Summary
Natural killer (NK) cells provide a major defence against human cytomegalovirus (HCMV) infection through the interaction of their surface receptors, including the activating and inhibitory killer immunoglobulin‐like receptors (KIRs), and human leucocyte antigen (HLA) class I molecules. Also γ marker (GM) allotypes, able to influence the NK antibody‐dependent cell‐mediated cytotoxicity, appear to be involved in the immunological control of virus infections, including HCMV. In some cases, their contribution requires epistatic interaction with other genes of the immune system, such as HLA. In the present report, with the aim of gaining insight into the immune mechanisms controlling HCMV, we have studied the possible associations among humoral and NK responses, and HCMV infections. In a previous study we assessed whether the KIR and HLA repertoire might influence the risk of developing symptomatic (n = 60) or asymptomatic (n = 60) disease after primary HCMV infection in the immunocompetent host. In the present study, the immunocompetent patients with primary symptomatic HCMV infection were genotyped for GM3/17 and GM23 allotypes, along with the 60 participants with a previous asymptomatic infection as controls. Notwithstanding the presence of missing data record, advanced missing data recovery techniques were able to show that individuals carrying the GM23 allotypes, both homozygous and heterozygous, GM17/17, HLA‐C2 and Bw4T KIR‐ligand groups are associated with the risk of developing symptomatic infection. Our findings on the role of both cellular and humoral immunity in the control of HCMV infection should be of value in guiding efforts to reduce HCMV‐associated health complications in the elderly, including immunosenescence, and in transplantation.
Keywords: γ marker, antibodies, human cytomegalovirus, human leucocyte antigen, killer immunoglobulin‐like receptor, natural killer
Abbreviations
- ADCC
antibody‐dependent cell‐mediated cytotoxicity
- FcγR
Fcγ receptors
- GM
γ marker
- HCMV
human cytomegalovirus
- HCV
hepatitis C virus
- HLA
human leucocyte antigens
- HSV‐1
herpes simplex virus type 1
- IFN
interferon
- Ig
immunoglobulin
- KIR
killer immunoglobulin‐like receptors
- MAR
missing at random
- MCAR
missing completely at random
- MICE
multivariate imputation by chained equations
- NK
natural killer
- OR
odds ratio
Introduction
Human cytomegalovirus (HCMV), a member of the Herpesviridae family, is a double‐stranded DNA virus that infects a high percentage of humans worldwide. As with the other herpesviruses, after recovery from acute HCMV infection the virus establishes a latent infection. Several cells are able to harbour HCMV in a slowly replicating or not replicating form. However, the exact mechanisms controlling latency are unclear. Immunosuppression, illness, or the use of chemotherapeutic agents can activate the virus from this latent state. Both innate and acquired immunity participate in protection against infection or re‐infection and resolution of established infection.1, 2
Human CMV is regarded as being the major cause of morbidity and mortality after transplantation as well as of lower graft survival. Translational research of HCMV infection performed in clinical conditions such as transplantation, cancer, immunodeficiency, and autoimmune and inflammatory diseases, strengthens the suggestion that HCMV can affect their evolution and prognosis through a process of “early” immunosenescence. HCMV infection has been associated with a variety of health problems and overall mortality in the elderly. Accordingly, recent data show that effective control of HCMV is impaired during healthy ageing, most probably due to loss of cellular control of early viral reactivation.3, 4 So, there is an urgent need for improved understanding of the virus–host balance during ageing and transplantation.
The cellular immune response is necessary to control latency and impede viral replication in latently infected individuals, and natural killer (NK) cells are essential in the control of HCMV, as demonstrated by lethal infections in adolescents lacking these cells. In fact, the most common clinical manifestation of NK cell dysfunction is recurrent and often severe herpes virus infections. The cellular immune response is necessary to control latency and impede viral replication in latently infected individuals, and NK cells are essential in the control of HCMV as demonstrated by lethal infections in adolescents lacking these cells. In fact, the most common clinical manifestation of NK cell dysfunction is recurrent, and often severe, herpesvirus infections.
Recently, the first experimental evidence has been provided that NK cells can efficiently control HCMV transmission in vitro both through soluble factors such as interferon‐γ (IFN‐γ) and by cell contact.5 The key role played by NK cells in the control of HCMV is further demonstrated by the existence in the HCMV genome of five viral genes capable of suppressing NK cell recognition of activating receptors.6 Furthermore, an HCMV‐infected young patient has been reported with severe combined immunodeficiency (T− B+ NK−) who recovered from HCMV disease without antiviral therapy after a significant expansion of IFN‐γ‐producing CD16+ CD94− NKG2C+ NK cells had occurred.7
Natural killer cells kill also by recognizing those coated with IgG via antibody‐dependent cell‐mediated cytotoxicity (ADCC). However, the exact role of HCMV antibodies in the control of infection in vivo is still unclear. In addition, although antibody plays an important role in protection against HCMV infection and disease, the level of protection is clearly incomplete.2, 8, 9, 10, 11
Several IgG allotypes have been identified in humans, determined by polymorphisms in the γ H chain and in the κ L chain.12 These allotypes have been shown to be involved in the immunological control of virus infections, such as hepatitis C virus (HCV)13 or herpes simplex virus type 1 (HSV‐1).14 Several mechanisms have been hypothesized to explain the role of γ marker (GM) allotypes in the control of virus infections, including their ability in modulating the strength of ADCC, so involving cells of the innate response such as NK cells. In addition, IgG allotypes may modulate the avidity of the FcγR–IgG interaction and could influence the efficacy of immune responses.
