Abstract
Background
: Noninvasive biomarkers to guide personalized treatment for castration‐resistant prostate cancer (CRPC) are needed. In this study, we analyzed hypermethylation patterns of two genes (GSTP1 and APC) in plasma cell‐free DNA (cfDNA) of CRPC patients. The aim of this study was to analyze the cfDNA concentrations and levels of the epigenetic markers and to assess the value of these biomarkers for prognosis.
Methods
: In this prospective study, patients were included before starting new treatment after developing CRPC. The blood samples were collected prior to start of the treatment and at three time points thereafter. cfDNA was extracted from 1.5 mL of plasma and before performing a methylation‐specific PCR, bisulfate modification was carried out.
Results
: The median levels of cfDNA, GSTP1, and APC copies in the baseline samples of CRPC patients (n = 47) were higher than in controls (n = 30). In the survival analysis, the group with baseline marker levels below median had significant less PCa‐related deaths (P‐values <0.02) and did not reach the median survival point. The survival distributions for the groups were statistically significant for the cfDNA concentration, GSTP1 and APC copies, as well as PSA combined with GSTP1 + APC (P‐values <0.03). Furthermore, there were strong positive correlations between PSA and marker response after starting treatment (P‐values <0.04).
Conclusions
: In conclusion, this study showed the kinetics of methylated cfDNA (GSTP1 and APC) in plasma of CRPC patients after starting treatment. Furthermore, the value of the markers before treatment is prognostic for overall survival. These results are promising for developing a test to guide treatment‐decision‐making for CRPC patients.
Keywords: APC, cell‐free DNA, epigenetics, GSTP1, hypermethylation, prostate cancer
Abbreviations
- ACTB
β‐actin
- ADT
androgen deprivation therapy
- APC
adenomatous polyposis coli
- AR
androgen receptor
- AR‐V7
androgen receptor splice variant 7
- CRPC
castration resistant prostate cancer
- cfDNA
cell‐free DNA
- CTC
circulating tumor cells
- GSTP1
gluthathione‐S‐tranfsferase π
- IQR
interquartile range
- PCa
prostate cancer
- PSA
prostate‐specific antigen
- RASSF1A
ras association domain family protein 1 isoform A
- RECIST
response evaluation criteria in solid tumors
1. INTRODUCTION
Most patients with advanced prostate cancer (PCa) will eventually develop castration‐resistant prostate cancer (CRPC). CRPC was accounting for over 26 000 deaths in the US in 2016.1 Significant improvements have been made in the treatment options for CRPC. Approved treatments are chemotherapy (docetaxel, cabazitaxel), androgen receptor (AR)‐targeted agents (enzalutamide and abiraterone acetate), and radiopharmaceuticals (Radium‐223).2, 3, 4, 5, 6, 7, 8 However, there are currently no biomarkers to guide personalized treatment decisions. Monitoring of treatment response is mostly done 3‐4 months after starting treatment, with measuring PSA levels and bone scintigraphy and/or other imaging modalities.9
Blood‐based biomarkers are promising to provide more molecular information on metastatic PCa than the primary tumor or a single metastasis alone.10 In the blood of advanced PCa patients circulating tumor cells (CTC) derived from the primary tumor and metastatic sites11 can be detected, as well as cell‐free, cancer‐derived nucleic acids like DNA and RNA.10 Circulating cell‐free DNA (cfDNA) is composed of small fragments of nucleic acids that are not associated with cells or cell fragments.12 Jung et al. showed that cfDNA levels were higher in metastatic PCa patients and were predictive for PCa‐specific survival.13 The detection of PCa‐specific genetic and epigenetic alterations within the cfDNA potentially is more useful.14, 15
The most common epigenetic alteration in PCa is DNA CpG island hypermethylation and previous studies showed detection of hypermethylated DNA in blood of PCa patients.16 GSTP1 hypermethylation was detected in 12% of patients with clinically localized PCa and 28% of patient with hormone refractory metastatic PCa.17 Furthermore, GSTP1 hypermethylation was shown to be the strongest predictor of PSA recurrence following radical prostatectomy17 and was correlated with the Gleason score and the extent of metastasis in patients with hormone‐refractory PCa.18 GSTP1 has been studied in combination with other hypermethylated genes, such as APC and RASSF1A. A recent meta‐analysis showed that a higher level of hypermethylated APC is associated with biochemical recurrence of patients with prostate cancer.19 The combination of these genes is used in a diagnostic tissue‐based test for PCa.20
We analyzed the hypermethylation patterns of CRPC patients in plasma cfDNA of two genes (GSTP1 and APC) and the housekeeping gene ACTB. The aim of this study was (i) to analyze the cfDNA concentrations and levels of methylated markers (GSTP1 and APC) in plasma of CRPC patients; (ii) to assess the value of the methylated markers as prognostic factor for overall survival; and (iii) to evaluate the association between the cfDNA and methylated markers changes in response to treatment.
