Brain metastases originate from cancer cells that have spread through the bloodstream and reached the brain from primary tumours in other organs. Metastatic masses are among the most common intracranial neoplasms, occurring in ∼25% of cancer patients. Most brain metastases derive from lung cancers (40–50% of cases), breast cancers (10–16%), melanoma (5–20%), kidney and ovarian cancers (5–10%) and more rarely colorectal cancers. Sometimes (5–10%) no primary site is detectable [1]. The onset of brain lesions from primary solid cancers is associated with poor prognosis with a median survival of 4–5 months [2]. Molecular mechanisms regulating metastatic spreading to the brain are largely unknown. RON (Recepteur d'Origine Nantais), also known as macrophage-stimulating receptor-1 (MSTR1) or stem cell derived tyrosine kinase (STK) in mice, belongs to the family of tyrosine kinase receptors of which MET is the prototype. It has been shown that RON regulates cellular proliferation, adhesion, motility and protection from apoptosis, all events resulting in the invasive growth genetic programme [3], which occurs in specific physiological conditions (i.e. embryonic development) and, when aberrantly regulated, contributes to tumour onset, progression and, above all, metastatic dissemination.
Short abstract
RON mutations might identify actionable targets in highly aggressive lung tumours http://ow.ly/RTUp30hSBX6
To the Editor:
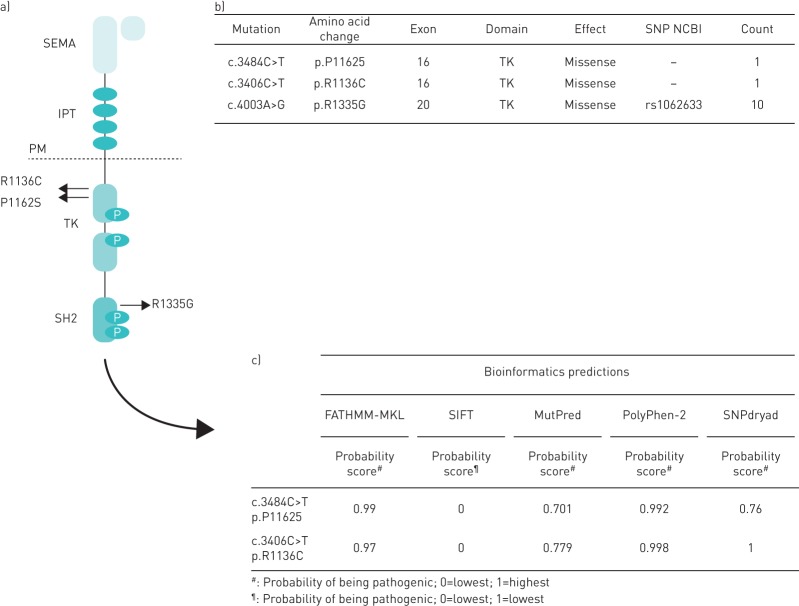
Brain metastases originate from cancer cells that have spread through the bloodstream and reached the brain from primary tumours in other organs. Metastatic masses are among the most common intracranial neoplasms, occurring in ∼25% of cancer patients. Most brain metastases derive from lung cancers (40–50% of cases), breast cancers (10–16%), melanoma (5–20%), kidney and ovarian cancers (5–10%) and more rarely colorectal cancers. Sometimes (5–10%) no primary site is detectable [1]. The onset of brain lesions from primary solid cancers is associated with poor prognosis with a median survival of 4–5 months [2]. Molecular mechanisms regulating metastatic spreading to the brain are largely unknown. RON (Recepteur d'Origine Nantais), also known as macrophage-stimulating receptor-1 (MSTR1) or stem cell derived tyrosine kinase (STK) in mice, belongs to the family of tyrosine kinase receptors of which MET is the prototype. It has been shown that RON regulates cellular proliferation, adhesion, motility and protection from apoptosis, all events resulting in the invasive growth genetic programme [3], which occurs in specific physiological conditions (i.e. embryonic development) and, when aberrantly regulated, contributes to tumour onset, progression and, above all, metastatic dissemination. RON aberrant activation in cancer cells is mainly due to receptor overexpression, achieved via gene amplification, enhanced transcription or post-transcriptional modifications, alternative splicing and protein truncation; point mutations and ligand activation loops, although rarer, might also occur. We had previously documented that the occurrence of MET somatic mutations in brain metastatic lesions from lung cancers (nonsmall cell lung cancers) is predictive of a lack of response to radiation therapy to the brain and significantly increased mortality [4]. Moreover, RON ligand, MSP (macrophage stimulating protein) is known to act as neurotrophic factor by modulating brain plasticity and regeneration [5]. The finding that RON is implicated in a number of human cancers categorises RON as a clinically relevant therapeutic target and led to the development of agents able to inhibit its function and activity. Various strategies are being investigated, the most promising being small molecular kinase inhibitors and neutralising antibodies against the receptors able to block the downstream signalling cascade. Yet strategies to target RON are further back than those for MET and drugs designed to specifically target RON are in early stages of development [6]. On this basis, this study aimed to evaluate the RON genetic profile in a cohort of surgically resected brain metastases from different solid cancers to