In a previous study we assessed whether the killer immunoglobulin‐like receptors (KIR) and HLA repertoire might influence the risk of developing symptomatic or asymptomatic disease after primary HCMV infection in the immunocompetent host. The frequency of the homozygous A haplotype (only KIR2DS4 as activating KIR) was significantly higher in symptomatic patients than in controls. By logistic regression, the risk of developing symptomatic disease was associated with the homozygous A haplotype and the HLABw4T allele. Combining the two independent variables, we found that 62% of symptomatic patients, but only 30% of controls, possessed the homozygous A haplotype or the HLABw4T allele with a highly significant odds ratio (OR) (3·75).15
In the present report, with the aim of gaining further insight into the immune mechanisms controlling HCMV, we have studied the possible interactive effects of humoral and NK responses to HCMV. For this purpose, we have analysed the distribution of GM allotypes, known to influence antibody responses to HCMV,16, 17, 18 in the cohorts of patients and controls described above.
Materials and methods
The characteristics of patients and controls have been described previously.15 Between November 2011 and May 2013, a total of 60 consecutive Caucasian patients with symptomatic acute HCMV infection were recruited from two Sicilian hospitals (University of Palermo Medical Centre and Cervello Hospital, Palermo, Italy). Only immunocompetent adults, with positive serological tests for acute HCMV infection (HCMV IgM‐positive/HCMV IgG‐negative test), with at least one of the following symptoms: fever, lymphadenopathy, splenomegaly, or clinical manifestations of hepatitis, encephalitis, or pneumonia were included. Patients with positive test for surface antigen of hepatitis B virus, anti‐human immunodeficiency virus, or anti‐HCV antibodies, or treated with immunosuppressant drugs were excluded. As a control, 60 Caucasian blood donors, matched to the study population by age (± 2 years) and sex, with a previous asymptomatic HCMV infection (HCMV IgM‐negative/HCMV IgG‐positive test, without any reported manifestations of acute HCMV infection as assessed by interview) were retrospectively enrolled. The population under study was homogeneous as both patients and controls were born in west Sicily from parents born in west Sicily from western Sicilian grandparents.
Of the 120 participants typed for KIR/HLA in the original study, only 98 patients (47 controls and 51 cases) were typed for GM17/3 and GM23 genotypes in the present study, owing to loss of the original DNA of 22 individuals – GM17/3 was not recorded for 17 patients and GM23 was missing for 21 patients (six patients missed both GM17/3 and GM23 records).
Given the pilot nature of this study and the long sample‐processing time interval, missing information was recovered as much as possible by using advanced missing data analysis techniques, as further detailed below.
DNA was obtained from peripheral blood leucocytes by the salting‐out technique.19 Using the PCR sequence‐specific primer technique, the DNA of cases and controls were genotyped for the presence of the three major KIR ligand groups, HLA‐C1, Cw alleles with asparagine at position 80 HLA‐C2, Cw alleles with lysine at position 80 and HLA‐Bw4, Bw4‐I, Bw alleles with isoleucine at position 80, Bw4‐T, Bw alleles with threonine at position 80. (Epitop‐TYPE kit; BAG Health Care GmbH, Lich, Germany). KIR genotyping was performed for both inhibitory and activating KIR using the KIR‐TYPE kit (BAG Health Care GmbH). KIR gene profiles were determined by the presence or absence of each KIR gene.15 For the determination of the IgG1 markers GM3 and GM17 (a G‐to‐A substitution determining an arginine‐to‐lysine change at residue 214 of the CH1 region), a TaqMan genotyping assay from Applied Biosystems (Foster City, CA) was used. In brief, the assay includes a PCR with primers 5′‐CCCAGACCTACATCTGCAACGTGA‐3′ (forward) and 5′‐CTGCCCTGGACTGGGACTGCAT‐3′ (reverse), which specifically amplify a 161‐bp fragment of the IGHG1 gene, as well as probes that discriminate the single nucleotide polymorphism VIC‐CTCTCACCAACTTTCTTGT‐NFQ (GM17‐specific) and FAM‐CTCTCACCAACTCTCTTGT‐NFQ (GM3‐specific).13 IgG2 markers GM23− and GM23+ (valine to methionine), were genotyped by a TaqMan® genotyping assay from Applied Biosystems Inc., employing the following primers and probes: forward primer: 5′‐CCCGAGGTCCAGTTCAACT‐3′; reverse primer: 5′‐CGTGGCTTTGTCTTGGCATTATG‐3′; reporter 1 (GM23– specific): VIC‐CACCTCCACGCCGTC‐NFQ; reporter 2 (GM23+ specific): FAM–CACCTCCATGCCGTC–NFQ.14 IgG3 markers GM5 and GM21 were not typed, because TaqMan genotyping assays for the IgG3 allotypes are not yet available. Because of almost absolute linkage disequilibrium at GM loci within an ethnic group, individuals positive for the IgG1 allotypes GM3 and GM17 are most likely positive for the IgG3 allotypes GM5 and GM21 (Pandey, unpublished observations).