2. MATERIALS AND METHODS
2.1. Patient population
In this prospective study, patients were enrolled prior to the start of chemotherapy (docetaxel or cabazitaxel) or androgen receptor signaling inhibitors (abiraterone, enzalutamide) after developing CRPC on androgen deprivation therapy (ADT). This was the primary treatment after developing CRPC. The subjects were included between February 2013 and March 2016, and treated at the Radboud university medical center (Nijmegen, The Netherlands). Exclusion criteria were a history of other malignancies in the last 5 years, baseline PSA level ≤5.0 ng/mL (to reduce the chance of non‐informative samples) and radiation therapy within 30 days of entry. Clinical data was extracted from the patients records. Approval was obtained from the Institutional Review Board and written informed consent was obtained from each patient. The control groups consisted of ten healthy age‐matched men with no evidence of PCa, ten men aged under 35 years of age and 10 healthy women.
2.2. Blood samples and cfDNA extraction
The blood samples were collected prior to the start of treatment and at week 15, week 27, and week 42. The blood collection was part of a scheduled follow‐up appointment, not focusing on the status of the disease. The blood samples were collected into an EDTA tube of 9 mL. The EDTA tubes were centrifuged at 1.300g for 10 min after which plasma was extracted and stored within two hours at −80°C. cfDNA was extracted from 1.5 mL of plasma using the Quick‐cfDNA Serum&Plasma kit (Zymo Research D4076). First, a centrifugation step of 16.000g for 10 min was performed to ensure efficient depletion of residual cells, cell debris, and genomic DNA.21 Supernatant was then transferred to a new 2.0 mL Eppendorf tube and extraction was performed following manufacturers protocol. The cfDNA was eluted in 27 μl of elution buffer. Size and yield of cfDNA was evaluated on a high sensitivity DNA chip of the Agilent 2100 Expert Bioanalyzer (Agilent Technologies Inc. Santa Clara, CA) and with the dsDNA High Sensitivity assay kit on a Qubit 3.0 fluorimeter (Thermo Fisher Scientific, Eugene, OR).
2.3. Bisulfite treatment
A total of 20 μl of cfDNA was used for the bisulphite modification using a commercially available kit (EZ DNA Methylation‐Lightning Kit, Zymo D5030, Zymo Research, Orange, CA). This reaction selectively deaminates unmethylated cytosine residues, resulting in a conversion to uracil, while 5‐methyl cytosine residues are not modified. The modified cfDNA was eluted into 25 μl EL‐Buffer (1 mM Tris‐HCl, pH8.0), then stored at −80°C before further processing.
2.4. Methylation‐specific PCR
A multiplex Taqman probe‐based approach was used to detect methylated copies of GSTP1 and APC (Table S1 in Appendix A−Supplementary data). ACTB was used as a housekeeping gene.20 A custom plasmid vector containing the relevant fragments of each of the three genes (IDT, Iowa) was included as a standard curve. Amplification of the multiplex methylation‐specific PCR assay was performed on the Lightcycler LC480 (Roche Ltd., Basel, Switzerland) with PerfeCTa multiplex qPCR ToughMix (Quantabio, Beverly, MA) in a total volume of 20 µL containing 1x buffer, 0.2 µmol/L of each primer and 0.2 μmol/L of each probe. As a positive control we used in vitro methylated DNA (Milipore). As a negative control we used DNA extracted from HCT116 cell line with knocked out methyltransferases. Methylation‐specific PCR conditions were: 95°C for 3 min to activate the Hotstart enzyme followed by 45 cycles of 95°C for 15 sec and 60°C for 1 min. Detection was performed with following filter combinations: FAM (465‐510), CFO 560 (533‐580), CFR 610 (533‐610), and Q670 (618‐660). Results were generated using the Lightcycler 480 software's (Roche) Abs quant/2nd derivate max and exported as Ct values (cycler number at which the amplification curves crossed the automatically set threshold value). These Ct values were used to calculate copy numbers based on a linear regression of the triplicate points of the standard curve of 8E + 06‐8 plasmid copies. A run was considered valid when R2 of at least five relevant data points of the standard curve was above 0.99; PCR efficiency was >80% and the no template control was negative. For the sample data, results were considered valid when the Ct of ACTB was detected within the standard curve (>80 copies).
