Abstract
Bioinks with shear-thinning/rapid solidification properties and strong mechanics are usually needed for the bioprinting of three-dimensional (3D) cell-laden constructs. As such, it remains challenging to generate soft constructs from bioinks at low concentrations that are favorable for cellular activities. Herein, we report a strategy to fabricate cell-laden constructs with tunable 3D microenvironments achieved by bioprinting of gelatin methacryloyl (GelMA)/alginate core/sheath microfibers, where the alginate sheath serves as a template to support and confine the GelMA pre-hydrogel in the core during the extrusion process, allowing for subsequent UV crosslinking. This novel strategy minimizes the bioprinting requirements for the core bioink, and facilitates the fabrication of cell-laden GelMA constructs at low concentrations. We showed the capability of generating various alginate hollow microfibrous constructs using a coaxial nozzle setup, and verified the diffusibility and perfusability of the bioprinted hollow structures which are important for the tissue-engineering applications. More importantly, the hollow alginate microfibers were used as templates for generating cell-laden GelMA constructs with soft microenvironments, by using GelMA pre-hydrogel as the bioink for the core phase during bioprinting. As such, GelMA constructs at extremely low concentrations (down to 1.5%) could be extruded to effectively support cellular activities including proliferation and spreading for various cell types. We believe that our strategy is likely to provide broad opportunities in bioprinting 3D constructs with cell-favorable microenvironments for applications in tissue engineering and pharmaceutical screening.
Keywords: Bioprinting, alginate, gelatin methacryloyl (GelMA), cell-laden, hydrogel, tissue engineering
1. Introduction
Extrusion bioprinting has attracted widespread attentions as it is a relatively simple technique that holds the potential to generate functional three-dimensional (3D) tissues or tissue models by providing precise spatial manipulation of various components such as living cells, supporting matrix, and functional molecules [1-5]. The bioprinting process usually involves layer-by-layer deposition of bioinks. To bioprint 3D cell-laden constructs with high structural fidelity, the bioinks should be designed with shear-thinning or rapid solidification properties [4, 6, 7]. In addition, the bioprinting process should be benign to ensure sufficient cell survival during bioprinting [8, 9]. Moreover, the bioprinted constructs should provide the encapsulated cells with favorable environments for proliferation, spreading, migration, and other normal cell activities [3, 4, 10].
Natural biomaterials are promising for the use in bioprinting due to their distinct cytocompatibility [11, 12]. To date, the natural biomaterials adapted for bioprinting include alginate [13-17], hyaluronic acid (HA) [6], collagen/gelatin [18] and their derivatives (e.g., gelatin methacryloyl (GelMA)) [19, 20], and fibrin [21], among others. For example, alginate has been bioprinted often through the use of core/sheath coaxial nozzle based on an ionic crosslinking mechanism [13-17]. HA has been modified into a shear-thinning and self-healing formulation using guest-host chemistry for bioprinting [6]. To improve cell adhesion and proliferation, both alginate and HA are usually further modified with the arginine-glycineaspartic acid (RGD) peptide sequence or blended with other functional components due to their lack of cell-adhesive moieties [6, 7, 22]. On the other hand, natural polymers that possess intrinsic cell-adhesion sites have also been directly used for bioprinting. In an example, bioprinting of collagen has been achieved by using a heated collector for thermal gelation and a biocompatible crosslinker of genipin for post-printing treatment [23]. In addition, collagen constructs have also been bioprinted using an aerosol crosslinking method via a core/sheath coaxial nozzle [24].
More recently, GelMA as a versatile chemically modified form of gelatin, has been widely used for extrusion bioprinted through in situ photocrosslinking or physical gelation [4, 10, 19, 20, 25]. However, the concentration of GelMA bioinks used for bioprinting is usually limited to relatively high values due to the requirement of rapid solidification for the formation of 3D constructs during the deposition process [10]. We have recently developed a bioink preparation method based on GelMA physical gels for direct GelMA bioprinting, but the concentration was still limited to approximately 3.0% [26]. Alternatively, a microfluidic bioprinting strategy has also demonstrated the possibility of extrusion of GelMA/alginate blend bioinks using a coaxial nozzle setup, where the alginate component could be immediately crosslinked with CaCl2 to achieve microfibrous structures followed by UV crosslinking of the GelMA component [7, 27, 28]. Nevertheless, the presence of alginate in the bioprinted microfibers inevitably affects the behavior of encapsulated cells, although it could be gradually released using a Ca2+-chelator.
In this study, we report a novel strategy to fabricate cell-laden constructs with tunable 3D microenvironments by bioprinting GelMA/alginate core/sheath microfibers using a coaxial nozzle setup, where the physically crosslinked alginate sheath serves as the template to confine the GelMA bioink in the core and to allow for subsequent UV crosslinking (Figure 1). This strategy minimizes the bioprinting requirements for the core bioink, facilitating the fabrication of cell-laden GelMA constructs at extremely low concentrations (<2.0%). We firstly examined the influence of the bioprinting parameters on the extrusion performance. To demonstrate potential applications, we then investigated the perfusion and diffusion properties of the bioprinted alginate hollow microfibrous constructs. We finally examined the cellular activities of various cell types in the GelMA cores of the bioprinted GelMA/alginate core/sheath microfibrous constructs.
Figure 1.
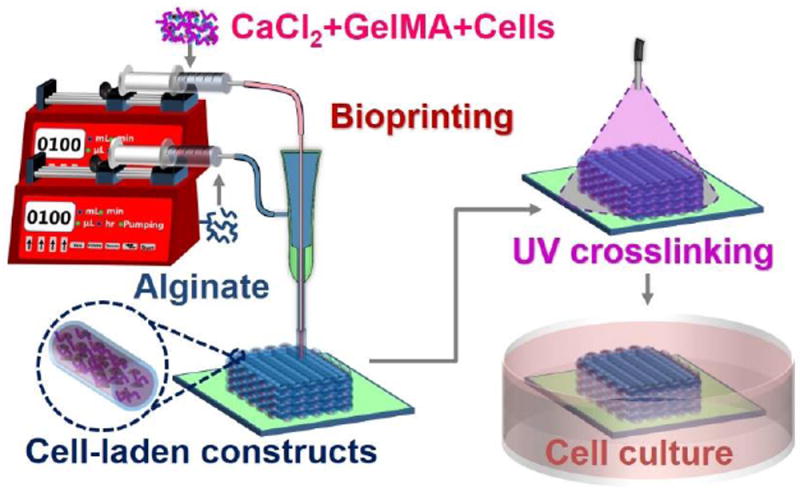
Schematic diagram showing the strategy of bioprinting GelMA/alginate core/sheath microfibers into 3D constructs with tunable microenvironments.