For statistical analysis, first, we eliminated all patients with one or more missing records on the KIR/HLA system. Statistical analyses performed on this data set, obtained after such list‐wise deletion of patients, will be referred to as complete case analyses. Crude complete case comparisons of gene frequencies were made using 2 × 2 contingency tables, analysed by the chi‐squared test or Fisher's exact test when mandatory. A complete case logistic regression model was also considered to estimate adjusted ORs. However, complete case analyses based on list‐wise deletion are wasteful and are often affected by serious biases.20 Results provided by Vach and Blettner21, 22 justify complete case logistic regression under the hypothesis that the missing data are confined to either a dichotomous outcome or to one or more predictor variables (but not to both), and some additional assumptions that are often difficult to test in practice. Moreover, our analysis includes some numerical variables (such as ‘age’), and so it is unclear whether the above‐mentioned results apply here.
For these reasons, we performed a confirmatory analysis using multiple imputations,20 imputing missing values m times to obtain m copies of a complete data set (not to be confused with complete case analysis), and then averaging logistic regression parameter estimates and P‐values for all m complete data sets,20 in order to have a single point estimate with associated uncertainty properly reflecting the presence of missing data. We used the chained equation imputation algorithm MICE described in van Buuren and Groothuis‐Oudshoorn.23 Under this general framework, four specific methods of imputation were used:20 (i) polytomous logistic regression; (ii) predictive mean matching; (iii) classification and regression tree; and (iv) random forest. Imputed values, and their ability to reproduce the distribution of the observed data, were compared with each other to assess their consistency, to minimize the risk of bias due to an inconsistent imputation of missing data due to the specific method used. It is also worth nothing that the risk of bias due to multiple imputations also depends on the reasons why the data are missing. Multiple imputations require that data are missing at random or MAR, that is the probability of an observation being missing depends only on observed measurements. However, when patients drop out of a study neither for health nor efficacy reasons (as in our case), missing patients can be considered a random sample from the total study population, and their characteristics are likely to be similar. In this situation, the probability of an observation being missing does not depend on observed or unobserved measurements, and data are missing completely at random (MCAR). However, MCAR data are also MAR,20 and hence such a technical requirement is very likely to be satisfied by our data.
Results
The univariate analysis did not show any significant association between the GM3/17 or GM23 polymorphisms and the risk of developing symptomatic HCMV infection, although a trend toward an effect of the GM23+/+ gene variant on the risk of CMV symptomatic infection was observed (Table 1). Hence, considering the whole original data set including all the KIR genes and their respective HLA ligands, with the addition of GM3/17 and GM23, by univariate analysis only KIR2DS2 and the HLA‐C2 ligand group, as well as the AA KIR haplotype, were significantly associated with the risk of HCMV symptomatic infection (P < 0·05).
Table 1.
Frequency of γ marker 3/17 (GM3/17) and GM23 in individuals with symptomatic and asymptomatic cytomegalovirus infection
| Genotypes | Frequency symptomatic infection n = 54 | Frequency asymptomatic infection n = 51 | Crude‐OR | P‐value |
|---|---|---|---|---|
| GM3/3 | 26 (48) | 23 (45) | ||
| GM3/17 | 21 (39) | 24 (47) | 0·78 1 | > 0·10 (P = 0·68) |
| GM17/17 | 7 (13) | 4 (8) | 1·54 2 | > 0·10 (P = 0·74)F |
| GM23−/− | 9 (17) | 15 (32) | ||
| GM23+/− | 28 (54) | 23 (49) | 2·01 3 | > 0·10 (P = 0·22) |
| GM23+/+ | 15 (29) | 9 (19) | 2·72 4 | > 0·10 (P = 0·15) |
Numbers in parenthesis refer to complete case sample sizes, after deleting patients with missing data. We tested whether frequency counts of GM3/3 and GM3/17 alleles are identically distributed across the two populations of asymptomatic and symptomatic patients.
1GM3/3 versus GM3/17; 2GM3/3 versus GM17/17; 3GM23−/− versus GM23+/−; 4GM23−/− versus GM23+/+; F: Fisher's exact test was used, because of the incorrect chi‐squared asymptotic approximation (the minimum expected count was < 5).
Then, we performed a multiple logistic regression analysis including GM3/17 and GM23 together with all the KIR and HLA gene variables that were both significant (P < 0·05) and not significant (P ≥ 0·05) in the univariate analysis. As mentioned above, owing to the high number of missing data in the GM3/17 and GM23 genes (a total of 22 patients), the complete case analysis included 98 patients (47 controls and 51 cases) without any missing data for the GM3/17 and GM23 genes. By multiple variable logistic regression GM3/17 (17/17), GM23 carriers (+/− and +/+), and KIR2DS5 were the only variables significantly associated with the risk of symptomatic HCMV infection (Table 2). A marginal statistical significance (P < 0·10) was reported for HLA‐C2 (P = 0·08) and HLA‐Bw4T (P = 0·05), whereas a marginal inverse correlation was found between KIR2DS2 and the risk of developing symptomatic infection (P = 0·07). In the analysis reported in the previous study, HLA‐C2 and HLA‐Bw4T were found to be independently associated (P < 0·05) with the outcome of the multiple logistic regression analysis, not taking into account GM3/17 and GM23.15 However, this analysis was performed on a data set of 98 observations and, as noted in the previous section, the exclusion of 22 patients from the analysis has the potential to alter estimates and effect sizes unpredictably.
Table 2.
