Abstract
Sea buckthorn is a plant of medicinal and nutritional importance owing in part to the high levels of essential fatty acids, linoleic (up to 42%) and α-linolenic (up to 39%) acids in the seed oil. Sea buckthorn can produce seeds either via the sexual pathway or by apomixis. The seed development and maturation programs are critically dependent on miRNAs. To understand miRNA-mediated regulation of sea buckthorn seed development, eight small RNA libraries were constructed for deep sequencing from developing seeds of a low oil content line ‘SJ1’ and a high oil content line ‘XE3’. High-throughput sequencing identified 137 known miRNA from 27 families and 264 novel miRNAs. The potential targets of the identified miRNAs were predicted based on sequence homology. Nineteen (four known and 15 novel) and 22 (six known and 16 novel) miRNAs were found to be involved in lipid biosynthesis and seed size, respectively. An integrated analysis of mRNA and miRNA transcriptome and qRT-PCR identified some key miRNAs and their targets (miR164d-ARF2, miR168b-Δ9D, novelmiRNA-108-ACC, novelmiRNA-23-GPD1, novelmiRNA-58-DGAT1, and novelmiRNA-191-DGAT2) potentially involved in seed size and lipid biosynthesis of sea buckthorn seed. These results indicate the potential importance of miRNAs in regulating lipid biosynthesis and seed size in sea buckthorn.
Introduction
Sea buckthorn (Hippophae L.) is one of the nutritionally and ecologically most important woody oil plants. Traditionally used for hundreds of years in China and Russia for health-related purposes, sea buckthorn has now gained popularity worldwide due to the unique composition of seed and pulp oil and the abundance of bioactive compounds in fruits, seeds, leaves and bark1. The red or orange berries of sea buckthorn contain fleshy pulp rich in vitamin C, carotenoids, flavonoids and monounsaturated palmitoleic acid (up to 39%), and a single seed with an oil content of 8–20%1,2, comprising high levels of linoleic acid (30–42%) and α-linolenic acid (20–39%)3, carotenoids, flavonol glycosides4, tocopherol and phytosterols5,6. The bioactive compounds in sea buckthorn products have clinically proven medicinal effects against tumor progression, inflammation, hypertension and gastric ulcers, and can promote wound healing and tissue regeneration7–11. The high levels of polyunsaturated fatty acids and the almost 1:1 ratio of linoleic acid to α-linolenic acid, which is considered to be beneficial for human health, makes sea buckthorn seed oil a niche product and warrants the study of seed development and oil biosynthesis pathways. Sea buckthorn sequences associated with fatty acid and triacylglycerol biosynthetic pathways were identified in the mature seed transcriptome and a comparison of gene expression and oil accumulation in seeds derived from four different developmental stages of fruit indicated that oil deposition begins very early in fruit development2. Recently, sea buckthorn was reported to produce seeds either sexually or occasionally by apomixis, with the latter ensuring reproduction in the absence of pollination12. Aside from these reports there is no other information available on sea buckthorn lipid biosynthesis and seed development pathways.
The seed development and maturation program is, to a major extent, regulated by microRNAs (miRNAs) and their targets encoding functional genes and transcription factors. miRNAs are endogenously encoded small RNAs that post-transcriptionally regulate gene expression. In plants, miRNAs play an essential role in numerous developmental and physiological processes, such as fatty acid biosynthesis13, growth and development14,15 and responses to various stresses16,17, and many miRNAs are conserved across species. The involvement of miRNAs in post-transcriptional regulation of seed or fruit development has been documented in apricot14, rice16, soybean18, peony19, Brassica napus13,20, and Cichorium intybus21. Examples of functional genes that are related to lipid biosynthesis and targeted by miRNAs, include 3-ketoacyl-ACP synthase (KAS) targeted by miR15913, 3-ketoacyl-ACP reductase (KAR) targeted by miR156 and miR602913, and stearoyl-acyl carrier protein Δ9-desaturase6 (SAD6) targeted by miR31922. Transcription factors play crucial roles in regulating lipid biosynthesis (WRINKLED123, LEAFY COTYLEDON24, and helix loop helix (bHLH)25) and seed size (ARF26, MYB27, and CNR28). For example, the Sesamum indicum bHLH transcription factor binds to E- or G-box elements in the FAD2 gene promoter and impacts lipid biosynthesis and accumulation during seed development25. The ARFs transcription factors involved in seed development are negatively regulated by miR16029,30. Accordingly, ARF10, ARF16, and ARF17 transcripts were highly increased in the miR160 foc mutant during early embryogenesis. miR167, which targets ARF6 and ARF8, is preferentially expressed during late embryogenesis26. A double mutation of miR159 (miR159ab), which enhances MYB33 and MYB65 expression leads to the formation of small seeds31. Additionally, the role of MYB56 in controlling seed size has been established in Arabidopsis26. Thus while miRNA-mediated regulatory networks controlling seed oil accumulation and seed development have been revealed in Arabidopsis, rice, and maize26, little is known in this area in woody oil crops.
miRNAs have been identified in woody oil plants such as olive, oil palm and peony19,32,33, but there are currently no reports on miRNAs present in developing seeds of sea buckthorn. To understand oil biosynthesis regulation in sea buckthorn seed, the present study was undertaken to identify miRNAs and their targets in developing seeds of sea buckthorn. Seeds of a high oil content sea buckthorn line ‘XE3’ and a low oil content line ‘SJ1’ were collected at four stages of fruit development: [green (G), green/yellow (G/Y), yellow/orange (Y/O) and orange/red (O/R) color stages], and used for small RNA (sRNA) sequencing. An integrated analysis of mRNA and miRNA transcriptome combined with qRT-PCR identified some novel and previously known miRNAs and their targets potentially involved in lipid biosynthesis. Expression of a subset of novel miRNAs was higher in the low oil content line ‘SJ1’ as compared to the high oil content line ‘XE3’, while the expression of miR164d with the target gene ARF2, which determines seed size, showed the opposite pattern. These results indicate the potential importance of miRNAs in regulating lipid biosynthesis and seed size of sea buckthorn via post-transcriptional mechanisms.
Results
Oil content, fatty acid composition and seed size of developing sea buckthorn seeds
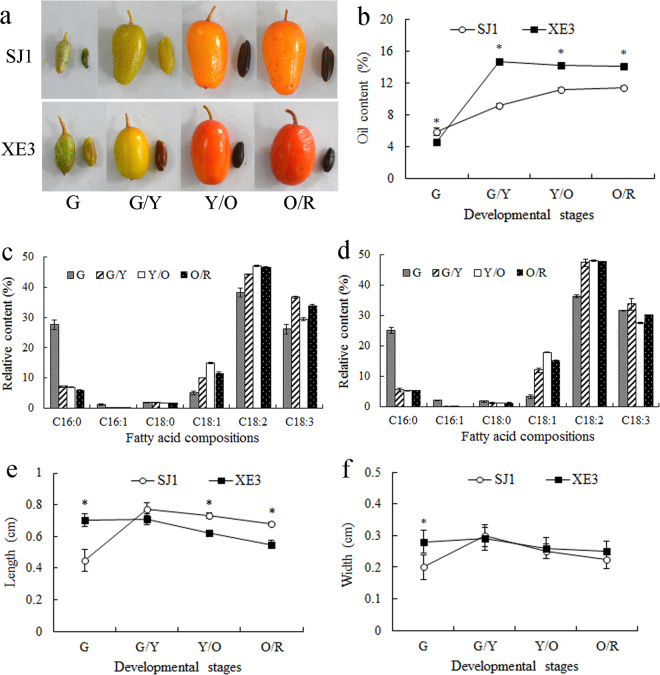
Sea buckthorn ‘SJ1’ and ‘XE3’ had previously been characterised as low and high oil content lines34, respectively. Here we explored oil accumulation, fatty acid composition and seed size across four developmental stages of the fruit from green (fruit swelling stage) to orange/red (fully matured stage). ‘XE3’ exhibited rapid accumulation (approximate 3.2-fold increase) of seed oil from 4.6% at G stage to 14.8% at G/Y stage (Fig. 1a,b), indicating a peak in oil accumulation between the early and the middle stages. Sea buckthorn seed oil has high concentrations of unsaturated fatty acids (>92%) such as palmitoleic (C16:1), oleic (C18:1), linoleic (C18:2) and linolenic (C18:3) acids (Fig. 1c,d). Six fatty acid components were measured in sea buckthorn seed oil. The relative content of palmitic (C16:0) and palmitoleic (C16:1) acids in line ‘SJ1’ (Fig. 1c) was higher than in line ‘XE3’ (Fig. 1d), but showed a decreasing trend with maturity in both lines. The relative contents of C18:1 and C18:2 first increased from G to Y/O stages and then decreased, and these two fatty acids contents in line ‘SJ1’ were lower than line ‘XE3’ from G/Y to O/R stages (Fig. 1c,d). The C18:2 levels peaked at the Y/O stage in both lines, while, C18:3 levels peaked in both lines at the G/Y stage.
Figure 1.
The oil content, fatty acid composition and size of developing sea buckthorn seeds. (a) Fruits and seeds of sea buckthorn at four development stages. G, G/Y, Y/O and O/R indicate the developmental stage of the fruit as indicated by color green, green/yellow, yellow/orange and orange/red, respectively. (b) Oil contents in seeds from lines ‘SJ1’ and ‘XE3’ at four development stages. (c) Fatty acid compositions in seeds from lines ‘SJ1’ at four development stages. C16:0, C16:1, C18:0, C18:1, C18:2 and C18:3 indicate palmitic, palmitoleic, stearic, oleic, linoleic and linolenic acids, respectively. (d) Fatty acid compositions in seeds from lines ‘XE3’ at four development stages. (e) Length of seeds from both lines at four development stages. (f) Width of seeds from both lines at four development stages. Error bars indicate standard deviations of three biological replicates. *Indicate significant differences of data between the two lines at the same developmental stage at the level of 0.05.
