CA204396 and CA193651, and P30 CA008748. A.G. and A.K. are Damon Runyon Clinical Investigators.
References
- Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, Bloomfield CD, Cazzola M, Vardiman JW. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- Balciunaite G, Ceredig R, Rolink AG. The earliest subpopulation of mouse thymocytes contains potent T, significant macrophage, and natural killer cell but no B-lymphocyte potential. Blood. 2005;105:1930–1936. doi: 10.1182/blood-2004-08-3087. [DOI] [PubMed] [Google Scholar]
- Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 2008;452:764–767. doi: 10.1038/nature06840. [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chonghaile TN, Roderick JE, Glenfield C, Ryan J, Sallan SE, Silverman LB, Loh ML, Hunger SP, Wood B, DeAngelo DJ, Stone R, Harris M, Gutierrez A, Kelliher MA, Letai A. Maturation stage of T-cell acute lymphoblastic leukemia determines BCL-2 versus BCL-XL dependence and sensitivity to ABT-199. Cancer Discov. 2014;4:1074–1087. doi: 10.1158/2159-8290.CD-14-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conter V, Valsecchi MG, Buldini B, Parasole R, Locatelli F, Colombini A, Rizzari C, Putti MC, Barisone E, Lo Nigro L, Santoro N, Ziino O, Pession A, Testi AM, Micalizzi C, Casale F, Pierani P, Cesaro S, Cellini M, Silvestri D, Cazzaniga G, Biondi A, Basso G. Early T-cell precursor acute lymphoblastic leukaemia in children treated in AIEOP centres with AIEOP-BFM protocols: a retrospective analysis. Lancet Haematol. 2016;3:e80–86. doi: 10.1016/S2352-3026(15)00254-9. [DOI] [PubMed] [Google Scholar]
- Coustan-Smith E, Mullighan CG, Onciu M, Behm FG, Raimondi SC, Pei D, Cheng C, Su X, Rubnitz JE, Basso G, Biondi A, Pui CH, Downing JR, Campana D. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. 2009;10:147–156. doi: 10.1016/S1470-2045(08)70314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij JD, Branstetter C, Ma J, Li Y, Walsh MP, Cheng J, Obulkasim A, Dang J, Easton J, Verboon LJ, Mulder HL, Zimmermann M, Koss C, Gupta P, Edmonson M, Rusch M, Lim JY, Reinhardt K, Pigazzi M, Song G, Yeoh AE, Shih LY, Liang DC, Halene S, Krause DS, Zhang J, Downing JR, Locatelli F, Reinhardt D, van den Heuvel-Eibrink MM, Zwaan CM, Fornerod M, Gruber TA. Pediatric non-Down syndrome acute megakaryoblastic leukemia is characterized by distinct genomic subsets with varying outcomes. Nat Genet. 2017;49:451–456. doi: 10.1038/ng.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de The H. Lessons taught by acute promyelocytic leukemia cure. Lancet. 2015;386:247–248. doi: 10.1016/S0140-6736(15)61278-8. [DOI] [PubMed] [Google Scholar]
- Della Gatta G, Palomero T, Perez-Garcia A, Ambesi-Impiombato A, Bansal M, Carpenter ZW, De Keersmaecker K, Sole X, Xu L, Paietta E, Racevskis J, Wiernik PH, Rowe JM, Meijerink JP, Califano A, Ferrando AA. Reverse engineering of TLX oncogenic transcriptional networks identifies RUNX1 as tumor suppressor in T-ALL. Nat Med. 2012;18:436–440. doi: 10.1038/nm.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD, McMichael JF, Wallis JW, Lu C, Shen D, Harris CC, Dooling DJ, Fulton RS, Fulton LL, Chen K, Schmidt H, Kalicki-Veizer J, Magrini VJ, Cook L, McGrath SD, Vickery TL, Wendl MC, Heath S, Watson MA, Link DC, Tomasson MH, Shannon WD, Payton JE, Kulkarni S, Westervelt P, Walter MJ, Graubert TA, Mardis ER, Wilson RK, DiPersio JF. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler HG, Sperling C, Ludwig WD. Terminal deoxynucleotidyl transferase (TdT) expression in acute myeloid leukemia. Leukemia. 1993;7:1142–1150. [PubMed] [Google Scholar]
- Dworzak MN, Buldini B, Gaipa G, Ratei R, Hrusak O, Luria D, Rosenthal E, Bourquin JP, Sartor M, Schumich A, Karawajew L, Mejstrikova E, Maglia O, Mann G, Ludwig WD, Biondi A, Schrappe M, Basso G. AIEOP-BFM Consensus Guidelines 2016 for flow cytometric immunophenotyping of Pediatric acute lymphoblastic leukemia. Cytometry Part B. 2017 doi: 10.1002/cyto.b.21518. [DOI] [PubMed] [Google Scholar]
