Abstract
Kinesin Family Member 3A (KIF3A) was recognized as a key factor of ciliogenesis and transport system of primary cilia in normal cells. However, its possible function on cancer cells has yet to be identified. In the present study, microarray tissue chips, including 230 breast cancer samples, were applied to determine the KIF3A expression pattern by immunological histological chemistry. Statistical analysis on the KIF3A expression level and the currently used clinicopathological characteristics of breast cancer patients was carried out. Follow-up data of these patients over 10 years were also used to evaluate the relationship between KIF3A and the survival rate. The expression levels of KIF3A were significantly higher in 140 breast cancer tissues than those of 90 para-carcinoma samples, which served as controls (P<0.001). In addition, in a further 70 paired samples, the same higher expression level was observed in cancer tissues compared with their self-paired controls (P<0.001). Furthermore, the high expression of KIF3A in breast cancer tissue correlated with the status of estrogen receptor, androgen receptor, epidermal growth factor receptor and Ki-67 of breast cancer patients, and were also related to their pathology grade and lymph node metastasis. The survival analysis showed a better survival rate in the patients with a higher expression level of KIF3A. Collectively, the triadic associations of KIF3A, the currently used clinicopathological parameters and survival rate suggest that KIF3A is involved in the tumorigenesis and progression of breast cancer. Thus, KIF3A could be considered a promising novel prognostic index in breast cancer.
Keywords: KIF3A, breast cancer, microarray tissue chip, primary cilia, clinical pathological characteristics, survival rate
Introduction
Kinesin Family Member 3A (KIF3A) is regarded as a motor protein, which is associated with the intraflagellar transport system of primary cilia and maintenance of ciliogenesis (1,2). In addition, KIF3A plays a role in primary cilia formation and in centriole cohesion and subdistal appendage organization and function (3). In 2013, Barakat et al reported that KIF3A is necessary for the initiation and maintenance of medulloblastoma for the first time (4). Liu et al also proved that KIF3A plays a critical role in prostate cancer (5). Recently, Kim et al found that KIF3A is a class of tumor suppressors in non-small cell lung cancer (6).
Previous findings showed that primary cilia decreased in breast cancer (7–9). In addition, the disrupted expression of KIF3A leads to ablate ciliogenesis and tumorigenesis in glioblastoma (10).
Thus, we hypothesized that KIF3A may affect the formation and/or pathological change of primary cilia in breast cancer, and subsequently on tumor progression. Therefore, the aim of this study was to explore the possible relationship of KIF3A and breast cancer progression, and by analyzing such a relationship to explore its possible clinical usage.
Materials and methods
Study subjects
The samples of tissue microarrays (Xinchao Biotechnology Company, Shanghai, China) were collected from 140 tissues of mammary carcinoma patients and 90 adjacent para-carcinoma tissues (2 cm from the tumor tissues) as controls. Within the total of 230 cases, 70 self-contrast tissues were included. A long-term follow-up was carried out to all the patients as long as 14 years, while the survival rate was measured up to 2013 and 2014, respectively. Details of the clinicopathological parameters are presented in the results.
Immunohistochemical detection of KIF3A
The expression level of KIF3A was detected by immunohistochemical staining, performed according to the instructions of the SP kit ZSGB-BIO, Beijing, China). The antibody for KIF3A was rabbit polyclonal anti-KIF3A (1:800; cat. no. K3513; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Results of the staining were evaluated separately by three pathologists under double-blind conditions and scored by the intensity of positive, the positive rates and final score (score of positive rates multiplied by score of intensity of positive). Detailed scoring system and criteria are presented in Table I.
Table I.