2. Materials and methods
2.1. Materials
Gelatin, methacrylic anhydride, 2-hydroxy-4’-(2-hydroxyethoxy)-2-methylpropiophenone (photoinitiator, PI), alginate, and CaCl2 were purchased from Sigma-Aldrich (St. Louis, MO, USA). GelMA molecules were synthesized according to our previously published protocol [25, 29], at a medium degree of methacryloyl substitution (53.8 ± 0.5%). Cell analysis and culture reagents including Live/Dead kit, PrestoBlue, Alexa 488-phalloidin, 4’,6-diamidino-2- phenylindole (DAPI), fetal bovine serum (FBS), penicillin/streptomycin (P/S), Hank’s balanced salt solution (HBSS), Dulbecco’s phosphate-buffered saline (DPBS), and Dulbecco’s modified eagle medium (DMEM) were purchased from ThermoFisher (Waltham, MA, USA). Endothelial growth medium (EGM)-2 was purchased from Lonza (Walkersville, MD, USA). All reagents were used without further purification. It should be noted that the concentration of GelMA, PI, CaCl2, and alginate was calculated by w/v (i.e., g mL-1), while the concentration of FBS and P/S was calculated by v/v.
2.2. Bioprinter
A commercial 3D printer (Lulzbot TAZ 4, Aleph Objects, Loveland, CO, USA) was modified to be capable of bioprinting hydrogel constructs. Specifically, we designed a printhead that allowed for easy installation and detachment of the custom-made core/sheath coaxial nozzle. The core/sheath coaxial nozzle was made by assembling a pair of 23G and 28G needles, where the core and the sheath were carefully aligned and bonded using epoxy glue. To obtain a stable nozzle with rigid connections, it was further heated in the oven (80 °C) for at least 3 h. Two individual syringe pumps were used to deliver the fluids for the core and sheath of the coaxial nozzle.
2.3. Bioprinting
For the bioprinting of alginate hollow microfibrous constructs, the core and sheath of the nozzle were fed with the CaCl2 solution and the alginate bioink, respectively, where the solvents were both deionized water. Unless otherwise noted, the concentration of the CaCl2 solution and the alginate bioink, the feeding rates of the CaCl2 solution and the alginate bioink (equal rates), and the nozzle moving speed were kept constant at 6.0% and 1.0%, 500 μL min-1, and 500 mm min-1, respectively.
For the bioprinting of GelMA/alginate core/sheath microfibrous constructs, the bioink for the core was 1.0%, 1.5%, or 2.0% GelMA containing 0.2% PI, 1.0% CaCl2, and cells at a density of 2 × 106 mL-1. The solvent of the GelMA bioinks was medium for the respective cell types. The bioink for the sheath was 1.0% alginate dissolved in DPBS. It should be noted that significant cell death was observed when the core GelMA biolinks contained 6.0% CaCl2, and therefore 1.0% CaCl2 was used instead of 6.0%. After mixing the cells in the GelMA bioink, the bioprinting process was initiated immediately and completed in approximately 10 min. After bioprinting, the GelMA bioink in the alginate sheath was further immediately crosslinked by UV illumination at a power of 3.95 W cm-2 (OmniCure S2000, Excelitas Technologies, Waltham, MA, USA), and washed with HBSS for 3 times of 5 min each to remove the residual CaCl2. The feeding rates of the CaCl2 solution and the alginate bioink (equal rates), and the nozzle moving speed were kept constant at 500 μL min-1, and 500 mm min-1, respectively.
2.4. Cell culture
Human umbilical vein endothelial cells (HUVECs, Angio-Proteomie, Boston, MA, USA) were cultured in EGM-2 supplemented with BulletKit and 1% P/S. MDA-MB-231 and MCF7 breast cancer cells, and NIH/3T3 mouse fibroblasts (American Type Culture Collection, Manassas, VA, USA) were cultured in DMEM supplemented with 10% FBS and 1% P/S. Medium was changed every other day. The culture environment was maintained in a 37°C incubator with 5% humidified atmosphere of CO2.
2.5. Characterization of bioprinted constructs
The compressive mechanical properties of the bioprinted constructs after UV crosslinking were measured using a mechanical testing machine (Instron 5943, Instron, Norwood, MA, USA). Scanning electron microscopy (SEM, JSM-5600LV, JEOL, Akishima, Tokyo, Japan) was used to image the freeze-dried constructs achieved by bioprinting of GelMA/alginate core/sheath microfibers. Live/Dead staining was conducted according to the manufacturer’s protocol (ThermoFisher), and imaged using an inverted fluorescence microscope (Zeiss Axio observer D1, Zeiss, Thornwood, NY, USA). Cell viability was quantified by counting the numbers of live and dead cells according to the images using ImageJ. In addition, the morphology of cells encapsulated within the bioprinted constructs was visualized using a laser scanning confocal microscope (IX83, Olympus, Shinjuku-ku, Tokyo, Japan) after F-actin and nuclei staining (phalloidin/DAPI). Cell proliferation was quantified using PrestoBlue by measuring the metabolic activity.
2.6. Fourier-transform infrared (FTIR) and nuclear magnetic resonance (NMR) analysis
Crosslinked and uncrosslinked GelMA samples (1.0%, 1.5%, and 2.0%) containing 0.2% PI and 1.0% CaCl2 were prepared by freeze-drying. The FTIR spectra of freeze-dried samples were measured with a Bruker Microspectrometer (Vertex 70, Bruker Optics Inc, Billerica, MA, USA). The spectra were recorded between 600 and 4000 cm-1 by collecting 128 scans at a 2-cm-1 resolution in the ATR mode. Proton NMR (1H NMR) spectra were measured using a 400-MHz NMR spectrometer (Unity Inova 400, Varian Associates, Varian, Palo Alto, USA) with automated sampling, locking, shimming, and tuning. Prior to data acquisition, freezedried samples were dissolved or homogenized (for crosslinked samples) in 1 mL of D2O at 50 °C. The spectra were processed using iNMR software (Nucleomatica, Molfetta, Italy). The free induction decays were Fourier-transformed using 8192 points and 5-Hz exponential broadening. An exponential window function was used to increase signal-to-noise ratio.
2.7. Statistical analysis
A comparison of values was performed by ANOVA and post-hoc test. Statistical analyses were conducted with at least 3 independent samples per experiment. Statistical significance was determined at p<0.05. Data were presented as average ± standard deviation.