Logistic regression model to predict the occurrence of symptomatic infection in the complete case analysis (n = 98, after list‐wise deletion of patients with one or more missing records on the KIR/HLA system)
| Variable | Code | Adjusted OR (95% CI) | P‐value |
|---|---|---|---|
| GM3/17 | 0: 3/3 (ref.) | ||
| 1: 3/17 | 1·00 (0·26–3·92) | 0·99 n.s. | |
| 2: 17/17 | 20·2 (1·13–359·8) | 0·04 1 | |
| GM23 | 0: −/− (ref.) | ||
| 1: −/+ | 14·53 (1·93–109·58) | 0·009 1 | |
| 2: +/+ | 19·35 (1·7–219·73) | 0·02 1 | |
| KIR2DS2 | 0: absent | ||
| 1: present | 0·14 (0·02–1·16) | 0·07 2 | |
| KIR2DS5 | 0: absent | ||
| 1: present | 8·89 (1·27–61·99) | 0·03 1 | |
| HLA‐C2 | 0: absent | ||
| 1: present | 3·22 (0·88–11·76) | 0·08 2 | |
| HLA‐Bw4T | 0: Absent | ||
| 1: Present | 4·43 (1–19·62) | 0·05 2 |
We report the following degree of evidence against the null hypothesis: 1statistical significance (P < 0·05); 2weak statistical significance (P < 0·10); n.s., not significant. TThreonine.
To overcome this limitation, four multiple imputation algorithms to replace missing data were used, enabling us to use a complete data set. For each method, we generated m = 100 imputations for each missing variable. The results of the four methods in terms of the frequency distribution of synthetic data are shown in Fig. 1. The imputation method that generates data with variability closer to the original data seems the random forest method (Fig. 1d), whereas a consistent bias is observed when imputations are generated by means of a polytomous logistic regression imputation model (Fig. 1a).
Figure 1.
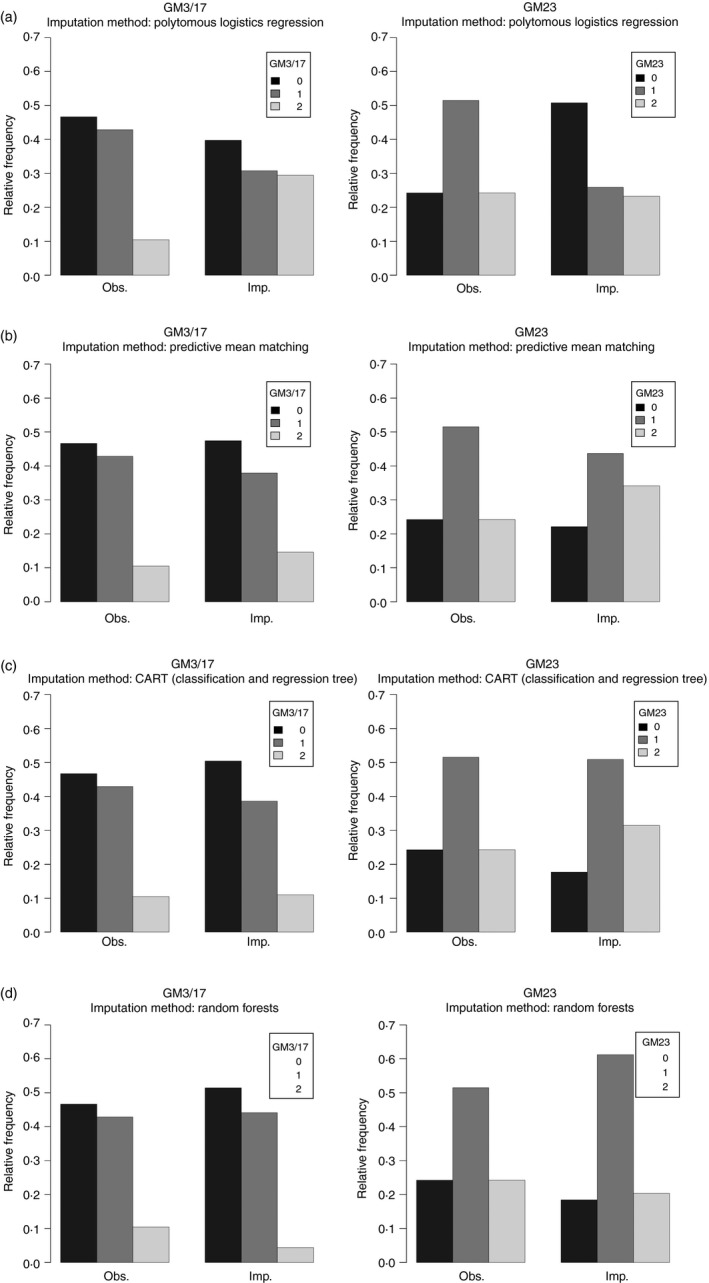
Comparison between observed and imputed relative frequency of γ marker 3/17 (GM3/17) and GM23 for each imputation method: (a) polytomous logistic regression; (b) predictive mean matching; (c) classification and regression tree; (d) random forest.
Table 3 reports all the significant (P < 0·05) or marginally significant (P < 0·10) variables shown by multiple variable logistic regression analyses performed with the original data set (complete case analysis, no imputation, 22 missing data) or the data sets with the imputed data (120 participants) according to the four imputation methods. The statistical significance levels reported by each individual method and the complete case analysis were consistent, confirming the validity of the findings obtained by the complete case analysis.
Table 3.
