Abstract
Aim:
Gene assessment of pancreatic adenocarcinoma disease via protein-protein interaction (PPI) Network Analysis.
Background:
Diagnosis, especially early detection of pancreatic adenocarcinoma as a lethal disease implies more investigation. PPI Network Analysis is a suitable tool to discover new aspects of molecular mechanism of diseases.
Methods:
In the present study the related genes to pancreatic adenocarcinoma are studied in the interactome unit and the key genes are highlighted. The significant clusters were introduced by Cluster-ONE application of Cytoscape software 3.4.0. The genes are retrieved from STRING date base and analyzed by Cytoscape software. The crucial genes based on analysis of central parameters were determined and enriched by ClueGO v2.3.5 via gene ontology.
Results:
The number of 24 key genes among 794 initial genes were highlighted as crucial nodes in relationship with pancreatic adenocarcinoma. All of the key genes were organized in a cluster including 216 nodes. The main related pathways and cancer diseases were determined.
Conclusion:
It was concluded that the introduced 24 genes are possible biomarker panel of pancreatic adenocarcinoma.
Key Words: pancreatic adenocarcinoma, Protein-Protein Interaction, biomarker panel, gene ontology, cluster
Introduction
Cancer is one of the most causes of deaths in the world. One of the most common cancer is pancreatic adenocarcinoma. Pancreatic cancer was ranked the fifth leading death of cancer in the world (1). Since late detection of pancreatic adenocarcinoma is accompanied with catastrophic situation, it is tried to find the new biomarker panel related to effective prognosis of pancreatic cancer (2). It is reported that seven miRNAs expression level in pancreatic cancer is altered (3). PPI network analysis to discover new biomarkers and assessment of different diseases, have attracted great attention of scientists (4). In this approach many genes, proteins, metabolites or RNAs which are related to a certain disease are collected and organized in an interactive organization as interactome (5-7). Each element (the node) plays an especial role in the network. In the scale free type of the networks a few number of the nodes which are characterized as central nodes play crucial roles such as control of the other nodes and facilitating speed of information circulation between the elements of the network (8, 9). The important features of central nodes are hub-nodes, bottleneck nodes and high value closeness nodes (10). The key nodes of the constructed network can be introduced as the informative biomarker panel (11). There are some dense parts (clusters) in a network that the nodes of these regions are interacted closely so they control similar pathways. The clusters form cores in center of the networks (12). Gene ontology (GO) that exposes molecular function, cellular components, biological processes and the biological pathways can be used as a useful tool to analysis of the role of a gene in biological environment (13). In the present study, pancreatic adenocarcinoma PPI network is constructed by Cytoscape software by using the genes of STRING data base. The candidate biomarker panel and the related biological pathways are introduced.
Methods
The significant related proteins to pancreatic adenocarcinoma are extracted from String data base (14). The 794 proteins correlated to pancreatic adenocarcinoma were found by disease query of STRING data base. This data base is one of the Cytoscape software 3.4.0 applications that provides interaction information from three different panels including disease query, protein query and PubMed query. The strength of protein interaction can be fitted for the network construction. The proteins were interacted via undirected edges and appear as an interctome unit by cytoscape software. The network was analyzed considering centrality parameters. The most important central parameter is degree value and the high value degree node is called hub-node. The hub-nodes were selected based on degree value more than mean+2SD (15). The numbers of 37 nodes were determined as hub-nodes. Two important central parameters are betweenness and closeness centralities. The 5% top nodes based on betweenness (16) and closeness values were selected as bottleneck nodes and high value closeness nodes. The common nodes between the selected three groups were identified as crucial nodes. The significant clusters (P-value≤0.001) of the network were determined by Cluster-ONE application of Cytoscape software (9). The elements of the main cluster enriched via gene ontology by ClueGO v2.3.5 plugin of Cytoscape (17). Chemical pathways were extracted from KEGG.
