Abstract
Antisense transcription is a widespread phenomenon in mammalian genomes, leading to production of RNAs molecules referred to as natural antisense transcripts (NATs). NATs apply diverse transcriptional and post‐transcriptional regulatory mechanisms to carry out a wide variety of biological roles that are important for the normal functioning of living cells, but their dysfunctions can be associated with human diseases. In this review, we attempt to provide a molecular basis for the involvement of NATs in the etiology of human disorders such as cancers and neurodegenerative and cardiovascular diseases. We also discuss the pros and cons of oligonucleotide‐based therapies targeted against NATs, and we comment on state‐of‐the‐art progress in this promising area of clinical research. WIREs RNA 2018, 9:e1461. doi: 10.1002/wrna.1461
This article is categorized under:
-
1
RNA in Disease and Development > RNA in Disease
-
2
Regulatory RNAs/RNAi/Riboswitches > Regulatory RNAs
-
3
RNA Interactions with Proteins and Other Molecules > Small Molecule–RNA Interactions
Keywords: antisense RNAs, antisense therapy, chromatin remodeling, NATs
1. INTRODUCTION
One of the most startling discoveries since the completion of the Human Genome Project (Lander et al., 2001) was that as many as 70% of genes show evidence of antisense transcription (Katayama et al., 2005; Lehner, Williams, Campbell, & Sanderson, 2002), producing the so‐called natural antisense transcripts (NATs). NATs overlap with sense genes, their promoters or other regulatory regions. Despite being processed mostly from protein‐coding loci, they frequently display distinct characteristics compared to their sense partners. For instance, many of them are not polyadenylated (Kung, Colognori, & Lee, 2013); they may be expressed at lower levels than sense transcripts (He, Vogelstein, Velculescu, Papadopoulos, & Kinzler, 2008); and they often display high tissue specificity (Faghihi et al., 2008; Modarresi et al., 2012). This prevalence and transcriptional diversity of antisense transcripts motivated a number of research projects aimed at deciphering their biological roles. Firstly, a knockdown of 797 evolutionarily conserved NATs demonstrated the existence of a regulatory relationship between the sense‐antisense partners (Faghihi, Kocerha, et al., 2010). It was later shown that NATs act by either repressing or promoting the expression of their target genes, referred to as discordant or concordant regulation, respectively (Faghihi & Wahlestedt, 2009). Although the regulation typically occurs in cis with the antisense transcript affecting its sense partner, trans action is also possible when they target transcripts from other genomic loci (Faghihi, Kocerha, et al., 2010; Guttman et al., 2011). From a mechanistic point of view, antisense transcripts modulate expression and processing of other RNAs in a number of ways (Figure 1). The best known and possibly the most frequent way involves recruitment of the gene‐silencing Polycomb Recessive Complex 2 (PRC2) that induces trimethylation of the lysine 27 residue in histone 2 (H3K27me3), indicating transcriptionally inactive genomic loci (Figure 1a). This cis effect is an example of discordant regulation, in which increased expression of NATs leads to inhibition of gene expression in sense orientation. Alternatively, antisense transcripts can affect the sense transcripts in a sequence‐specific manner by coming into direct RNA:RNA interactions, although here also interactions in trans are possible. In a duplex with mated pre‐mRNA molecules, NATs cause alternative splicing events (Hu, Wang, & Shan, 2016) by masking splice sites and/or other splicing signals, or they may lead to reduction of transcript availability through nuclear retention (Kumar & Carmichael, 1997) (Figure 1b). Furthermore, formation of RNA duplexes might trigger editing events by providing ADARs (adenosine deaminases acting on RNA) with double‐stranded RNA substrates. Other examples include affecting transcript stability by guiding protein‐coding transcripts to degradation within a Staufen‐mediated decay pathway or by abrogation of miRNA functions through masking miRNA binding sites in the 3′ untranslated regions (Szczesniak & Makalowska, 2016) (Figure 1d). Some NATs also function as microRNA sponges, that is, they contain multiple binding sites that associate with specific miRNAs to prevent them from interacting with other RNAs (Figure 1c). There are also other known NATs‐mediated mechanisms such as transcriptional interference (Figure 1e) or entering into interactions with splicing factors; these and other functions of NATs are reviewed elsewhere (Rosikiewicz & Makalowska, 2016; Wight & Werner, 2013). Below we provide a more detailed insight into their selected modes of action, focusing on those involved in the etiology of human diseases or those that are otherwise important from the human health perspective (Table 1). Our intention is to put most emphasis on the so called cis‐NATs, that is, such NATs that have their sense partners in the same genomic locus. We then describe current strategies aimed at harnessing this yet‐unexplored regulatory potential of NATs to cure human diseases; finally, we discuss the current progress and future prospects.
Figure 1.
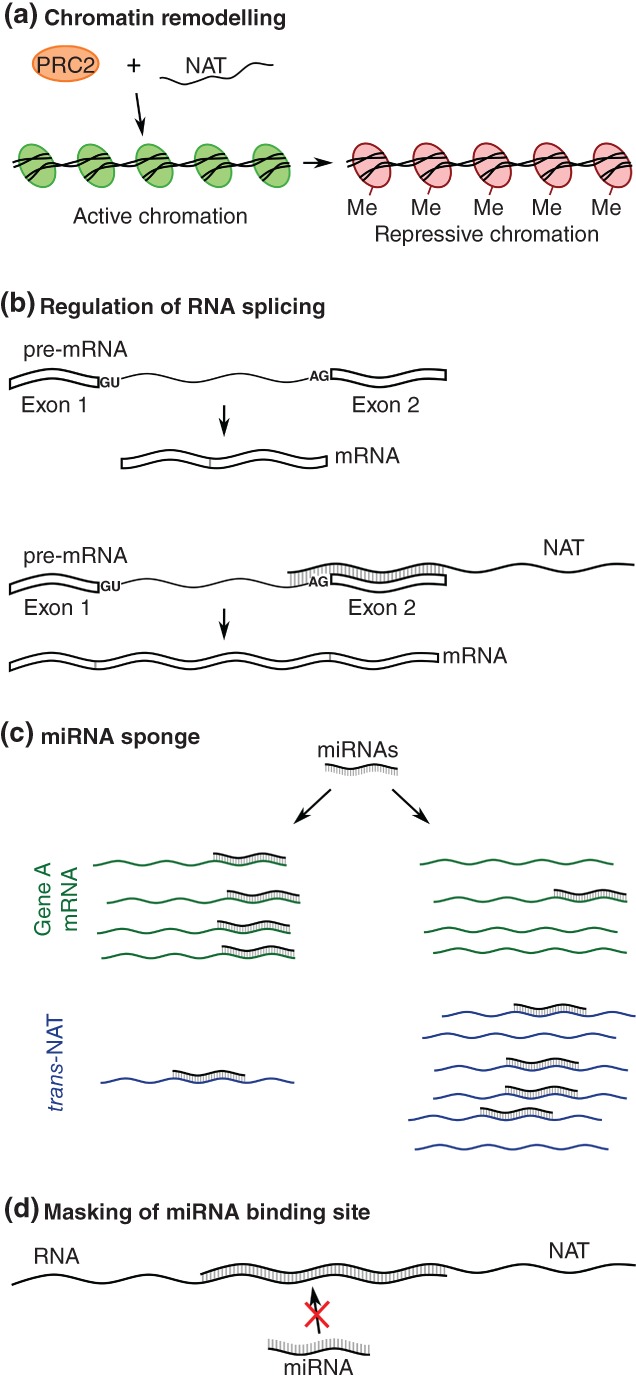
Functions of NATs. (a) Chromatin remodeling by the PRC1 or PRC2 complexes recruited by the NAT. Me, H3K27me3 histone modification. (b) Regulation of the alternative splicing by the RNA:RNA interaction between pre‐mRNA and NAT within the 3′ end of the pre‐mRNA’s retained intron. (c) miRNA sponge formed by the increased amount of trans‐NATs possessing miRNA target sites. (d) miRNA masking by the RNA:RNA interaction between RNA and NAT within the miRNA target sequence
Table 1.
A list of natural antisense transcripts and their mechanisms of action in human diseases that are mentioned in this review
| Mechanism | Natural antisense transcript | Sense gene | NAT localization relative to a sense gene | Example of disease | References |
|---|---|---|---|---|---|
| Histone modification | HOTAIR | HOXC11 | 5′ end overlap | Breast cancer | Gupta et al. (2010) |
| ANRIL | CDKN2B | full overlap | Hepatocellular carcinoma | Pasmant et al. (2007) and Huang et al. (2015) | |
| TUG1 | MORC2 | 5′ end overlap | Small Cell Lung cancer | Xu et al. (2015), Ma et al. (2016) and Niu et al. (2017) | |
| APOA1‐AS | APOA1 | full overlap | Cardiovascular diseases | Rader (2002) and Halley et al. (2014) | |
| Methylation of CpG islands | Aberrant LUC7L | HBA2 | 3′ end overlap | Alpha‐thalassemia | Tufarelli, Frischauf, Hardison, Flint, and Higgs (2001) and Tufarelli et al. (2003) |
| Genomic imprinting | UBE3A‐AS | UBE3A | 3′ end overlap | Angelman syndrome | Rougeulle, Glatt, and Lalande (1997), Chamberlain and Brannan (2001), Johnstone et al. (2006), Meng, Person, and Beaudet (2012) |
| KCNQ1OT1 | KCNQ1 | 3′ end overlap | Beckwith‐Wiedemann syndrome | Caspary, Cleary, Baker, Guan, and Tilghman (1998), Thakur et al. (2004), Nakano et al. (2006), Pandey et al. (2008), Robbins, Chen, Wells, and Rivera (2012), and Zhang et al. (2014) | |
| Modulation of alternative splicing | UXT‐AS1 | UXT | 5′ end overlap | Colorectal cancer | Yin et al. (2017) |
| ZEB2‐AS1 | ZEB2 | 5′ end overlap | Hepatocellular carcinoma | Beltran et al. (2008), Petrova, Schecterson, and Gumbiner (2016) | |
| 17A | GABBR2 | full overlap | Alzheimer’s disease | Massone et al., 2011 | |
| AC021224.1–201 | retro_hsap_1933 | HIV | Mayeda, Munroe, Caceres, and Krainer (1994), Tange, Damgaard, Guth, Valcarcel, and Kjems (2001), Damgaard, Tange, and Kjems (2002) and Bryzghalov, Szczesniak, and Makalowska (2016) | ||
| ATX8OS | ATX8 | 5′ end overlap | Spinocerebellar Ataxia Type 8 | Moseley et al. (2006) and Daughters et al. (2009) | |
| miRNA sponges | PTENpg1 antisense β | PTENpg1 | 5′ end overlap | Clear‐Cell Renal Cell carcinoma | Poliseno et al. (2010), Johnsson et al. (2013), and Grander and Johnsson (2016) |
| ZFAS1 | ZNFX1 | 5′ end overlap | Hepatocellular carcinoma | Li et al. (2015) and Thorenoor et al. (2016) | |
| TUG1 | MORC2 | 5′ end overlap | Endometrial cancer | Liu et al., 2017 | |
| Masking miRNA binding sites | BACE1‐AS | BACE1 | full overlap | Alzheimer’s disease | Fukumoto, Cheung, Hyman, and Irizarry (2002), Holsinger, McLean, Beyreuther, Masters, and Evin (2002), Faghihi et al. (2008), Faghihi, Kocerha, et al. (2010), Faghihi, Zhang, et al. (2010), and Yan, Fan, Zhou, and Vassar (2016) |
2. NATURAL ANTISENSE TRANSCRIPTS IN HUMAN DISEASES: MOLECULAR BASIS
2.1. Histone modifications
The NATs are well known to regulate gene expression by recruitment of complex epigenetic machinery, resulting in histone modifications and transcriptional deregulation of target genes (Kaikkonen, Lam, & Glass, 2011). Polycomb Repressive Complex 1 (PRC21) and 2 (PRC2) represent the two best‐described chromatin remodeling complexes, leading to mono‐ubiquitinylation of lysine 119 of Histone 2A (H2AK119ub) and methylation of histone H3 at lysine 27 (H3K27me3), respectively (Cao et al., 2002; Wang et al., 2004) (Figure 1a). Other proteins involved in chromatin reorganization include Lysine Specific Demethylase 1 (LSD1) and G9a Methyltransferase. LSD1 demethylates histone H3 at lysine 4 (H3K4) (Shi et al., 2004), while G9a is responsible for di‐ and tri‐methylation of Histone H3 at lysine 9 (Casciello, Windloch, Gannon, & Lee, 2015). A prominent example of a NAT employing this mechanism is the HOX Transcript Antisense Intergenic RNA (HOTAIR) that promotes HOX gene silencing by interacting with PRC2 and LSD1 and recruiting them to defined genomic loci. HOTAIR is upregulated in a variety of cancers (Gupta et al., 2010), contributing to their etiology. In breast cancer, one of the most common cancers in women, HOTAIR triggers silencing of metastasis suppressor genes including Protocadherin 10 (PCDH10), Protocadherin Beta 5 (PCDHB5) and Junctional Adhesion Molecule 2 (JAM2) (Croce, 2010).