3. Results and Discussion
3.1. Bioprinting of perfusable alginate microfibrous hollow constructs
Attributing to the rapid gelation property upon contact with CaCl2 and superior biocompatibility, alginate has been widely used as a promising bioink for bioprinting [7, 30]. In this study, alginate hollow microfibers were generated and deposited into 3D constructs using an extrusion bioprinter via a core/sheath coaxial nozzle, where the core and sheath channels of the nozzle were fed with a CaCl2 solution and an alginate bioink, respectively (Figure S1). To facilitate the bioprinting process, a modified printhead was designed, which allowed for easy installation and detachment of the custom-made core/sheath coaxial nozzle (Figure S2). The core/sheath coaxial nozzle was assembled by inserting a 27G needle into an 18G needle and fixing the connections using epoxy glue [7, 27]. It should be noted that the fabrication of a concentric and stable core/sheath coaxial nozzle was the key to the continuous generation of hollow alginate microfibers. Therefore, the two layers of the nozzle were carefully aligned during the assembling process, and the nozzle was further heated in an oven (80 °C) for at least 3 h to stabilize the connections.
The generation of the hollow alginate microfibers was initiated prior to the bioprinting, where the feeding rates of the CaCl2 solution and the alginate bioink should match each other within a certain range (Figure 2A). Otherwise, the hollow alginate microfibers could not be continuously produced due to needle clogging, making the bioprinting impossible. To further ensure successful bioprinting, the extrusion rate of the alginate microfiber and the nozzle moving speed should also match each other. For example, when the extrusion rate of the alginate microfiber was higher than the nozzle moving speed, hollow microfibrous constructs with curvy channels were obtained (Figure 2B-E). Although curvy microfibers were reported before [14], those with hollow interiors have not been demonstrated. While irrelevant in the current work, these hollow and perfusable microfibrous constructs with curvy channels might be useful for certain applications such as mimicking the tubules in the nephron [31, 32]. When the two speeds were equal, the hollow microfibrous constructs with straight channels could be generated (Figure 2F,G and Movie S1), and these conditions were chosen in our subsequent studies. Further enlarging the difference of these two speeds resulted in the failure of bioprinting because of needle clogging or the dragging of the deposited microfibers.
Figure 2.
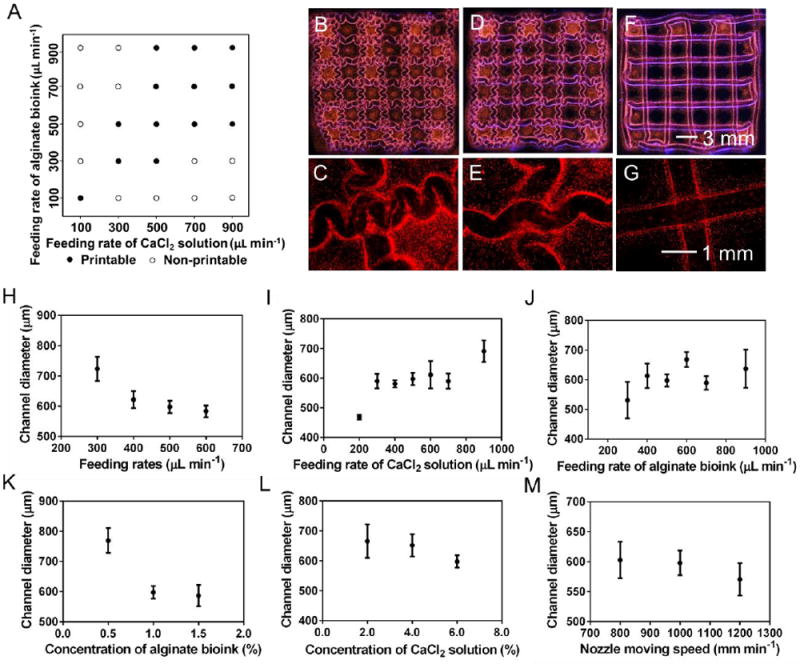
Bioprinting performance of alginate bioinks. A) Printability map showing the effect of feeding rates of the CaCl2 solution and the alginate bioink. B-G) Photographs showing bioprinted hollow constructs with curved and straight channels, the feeding rates of the CaCl2 solution and the alginate bioink (equal rates) were (B, C) 900 μL min-1, (D, E) 700 μL min-1, and (F, G) 400 μL min-1. H-M) Channel diameter of the hollow constructs as a function of (H) feeding rates of the CaCl2 solution and the alginate bioink (equal rates), (I) feeding rate of the CaCl2 solution, (J) feeding rate of the alginate, (K) concentration of the alginate bioink, (L) concentration of the CaCl2 solution, and (M) nozzle moving speed. Unless otherwise noted, the concentration of the CaCl2 solution and the alginate bioink, the feeding rates of the CaCl2 solution and the alginate bioink (equal rates), and the nozzle moving speed were kept constant at 6.0% and 1.0%, 500 μL min-1 and 500 mm min-1, respectively. The core-sheath nozzle was made using 23G/28G needles.
We then systematically investigated the influence of bioprinting parameters on the channel diameter of the alginate hollow microfibers. When the feeding rates of the CaCl2 solution and the alginate bioink (equal rates) were increased from 300 to 600 μL min-1, the channel diameter was decreased from 723 ± 40 to 583 ± 19 μm (Figure 2H). In addition, the channel diameter was increased from 469 ± 8 to 691 ± 37 μm by elevating the feeding rate of the CaCl2 solution from 200 to 900 μL min-1 when the feeding rate of the alginate bioink was 500 μL min-1 (Figure 2I), while the channel diameter was increased from 531 ± 61 to 637 ± 64 μm by raising the feeding rate of the alginate bioink from 300 to 900 μL min-1 when the feeding rate of the CaCl2 solution was 500 μL min-1 (Figure 2J). Results also indicated that lower concentration of alginate led to larger channel diameters (Figure 2K), while the concentration of the CaCl2 solution and the nozzle moving speed did not exert noticeable influence on the channel diameter in the explored ranges (Figure 2L,M), at feeding rates of 500 mm min-1. Moreover, the thickness of the channel wall was around 100 μm, which did not significantly change when the bioprinting parameters were tuned. Our results indicated similar trends but different absolute values with those reported in the literature, which is reasonable due to the size difference of the core/sheath coaxial nozzles used in these studies [14, 33, 34].
We next bioprinted a 10-layer construct, in which the distance between the contiguous microfibers was around 2.3 mm (Figure 3A-C). By reducing the distance to 0.0 mm, an integral piece of construct was bioprinted (Figure 3D-F), in which adjacent microchannels shared single walls (Figure 3F), indicating that the individual microfibers in the construct were bonded together. The integration should be attributed to the delayed crosslinking of the external surface of the microfibers as the calcium ions was gradually diffused from the interior, thus facilitating the bonding of adjacent microfibers through crosslinking of alginate. It should be noted that both types of the 10-layer constructs were perfusable (Movies S2 and S3), but slight deformation of the 10-layer constructs was observed due to the soft nature of alginate hydrogels that could not act against the gravity. Much thicker constructs are achievable by further increasing the number of layers, but perfusion is challenging for these constructs with more than 10 layers because of increased quantity of sharply curved channels and thus higher hydraulic resistance. Moreover, a perfusable “MIT”-patterned construct was bioprinted (Figure 3G and Movie S4), indicating the versatility of the bioprinting process.