Methods of scoring system and criteria for immunohistochemical results.
| Score | Criteria |
|---|---|
| Intensity of positive | |
| 0 | Negative (−) |
| 1 | Weakly positive (+) |
| 2 | Medium positive (++) |
| 3 | Strong positive (+++) |
| Positive rate | |
| 0 | No or 0% nuclear and cytoplasm staining (−) |
| 1 | <15% or occasional nuclear staining (1–15%) (+) |
| 2 | >15 to 75% clear positive nuclear staining (++) |
| 3 | >75% positive staining (+++) |
| Final score | |
| 0 | Total score 0–2 |
| 1 | Total score 3–5 |
| 2 | Total score 6–8 |
| 3 | Total score 9–11 |
Statistical analysis
The Chi-square test was performed to analyze the differences in the expression of KIF3A between the 140 tumor tissues and 90 adjacent para-carcinoma tissues, as well as the correlations of clinicopathological parameters of corresponding patients. Kaplan-Meier survival values were calculated to evaluate the connection between the expression level of KIF3A and the survival rate. Survival between the groups was compared using the log-rank test. Statistical significance was set at P<0.05, while P<0.001 indicated extremely statistically significant. The software used was SPSS Version 21.0 (SPSS, Inc., Chicago, IL, USA).
Results
Expression pattern of KIF3A in breast cancer
Tissue microarrays were used to detect the expression status of KIF3A in 230 cases. The intensity of positive, the positive rate and the final score were measured for further statistical analysis, respectively (Tables II and III).
Table II.
Expression level of KIF3A in 140 cases breast cancer patients and 90 para-carcinoma tissues.
| Score | ||||||
|---|---|---|---|---|---|---|
| Item | 0 | 1 | 2 | 3 | No. | P-value |
| Intensity of positive | <0.001a | |||||
| Cancer | 4 | 22 | 56 | 58 | 140 | |
| Formal | 24 | 31 | 31 | 4 | 90 | |
| No. | 28 | 53 | 87 | 62 | 230 | |
| Positive rate | <0.001a | |||||
| Cancer | 8 | 21 | 66 | 45 | 140 | |
| Formal | 23 | 16 | 46 | 5 | 90 | |
| No. | 31 | 37 | 112 | 50 | 230 | |
| Final score | <0.001a | |||||
| Cancer | 8 | 32 | 52 | 48 | 140 | |
| Formal | 25 | 30 | 31 | 4 | 90 | |
| No. | 33 | 62 | 83 | 52 | 230 | |
P<0.001. KIF3A, Kinesin Family Member 3A.
Table III.
The expression level of KIF3A in 70 self-contrast patients.
| Formal | ||||||
|---|---|---|---|---|---|---|
| Item | 0 | 1 | 2 | 3 | No. | P-value |
| Intensity of positive | <0.001 | |||||
| Cancer | ||||||
| 0 | 1 | 3 | 7 | 6 | 17 | |
| 1 | 0 | 5 | 11 | 10 | 26 | |
| 2 | 1 | 2 | 9 | 11 | 23 | |
| 3 | 0 | 0 | 2 | 2 | 4 | |
| No. | 2 | 10 | 29 | 29 | 70 | |
| Positive rate | <0.001 | |||||
| Cancer | ||||||
| 0 | 1 | 3 | 8 | 4 | 16 | |
| 1 | 2 | 1 | 6 | 5 | 14 | |
| 2 | 0 | 5 | 18 | 14 | 37 | |
| 3 | 0 | 0 | 2 | 1 | 3 | |
| No. | 3 | 9 | 34 | 24 | 70 | |
| Final score | <0.001 | |||||
| Cancer | ||||||
| 0 | 1 | 5 | 6 | 5 | 17 | |
| 1 | 2 | 7 | 6 | 9 | 24 | |
| 2 | 0 | 5 | 11 | 10 | 25 | |
| 3 | 0 | 0 | 2 | 1 | 3 | |
| No. | 3 | 17 | 25 | 25 | 70 | |
P<0.001. KIF3A, Kinesin Family Member 3A.