A similar mechanism was proposed for Antisense Noncoding RNA in the INK4 Locus (ANRIL) initially identified in melanoma patients (Pasmant et al., 2007). ANRIL is overexpressed in a variety of human cancers, including breast, gastric, liver, lung, melanoma, and prostate carcinomas. The data accumulated so far implicate ANRIL in chromatin reorganization and silencing of the tumor suppressor locus INK4b‐ARF‐INK4a by interacting with Chromobox 7 (CBX7) and Suppressor of Zeste 12 Protein Homologue (SUZ12), which are components of PRC1 and PRC2, respectively. Moreover, Huang et al. (2015) found increased expression of ANRIL in hepatocellular carcinoma (HCC) and noted its association with tumor size. In HCC, ANRIL recruits Enhancer of Zeste Homologue 2 (EZH2), a component of PRC2, to the Kruppel‐Like Factor 2 (KLF2) promoter, triggering epigenetic silencing of this gene. Similarly, Taurine Upregulated Gene (TUG1) enters into interactions with the EZH2 component of PRC2. TUG1, a NAT for MORC Family CW‐Type Zinc Finger 2 (MORC2), is associated with poor prognosis in patients with cancers (Ma et al., 2016; Niu et al., 2017), and its high expression level correlates with increased cancer cell proliferation and metastasis (Xu et al., 2015). Niu et al. found that in small‐cell lung cancer (SCLC), TUG1 interacts with EZH2, leading to deregulation of LIMK2b Kinase, which is implicated in tumor growth. This results in enhanced chemoresistance and growth of SCLC cells (Niu et al., 2017).
Apolipoprotein A1 Antisense Transcript (APOA1‐AS) and its role in the cholesterol pathway can be mentioned as another example of a NAT involved in epigenetic regulation and disease development. The APOA1‐AS regulates Apolipoprotein A1 (APOA1), the major protein component of high‐density lipoprotein (HDL) in plasma. Importantly, low levels of HDL cholesterol are a major risk factor of cardiovascular diseases (Rader, 2002). APOA1‐AS overlaps the fourth exon of APOA1 and impedes its transcription, most likely by promoting LSD1 assembly to an APOA1 locus (Halley et al., 2014). LSD1, also known as KDM1A, catalyses demethylation of histone lysine marks from transcriptionally active promoters (reviewed in Shi, 2007). Interestingly, targeting APOA1‐AS with a siRNA leads to increased H3K4 trimethylation in the APOA1 region and enhanced gene expression, suggesting that inhibitory interaction between APOA1‐AS and LSD1 was interrupted (Halley et al., 2014).
2.2. Methylation of CpG islands
Vertebrate promoters are often associated with regions rich in CG dinucleotides, referred to as CpG islands. In the human genome, the CpG islands are found in approximately 70% of annotated gene promoters (Saxonov, Berg, & Brutlag, 2006). Their methylation leads to decreased transcription by reducing accessibility of DNA regions to transcription machinery (Saxonov et al., 2006), and some antisense transcripts are implicated in this kind of regulation. An example can be observed in α‐thalassemia—an inherited form of anemia caused by disruption in α‐Globin synthesis (Muncie & Campbell, 2009; Tufarelli et al., 2003). Although the abnormal gene expression in α‐thalassemia patients is mostly due to mutations in the α‐Globin (HBA2) gene sequence, the methylation status of α‐Globin CpG islands is changed in the disease as well (Tufarelli et al., 2003). This phenomenon could be attributed to an α‐Globin antisense transcript that arises from a disrupted LUC7‐Like (LUC7L) gene localized on the opposite DNA strand in the convergent orientation to the α‐Globin cluster. It was proposed that transcription of LUC7L represses expression of HBA2 by triggering methylation of its CpG islands (Tufarelli et al., 2001, 2003). In agreement with this hypothesis, interesting insights have come from the analysis of patients with a deletion of the 3′ region of the α‐Globin locus that leaves the 5′ part of the HBA2 gene intact. The deletion also affects LUC7L by removing the transcription termination signal, resulting in generation of a long antisense transcript stretching up to the HBA2 promoter. A growing body of evidence shows that methylation of CpG islands of HBA2 is triggered by transcription of this aberrant antisense transcript (Tufarelli et al., 2001, 2003). Moreover, replacement of LUC7L with the Human Ubiquitin C promoter leads to methylation of the HBA2 promoter, suggesting that antisense transcription itself is required rather than the action of the aberrant antisense RNA transcript (Tufarelli et al., 2003).
2.3. Genomic imprinting
In diploid organisms, the majority of autosomal genes are expressed from both chromosomes, yet a limited subset of genes display parental‐specific expression (Barlow & Bartolomei, 2014). The monoallelic gene expression is achieved through the so‐called genomic imprinting, an epigenetic phenomenon that causes modifications at the DNA or histone level during gametogenesis (Barlow & Bartolomei, 2014). Imprinted genes are often organized into clusters regulated by the imprinting control center (Barlow & Bartolomei, 2014; Delaval & Feil, 2004). NATs are among the key players in genomic imprinting. They act by formation of NAT:mRNA duplexes that later can either induce RNA interference (RNAi) or coat the chromosomal region and recruit repressive chromatin proteins to target DNA sequences (Barlow & Bartolomei, 2014).
An example of a disorder caused by disruption in imprinting is Beckwith–Wiedemann syndrome (BWS) (Lee et al., 1999; Smilinich et al., 1999). Analyses of BWS patients have shown abnormal biallelic gene expression as a result of the loss of maternal‐specific methylation in the KvDMR1 cluster at the 11p15.5 chromosomal region (Lee et al., 1999; Smilinich et al., 1999). Interestingly, in the majority of patients the expression of otherwise transcriptionally silenced KCNQ1OT1 antisense transcript was found. As the orthologous imprinted cluster has been found at the distal end of mouse chromosome 7 (Caspary et al., 1998), mouse models were used to analyze KCNQ1OT1 and shed light on the mechanism of NAT action. Experiments have confirmed that Kcnq1ot1 RNA does not form a double‐stranded RNA and therefore does not take part in dsRNA‐mediated silencing (Thakur et al., 2004). A recent study by Zhang et al. (2014) has demonstrated the role of Kcnq1ot1 in long‐range chromatin interactions. They used a novel RNA‐guided chromosome conformation capture approach and showed that Kcnq1ot1 RNA acts as a scaffold for a long‐range intrachromosomal loop formation between the KvDMR1 cluster and promoter region. Furthermore, Kcnq1ot1 RNA recruits chromatin modifiers such as PRC1, PRC2 and Euchromatic Histone‐Lysine N‐Methyltransferase 2 (EHMT2) to maintain monoallelic expression of the KvDMR1 cluster (Pandey et al., 2008; Robbins et al., 2012; Zhang et al., 2014). Interestingly, KCNQ1OT1 is upregulated in a variety of carcinomas including colorectal cancer (CRC; Nakano et al., 2006), yet its mode of action there remains to be elucidated.
Ubiquitin‐protein ligase E3A (UBE3A) encodes a component of the ubiquitin protein degradation system and plays a crucial role in the development and function of the nervous system (Greer et al., 2010; Kishino, Lalande, & Wagstaff, 1997; Scheffner, Huibregtse, Vierstra, & Howley, 1993). The UBE3A gene is biallelically expressed in most human tissues except for brain, where it shows maternal‐specific expression (Matsuura et al., 1997; Rougeulle et al., 1997; Vu & Hoffman, 1997). It is localized in a region of chromosome 15q11‐q13, and disruptions in the inheritance of paternal or maternal imprinting in this region are associated with two neurological disorders: Prader–Willi syndrome (PWS) and Angelman syndrome (AS), respectively (Chamberlain & Lalande, 2010; Matsuura et al., 1997). Genes involved in development of these disorders are under control of the imprinting control center, either the Prader–Willi syndrome imprinting center (PWS‐IC) or the Angelman syndrome imprinting center (AS‐IC) (Kantor, Makedonski, Green‐Finberg, Shemer, & Razin, 2004). The UBE3A gene is adjacent to a paternally expressed cluster of genes positively regulated by the PWS‐IC. This gene was also shown to be downregulated in cis by the antisense transcript known as UBE3A‐AS (Rougeulle et al., 1997). Just as in the case of KCNQ1OT1, and due to the fact that the orthologous region on mouse chromosome 7C is conserved in terms of gene content and PWS‐IC function (Yang et al., 1998), a mouse model was used to test the molecular basis of the diseases, leading to the conclusion that PWS‐IC negatively regulates parental Ube3a gene expression but activates Ube3a‐as (Chamberlain & Brannan, 2001). Moreover, the deletion of the Ube3a‐as promoter region as well as Ube3a‐as depletion by early termination of its expression results in activation of the paternal Ube3a gene in mutant mice (Meng et al., 2012). However, the molecular background of the cis silencing of Ube3a by Ube3a‐as is unclear. It can be caused by the presence of the antisense RNA and/or by the transcription event itself. Importantly, the expression of Ube3a‐as was also related with maternal imprinting defects (Johnstone et al., 2006), further supporting the hypothesis that Ube3a‐as mediates silencing of the Ube3a gene.
2.4. Modulation of alternative splicing
Alternative splicing of pre‐mRNA molecules is a posttranscriptional process that leads to generation of multiple transcripts from a single gene, thus contributing to the observed diversity at transcriptional and protein levels in eukaryotic organisms. UXT‐AS1 is an antisense long noncoding RNA that can coordinate alternative splicing of the Ubiquitously Expressed Prefoldin‐Like Chaperone (UXT) gene and thereby promote the progression of CRC (Figure 1b). The UXT gene has two alternatively spliced transcripts, UXT1 and UXT2, that are down‐ and upregulated in CRC, respectively. Yin et al. (2017) demonstrated that increased expression of UXT‐AS1 in CRC cells results in a shift of the splicing pattern from UXT1 to UXT2. UXT‐AS1 is supposed to achieve this by base‐pairing with UXT pre‐mRNA.
Another example is ZEB2 Antisense RNA 1 (ZEB2‐AS1), which modulates alternative splicing of the Zinc Finger E‐Box Binding Homeobox 2 (ZEB2) gene. This gene encodes a transcriptional repressor of E‐Cadherin and is associated with tumorigenesis (Beltran et al., 2008; Petrova et al., 2016). As with UXT‐AS1, ZEB2‐AS1 achieves this splicing regulatory function by entering into direct RNA:RNA interactions. ZEB2‐AS1 binds to the 5′ splice site of the first intron in ZEB2 pre‐mRNA, which leads to intron retention. Interestingly, the retained intron contains an internal Ribosome Entry Site (IRES) necessary for expanded expression of the ZEB2 protein, while the forms with the intron spliced out have no protein coding capacity. The ZEB2 translation is dependent on the IRES. Thus, the NAT expression indirectly facilitates expression of the ZEB2 protein.
NATs are known to contribute to alternative splicing modulation in the brain, which is known for frequent occurrence of tissue‐specific alternative splicing isoforms (Chen & Manley, 2009). For instance, 17A ncRNA is considered as a splicing regulator of Gamma‐Aminobutyric Acid Type B Receptor Subunit 2 (GABBR2, also known as GPR51). 17A is overexpressed in cerebral cortices of patients with Alzheimer’s disease. From studies of neuroblastoma cells, it is known that this NAT induces the synthesis of alternative splice isoforms of GABBR2 and therefore abolishes intracellular signaling, leading to overproduction of neurotoxic β‐Amyloid peptide (Massone et al., 2011). GABBR2 is a component of a heterodimeric G‐protein coupled receptor for gamma‐aminobutyric acid (GABA); among other functions, it takes part in the regulation of neurotransmitter release (Geng et al., 2012; White et al., 1998). The 17A masks two putative splicing‐associated sequences, including a branch point and a polypyrimidine tract. As a result, a shorter alternative isoform—lacking the signal peptide—is expressed rather than the canonical full‐length splice variant. The signal peptide is required for the intramembrane localization of the GABBR2 protein, leaving the truncated isoform dysfunctional.