2.5. Statistical analysis
The study must be considered of exploratory nature and hypothesis‐generating. Hypermethylated GSTP1 and APC copies were combined as one factor with the principal component analysis, as well as the combination of the methylated markers with PSA. Differences in levels of cfDNA and methylated markers between controls and CRPC patients were analyzed using the Mann–Whitney U test. For survival analysis Kaplan–Meier curves and the log rank test was used. To evaluate the correlation between PSA response and marker responses after starting treatment Spearman's correlation coefficient (rs) was determined. Significance levels of P < 0.05 were concluded statistically significant. Statistical tests were performed using SPSS version 22.0 (IBM Corp., Armonk, NY).
3. RESULTS
3.1. Study population
In this prospective study 50 CRPC patients were included. Three subjects were excluded because one patient had been included twice and two patients had no biomarker levels in the baseline blood samples. Furthermore, 10 age‐matched man (median age 64 years [interquartile range IQR 58‐67]), 10 men younger than 35 years old (median age 27 years [IQR 26‐29]), and 10 women (median age 55 years [IQR 35‐66]) were sampled as controls. The patient characteristics are summarized in Table 1. The median age of patients was 68 years (IQR 60‐71 years) and median baseline PSA was 108 ng/mL.
Table 1.
Patient characteristics
| n = 47 (IQR/%) | |
|---|---|
| Age (median, yrs) | 68 (60‐71) |
| Metastasize status, n | |
| Primary M+ diagnosis | 30 (64%) |
| M+ after primary treatment | 13 (28%) |
| Unknown | 4 (8%) |
| Primary treatment, n | |
| Radical prostatectomy | 13 (28%) |
| External radiation therapy | 14 (30%) |
| Brachytherapy | 1 (2%) |
| Prior systemic treatment, n | |
| Androgen deprivation | 47 (100%) |
| Chemotherapy | 8 (17%) |
| Treatment at inclusion, n | |
| Docetaxel | 22 (47%) |
| Cabazitaxal | 2 (4%) |
| Abiraterone | 16 (34%) |
| Enzalutamide | 7 (15%) |
| Samples available for analysis, n | |
| Baseline | 47 (100%) |
| 2nd timepoint | 47 (100%) |
| 3rd timepoint | 37 (79%) |
| 4th timepoint | 25 (53%) |
| sPSA, ng/mL (median) | |
| Baseline PSA | 108 (35.0‐220) |
| 2nd timepoint | 38.1 (3.6‐140.0) |
| 3rd timepoint | 34.0 (3.5‐76.2) |
| 4th timepoint | 19.0 (2.9‐155.0) |
| Follow‐up, months (median) | 18 (11‐26) |
| Patients died of PCa, n | 18 (38%) |
3.2. Detection of cfDNA and hypermethylation markers in plasma of CRPC patients
The median levels of cfDNA in the baseline samples of the CRPC patients were significantly higher than in the age‐matched controls and men <35 years (P < 0.01 and P < 0.01, respectively) (Table 2 and Figure 1). The median level of cfDNA in the female controls was lower than in the CRPC patients, although not statistically significant (P = 0.07). Hypermethylation of GSTP1 and APC could not be measured in all samples probably because the copy numbers were below the detection limit (Table 2). Hypermethylation of GSTP1 was observed in 91% of CRPC patients at baseline with a median of 32 copies. This was significantly higher than in the 5 (71%) age‐matched controls (median 2 copies; P < 0.01) and in 6 (86%) men <35 years (median 14 copies; P = 0.02). There was no statistical difference with the 9 (100%) women (median 14 copies; P = 0.13). APC was statistically significant lower in all control groups (age‐matched men P < 0.01, men <35 years P < 0.01, and women P < 0.01) compared to the CRPC patients (median of 59 copies) (Table 2).