Figure 3.
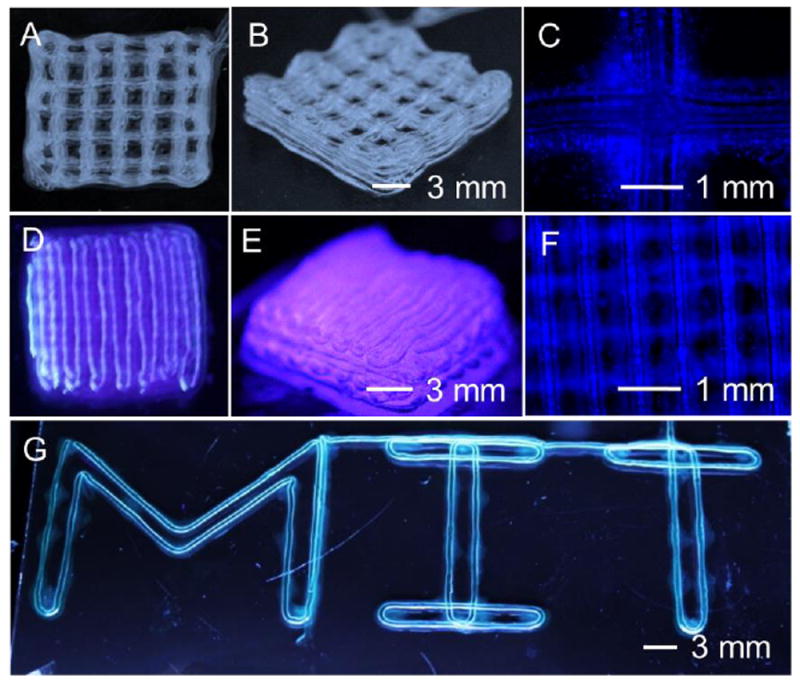
Bioprinting of 3D hollow microfibrous constructs. A-F) A 10-layer construct with a distance between the contiguous microfibers of (A-C) 2.3 mm, and (D-F) 0.0 mm. G) A perfusable “MIT”-patterned construct.
More importantly, we found that, by using a customized printhead and nozzle we were able to remove the nozzle and bioprinted hollow microfibrous construct together upon completion of the bioprinting process while keeping them still connected, and transfer them into a polydimethylsiloxane (PDMS) device to enable perfusion by directly fitting the nozzle onto a syringe (Figures S2 and S3A,B). It should be noted that the connection between the extruded alginate microfiber and the coaxial nozzle was not strong and was solely dependent on the residual alginate within the sheath of the nozzle, and thus a movable supporting device was installed around the middle section of the nozzle and was carefully moved down to the tip to protect the connection immediately after bioprinting (Figure S4), to ensure that no force was exerted during the transfer process. In addition, to achieve better connection, the two syringe pumps should be stopped slightly earlier prior to finishing the bioprinting process to prevent fluid overflow.
Although perfusion of bioprinted alginate microfibrous hollow constructs has been achieved before, the connections between the microfibers and the perfusion devices are typically created by using a micropositioning system, limiting the concentration of alginate bioinks to 2.0% as lower concentrations would result in hollow microfibers that were too weak to connect [14, 33, 34]. In comparison, our results indicated that as low as 1.0% alginate bioink could be bioprinted into perfuable hollow microfibrous constructs, enabling the fabrication of structures with higher wall porosity, which is crucial for the transport of nutrients, oxygen, and waste during cell cultures or perfusion studies. Together with the customized printhead and nozzle, the connection between the bioprinted microfiber and the perfusion device (i.e., the core/sheath coaxial nozzle used for bioprinting) was formed automatically during a bioprinting process. This connection could be subsequently protected by the movable supporting device after bioprinting, allowing for convenient transfer of the perfusable alginate hollow microfibrous construct into an additional device such as a bioreactor, potentially serving as a platform for engineering tissues containing perfusion vascular patterns.
We next conducted perfusion tests of the bioprinted hollow microfibrous constructs (4 layers) using a food dye at a flow rate of 200 μL min-1. As expected, the food dye not only flowed out from the end of the continuous microchannel of the construct, but also diffused through the microchannel wall due to the relatively high porosity of the 1.0% alginate bioink used for the bioprinting. The diffusion process was quantified by calculating the normalized gray-scale intensity of the areas within the enclosing microfibers using ImageJ, as a function of perfusion length (the distance that food dye solution has traveled), which was captured from the recorded perfusion videos (Movie S5). The normalized gray-scale intensity increased from 1.00 ± 0.04 to 1.23 ± 0.04 when the perfusion length increased from 0.0 to 63.0 cm (Figure S3C,D and Movie S5), confirming the existence of diffusion during the perfusion process, which should be attributed to the porous structure of the alginate hydrogel [35] and the thin thickness of the microchannel wall. It should be noted that, the diffusion property of these bioprinted hollow microfibrous constructs is critical for their extended applications in generating vascularized tissues, due to the ability of nutrients, oxygen, and waste to exchange through the porous walls of the microfibers [35].
3.2. Bioprinting of cell-laden constructs with tunable 3D microenvironments
After characterizing the bioprinting of hollow microfibrous constructs with optimized parameters, we next investigated the use of the alginate hollow microfibers as templates for building cell-laden constructs with soft microenvironments. The main change here lies in that the core of the nozzle was loaded with a GelMA bioink containing CaCl2 instead of a pure CaCl2 solution (Figures 1 and S5), while the sheath was still fed with the alginate bioink. Attributing to the good printability, the alginate microfibers could function to support and confine the GelMA bioink during the extrusion process, allowing for the subsequent UV crosslinking to obtain stable cell-laden microfibrous structures. Such a strategy minimizes the bioprinting requirements for the cell-laden bioinks, making it possible to generate soft microenvironments by using GelMA bioinks at extremely low concentrations of down to 1.5%. To the best of our knowledge, such a pure GelMA microenvironment of 1.5% is so far the lowest-attainable using extrusion bioprinting. Previously, the lowest concentrations of GelMA bioinks achievable through extrusion bioprinting have been limited to 5.0% and 3.0%, as reported by the Burdick group and ourselves, respectively [10, 26].