Results
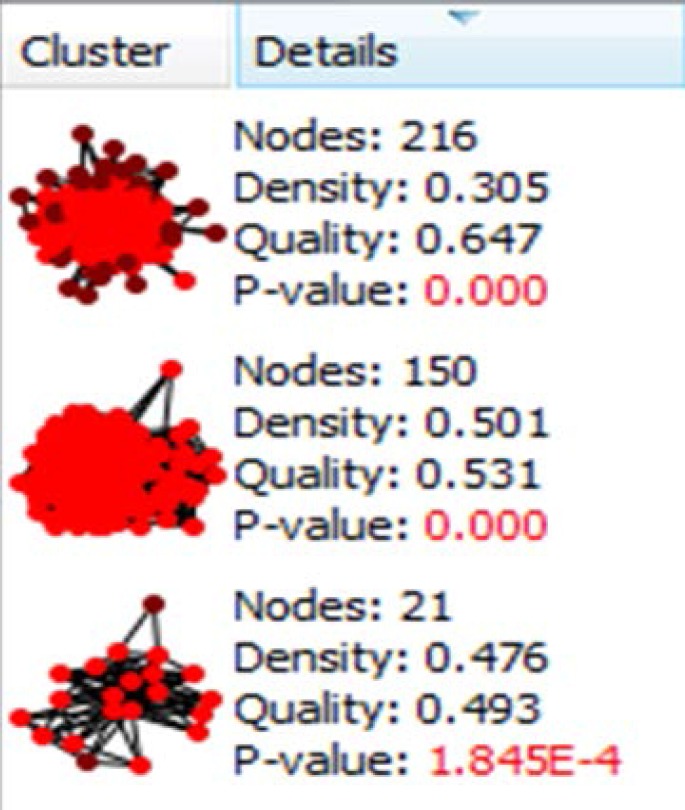
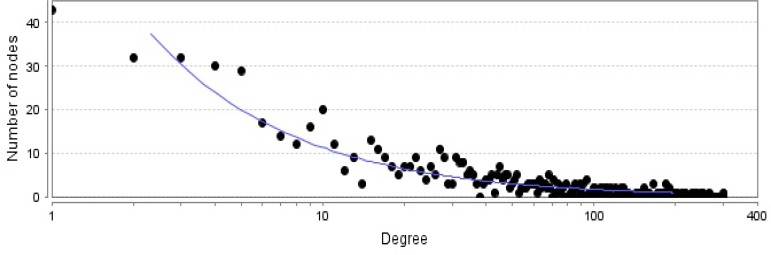
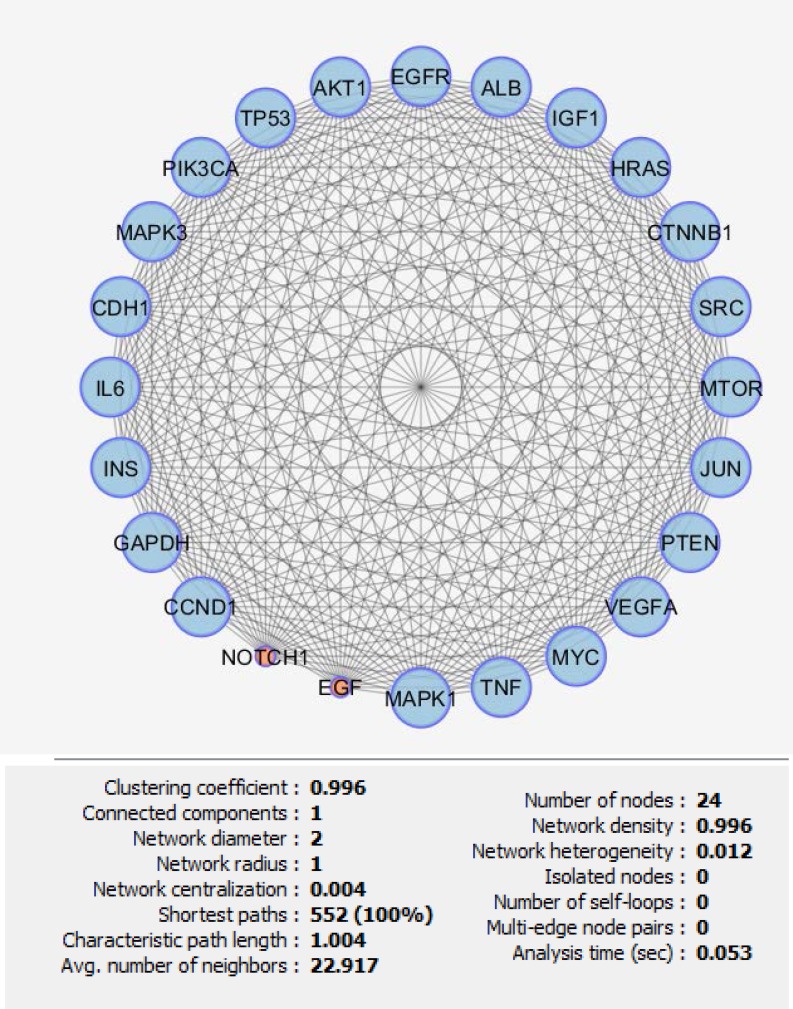
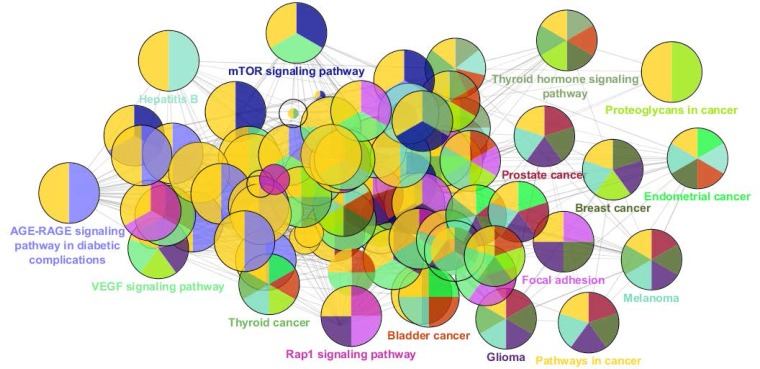
There are certain genes related to diseases in STRING database. The number of 794 genes were retried for pancreatic adenocarcinoma. The constructed network was included 145 isolated and one paired nodes and a main connected component specified with 647 nodes. Network density and cluster coefficient were 0.040 and 0.397 respectively. It is corresponded to a relatively uncondensed network. However, cluster analysis showed that there are three significant clusters in the network (see figure 1). The network was analyzed based on centrality parameters. The crucial nodes (number of 24 key genes) as are described in material and methods, were determined and accompanied with their centrality parameters such as degree, betweenness centrality and closeness centrality tabulated in table 1. Surprisingly, all off the 24 important nodes are presented in cluster-1 and cluster-2. As it is depicted in figure 1 these clusters include 216 and 150 nodes respectively. It seems that these clusters are the main functional and structural parts of the network and cluster 2 are like a subcluster of cluster-1. In the other hand, presence of the 24 crucial node in the cluster-1 indicates that these nodes play main role in controlling of the network. The degree distribution curve (see figure 2) is corresponded to scale free network. In this type of the networks the most nodes have low amounts of degree and just a few nodes are characterized with high values of degree. Since the 24 introduced key genes are the main elements in the network, their connections as a subinteractome is shown in figure 3 and the related biological pathways were assessed via gene ontology enrichment analysis (see figure 4).
Figure 1.