evaluate mutational frequency. A total of 57 formalin-fixed paraffin-embedded samples of brain metastases were retrieved from the archives of the Pathology Division of the University of Turin at the Città della Salute e della Scienza, Molinette Hospital. The study received ethical approval from the local institutional review board (the Local Ethics Committee of Città della Salute e della Scienza, Molinette Hospital, Turin, Italy). Of the 57 cases, 31 were female (54.4%) and 26 (45.6%) were male; the mean age at diagnosis was 60.4±12.19 years. The primary site of origin of the metastastic samples analysed was distributed, coherently with the epidemiological data, as follows: 36.8% (21 cases) derived from lung cancers; 33.3% (19 cases) from breast cancer, 14% from melanoma (eight cases), 8.8% (five cases) from colorectal cancers and 7% (four cases) from ovarian tumours. For each sample, the entire RON coding sequence was analysed. Mutational analysis allowed the identification of somatic mutations in two out of the 57 evaluated patients/samples. The lineage of origin of the RON-mutated lesions was the lung in both cases, and interestingly, both mutations clustered in the tyrosine-kinase portion of the receptor (figure 1). The first mutation identified, R1136C, was a novel change; while the second P1162S has already been reported in one melanoma sample [7], but no data are available about its effect on protein functions. We thus moved to assess the possible pathogenic impact of these substitutions by submitting them to a panel of in silico bioinformatics programmes, able to predict the effect of coding variants on protein function and disease onset. In detail, we surveyed the following five tools: 1) FATHMM-MKL [8], which predicts the functional, molecular and phenotypic consequences of protein missense variants using hidden Markov models; 2) SIFT [9], which predicts whether an amino acid substitution affects protein function based on the degree of conservation of amino acid residues in sequence alignments derived from closely related sequences; 3) MutPred [10], which classifies an amino acid substitution as disease-associated or neutral in humans; 4) PolyPhen-2 [11], which predicts the possible impact of an amino acid substitution on the structure and function of a human protein using physical and comparative considerations; and 5) SNPdryad [12], which predicts the deleterious effect of amino acid substitutions occurred in human proteins. For each algorithm, the results interpretation, based on output scores, allowed us to predict a damaging or deleterious effect of the two identified changes. In addition, ten out of the 57 analysed cases (17.5%) harboured the R1335G polymorphism (NCBI rs1062633) which has been previously reported to predispose to gastro-oesophageal cancer [13]. In the present cohort, the single nucleotide polymorphism was present in metastatic lesions from different lineages of origin: lung (0.3%), breast (5.3%), melanoma (5.3%) and ovary (1.8%). This discovery allows us to hypothesise that R1335G has a role in influencing the spread of many cancer types other than gastro-oesophageal tumours.
FIGURE 1.
a) RON tyrosine kinase receptor structure; highlighted are the two somatic mutations detected in brain secondary lesions from lung cancers (R1136C and P1162S) and R1335G polymorphism (NCBI rs1062633). b) Details on somatic mutations and recurrent single nucleotide polymorphism (SNP). c) Bioinformatics prediction of pathogenic role of the changes found. IPT: immunoglobulin-plexin-transcription; PM: plasma membrane; SEMA: semaphorin; TK: tyrosine kinase.
From the data obtained in the present study, the mutational frequency of RON in brain metastases from solid cancers is 3.5%. This result acquires relevance when compared to the RON mutational frequency in an unselected neoplastic population, which is known to be ∼1% (data obtained from COSMIC database). The RON gene mutational incidence becomes significantly higher (9.5%) among brain lesions derived from the lung. Notably the RON receptor is involved in the development of epithelial tissues, including the lungs [14], and is often overexpressed in lung cancers [15].