Table 2.
cfDNA and biomarker levels in PCa patients versus controls
| PCa patients | Age‐matched men | Men aged <35 yrs | Women | ||||
|---|---|---|---|---|---|---|---|
| Time point | 1 | 2 | 3 | 4 | ‐ | ‐ | ‐ |
| No. patients | 47 | 47 | 37 | 25 | 10 | 10 | 10 |
| Exclusion ‡ | 0 | 2 (4%) | 2 (4%) | 0 | 3 (30%) | 3 (30%) | 1 (10%) |
| Yield bioanalyzer (median [IQR]) | 5.0 (3.4‐9.6) | 6.9 (3.7‐10.2) | 6.9 (4.0‐12.7) | 5.7 (3.6‐8.7) | 2.6 (2.0‐4.0) P < 0.01 * | 1.9 (1.4‐2.6) P < 0.01 * | 2.7 (2.4‐7.4) P = 0.07 |
| cfDNA ng/mL bioanalyzer | 3.4 (2.2‐6.4) | 4.6 (2.4‐6.8) | 4.6 (2.6‐8.5) | 3.8 (2.4‐5.8) | 1.7 (1.3‐2.7) P < 0.01 * | 1.2 (0.9‐1.7) P < 0.01 * | 1.8 (1.6‐4.9) P = 0.07 |
| ACTB (evaluable samples) Copies (median [IQR]) | 47 (100%) 330 (2‐764) | 45 (100%) 258 (2‐669) | 35 (100%) 27 (2‐764) | 25 (100%) 219 (2‐725) | 7 (100%) 286 (232‐531) P = 0.20 | 7 (100%) 239 (200‐274) P = 0.09 | 9 (100%) 489 (306‐1407) P = 0.15 |
| GSTP1 (evaluable samples) Copies (median [IQR]) | 43 (91%) 32 (7‐83) | 38 (84%) 11 (2‐46) | 25 (71%) 16 (10‐147) | 22 (88%) 68 (7‐167) | 5 (71%) 2 (2‐16) P < 0.01 * | 6 (86%) 14 (2‐87) P = 0.02 * | 9 (100%) 14 (2‐37) P = 0.13 |
| APC (evaluable samples) Copies (median [IQR]) | 37 (79%) 59 (14‐138) | 25 (56%) 31 (12‐172) | 26 (74%) 26 (5‐88) | 24 (96%) 50 (13‐183) | 3 (43%) 12 (10‐12) P < 0.01 * | 1 (14%) 10 P <0.01 * | 4 (44%) 15 (7‐23) P < 0.01 * |
Exclusion samples with ACTB calculated copies below the detection limit (<80 copies ACTB).
Statistically significant different from patient baseline samples (Mann–Whitney U test, significance level P < 0.05).
Figure 1.
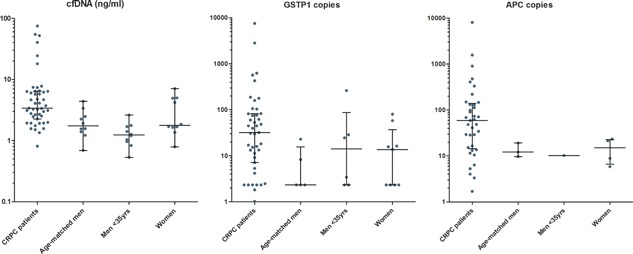
cfDNA concentrations and methylated GSTP1 and APC copies in patients versus controls
3.3. Prognostic value of baseline cfDNA and epigenetic markers
At the time of analysis 18 (38%) of the patients had died from PCa. The median follow‐up was 18 months (IQR 11‐26 months). To assess the prognostic value of the baseline levels of cfDNA and methylated markers, survival analysis was performed. The patients were divided into two groups: a group with the baseline level below the median and a group with a baseline level higher than the median. Figure 2 shows the Kaplan–Meier analysis of overall survival in association with the cfDNA concentration, level of the methylated markers and the combined factors GSTP1+APC and PSA+GSTP1+APC. For all markers, the group with baseline levels below median did not reach the median survival point. Patients with a cfDNA concentration below the median of 3.4 ng/mL had a significant lower number of deaths than the patients with a baseline value >3.4 ng/mL (21% vs 57%, P = 0.02) and a better overall survival (log rank P = 0.01). There was also a significant difference in overall survival for methylated GSTP1 baseline level below median compared to a higher GSTP1 Level (P = 0.02) and for methylated APC baseline levels (P = 0.03). When using the combined factor GSTP1+APC, there were significant less deaths in the group with the baseline value below the median (21% vs 57%, P = 0.02). Furthermore, survival analysis showed a trend to better overall survival in the low level group, although not statistically significant (P = 0.07). De combination of PSA with GSTP1 and APC resulted in a significant better overall survival for patients with a baseline value below median (P = 0.01).