The three significant clusters related to the PPI network of pancreatic adenocarcinoma and their properties are presented
Table 1.
The 24 crucial nodes related to the PPI network of pancreatic adenocarcinoma are presented. D, BC and CC are abbreviations of degree, betweenness centrality and closeness centrality respectively
| Name | Description | D | BC | CC |
|---|---|---|---|---|
| TP53 | tumor protein p53 | 301 | 1 | 0.63 |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase | 266 | 0.29 | 0.61 |
| VEGFA | vascular endothelial growth factor A | 253 | 0.29 | 0.59 |
| AKT1 | v-akt murine thymoma viral oncogene homolog 1 | 250 | 0.29 | 0.59 |
| ALB | albumin | 249 | 0.43 | 0.59 |
| EGFR | epidermal growth factor receptor | 239 | 0.29 | 0.59 |
| MYC | v-myc myelocytomatosis viral oncogene homolog (avian) | 232 | 0.29 | 0.58 |
| EGF | epidermal growth factor | 230 | 0.14 | 0.58 |
| INS | Insulin | 218 | 0.29 | 0.57 |
| PIK3CA | phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha | 212 | 0.14 | 0.57 |
| JUN | jun proto-oncogene | 203 | 0.07 | 0.56 |
| IL6 | interleukin 6 (interferon, beta 2) | 201 | 0.14 | 0.55 |
| SRC | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | 199 | 0.29 | 0.56 |
| HRAS | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | 192 | 0.14 | 0.55 |
| MAPK1 | mitogen-activated protein kinase 1 | 190 | 0.14 | 0.56 |
| CCND1 | cyclin D1 | 190 | 0.14 | 0.55 |
| CDH1 | cadherin 1, type 1, E-cadherin (epithelial) | 184 | 0.14 | 0.55 |
| TNF | tumor necrosis factor | 184 | 0.07 | 0.55 |
| MAPK3 | mitogen-activated protein kinase 3 | 182 | 0.07 | 0.55 |
| IGF1 | insulin-like growth factor 1 (somatomedin C) | 176 | 0.07 | 0.54 |
| PTEN | phosphatase and tensin homolog | 175 | 0.07 | 0.54 |
| CTNNB1 | catenin (cadherin-associated protein), beta 1, 88kDa | 173 | 0.14 | 0.54 |
| NOTCH1 | notch 1 | 172 | 0.14 | 0.54 |
| MTOR | mechanistic target of rapamycin (serine/threonine kinase) | 155 | 0.07 | 0.54 |
Figure 2.