Comparisons between the statistically significant variables (P < 0·05 or P < 0·10) independently associated with the risk of developing symptomatic cytomegalovirus infection in the multiple variable logistic regressions performed in the complete case database (n = 98) and after imputing missing values according to the four different imputation methods used (n = 120)
| No imputation (P) | Method 1 (P) | Method 2 (P) | Method 3 (P) | Method 4 (P) | Overall statistically significant | |
|---|---|---|---|---|---|---|
| GM3/17 | 0·99 n.s. | 0·8 n.s. | 0·4 n.s. | 0·6 n.s. | 0·7 n.s. | 0/5 |
| GM17/17 | 0·04 1 | 0·01 1 | 0·05 2 | 0·08 2 | 0·08 2 | 4/5 |
| GM23−/+ | 0·009 1 | 0·004 1 | 0·01 1 | 0·04 1 | 0·04 1 | 5/5 |
| GM23+/+ | 0·01 1 | 0·009 1 | 0·01 1 | 0·06 2 | 0·05 2 | 4/5 |
| KIR2DS2 | 0·06 2 | 0·01 1 | 0·01 1 | 0·02 1 | 0·02 1 | 5/5 |
| KIR2DS5 | 0·02 1 | 0·01 1 | 0·02 1 | 0·06 2 | 0·05 2 | 5/5 |
| HLA‐C2 | 0·07 2 | 0·02 1 | 0·02 1 | 0·03 1 | 0·03 1 | 5/5 |
| HLA‐Bw4T | 0·05 2 | 0·03 1 | 0·05 2 | 0·05 2 | 0·07 2 | 5/5 |
| Haplotype AA | 0·47 n.s. | 0·45 n.s. | 0·20 n.s. | 0·16 n.s. | 0·18 n.s. | 0/5 |
We report the following degree of evidence against the null hypothesis: 1statistical significance (P < 0·05); 2weak statistical significance (P < 0·10); n.s., not significant. TThreonine.
The GM23‐carrying genotypes were shown to be significantly or marginally associated with the risk of symptomatic infection by all the analyses, whereas only the GM17/17 genotype showed a significant association. Notably, in the multiple imputation method used, the missing data were imputed 100 times, and the reported ORs are the mean of these imputations. Hence, these methods are very conservative, with a consequent reduction of the statistical significance. Under these conditions, the reports of weak (marginal) significance by some of these methods are considered a valuable result, when others report significant results, in the case of GM23+/+ (statistically significant in three cases, marginal in two) (Table 3).
Furthermore, HLA‐C2 and HLA‐Bw4T were shown to be significant. These gene variants were already shown to be significant by the multivariate analysis, not taking into account GM23 and GM3/17. These results confirm that the imputation methods did not significantly alter the original variability of the results, supporting the reliability of these findings. The only variable no longer significant after the inclusion in the model of GM23 and GM3/17 was the KIR haplotype (A/A), which was significantly associated with the outcome in the original study.15 However, it is worth noting that KIR2DS2 was found to be independently associated with the outcome (negatively), being less frequent in symptomatic patients, supporting the role of activating KIRs in the control of the infection.
The multiple logistic regression analysis reporting the ORs performed with the complete data set obtained using the random forest imputation method is shown in Table 4. This analysis confirms that HLA‐C2 and GM23 in homo‐ and heterozygosity showed the strongest association with the risk of symptomatic infection, whereas KIR2DS2 seems to be protective, being more frequent in the controls.
Table 4.
Logistic regression model to predict the occurrence of symptomatic infection in the database after imputing missing values according to the “random forest” method (n = 120)
| Variable | Code | Adjusted OR (95% CI) | P‐value |
|---|---|---|---|
| GM3/17 | 0: 3/3 (ref.) | ||
| 1: 3/17 | 1·28 (0·34–4·74) | 0·71 n.s. | |
| 2: 17/17 | 11·88 (0·70–202·3) | 0·08 2 | |
| GM23 | 0: −/− (ref.) | ||
| 1: −/+ | 8·32 (1·02–67·58) | 0·04 1 | |
| 2: +/+ | 10·85 (0·97–121·57) | 0·05 2 | |
| KIR2DS2 | 0: absent | ||
| 1: present | 0·13 (0·02–0·77) | 0·02 1 | |
| KIR2DS5 | 0: absent | ||
| 1: present | 4·99 (0·99–25·15) | 0·05 2 | |
| HLA‐C2 | 0: absent | ||
| 1: present | 3·57 (1·13–11·30) | 0·03 1 | |
| HLA‐Bw4T | 0: Absent | ||
| 1: Present | 3·42 (0·88–13·20) | 0·07 2 |
We report the following degree of evidence against the null hypothesis: 1statistical significance (P < 0·05); 2weak statistical significance (P < 0·10); n.s., not significant. TThreonine.
Finally, 32 out of 51 symptomatic patients (63%) but only 18 out of 47 controls (38%) possess both the GM23 allotypes and HLA‐C2 gene group with a highly significant OR (OR = 2·71; P = 0·01).
Discussion
Several studies support the hypothesis that polygenic inheritance controls common variations in the clinical course of HCMV infection.24 We reported that a series of polymorphic key regulators at the interface of innate and acquired immunity (i.e. HLA class I molecules, KIR and GM3/17 and GM23 allotypes) are associated with this clinical variability, strengthening the essential role of both kinds of immunity in the control of HCMV infection.