A recent bioinformatics analysis pinpointed the possibility that antisense transcripts derived from retrotransposed genes are affecting their progenitor genes by means of full or partial RNA:RNA complementarity, leading to a number of possible regulatory mechanisms in trans (Bryzghalov et al., 2016). For instance, a transcript transcribed in antisense to human retrocopy retro_hsap_1933 (RetrogeneDB, Kabza, Ciomborowska, & Makalowska, 2014), referred to as AC021224.1–201 or NONHSAT058863.2, could be involved in splicing regulation of its progenitor gene, Nuclear Ribonucleoprotein Particle A1 Protein (hnRNPA1) (Bryzghalov et al., 2016). The NAT is able to mask the 5′ splice site in the sixth intron of hnRNPA1, resulting in production of a longer isoform, while lack of the NAT: pre‐mRNA interaction gives rise to a shorter isoform. The shorter isoform was found to play regulatory roles in human immunodeficiency (HIV‐1) virus splicing and replication (Bryzghalov et al., 2016), as it blocks binding of the U2 spliceosomal RNA to the branch points of an intron in the viral tat gene (Damgaard et al., 2002; Tange et al., 2001). On the other hand, this isoform lacks protein domains required for canonical functions of the hnRNPA1 protein, that is, alternative splicing activity, stable binding of RNAs and optimal RNA annealing (Bryzghalov et al., 2016; Mayeda et al., 1994). Therefore, the NAT‐mediated modulation of alternative splicing leads to a critical switch in biological roles played by hnRNPA1.
In addition to direct masking of splice sites, NATs can also influence splicing indirectly by altering the activities of splicing factors such as Muscleblind‐Like Splicing Regulator 1 (MBLN1) and CUGBP Elav‐Like Family Member 1 (CELF1) (Daughters et al., 2009; Wang et al., 2015). The brain‐expressed ATXN8 Opposite Strand (ATX8OS) gene is a natural antisense noncoding RNA overlapping the first exon of the Kelch‐Like Family Member 1 gene (KLHL1). In Spinocerebellar Ataxia Type 8 (SCA8), CUG trinucleotide expansions from ATX8OS are thought to mediate sequestration of the MBLN1 protein, leading to a shift in splicing pattern in a number of genes including GABA‐A Transporter 4 RNA (GABT4) (Daughters et al., 2009; Moseley et al., 2006). GABT4 encodes a sodium‐dependent transporter of GABA that maintains low extracellular levels of GABA (Borden et al., 1994). ATX8OS was found to deregulate splicing of the seventh exon in GABT4 in human frontal lobe tissue, which is mediated by both sequestration of MBNL1 and enhanced expression or activity of CELF1 (Daughters et al., 2009).
2.5. miRNA sponges
A number of NATs act in trans as competitive endogenous RNAs (ceRNAs, miRNA sponges) (Figure 1c). They possess multiple binding sites by which they associate with miRNAs and prevent them from interaction with mRNAs. As a result, ceRNAs modulate expression levels of accessible miRNAs and mRNAs under their control (Sen, Ghosal, Das, Balti, & Chakrabarti, 2014).
One of the first described miRNA sponges is Phosphatase and Tensin Homologue Pseudogene 1 (PTENP1, also known as PTENpg1) (Poliseno et al., 2010). The PTENpg1 locus encodes three functional transcripts: PTENpg1 sense, PTENpg1 antisense α and PTENpg1 antisense β, which regulate expression of the Phosphatase and Tensin Homologue (PTEN) tumor suppressor gene. By competing for miR21, PTENpg1 reduces interaction between PTEN and miR21, thus suppressing cancer progression (Yu et al., 2014). This sponge activity of PTENpg1 is made possible by RNA:RNA interaction with PTENpg1 antisense β transcript, which provides the PTENpg1 sense transcript with a polyA tail that is essential for nuclear export and cytoplasmic function of this ceRNA molecule (Grander & Johnsson, 2016; Johnsson et al., 2013). Interestingly, PTENpg1 α, another antisense splice form, recruits chromatin remodeling complexes to the PTEN promoter, resulting in transcriptional silencing of PTEN and further contributing to this elegant regulatory pathway (Grander & Johnsson, 2016).
There are more NATs acting as ceRNAs and associated with cancer development and progression. The gene mir‐150 is a known tumor suppressor for a variety of carcinomas including CRC (Feng et al., 2014) and osteosarcoma (Qu, Pan, Kang, Dong, & Zhao, 2016). In HCC, mir‐150 suppresses Zinc Finger E‐box Binding Homeobox 1 (ZEB1) and thereby inhibits cell invasion and metastasis. An antisense transcript, ZFAS1, is a sponge for mir‐150 and is considered as an oncogene in HCC (Li et al., 2015). Interestingly, bioinformatic analysis suggests that ZFAS1 has the potential to sponge mir‐590‐3p in CRC tissues (Thorenoor et al., 2016), yet no experimental validation exists.
TUG1, another antisense lncRNA acting as a competitive endogenous RNA in cancer, is upregulated in endometrial cancer. By miR‐299 and miR‐34a‐5p sponging, TUG1 regulates expression levels of Vascular Endothelial Growth Factor A (VEGFA), which is considered to be the main factor responsible for tumor angiogenesis in human cancers (Liu et al., 2017).
2.6. Masking miRNA binding sites
Antisense transcripts can bind to mRNA molecules and mask miRNA binding sites, leading to increased stability of mRNAs that would otherwise be targeted by miRNAs (Figure 1d). This mechanism has been proposed as an important player in regulation of β‐Secretase 1 (BACE1) expression, a crucial enzyme in Alzheimer’s disease pathophysiology (Faghihi et al., 2008; Faghihi, Zhang, et al., 2010). BACE1 can be involved in generation of neurotoxic β‐Amyloid peptides by sequential proteolytic cleavages of amyloid precursor protein (Yan et al., 2016). Previous studies have shown that concentration as well as enzymatic activity of BACE1 is increased in Alzheimer’s disease brains (Fukumoto et al., 2002; Holsinger et al., 2002). It was reported that the BACE1 Antisense Transcript (BACE1‐AS) enhances the stability of BACE1 and therefore may be involved in disease development (Faghihi et al., 2008). Furthermore, an interplay between miR‐485‐5p and BACE1‐AS was suggested as a potential mechanism involved in this dysregulation (Faghihi, Zhang, et al., 2010). BACE1‐AS overlaps the sixth exon of the BACE1 gene where the miR‐485‐5p binding site is localized. In line with this hypothesis, in Alzheimer’s disease brains, upregulation of BACE1‐AS was correlated with downregulation of miR‐485‐5p. Moreover, overexpression of these two noncoding RNAs is associated with downregulation of BACE1. Finally, further experiments have established that BACE1 is stabilized by its antisense transcript by means of RNA:RNA interactions that lead to masking of a miRNA binding site in BACE1 (Faghihi, Zhang, et al., 2010).
2.7. Dicer‐dependent mechanisms
c‐MYC is an oncogene deregulated in various human cancers that promotes cell growth and proliferation (Stine, Walton, Altman, Hsieh, & Dang, 2015). Napoli, Piccinelli, Mapelli, Pisignano, and Catapano (2017) identified several NATs overlapping the c‐MYC gene and its 3′ distal region and described the complex network of interactions involved in c‐MYC expression regulation. One of the NATs, referred to as NAT6531, was found to form a stem‐loop, recognized by Dicer, an enzyme that cuts dsRNAs, leading to production of microRNAs or endo‐siRNAs (Kim, Han, & Siomi, 2009). The NAT6531‐derived small RNAs are believed to target the promoter and intron 1 sequences of c‐MYC (Napoli et al., 2017). As suggested by Napoli et al., this regulatory mechanism, based on the interplay between NATs and NAT‐derived small RNAs, may represent an important layer of transcriptome tuning that goes beyond c‐MYC locus. Intriguingly, the antisense‐mediated regulation is not limited to NAT6531 there. Another antisense transcript, NAT7281, is associated with repression of c‐MYC transcription and shutdown of NAT6531 and NAT6531‐derived small RNAs, contributing a lot to the observed regulatory complexity in the c‐Myc locus (Napoli et al., 2017).
A dicer‐dependent mechanism was also suggested in Huntington’s disease (Chung, Rudnicki, Yu, & Margolis, 2011), a neurodegenerative disorder caused by CAG trinucleotide repeat expansion within the first exon of the Huntingtin (HTT) gene. Huntingtin Natural Antisense Transcript (HTTAS) seems to play a regulatory role on the HHT gene. Chung et al. (2011) used the mouse Dicer‐null and wild‐type embryonic stem cells as well as different HTTAS constructs to investigate the possible roles of Dicer in silencing of HTT. In line with previous experiment conducted on human embryonic kidney cells, high levels of one isoform of HTTAS, referred to as HTTAS_v1, lead to reduced expression of HTT in mouse wild‐type embryonic cells. Contrary to wild‐type cells, the expression of HTT in Dicer‐null lines significantly increased in spite of the presence of HTTAS_v1. Both observation strongly support the involvement of Dicer in regulation of HTT by NATs.
2.8. Other associations with diseases
In addition to NATs for which mechanisms of action in diseases are quite well established, the exact molecular functions of most NATs are not fully understood. This is because of a lack of experimental evidence as well as the complexity of the underlying phenomena, leading to ambiguity in interpretation of results, especially now that multiple NATs are believed to be implicated in more than one mode of action. Some examples were provided above. Another such antisense noncoding RNA is TRAF3IP2‐AS1, for which at least two different mechanisms have been proposed. Firstly, it can be involved in the epigenetic modulation of the TRAF3 Interacting Protein 2 gene (TRAF3IP2) through alterations of the chromatin state (Bannon et al., 2015). In line with this, overexpression of TRAF3IP2‐AS1 leads to significantly decreased expression of at least one of the TRAF3IP2 transcripts. Secondly, it was proposed that TRAF3IP2‐AS1 can be involved in the post‐transcriptional control of TRAF3IP2 expression by formation of RNA:RNA duplexes (Morelli, Magnanini, Mungall, Negrini, & Barbanti‐Brodano, 2000), as the NAT overlaps the second exon of the TRAF3IP2 gene where the start codon of one of protein‐coding isoforms is located. TRAF3IP2‐AS1 seems to be present exclusively in the nucleus, compared to the global cellular distribution of TRAF3IP2, reinforcing possible regulatory functions of this NAT (Bannon et al., 2015). Further examples of NATs with not yet fully resolved modes of action come from studies on NAT: protein interactions. High‐Mobility Group Box 2 (Hmgb2) is a multifunctional protein participating in regulation of gene transcription by modulating transcription factor activity (Reeves, Langan, & Nissen, 1991; Wanschura et al., 1996; Yamanaka et al., 2015). An interplay between this protein and an antisense transcript called LPR1‐AS was suggested to be relevant in Alzheimer’s disease (Yamanaka et al., 2015). LPR1‐AS is an antisense transcript of Low Density Lipoprotein Receptor‐Related Protein 1 (LRP1)—a large multifunctional endocytic receptor implicated in a variety of physiological processes (Lillis, Van Duyn, Murphy‐Ullrich, & Strickland, 2008). Among its functions, LPR1 is involved in removal of soluble β‐Amyloid (Kang et al., 2000; Shibata et al., 2000). A further analysis confirmed that Lrp1‐AS binds to Hmgb2 (Yamanaka et al., 2015). In turn, Hmgb2 can interact with Sterol Regulatory Element‐Binding Protein 1a (Srebp1a)—a transcription factor of Lrp1 (Bown et al., 2011). As a consequence of Lrp1‐AS binding to Hmgb2, the inhibition of Srebp1a transcriptional activity occurs, and expression of Lrp1 is decreased. Intriguingly, RNA:RNA base‐pairing with Lrp1 mRNA prevents the interaction between Lrp1‐AS and Hmgb2 (Yamanaka et al., 2015).
Although in many cases the mechanism behind a NAT’s action has not been resolved, a significant portion of them represent promising molecular markers for human disorders. Above all, differentially expressed NATs are prognostic and diagnostic markers in a variety of cancers. A prominent example is HOTAIR, which is significantly overexpressed in multiple tumors including breast (Gupta et al., 2010), colorectal (Kogo et al., 2011), hepatocellular (Yang et al., 2011), and pancreatic (Kim et al., 2013) carcinomas; its upregulation is associated with poor prognosis among patients. Importantly, besides being dysregulated in a disease, the optimal markers should be stable and easily detectable in body fluids. For example, Prostate Cancer Associated 3 (PCA3) is significantly upregulated in prostate cancer (PC; Bussemakers et al., 1999). PCA3 is more specific than prostate‐specific antigen (PSA), which is commonly used to predict the risk of PC occurrence. Moreover, the PCA3 test is a noninvasive assay, allowing the measurement of PCA3 expression in patient urine samples (Bussemakers et al., 1999).