Degree distribution curve of PPI network of pancreatic adenocarcinoma is illustrated. The statistical parameters are determined as: fitted power law; y=74.353x-0.816, correlation; 0.910 and R-square on logarithmized value; 0.804
Figure 3.
The sub network including 24 crucial nodes of pancreatic PPI network is represented. All of these genes are included in cluster-1.
Figure 4.
The biological pathways related to the 24 crucial nodes of pancreatic adenocarcinoma PPI network are extracted from KEGG_01.03.2017:7194. The details and statistical parameters are presented as: final kappa score groups = 60, final group size after merging: 16, GO terms: 85, GO term connections: 471. The network was constructed by ClueGO v2.3.5. The main pathways are represented but the associated pathways are hidden. The colors are corresponded on the pathways or diseases, For example the “yellow color” refers to “pathways in cancer” and “purple color” to glioma
Discussion
Analysis of different diseases has attracted attention of researchers and scientists in the biology and medicine fields. Some gastrohepato diseases are targeted by the experts in bioinformatics and medical informatics (18, 19). There are several molecular investigations especially genetic approaches about pancreatic adenocarcinoma. However, there is a report about switching angiogenic to pancreatic cancer but there is no complete gene analysis about pancreatic adenocarcinoma (20, 21). In current research the scale free PPI network of pancreatic adenocarcinoma is constructed and was analyzed.
The number of 24 crucial nodes which are organized in dense part of the network were introduced as the key elements of the network. Presence of all the key nodes in the 2 significant clusters indicates that the crucial nodes are selected in a right method. In the other hand, the compressed linkages between the key nodes (as depicted in figure 3) are corresponded to interactive role of each one to control the network integrity. TP53, AKT1, EGFR, EGF, MYC and HRAS are the six well known genes that their expression level alterations in various cancers are reported frequently. There are some evidence that indicates the coloration between these genes expression changes and hepatogastro diseases (22, 23). Cowgill et al. reported mutations in TP53, KRAS, SMAD4 (DPC4) and P16 (CDKN2) genes in pancreatic cancer patients (24).
Expression change of ALB in several cancers is highlighted. However, ALB is a blood carrier which mostly is involved in molecular transport (25). C-reactive protein/ALB ratio was used as a pancreatic cancer index by Haruki et al. (26). The relationship between GAPDH and colorectal cancers (CRCs) is investigated in the mutant cells which are attributed to CRCs. In the mutant cells GAPDH was affected ;however, the normal cells was not (27). VEGFA mediated inhibition of several cytokines in the cultured pancreatic cancer cell line is discussed in details (28).
Insulin, the main product of pancreas is well known hormone with central role in body metabolism. Insulin expression change is main feature of diabetes (29). There is a potent relationship between pancreatic ductal adenocarcinoma and diabetes. This disease is the most common and lethal feature of pancreatic cancer (30).
It is reported that PIK3CA mutations can lead to pancreatic tumor beginning (31). The role of JUN in cell proliferation is confirmed (32). Tessari et al. were shown that the proto-oncogene C-Jun staining is a suitable approach in pancreatic cancer study (33). Rezaei-Tavirani et al. were shown TP53, EGFR, AKT1 and CTNNB1 are the main part of gastric adenocarcinoma biomarker panel (34). The effective role of CCND1 and MYC in 31 pancreatic cancer cell lines are assessed and emphasized (35).