Present results extend our previous findings on the role of KIR/HLA genetic variants in the control of primary HCMV infection in immunocompetent hosts by showing an additive effect of GM allotypes. Particularly, GM17/17 was shown to be associated with the risk of developing symptomatic infection only when present in homozygosity, but not as the heterozygous GM3/17 genotype, suggesting a gene–dose effect. Moreover, an association with the risk of symptomatic infection was also observed for GM23 carriers (GM23+/− and GM23+/+), with no evidence of a gene–dose effect.
The GM allotypes have been shown to be involved in the immunological control of virus infections, such as HCV13 or HSV‐1.14 In some cases, their contribution requires epistatic interaction with other genes of the immune system such as HLA genes.14, 25 These findings further underscore the importance of epistasis.
Our data show that besides GM17/17 and GM23 carriers, HLA‐C2 and HLA‐Bw4 are independently associated with the risk of HCMV symptomatic infection in a multiple logistic regression analysis. Therefore, besides a possible epistatic interaction of GM allotypes with HLA gene variants, an independent effect of these GM allotypes can be hypothesized on the basis of our results. This genetic effect together with that of HLA and KIR gene variants contribute different inherited genotypes with variable ability to influence the final outcome of HCMV infection, by modulating the immune response at different levels, both innate and acquired.
The mechanism of GM gene involvement in response to HCMV could involve the host immunosurveillance mechanism against virally infected cells, i.e. ADCC. IgG antibody‐mediated ADCC is, in fact, triggered upon ligation of FcγR to the Fc region of IgG, where the majority of GM alleles are expressed. Hence, genetic variation might contribute to the differences in the magnitude of ADCC.26 GM allotypes are expressed in the constant region of γ chains, but they can influence antibody specificity and affinity by changing the conformation of the variable region, hence modulating the kinetic competence of antigen‐binding sites.27
Furthermore, it has been shown that two HCMV‐encoded FcγR, which the virus uses to evade Fcγ‐mediated effector functions, differentially bind to non‐immune IgG1 expressing different GM allotypes.28, 29, 30, 31 Although viral FcγRs have been shown to bind the CH2–CH3 interface of IgG γ‐chains, as stated above, amino acid substitutions distant from the binding site could influence the protein conformation and so indirectly affect its binding affinity. The importance of GM allotypes expressed in the CH1 region of the γ1 chain for the viral FcγR binding has been conclusively shown for HSV‐1.32 In particular, higher affinity of GM17‐expressing IgG1 to the viral FcγR would imply that individuals with the GM17 allotype would be more likely to have the Fcγ domains of their anti‐HCMV IgG antibodies scavenged, hence decreasing their immunological competence to eliminate the virus through Fcγ‐mediated effector mechanisms, such as ADCC. Consequently, individuals possessing the GM17 allotype would be expected to be at an increased risk of developing HCMV‐associated diseases, such as symptomatic infection, whereas those carrying the GM3 allotype, which presents a lower affinity to the viral FcγR, would be expected to have a reduced risk of symptomatic infection.
There is a strong biological rationale for the involvement of GM allotypes and KIR/HLA variants in the outcome of HCMV infections based on their immunological properties and previously published reports. However, repeating these studies using an independent study population is the best way to determine whether our observations are not a result of chance.
The results of the present analysis show that AA haplotype is no longer associated with the risk of symptomatic infection, as was conversely reported in the previous manuscript. The lack of significance of the AA haplotype was unexpected, because the correlation between AA haplotype and the risk of CMV infection/reactivation was replicated in several studies, besides ours. This may suggest that the role of AA haplotype is not that important, as previously shown; however, an unexplained statistical segregation of the data, after the introduction of new variables in the model (GM23/23 and GM3/17) might explain this unexpected result, as frequently occurs, as the new variables may subsume the role of previous important variables for reasons that are sometimes difficult to explain on biological grounds.
In contrast, it is important to underline that variables identified in our previous study as being significantly associated with the risk of HCMV symptomatic infection, such as HLA‐C2 and HLA‐Bw4T, were also significantly (or marginally) associated with the risk of HCMV symptomatic infection in the present analyses after the introduction of GM allotypes in the multivariable regression analysis. This observation strengthens the association between these variables and the outcome, which is consistent in all the multiple imputation models used, despite the differences in the imputed data for GM3/17 and GM23 generated by each model. In particular, GM23, both homozygous and heterozygous, and HLA‐C2 are the variables that were shown to be consistently associated with the outcome in all the imputation models used. When we combined these two variables, the risk of symptomatic HCMV infection was about three times more frequent in individuals bearing the GM23 allele and HLA‐C2 (53·3%) than in those not bearing these gene variants (30%) (OR 2·71; P = 0·01).
With regard to the KIR2DS5, it is true that it was shown to be associated with the risk of symptomatic infection, and this is apparently in contrast to the findings of the previous paper,15 supporting the role of activating receptors in the control of the infection. However, the present analysis showed that another activating receptor, KIR2DS2, is negatively associated with the risk of infection, being more frequent in controls than in cases (OR = 0·14). Therefore, the two activating receptors independently associated with the risk of symptomatic infection have opposite roles, one detrimental (KIR2DS5), the other protective (KIR2DS2). This confirms the analysis of the previous paper15 showing no difference in the number of activating receptors between cases and controls.
Linkage disequilibrium in the GM gene complex is very strong. It is possible that there are other genes in and around this complex (on chromosome 14) that influence the outcome of HCMV infection by as yet unknown mechanisms (other than ADCC). Significant linkage disequilibrium between the alleles of these putative genes and GM23 and HLA‐C2 alleles could also give rise to the associations observed in this investigation.