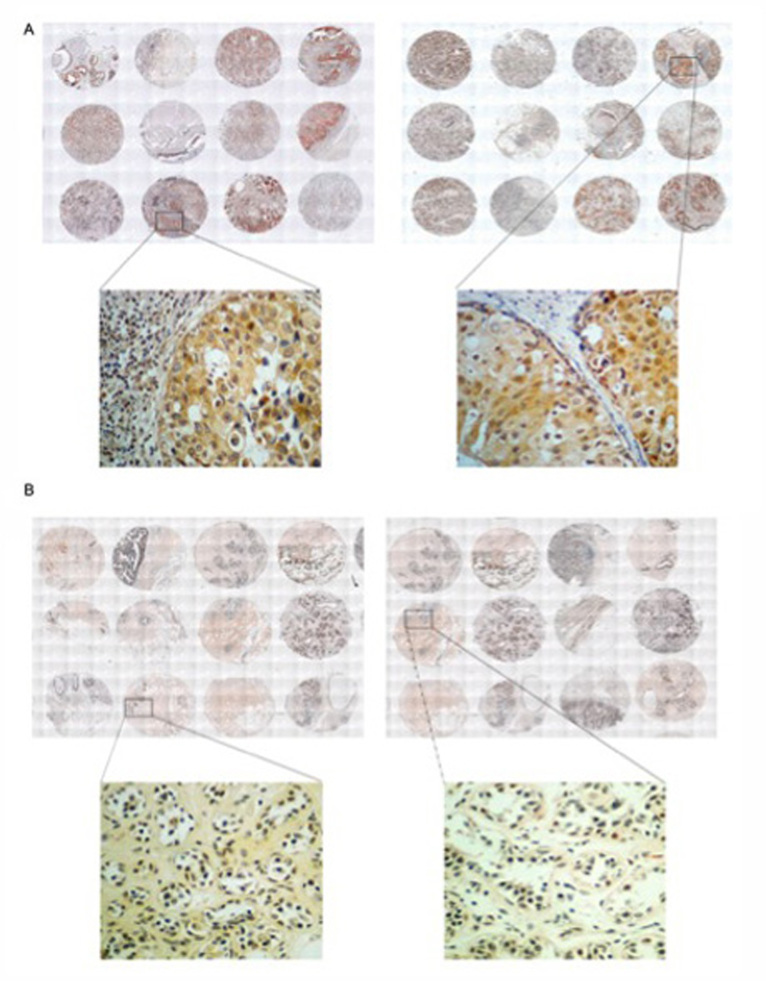
The Chi-square test on the scores of intensity of positive, positive rate and the final score, showed KIF3A expression levels were significantly higher in 140 breast cancer tissues than those in adjacent para-carcinoma tissues (P<0.01). The same significant difference was also observed in the 70 self-contrast cases (Fig. 1).
Figure 1.
IHC of KIF3A in human breast cancer and para-carcinoma tissues microarrays. IHC of KIF3A in (A) human breast cancer and (B) para-carcinoma tissues (magnification, ×400). IHC, immunological histological chemistry; KIF3A, Kinesin Family Member 3A.
Expression of KIF3A and clinicopathological parameters
Currently used clinical pathological parameters of the 230 breast cancer cases in this study are shown in Table IV. The average age of the patients was 53 (from 31 to 83 years). Pathological grade was categorized as grade I, II and III.
Table IV.
Regular clinicopathological parameters of breast cancer.
| Characteristics | No. |
|---|---|
| Age | |
| ≤53 | 80 |
| >53 | 60 |
| Pathology grade | |
| I | 12 |
| I–II | 21 |
| II | 95 |
| III | 7 |
| ER | |
| Positive | 88 |
| Negative | 42 |
| PR | |
| Positive | 62 |
| Negative | 51 |
| AR | |
| Positive | 101 |
| Negative | 40 |
| HER2 | |
| Positive | 42 |
| Negative | 89 |
| Lymph node metastasis | |
| TnN0 | 86 |
| TnNn | 46 |
| P53 | |
| Positive | 86 |
| Negative | 47 |
| Ki-67 | |
| − | 24 |
| + | 75 |
| ++ | 22 |
| +++ | 10 |
| Ck56 | |
| Positive | 19 |
| Negative | 112 |
| EGFR | |
| Positive | 38 |
| Negative | 102 |
| TN | |
| TNBC | 18 |
| NTNBC | 103 |
ER, estrogen receptor; PR, progesterone receptor; AR, androgen receptor; HER2, human epidermal growth factor receptor 2; EGFR, epidermal growth factor receptor; TN, triple-negative; TNBC, triple-negative breast cancer; NTNBC, non-triple-negative breast cancer.