Elevation of serum level of cytokines TNF and Il6 in pancreatic cancer patients were assessed by Falconer et al. (36). Since SRC is a suitable target in chemotherapy, its overexpression in pancreatic cancer refers to the crucial role of SRC in patients (37). Investigation showed that MAPK1 expression reduces in primary stage of disease (38).
Assessment of CDH1 together 11 genes in 15 types of cancer including pancreas cancer was corresponded to effect on several biological processes such as apoptosis, cell cycle regulation (39). The finding indicates that loss of ErbB2 leads to decrement of MAPK activation of the cells of pancreatic adenocarcinoma patients (40). The previous studies indicate that loss of PTEN elevates cell proliferation and cell invasion in pancreatic adenocarcinoma. In this pathway PTEN increases Phospho-AKT and Phospho-mTOR (41).
The IGF-1/PI3K/PTEN/Akt/NF-кB cascade was introduced as a main process in five pancreatic cancer cell line which was regulated by PTEN (42). Inhibitory role of NOTCH1 on pancreatic cancer cell invasion is reported by Wang et al. (43).
However, the positive pieces of evidence are corresponded to the role of all 24 introduced genes in pancreatic cancer but there are some documents about relationship between these markers and the other diseases especially different types of cancers. For example expression change of EGFR in lung cancer and EGF and TP53 in gliosarcoma is reported (44-46).
In the report of Bloomston et al. 25 microRNAs related to pancreas cancer are introduced which 21 numbers of them were overexpressed (47). It is a useful approach if the target genes of the detected microRNAs be analyzed.
The highlighted pathways and the related diseases in figure 4 are a clear schema of relationship between these genes and the various types of cancers. Thyroid, prostate, bladder, breast and endometrial cancers accompanied with melanoma and glioma are presented in the figure 4. Yellow color which refers to “pathways in cancers” is seen in almost all of the represented circles. The significant pathways including VEGF signaling pathway, MTOR signaling pathway and Rap1 signaling pathway are attributed to the 24 crucial genes. VEGFA is the gene of row-3 in table 1. It can be proposed that a combination of a few number of these genes in a feature of a suitable biomarker panel is a useful tool in diagnosis and prognosis of pancreatic adenocarcinoma.
In conclusion 24 genes were introduced as possible biomarker panel related to pancreatic adenocarcinoma. However, each one of them separately or in combination (68) with the other are related to the other cancers, more investigation in the field can lead to representation of a suitable biomarker panel among them.
Acknowledgment
This project is supported by Proteomics Research Center, Shahid Beheshti University of Medical Sciences.
Conflict of interests
The authors declare that they have no conflict of interest.
References
- 1.Yadav D, Lowenfels AB. The epidemiology of pancreatitis and pancreatic cancer. Gastroenterology. 2013;144:1252–61. doi: 10.1053/j.gastro.2013.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ranjzad F, Mahban A, Shemirani AI, Mahmoudi T, Vahedi M, Nikzamir A, et al. Influence of gene variants related to calcium homeostasis on biochemical parameters of women with polycystic ovary syndrome. J Assist Reprod Genet. 2011;28:225–32. doi: 10.1007/s10815-010-9506-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Safaee A, Fatemi SR, Ashtari S, Vahedi M, Moghimi-Dehkordi B, Zali MR. Four years incidence rate of colorectal cancer in Iran: a survey of national cancer registry data - implications for screening. Asian Pac J Cancer Prev. 2012;13:2695–98. doi: 10.7314/apjcp.2012.13.6.2695. [DOI] [PubMed] [Google Scholar]