We next investigated the mechanical properties of the bioprinted 4-layer GelMAembedded constructs (12 × 12 mm2). Specifically, the Young’s modulus increased from 1.17 ± 0.04 to 2.40 ± 0.36 kPa when the concentration of the GelMA bioinks was elevated from 1.0% to 2.0% (Figure 4A,B), which were comparable to the pure GelMA constructs we reported before [26], indicating that the structures were relatively stable. The SEM analyses were additionally performed to study the fine structure of the bioprinted microfibrous constructs. Images indicated that the 1.0% GelMA constructs only had the surrounding alginate wall with a very dilute, almost non-existent GelMA network in the interior (Figure 4C), indicating that the UV crosslinking did not generate measurable GelMA structures in this case. By contrast, the 1.5% and 2.0% GelMA constructs owned porous structures in the alginate hollow microfibers (Figure 4D,E). These results are comparable with our previous reports that lower concentrations of GelMA resulted in hydrogels with larger pores [26].
Figure 4.
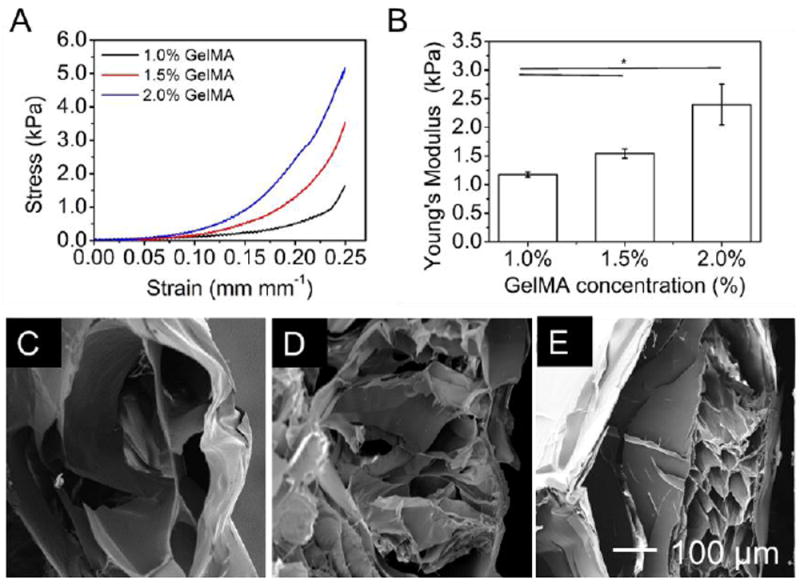
Bioprinting of GelMA/alginate core/sheath constructs with tunable microenvironments. A) Stress-strain curves of the bioprinted constructs with different concentrations of GelMA. B) Young’s modulus of the bioprinted constructs. C-E) SEM images of the bioprinted GelMA/alginate constructs after freeze-drying showing the porosity of the cores: (A) 1.0% GelMA; (B) 1.5% GelMA; (C) 2.0% GelMA.
To encapsulate cells and initiate the ionic crosslinking of alginate during bioprinting, the GelMA bioinks in the core of the nozzle were supplied with 1.0% CaCl2, which may exert negative effects on the encapsulated cells, although GelMA itself has been proven compatible with various cell types [5, 7]. Therefore, we investigated the influence of CaCl2 on the viability of different cells, including MDA-MB-231 and MCF7 breast cancer cells, NIH/3T3 mouse fibroblasts, and HUVECs in 24-well plates with media containing the same concentration of CaCl2 for up to 20 min (control samples). Results indicated that CaCl2 did not show significant impact on the viability of MDA-MB-231 cells over the 20-min treatment, all at above 92.6% ± 2.5% (Figures 5A and S6A-E). In comparison, after the 20-min treatment, the viabilities of MCF7, NIH/3T3, and HUVECs decreased from 94.2.0% ± 2.8% to 80.4% ± 0.7%, from 96.2.0% ± 1.5% to 77.4% ± 3.9%, and from 98.4% ± 1.4% to 87.3% ± 5.1.0%, respectively (Figures 5A and S6F-T). These results were in the normal range and acceptable for various biofabrication techniques [7, 10, 19], showing that our strategy could be feasible for the bioprinting of soft cell-laden constructs.
Figure 5.
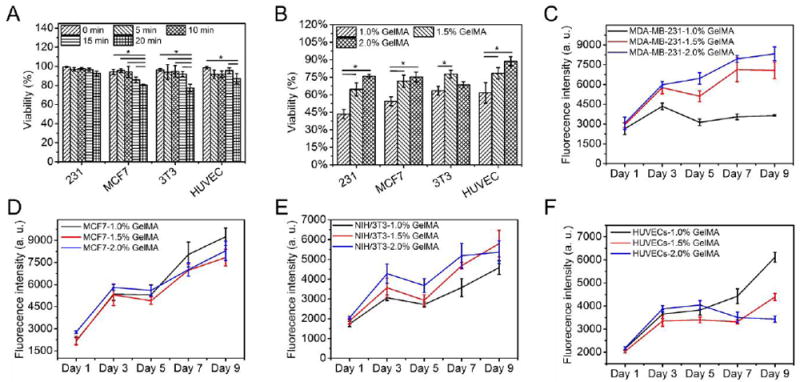
Cell viability and proliferation. A,B) Quantified viability of cells (A) cultured in 24- well plates over a 20-min incubation using media containing 1.0% CaCl2 and (B) in bioprinted constructs (Day 1). C-F) Quantified proliferation of cells over a 9-day culture period, for (C) MDA-MB-231, (D) MCF7, (E) NIH/3T3, and (F) HUVECs.
To minimize the exposure time of cells in the presence of CaCl2, the bioprinted constructs were washed with HBSS immediately after UV crosslinking, and the entire bioprinting process was completed within approximately 10 min. The cell viabilities post-bioprinting at Day 1 were characterized by Live/Dead staining, which were in general lower than those of the control samples. We noticed that the cell viabilities were quite low (from 43.4% ± 3.9% to 63.4% ± 3.8% for MDA-MB-231, MCF7, NIH/3T3, and HUVECs) when the cores were fed with 1.0% GelMA bioink, which could be ascribed to the lack of cell-anchoring points as the 1.0% GelMA bioink could be barely crosslinked (Figures 5B and S7A-D). In contrast, the cell viabilities were significantly enhanced to the range between 64.6% ± 5.6% and 88.7% ± 4.0% for MDA-MB-231, MCF7, NIH/3T3, and HUVECs when the cores were fed with 1.5% and 2.0% GelMA bioinks (Figures 5B and S7E-L). It should be noted that the 1.0% GelMA bioink could barely form hydrogel through UV crosslinking, while the 1.5% and 2.0% GelMA bioinks resulted in successful formation of hydrogels, which could be indicated by the aggregation of the encapsulated cells (Figure S7A-D) and the uniform cell distributions (Figure S7E-L), respectively. According to the FTIR spectra, no significant difference of crosslinking efficiency was observed among the different concentrations investigated (Figure S8), while the 1H NMR spectra indicated that the crosslinking efficiency was quite high for the 1% GelMA bioink (Figure S9). Therefore, we believe that the failure of hydrogel formation through UV crosslinking in the case of 1% GelMA bioink should be attributed to the insufficient density of the crosslinked polymer network but not the crosslinking efficiency. Furthermore, we quantified the proliferation of the laden cells by measuring the metabolic activities using Prestoblue. Overall, the proliferation of most cells in the bioprinted constructs remained an increasing trend (Figure 5C-F). Exceptions include the 1.0% GelMA constructs with MDA-MB-231 cells, and 2.0% GelMA constructs with HUVECs, potentially indicating that different cells may have their preferred microenvironments and stiffness [36, 37].