2.9. Related databases
Data from high‐throughput experiments provide more and more information about NATs that are potentially associated with development of disease or that can be used as promising molecular markers. Despite this fact, there is lack of frequently updated and well‐maintained human NAT‐oriented databases. However, a variety of information may be taken from wider data collections. For example, the basic characteristic of NATs, including genomic location, gene structure and sequence, can be established based on the Ensembl Genome Browser (Yates et al., 2016). In this database, 11,387 antisense transcripts assigned to 5,895 human genes (GRCh38.p10 human genome assembly) are annotated. According to Transcription Support Level (TSL) provided by GENECODE (Harrow et al., 2012), 965 of these are supported by at least one nonsuspect mRNA, whereas 2,088 and 3,103 are supported by multiple or single Expressed Sequence Tags (EST), respectively. Gathered together, approximately 54% of antisense transcripts from Ensembl are quite well‐supported in terms of structural models and may be considered as not speculative.
As the application of long noncoding RNAs as disease markers (including NATs) is now a field of intensive research, hundreds of related publications exist in the public domain, and this has triggered development of very specialized databases. A noteworthy example is the Lnc2Cancer (Ning et al., 2016) database that collects experimentally supported associations between human lncRNA and cancer. Importantly, the data are manually curated and include as many as 666 lncRNAs in 97 cancer types along with their functional description, expression patterns and references in the published literature. According to Ensembl annotations, at least 79 of those lncRNAs represent genes encoding antisense transcripts. As a further example, LncRNADisease (Chen et al., 2013) is a platform for searching and analyzing experimentally supported lncRNA‐disease association data from 166 diseases. This online resource also implements a bioinformatics tool designed to predict novel lncRNA‐disease associations based on the genomic context of a given lncRNA. A 120 of stored lncRNAs are annotated in ENSEMBL or NCBI as antisense transcripts. Another useful yet highly specialized database is KTCNlncDB, which focuses on lncRNAs expressed in human keratoconus and nonkeratoconus corneas (Szczesniak et al., 2017). The database contains a collection of 16,331 lncRNAs, including 2,222 NATs, obtained from the analysis of RNA‐Seq experiments conducted on 50 samples from patients with disease or from healthy controls. In addition to lncRNA characteristics, the database provides information about predicted functional lncRNA–RNA interactions. From the remaining resources for NATs‐related data, at least two additional databases deserve mentioning: the LincSNP 2.0 (Ning et al., 2017) database and the lncRNASNP (Gong, Liu, Zhang, Miao, & Guo, 2015) database. Both store data on disease‐associated single nucleotide polymorphisms (SNPs) in genes.
3. THERAPEUTIC APPLICATIONS
As discussed above, NATs employ a number of mechanisms, frequently leading to changes in the expression of their mate protein‐coding genes that are transcribed in sense orientation. Although there are reports of NATs showing concordant regulation whereby inhibiting NATs leads to decreases in levels of sense transcripts, most of the functionally described NATs appear to repress the expression of protein coding genes; this is referred to as discordant regulation. Keeping in mind that as many as 70% of mammalian genes have antisense partners (Katayama et al., 2005), inhibiting NATs could be widely applied to enhance expression of other genes (Wahlestedt, 2006), providing a potent tool to develop molecular and pharmacological strategies. In fact, this approach was first published a long time ago (Katayama et al., 2005), but only now has technology allowed successful applications, including treatment of hitherto incurable diseases. Importantly, there are several features of NATs that make them excellent targets for therapy. First, these noncoding RNAs typically act in cis, and therefore only the genes of interest are affected (Modarresi et al., 2012); this gives hope for development of highly targeted therapeutic drugs. Modulating NATs will only have an effect in cells that express the targeted RNA, leaving other cells (e.g., healthy ones) unaffected. In addition, it often appears difficult or even impossible to design a small molecule specific enough to target only a given protein, especially if similar, related proteins exist. There is also a problem with complex proteins in that it is challenging to block all key functions or only selected ones.
On the other hand, antisense transcripts produce no proteins (Khorkova, Myers, Hsiao, & Wahlestedt, 2014), leaving most classical approaches that use small molecule drugs ineffective. Therefore, to affect these RNA molecules—either their expression or processing—oligonucleotide‐based strategies have been developed. Chemically, these oligonucleotides can be divided into two categories: antisense DNA oligonucleotides (ASOs) and duplex RNAs. The two types act via different mechanisms (Figure 2). ASOs themselves are functionally and structurally diversified. Gapmers are ASOs that consist of a DNA gap flanked by chemically modified bases that increase its binding to complementary RNAs and provide higher resistance against nucleases (Deleavey & Damha, 2012; Watts & Corey, 2012). Gapmers form a heteroduplex with mate RNAs that is then recognized by RNase H1, a nonsequence‐specific endonuclease that specifically cleaves the RNA strand (Stein & Hausen, 1969) (Figure 2a). Another class of ASOs (lacking a central gap) bind to RNA molecules and block access for splicing factors, thus affecting splicing pattern of the RNA molecule (Deleavey & Damha, 2012; Watts & Corey, 2012).
Figure 2.
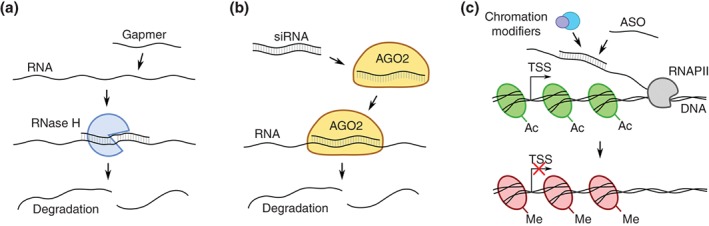
Regulation of RNA levels using antisense oligonucleotides (ASO). (a) RNA level downregulation by the ASO gapmer. The RNA:gapmer duplex is recognized by the RNase H, which leads to transcript cleavage and degradation. (b) RNA level downregulation by small interfering RNA (siRNA). The siRNA is loaded into an RNA‐induced silencing complex with argonaute‐2 protein (AGO2), which leads to target transcript cleavage and degradation within the RNAi pathway. (c) Modulation of repressive epigenetic changes by the chromatin modifiers recognizing the ASO:RNA duplex. TSS, transcription start site; Ac, H3K27ac histone modification; Me, H3K27me3 histone modification
Duplex RNAs, on the other hand, function through RNAi mechanisms, where they cleave the targeted RNA molecules much like endogenous siRNAs (Wilson & Doudna, 2013) (Figure 2b). However, alternative modes of action are possible. For example, like ASOs, they can induce chromatin modifications and modulate transcription by targeting promoter‐associated RNAs (Figure 2c). It is difficult to say whether ASOs or duplex RNAs are superior, but their choice might be in large part dictated by cellular localization of the target antisense transcripts (Lennox & Behlke, 2016). Generally, ASOs seem to be more reliable in nuclei (Lennox & Behlke, 2016), so they might be preferentially used to modulate expression of nuclear antisense RNAs, while duplex RNAs might be oligonucleotides of choice when a target is believed to function in the cytoplasm.
The best‐studied therapeutic oligonucleotides are those designed against mRNAs and miRNAs. Six of these provide clear clinical benefits and are Food and Drug Administration (FDA)‐approved for the treatment of conditions such as macular degeneration (Stein & Castanotto, 2017), and many more are currently being clinically tested (Matsui & Corey, 2017). Therefore, although still at its infancy, designing oligonucleotides targeting antisense lncRNAs can take advantage of previous experience with other classes of RNAs. However, application of NATs as targets for oligonucleotide‐based therapies has plenty of difficulties. First, even though it is generally easier to design an oligonucleotide complementary to RNA of interest than to design a small molecule targeting proteins, the oligonucleotides often cause the so‐called off‐target effect, that is, other than intended interactions occur, frequently leading to adverse phenotypic effects. The off‐target effect can be provoked in several ways. First, oligonucleotides might bind to proteins and induce a nonspecific reaction such as the interferon response (Krieg, 2012). They can also show partial complementarity to unintended targets, especially given that even partially complementary nucleic acids can interact. There are also toxicological challenges including proinflammatory effects, nephrotoxicity, thrombocytopenia, and hepatotoxicity (Frazier, 2015). These and other adverse effects have been previously reviewed and discussed (Engelhardt et al., 2015; Frazier, 2015). There are, however, ways to minimize the consequences of the off‐target effect such as designing the antisense drugs while considering a detailed bioinformatics analysis for identification of potential targets showing both full and partial complementarity (Bennett & Swayze, 2010). Additionally, it is generally advisable to test multiple oligonucleotides per target gene and consider alternative hypotheses to explain the observed results (Matsui & Corey, 2017).
The mechanisms of action for antisense lncRNAs are often obscure. This can raise problems, as in the case of Genasence, an oligonucleotide‐based drug that initially yielded promising results. Clinical trials showed almost no beneficial effect in patients (Bedikian, Garbe, Conry, Lebbe, & Grob, 2014; Lai et al., 2003). It was later demonstrated that the observations from initial studies were most likely triggered by sequence‐independent processes and resulted from the off‐target effect (Lai et al., 2003; Winkler, Stessl, Amartey, & Noe, 2010). Another example involves attempts to restore the correct splicing pattern in an IDS gene associated with mucopolysaccharidosis II. Oligonucleotides expected to restore the right splicing pattern triggered production of yet another aberrant splice form. It later appeared that the oligonucleotides used were masking a cis‐acting element essential for 5′ splice site recognition, once again highlighting the importance of functional studies aimed at understanding the mechanism of action for both oligonucleotides and the NAT before attempting clinical trials. Similarly, it should be resolved whether the NATs of interest act in the nucleus or in the cytoplasm; this is associated with both proper understanding of the underlying mode of action and choice of the targeting strategy (Lennox & Behlke, 2016).
Last but not least, one of the most serious obstacles en route to the clinical translation of oligonucleotide‐based approaches is their relatively poor delivery to target organs and tissues after systemic delivery. It is assumed that on average < 1% of oligonucleotides reach the correct cellular compartment (Godfrey et al., 2017), and further progress is dependent on better understanding of the mechanisms underlying cellular uptake, transport and metabolism. Increasing the intravenously administered doses in order to deliver sufficient amounts of therapeutic oligonucleotides to targeted cells is restricted by their associated toxicity. Furthermore, their circulation is restricted due to the natural tissue barriers such as the blood–brain barrier: direct delivery to the central nervous system is the most commonly used method and is achieved through intracerebroventricular or intrathecal injection. However, this kind of therapy is relatively expensive and requires specialist expertise as well as hospital visits.
Despite the abovementioned barriers to developing NAT‐based therapies, substantial progress has been achieved in recent years—first due to advancements in chemistry and technology, and second thanks to our better understanding of the biology behind antisense transcription and its RNA products. As a result, several successful research projects have been performed, and some therapies have reached clinical phases. Selected cases are now being thoroughly investigated and augur efficient therapies for thus far incurable conditions. Some examples are described below.
3.1. UBE3A‐ATS: Angelman syndrome
Oligonucleotide strategies to activate gene expression—by means of targeting the inhibitory NATs—could be beneficial in the treatment of diseases arising from haploinsufficiency such as Angelman syndrome. Angelman syndrome is a neurodevelopmental disorder with no known curative treatments. It results from lack of function of the maternal gene UBE3A (encoding E3 Ubiquitin Protein Ligase) as a consequence of a mutation, usually a deletion, that affects this gene. As the paternal version is genetically imprinted, that is, silenced in cis by its antisense transcript, no active copy of the gene remains (Meng et al., 2012; Rougeulle, Cardoso, Fontes, Colleaux, & Lalande, 1998). Inactivation of the antisense transcript should therefore lead to derepression of the parental gene and compensate for the lack of expression from the maternal allele. Indeed, in mice the reduction of antisense RNA (Ube3a‐ATS) expression with two ASO gapmers triggers substantial increase of paternal allele expression (Ube3a) (Meng et al., 2015). Administration of ASOs partially reversed some cognitive defects in mice. Importantly, the expression of other genes expressed from the same locus (Snrpn, Snord115, and Snord116) was not affected.
3.2. APOA1‐AS: cholesterol metabolism
APOA1, the major protein component of HDL in plasma (Barbaras, Puchois, Fruchart, & Ailhaud, 1987), has an antisense transcript, APOA1‐AS, that acts as a negative transcriptional regulator of APOA1 both in vitro and in vivo (Halley et al., 2014). As a consequence, inhibition of APOA1‐AS in cultured cells results in the increased expression of APOA1. Chromatin immunoprecipitation analyses indicate that APOA1‐AS can modulate histone methylation patterns that mark active and/or inactive gene expression (Figure 2c). Finally, targeting APOA1‐AS using ASOs also leads to an increase in APOA1 expression in human and monkey liver cells. Altogether, these findings promise a strategy to control cholesterol concentration. This is especially important in that low HDL cholesterol concentration reflects increased susceptibility to cardiovascular diseases, and raising HDL pharmacologically remains a proposed strategy to reduce the occurrence of cardiovascular diseases (Rader, 2002).