According to the intensity of positive, the positive rate and the final score, the expression pattern of KIF3A and the clinicopathological parameters in the 140 cases were evaluated, respectively (Tables V–VII).
Table V.
Relationship between KIF3A expression and clinicopathological parameters in the140 cases by intensity of positive.
| Intensity of positive | ||||||
|---|---|---|---|---|---|---|
| Item | 0 | 1 | 2 | 3 | No. | P-value |
| Age | 0.728 | |||||
| ≤53 | 2 | 15 | 31 | 32 | 80 | |
| >53 | 2 | 7 | 25 | 26 | 60 | |
| Pathology grade | 0.000b | |||||
| I | 1 | 7 | 4 | 0 | 12 | |
| I–II | 0 | 2 | 12 | 8 | 22 | |
| II | 2 | 13 | 37 | 43 | 95 | |
| III | 0 | 0 | 2 | 5 | 7 | |
| ER | ||||||
| Positive | 2 | 12 | 35 | 39 | 88 | 0.463 |
| Negative | 1 | 10 | 16 | 15 | 42 | |
| PR | ||||||
| Positive | 2 | 10 | 32 | 34 | 78 | 0.475 |
| Negative | 0 | 11 | 20 | 20 | 51 | |
| AR | ||||||
| Positive | 4 | 12 | 36 | 48 | 100 | 0.021a |
| Negative | 0 | 10 | 20 | 10 | 40 | |
| HER2 | ||||||
| Positive | 1 | 5 | 13 | 23 | 42 | 0.211 |
| Negative | 2 | 17 | 38 | 32 | 89 | |
| Lymph node | ||||||
| metastasis | ||||||
| TnN0 | 4 | 4 | 23 | 37 | 86 | 0.013a |
| TnNn | 0 | 18 | 21 | 16 | 46 | |
| P53 | ||||||
| Positive | 1 | 15 | 33 | 37 | 86 | 0.495 |
| Negative | 2 | 7 | 21 | 16 | 46 | |
| Ki-67 | ||||||
| − | 1 | 6 | 11 | 6 | 24 | 0.320 |
| + | 2 | 11 | 32 | 30 | 75 | |
| ++ | 0 | 4 | 7 | 11 | 22 | |
| +++ | 0 | 1 | 3 | 6 | 10 | |
| Ck56 | ||||||
| Positive | 0 | 2 | 10 | 7 | 19 | 0.757 |
| Negative | 3 | 20 | 43 | 46 | 112 | |
| EGFR | ||||||
| Positive | 4 | 19 | 43 | 36 | 102 | 0.023a |
| Negative | 0 | 3 | 13 | 21 | 37 | |
| TN | ||||||
| TNBC | 0 | 6 | 5 | 7 | 18 | 0.046 |
| NTNBC | 6 | 23 | 43 | 31 | 103 | |
P<0.05
P<0.001. KIF3A, Kinesin Family Member 3A; ER, estrogen receptor; PR, progesterone receptor; AR, androgen receptor; HER2, human epidermal growth factor receptor 2; EGFR, epidermal growth factor receptor; TN, triple-negative; TNBC, triple-negative breast cancer; NTNBC, non-triple-negative breast cancer.
Table VII.
Relationship between KIF3A expression and clinical pathological parameters in the140 cases by final score.