Morphological analyses were next performed. Without a sufficiently crosslinked network of GelMA at 1.0% concentration, the encapsulated cells could barely attach (Figure 6A). In comparison, higher concentrations (1.5% and 2.0%) of GelMA provided favorable 3D microenvironments for the cells to adhere, spread, proliferate, and interact (Figure 6B,C). Confocal images indicated that MDA-MB-231 cells tended to aggregate in the 1.0% and 1.5% GelMA constructs (Figure 6D-G), and smaller aggregates were found in the 2.0% GelMA constructs (Figure 6H,I). MCF7 cells exhibited spreading in the 1.0% and 1.5% GelMA constructs but much less spreading in the 2.0% GelMA constructs (Figure S10A-C). For NIH/3T3 fibroblasts and HUVECs, considerable cell spreading only appeared in the 1.5% GelMA constructs (Figure S10D-I). In summary, all the cells displayed normal spreading in the microenvironments with intermediate concentration of 1.5% GelMA. While only cancer cells of MDA-MB-231 and MCF7 showed spreading in the 1.0% GelMA constructs, probably attributed to their higher resistance to harsh environments than normal cells. In contrast, cell spreading was limited in microenvironment of 2.0% GelMA with relatively strong mechanics. These observations on the cell morphology were in good agreement with the viability and proliferation data (Figure 5B-F), again suggesting the varying microenvironmental effects on different cell types.
Figure 6.
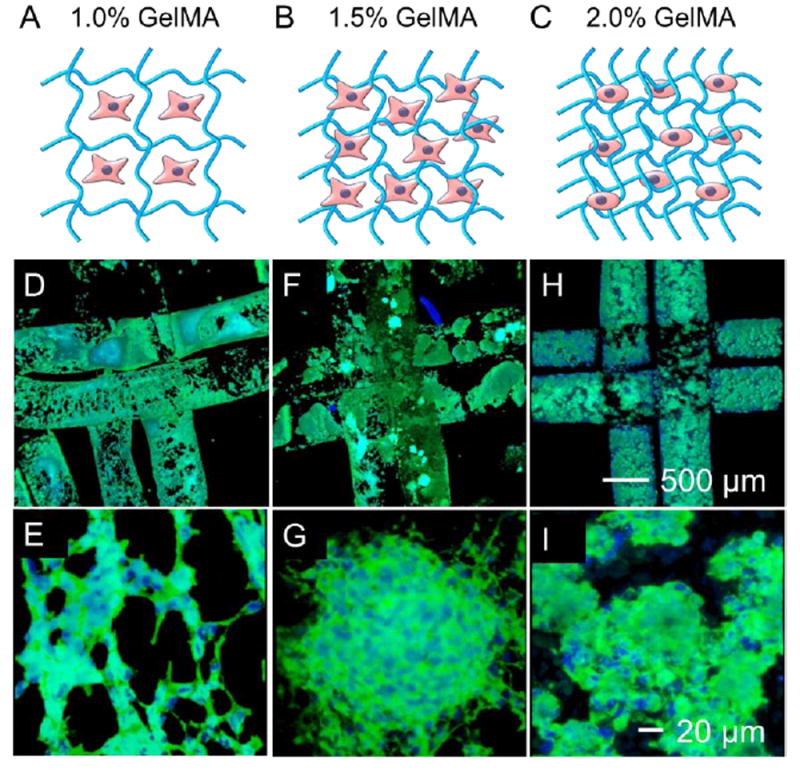
Cell spreading in the bioprinted constructs. A-C) Schematic diagram showing the structures of (A) 1.0% GelMA constructs, (B) 1.5% GelMA constructs, and (C) 2.0% GelMA constructs. D-I) Fluorescence images showing MDA-MB-231 cells stained for F-actin and nuclei at Day 9, in bioprinted constructs of (D,E) 1.0% GelMA, (F,G) 1.5% GelMA, and (H,I) 2.0% GelMA.
To enable sufficient cell spreading, microenvironments tunable towards low stiffness and containing biologically active motifs are usually desired, which seldom satisfy the requirements for conventional 3D bioprinting due to the weak mechanical properties of these hydrogels in serving as bioinks. In addressing such a challenge, templating strategies have been employed for the bioprinting of cell-laden constructs with soft microenvironments [2, 3, 24]. In a recent work, collagen/alginate constructs were bioprinted via an aerosol crosslinking strategy [24]. However, since the aerosol crosslinking process was relatively slow, this strategy necessitated pre-crosslinking of the alginate bioink by mixing it with CaCl2 prior to bioprinting to achieve structural fidelity, making it more complicated comparing to our approach. As such, the concentration of the alginate bioink was limited to 4.0%, which could inhibit the diffusion rate and thus the transport of nutrients, oxygen, and wastes through the alginate sheath. Indeed, although proliferation of osteoblast-like cells and human adiposederived stem cells (hASCs) were demonstrated, spreading only occurred in some cells after a 20-day culture. In comparison, our bioprinting strategy with low-stiffness GelMA interiors and low-concentration alginate sheaths facilitated the spreading of the embedded cells within relatively short periods of time.
To this end, our strategy could open an alternative way to bioprint cell-laden constructs that allow for enhanced cellular activities such as proliferation and spreading. The results indicated that the bioprinted constructs may be suitable for encapsulating various cell types while maintaining an integral structure due to the existence of the alginate sheaths surrounding the microfibers as templates, which could be rarely achieved through other reported strategies [10, 19]. It should be noted that, the bioprinted GelMA microfibrous constructs were surrounded and thus separated by alginate sheaths, which would collapse without the sheaths. Therefore, the alginate templates were not removed in any case in our study to support the structures of the bioprinted constructs. While GelMA was used to demonstrate the concept, it is further possible to adapt other hydrogel materials, such as collagen [24], fibrin, and Matrigel as the internal bioink for desired applications.