3.3. SCN1ANAT: Dravet syndrome
Another example of discordant regulation is SCN1A. Depleting its antisense transcript (SCN1ANAT, also known as AC010127.3) or blocking the antisense transcript’s interactions with epigenetic proteins by means of oligonucleotides leads to upregulation of this protein‐coding gene (Li & Rana, 2014; Stein et al., 2010). It was demonstrated that ASOs are able to partially reverse the disease phenotype in a mouse model of Dravet syndrome (Kaboli, Rahmat, Ismail, & Ling, 2015).
3.4. BDNF‐AS: neurodegenerative disorders
BDNF (brain‐derived neurotrophic factor) belongs to a class of neurotrophins that function as growth factors and are essential in different aspects of neuronal biology, including their growth, differentiation and maintenance (Hasbi et al., 2009; Yoshimura, Ito, & Endo, 2009). Some psychiatric and neurodegenerative disorders are associated with impaired expression levels of neurotrophins (Dell'Osso et al., 2010; Frade & Lopez‐Sanchez, 2010; Laske et al., 2011). Therefore, their upregulation is expected to have beneficial effects in at least some conditions. BDNF has its antisense partner (BDNF‐AS or BDNF‐OS), downregulating expression levels of BDNF in vivo and in vitro as in the three cases described above. Additionally, upregulation of BDNF has been achieved in a locus‐specific manner by inhibiting BDNF‐AS transcripts with ASOs (Modarresi et al., 2012) (Figure 2a).
3.5. BACE1‐AS: Alzheimer’s disease
The Beta‐Site Cleavage Enzyme 1 (BACE1) and its antisense transcript (BACE1‐AS) are examples of concordant regulation whereby the NAT stabilizes BACE1 by masking microRNA binding sites (Figure 1d). Both BACE1 and BACE1‐AS expression are elevated in patients with Alzheimer’s disease (AD). BACE1 plays a rate‐limiting role in production of β‐Amyloid, a peptide fragment that is pathogenic in AD, making it an attractive target for pharmaceutical companies. Unfortunately, blocking BACE1 activity with conventional drugs leads to adverse effects that are most likely associated with other molecular functions of the enzyme (Willem, Lammich, & Haass, 2009). It was reported, however, that infusion of siRNAs against BACE1‐AS into mouse brains leads to decreased levels of BACE1‐AS and its sense partner BACE1 at mRNA and protein levels (Faghihi et al., 2008) (Figure 2b). As a result, reduced soluble levels and aggregation of β‐amyloid were observed in vivo. Therefore, indirect reduction of BACE1 cellular levels by means of targeting its antisense transcript, rather than switching the gene completely off, represents an opportunity to pharmacologically control AD‐associated aggregation of β‐Amyloid when traditional approaches are not effective.
In spite of the fact that oligonucleotide‐based therapies, with all their pros and cons, seem to be the most promising and clinically advanced approach for treatment of NAT‐associated diseases, other strategies are possible as well, in particular a CRISPR‐Cas9 system (clustered regularly interspaced short palindromic repeats‐associated nuclease 9). For example, recent reports have shown that CRISPR‐Cas9 components are able to in vivo correct a gene mutation in mouse models of Duchenne muscular dystrophy and rescue the disease phenotype (Long et al., 2016; Nelson et al., 2016; Tabebordbar et al., 2016). Although this is a significant step forward on the way to bring CRISPR‐Cas9 to the clinic, there are several major difficulties there, such as off target effects, poor fitness of edited cells or immunogenicity of CRISPR‐Cas9 components (reviewed in Dai et al., 2016). Application of the system in the area of antisense RNAs might be even more challenging than that, as cis‐NATs share genomic regions with their mate genes, making it harder to avoid the off target effect.
4. CONCLUSION
NATs are now recognized as an essential component of the human transcriptome, though we are only beginning to explore their biological relevance and their regulatory mechanisms. The fact that they are required for metabolism of healthy cells as well as blamed for causing human diseases opens up the possibility of using them in medical research and clinical applications. First, they can serve as biomarkers for diseases, especially since—as opposed to mRNAs—they are functional at the RNA level; hence their expression is expected to be a better indicator of disease than protein‐coding transcripts. Additionally, they represent promising targets for new therapeutic strategies. Successful manipulation of NATs and other long noncoding RNAs in vitro and in vivo with oligonucleotide technology triggered the interest of pharmaceutical companies such as OPCO (http://www.opko.com/), which is developing a CURNA platform utilizing antagoNATs (ASOs against NAT transcripts) to suppress the activity of disease‐causing NATs. The company is currently screening over 400 gene targets and is conducting preclinical studies for proteins associated mostly with orphan diseases, that is, conditions affecting less than 200,000 American citizens (Orphan Drug Act, FDA) including Dravet syndrome, Rett syndrome, and mucopolysaccharidosis I. It therefore appears imminent that in spite of the abovementioned technical impediments, NATs will become targets for ncRNA‐based strategies against diverse human conditions including cancer, cardiovascular, neurological, and muscular diseases.
CONFLICT OF INTEREST
The authors have declared no conflicts of interest for this article.
RELATED WIREs ARTICLES
Targeting RNA splicing for disease therapy
Long noncoding RNAs in mammalian cells: What, where, and why?
Chromatin‐associated noncoding RNAs in development and inheritance
Current progress on microRNAs‐based therapeutics in neurodegenerative diseases
Modulating splicing with small molecular inhibitors of the spliceosome
ACKNOWLEDGMENTS
This work is supported by the Polish Ministry of Science and Higher Education (decision No. 1268/MOB/IV/2015/0 to M.W.S.—Mobility Plus project), the National Science Centre (grant No. 2014/15/D/NZ2/00525 to M.W.S. and grant No. 2013/11/N/NZ2/02524 to W.R.), and the KNOW Poznan RNA Centre (grant No. 01/KNOW2/2014). A part of the graphical abstract’s figure came from http://www.freepik.com.
Wanowska E, Kubiak MR, Rosikiewicz W, Makałowska I, Szcześniak MW. Natural antisense transcripts in diseases: From modes of action to targeted therapies. WIREs RNA. 2018;9:e1461. https://doi.org/10.1002/wrna.1461
Funding information KNOW Poznan RNA Centre, Grant/Award number: 01/KNOW2/2014; National Science Centre, Grant/Award numbers: 2013/11/N/NZ2/02524, 2014/15/D/NZ2/00525; Ministry of Science and Higher Education
References
REFERENCES
- Bannon, M. J. , Savonen, C. L. , Jia, H. , Dachet, F. , Halter, S. D. , Schmidt, C. J. , … Kapatos, G. (2015). Identification of long noncoding RNAs dysregulated in the midbrain of human cocaine abusers. Journal of Neurochemistry, 135, 50–59. https://doi.org/10.1111/jnc.13255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbaras, R. , Puchois, P. , Fruchart, J. C. , & Ailhaud, G. (1987). Cholesterol efflux from cultured adipose cells is mediated by LpAI particles but not by LpAI:AII particles. Biochemical and Biophysical Research Communications, 142, 63–69. https://doi.org/0006-291X(87)90451-7 [DOI] [PubMed] [Google Scholar]
- Barlow, D. P. , & Bartolomei, M. S. (2014). Genomic imprinting in mammals. Cold Spring Harb Perspect Biol, 6, a01838 https://doi.org/10.1101/cshperspect.a018382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedikian, A. Y. , Garbe, C. , Conry, R. , Lebbe, C. , & Grob, J. J. (2014). Dacarbazine with or without oblimersen (a Bcl‐2 antisense oligonucleotide) in chemotherapy‐naive patients with advanced melanoma and low‐normal serum lactate dehydrogenase: 'The AGENDA trial'. Melanoma Research, 24, 237–243. https://doi.org/10.1097/CMR.0000000000000056 [DOI] [PubMed] [Google Scholar]
- Beltran, M. , Puig, I. , Pena, C. , Garcia, J. M. , Alvarez, A. B. , Pena, R. , … de Herreros, A. G. (2008). A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1‐induced epithelial‐mesenchymal transition. Genes & Development, 22, 756–769. https://doi.org/10.1101/gad.455708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, C. F. , & Swayze, E. E. (2010). RNA targeting therapeutics: Molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annual Review of Pharmacology and Toxicology, 50, 259–293. https://doi.org/10.1146/annurev.pharmtox.010909.105654 [DOI] [PubMed] [Google Scholar]
- Borden, L. A. , Dhar, T. G. , Smith, K. E. , Branchek, T. A. , Gluchowski, C. , & Weinshank, R. L. (1994). Cloning of the human homologue of the GABA transporter GAT‐3 and identification of a novel inhibitor with selectivity for this site. Receptors & Channels, 2, 207–213. [PubMed] [Google Scholar]
- Bown, M. J. , Jones, G. T. , Harrison, S. C. , Wright, B. J. , Bumpstead, S. , Baas, A. F. , … Samani, N. J. (2011). Abdominal aortic aneurysm is associated with a variant in low‐density lipoprotein receptor‐related protein 1. American Journal of Human Genetics, 89, 619–627. https://doi.org/10.1016/j.ajhg.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryzghalov, O. , Szczesniak, M. W. , & Makalowska, I. (2016). Retroposition as a source of antisense long non‐coding RNAs with possible regulatory functions. Acta Biochimica Polonica, 63, 825–833. https://doi.org/10.18388/abp.2016_1354 [DOI] [PubMed] [Google Scholar]
- Bussemakers, M. J. , van Bokhoven, A. , Verhaegh, G. W. , Smit, F. P. , Karthaus, H. F. , Schalken, J. A. , … Isaacs, W. B. (1999). DD3: A new prostate‐specific gene, highly overexpressed in prostate cancer. Cancer Research, 59, 5975–5979. [PubMed] [Google Scholar]
- Cao, R. , Wang, L. , Wang, H. , Xia, L. , Erdjument‐Bromage, H. , Tempst, P. , … Zhang, Y. (2002). Role of histone H3 lysine 27 methylation in Polycomb‐group silencing. Science, 298, 1039–1043. https://doi.org/1076997 [DOI] [PubMed] [Google Scholar]
- Casciello, F. , Windloch, K. , Gannon, F. , & Lee, J. S. (2015). Functional role of G9a histone methyltransferase in cancer. Frontiers in Immunology, 6(487). https://doi.org/10.3389/fimmu.2015.00487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary, T. , Cleary, M. A. , Baker, C. C. , Guan, X. J. , & Tilghman, S. M. (1998). Multiple mechanisms regulate imprinting of the mouse distal chromosome 7 gene cluster. Molecular and Cellular Biology, 18, 3466–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain, S. J. , & Brannan, C. I. (2001). The Prader‐Willi syndrome imprinting center activates the paternally expressed murine Ube3a antisense transcript but represses paternal Ube3a. Genomics, 73, 316–322. https://doi.org/S0888-7543(01)96543-7 [DOI] [PubMed] [Google Scholar]
- Chamberlain, S. J. , & Lalande, M. (2010). Angelman syndrome, a genomic imprinting disorder of the brain. The Journal of Neuroscience, 30, 9958–9963. https://doi.org/10.1523/JNEUROSCI.1728-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, G. , Wang, Z. , Wang, D. , Qiu, C. , Liu, M. , Chen, X. , … Cui, Q. (2013). LncRNADisease: A database for long‐non‐coding RNA‐associated diseases. Nucleic Acids Research, 41, D983–D986. https://doi.org/10.1093/nar/gks1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M. , & Manley, J. L. (2009). Mechanisms of alternative splicing regulation: Insights from molecular and genomics approaches. Nature Reviews. Molecular Cell Biology, 10, 741–754. https://doi.org/10.1038/nrm2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, D. W. , Rudnicki, D. D. , Yu, L. , & Margolis, R. L. (2011). A natural antisense transcript at the Huntington's disease repeat locus regulates HTT expression. Human Molecular Genetics, 20, 3467–3477. https://doi.org/10.1093/hmg/ddr263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce, C. M. (2010). LINCing chromatin remodeling to metastasis. Nature Biotechnology, 28, 931–932. https://doi.org/10.1038/nbt0910-931 [DOI] [PubMed] [Google Scholar]