Seed length in ‘SJ1’ was approximately 1.7-fold higher at the G/Y stage compared to the G stage. Interestingly seed lengths in ‘SJ1’ and ‘XE3’ decreased by 11.7% and 22.5%, respectively, from the G/Y to the O/R stage (Fig. 1e). The pattern for the width of seeds matched that of the length (Fig. 1f), indicating that growth of sea buckthorn seeds takes place mainly at early stages (G–G/Y). Thus, lipid biosynthesis and seed size in sea buckthorn is developmentally controlled.
Identification of functional genes involved in lipid biosynthesis and seed size
To explore unigenes involved in lipid biosynthesis and seed size, eight mRNA libraries from seeds extracted from four developmental stages (G, G/Y, Y/O, and O/R) were constructed by Illumina Hiseq2500, and 161,739,044 and 182,289,308 clean reads was generated for lines ‘SJ1’ and ‘XE3’ seeds, respectively (Supplementary Table S1). In total, 323,881 unigenes (≥200 bp) were assembled and used as reference for the discovery of pre-miRNA and miRNA sequences. Functional annotation revealed 79,413, 69,924,99,916 and 28,579 unigenes with alignments to the Nr (Non-redundant protein database), COG (Clusters of orthologous groups of protein), Swiss-Prot (Annotated protein sequence database) and KEGG (Kyoto encyclopedia of genes and genomes) databases, respectively. The GO (Gene ontology) database provides functional terms for genes across all species. In sea buckthorn transcriptome library, 167 and 520 functional unigenes were grouped into “developmental process” and “metabolic process”, respectively (Supplementary Table S2). In total, 3153 unigenes were assigned to “Lipid transport and metabolism” based on COG classifications according to phylogenetic relationships (Supplementary Table S2). We used KEGG pathway database with KAAS (KEGG Automatic Annotation Server) to predict the lipid biosynthesis network of sea buckthorn. The “lipid metabolism” category containing 1737 unigenes were grouped into 17 pathways (Fig. 2 and Supplementary Table S3). A maximum of 342 unigenes were involved in glycerophospholipid metabolism (ko00564) followed by 222 unigenes involved in glycerolipid metabolism (ko00561), and 141 unigenes involved in fatty acid biosynthesis (ko00061).
Figure 2.
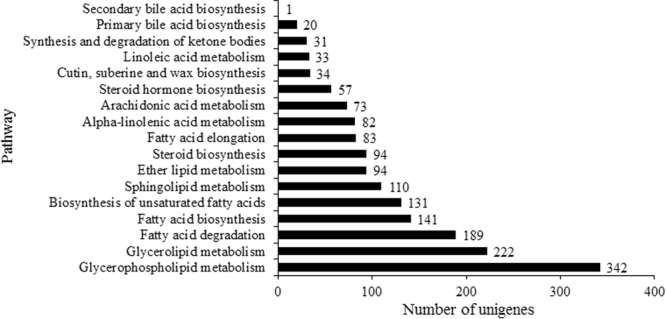
Distribution of annotated unigenes involved in lipid metabolism based on KEGG pathway analysis.
Identification of known and novel miRNAs in developing sea buckthorn seeds
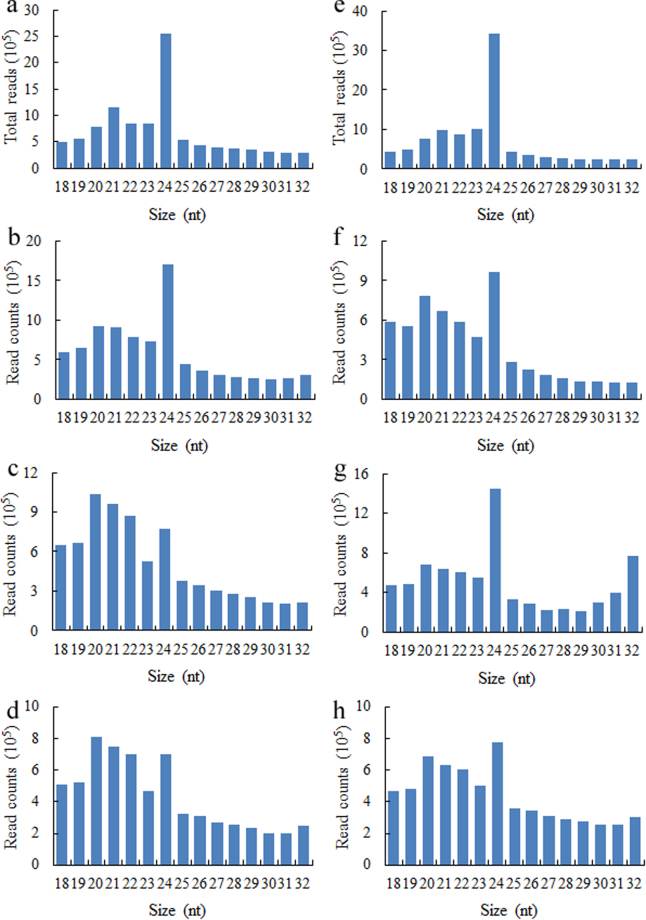
To identify miRNA-mediated regulation that may be involved in sea buckthorn lipid biosynthesis and seed size, eight sRNA libraries from the same eight samples as above were analyzed by Illumina Hiseq2500. Deep sequencing of all eight sRNA libraries yielded 33,184,369 and 30,410,875 clean reads in lines ‘SJ1’ and ‘XE3’, respectively (Table 1). Further quality evaluation of the sRNA sequencing data by the FastQC software identified high quality clean reads for sea buckthorn seed miRNAs (Supplementary Fig. S1). Within this dataset, 20 to 24 nts long sequences were the most abundant, with the 24 nts long sRNAs as most abundant in six libraries (G and G/Y stages in line ‘SJ1’, all four stages in line ‘XE3’) (Fig. 3a,b,e,f,g,h). This length distribution pattern of sRNAs in sea buckthorn is consistent with that in other plant species13,35–37. However, in the Y/O and O/R stages of line ‘SJ1’ seed datasets the number of 20 to 22 nts long sRNAs exceeded the number of 24 nts long sRNAs (Fig. 3c,d). The 20–22 nts plant sRNA mediate target gene cleavage or inhibition of protein translation, while the 24 nt sRNA class affects chromatin modelling of target genes38.
Table 1.
Summary of sequencing data of eight sRNA libraries.
| Line ‘SJ1’ seeds | Line ‘XE3’ seeds | |||||||
|---|---|---|---|---|---|---|---|---|
| G | G/Y | Y/O | O/R | G | G/Y | Y/O | O/R | |
| Raw reads* | 14,468,271 | 14,339,275 | 13,300,277 | 10,891,810 | 14,025,713 | 11,691,931 | 13,401,496 | 11,479,969 |
| Clean reads** | 10,246,589 | 8,742,899 | 7,693,414 | 6,501,467 | 10,250,860 | 5,974,796 | 7,632,683 | 6,552,536 |
| Q20 of clean read (%) | 99.76 | 99.81 | 99.83 | 99.77 | 99.80 | 99.84 | 99.81 | 99.84 |
| Q30 of clean read (%) | 98.74 | 98.90 | 99.13 | 99.00 | 98.90 | 99.17 | 99.12 | 99.16 |
| GC (%) | 45.15 | 46.13 | 47.96 | 48.39 | 43.54 | 46.82 | 47.59 | 48.34 |
*The raw reads with lengths of 1 to 51 nts; **Clean reads with lengths of 18 to 32 nts; G, G/Y, Y/O and O/R indicate the developmental stage of the fruit as indicated by color green, green/yellow, yellow/orange and orange/red, respectively; Q20 and Q30 indicate the percentage of bases with Phred values >20 and >30, respectively; GC indicates the GC ratio of total base number.
Figure 3.
Length distribution of clean reads of eight small RNA libraries in sea buckthorn seeds. (a,b,c and d) Indicate clean reads of seeds from line ‘SJ1’ at four developmental stages G, G/Y, Y/O and O/R, respectively. (e,f,g and h) indicate clean reads of seeds from line ‘XE3’ at four developmental stages G, G/Y, Y/O and O/R, respectively. G, G/Y, Y/O and O/R indicate the developmental stage of the fruit as indicated by color green, green/yellow, yellow/orange and orange/red, respectively.
Although these data suggest that different mechanisms may operate in the two lines during seed development, further studies will be required to clarify the significance of the abundant 20–22 nts class in line ‘SJ1’.
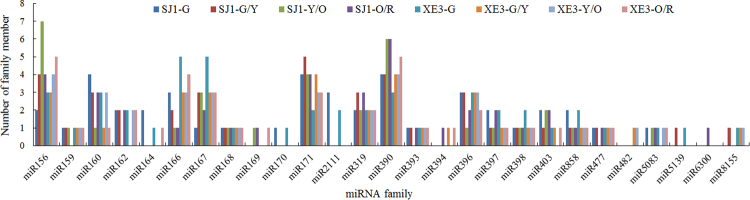
The unique sRNA sequences were mapped against the miRBase sequence database (Release 20) allowing zero mismatches. A total of 137 known miRNAs sequences and pre-miRNAs were identified (Fig. 4 and Supplementary Table S4), of which 51 miRNAs were shared in both lines across the four seed developmental stages, and 47 and 39 miRNAs were identified to be development-specific in lines ‘SJ1’ and ‘XE3’, respectively. All miRNA family members and normalized reads are shown in Supplementary Table S5. All known miRNAs were clustered into 27 miRNA families (Fig. 4), among which 21 miRNA families (miR156, miR159, miR160, miR162, miR164, miR166, miR167, miR168, miR169, miR170, miR171, miR2111, miR319, miR390, miR393, miR394, miR396, miR397, miR398, miR403, and miR858) are highly conserved in Arabidopsis15,39, indicating that these conserved miRNAs may have the fundamental regulatory roles in developing sea buckthorn seed.
Figure 4.
Distribution of known miRNA family size in sea buckthorn seed. SJ1-G, SJ1-G/Y, SJ1-Y/O and SJ1-O/R indicate the developmental stage of the line ‘SJ1’ fruit as indicated by color green, green/yellow, yellow/orange and orange/red, respectively; XE3-G, XE3-G/Y, XE3-Y/O and XE3-O/R indicate the developmental stage of the line ‘XE3’ fruit as indicated by color green, green/yellow, yellow/orange and orange/red, respectively.