- Goyama S, Schibler J, Cunningham L, Zhang Y, Rao Y, Nishimoto N, Nakagawa M, Olsson A, Wunderlich M, Link KA, Mizukawa B, Grimes HL, Kurokawa M, Liu PP, Huang G, Mulloy JC. Transcription factor RUNX1 promotes survival of acute myeloid leukemia cells. J Clin Invest. 2013;123:3876–3888. doi: 10.1172/JCI68557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez A, Dahlberg SE, Neuberg DS, Zhang J, Grebliunaite R, Sanda T, Protopopov A, Tosello V, Kutok J, Larson RS, Borowitz MJ, Loh ML, Ferrando AA, Winter SS, Mullighan CG, Silverman LB, Chin L, Hunger SP, Sallan SE, Look AT. Absence of biallelic TCRgamma deletion predicts early treatment failure in pediatric T-cell acute lymphoblastic leukemia. J Clin Oncol. 2010;28:3816–3823. doi: 10.1200/JCO.2010.28.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homminga I, Pieters R, Langerak AW, de Rooi JJ, Stubbs A, Verstegen M, Vuerhard M, Buijs-Gladdines J, Kooi C, Klous P, van Vlierberghe P, Ferrando AA, Cayuela JM, Verhaaf B, Beverloo HB, Horstmann M, de Haas V, Wiekmeijer AS, Pike-Overzet K, Staal FJ, de Laat W, Soulier J, Sigaux F, Meijerink JP. Integrated transcript and genome analyses reveal NKX2-1 and MEF2C as potential oncogenes in T cell acute lymphoblastic leukemia. Cancer Cell. 2011;19:484–497. doi: 10.1016/j.ccr.2011.02.008. [DOI] [PubMed] [Google Scholar]
- Hosokawa H, Rothenberg EV. Cytokines, Transcription Factors, and the Initiation of T-Cell Development. Cold Spring Harb Perspect Biol. 2017 Jul 17; doi: 10.1101/cshperspect.a028621. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikawa T, Hirose S, Masuda K, Kakugawa K, Satoh R, Shibano-Satoh A, Kominami R, Katsura Y, Kawamoto H. An essential developmental checkpoint for production of the T cell lineage. Science. 2010;329:93–96. doi: 10.1126/science.1188995. [DOI] [PubMed] [Google Scholar]
- Kawamoto H, Ikawa T, Masuda K, Wada H, Katsura Y. A map for lineage restriction of progenitors during hematopoiesis: the essence of the myeloid-based model. Immunol Rev. 2010;238:23–36. doi: 10.1111/j.1600-065X.2010.00959.x. [DOI] [PubMed] [Google Scholar]
- Kim KH, Kim W, Howard TP, Vazquez F, Tsherniak A, Wu JN, Wang W, Haswell JR, Walensky LD, Hahn WC, Orkin SH, Roberts CW. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nat Med. 2015;21:1491–1496. doi: 10.1038/nm.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klco JM, Miller CA, Griffith M, Petti A, Spencer DH, Ketkar-Kulkarni S, Wartman LD, Christopher M, Lamprecht TL, Helton NM, Duncavage EJ, Payton JE, Baty J, Heath SE, Griffith OL, Shen D, Hundal J, Chang GS, Fulton R, O'Laughlin M, Fronick C, Magrini V, Demeter RT, Larson DE, Kulkarni S, Ozenberger BA, Welch JS, Walter MJ, Graubert TA, Westervelt P, Radich JP, Link DC, Mardis ER, DiPersio JF, Wilson RK, Ley TJ. Association Between Mutation Clearance After Induction Therapy and Outcomes in Acute Myeloid Leukemia. JAMA. 2015;314:811–822. doi: 10.1001/jama.2015.9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Leid M, Rothenberg EV. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science. 2010;329:89–93. doi: 10.1126/science.1188989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo-Coco F, Avvisati G, Vignetti M, Thiede C, Orlando SM, Iacobelli S, Ferrara F, Fazi P, Cicconi L, Di Bona E, Specchia G, Sica S, Divona M, Levis A, Fiedler W, Cerqui E, Breccia M, Fioritoni G, Salih HR, Cazzola M, Melillo L, Carella AM, Brandts CH, Morra E, von Lilienfeld-Toal M, Hertenstein B, Wattad M, Lubbert M, Hanel M, Schmitz N, Link H, Kropp MG, Rambaldi A, La Nasa G, Luppi M, Ciceri F, Finizio O, Venditti A, Fabbiano F, Dohner K, Sauer M, Ganser A, Amadori S, Mandelli F, Dohner H, Ehninger G, Schlenk RF, Platzbecker U, Gruppo Italiano Malattie Ematologiche dA, German-Austrian Acute Myeloid Leukemia Study G, Study Alliance L. Retinoic acid and arsenic trioxide for acute promyelocytic leukemia. N Engl J Med. 2013;369:111–121. doi: 10.1056/NEJMoa1300874. [DOI] [PubMed] [Google Scholar]