The HLA genes investigated in this report should influence the outcome of HCMV infection by regulating the activity of NK cells, key players in anti‐viral immunity (see Introduction). The KIRs on NK cells bind their HLA ligands on target cells with differential affinity. Particular allelic combinations of KIR and HLA genes have been reported to be associated with the resolution to virus infection and/or susceptibility. In this regard, it is interesting to note that it also concerns autoimmune diseases. In fact, HLA‐B27 interactions with KIR have been implicated in the pathogenesis of ankylosing spondylitis.33, 34
Mechanisms underlying the epistatic interaction between GM and HLA alleles in the outcome of HCMV infection could also involve the recognition of HCMV antigens by the B‐cell membrane‐bound IgG receptors expressing different GM alleles, followed by processing and presentation to the peptide‐binding groove of at‐risk HLA alleles.
Collectively these data suggest that genetic variations in the innate immunity cells, such as NK cells and their KIR repertoire, or molecules, such as IgG genes (GM variants), play a crucial role in the control of HCMV infection.
However, it is important to delineate the mechanisms underlying the involvement of GM allotypes in the outcome of HCMV infection. Such studies may lead to the identification of novel pathways of host immune response that may be helpful in the designing of vaccines for protection against this infection. Some progress toward this end has been made,2 but more studies are needed. Our information on the role of both cellular and humoral roles in control of HCMV infection should be of value in guiding efforts to reduce HCMV‐associated health complications in the elderly, including immunosenescence,35 and in transplantation.
Disclosure
The authors state that they have no no conflicts of interest to declare.
Acknowledgements
CCO and DDB collected the data; JPP, AA and GA performed the experiments and compiled the data for the summation and analysis; MB performed statistical analysis. DDB, JPP and CCA designed the study. DDB, MB and CC wrote the paper. All authors analysed the data, reviewed the paper, approved the final version and agreed to submit the paper. This work was supported in part by a grant from the Ministry of University (PRIN: progetti di ricerca di rilevante interesse nazionale – Bando 2015 Prot. 20157ATSLF ‘Discovery of molecular and genetic/epigenetic signatures underlying resistance to age‐related diseases and comorbidities’) to CCA, GC and DDB; GA and AA are fellows of this project. We thank Dr Sarumathi Mohan for assistance in GM genotyping.
References
- 1. Cannon MJ, Schmid DS, Hyde TB. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev Med Virol 2010; 20:202–13. [DOI] [PubMed] [Google Scholar]
- 2. Hanley PJ, Bollard CM. Controlling cytomegalovirus: helping the immune system take the lead. Viruses 2014; 6:2242–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Solana R, Tarazona R, Aiello AE, Akbar AN, Appay V, Beswick M et al CMV and immunosenescence: from basics to clinics. Immun Ageing 2012; 9:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Parry HM, Zuo J, Frumento G, Mirajkar N, Inman C, Edwards E et al Cytomegalovirus viral load within blood increases markedly in healthy people over the age of 70 years. Immun Ageing 2016; 13:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wu Z, Sinzger C, Reichel JJ, Just M, Mertens T. Natural killer cells can inhibit the transmission of human cytomegalovirus in cell culture by using mechanisms from innate and adaptive immune responses. J Virol 2015; 89:2906–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wilkinson GW, Tomasec P, Stanton RJ, Armstrong M, Prod'homme V, Aicheler R et al Modulation of natural killer cells by human cytomegalovirus. J Clin Virol 2008; 41:206–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kuijpers TW, Baars PA, Dantin C, van den Burg M, van Lier RA, Roosnek E. Human NK cells can control CMV infection in the absence of T cells. Blood 2008; 112:914–5. [DOI] [PubMed] [Google Scholar]
- 8. Wu Z, Sinzger C, Frascaroli G, Reichel J, Bayer C, Wang L et al Human cytomegalovirus‐induced NKG2Chi CD57hi natural killer cells are effectors dependent on humoral antiviral immunity. J Virol 2013; 87:7717–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moss P, Rickinson A. Cellular immunotherapy for viral infection after HSC transplantation. Nat Rev Immunol 2005; 5:9–20. [DOI] [PubMed] [Google Scholar]
- 10. Biron CA, Byron KS, Sullivan JL. Severe herpesvirus infections in an adolescent without natural killer cells. N Engl J Med 1989; 320:1731–5. [DOI] [PubMed] [Google Scholar]
- 11. Parikh Bijal A, Piersma Sytse J, Yokoyama Wayne M. NK cells in antiviral defense In: Ratcliffe MJH, ed. Encyclopedia of Immunobiology. Oxford: Academic Press, 2016: 243–52. [Google Scholar]
- 12. Oxelius VA, Pandey JP. Human immunoglobulin constant heavy G chain (IGHG) (Fcγ) (GM) genes, defining innate variants of IgG molecules and B cells, have impact on disease and therapy. Clin Immunol 2013; 149:475–86. [DOI] [PubMed] [Google Scholar]