All of the non-annotated reads from the eight sRNA libraries were aligned with the miRBase sequence database (Release 20), and the miRDeep software was used to predict potential novel miRNAs40. A total of 264 potential novel miRNAs (designated as novelmiRNA-1 to novelmiRNA-264) were predicted (Supplementary Table S6). Four miRNAs (novelmiRNA-5, novelmiRNA-27, novelmiRNA-67, and novelmiRNA-74) in line ‘SJ1’, and six miRNAs (novelmiRNA-27, novelmiRNA-37, novelmiRNA-44, novelmiRNA-74, novelmiRNA-149, and novelmiRNA-261) in line ‘XE3’ were expressed at all four developmental stages (Supplementary Table S7). To determine miRNAs that were expressed specifically at any developmental stage, the significantly differentially expressed miRNAs were compared between libraries of two different developmental stages (16 pairwise comparison groups in Supplementary Table S8). A total of 177 down-regulated and 175 up-regulated miRNAs were identified among the 16 pairwise comparison groups (Table 2). These results suggest that miRNA-mediated regulatory mechanisms may have a significant role in sea buckthorn seed development and oil accumulation.
Table 2.
Numbers of the significantly differential expression miRNAs for each of 16 pairwise comparison groups.
| Pairwise comparison groups | Number of up-regulation miRNA | Number of down-regulation miRNA |
|---|---|---|
| SJ1-G vs. SJ1-G/Y | 7 | 4 |
| SJ1-G vs. SJ1-Y/O | 5 | 0 |
| SJ1-G vs. SJ1-O/R | 7 | 9 |
| SJ1-G/Y vs. SJ1-Y/O | 5 | 4 |
| SJ1-G/Y vs. SJ1-O/R | 8 | 4 |
| SJ1-Y/O vs. SJ1-O/R | 2 | 9 |
| XE3-G vs. XE3-G/Y | 4 | 9 |
| XE3-G vs. XE3-Y/O | 9 | 6 |
| XE3-G vs. XE3-O/R | 9 | 13 |
| XE3-G/Y vs. XE3-Y/O | 20 | 15 |
| XE3-G/Y vs. XE3-O/R | 18 | 9 |
| XE3-Y/O vs. XE3-O/R | 14 | 7 |
| SJ1-G vs. XE3-G | 26 | 13 |
| SJ1-G/Y vs. XE3-G/Y | 16 | 20 |
| SJ1-Y/O vs. XE3-Y/O | 9 | 22 |
| SJ1-O/R vs. XE3-O/R | 18 | 31 |
SJ1-G, SJ1-G/Y, SJ1-Y/O and SJ1-O/R indicate the developmental stage of line ‘SJ1’ fruit as indicated by color green, green/yellow, yellow/orange and orange/red, respectively; XE3-G, XE3-G/Y, XE3-Y/O and XE3-O/R indicate the developmental stage of line ‘XE3’ fruit as indicated by color green, green/yellow, yellow/orange and orange/red, respectively.
Identification of differentially expressed miRNAs and their target genes involved in lipid biosynthesis and seed size
Since the sea buckthorn genome has not as yet been published, we determined the de novo mRNA transcriptome of seeds from four developmental stages of ‘SJ1’ and ‘XE3’ for identification of miRNA target genes41. Using psRNATarget (plant miRNA target prediction server), 3074 gene targets were predicted. Some genes were targeted with multiple miRNA, leading to a total of 5594 putative miRNA-target interactions. The identified gene targets were aligned using BLASTX against the protein databases Nr, COG, and Uniprot, followed by GO and KEGG analysis (Supplementary Table S9).
The putative gene targets of known and novel miRNAs identified in this study were subjected to GO analysis to investigate gene ontology. One hundred and seven genes (targeted by 91 miRNAs) were involved in seven different cellular components, 197 genes (targeted by 148 miRNAs) took part in seven molecular functions, and 159 genes (targeted by 98 miRNAs) participated in nine biological processes. We followed the GO term “metabolic processes” containing 56 target genes (Supplementary Table S10); as examples acetyl-CoA carboxylase (ACC, c103701_g1_i1) and delta-9-desaturase (Δ9D, c119361_g2_i1) were grouped into subterm fatty acid biosynthetic process (GO: 0006633), while GPD1 (c138230_g1_i3) was grouped into subterm carbohydrate metabolic process (GO: 0005975).
Based on the predicted miRNA-target interaction and functional annotation, we focussed on the miRNAs and their target genes involved in lipid biosynthesis. Four known (aly-miR170-5p, gma-miR168b, gma-miR164d, and zma-miR159i-3p) and 15 novel miRNAs were found to be involved in lipid biosynthesis (Table 3). Δ9D (c119361_g2_i1) targeted by gma-miR168b, ACC (c103701_g1_i1) targeted by novelmiRNA-108, glycerol-3-phosphate dehydrogenase (GPD1, c138230_g1_i3) targeted by novelmiRNA-23, diacylglycerol O-acyltransferase1 (DGAT1, c144982_g1_i2) targeted by novelmiRNA-58, and diacylglycerol O-acyltransferase 2 (DGAT2, c220405_g1_i1) targeted by novelmiRNA-191 were identified. The read counts of several novel miRNA, with potential target genes involved in lipid biosynthesis, were significantly higher in the low oil content line ‘SJ1’ as compared to the high oil content line ‘XE3’ (Supplementary Table S8). Based on these results it can be speculated that these miRNAs play an important role in regulating lipid biosynthesis.
Table 3.
Predicted targets involved in lipid metabolism for miRNAs.
| KEGG Pathway | miRNA name | Target ID | Annotation for targets | Gene name |
|---|---|---|---|---|
| Fatty acid biosynthesis | novelmiRNA-2 | c141756_g2_i2 | long-chain acyl-CoA synthetase | ACSL |
| novelmiRNA-108 | c103701_g1_i1 | acetyl-CoA carboxylase carboxyl transferase | ACC | |
| novelmiRNA-110 | c141756_g2_i2 | long-chain acyl-CoA synthetase | ACSL | |
| Fatty acid elongation | novelmiRNA-170 | c168561_g1_i1 | enoyl-CoA hydratase | ECHS |
| Fatty acid degradation | aly-miR170-5p | c60336_g1_i1 | alcohol dehydrogenase | adh |
| novelmiRNA-2 | c141756_g2_i2 | long-chain acyl-CoA synthetase | ACSL | |
| novelmiRNA-110 | c141756_g2_i2 | long-chain acyl-CoA synthetase | ACSL | |
| novelmiRNA-170 | c168561_g1_i1 | enoyl-CoA hydratase | ECHS | |
| Biosynthesis of unsaturated fatty acids | gma-miR168b | c119361_g2_i1 | delta-9-desaturase | Δ9D |
| novelmiRNA-58 | c145891_g1_i4 | helix loop helix transcription factor | HLH | |
| novelmiRNA-77 | c142283_g2_i1 | helix loop helix transcription factor | HLH | |
| Glycerolipid metabolism | gma-miR164d | c154991_g1_i1 | dihydroxyacetone kinase | DAK |
| novelmiRNA-11 | c192054_g1_i1 | phosphatidate phosphatase | LPIN | |
| novelmiRNA-23 | c133634_g3_i5 | phospholipid:diacylglycerol acyltransferase | PDAT | |
| novelmiRNA-58 | c144982_g1_i2 | diacylglycerol O-acyltransferase 1 | DGAT1 | |
| novelmiRNA-191 | c220405_g1_i1 | diacylglycerol O-acyltransferase 2 | DGAT2 | |
| Glycero- phospholipid metabolism | novelmiRNA-11 | c192054_g1_i1 | phosphatidate phosphatase | LPIN |
| novelmiRNA-23 | c138230_g1_i3 | glycerol-3-phosphate dehydrogenase | GPD1 | |
| novelmiRNA-64 | c176644_g1_i1 | lysophospholipid hydrolase | NTE | |
| Steroid biosynthesis | novelmiRNA-10 | c81203_g2_i1 | cycloartenol synthase | CAS |
| novelmiRNA-58 | c131674_g1_i1 | sterol-4-alpha-methyl oxidase | SMO2 | |
| novelmiRNA-58 | c144982_g1_i3 | sterol O-acyltransferase | SOAT | |
| novelmiRNA-179 | c81203_g2_i1 | cycloartenol synthase | CAS | |
| novelmiRNA-224 | c137936_g1_i1 | cycloartenol synthase | CAS | |
| novelmiRNA-224 | c137936_g1_i2 | lanosterol synthase | LSS | |
| novelmiRNA-232 | c81203_g2_i1 | cycloartenol synthase | CAS | |
| Sphingolipid metabolism | zma-miR159i-3p | c262907_g1_i1 | beta-galactosidase | lacZ |
| novelmiRNA-23 | c130447_g1_i3 | neutral ceramidase | ASAH2 | |
| novelmiRNA-108 | c167130_g1_i1 | sphingolipid delta-4 desaturase | DEGS | |
| novelmiRNA-151 | c209847_g1_i1 | beta-galactosidase | GLB1 | |
| novelmiRNA-170 | c199679_g1_i1 | sphingomyelin phosphodiesterase 2 | SMPD2 |
Many transcription factors play an essential role in controlling seed size26,42, however, the transcription factor regulatory network controlling seed size in woody oil crops remains largely unknown. Previous studies showed that ARF, MYB, CNR (cell number regulator) transcription factors and the MED gene play key role in controlling seed development and seed size in model plants28–31,43. In developing sea buckthorn seed, the gene targets of several miRNAs encode ARF, MYB, and CNR transcription factors (Table 4), including ARF2 (c88882_g1_i1 and c145594_g1_i3) targeted by gma-miR164d; ARF18 (c145777_g1_i1) targeted by gma-miR160b, gma-miR160c, gma-miR160d, and gma-miR160e; MYB (c133448_g2_i1) targeted by aly-miR166e-5p; and CNR13 (c37818_g1_i1) targeted by novelmiRNA-60, novelmiRNA-86, novelmiRNA-93, and novelmiRNA-196 (Table 4). CNR transcription factor are considered as general regulators of plant cell number and organ size. In sea buckthorn seed, MED12 (c180446_g1_i1), MED16 (c172958_g1_i1), and MED30 (c129331_g2_i1) were targeted by novelmiRNA-98, novelmiRNA-28, and novelmiRNA-23, respectively. Whether these genes control seed size in sea buckthorn remain to be determined.