- 4.Li Z, Ivanov AA, Su R, Gonzalez-Pecchi V, Qi Q, Liu S, et al. The OncoPPi network of cancer-focused protein–protein interactions to inform biological insights and therapeutic strategies. Nat Commun. 2017;8:14356. doi: 10.1038/ncomms14356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peyvandi AA, Khoshsirat S, Safaei A, Rezaei-Tavirani M, Azodi-Zamanian M. Interactome Analysis of 11-Dehydrosinulariolide-Treated Oral Carcinoma Cell Lines Such as Ca9-22 and CAL-27 and Melanoma Cell Line. Int J Cancer Manag. 2017;10:10096. [Google Scholar]
- 6.Safaei A, Rezaei-Tavirani M, Oskouie AA, Mohebbi SR, Shabani M, Sharifian A. Serum Metabolic Profiling of Advanced Cirrhosis Based on HCV. Hepat Mon. 2017;17:44431. [Google Scholar]
- 7.Izadi F, Zamanian-azodi M, Mansouri V, Khodadoostan M, Naderi N. Exploring conserved mRNA-miRNA interactions in colon and lung cancers. Gastroenterol Hepatol Bed Bench. 2017;10:184–93. [PMC free article] [PubMed] [Google Scholar]
- 8.Maghvan PV, Rezaei–Tavirani M, Zali H, Nikzamir A, Rostami-Nejad M, Khodadoostan M, et al. Network analysis of common genes related to esophageal, gastric, and colon cancers. Gastroenterol Hepatol Bed Bench. 2017:10. [PMC free article] [PubMed] [Google Scholar]
- 9.Abbaszadeh HA, Peyvandi AA, Sadeghi Y, Safaei A, Zamanian-Azodi M, Khoramgah MS, et al. Er: YAG laser and cyclosporin A effect on cell cycle regulation of human gingival fibroblast cells. J Lasers Med Sci. 2017;8:143–9. doi: 10.15171/jlms.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khayyer N, Azodi MZ, Mansouri V, Ghssemi-Broumand M, Rezaei-Tavirani M, Heidari MH, et al. Oral Squamous Cell Cancer protein-protein interaction network interpretation in comparison to esophagus adenocarcinoma. Gastroenterol Hepatol Bed Bench. 2017;10:118–24. [PMC free article] [PubMed] [Google Scholar]
- 11.Rezaei-Tavirani M, Okhovatian F, Azodi MZ, Tavirani MR. Duchenne muscular dystrophy (DMD) protein-protein interaction mapping. Iran J Child Neurol. 2017;11:7–14. [PMC free article] [PubMed] [Google Scholar]
- 12.Safaei A, Tavirani MR, Azodi MZ, Lashay A, Mohammadi SF, Broumand MG, et al. Diabetic Retinopathy and Laser Therapy in Rats: A Protein-Protein Interaction Network Analysis. J Lasers Med Sci. 2017;8:s20–27. doi: 10.15171/jlms.2017.s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karbalaei R, Piran M, Rezaei-Tavirani M, Asadzadeh-Aghdaei H, Heidari MH. One systems biology analysis protein-protein interaction of NASH and IBD based on comprehensive gene information. Gastroenterol Hepatol Bed Bench. 2017;10:194–201. [PMC free article] [PubMed] [Google Scholar]
- 14.Baghaei K, Shokrzadeh L, Jafari F, Dabiri H, Yamaoka Y, Bolfion M, et al. Determination of Helicobacter pylori virulence by analysis of the cag pathogenicity island isolated from Iranian patients. Dig Liver Dis. 2009;41:634–38. doi: 10.1016/j.dld.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Safari‐Alighiarloo N, Taghizadeh M, Tabatabaei SM, Shahsavari S, Namaki S, Khodakarim S, et al. Identification of new key genes for type 1 diabetes through construction and analysis of protein–protein interaction networks based on blood and pancreatic islet transcriptomes. J Diabetes. 2017;9:764–77. doi: 10.1111/1753-0407.12483. [DOI] [PubMed] [Google Scholar]
- 16.Rezaei-Tavirani M, Zamanian-Azodi M, Rajabi S, Masoudi-Nejad A, Rostami-Nejad M, Rahmatirad S. Protein Clustering and Interactome Analysis in Parkinson and Alzheimer's Diseases. Arch Iran Med. 2016;19:101–109. [PubMed] [Google Scholar]