| Final score | ||||||
|---|---|---|---|---|---|---|
| Item | 0 | 1 | 2 | 3 | No. | P-value |
| Age | 0.465 | |||||
| ≤53 | 6 | 18 | 26 | 30 | 80 | |
| >53 | 2 | 14 | 26 | 18 | 60 | |
| Pathology grade | 0.001a | |||||
| G I | 3 | 6 | 3 | 0 | 12 | |
| G I–II | 0 | 4 | 13 | 5 | 22 | |
| G II | 4 | 21 | 32 | 38 | 95 | |
| G III | 0 | 1 | 3 | 3 | 7 | |
| ER | 0.008a | |||||
| Positive | 5 | 15 | 39 | 24 | 83 | |
| Negative | 2 | 15 | 9 | 21 | 47 | |
| PR | 0.135 | |||||
| Positive | 3 | 10 | 29 | 20 | 62 | |
| Negative | 2 | 20 | 20 | 25 | 67 | |
| AR | 0.309 | |||||
| Positive | 7 | 19 | 37 | 37 | 100 | |
| Negative | 1 | 13 | 15 | 11 | 40 | |
| HER2 | 0.344 | |||||
| Positive | 2 | 7 | 14 | 19 | 42 | |
| Negative | 5 | 23 | 35 | 26 | 89 | |
| Lymph node metastases | 0.218a | |||||
| TnN0 | 5 | 8 | 21 | 20 | 54 | |
| TnNn | 3 | 23 | 31 | 27 | 84 | |
| P53 | 0.125 | |||||
| Positive | 2 | 22 | 29 | 33 | 86 | |
| Negative | 5 | 9 | 19 | 13 | 46 | |
| Ki-67 | 0.023a | |||||
| − | 2 | 6 | 10 | 6 | 24 | |
| + | 4 | 19 | 29 | 23 | 75 | |
| ++ | 1 | 3 | 8 | 10 | 22 | |
| +++ | 0 | 2 | 1 | 7 | 10 | |
| Ck56 | 0.872 | |||||
| Positive | 0 | 4 | 8 | 7 | 19 | |
| Negative | 7 | 26 | 40 | 39 | 112 | |
| EGFR | 0.054 | |||||
| Positive | 6 | 28 | 38 | 30 | 102 | |
| Negative | 2 | 4 | 14 | 17 | 37 | |
| TN | 0.873 | |||||
| TNBC | 0 | 3 | 8 | 7 | 18 | |
| NTNBC | 3 | 17 | 43 | 40 | 103 | |
P<0.05
P<0.001. KIF3A, Kinesin Family Member 3A; ER, estrogen receptor; PR, progesterone receptor; AR, androgen receptor; HER2, human epidermal growth factor receptor 2; EGFR, epidermal growth factor receptor; TN, triple-negative; TNBC, triple-negative breast cancer; NTNBC, non-triple-negative breast cancer.
In terms of the intensity of positive in the 140 breast cancer patients, the statistical analysis revealed that the higher level expression of KIF3A was correlated with the status of lymph node metastasis, pathological grade, and the expression of androgen receptor (AR) and epidermal growth factor receptor (EGFR). In addition, the Chi-square test on both the positive rate and final score indicated that a higher level of expression of KIF3A was correlated with higher pathology grade and the expression of estrogen receptor (ER) and Ki-67.
Expression of KIF3A and prognosis in breast cancer patients
Kaplan-Meier analysis of the intensity of positive, the positive rate and the final score was applied to explore the association between KIF3A expression and the survival rate of 140 patients followed up to the year of 2013 and 2014.
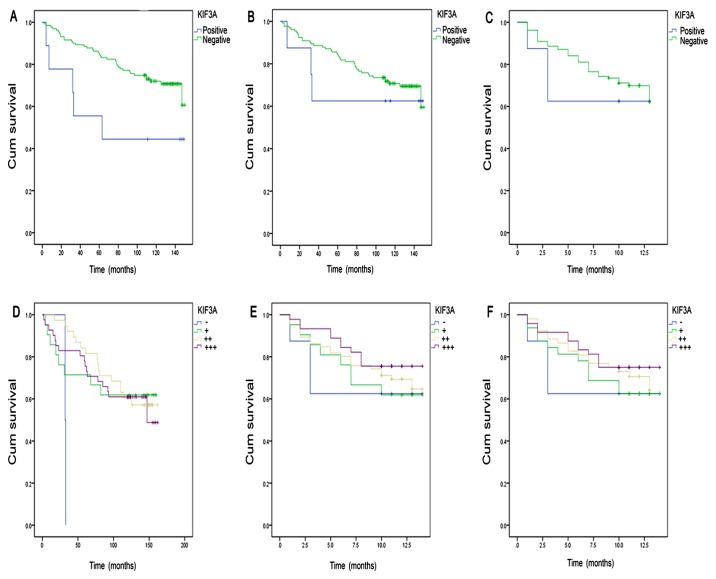
For the cases followed up to 2013, grouped as KIF3A positive (+, ++, +++) and negative (−), a statistical significance was only identified between KIF3A expression and survival rate (P=0.047) when evaluated by intensity of positive (Fig. 2 and Table VIII).