Table 4.
Predicted targets involved in seed size for miRNAs.
| miRNA name | Target ID | Annotation for targets | Gene name |
|---|---|---|---|
| gma-miR164d | c88882_g1_i1 | auxin response factor 2 | ARF2 |
| gma-miR164d | c145594_g1_i3 | auxin response factor 2 | ARF2 |
| novelmiRNA-167 | c146624_g3_i1 | auxin response factor 2 | ARF2 |
| gma-miR160d | c145777_g1_i1 | auxin response factor 18 | ARF18 |
| gma-miR160b | c145777_g1_i1 | auxin response factor 18 | ARF18 |
| gma-miR160c | c145777_g1_i1 | auxin response factor 18 | ARF18 |
| gma-miR160e | c145777_g1_i1 | auxin response factor 18 | ARF18 |
| novelmiRNA-204 | c145777_g1_i1 | auxin response factor 18 | ARF18 |
| aly-miR166e-5p | c133448_g2_i1 | MYB domain-containing protein | MYB |
| novelmiRNA-48 | c146969_g1_i2 | Myb-like protein L | MYB |
| novelmiRNA-76 | c146969_g1_i2 | Myb-like protein L | MYB |
| novelmiRNA-122 | c146969_g1_i2 | Myb-like protein L | MYB |
| novelmiRNA-128 | c146969_g1_i2 | Myb-like protein L | MYB |
| novelmiRNA-174 | c147552_g2_i2 | Myb-related protein | MYB |
| novelmiRNA-201 | c146969_g1_i2 | Myb-like protein L | MYB |
| novelmiRNA-234 | c146969_g1_i2 | Myb-like protein L | MYB |
| novelmiRNA-23 | c129331_g2_i1 | mediator of RNA polymerase II transcription subunit 30 | MED30 |
| novelmiRNA-28 | c172958_g1_i1 | mediator of RNA polymerase II transcription subunit 16 | MED16 |
| novelmiRNA-98 | c180446_g1_i1 | mediator of RNA polymerase II transcription subunit 12 | MED12 |
| novelmiRNA-60 | c37818_g1_i1 | cell number regulator 13 | CNR13 |
| novelmiRNA-86 | c37818_g1_i1 | cell number regulator 13 | CNR13 |
| novelmiRNA-93 | c37818_g1_i1 | cell number regulator 13 | CNR13 |
| novelmiRNA-196 | c37818_g1_i1 | cell number regulator 13 | CNR13 |
qRT-PCR validation of miRNAs and corresponding target genes
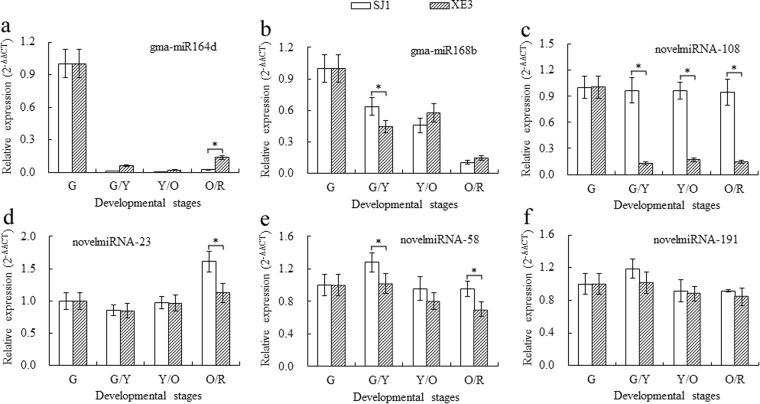
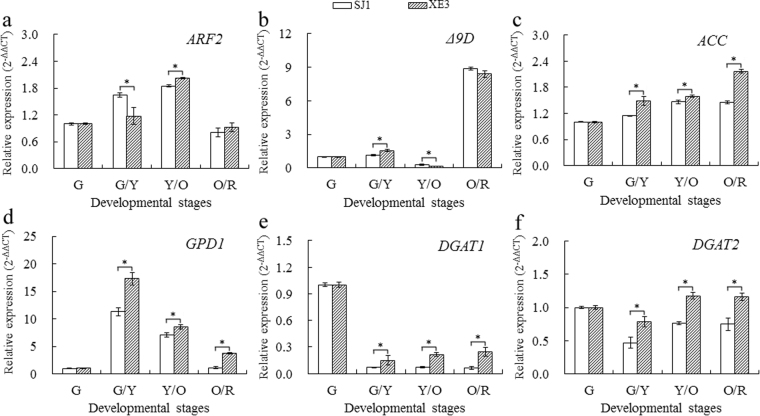
An integrated analysis of mRNA and miRNA transcriptome and qRT-PCR identified some miRNAs and their targets (miR164d-ARF2, miR168b-Δ9D, novelmiRNA-108-ACC, novelmiRNA-23-GPD1, novelmiRNA-58-DGAT1, and novelmiRNA-191-DGAT2) potentially involved in seed size and lipid biosynthesis (Figs 5 and 6). The qRT-PCR data of miRNAs matched the expression profiles obtained by sRNA sequencing of the G, G/Y, Y/O and O/R libraries, and the qRT-PCR data of the corresponding target gene showed a trend opposite to that of miRNA expression (Fig. 6). Relative expression levels of gma-miR164d, gma-miR168b, and novelmiRNA-108 decreased in both lines from G to G/Y stages (Fig. 5a,b,c), but gma-miR164d and gma-miR168b expression levels were up-regulated at O/R and Y/O stages, respectively, in seed of line ‘XE3’. By contrast, ARF2 (c145594_g1_i3), a target of gma-miR164d, was up-regulated from G to Y/O stages and then down-regulated at O/R stage (Fig. 6a). Δ9D gene (c119361_g2_i1) targeted by gma-miR168b was up-regulated at G/Y stage and then down-regulated at Y/O stage followed by a sharp increase at the O/R stage. Δ9D expression in ‘SJ1’ seed was higher than in ‘XE3’ seed (Fig. 6b). ACC (c103701_g1_i1) targeted by novelmiRNA-108 was up-regulated in both lines with higher expression in ‘XE3’ seed (Fig. 6c).
Figure 5.
qRT-PCR validation of select miRNAs putatively related to lipid biosynthesis and seed size in sea buckthorn. (a) Gma-miR164d expression. (b) Gma-miR168b expression. (c) NovelmiRNA-108 expression. (d) NovelmiRNA-23 expression. (e) NovelmiRNA-58 expression. (f) NovelmiRNA-191 expression. G, G/Y, Y/O and O/R indicate the developmental stage of the fruit as indicated by color green, green/yellow, yellow/orange and orange/red, respectively. *Indicate significant differences of gene relative expression level between the two lines at the same developmental stages, at the level of 0.05.
Figure 6.
qRT-PCR validation of target genes related to lipid biosynthesis and seed size in sea buckthorn. (a) ARF2 expression. (b) Δ9D expression. (c) GPD1 expression. (d) DGAT1 expression. (e) DGAT2 expression. (f) NovelmiRNA-191 expression. G, G/Y, Y/O and O/R indicate the developmental stage of the fruit as indicated by color green, green/yellow, yellow/orange and orange/red, respectively. *Indicate significant differences of gene relative expression level between the two lines at the same developmental stages, at the level of 0.05.
The novelmiRNA-23 was first down-regulated at G/Y stage and then up-regulated from Y/O to O/R stages in both lines (Fig. 5d). The novelmiRNA-58 and novelmiRNA-191 were first up-regulated and then down-regulated (Fig. 5e,f), and their expression levels in ‘SJ1’ seed were higher than in ‘XE3’ from G/Y to O/R stages. On the contrary, GPD1 (c138230_g1_i3) targeted by novelmiRNA-23 first increased at G/Y stage and then declined from Y/O to O/R stages (Fig. 6d). DGAT1 (c144982_g1_i2) and DGAT2 (c220405_g1_i1) targeted by novelmiRNA-58 and novelmiRNA-191, respectively, first declined at G/Y stage and then increased from Y/O to O/R stages. The expression levels of GPD1, DGAT1, and DGAT2 in ‘XE3’ seeds were significantly higher than in ‘SJ1’ seeds from G/Y to O/R stages (Fig. 6e,f).
Regulation of Δ9D by miR168b, as determined by luciferase activity assays
Target prediction analysis server psRNAtarget was used to assess the complementarity between miR168b and the target site. miR168b was predicted to have the potential to target Δ9D 3′UTR (Fig. 7a). Luciferase activity in 293 cells co-transfected with the miR168b recombinant expression vector and the expression vector containing the 3′UTR of Δ9D fused with the reporter gene was decreased by nearly 26.9% (p < 0.05) compared to that in the control group (Fig. 7b). These results indicate that Δ9D is one of the target genes of miR168b.
Figure 7.
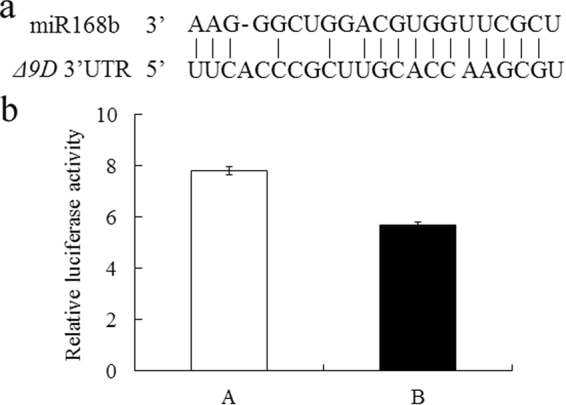
The validation of miR168b and target Δ9D. (a) Prediction of the binding sites of miR168b in Δ9D mRNA using psRNATarget. (b) Effect of miR168b expression on luciferase activity in transfected cells: (A) pCDNA3.1 + pmirGLO-Δ9D, (B) pCDNA3.1-miR168b + pmirGLO-Δ9D. Data are represented as the mean ± standard deviation (SD) from three independent experiments.