- 17.Chaiyadet S, Smout M, Laha T, Sripa B, Loukas A, Sotillo J. Proteomic characterization of the internalization of Opisthorchis viverrini excretory/secretory products in human cells. Parasitol Int. 2017;66:494–502. doi: 10.1016/j.parint.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donaldson EF, Harrington PR, O'Rear JJ, Naeger LK. Clinical evidence and bioinformatics characterization of potential hepatitis C virus resistance pathways for sofosbuvir. Hepatology. 2015;61:56–65. doi: 10.1002/hep.27375. [DOI] [PubMed] [Google Scholar]
- 19.Poon TC, Wong N, Lai PB, Rattray M, Johnson PJ, Sung JJ. A tumor progression model for hepatocellular carcinoma: bioinformatic analysis of genomic data. Gastroenterology. 2006;131:1262–70. doi: 10.1053/j.gastro.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 20.Abdollahi A, Schwager C, Kleeff J, Esposito I, Domhan S, Peschke P, et al. Transcriptional network governing the angiogenic switch in human pancreatic cancer. Proc Natl Acad Sci U S A. 2007;104:12890–5. doi: 10.1073/pnas.0705505104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 22.Suda K, Tomizawa K, Mitsudomi T. Biological and clinical significance of KRAS mutations in lung cancer: an oncogenic driver that contrasts with EGFR mutation. Cancer Metastasis Rev. 2010;29:49–60. doi: 10.1007/s10555-010-9209-4. [DOI] [PubMed] [Google Scholar]
- 23.Lu SH, Hsieh LL, Luo FC, Weinstein I. Amplification of the EGF receptor and c‐myc genes in human esophageal cancers. Int J Cancer. 1988;42:502–5. doi: 10.1002/ijc.2910420406. [DOI] [PubMed] [Google Scholar]
- 24.Cowgill SM, Muscarella P. The genetics of pancreatic cancer. Am J Surg. 2003;186:279–86. doi: 10.1016/s0002-9610(03)00226-5. [DOI] [PubMed] [Google Scholar]
- 25.Rezaei-Tavirani M, Tadayon R, Mortazavi SA, Medhet A, Namaki S, Kalantari S, et al. Fluoxetine competes with cortisol for binding to human serum albumin. Iran J Pharm Res. 2012;11:325. [PMC free article] [PubMed] [Google Scholar]
- 26.Haruki K, Shiba H, Shirai Y, Horiuchi T, Iwase R, Fujiwara Y, et al. The C-reactive protein to albumin ratio predicts long-term outcomes in patients with pancreatic cancer after pancreatic resection. World J Surg. 2016;40:2254–60. doi: 10.1007/s00268-016-3491-4. [DOI] [PubMed] [Google Scholar]
- 27.Yun J, Mullarky E, Lu C, Bosch KN, Kavalier A, Rivera K, et al. Vitamin C selectively kills KRAS and BRAF mutant colorectal cancer cells by targeting GAPDH. Science. 2015;350:1391–6. doi: 10.1126/science.aaa5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Y, Antony S, Meitzler JL, Lu J, Juhasz A, Jiang G, et al. Dexamethasone suppresses cytokine-induced dual oxidase 2 (Duox2) and VEGF-A expression in human pancreatic cancer cells in vitro and pancreatic cancer growth in xenografts. AACR. 2016;76:1456. [Google Scholar]
- 29.Baghestani AR, Daneshva T, Pourhoseingholi MA, Asadzadeh H. Survival of Colorectal Cancer in the Presence of Competing- Risks - Modeling by Weibull Distribution. Asian Pac J Cancer Prev. 2016;17:1193–96. [PubMed] [Google Scholar]
- 30.Andersen DK, Korc M, Petersen GM, Eibl G, Li D, Rickels MR, et al. Diabetes, Pancreatogenic Diabetes, and Pancreatic Cancer. Diabetes. 2017;66:1103–10. doi: 10.2337/db16-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Payne S, Maher M, Tran N, Van De Hey D, Foley T, Yueh A, et al. PIK3CA mutations can initiate pancreatic tumorigenesis and are targetable with PI3K inhibitors. Oncogenesis. 2015;4:169. doi: 10.1038/oncsis.2015.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bai L, Mao R, Wang J, Ding L, Jiang S, Gao C, et al. ERK1/2 promoted proliferation and inhibited apoptosis of human cervical cancer cells and regulated the expression of c-Fos and c-Jun proteins. Med Oncol. 2015;32:57. doi: 10.1007/s12032-015-0490-5. [DOI] [PubMed] [Google Scholar]
- 33.Tessari G, Ferrara C, Poletti A, Dubrovich A, Corsini A, Del Favero G, et al. The expression of proto-oncogene c-jun in human pancreatic cancer. Anticancer Res. 1999;19:863–7. [PubMed] [Google Scholar]