- Dai, W. J. , Zhu, L. Y. , Yan, Z. Y. , Xu, Y. , Wang, Q. L. , & Lu, X. J . (2016). CRISPR‐Cas9 for in vivo gene therapy: Promise and hurdles. Molecular Therapy–Nucleic Acids, 5, e349 https://doi.org/10.1038/mtna.2016.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard, C. K. , Tange, T. O. , & Kjems, J. (2002). hnRNP A1 controls HIV‐1 mRNA splicing through cooperative binding to intron and exon splicing silencers in the context of a conserved secondary structure. RNA, 8, 1401–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughters, R. S. , Tuttle, D. L. , Gao, W. , Ikeda, Y. , Moseley, M. L. , Ebner, T. J. , … Ranum, L. P. (2009). RNA gain‐of‐function in spinocerebellar ataxia type 8. PLoS Genetics, 5, e1000600 https://doi.org/10.1371/journal.pgen.1000600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaval, K. , & Feil, R. (2004). Epigenetic regulation of mammalian genomic imprinting. Current Opinion in Genetics & Development, 14, 188–195. https://doi.org/S0959437X04000206 [DOI] [PubMed] [Google Scholar]
- Deleavey, G. F. , & Damha, M. J. (2012). Designing chemically modified oligonucleotides for targeted gene silencing. Chemistry & Biology, 19, 937–954. https://doi.org/10.1016/j.chembiol.2012.07.011 [DOI] [PubMed] [Google Scholar]
- Dell'Osso, L. , Del Debbio, A. , Veltri, A. , Bianchi, C. , Roncaglia, I. , Carlini, M. , … Piccinni, A. (2010). Associations between brain‐derived neurotrophic factor plasma levels and severity of the illness, recurrence and symptoms in depressed patients. Neuropsychobiology, 62, 207–212. https://doi.org/10.1159/000319946 [DOI] [PubMed] [Google Scholar]
- Engelhardt, J. A. , Fant, P. , Guionaud, S. , Henry, S. P. , Leach, M. W. , Louden, C. , … Frazier, K. S. (2015). Scientific and regulatory policy committee points‐to‐consider paper*: Drug‐induced vascular injury associated with nonsmall molecule therapeutics in preclinical development: Part 2. Antisense oligonucleotides. Toxicologic Pathology, 43, 935–944. https://doi.org/10.1177/0192623315570341 [DOI] [PubMed] [Google Scholar]
- Faghihi, M. A. , Kocerha, J. , Modarresi, F. , Engstrom, P. G. , Chalk, A. M. , Brothers, S. P. , … Wahlestedt, C. (2010). RNAi screen indicates widespread biological function for human natural antisense transcripts. PLoS ONE, 5, e13177 https://doi.org/10.1371/journal.pone.0013177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi, M. A. , Modarresi, F. , Khalil, A. M. , Wood, D. E. , Sahagan, B. G. , Morgan, T. E. , … Wahlestedt, C. (2008). Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed‐forward regulation of beta‐secretase. Nature Medicine, 14, 723–730. https://doi.org/10.1038/nm1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi, M. A. , & Wahlestedt, C. (2009). Regulatory roles of natural antisense transcripts. Nature Reviews. Molecular Cell Biology, 10, 637–643. https://doi.org/10.1038/nrm2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi, M. A. , Zhang, M. , Huang, J. , Modarresi, F. , Van der Brug, M. P. , Nalls, M. A. , … Wahlestedt, C. (2010). Evidence for natural antisense transcript‐mediated inhibition of microRNA function. Genome Biology, 11, R56 https://doi.org/10.1186/gb-2010-11-5-r56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, J. , Yang, Y. , Zhang, P. , Wang, F. , Ma, Y. , Qin, H. , & Wang, Y. (2014). miR‐150 functions as a tumour suppressor in human colorectal cancer by targeting c‐Myb. Journal of Cellular and Molecular Medicine, 18, 2125–2134. https://doi.org/10.1111/jcmm.12398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frade, J. M. , & Lopez‐Sanchez, N. (2010). A novel hypothesis for Alzheimer disease based on neuronal tetraploidy induced by p75 (NTR). Cell Cycle, 9, 1934–1941. https://doi.org/10.4161/cc.9.10.11582 [DOI] [PubMed] [Google Scholar]
- Frazier, K. S. (2015). Antisense oligonucleotide therapies: The promise and the challenges from a toxicologic pathologist's perspective. Toxicologic Pathology, 43, 78–89. https://doi.org/10.1177/0192623314551840 [DOI] [PubMed] [Google Scholar]
- Fukumoto, H. , Cheung, B. S. , Hyman, B. T. , & Irizarry, M. C. (2002). Beta‐secretase protein and activity are increased in the neocortex in Alzheimer disease. Archives of Neurology, 59, 1381–1389. https://doi.org/noc20210 [DOI] [PubMed] [Google Scholar]
- Geng, Y. , Xiong, D. , Mosyak, L. , Malito, D. L. , Kniazeff, J. , Chen, Y. , … Fan, Q. R. (2012). Structure and functional interaction of the extracellular domain of human GABA(B) receptor GBR2. Nature Neuroscience, 15, 970–978. https://doi.org/10.1038/nn.3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey, C. , Desviat, L. R. , Smedsrod, B. , Pietri‐Rouxel, F. , Denti, M. A. , Disterer, P. , … Arechavala‐Gomeza, V. (2017). Delivery is key: Lessons learnt from developing splice‐switching antisense therapies. EMBO Molecular Medicine, 9, 545–557. https://doi.org/10.15252/emmm.201607199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, J. , Liu, W. , Zhang, J. , Miao, X. , & Guo, A. Y. (2015). lncRNASNP: A database of SNPs in lncRNAs and their potential functions in human and mouse. Nucleic Acids Research, 43, D181–D186. https://doi.org/10.1093/nar/gku1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grander, D. , & Johnsson, P. (2016). Pseudogene‐expressed RNAs: Emerging roles in gene regulation and disease. Current Topics in Microbiology and Immunology, 394, 111–126. https://doi.org/10.1007/82_2015_442 [DOI] [PubMed] [Google Scholar]
- Greer, P. L. , Hanayama, R. , Bloodgood, B. L. , Mardinly, A. R. , Lipton, D. M. , Flavell, S. W. , … Greenberg, M. E. (2010). The Angelman syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell, 140, 704–716. https://doi.org/10.1016/j.cell.2010.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, R. A. , Shah, N. , Wang, K. C. , Kim, J. , Horlings, H. M. , Wong, D. J. , … Chang, H. Y. (2010). Long non‐coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature, 464, 1071–1076. https://doi.org/10.1038/nature08975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman, M. , Donaghey, J. , Carey, B. W. , Garber, M. , Grenier, J. K. , Munson, G. , … Lander, E. S. (2011). lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature, 477, 295–300. https://doi.org/10.1038/nature10398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halley, P. , Kadakkuzha, B. M. , Faghihi, M. A. , Magistri, M. , Zeier, Z. , Khorkova, O. , … Wahlestedt, C. (2014). Regulation of the apolipoprotein gene cluster by a long noncoding RNA. Cell Reports, 6, 222–230. https://doi.org/10.1016/j.celrep.2013.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow, J. , Frankish, A. , Gonzalez, J. M. , Tapanari, E. , Diekhans, M. , Kokocinski, F. , … Hubbard, T. J. (2012). GENCODE: The reference human genome annotation for the ENCODE project. Genome Research, 22, 1760–1774. https://doi.org/10.1101/gr.135350.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasbi, A. , Fan, T. , Alijaniaram, M. , Nguyen, T. , Perreault, M. L. , O'Dowd, B. F. , & George, S. R. (2009). Calcium signaling cascade links dopamine D1‐D2 receptor heteromer to striatal BDNF production and neuronal growth. Proceedings of the National Academy of Sciences of the United States of America, 106, 21377–21382. https://doi.org/10.1073/pnas.0903676106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y. , Vogelstein, B. , Velculescu, V. E. , Papadopoulos, N. , & Kinzler, K. W. (2008). The antisense transcriptomes of human cells. Science, 322, 1855–1857. https://doi.org/10.1126/science.1163853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsinger, R. M. , McLean, C. A. , Beyreuther, K. , Masters, C. L. , & Evin, G. (2002). Increased expression of the amyloid precursor beta‐secretase in Alzheimer's disease. Annals of Neurology, 51(6), 783–786. https://doi.org/10.1002/ana.10208 [DOI] [PubMed] [Google Scholar]
- Hu, S. , Wang, X. , & Shan, G. (2016). Insertion of an Alu element in a lncRNA leads to primate‐specific modulation of alternative splicing. Nature Structural & Molecular Biology, 23, 1011–1019. https://doi.org/10.1038/nsmb.3302 [DOI] [PubMed] [Google Scholar]
- Huang, M. D. , Chen, W. M. , Qi, F. Z. , Xia, R. , Sun, M. , TP, X. , … Shu, Y. Q. (2015). Long non‐coding RNA ANRIL is upregulated in hepatocellular carcinoma and regulates cell apoptosis by epigenetic silencing of KLF2. Journal of Hematology & Oncology, 8(50). https://doi.org/10.1186/s13045-015-0146-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson, P. , Ackley, A. , Vidarsdottir, L. , Lui, W. O. , Corcoran, M. , Grander, D. , & Morris, K. V. (2013). A pseudogene long‐noncoding‐RNA network regulates PTEN transcription and translation in human cells. Nature Structural & Molecular Biology, 20, 440–446. https://doi.org/10.1038/nsmb.2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone, K. A. , DuBose, A. J. , Futtner, C. R. , Elmore, M. D. , Brannan, C. I. , & Resnick, J. L. (2006). A human imprinting centre demonstrates conserved acquisition but diverged maintenance of imprinting in a mouse model for Angelman syndrome imprinting defects. Human Molecular Genetics, 15, 393–404. https://doi.org/10.1093/hmg/ddi456 [DOI] [PubMed] [Google Scholar]
- Kaboli, P. J. , Rahmat, A. , Ismail, P. , & Ling, K. H. (2015). MicroRNA‐based therapy and breast cancer: A comprehensive review of novel therapeutic strategies from diagnosis to treatment. Pharmacological Research, 97, 104–121. https://doi.org/10.1016/j.phrs.2015.04.015 [DOI] [PubMed] [Google Scholar]
- Kabza, M. , Ciomborowska, J. , & Makalowska, I. (2014). RetrogeneDB‐‐a database of animal retrogenes. Molecular Biology and Evolution, 31, 1646–1648. https://doi.org/10.1093/molbev/msu139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaikkonen, M. U. , Lam, M. T. , & Glass, C. K. (2011). Non‐coding RNAs as regulators of gene expression and epigenetics. Cardiovascular Research, 90, 430–440. https://doi.org/10.1093/cvr/cvr097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, D. E. , Pietrzik, C. U. , Baum, L. , Chevallier, N. , Merriam, D. E. , Kounnas, M. Z. , … Koo, E. H. (2000). Modulation of amyloid beta‐protein clearance and Alzheimer's disease susceptibility by the LDL receptor‐related protein pathway. The Journal of Clinical Investigation, 106, 1159–1166. https://doi.org/10.1172/JCI11013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor, B. , Makedonski, K. , Green‐Finberg, Y. , Shemer, R. , & Razin, A. (2004). Control elements within the PWS/AS imprinting box and their function in the imprinting process. Human Molecular Genetics, 13, 751–762. https://doi.org/ddh085 [DOI] [PubMed] [Google Scholar]
- Katayama, S. , Tomaru, Y. , Kasukawa, T. , Waki, K. , Nakanishi, M. , Nakamura, M. , … FANTOM Consortium (2005). Antisense transcription in the mammalian transcriptome. Science, 309, 1564–1566. https://doi.org/10.1126/science.1112009 [DOI] [PubMed] [Google Scholar]
- Khorkova, O. , Myers, A. J. , Hsiao, J. , & Wahlestedt, C. (2014). Natural antisense transcripts. Human Molecular Genetics, 23, R54–R63. https://doi.org/10.1093/hmg/ddu207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K. , Jutooru, I. , Chadalapaka, G. , Johnson, G. , Frank, J. , Burghardt, R. , … Safe, S. (2013). HOTAIR is a negative prognostic factor and exhibits pro‐oncogenic activity in pancreatic cancer. Oncogene, 32, 1616–1625. https://doi.org/10.1038/onc.2012.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, V. N. , Han, J. , & Siomi, M. C. (2009). Biogenesis of small RNAs in animals. Nature Reviews. Molecular Cell Biology, 10, 126–139. https://doi.org/10.1038/nrm2632 [DOI] [PubMed] [Google Scholar]