Discussion
Sea buckthorn (Hippophae L.) is a nutritionally and ecologically important woody plant known for the unique composition of its seed and fruit oil1. Although genes related to oil biosynthesis have been identified in sea buckthorn in previous2,44 and present studies, the regulatory mechanism of sea buckthorn oil accumulation is poorly understood. To explore if miRNA-mediated post-transcriptional regulation is involved in controlling oil accumulation and seed size in sea buckthorn, we analyzed the dynamic patterns of oil content, fatty acids composition, and seed size of developing seeds of lines ‘SJ1’ and ‘XE3’ (Fig. 1). Line ‘SJ1’ has low seed oil content and bigger seeds, and line ‘XE3’ has high seed oil content and smaller seeds34. We carried out comparative deep miRNA transcriptomic analysis in the two lines at four developmental stages of seeds, which generated 33,184,369 and 30,410,875 clean reads for lines ‘SJ1’ and ‘XE3’, respectively (Table 1). Bioinformatics analysis identified a total of 137 known and 264 novel miRNAs in developing sea buckthorn seeds (Supplementary Table S4 and Supplementary Table S6). Nineteen (four known and 15 novel) and 22 (six known and 16 novel) miRNAs were found to be involved in lipid biosynthesis and seed size, respectively, suggesting that miRNAs regulate oil accumulation and seed size in sea buckthorn seeds.
miRNA and their target genes involved in fatty acid biosynthesis in sea buckthorn
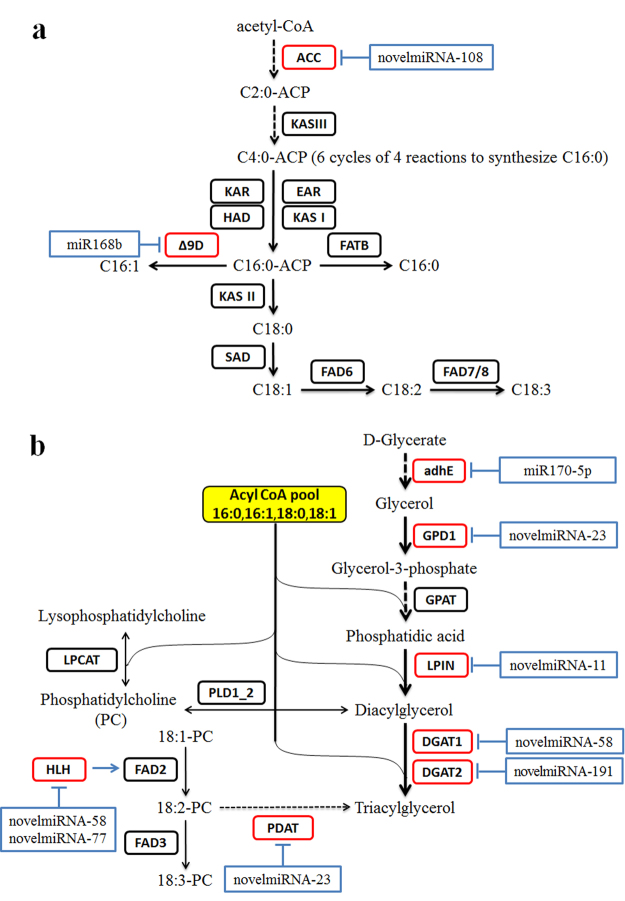
Eight miRNAs associated with fatty acid metabolic pathways were identified (Table 3). The expression of Δ9D gene that is involved in the conversion of C16:0 to C16:144 (Fig. 8a) was up-regulated at G/Y stage, down-regulated at Y/O stage, and sharply increased at O/R stage (Fig. 6b). Δ9D is a direct target of gma-miR168b, and its expression was significantly down-regulated in gma-miR168b transduced cells (Fig. 7b). As would be expected the expression trends of Δ9D and gma-miR168b were opposite of each other (Figs 5b and 6b), thus confirming that the gma-miR168b-target Δ9D relationship in conserved in sea buckthorn. Interestingly, while the expression of Δ9D was sharply up-regulated at the O/R stage, the C16:1 levels in both lines remained very low (Fig. 1c,d). This could be due to further regulation of Δ9D at the post-transcriptional level or lack of enough precursor C16:0-ACP. The down-regulation of Δ9D at Y/O stage was coordinated with high expressions of ketoacyl-ACP reductase (KAR) and ketoacyl-ACP synthase II (KAS II) and increased and decreased levels of C18:0 and C16:1, respectively (Fig. 8a), indicating higher precursor availability for the synthesis of C18 unsaturated fatty acids45. It is possible that inhibition of Δ9D by gma-miR168b in sea buckthorn seed directs conversion of C16:0 to higher levels of C18:2 and C18:3. It would be interesting to study the regulation of miR168b in sea buckthorn fruit pulp which, unlike the seed, contains high levels of C16:1 that is important for human health, but is also a valuable renewable source for industrial chemicals and biodiesel46. miR168 is also stress- and abscisic acid- inducible47. Furthermore, in mice fed on rice grain containing high levels of miR168a, the rice miR168a could bind to mRNA encoding human/mouse low-density lipoprotein receptor adapter protein 1 (LDLRAP1), thereby reducing its expression in the liver and increasing LDL levels in mouse plasma48.
Figure 8.
Sea buckthorn sequences associated with fatty acid (a) and TAG (b) biosynthetsis pathways. The miRNAs and its putative target genes are shown in blue and red boxes, respectively. Acetyl-CoA carboxylase (ACC); 3-oxoacyl-ACP synthase I/II/III (KAS I/II/III); 3-oxoacyl-ACP reductase (KAR); enoyl-acyl-ACP reductase (EAR); 3-hydroxyacyl-ACP dehydratase (HAD); delta-9-desaturase (Δ9D); fatty acyl-ACP thioesterases B (FATB); stearoyl-ACP desaturase (SAD); fatty acid desaturase (FAD); aldehyde dehydrogenases (adhE), glycerol-3-phosphate dehydrogenase (GPD1); glycerol-3-phosphate acyltransferase (GPAT); diacylglycerol O-acyltransferase (DGAT); phosphatidate phosphatase (LPIN); lysophosphatidylcholine acyltransferase (LPCAT); phospholipase D (PLD1_2); helix loop helix (HLH); phospholipid diacylglycerol acyltransferase (PDAT).
Expression of novelmiRNA-108 increased steadily with seed development in line ‘SJ1’, but declined sharply at the G/Y stage in ‘XE3’, which had significantly lower levels of this miRNA than in ‘SJ1’ (Fig. 5c). One of the putative targets of this miRNA is the ACC gene, which encodes the first committed enzyme that controls the flux of carbon into fatty acids49 (Fig. 8a). ACC was up-regulated in both lines from G/Y to O/R stages and its expression was significantly higher in ‘XE3’ as compared to ‘SJ1’ (Fig. 6c), which agree with the higher oil content in ‘XE3’ (Fig. 1b).
The putative target of novelmiRNA-58 is an HLH transcription factor. This protein binds as a homodimers or as heterodimers to specific target sequences in the FAD2 gene promoter25. FAD2 catalyzes the first extra-plastidial desaturation in plants, converting oleic acid to linoleic acid. The expressions of novelmiRNA-58 was higher in ‘SJ1’ than in ‘XE3’, and it increased at the G/Y stage in ‘SJ1’ and then decreased from G/Y to O/R stages (Fig. 5e). Interestingly, the level of C18:2 in ‘SJ1’ seeds were slightly lower than in ‘XE3’ seeds from G/Y to O/R stages (Fig. 1c,d). Further validation in required of the roles of novelmiRNA-108 and novelmiRNA-58 in fatty acids biosynthesis.
The TAG biosynthesis miRNA and their targets in sea buckthorn
A primary substrate for TAG biosynthesis is glycerol-3-phosphate (G3P) and its levels directly limit TAG biosynthesis in seeds50. G3P can be catalysed by glycerol-3-phosphate dehydrogenase (GPD1) from glycerol51 (Fig. 8b). GPD1, a rate-limiting enzyme of lipid synthesis52, plays a key role in carbohydrate and lipid metabolism53. A twofold increase in GPD activity led to a three to four -fold increase in the level of G3P in transgenic oil-seed rape, resulting in a 40% increase in the final oil content of the seed50.
In the Kennedy pathway, the enzyme glycerol-3-phosphate acyltransferase (GPAT) utilizes G3P and acyl-CoA as substrates to form lysophosphatidic acid (Lyso-PA), which is acylated by 1-acyl-sn-glycerol-3-phosphate acyltransferase (LPAAT) to form phosphatidic acid (PA). Next, PA is dephosphorylated by phosphatidate phosphatase (LPIN) to form diacylglycerol (DAG), which is finally converted to TAG by diacylglycerol O-acyltransferase (DGAT1 and DGAT2)44,49 (Fig. 8b). It is well known that the DGAT enzyme catalyzes a rate-limiting reaction in TAG bioassembly, and DGAT1 and DGAT2 genes are responsible for the progress of this reaction54. Seed-specific overexpression of AtDGAT1 increased the seed oil content by up to 8.3% in transgenic Brassica juncea as compared to wild type plants55. The expression patterns of both XsDGAT1 and XsDGAT2 correlated with oil accumulation in developing Xanthoceras sorbifolia embryos, and overexpression of these genes increased total seed oil content in transgenic plants as compared to wild-type plants54. Furthermore, we revealed in sea buckthorn that the high coordinated expression of source ‘GPD1’ and sink ‘DGAT1 and DGAT2’ genes results in oil accumulation in the pulp56,57.