Figure 2.
Kaplan-Meier survival curves of KIF3A expression and survival rate of 140 breast cancer cases in 2013. (A) Kaplan-Meier survival analysis basing on KIF3A intensity of positive (P=0.047). (B) Kaplan-Meier survival analysis basing on KIF3A-positive rate (P=0.068). (C) Kaplan-Meier survival analysis basing on KIF3A final score (P=0.635). (D) Kaplan-Meier survival of the intensity of positive (P=0.045). (E) Kaplan-Meier survival of the positive rate (P=0.217). (F) Kaplan-Meier survival of the final score (P=0.223). KIF3A, Kinesin Family Member 3A.
Table VIII.
Correlation of KIF3A expression and the survival rate of 140 breast cancer cases followed up to 2013.
| Item | No. | P-value |
|---|---|---|
| Intensity of positive | 0.047a | |
| Positive | 136 | |
| Negative | 4 | |
| Positive rate | 0.628 | |
| Positive | 132 | |
| Negative | 8 | |
| Final score | 0.635 | |
| Positive | 132 | |
| Negative | 8 |
P<0.05. KIF3A, Kinesin Family Member 3A.
For the cases up to 2014, positive (++) patients showed an improved prognosis (P=0.045; Fig.2 and Table IX).
Table IX.
Correlation of KIF3A expression and the survival rate of 140 breast cancer cases followed up to 2014.
| Item | No. | P-value |
|---|---|---|
| Intensity of positive | 0.045a | |
| 0 | 4 | |
| 1 | 22 | |
| 2 | 56 | |
| 3 | 58 | |
| Positive rate | 0.217 | |
| 0 | 8 | |
| 1 | 21 | |
| 2 | 66 | |
| 3 | 45 | |
| Final score | 0.223 | |
| 0 | 8 | |
| 1 | 32 | |
| 2 | 52 | |
| 3 | 48 |
P<0.05. KIF3A, Kinesin Family Member 3A.
Discussion
KIF3 is a heterotrimeric complex that consists of KIF3A, KIF3B, and kinesis-associated protein 3 (KAP3) (11), the complex is considered as microtubule (MT)-dependent molecular motors that function in intracellular transport (12), which is expressed ubiquitously. KIF3A is involved in the anterograde transport of membranous organelles, distinct from synaptic vesicle precursors and from vesicles, also required for ciliary basal feet formation and MT anchoring to mother centriole (3). Thus, KIF3A plays an important role in the procedure of ciliogenesis.
KIF3A was also reported to be associated with certain pathological processes. It has been reported that KIF3A is involved in forming defective bone formation and osteopenia (13) and defective osteoblastic differentiation in dental mesenchymal stem/precursor cells (14). Since 2013, the expression and function of KIF3A has been regarded as statistically significantly and correlated with several tumors such as glioblastoma, prostate cancer and medulloblastoma (4,5,10). It has been demonstrated that disruption of the expression of KIF3A leads to ablate ciliogenesis and tumorigenesis in glioblastoma (10).
Since KIF3A is associated with ciliogenesis and primary cilia decrease in breast cancer (7–9), identifying the association of KIF3A and breast cancer progression is of great importance. Our results showed that the expression of KIF3A is extremely higher in breast cancer tissues than that in para-carcinoma tissues, and this difference was confirmed by the 70 self-contrast tissues. Barakat et al have reported that the difference of KIF3A expression has the same relationship with primary cilia (4). Based on our results, KIF3A is associated with progression of breast cancer.
No reports have previously focused on the relationship between KIF3A expression and breast cancer progression. Thus, we statistically analyzed the clinical mainstream clinicopathological parameters. Our data suggested that the high expression of KIF3A was correlated with clinical diagnosis and prognosis including: lymph node metastasis, pathological grade, AR, ER, EGFR and Ki-67. In clinic, these parameters are not sufficient for the accurate diagnosis and prognosis of breast cancer, particularly triple negative breast cancer. Thus, it is imperative to add new parameters for breast cancer, and according to findings of the present study, we suggest that KIF3A be a new candidate parameter of breast cancer.