- Macdonald D, Reiter A, Cross NC. The 8p11 myeloproliferative syndrome: a distinct clinical entity caused by constitutive activation of FGFR1. Acta Haematol. 2002;107:101–107. doi: 10.1159/000046639. [DOI] [PubMed] [Google Scholar]
- Matutes E, Pickl WF, Van't Veer M, Morilla R, Swansbury J, Strobl H, Attarbaschi A, Hopfinger G, Ashley S, Bene MC, Porwit A, Orfao A, Lemez P, Schabath R, Ludwig WD. Mixed-phenotype acute leukemia: clinical and laboratory features and outcome in 100 patients defined according to the WHO 2008 classification. Blood. 2011;117:3163–3171. doi: 10.1182/blood-2010-10-314682. [DOI] [PubMed] [Google Scholar]
- Norton JD, Campana D, Hoffbrand AV, Janossy G, Coustan-Smith E, Jani H, Yaxley JC, Prentice HG. Rearrangement of immunoglobulin and T cell antigen receptor genes in acute myeloid leukemia with lymphoid-associated markers. Leukemia. 1987;1:757–761. [PubMed] [Google Scholar]
- Ogiwara H, Sasaki M, Mitachi T, Oike T, Higuchi S, Tominaga Y, Kohno T. Targeting p300 Addiction in CBP-Deficient Cancers Causes Synthetic Lethality by Apoptotic Cell Death due to Abrogation of MYC Expression. Cancer Discov. 2016;6:430–445. doi: 10.1158/2159-8290.CD-15-0754. [DOI] [PubMed] [Google Scholar]
- Pan R, Hogdal LJ, Benito JM, Bucci D, Han L, Borthakur G, Cortes J, DeAngelo DJ, Debose L, Mu H, Dohner H, Gaidzik VI, Galinsky I, Golfman LS, Haferlach T, Harutyunyan KG, Hu J, Leverson JD, Marcucci G, Muschen M, Newman R, Park E, Ruvolo PP, Ruvolo V, Ryan J, Schindela S, Zweidler-McKay P, Stone RM, Kantarjian H, Andreeff M, Konopleva M, Letai AG. Selective BCL-2 inhibition by ABT-199 causes on-target cell death in acute myeloid leukemia. Cancer Discov. 2014;4:362–375. doi: 10.1158/2159-8290.CD-13-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parreira L, Carvalho C, Moura H, Melo A, Santos P, Guimaraes JE, Parreira A. Configuration of immunoglobulin and T cell receptor beta and gamma genes in acute myeloid leukaemia: pitfalls in the analysis of 40 cases. J Clin Pathol. 1992;45:193–200. doi: 10.1136/jcp.45.3.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel KP, Khokhar FA, Muzzafar T, James You M, Bueso-Ramos CE, Ravandi F, Pierce S, Medeiros LJ. TdT expression in acute myeloid leukemia with minimal differentiation is associated with distinctive clinicopathological features and better overall survival following stem cell transplantation. Mod Pathol. 2013;26:195–203. doi: 10.1038/modpathol.2012.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick K, Wade R, Goulden N, Mitchell C, Moorman AV, Rowntree C, Jenkinson S, Hough R, Vora A. Outcome for children and young people with Early T-cell precursor acute lymphoblastic leukaemia treated on a contemporary protocol, UKALL 2003. Br J Haematol. 2014;166:421–424. doi: 10.1111/bjh.12882. [DOI] [PubMed] [Google Scholar]
- Pui JC, Allman D, Xu L, DeRocco S, Karnell FG, Bakkour S, Lee JY, Kadesch T, Hardy RR, Aster JC, Pear WS. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11:299–308. doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- Riemke P, Czeh M, Fischer J, Walter C, Ghani S, Zepper M, Agelopoulos K, Lettermann S, Gebhardt ML, Mah N, Weilemann A, Grau M, Groning V, Haferlach T, Lenze D, Delwel R, Prinz M, Andrade-Navarro MA, Lenz G, Dugas M, Muller-Tidow C, Rosenbauer F. Myeloid leukemia with transdifferentiation plasticity developing from T-cell progenitors. EMBO J. 2016;35:2399–2416. doi: 10.15252/embj.201693927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt CA, Oettle H, Neubauer A, Seeger K, Thiel E, Huhn D, Siegert W, Ludwig WD. Rearrangements of T-cell receptor delta, gamma and beta genes in acute myeloid leukemia coexpressing T-lymphoid features. Leukemia. 1992;6:1263–1267. [PubMed] [Google Scholar]
- Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW. WHO classification of tumours of haematopoietic and lymphoid tissues. 4. International Agency for Research on Cancer Press; Lyon, France: 2008. [Google Scholar]
- The Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosello V, Mansour MR, Barnes K, Paganin M, Sulis ML, Jenkinson S, Allen CG, Gale RE, Linch DC, Palomero T, Real P, Murty V, Yao X, Richards SM, Goldstone A, Rowe J, Basso G, Wiernik PH, Paietta E, Pieters R, Horstmann M, Meijerink JP, Ferrando AA. WT1 mutations in T-ALL. Blood. 2009;114:1038–1045. doi: 10.1182/blood-2008-12-192039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vlierberghe P, Palomero T, Khiabanian H, Van der Meulen J, Castillo M, Van Roy N, De Moerloose B, Philippe J, Gonzalez-Garcia S, Toribio ML, Taghon T, Zuurbier L, Cauwelier B, Harrison CJ, Schwab C, Pisecker M, Strehl S, Langerak AW, Gecz J, Sonneveld E, Pieters R, Paietta E, Rowe JM, Wiernik PH, Benoit Y, Soulier J, Poppe B, Yao X, Cordon-Cardo C, Meijerink J, Rabadan R, Speleman F, Ferrando A. PHF6 mutations in T-cell acute lymphoblastic leukemia. Nat Genet. 2010;42:338–342. doi: 10.1038/ng.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vlierberghe P, Ambesi-Impiombato A, Perez-Garcia A, Haydu JE, Rigo I, Hadler M, Tosello V, Della Gatta G, Paietta E, Racevskis J, Wiernik PH, Luger SM, Rowe JM, Rue M, Ferrando AA. ETV6 mutations in early immature human T cell leukemias. J Exp Med. 2011a;208:2571–2579. doi: 10.1084/jem.20112239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vlierberghe P, Patel J, Abdel-Wahab O, Lobry C, Hedvat CV, Balbin M, Nicolas C, Payer AR, Fernandez HF, Tallman MS, Paietta E, Melnick A, Vandenberghe P, Speleman F, Aifantis I, Cools J, Levine R, Ferrando A. PHF6 mutations in adult acute myeloid leukemia. Leukemia. 2011b;25:130–134. doi: 10.1038/leu.2010.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H, Masuda K, Satoh R, Kakugawa K, Ikawa T, Katsura Y, Kawamoto H. Adult T-cell progenitors retain myeloid potential. Nature. 2008;452:768–772. doi: 10.1038/nature06839. [DOI] [PubMed] [Google Scholar]
- Walter K, Cockerill PN, Barlow R, Clarke D, Hoogenkamp M, Follows GA, Richards SJ, Cullen MJ, Bonifer C, Tagoh H. Aberrant expression of CD19 in AML with t(8;21) involves a poised chromatin structure and PAX5. Oncogene. 2010;29:2927–2937. doi: 10.1038/onc.2010.56. [DOI] [PubMed] [Google Scholar]
- West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest. 2014;124:30–39. doi: 10.1172/JCI69738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, Lu C, Chen SC, Wei L, Collins-Underwood JR, Ma J, Roberts KG, Pounds SB, Ulyanov A, Becksfort J, Gupta P, Huether R, Kriwacki RW, Parker M, McGoldrick DJ, Zhao D, Alford D, Espy S, Bobba KC, Song G, Pei D, Cheng C, Roberts S, Barbato MI, Campana D, Coustan-Smith E, Shurtleff SA, Raimondi SC, Kleppe M, Cools J, Shimano KA, Hermiston ML, Doulatov S, Eppert K, Laurenti E, Notta F, Dick JE, Basso G, Hunger SP, Loh ML, Devidas M, Wood B, Winter S, Dunsmore KP, Fulton RS, Fulton LL, Hong X, Harris CC, Dooling DJ, Ochoa K, Johnson KJ, Obenauer JC, Evans WE, Pui CH, Naeve CW, Ley TJ, Mardis ER, Wilson RK, Downing JR, Mullighan CG. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuurbier L, Gutierrez A, Mullighan CG, Cante-Barrett K, Gevaert AO, de Rooi J, Li Y, Smits WK, Buijs-Gladdines JG, Sonneveld E, Look AT, Horstmann M, Pieters R, Meijerink JP. Immature MEF2C-dysregulated T-cell leukemia patients have an early T-cell precursor acute lymphoblastic leukemia gene signature and typically have non-rearranged T-cell receptors. Haematologica. 2014;99:94–102. doi: 10.3324/haematol.2013.090233. [DOI] [PMC free article] [PubMed] [Google Scholar]