In the present study, sea buckthorn DGAT1 and DGAT2 were targeted by novelmiRNA-58 and novelmiRNA-191, respectively. The expression of novelmiRNA-58 and novelmiRNA-191 peaked at the G/Y stage in both lines (Fig. 5e,f) and then decreased. The target genes DGAT1 and DGAT2 showed a trend opposite to those of miRNAs. Also DGAT1 and DGAT2 expression was lower in ‘SJ1’ as compared to ‘XE3’ (Fig. 6e,f), while the expression of novelmiRNA-58 and novelmiRNA-191 was always higher in ‘SJ1’ compared to ‘XE3’ (Fig. 5e,f). These results suggest that DGAT1 and DGAT2 genes are regulated by the novel miRNAs but further validation is required.
We identified the putative target of novelmiRNA-23 as GPD1, which was expressed at higher levels in ‘XE3’ as compared to ‘SJ1’, and in both lines it was first up-regulated at G/Y stage and then down-regulated (Fig. 6d). The gene expression changes correlated with oil content changes in both lines (Fig. 1b), and, as would be expected, were opposite to the expression trends of the miRNAs (Fig. 5d). In addition to establishing a putative miRNA-target gene relationship, these results clearly indicate that most TAG biosynthesis occurrs at the early to -mid stage of sea buckthorn seed development (Fig. 1b). Earlier GPD1 was identified as the target of jcu_MIR403 in Jatropha, and of oco-miR044 in Oryza coarctata seeds, and GPD1 expression was correlated to increased carbon flux and TAG biosynthesis58,59. Future studies will reveal how important the role of novelmiRNA-23 is in regulating GPD1 and consequently TAG biosynthesis in sea buckthorn seeds.
The seed size miRNAs and their targets in sea buckthorn
ARFs are transcription factors involved in auxin signal transduction during many stages of plant growth development via regulation of auxin response genes26. The crucial roles of ARFs in distinct biological processes and tissues are well understood in Arabidopsis. For example, mutation in ARF5 impairs the initiation of the body axis. Multiple ARF family members act as targets of conserved plant miRNA families including miR160, miR167 and miR390, and ARF regulate miRNA expression60,61. For example, ARF10, ARF16 and ARF17 transcript levels were highly increased in the miR160 foc mutant during early embryogenesis, and miR167, which targets ARF6 and ARF8, is preferentially expressed in rice seed, suggesting its involvement during late embryogenesis (seed maturation)26.
In this study, gma-miR164d was predicted to target ARF2, which is a repressor of cell division and organ growth, and determines the final size of the seed62. gma-miR164d showed very high expression at G stage, then relatively low expression at G/Y and Y/O stages, and then again up-regulated at the O/R stage (Fig. 5a). As would be expected, ARF2 was up-regulated during the G/Y and Y/O stages (Fig. 6a), when the length and width of seeds decreased (Fig. 1e,f). While the change in seed size cannot be fully explained by the expression patterns of miR164d (Fig. 1e,f), the possibility of regulation of ARF2 by miR164d cannot be ruled out at all seed developmental stages.
Two key determinants of organ size are cell number and cell size, and altering either one may affect the plant organ size. CNR and MED are considered as general regulators of plant cell number, final organ size, and fruit growth28,43. CNR1 reduced maize ear length and overall plant size when overexpressed, and the ear length increased when its expression was silenced28. Overexpression of MED25 produced small organs owing to decreases in both cell number and cell size in Arabidopsis43. In this study, CNR13, MED12, MED16, and MED30 were targeted by novelmiRNA-60, novelmiRNA-98, novelmiRNA-28, and novelmiRNA-23, respectively, indicating that several novel miRNAs may be involved in controlling seed size in sea buckthorn.
In conclusion, conserved and novel miRNAs were identified in two sea buckthorn lines. The putative identities of target genes of some miRNAs indicate that lipid biosynthesis and seed size, among other physiological processes, may be regulated by miRNAs in sea buckthorn.
Materials and Methods
Plant materials
Sea buckthorn lines ‘SJ1’ and ‘XE3’ belonged to H. rhamnoides ssp. mongolica grew at the Institute of Berries, Heilongjiang Academy of Agricultural Sciences in Suiling county, Heilongjiang Province, China (47°14′12.3″ northern latitude, 127°05′39.9″ east longitude). The orchard had a mean annual rainfall of 570.6 mm, mean annual temperature of 2.0 °C, mean annual evaporation capacity of 1242.5 mm and effective accumulative temperature of 2460.4 °C63. Line ‘SJ1’ was selected from seedlings of cultivar ‘Wulangemu’ (H. rhamnoides ssp. mongolica) in 1990s, and line ‘XE3’ was selected from seedlings of Russia cultivars (H. rhamnoides ssp. mongolica) in 2000s. Only one pollinate tree was cultivar ‘Wucixiong’ (ssp. mongolica.). The three trees of line ‘SJ1’ for collecting fruits were all cutting seedlings for three biological repetitions; the same was for line ‘XE3’. So the lines are stable and conserved over the generation. The molecular marker-based genetic similarity of these two lines is 0.76164. Fruits of both lines at four developmental stages described as green (G), green/yellow (G/Y), yellow/orange (Y/O) and orange/red (O/R)2 were harvested in 2015. The fruits of line ‘SJ1’ were collected on 25 June (G), 17 July (G/Y), 8 August (Y/O) and 30 August (O/R); the fruits of line ‘XE3’ were collected on 6 July (G), 28 July (G/Y), 19 August (Y/O) and 10 September (O/R). The samples were immediately frozen in liquid nitrogen and stored at −80 °C.
Oil content and fatty acid composition analysis
Sea buckthorn seed oil was isolated using a methanol-chloroform extraction procedure65,66. Seed sample powder (300 mg) was homogenised in methanol (2 ml) for 1 min, and after adding chloroform (4 ml) homogenization was continued for a further 2 min. The mixtures were sonicated in an ultrasonic bath for 30 min, centrifuged and filtered. The solid residues were re-suspended in chloroform/methanol (2:1, v/v, 4 ml) and homogenised again for 3 min and filtered. The volume of 1 ml of 0.88% KCl solution was added to the combined filtrates, and the mixtures were mixed thoroughly by vortexing and then centrifuged. The lower phase containing the purified oils was collected and evaporated to dryness under nitrogen.
Fatty acid composition was determined as fatty acid methyl esters (FAMEs) based on the boron trifluoride in methanol catalysis67. GC–TOF/MS analysis of FAMEs was performed on a Clarus 680 GC coupled with AxION iQT TOF/MS system (PerkinElmer, Shelton, USA). The system was equipped with Agilent J&W DB-23 capillary column (60 m × 0.25 mm × 0.25 μm). Fatty acid composition was measured and expressed as weight percentage of each fatty acid to the total fatty acids according to our previous study45. The analyses were conducted in three replicates.
mRNA and sRNA library construction and Illumina high-throughput sequencing
Total RNA was isolated from four developmental stages of lines ‘SJ1’ and ‘XE3’ seeds, using TRIzol RNA Extraction Kit (Invitrogen, Carlsbad, CA, USA). Total RNA was quantified and qualified by Agilent 2100 Bioanalyzer (1.9 < A260/A280 < 2.1, 2.0 < A260/A230 < 2.5 and RNA Integrity Number value ≥ 8.0). Next generation sequencing libraries were constructed according to the manufacturer’s protocol (NEBNext® Ultra™ RNA Library Prep Kit for Illumina). Enrichment of mRNA, fragment interruption, addition of adapters, size selection and PCR amplification were performed by GENEWIZ Inc. (Suzhou, China), and RNA-Seq was conducted by Illumina HiSeq2500 System. De novo assembly of the clean reads after removal of ambiguous nucleotides (N) and low-quality bases of raw reads based on Q20, Q30, N and GC parentages, was performed using Trinity software r2013-02-2568. All unigenes were aligned using Blastx algorithms (E-value ≤ 10−5) to identify homologous genes, and to the Nr, COG, Swiss-Prot and KEGG databases69. The unigenes were mapped to the KEGG metabolic pathway database to elucidate the complex biological behaviors of unigenes using KAAS70. Orthologous gene products were classified and the functions of unigenes was predicted using the COG database. GO classifications of unigenes were obtained using WEGO software71 after annotation by the Blast2GO program72 to elucidate the distribution of gene functions.
To identify miRNA involved in regulating lipid biosynthesis and seed size in sea buckthorn seeds, the RNA samples from four developmental stages of lines ‘SJ1’ and ‘XE3’ were used for small RNA sequences. Eight cDNA libraries were constructed according to the manufacturer’s protocol (NEBNext® Multiplex Small RNA Library Prep Set Kit for Illumina). The libraries were multiplexed and loaded on an Illumina HiSeq2500 instrument according to manufacturer’s instructions. The sequences were processed and analyzed by GENEWIZ Inc. (Suzhou, China).
Identification and analysis of known and novel miRNAs
The overall procedure for analyzing sRNA libraries is shown in Supplementary Fig. S1. All low-quality reads were removed, and 5′ and 3′ adapter sequences were trimmed using Genome Analyzer Pipeline v1.9. The remaining low -quality reads with ‘n’ were removed using Trimmomatic v0.3. Sequences shorter than 18 nts and longer than 32 nts were excluded from further analysis. Small RNAs were identified by mapping with miRDeep2 software and excluded from further miRNA predictions and analyses.
To identify conserved plant miRNAs in sea buckthorn, sRNA sequences were aligned with known plant miRNAs (Viridiplantae) in the miRBase database (Release 20) using miRDeep2 software. Complete alignment of the sequences was required and zero mismatches were allowed. To search for novel miRNAs, small RNA sequences were matched against assembled mRNA-seq contigs using the MiRanda software. As miRNA precursors have a characteristic hairpin structure, the next step to select candidate sequences was secondary structure analysis using MiRDeep2. Stem-loop structures should have the miRNA sequence at one arm of the stem and a corresponding antisense sequence at the opposite arm. Finally, precursor candidate sequences were checked using the BLASTn algorithm from the miRBase (www.miRBase.org) and NCBI databases.