To the best of our knowledge this is the first report of RON somatic mutations with deleterious effects in brain lesions from lung cancers. It should be noted that although bioinformatics tools can predict the molecular effect, this might not result in a full pathological phenotype, and in vitro and in vivo studies are required to further support this preliminary in silico evidence. Nevertheless, some hypothesises are allowed regarding the putative mechanisms of RON-mediated brain metastasis. Previous data showed that in breast cancers RON activation promotes metastases dissemination by triggering aberrant DNA methylation and suggested an association with worse outcomes for patients [16]. Here we point out the occurrence of genetic lesions affecting RON associated with lung cancer distant dissemination. Overall these findings converge on the important role of RON in metastatic spread, in both breast and lung cancers. On one hand, in the case of breast cancer the receptor activated by its ligand controls epigenetic events, on the other hand, as reported here in lung cancer, the receptor is activated as a consequence of genetic alterations. The two mechanisms might reflect functional differences and molecular heterogeneity between the primary tumours. In conclusion, the identified RON mutations might have clinical and prognostic implications: 1) as functional markers of highly aggressive lung tumours; and 2) actionable for an approach targeted to blocking the RON receptor in patients suffering from advanced diseases with cerebral metastatic dissemination.
Footnotes
Conflict of interest: None declared.
Funding: AIRC IG, Project no 15572-AIRC Special Program 5xMille 2010 MCO, Project no 9970. Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1.Lagerwaard FJ, Levendag PC, Nowak PJ, et al. . Identification of prognostic factors in patients with brain metastastes: a review of 1292 patients. Int J Radiat Oncol Biol Phys 1999; 43: 795–803. [DOI] [PubMed] [Google Scholar]
- 2.Wong E, Rowbottom L, Taso M, et al. . Correlating symptoms and their changes with survival in patients with brain metastases. Ann Palliat Med 2016; 5: 253–266. [DOI] [PubMed] [Google Scholar]
- 3.Benvenuti S, Comoglio PM. The MET receptor tyrosine kinase in invasion and metastasis. J Cell Physiol 2007; 213: 316–325. [DOI] [PubMed] [Google Scholar]
- 4.Stella GM, Senetta R, Inghilleri S, et al. . MET mutations are associated with aggressive and radioresistant brain metastatic non-small-cell lung cancer. Neuro Oncol 2016; 18: 598–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stella MC, Vercelli A, Repici M, et al. . Macrophage stimulating protein is a novel neurotrophic factor. Mol Biol Cell 2001; 12: 1341–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zarei O, Benvenuti S, Ustun-Alkan F, et al. . Strategies of targeting the extracellular domain of RON tyrosine kinase receptor for cancer therapy and drug delivery. J Cancer Res Clin Oncol 2016; 142: 2429–2446. [DOI] [PubMed] [Google Scholar]
- 7.Krauthammer M, Kong Y, Bacchiocchi A, et al. . Exome sequencing identifies recurrent mutations in NF1 and RASopathy genes in sun-exposed melanoma. Nat Genet 2015; 47: 996–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shihab HA, Rogers MF, Gough J, et al. . An integrative approach to predicting the functional consequences of non-coding and coding sequence variation. Bioinformatics 2015; 31: 1536–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu Rev Genomics Hum Genet 2006; 7: 61–80. [DOI] [PubMed] [Google Scholar]
- 10.Li B, Krishnan VG, Mort ME, et al. . Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics 2009; 25: 2744–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adzhubei IA, Schmidt S, Peshkin L, et al. . A method and server for predicting damaging missense mutations. Nat Methods 2010; 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong KC, Zhang Z. SNPdryad: predicting deleterious non-synonymous human SNPs using only orthologous protein sequences. Bioioformatics 2014; 30: 1112–1119. [DOI] [PubMed] [Google Scholar]
- 13.Catenacci DV, Cervantes G, Yala S, et al. . RON (MST1R) is a novel prognostic marker and therapeutic target for gastroesophageal adenocarcinoma. Cancer Biol Ther 2011; 12: 9–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaudino G, Avantaggiato V, Follenzi A, et al. . The proto-oncogene RON is involved in development of epithelial, bone and neuro-endocrine tissues. Oncogene 1995; 11: 2627–2637. [PubMed] [Google Scholar]
- 15.Kanteti R, Krishnaswamy S, Catenacci D, et al. . Differential expression of RON in small and non-small cell lung cancers. Genes Chromosomes Cancer 2012; 51: 841–851. [DOI] [PubMed] [Google Scholar]
- 16.Cunha S, Lin YC, Goossen EA, et al. . The Ron receptor tyrosine kinase promotes metastasis by triggering MBD4-dependent DNA methylation reprogramming. Cell Reports 2014; 6: 141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]