One main limitation of the technology lies in the fact that, the cells have to be dispersed in bioinks containing 1.0% CaCl2, which may exert negative effects on the cells, necessitating fast completion of the bioprinting process followed by extensive wash with buffer solutions. In addition, generation of dendroid structures using such co-axial extrusion bioprinting strategies has been historically challenging, which is a limitation in our approach as well. Nevertheless, we foresee that with additional optimization, our strategy could be generalized to bioprint 3D constructs with cell-favorable microenvironments for widespread applications in tissue engineering and pharmaceutical screening.
4. Conclusions
We have developed a strategy to generate cell-laden GelMA constructs with tunable and favorable microenvironments for various cell types by coaxial extrusion bioprinting of GelMA/alginate core/sheath microfibers, where the alginate sheath serves as a template to support and confine the GelMA hydrogel in the core to allow for subsequent UV crosslinking. This strategy could minimize the bioprinting requirements for the bioinks used in the core, facilitating the fabrication of cell-laden GelMA constructs at extremely low concentrations (<2.0%), which is challenge using conventional bioprinting strategies. Indeed, such constructs based on GelMA alone are not structurally stable and have not been possible to bioprint to date as they are too soft to overcome their gravity [10, 19]. It is further worth noting that this strategy enables a high degree of control over the 3D microenvironments for the encapsulated cells of various types, which has been difficult to achieve with conventional bioprinting strategies [7, 10, 19].
Supplementary Material
Acknowledgments
The authors acknowledge funding from the National Institutes of Health (AR057837, DE021468, AR068258, AR066193, EB022403, EB021148) and the Presidential Early Career Award for Scientists and Engineers (PECASE) and the Fundamental Research Funds for the Central Universities from China (No. 14D310106). W.L. acknowledges the financial support from the program of China Scholarships Council (No. 201406630041). Y.S.Z. acknowledges the National Cancer Institute of the National Institutes of Health Pathway to Independence Award (K99CA201603). We acknowledges the help of data analysis from Dr. Samson Afewerki and Dr. Mingyu Qiao.
Footnotes
Supplementary material for this article is available online.
References
- 1.Derby B. Printing and prototyping of tissues and scaffolds. Science. 2012;338:921–6. doi: 10.1126/science.1226340. [DOI] [PubMed] [Google Scholar]
- 2.Kang H-W, Lee SJ, Ko IK, Kengla C, Yoo JJ, Atala A. A 3D bioprinting system to produce human-scale tissue constructs with structural integrity. Nat Biotechnol. 2016;34:312–9. doi: 10.1038/nbt.3413. [DOI] [PubMed] [Google Scholar]
- 3.Kolesky DB, Homan KA, Skylar-Scott MA, Lewis JA. Three-dimensional bioprinting of thick vascularized tissues. Proc Natl Acad Sci U S A. 2016;113:3179–84. doi: 10.1073/pnas.1521342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kolesky DB, Truby RL, Gladman A, Busbee TA, Homan KA, Lewis JA. 3D bioprinting of vascularized, heterogeneous cell-laden tissue constructs. Adv Mater. 2014;26:3124–30. doi: 10.1002/adma.201305506. [DOI] [PubMed] [Google Scholar]
- 5.Liu W, Zhang YS, Heinrich MA, De Ferrari F, Jang HL, Bakht SM, Alvarez MM, Yang J, Li Y-C, Trujillo-de Santiago G, Miri AK, Zhu K, Khoshakhlagh P, Prakash G, Cheng H, Guan X, Zhong Z, Ju J, Zhu GH, Jin X, Shin SR, Dokmeci MR, Khademhosseini A. Rapid Continuous Multimaterial Extrusion Bioprinting. Adv Mater. 2017;29:1604630. doi: 10.1002/adma.201604630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ouyang L, Highley CB, Rodell CB, Sun W, Burdick JA. 3D Printing of shear-thinning hyaluronic acid hydrogels with secondary crosslinking. ACS Biomater Sci Eng. 2016 doi: 10.1021/acsbiomaterials.6b00158. [DOI] [PubMed] [Google Scholar]
- 7.Jia W, Gungor-Ozkerim PS, Zhang YS, Yue K, Zhu K, Liu W, Pi Q, Byambaa B, Dokmeci MR, Shin SR, Khademhosseini A. Direct 3D bioprinting of perfusable vascular constructs using a blend bioink. Biomaterials. 2016;106:58–68. doi: 10.1016/j.biomaterials.2016.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blaeser A, Duarte Campos DF, Puster U, Richtering W, Stevens MM, Fischer H. Controlling shear stress in 3D bioprinting is a key factor to balance printing resolution and stem cell integrity. Adv Healthc Mate. 2016;5:326–33. doi: 10.1002/adhm.201500677. [DOI] [PubMed] [Google Scholar]
- 9.Murphy SV, Atala A. 3D bioprinting of tissues and organs. Nat Biotechnol. 2014;32:773–85. doi: 10.1038/nbt.2958. [DOI] [PubMed] [Google Scholar]
- 10.Ouyang L, Highley CB, Sun W, Burdick JA. A Generalizable Strategy for the 3D Bioprinting of Hydrogels from Nonviscous Photo-crosslinkable Inks. Adv Mater. 2016;29:1604983. doi: 10.1002/adma.201604983. [DOI] [PubMed] [Google Scholar]
- 11.Carrow JK, Kerativitayanan P, Jaiswal MK, Lokhande G, Gaharwar AK. Essentials of 3D Biofabrication and Translation. USA: Academic Press; 2015. pp. 229–48. [Google Scholar]
- 12.Melchels FPW, Domingos MAN, Klein TJ, Malda J, Bartolo PJ, Hutmacher DW. Additive manufacturing of tissues and organs. Prog Polym Sci. 2012;37:1079–104. [Google Scholar]
- 13.Tamayol A, Najafabadi AH, Aliakbarian B, Arab-Tehrany E, Akbari M, Annabi N, Juncker D, Khademhosseini A. Hydrogel Templates for Rapid Manufacturing of Bioactive Fibers and 3D Constructs. Adv Healthc Mate. 2015;4:2146–53. doi: 10.1002/adhm.201500492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao Q, He Y, Fu J-z, Liu A, Ma L. Coaxial nozzle-assisted 3D bioprinting with built-in microchannels for nutrients delivery. Biomaterials. 2015;61:203–15. doi: 10.1016/j.biomaterials.2015.05.031. [DOI] [PubMed] [Google Scholar]
- 15.Luo Y, Lode A, Gelinsky M. Direct Plotting of Three-Dimensional Hollow Fiber Scaffolds Based on Concentrated Alginate Pastes for Tissue Engineering. Adv Healthc Mate. 2013;2:777–83. doi: 10.1002/adhm.201200303. [DOI] [PubMed] [Google Scholar]