The expression of KIF3A was associated with survival in breast cancer patients up to 2013. Furthermore, Kaplan-Meier survival curves showed positive (++) KIF3A combined with longer survival according to the data of 2014. Based on the statistical analysis on the relationship with existing parameters and KIF3A, the high expression of KIF3A is associated with ER, AR, EGFR and Ki-67. Concerning the associated parameters, ER, AR and Ki-67 are regarded as a reference index to evaluate prognosis status. Of these, ER and AR are selected from hormonic effect and Ki-67 is based on cell cycle, while KIF3A is associated with the pathological change of primary cilia. Therefore, we suggest KIF3A can be used as a new parameter to evaluate prognosis in a novel way.
In conclusion, the high expression of KIF3A is associated with the progression of breast cancer. Furthermore, its high expression is also associated with breast cancer prognosis parameters ER, AR, EGFR and Ki-67. These results indicate that KIF3A can be used as a diagnostic indicator, and also as a new prognosis parameter to evaluate breast cancer considering its particular function on the pathological change of primary cilia.
Table VI.
Relationship between KIF3A expression and clinicopathological parameters in the140 cases by positive rate.
| Positive rate | ||||||
|---|---|---|---|---|---|---|
| Item | 0 | 1 | 2 | 3 | No. | P-value |
| Age | 0.358 | |||||
| ≤53 | 6 | 12 | 33 | 29 | 80 | |
| >53 | 2 | 9 | 33 | 16 | 60 | |
| Pathology grade | 0.000b | |||||
| I | 3 | 4 | 5 | 0 | 12 | |
| I–II | 0 | 2 | 16 | 4 | 22 | |
| II | 4 | 15 | 40 | 36 | 95 | |
| III | 0 | 0 | 4 | 3 | 7 | |
| ER | 0.031a | |||||
| Positive | 5 | 9 | 47 | 27 | 83 | |
| Negative | 2 | 10 | 15 | 20 | 47 | |
| PR | 0.184 | |||||
| Positive | 3 | 5 | 34 | 20 | 62 | |
| Negative | 2 | 14 | 29 | 22 | 67 | |
| AR | 0.842 | |||||
| Positive | 7 | 12 | 48 | 33 | 100 | |
| Negative | 1 | 9 | 18 | 12 | 40 | |
| HER2 | 0.579 | |||||
| Positive | 2 | 5 | 18 | 17 | 42 | |
| Negative | 5 | 14 | 45 | 25 | 89 | |
| Lymph node metastasis | 0.135 | |||||
| TnN0 | 5 | 5 | 23 | 21 | 54 | |
| TnNn | 3 | 16 | 42 | 23 | 84 | |
| P53 | 0.184 | |||||
| Positive | 2 | 13 | 40 | 31 | 86 | |
| Negative | 5 | 7 | 22 | 12 | 46 | |
| Ki-67 | 0.009a | |||||
| − | 2 | 5 | 12 | 5 | 24 | |
| + | 4 | 11 | 39 | 21 | 75 | |
| ++ | 1 | 2 | 8 | 11 | 22 | |
| +++ | 0 | 1 | 3 | 6 | 10 | |
| Ck56 | 0.873 | |||||
| Positive | 0 | 3 | 9 | 7 | 19 | |
| Negative | 7 | 16 | 53 | 36 | 112 | |
| EGFR | 0.109 | |||||
| Positive | 6 | 19 | 48 | 29 | 102 | |
| Negative | 2 | 2 | 18 | 15 | 37 | |
| TN | 0.657 | |||||
| TNBC | 0 | 4 | 8 | 6 | 18 | |
| NTNBC | 6 | 14 | 52 | 31 | 103 | |
P<0.05
P<0.001. KIF3A, Kinesin Family Member 3A; ER, estrogen receptor; PR, progesterone receptor; AR, androgen receptor; HER2, human epidermal growth factor receptor 2; EGFR, epidermal growth factor receptor; TN, triple-negative; TNBC, triple-negative breast cancer; NTNBC, non-triple-negative breast cancer.