For the frequency analysis of all identified miRNAs, sRNA reads were aligned in MiRDeep2 software. As reference, we used both previously annotated pre-miRNAs from miRBase and the putative pre-miRNAs identified in this work. The read counts of identified miRNAs in eight libraries were normalized as transcripts per million (TPM) according to the formula: Normalized expression = actual miRNA count/total count of clean reads × 1,000,00014. To assess whether the miRNA was differentially expressed, we independently used the R package EdgeR73. We considered miRNAs to be significantly differential expression if they had p value ≤ 0.05 and log2 (fold change) ≥2.
Prediction of miRNA targets
Prediction of target genes of novel miRNAs was performed against assembled RNA-seq contigs using psRNAtarget74, with the default parameters and a maximum expectation value of 4 (number of mismatches allowed). Function of each targeted gene was identified based on in-house sea buckthorn fruits mRNA-seq data; this analysis was conducted using the blast2GO v2.3.5 software75. Annotation was improved by analyzing conserved domains/families using the InterProScan tool. Orientation of the transcripts was obtained from BLAST annotations.
qRT-PCR analysis of miRNAs and their target genes
The RNA samples used for qRT-PCR analysis were the same as those for mRNA and miRNA sequence experiments. RNA was extracted as described under sRNA library preparation. First-strand cDNA synthesis was carried out using the Mir-XTM miRNA first-strand cDNA synthesis kit (TaKaRa), and qRT-PCR of miRNAs was performed using the Mir-XTM miRNA qRT-PCR SYBR kit (TaKaRa) and miRNA-specific primers in an ABI 7500 Real-Time PCR system (Applied Biosystems, Foster, USA). Small nuclear RNA U6 was used as an internal control. All samples included three technical repetitions. The primer sequences are shown in Supplementary Table S11.
Predicted target genes were validated by qRT-PCR using specific primers designed with Primer Premier 5.0. The first strand cDNA was synthesized from the RNAs using a PrimeScriptTM RT reagent Kit with gDNA Eraser (TaKaRa, Dalian, China). qRT-PCR was performed in an ABI 7500 Real-Time PCR system (Applied Biosystems, Foster, USA) using the SYBR Premix Ex TaqTM II Kit (TaKaRa, Dalian, China) according to the manufacturer’s instructions. The relative gene expression for qRT-PCR data was calculated by the 2−ΔΔCt method. All of the analyzed target genes were tested with three replicates. The primer sequences are shown in Supplementary Table S12.
Dual-luciferase reporter assay
Fragments from the 3′UTR of Δ9D containing the predicted binding sequences for miR168b were amplified and sub-cloned into pmirGLO luciferase promoter vector. The pCDNA3.1 plasmid was used as the template vector. The fragment containing the nucleotide sequences of precursor of the miR168b were cloned into the vector to construct the recombinant vector expressing miR168b pCDNA3.1 as described earlier. The pmirGLO vector containing 3′UTR of Δ9D were co-transfected with pCDNA3.1 or pCDNA3.1 containing pre-miR168b using lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol and previous report76–78. Forty-eight h after treatment, the expressed luciferase firefly and renilla activity was measured using a luciferase reporter assay kit (BioVision, Inc., CA, USA). Renilla was used as a transfection control.
Electronic supplementary material
Acknowledgements
This research was supported by the National Natural Science Foundation of China (No. 31570681). The authors acknowledge Professor Hilde Nybom for her help in editing the manuscript.
Author Contributions
J.D. and Y.G. designed and carried out the experiment of this study, C.R. conceived the study and C.R. and K.P. drafted and revised the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-22464-w.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ruan CJ, Rumpunen K, Nybom H. Advances in improvement of quality and resistance in a multipurpose crop: sea buckthorn. Crit. Rev. Biotechnol. 2013;33:126–144. doi: 10.3109/07388551.2012.676024. [DOI] [PubMed] [Google Scholar]
- 2.Fatima T, et al. Fatty acid composition of developing sea buckthorn (Hippophae rhamnoides L.) berry and the transcriptome of the mature seed. PloS ONE. 2012;7:e34099. doi: 10.1371/journal.pone.0034099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang BR, Kallio HP. Composition and physiological effect of sea buckthorn (Hippophae) lipids. Trends Food Sci. Tech. 2002;13:160–167. doi: 10.1016/S0924-2244(02)00136-X. [DOI] [Google Scholar]
- 4.Li W, Ruan CJ, Teixeira da Silva JA, Guo H, Zhao CE. NMR metabolomics of berry quality in sea buckthorn (Hippophae L.) Mol. Breeding. 2013;31:57–67. doi: 10.1007/s11032-012-9768-x. [DOI] [Google Scholar]
- 5.Gornas P, Misina I, Krasnova I, Seglina D. Tocopherol and tocotrienol contents in the sea buckthorn berry beverages in Baltic countries: Impact of the cultivar. Fruits. 2016;71:399–405. doi: 10.1051/fruits/2016030. [DOI] [Google Scholar]
- 6.Teleszko M, Wojdylo A, Rudzinska M, Oszmianski J, Golis T. Analysis of lipophilic and hydrophilic bioactive compounds content in sea buckthorn (Hippophae rhamnoides L.) berries. J. Agr. Food Chem. 2015;63:4120–4129. doi: 10.1021/acs.jafc.5b00564. [DOI] [PubMed] [Google Scholar]
- 7.Grey C, Widén C, Adlercreutz P, Rumpunen K, Duan RD. Antiproliferative effects of sea buckthorn (Hippophae rhamnoides L.) extracts on human colon and liver cancer cell lines. Food Chem. 2010;120:1004–1010. doi: 10.1016/j.foodchem.2009.11.039. [DOI] [Google Scholar]
- 8.Koyama T, Taka A, Togashi H. Effects of a herbal medicine, Hippophae rhamnoides, on cardiovascular functions and coronary microvessels in the spontaneously hypertensive stroke-prone rat. Clin. Hemorheol. Micro. 2009;41:17–26. doi: 10.3233/CH-2009-1148. [DOI] [PubMed] [Google Scholar]
- 9.Basu M, et al. Anti-atherogenic effects of seabuckthorn (Hippophaea rhamnoides) seed oil. Phytomedicine. 2007;14:770–777. doi: 10.1016/j.phymed.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 10.Olas B. Sea buckthorn as a source of important bioactive compounds in cardiovascular diseases. Food Chem. Toxicol. 2016;97:199–204. doi: 10.1016/j.fct.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 11.Krejcarova J, Strakova E, Suchy P, Herzig I, Karaskova K. Sea buckthorn (Hippophae rhamnoides L.) as a potential source of nutraceutics and its therapeutic possibilities - a review. Acta. Vet. Brno. 2015;84:257–268. doi: 10.2754/avb201584030257. [DOI] [Google Scholar]
- 12.Mangla Y, et al. Facultative apomixis and development of fruit in a deciduous shrub with medicinal and nutritional uses. Aob. Plants. 2015;7:plv098. doi: 10.1093/aobpla/plv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, et al. Identification of microRNAs actively involved in fatty acid biosynthesis in developing Brassica napus seeds using high-throughput sequencing. Front. Plant Sci. 2016;7:1570. doi: 10.3389/fpls.2016.01570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niu J, et al. Integrated mRNA and miRNA transcriptome reveal a cross-talk between developing response and hormone signaling for the seed kernels of Siberian apricot. Sci. Rep. 2016;6:35675. doi: 10.1038/srep35675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 16.Mutum DR, et al. Identification of novel miRNAs from drought tolerant rice variety Nagina 22. Sci. Rep. 2016;6:30786. doi: 10.1038/srep30786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khraiwesh B, Zhu JK, Zhu J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta. 2012;1819:137–148. doi: 10.1016/j.bbagrm.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song QX, et al. Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant Biol. 2011;11:5. doi: 10.1186/1471-2229-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin QJ, et al. Identification and characterization of microRNAs from tree peony (Paeonia ostii) and their response to copper stress. PloS ONE. 2015;10:e0117584. doi: 10.1371/journal.pone.0117584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korbes AP, et al. Identifying conserved and novel microRNAs in developing seeds of Brassica napus using deep sequencing. PloS ONE. 2012;7:e50663. doi: 10.1371/journal.pone.0050663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Srivastava S, Singh N, Srivastava G, Sharma A. MiRNA mediated gene regulatory network analysis of Cichorium intybus (chicory). Agri. Gene. 2017;3:37–45. [Google Scholar]
- 22.Klinkenberg J, et al. Two fatty acid desaturases, stearoyl-acyl carrier protein Δ9-desaturase6 and fatty aciddesaturase3, are involved in drought and hypoxia stress signaling in Arabidopsis crown galls. Plant Physiol. 2014;164:570–583. doi: 10.1104/pp.113.230326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adhikari ND, Bates PD, Browse J. WRINKLED1 rescues feedback inhibition of fatty acid synthesis in hydroxylase-expressing seeds. Plant Physiol. 2016;171:179–191. doi: 10.1104/pp.15.01906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manan S, et al. Soybean LEC2 regulates subsets of genes involved in controlling the biosynthesis and catabolism of seed storage substances and seed development. Front. Plant Sci. 2017;8:604. doi: 10.3389/fpls.2017.01604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim MJ, Kim J, Shin JS, Suh MC. The SebHLH transcription factor mediates trans-activation of the SeFAD2 gene promoter through binding to E- and G-box elements. Plant Mol. Biol. 2007;64:453–466. doi: 10.1007/s11103-007-9165-8. [DOI] [PubMed] [Google Scholar]
- 26.Gupta M, Bhaskar PB, Sriram S, Wang PH. Integration of omics approaches to understand oil/protein content during seed development in oilseed crops. Plant Cell Rep. 2017;36:637–652. doi: 10.1007/s00299-016-2064-1. [DOI] [PubMed] [Google Scholar]
- 27.Wu G, et al. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009;138:750–759. doi: 10.1016/j.cell.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo M, Simmons CR. Cell number counts–thefw2.2 and CNR genes and implications for controlling plant fruit and organ size. Plant Sci. 2011;181:1–7. doi: 10.1016/j.plantsci.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 29.Liu PP, et al. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 2007;52:133–146. doi: 10.1111/j.1365-313X.2007.03218.x. [DOI] [PubMed] [Google Scholar]