- 34.Rezaei-Tavirani M, Rezaei-Tavirani M, Mansouri V, Mahdavi SM, Valizadeh R, Rostami-Nejad M, et al. Introducing crucial protein panel of gastric adenocarcinoma disease. Gastroenterol Hepatol Bed Bench. 2017;10:21. [PMC free article] [PubMed] [Google Scholar]
- 35.Mahlamäki EH, Bärlund M, Tanner M, Gorunova L, Höglund M, Karhu R, et al. Frequent amplification of 8q24, 11q, 17q, and 20q‐specific genes in pancreatic cancer. Genes Chromosomes Cancer. 2002;35:353–8. doi: 10.1002/gcc.10122. [DOI] [PubMed] [Google Scholar]
- 36.Falconer JS, Fearon K, Plester CE, Ross JA, Carter DC. Cytokines, the acute-phase response, and resting energy expenditure in cachectic patients with pancreatic cancer. Ann Surg. 1994;219:325. doi: 10.1097/00000658-199404000-00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Messersmith WA, Rajeshkumar N, Tan AC, Wang XF, Diesl V, Choe SE, et al. Efficacy and pharmacodynamic effects of bosutinib (SKI-606), a Src/Abl inhibitor, in freshly generated human pancreas cancer xenografts. Mol Cancer Ther . 2009;8:1484–93. doi: 10.1158/1535-7163.MCT-09-0075. [DOI] [PubMed] [Google Scholar]
- 38.Furukawa T, Kanai N, Shiwaku H, Soga N, Uehara A, Horii A. AURKA is one of the downstream targets of MAPK1/ERK2 in pancreatic cancer. Oncogene. 2006;25:4831–9. doi: 10.1038/sj.onc.1209494. [DOI] [PubMed] [Google Scholar]
- 39.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–9. [PubMed] [Google Scholar]
- 40.Skrypek N, Vasseur R, Vincent A, Duchêne B, Van Seuningen I, Jonckheere N. The oncogenic receptor ErbB2 modulates gemcitabine and irinotecan/SN-38 chemoresistance of human pancreatic cancer cells via hCNT1 transporter and multidrug-resistance associated protein MRP-2. Oncotarget. 2015;6:10853. doi: 10.18632/oncotarget.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Zhang J, Xu K, Xiao Z, Sun J, Xu J, et al. PTEN/PI3K/mTOR/B7-H1 signaling pathway regulates cell progression and immuno-resistance in pancreatic cancer. Hepatogastroenterology. 2013;60:1766–72. [PubMed] [Google Scholar]
- 42.Ma J, Sawai H, Matsuo Y, Ochi N, Yasuda A, Takahashi H, et al. IGF-1 mediates PTEN suppression and enhances cell invasion and proliferation via activation of the IGF-1/PI3K/Akt signaling pathway in pancreatic cancer cells. J Surg Res. 2010;160:90–101. doi: 10.1016/j.jss.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 43.Rostami-Nejad M, Villanacci V, Hogg-Kollars S, Volta U, Manenti S, Reza-Zali M, et al. Endoscopic and histological pitfalls in the diagnosis of celiac disease: A multicentre study assessing the current practice. Rev Esp Enferm Dig. 2013;105:326–33. doi: 10.4321/s1130-01082013000600003. [DOI] [PubMed] [Google Scholar]
- 44.Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 45.Reis RM, Könü-Lebleblicioglu D, Lopes JM, Kleihues P, Ohgaki H. Genetic profile of gliosarcomas. Am J Pathol. 2000;156:425–32. doi: 10.1016/S0002-9440(10)64746-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Biernat W, Aguzzi A, Sure U, Grant JW, Kleihues P, Hegi ME. Identical mutations of the p53 tumor suppressor gene in the gliomatous and the sarcomatous components of gliosarcomas suggest a common origin from glial cells. J Neuropathol Exp Neurol. 1995;54:651–6. doi: 10.1097/00005072-199509000-00006. [DOI] [PubMed] [Google Scholar]
- 47.Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–8. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 48.Nazemalhosseini-Mojarad E, Haghighi A, Taghipour N, Keshavarz A, Mohebi SR, Zali MR, et al. Subtype analysis of Cryptosporidium parvum and Cryptosporidium hominis isolates from humans and cattle in Iran. Vet Parasitol. 2011;179:250–52. doi: 10.1016/j.vetpar.2011.01.051. [DOI] [PubMed] [Google Scholar]