- Kishino, T. , Lalande, M. , & Wagstaff, J. (1997). UBE3A/E6‐AP mutations cause Angelman syndrome. Nature Genetics, 15, 70–73. https://doi.org/10.1038/ng0197-70 [DOI] [PubMed] [Google Scholar]
- Kogo, R. , Shimamura, T. , Mimori, K. , Kawahara, K. , Imoto, S. , Sudo, T. , … Mori, M. (2011). Long noncoding RNA HOTAIR regulates polycomb‐dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Research, 71, 6320–6326. https://doi.org/10.1158/0008-5472.CAN-11-1021 [DOI] [PubMed] [Google Scholar]
- Krieg, A. M. (2012). CpG still rocks! Update on an accidental drug. Nucleic Acid Therapeutics, 22, 77–89. https://doi.org/10.1089/nat.2012.0340 [DOI] [PubMed] [Google Scholar]
- Kumar, M. , & Carmichael, G. G. (1997). Nuclear antisense RNA induces extensive adenosine modifications and nuclear retention of target transcripts. Proceedings of the National Academy of Sciences of the United States of America, 94, 3542–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung, J. T. , Colognori, D. , & Lee, J. T. (2013). Long noncoding RNAs: Past, present, and future. Genetics, 193, 651–669. https://doi.org/10.1534/genetics.112.146704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, J. C. , Benimetskaya, L. , Santella, R. M. , Wang, Q. , Miller, P. S. , & Stein, C. A. (2003). G3139 (oblimersen) may inhibit prostate cancer cell growth in a partially bis‐CpG‐dependent non‐antisense manner. Molecular Cancer Therapeutics, 2, 1031–1043. [PubMed] [Google Scholar]
- Lander, E. S. , Linton, L. M. , Birren, B. , Nusbaum, C. , Zody, M. C. , Baldwin, J. , … International Human Genome Sequencing Consortium (2001). Initial sequencing and analysis of the human genome. Nature, 409, 860–921. https://doi.org/10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Laske, C. , Stellos, K. , Hoffmann, N. , Stransky, E. , Straten, G. , Eschweiler, G. W. , & Leyhe, T. (2011). Higher BDNF serum levels predict slower cognitive decline in Alzheimer's disease patients. The International Journal of Neuropsychopharmacology, 14, 399–404. https://doi.org/10.1017/S1461145710001008 [DOI] [PubMed] [Google Scholar]
- Lee, M. P. , DeBaun, M. R. , Mitsuya, K. , Galonek, H. L. , Brandenburg, S. , Oshimura, M. , & Feinberg, A. P. (1999). Loss of imprinting of a paternally expressed transcript, with antisense orientation to KVLQT1, occurs frequently in Beckwith‐Wiedemann syndrome and is independent of insulin‐like growth factor II imprinting. Proceedings of the National Academy of Sciences of the United States of America, 96, 5203–5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner, B. , Williams, G. , Campbell, R. D. , & Sanderson, C. M. (2002). Antisense transcripts in the human genome. Trends in Genetics, 18, 63–65. https://doi.org/S0168-9525(02)02598-2 [DOI] [PubMed] [Google Scholar]
- Lennox, K. A. , & Behlke, M. A. (2016). Cellular localization of long non‐coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Research, 44, 863–877. https://doi.org/10.1093/nar/gkv1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T. , Xie, J. , Shen, C. , Cheng, D. , Shi, Y. , Wu, Z. , … Zhu, Z. (2015). Amplification of long noncoding RNA ZFAS1 promotes metastasis in hepatocellular carcinoma. Cancer Research, 75, 3181–3191. https://doi.org/10.1158/0008-5472.CAN-14-3721 [DOI] [PubMed] [Google Scholar]
- Li, Z. , & Rana, T. M. (2014). Therapeutic targeting of microRNAs: Current status and future challenges. Nature Reviews. Drug Discovery, 13, 622–638. https://doi.org/10.1038/nrd4359 [DOI] [PubMed] [Google Scholar]
- Lillis, A. P. , Van Duyn, L. B. , Murphy‐Ullrich, J. E. , & Strickland, D. K. (2008). LDL receptor‐related protein 1: Unique tissue‐specific functions revealed by selective gene knockout studies. Physiological Reviews, 88, 887–918. https://doi.org/10.1152/physrev.00033.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L. , Chen, X. , Zhang, Y. , Hu, Y. , Shen, X. , & Zhu, W. (2017). Long non‐coding RNA TUG1 promotes endometrial cancer development via inhibiting miR‐299 and miR‐34a‐5p. Oncotarget, 8, 31386–31394. https://doi.org/10.18632/oncotarget.15607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, C. , Amoasii, L. , Mireault, A. A. , McAnally, J. R. , Li, H. , Sanchez‐Ortiz, E. , … Olson, E. N. (2016). Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science, 351, 400–403. https://doi.org/10.1126/science.aad5725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, B. , Li, M. , Zhang, L. , Huang, M. , Lei, J. B. , GH, F. , … Wang, Y. L. (2016). Upregulation of long non‐coding RNA TUG1 correlates with poor prognosis and disease status in osteosarcoma. Tumour Biology, 37, 4445–4455. https://doi.org/10.1007/s13277-015-4301-6 [DOI] [PubMed] [Google Scholar]
- Massone, S. , Vassallo, I. , Fiorino, G. , Castelnuovo, M. , Barbieri, F. , Borghi, R. , … Pagano, A. (2011). 17A, a novel non‐coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiology of Disease, 41, 308–317. https://doi.org/10.1016/j.nbd.2010.09.019 [DOI] [PubMed] [Google Scholar]
- Matsui, M. , & Corey, D. R. (2017). Non‐coding RNAs as drug targets. Nature Reviews. Drug Discovery, 16, 167–179. https://doi.org/10.1038/nrd.2016.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura, T. , Sutcliffe, J. S. , Fang, P. , Galjaard, R. J. , Jiang, Y. H. , Benton, C. S. , … Beaudet, A. L. (1997). De novo truncating mutations in E6‐AP ubiquitin‐protein ligase gene (UBE3A) in Angelman syndrome. Nature Genetics, 15, 74–77. https://doi.org/10.1038/ng0197-74 [DOI] [PubMed] [Google Scholar]
- Mayeda, A. , Munroe, S. H. , Caceres, J. F. , & Krainer, A. R. (1994). Function of conserved domains of hnRNP A1 and other hnRNP a/B proteins. The EMBO Journal, 13, 5483–5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, L. , Person, R. E. , & Beaudet, A. L. (2012). Ube3a‐ATS is an atypical RNA polymerase II transcript that represses the paternal expression of Ube3a. Human Molecular Genetics, 21, 3001–3012. https://doi.org/10.1093/hmg/dds130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, L. , Ward, A. J. , Chun, S. , Bennett, C. F. , Beaudet, A. L. , & Rigo, F. (2015). Towards a therapy for Angelman syndrome by targeting a long non‐coding RNA. Nature, 518, 409–412. https://doi.org/10.1038/nature13975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modarresi, F. , Faghihi, M. A. , Lopez‐Toledano, M. A. , Fatemi, R. P. , Magistri, M. , Brothers, S. P. , … Wahlestedt, C. (2012). Inhibition of natural antisense transcripts in vivo results in gene‐specific transcriptional upregulation. Nature Biotechnology, 30, 453–459. https://doi.org/10.1038/nbt.2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli, C. , Magnanini, C. , Mungall, A. J. , Negrini, M. , & Barbanti‐Brodano, G. (2000). Cloning and characterization of two overlapping genes in a subregion at 6q21 involved in replicative senescence and schizophrenia. Gene, 252, 217–225. https://doi.org/S0378-1119(00)00231-6 [DOI] [PubMed] [Google Scholar]
- Moseley, M. L. , Zu, T. , Ikeda, Y. , Gao, W. , Mosemiller, A. K. , Daughters, R. S. , … Ranum, L. P. W. (2006). Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nature Genetics, 38, 758–769. https://doi.org/10.1038/ng1827 [DOI] [PubMed] [Google Scholar]
- Muncie Jr., H. L. , & Campbell, J. (2009). Alpha and beta thalassemia. American Family Physician, 80, 339–344. [PubMed] [Google Scholar]
- Nakano, S. , Murakami, K. , Meguro, M. , Soejima, H. , Higashimoto, K. , Urano, T. , … Oshimura, M. (2006). Expression profile of LIT1/KCNQ1OT1 and epigenetic status at the KvDMR1 in colorectal cancers. Cancer Science, 97, 1147–1154. https://doi.org/10.1111/j.1349-7006.2006.00305.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli, S. , Piccinelli, V. , Mapelli, S. N. , Pisignano, G. , & Catapano, C. V. (2017). Natural antisense transcripts drive a regulatory cascade controlling c‐MYC transcription. RNA Biology, 14 (12), 1742–1755. https://doi.org/10.1080/15476286.2017.1356564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, C. E. , Hakim, C. H. , Ousterout, D. G. , Thakore, P. I. , Moreb, E. A. , Castellanos Rivera, R. M. , … Gersbach, C. A. (2016). In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science, 351, 403–407. https://doi.org/10.1126/science.aad5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning, S. , Yue, M. , Wang, P. , Liu, Y. , Zhi, H. , Zhang, Y. , … Li, X. (2017). LincSNP 2.0: An updated database for linking disease‐associated SNPs to human long non‐coding RNAs and their TFBSs. Nucleic Acids Research, 45, D74–D78. https://doi.org/10.1093/nar/gkw945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning, S. , Zhang, J. , Wang, P. , Zhi, H. , Wang, J. , Liu, Y. , … Li, X. (2016). Lnc2Cancer: A manually curated database of experimentally supported lncRNAs associated with various human cancers. Nucleic Acids Research, 44, D980–D985. https://doi.org/10.1093/nar/gkv1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu, Y. , Ma, F. , Huang, W. , Fang, S. , Li, M. , Wei, T. , & Guo, L. (2017). Long non‐coding RNA TUG1 is involved in cell growth and chemoresistance of small cell lung cancer by regulating LIMK2b via EZH2. Molecular Cancer, 16(5), 5 https://doi.org/10.1186/s12943-016-0575-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, R. R. , Mondal, T. , Mohammad, F. , Enroth, S. , Redrup, L. , Komorowski, J. , … Kanduri, C. (2008). Kcnq1ot1 antisense noncoding RNA mediates lineage‐specific transcriptional silencing through chromatin‐level regulation. Molecular Cell, 32, 232–246. https://doi.org/10.1016/j.molcel.2008.08.022 [DOI] [PubMed] [Google Scholar]
- Pasmant, E. , Laurendeau, I. , Heron, D. , Vidaud, M. , Vidaud, D. , & Bieche, I. (2007). Characterization of a germ‐line deletion, including the entire INK4/ARF locus, in a melanoma‐neural system tumor family: Identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Research, 67, 3963–3969. https://doi.org/10.1158/0008-5472.CAN-06-2004 [DOI] [PubMed] [Google Scholar]
- Petrova, Y. I. , Schecterson, L. , & Gumbiner, B. M. (2016). Roles for E‐cadherin cell surface regulation in cancer. Molecular Biology of the Cell, 27, 3233–3244. https://doi.org/10.1091/mbc.E16-01-0058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno, L. , Salmena, L. , Zhang, J. , Carver, B. , Haveman, W. J. , & Pandolfi, P. P. (2010). A coding‐independent function of gene and pseudogene mRNAs regulates tumour biology. Nature, 465, 1033–1038. https://doi.org/10.1038/nature09144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, Y. , Pan, S. , Kang, M. , Dong, R. , & Zhao, J. (2016). MicroRNA‐150 functions as a tumor suppressor in osteosarcoma by targeting IGF2BP1. Tumour Biology, 37, 5275–5284. https://doi.org/10.1007/s13277-015-4389-8 [DOI] [PubMed] [Google Scholar]
- Rader, D. J. (2002). High‐density lipoproteins and atherosclerosis. The American Journal of Cardiology, 90, 62i–70i. https://doi.org/S0002914902026358 [DOI] [PubMed] [Google Scholar]
- Reeves, R. , Langan, T. A. , & Nissen, M. S. (1991). Phosphorylation of the DNA‐binding domain of nonhistone high‐mobility group I protein by cdc2 kinase: Reduction of binding affinity. Proceedings of the National Academy of Sciences of the United States of America, 88, 1671–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins, K. M. , Chen, Z. , Wells, K. D. , & Rivera, R. M. (2012). Expression of KCNQ1OT1, CDKN1C, H19, and PLAGL1 and the methylation patterns at the KvDMR1 and H19/IGF2 imprinting control regions is conserved between human and bovine. Journal of Biomedical Science, 19, 95 https://doi.org/10.1186/1423-0127-19-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosikiewicz, W. , & Makalowska, I. (2016). Biological functions of natural antisense transcripts. Acta Biochimica Polonica, 63, 665–673. https://doi.org/10.18388/abp.2016_1350 [DOI] [PubMed] [Google Scholar]