- 30.Mallory AC, Bartel DP, Bartel B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell. 2005;17:1360–1375. doi: 10.1105/tpc.105.031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allen RS, et al. Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proc. Natl. Acad. Sci. USA. 2007;104:16371–16376. doi: 10.1073/pnas.0707653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nasaruddin NM, Harikrishna K, Othman RY, Hoon LS, Harikrishna A. Computational prediction of microRNAs from oil palm (Elaeis guineensis Jacq.) expressed sequence tags. Asia Pac. J. Mol. Biol. Biotechnol. 2007;15:107–113. [Google Scholar]
- 33.Donaire L, Pedrola L, Rosa R, Llave C. High-throughput sequencing of RNA silencing-associated small RNAs in olive (Olea europaea L.) PloS ONE. 2011;6:e27916. doi: 10.1371/journal.pone.0027916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding J, et al. Characterization and identification of ISSR markers associated with oil content in sea buckthorn berries. Genet. Mol. Res. 2016;15:gmr15038278. doi: 10.4238/gmr.15038278. [DOI] [PubMed] [Google Scholar]
- 35.Xu W, Cui QH, Li F, Liu AZ. Transcriptome-wide identification and characterization of microRNAs from castor bean (Ricinus communis L.) PloS ONE. 2013;8:e69995. doi: 10.1371/journal.pone.0069995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of micro-RNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y, et al. Identification of miRNA from eggplant (Solanum melongena L.) by small RNA deep sequencing and their response to Verticillium dahliae infection. PloS ONE. 2013;8:e72840. doi: 10.1371/journal.pone.0072840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y, El-Kassaby YA. Landscape of fluid sets of hairpin-derived 21-/24-nt-log small RNAs at seed set uncovers special epigenetic features in Picea glauca. Genome Biol. Evol. 2017;9:82–92. doi: 10.1093/gbe/evw283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fahlgren N, et al. Computational and analytical framework for small RNA profiling by high-throughput sequencing. RNA. 2009;15:992–1002. doi: 10.1261/rna.1473809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friedlander MR, et al. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 41.Zhao H, Chen D, Peng Z, Wang L, Gao Z. Identification and characterization of microRNAs in the leaf of ma bamboo (Dendrocalamus latiflorus) by deep sequencing. PLoS ONE. 2013;8:e78755. doi: 10.1371/journal.pone.0078755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang CH, Yang CC. Identification of ICE1 as a negative regulator of ABA-dependent pathways in seeds and seedlings of Arabidopsis. Plant Mol. Biol. 2015;88:459–470. doi: 10.1007/s11103-015-0335-9. [DOI] [PubMed] [Google Scholar]
- 43.Xu R, Li YH. Control of final organ size by Mediator complex subunit 25 in Arabidopsis thaliana. Development. 2011;138:4545–4554. doi: 10.1242/dev.071423. [DOI] [PubMed] [Google Scholar]
- 44.Ding J, Wang L, Ruan CJ. Comparative transcriptome analysis of lipid biosynthesis in seeds and non-seed tissues of sea buckthorn. Genes Genom. 2017;39:1021–1033. doi: 10.1007/s13258-017-0564-1. [DOI] [Google Scholar]
- 45.Ding J, Ruan CJ, Guan Y, Wu LR, Shan JY. Coordinated regulation mechanism of multigenes involved in high accumulation of C18 unsaturated fatty acids in sea buckthorn seed. Acta Bot. Boreal. –Occident. Sin. 2017;37:1080–1089. [Google Scholar]
- 46.Wu YM, Li RZ, Hildebrand DF. Biosynthesis and metabolic engineering of palmitoleate production, an important contributor to human health and sustainable industry. Prog. Lipid Res. 2012;51:340–349. doi: 10.1016/j.plipres.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 47.Li W, et al. Transcriptional regulation of Arabidopsis MIR168a and ARGONAUTE1 homeostasis in abscisic acid and abiotic stress responses. Plant Physiol. 2012;158:1279–1292. doi: 10.1104/pp.111.188789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang L, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 2012;22:107–126. doi: 10.1038/cr.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang JX, et al. Comparative transcriptomic analysis of two Brassica napus near-isogenic lines reveals a network of genes that influences seed oil accumulation. Front. Plant Sci. 2016;7:1498. doi: 10.3389/fpls.2016.01498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vigeolas H, Waldeck P, Zank T, Geigenberger P. Increasing seed oil content in oil-seed rape (Brassica napus L.) by over-expression of a yeast glycerol-3-phosphate dehydrogenase under the control of a seed-specific promoter. Plant Biotechnol. J. 2007;5:431–441. doi: 10.1111/j.1467-7652.2007.00252.x. [DOI] [PubMed] [Google Scholar]
- 51.Vigeolas H, Geigenberger P. Increased levels of glycerol-3-phosphate lead to a stimulation of flux into triacylglycerol synthesis after supplying glycerol to developing seeds of Brassica napus L. in planta. Planta. 2004;219:827–835. doi: 10.1007/s00425-004-1273-y. [DOI] [PubMed] [Google Scholar]
- 52.Remize F, Barnavon L, Dequin S. Glycerol export and glycerol-3-phosphate dehydrogenase, but not glycerol phosphatase, are rate limiting for glycerol production in Saccharomyces cerevisiae. Metab. Eng. 2001;3:301–312. doi: 10.1006/mben.2001.0197. [DOI] [PubMed] [Google Scholar]
- 53.Zhang X, Zhang H, Zhang FC. Subcellular localization and expression of glyceraldehyde-3-phosphate dehydrogenase gene from Halostachys caspica under salt stress. Acta Bot. Boreal.-Occident. Sin. 2015;35:1283–1288. [Google Scholar]
- 54.Guo HH, et al. Two novel diacylglycerol acyltransferase genes from Xanthoceras sorbifolia are responsible for its seed oil content. Gene. 2013;527:266–274. doi: 10.1016/j.gene.2013.05.076. [DOI] [PubMed] [Google Scholar]
- 55.Savadi S, Naresh V, Kumar V, Bhat SR. Seed-specific overexpression of Arabidopsis DGAT1 in Indian mustard (Brassica juncea) increases seed oil content and seed weight. Botany. 2015;94:177–184. doi: 10.1139/cjb-2015-0218. [DOI] [Google Scholar]
- 56.Ding J, Ruan CJ, Shan JY, Guan Y. Expression of key genes involved in lipid biosynthesis and accumulation during seeds formation and development in Hippophae rhamnoides. Acta Bot. Boreal.-Occident. Sin. 2016;36:1642–1647. [Google Scholar]
- 57.Ding J, et al. Coordinated expression of source and sink genes involved in lipid biosynthesis and accumulation during sea buckthorn pulp development. Forest Res. 2017;30:902–907. [Google Scholar]
- 58.Galli V, et al. Identifying microRNAs and transcript targets in Jatropha seeds. PloS ONE. 2014;9:e83727. doi: 10.1371/journal.pone.0083727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mondal TK, Ganie SA, Debnath AB. Identification of novel and conserved miRNAs from extreme halophyte, Oryza coarctata, a wild relative of rice. PloS ONE. 2015;10:e0140675. doi: 10.1371/journal.pone.0140675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xie ZX, Khanna K, Ruan SL. Expression of microRNAs and its regulation in plants. Semin. Cell Dev. Biol. 2010;21:790–797. doi: 10.1016/j.semcdb.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Curaba J, Spriggs A, Taylor J, Li ZY, Helliwell C. miRNA regulation in the early development of barley seed. BMC Plant Biol. 2012;12:120. doi: 10.1186/1471-2229-12-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schruff MC, et al. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development. 2006;133:251–261. doi: 10.1242/dev.02194. [DOI] [PubMed] [Google Scholar]
- 63.Shan JY, Ding J, Wu YX. The evaluation for introduced sea buckthorn adaptation of the third Russia’s generation in northeast black soil area. Global Seabuckthorn Res. Dev. 2014;4:5–9. [Google Scholar]
- 64.Ding J, et al. Analysis of genetic relationships in sea buckthorn (Hippophae rhamnoides) germplasm from China and other countries using ISSR markers. J. Hortic. Sci. Biotech. 2015;90:599–606. doi: 10.1080/14620316.2015.11668721. [DOI] [Google Scholar]
- 65.Yang BR, Kallio H. Fatty acid composition of lipids in sea buckthorn (Hippophae rhamnoides L.) berries of different origins. J. Agr. Food Chem. 2001;49:1939–1947. doi: 10.1021/jf001059s. [DOI] [PubMed] [Google Scholar]
- 66.Vuorinen AL, et al. Effect of growth environment on the gene expression and lipids related to triacylglycerol biosynthesis in sea buckthorn (Hippophae rhamnoides) berries. Food Res. Int. 2015;77:608–619. doi: 10.1016/j.foodres.2015.08.023. [DOI] [Google Scholar]
- 67.Sanchez-Salcedo EM, Sendra E, Carbonell-Barrachina AA, Martinez JJ, Hernandez F. Fatty acids composition of Spanish black (Morus nigra L.) and white (Morus alba L.) mulberries. Food Chem. 2016;190:566–571. doi: 10.1016/j.foodchem.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 68.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tatusov RL, et al. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:277–280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ye J, et al. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006;34:293–297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Conesa A, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 73.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dai X, Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 2011;39:155–159. doi: 10.1093/nar/gkr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Conesa A, Gotz S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics. 2008;2008:1–13. doi: 10.1155/2008/619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Agatheeswaran S, Pattnayak NC, Chakraborty S. Identification and functional characterization of the miRNA-gene regulatory network in chronic myeloid leukemia lineage negative cells. Sci. Rep. 2016;6:32493. doi: 10.1038/srep32493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kumari B, et al. Dynamic changes in global microRNAome and transcriptome reveal complex miRNA-mRNA regulated host response to Japanese Encephalitis Virus in microglial cells. Sci. Rep. 2016;6:20263. doi: 10.1038/srep20263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Qian P, et al. bmo-miR-275 down-regulates expression of Bombyx mori sericin gene 2 in vitro. PloS ONE. 2018;13:e0190464. doi: 10.1371/journal.pone.0190464. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.