- 16.Ozbolat IT, Hospodiuk M. Current advances and future perspectives in extrusion-based bioprinting. Biomaterials. 2016;76:321–43. doi: 10.1016/j.biomaterials.2015.10.076. [DOI] [PubMed] [Google Scholar]
- 17.Yu Y, Moncal KK, Li J, Peng W, Rivero I, Martin JA, Ozbolat IT. Three-dimensional bioprinting using self-Assembling scalable scaffold-free “tissue strands” as a new bioink. Scientific Reports. 2016;6:28714. doi: 10.1038/srep28714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith CM, Stone AL, Parkhill RL, Stewart RL, Simpkins MW, Kachurin AM, Warren WL, Williams SK. Three-dimensional bioassembly tool for generating viable tissue-engineered constructs. Tissue Eng. 2004;10:1566–76. doi: 10.1089/ten.2004.10.1566. [DOI] [PubMed] [Google Scholar]
- 19.Billiet T, Gevaert E, De Schryver T, Cornelissen M, Dubruel P. The 3D printing of gelatin methacrylamide cell-laden tissue-engineered constructs with high cell viability. Biomaterials. 2014;35:49–62. doi: 10.1016/j.biomaterials.2013.09.078. [DOI] [PubMed] [Google Scholar]
- 20.Luiz EB, Juliana CC, Vijayan M, Ana LC, Nupura SB, Wesleyan AA, Pinar Z, Nihal EV, Amir MG, Mehmet RD, Ali K. Direct-write bioprinting of cell-laden methacrylated gelatin hydrogels. Biofabrication. 2014;6:024105. doi: 10.1088/1758-5082/6/2/024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamura M, Iwanaga S, Henmi C, Arai K, Nishiyama Y. Biomatrices and biomaterials for future developments of bioprinting and biofabrication. Biofabrication. 2010;2:014110. doi: 10.1088/1758-5082/2/1/014110. [DOI] [PubMed] [Google Scholar]
- 22.Jia J, Richards DJ, Pollard S, Tan Y, Rodriguez J, Visconti RP, Trusk TC, Yost MJ, Yao H, Markwald RR. Engineering alginate as bioink for bioprinting. Acta Biomater. 2014;10:4323–31. doi: 10.1016/j.actbio.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim YB, Lee H, Kim GH. Strategy to Achieve Highly Porous/Biocompatible Macroscale Cell Blocks, Using a Collagen/Genipin-bioink and an Optimal 3D Printing Process. ACS Appl Mat Interfaces. 2016;8:32230–40. doi: 10.1021/acsami.6b11669. [DOI] [PubMed] [Google Scholar]
- 24.Yeo M, Lee J-S, Chun W, Kim GH. An Innovative Collagen-Based Cell-Printing Method for Obtaining Human Adipose Stem Cell-Laden Structures Consisting of Core–Sheath Structures for Tissue Engineering. Biomacromolecules. 2016;17:1365–75. doi: 10.1021/acs.biomac.5b01764. [DOI] [PubMed] [Google Scholar]
- 25.Loessner D, Meinert C, Kaemmerer E, Martine LC, Yue K, Levett PA, Klein TJ, Melchels FP, Khademhosseini A, Hutmacher DW. Functionalization, preparation and use of cell-laden gelatin methacryloyl-based hydrogels as modular tissue culture platforms. Nat Protoc. 2016;11:727–46. doi: 10.1038/nprot.2016.037. [DOI] [PubMed] [Google Scholar]
- 26.Liu W, Heinrich MA, Zhou Y, Akpek A, Hu N, Liu X, Guan X, Zhong Z, Jin X, Khademhosseini A, Zhang YS. Extrusion Bioprinting of Shear-Thinning Gelatin Methacryloyl Bioinks. Adv Healthc Mate. 2017:1601451. doi: 10.1002/adhm.201601451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang YS, Arneri A, Bersini S, Shin S-R, Zhu K, Goli-Malekabadi Z, Aleman J, Colosi C, Busignani F, Dell’Erba V. Bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium and heart-on-a-chip. Biomaterials. 2016;110:45–59. doi: 10.1016/j.biomaterials.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colosi C, Shin SR, Manoharan V, Massa S, Costantini M, Barbetta A, Dokmeci MR, Dentini M, Khademhosseini A. Microfluidic Bioprinting of Heterogeneous 3D Tissue Constructs Using Low-Viscosity Bioink. Adv Mater. 2015;28:677–84. doi: 10.1002/adma.201503310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nichol JW, Koshy ST, Bae H, Hwang CM, Yamanlar S, Khademhosseini A. Cell-laden microengineered gelatin methacrylate hydrogels. Biomaterials. 2010;31:5536–44. doi: 10.1016/j.biomaterials.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu K, Shin SR, van Kempen T, Li Y-C, Ponraj V, Nasajpour A, Mandla S, Hu N, Liu X, Leijten J, Lin Y-D, Hussain MA, Zhang YS, Tamayol A, Khademhosseini A. Gold Nanocomposite Bioink for Printing 3D Cardiac Constructs. Adv Funct Mater. 2017;27:1605352. doi: 10.1002/adfm.201605352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Homan KA, Kolesky DB, Skylar-Scott MA, Herrmann J, Obuobi H, Moisan A, Lewis JA. Bioprinting of 3D convoluted renal proximal tubules on perfusable chips. Scientific reports. 2016;6 doi: 10.1038/srep34845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grolman JM, Zhang D, Smith AM, Moore JS, Kilian KA. Rapid 3D extrusion of synthetic tumor microenvironments. Adv Mater. 2015;27:5512–7. doi: 10.1002/adma.201501729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Yu Y, Ozbolat IT. Direct bioprinting of vessel-like tubular microfluidic channels. Journal of nanotechnology in engineering and medicine. 2013;4:020902. [Google Scholar]
- 34.Zhang Y, Yu Y, Chen H, Ozbolat IT. Characterization of printable cellular micro-fluidic channels for tissue engineering. Biofabrication. 2013;5:025004. doi: 10.1088/1758-5082/5/2/025004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amsden B, Turner N. Diffusion characteristics of calcium alginate gels. Biotechnol Bioeng. 1999;65:605–10. doi: 10.1002/(sici)1097-0290(19991205)65:5<605::aid-bit14>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 36.Chaudhuri O, Gu L, Klumpers D, Darnell M, Bencherif SA, Weaver JC, Huebsch N, Lee H-p, Lippens E, Duda GN, Mooney DJ. Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat Mater. 2016;15:326–34. doi: 10.1038/nmat4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baker BM, Trappmann B, Wang WY, Sakar MS, Kim IL, Shenoy VB, Burdick JA, Chen CS. Cell-mediated fibre recruitment drives extracellular matrix mechanosensing in engineered fibrillar microenvironments. Nat Mater. 2015;14:1262–8. doi: 10.1038/nmat4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