- 13. Pandey JP, Namboodiri AM, Luo Y, Wu Y, Elston RC, Thomas DL et al Genetic markers of IgG influence the outcome of infection with hepatitis C virus. J Infect Dis 2008; 198:1334–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Moraru M, Black LE, Muntasell A, Portero F, López‐Botet M, Reyburn HT et al NK Cell and Ig interplay in defense against herpes simplex virus Type 1: epistatic interaction of CD16A and IgG1 allotypes of variable affinities modulates antibody‐dependent cellular cytotoxicity and susceptibility to clinical reactivation. J Immunol 2015; 195:1676–84. [DOI] [PubMed] [Google Scholar]
- 15. Di Bona D, Scafidi V, Plaia A, Colomba C, Nuzzo D, Occhino C et al HLA and killer cell immunoglobulin‐like receptors influence the natural course of CMV infection. J Infect Dis 2014; 210:1083–9. [DOI] [PubMed] [Google Scholar]
- 16. Pandey JP, Kistner‐Griffin E, Radwan FF, Kaur N, Namboodiri AM, Black L et al Immunoglobulin genes influence the magnitude of humoral immunity to cytomegalovirus glycoprotein B. J Infect Dis 2014; 210:1823–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pandey JP, Namboodiri AM, Mohan S, Nietert PJ, Peterson L. Genetic markers of immunoglobulin G and immunity to cytomegalovirus in patients with breast cancer. Cell Immunol 2017; 312:67–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pandey JP, Kaur N, Costa S, Amorim J, Nabico R, Linhares P et al Immunoglobulin genes implicated in glioma risk. Oncoimmunology 2014; 3:e28609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988; 16:1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Van Buuren S. Flexible Imputation of Missing Data. Boca Raton, FL: Chapman & Hall/CRC Press, 2012: 342. CRC Press, Amazon, eBook [Google Scholar]
- 21. Vach W, Blettner M. Biased estimation of the odds ratio in case–control studies due to the use of ad hoc methods of correcting for missing values for confounding variables. Am J Epidemiol 1991; 134:895–907. [DOI] [PubMed] [Google Scholar]
- 22. Vach W, Blettner M. Logistic regression with incompletely observed categorical covariates – investigating the sensitivity against violation of the missing at random assumption. Stat Med 1995; 14:1315–29. [DOI] [PubMed] [Google Scholar]
- 23. van Buuren S, Groothuis‐Oudshoorn K. Mice: multivariate imputation by chained equations in R. J Stat Softw 2011; 45:1–67. [Google Scholar]
- 24. Gelemanovic A, Dobberpuhl K, Krakar G, Patarcic I, Kolcic I, Polasek O. Host genetics and susceptibility to congenital and childhood cytomegalovirus infection: a systematic review. Croat Med J 2016; 57:321–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pandey JP, Montes‐Cano MA, Aguilar‐Reina J, Gonzalez‐Escribano MF. Interactive effects of immunoglobulin γ and human leucocyte antigen genotypes on clearance and persistence of infection with hepatitis C virus. Clin Exp Immunol 2007; 150:518–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pandey JP. Immunoglobulin GM genes, cytomegalovirus immunoevasion, and the risk of glioma, neuroblastoma, and breast cancer. Front Oncol 2014; 4:236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Torres M, Casadevall A. The immunoglobulin constant region contributes to affinity and specificity. Trends Immunol 2008; 29:91–7. [DOI] [PubMed] [Google Scholar]
- 28. Atalay R, Zimmermann A, Wagner M, Borst E, Benz C, Messerle M et al Identification and expression of human cytomegalovirus transcription units coding for two distinct Fcγ receptor homologs. J Virol 2002; 76:8596–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Corrales‐Aguilar E, Trilling M, Hunold K, Fiedler M, Le VT, Reinhard H et al Human cytomegalovirus Fcγ binding proteins gp34 and gp68 antagonize Fcγ receptors I, II and III. PLoS Pathog 2014; 10:e1004131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Namboodiri A, Pandey JP. The human cytomegalovirus TRL11/IRL11‐encoded FcγR binds differentially to allelic variants of immunoglobulin G1 Arch. Virol 2011; 156:907–10. [DOI] [PubMed] [Google Scholar]
- 31. Pandey JP, Namboodiri AM, Radwan FF, Nietert PJ. The decoy Fcγ receptor encoded by the cytomegalovirus UL119‐UL118 gene has differential affinity to IgG proteins expressing different GM allotypes. Hum Immunol 2015; 76:591–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Atherton A, Armour KL, Bell S, Minson AC, Clark MR. The herpes simplex virus type 1 Fc receptor discriminates between IgG1 allotypes. Eur J Immunol 2000; 30:2540–7. [DOI] [PubMed] [Google Scholar]
- 33. Aiello A, Accardi G, Candore G, Gambino CM, Caruso C, Di Bona D. The importance of the interactions between KIRs and HLA ligands in the development of human autoimmune and viral diseases In: Accardi G, Caruso C, eds. Updates in Pathobiology: Causality and Chance in Ageing, Age‐Related Diseases and Longevity. Palermo: Palermo University Press, 2017: 91–110. [Google Scholar]
- 34. Cauli A, Piga M, Dessole G, Porru G, Floris A, Vacca A et al Killer‐cell immunoglobulin‐like receptors (KIR) and HLA‐class I heavy chains in ankylosing spondylitis. Drug Dev Res 2014; 75(Suppl 1):S15–9. [DOI] [PubMed] [Google Scholar]
- 35. Caruso C, Vasto S. Immunity and aging In: Ratcliffe MJH, ed. Encyclopedia of Immunobiology. Oxford: Academic Press, 2016: 127–32. [Google Scholar]