Acknowledgements
We gratefully thank Professor Tao Yi for providing technical support.
Funding
This study was supported by the National Major Scientific and Technological Special Project for ‘Significant New Drugs Development’ (2013zx09301304001).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
PX and SC carried out most of the experimental work, performed the immunohistochemical staining of tissue chip. SC analyzed the data. PX wrote the paper. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was based on the samples of tissue microarrays brought from Xinchao, Shanghai.
Consent for publication
All authors have agreed to submit the manuscript.
Competing interests
All authors declares that they have no conflict of interest.
References
- 1.Kolpakova-Hart E, Jinnin M, Hou B, Fukai N, Olsen BR. Kinesin-2 controls development and patterning of the vertebrate skeleton by Hedgehog- and Gli3-dependent mechanisms. Dev Biol. 2007;309:273–284. doi: 10.1016/j.ydbio.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 3.Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279:519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- 4.Barakat MT, Humke EW, Scott MP. Kif3a is necessary for initiation and maintenance of medulloblastoma. Carcinogenesis. 2013;34:1382–1392. doi: 10.1093/carcin/bgt041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Z, Rebowe RE, Wang Z, Li Y, Wang Z, DePaolo JS, Guo J, Qian C, Liu W. KIF3a promotes proliferation and invasion via Wnt signaling in advanced prostate cancer. Mol Cancer Res. 2014;12:491–503. doi: 10.1158/1541-7786.MCR-13-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim M, Suh YA, Oh JH, Lee BR, Kim J, Jang SJ. KIF3A binds to β-arrestin for suppressing Wnt/β-catenin signalling independently of primary cilia in lung cancer. Sci Rep. 2016;6:32770. doi: 10.1038/srep32770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDermott KM, Liu BY, Tlsty TD, Pazour GJ. Primary cilia regulate branching morphogenesis during mammary gland development. Curr Biol. 2010;20:731–737. doi: 10.1016/j.cub.2010.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Menzl I, Lebeau L, Pandey R, Hassounah NB, Li FW, Nagle R, Weihs K, McDermott KM. Loss of primary cilia occurs early in breast cancer development. Cilia. 2014;3:7. doi: 10.1186/2046-2530-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan K, Frolova N, Xie Y, Wang D, Cook L, Kwon YJ, Steg AD, Serra R, Frost AR. Primary cilia are decreased in breast cancer: Analysis of a collection of human breast cancer cell lines and tissues. J Histochem Cytochem. 2010;58:857–870. doi: 10.1369/jhc.2010.955856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoang-Minh LB, Deleyrolle LP, Siebzehnrubl D, Ugartemendia G, Futch H, Griffith B, Breunig JJ, De Leon G, Mitchell DA, Semple-Rowland S, et al. Disruption of KIF3A in patient-derived glioblastoma cells: Effects on ciliogenesis, hedgehog sensitivity, and tumorigenesis. Oncotarget. 2016;7:7029–7043. doi: 10.18632/oncotarget.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo X, Macleod GT, Wellington A, Hu F, Panchumarthi S, Schoenfield M, Marin L, Charlton MP, Atwood HL, Zinsmaier KE. The GTPase dMiro is required for axonal transport of mitochondria to Drosophila synapses. Neuron. 2005;47:379–393. doi: 10.1016/j.neuron.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 12.Ichinose S, Ogawa T, Hirokawa N. Mechanism of Activity-Dependent Cargo Loading via the Phosphorylation of KIF3A by PKA and CaMKIIa. Neuron. 2015;87:1022–1035. doi: 10.1016/j.neuron.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 13.Qiu N, Xiao Z, Cao L, Buechel MM, David V, Roan E, Quarles LD. Disruption of Kif3a in osteoblasts results in defective bone formation and osteopenia. J Cell Sci. 2012;125:1945–1957. doi: 10.1242/jcs.095893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang S, Chen G, Feng L, Jiang Z, Yu M, Bao J, Tian W. Disruption of kif3a results in defective osteoblastic differentiation in dental mesenchymal stem/precursor cells via the Wnt signaling pathway. Mol Med Rep. 2016;14:1891–1900. doi: 10.3892/mmr.2016.5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.