- Rougeulle, C. , Cardoso, C. , Fontes, M. , Colleaux, L. , & Lalande, M. (1998). An imprinted antisense RNA overlaps UBE3A and a second maternally expressed transcript. Nature Genetics, 19, 15–16. https://doi.org/10.1038/ng0598-15 [DOI] [PubMed] [Google Scholar]
- Rougeulle, C. , Glatt, H. , & Lalande, M. (1997). The Angelman syndrome candidate gene, UBE3A/E6‐AP, is imprinted in brain. Nature Genetics, 17, 14–15. https://doi.org/10.1038/ng0997-14 [DOI] [PubMed] [Google Scholar]
- Saxonov, S. , Berg, P. , & Brutlag, D. L. (2006). A genome‐wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proceedings of the National Academy of Sciences of the United States of America, 103, 1412–1417. https://doi.org/10.1073/pnas.0510310103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner, M. , Huibregtse, J. M. , Vierstra, R. D. , & Howley, P. M. (1993). The HPV‐16 E6 and E6‐AP complex functions as a ubiquitin‐protein ligase in the ubiquitination of p53. Cell, 75, 495–505. https://doi.org/0092-8674(93)90384-3 [DOI] [PubMed] [Google Scholar]
- Sen, R. , Ghosal, S. , Das, S. , Balti, S. , & Chakrabarti, J. (2014). Competing endogenous RNA: The key to posttranscriptional regulation. ScientificWorldJournal, 2014(896206), 1–6. https://doi.org/10.1155/2014/896206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Y. (2007). Histone lysine demethylases: Emerging roles in development, physiology and disease. Nature Reviews. Genetics, 8, 829–833. https://doi.org/10.1038/nrg2218 [DOI] [PubMed] [Google Scholar]
- Shi, Y. , Lan, F. , Matson, C. , Mulligan, P. , Whetstine, J. R. , Cole, P. A. , … Shi, Y. (2004). Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell, 119, 941–953. https://doi.org/10.1016/j.cell.2004.12.012 [DOI] [PubMed] [Google Scholar]
- Shibata, M. , Yamada, S. , Kumar, S. R. , Calero, M. , Bading, J. , Frangione, B. , … Zlokovic, B. V. (2000). Clearance of Alzheimer's amyloid‐ss(1‐40) peptide from brain by LDL receptor‐related protein‐1 at the blood‐brain barrier. The Journal of Clinical Investigation, 106, 1489–1499. https://doi.org/10.1172/JCI10498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smilinich, N. J. , Day, C. D. , Fitzpatrick, G. V. , Caldwell, G. M. , Lossie, A. C. , Cooper, P. R. , … Higgins, M. J. (1999). A maternally methylated CpG island in KvLQT1 is associated with an antisense paternal transcript and loss of imprinting in Beckwith‐Wiedemann syndrome. Proceedings of the National Academy of Sciences of the United States of America, 96, 8064–8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, C. A. , & Castanotto, D. (2017). FDA‐approved oligonucleotide therapies in 2017. Molecular Therapy, 25, 1069–1075. https://doi.org/10.1016/j.ymthe.2017.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, C. A. , Hansen, J. B. , Lai, J. , Wu, S. , Voskresenskiy, A. , Hog, A. , … Koch, T. (2010). Efficient gene silencing by delivery of locked nucleic acid antisense oligonucleotides, unassisted by transfection reagents. Nucleic Acids Research, 38, e3 https://doi.org/10.1093/nar/gkp841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, H. , & Hausen, P. (1969). Enzyme from calf thymus degrading the RNA moiety of DNA‐RNA hybrids: Effect on DNA‐dependent RNA polymerase. Science, 166, 393–395. [DOI] [PubMed] [Google Scholar]
- Stine, Z. E. , Walton, Z. E. , Altman, B. J. , Hsieh, A. L. , & Dang, C. V. (2015). MYC, metabolism, and cancer. Cancer Discovery, 5, 1024–1039. https://doi.org/10.1158/2159-8290.CD-15-0507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesniak, M. W. , Kabza, M. , Karolak, J. A. , Rydzanicz, M. , Nowak, D. M. , Ginter‐Matuszewska, B. , … Gajecka, M. (2017). KTCNlncDB‐a first platform to investigate lncRNAs expressed in human keratoconus and non‐keratoconus corneas. Database: The Journal of Biological Databases and Curation, 2017 https://doi.org/10.1093/database/baw168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesniak, M. W. , & Makalowska, I. (2016). lncRNA‐RNA interactions across the human transcriptome. PLoS ONE, 11, e0150353 https://doi.org/PONE-D-15-35227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabebordbar, M. , Zhu, K. , Cheng, J. K. W. , Chew, W. L. , Widrick, J. J. , Yan, W. X. , … Wagers, A. J. (2016). In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science, 351, 407–411. https://doi.org/10.1126/science.aad5177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange, T. O. , Damgaard, C. K. , Guth, S. , Valcarcel, J. , & Kjems, J. (2001). The hnRNP A1 protein regulates HIV‐1 tat splicing via a novel intron silencer element. The EMBO Journal, 20, 5748–5758. https://doi.org/10.1093/emboj/20.20.5748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur, N. , Tiwari, V. K. , Thomassin, H. , Pandey, R. R. , Kanduri, M. , Gondor, A. , … Kanduri, C. (2004). An antisense RNA regulates the bidirectional silencing property of the Kcnq1 imprinting control region. Molecular and Cellular Biology, 24, 7855–7862. https://doi.org/24/18/7855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorenoor, N. , Faltejskova‐Vychytilova, P. , Hombach, S. , Mlcochova, J. , Kretz, M. , Svoboda, M. , & Slaby, O. (2016). Long non‐coding RNA ZFAS1 interacts with CDK1 and is involved in p53‐dependent cell cycle control and apoptosis in colorectal cancer. Oncotarget, 7, 622–637. https://doi.org/10.18632/oncotarget.5807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufarelli, C. , Frischauf, A. M. , Hardison, R. , Flint, J. , & Higgs, D. R. (2001). Characterization of a widely expressed gene (LUC7‐LIKE; LUC7L) defining the centromeric boundary of the human alpha‐globin domain. Genomics, 71, 307–314. https://doi.org/S0888-7543(00)96394-8 [DOI] [PubMed] [Google Scholar]
- Tufarelli, C. , Stanley, J. A. , Garrick, D. , Sharpe, J. A. , Ayyub, H. , Wood, W. G. , & Higgs, D. R. (2003). Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nature Genetics, 34, 157–165. https://doi.org/ng1157 [DOI] [PubMed] [Google Scholar]
- Vu, T. H. , & Hoffman, A. R. (1997). Imprinting of the Angelman syndrome gene, UBE3A, is restricted to brain. Nature Genetics, 17, 12–13. https://doi.org/10.1038/ng0997-12 [DOI] [PubMed] [Google Scholar]
- Wahlestedt, C. (2006). Natural antisense and noncoding RNA transcripts as potential drug targets. Drug Discovery Today, 11, 503–508. https://doi.org/10.1016/j.drudis.2006.04.013 [DOI] [PubMed] [Google Scholar]
- Wang, E. T. , Ward, A. J. , Cherone, J. M. , Giudice, J. , Wang, T. T. , Treacy, D. J. , … Burge, C. B. (2015). Antagonistic regulation of mRNA expression and splicing by CELF and MBNL proteins. Genome Research, 25, 858–871. https://doi.org/10.1101/gr.184390.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Wang, L. , Erdjument‐Bromage, H. , Vidal, M. , Tempst, P. , Jones, R. S. , & Zhang, Y. (2004). Role of histone H2A ubiquitination in Polycomb silencing. Nature, 431, 873–878. https://doi.org/nature02985 [DOI] [PubMed] [Google Scholar]
- Wanschura, S. , Schoenmakers, E. F. , Huysmans, C. , Bartnitzke, S. , Van de Ven, W. J. , & Bullrdiek, J. (1996). Mapping of the human HMG2 gene to 4q31. Genomics, 31, 264–265. https://doi.org/S0888754396900464 [DOI] [PubMed] [Google Scholar]
- Watts, J. K. , & Corey, D. R. (2012). Silencing disease genes in the laboratory and the clinic. The Journal of Pathology, 226, 365–379. https://doi.org/10.1002/path.2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, J. H. , Wise, A. , Main, M. J. , Green, A. , Fraser, N. J. , Disney, G. H. , … Marshall, F. H. (1998). Heterodimerization is required for the formation of a functional GABA(B) receptor. Nature, 396, 679–682. https://doi.org/10.1038/25354 [DOI] [PubMed] [Google Scholar]
- Wight, M. , & Werner, A. (2013). The functions of natural antisense transcripts. Essays in Biochemistry, 54, 91–101. https://doi.org/10.1042/bse0540091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willem, M. , Lammich, S. , & Haass, C. (2009). Function, regulation and therapeutic properties of beta‐secretase (BACE1). Seminars in Cell & Developmental Biology, 20, 175–182. https://doi.org/10.1016/j.semcdb.2009.01.003 [DOI] [PubMed] [Google Scholar]
- Wilson, R. C. , & Doudna, J. A. (2013). Molecular mechanisms of RNA interference. Annual Review of Biophysics, 42, 217–239. https://doi.org/10.1146/annurev-biophys-083012-130404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, J. , Stessl, M. , Amartey, J. , & Noe, C. R. (2010). Off‐target effects related to the phosphorothioate modification of nucleic acids. ChemMedChem, 5, 1344–1352. https://doi.org/10.1002/cmdc.201000156 [DOI] [PubMed] [Google Scholar]
- Xu, Y. , Wang, J. , Qiu, M. , Xu, L. , Li, M. , Jiang, F. , … Xu, L. (2015). Upregulation of the long noncoding RNA TUG1 promotes proliferation and migration of esophageal squamous cell carcinoma. Tumour Biology, 36, 1643–1651. https://doi.org/10.1007/s13277-014-2763-6 [DOI] [PubMed] [Google Scholar]
- Yamanaka, Y. , Faghihi, M. A. , Magistri, M. , Alvarez‐Garcia, O. , Lotz, M. , & Wahlestedt, C. (2015). Antisense RNA controls LRP1 sense transcript expression through interaction with a chromatin‐associated protein, HMGB2. Cell Reports, 11, 967–976. https://doi.org/10.1016/j.celrep.2015.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, R. , Fan, Q. , Zhou, J. , & Vassar, R. (2016). Inhibiting BACE1 to reverse synaptic dysfunctions in Alzheimer's disease. Neuroscience and Biobehavioral Reviews, 65, 326–340. https://doi.org/10.1016/j.neubiorev.2016.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, T. , Adamson, T. E. , Resnick, J. L. , Leff, S. , Wevrick, R. , Francke, U. , … Brannan, C. I. (1998). A mouse model for Prader‐Willi syndrome imprinting‐centre mutations. Nature Genetics, 19, 25–31. https://doi.org/10.1038/ng0598-25 [DOI] [PubMed] [Google Scholar]
- Yang, Z. , Zhou, L. , LM, W. , Lai, M. C. , Xie, H. Y. , Zhang, F. , & Zheng, S. S. (2011). Overexpression of long non‐coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Annals of Surgical Oncology, 18, 1243–1250. https://doi.org/10.1245/s10434-011-1581-y [DOI] [PubMed] [Google Scholar]
- Yates, A. , Akanni, W. , Amode, M. R. , Barrell, D. , Billis, K. , Carvalho‐Silva, D. , … Flicek, P. (2016). Ensembl 2016. Nucleic Acids Research, 44, D710–D716. https://doi.org/10.1093/nar/gkv1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, J. , Luo, W. , Zeng, X. , Zeng, L. , Li, Z. , Deng, X. , … Hu, W. (2017). UXT‐AS1‐induced alternative splicing of UXT is associated with tumor progression in colorectal cancer. American Journal of Cancer Research, 7, 462–472. [PMC free article] [PubMed] [Google Scholar]
- Yoshimura, R. , Ito, K. , & Endo, Y. (2009). Differentiation/maturation of neuropeptide Y neurons in the corpus callosum is promoted by brain‐derived neurotrophic factor in mouse brain slice cultures. Neuroscience Letters, 450, 262–265. https://doi.org/10.1016/j.neulet.2008.12.010 [DOI] [PubMed] [Google Scholar]
- Yu, G. , Yao, W. , Gumireddy, K. , Li, A. , Wang, J. , Xiao, W. , … Xu, H. (2014). Pseudogene PTENP1 functions as a competing endogenous RNA to suppress clear‐cell renal cell carcinoma progression. Molecular Cancer Therapeutics, 13, 3086–3097. https://doi.org/10.1158/1535-7163.MCT-14-0245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Zeitz, M. J. , Wang, H. , Niu, B. , Ge, S. , Li, W. , … Hu, J. F. (2014). Long noncoding RNA‐mediated intrachromosomal interactions promote imprinting at the Kcnq1 locus. The Journal of Cell Biology, 204, 61–75. https://doi.org/10.1083/jcb.201304152 [DOI] [PMC free article] [PubMed] [Google Scholar]