Figure 2.
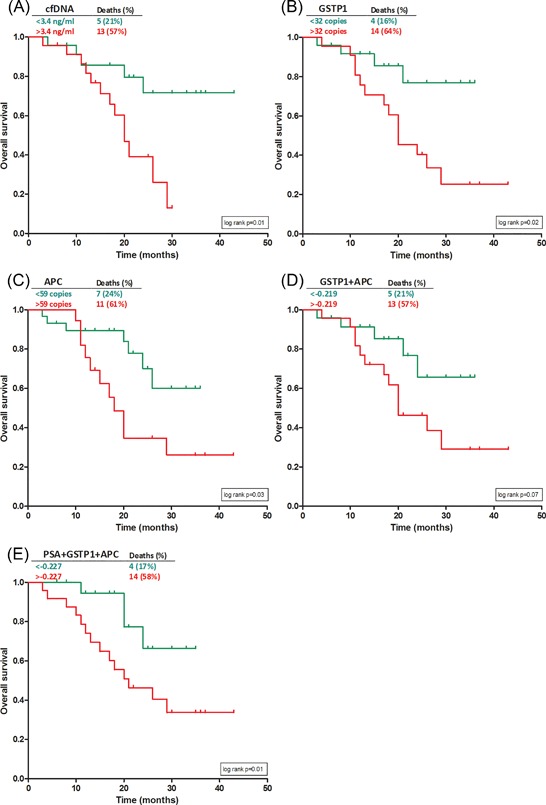
Kaplan–Meier analysis of overall survival according to baseline values of cfDNA (A) and methylated markers (B‐D) and the combination of PSA with GSTP1+APC (E)
3.4. cfDNA and methylated markers response after treatment as survival predictors
To evaluate the association with overall survival, patients were stratified into four groups with the median of the baseline sample used as cutoff‐point: (i) both samples below baseline median; (ii) first sample below baseline median and second sample higher; (iii) both samples higher than baseline median; and (iv) first sample higher and second sample below baseline median. There were statistically significant less PCa‐related deaths in the group with cfDNA and markers below median at baseline and the first timepoint (P‐values <0.02). A Log rank test was run to determine if there were differences in the survival distributions for the four patient groups. The patient group with both samples below baseline median had significantly higher overall survival for all markers (Figure S1, Appendix A−Supplementary data).
4. DISCUSSION
To guide personalized treatment decisions in advanced PCa, reliable markers are needed to monitor disease activity and as predictors for survival. In this study, we found higher concentrations of cfDNA and higher levels of methylated markers in blood plasma of CRPC patients compared to healthy controls. Furthermore, the baseline levels of cfDNA and GSTP1/APC before starting treatment were prognostic for overall survival.
In this study, blood plasma was used to extract cfDNA. Serum contains higher cfDNA concentrations, however, serum is known to be contaminated by genomic DNA.21 Several studies have demonstrated that the increased level of cfDNA in serum is mainly due to lysis of the clotting white blood cells in the collection tubes. Thijssen et al22 showed that plasma DNA better reflects the in vivo concentrations of cfDNA than serum DNA. The measured level of cfDNA has been shown to be greatly influenced by blood‐sampling, processing and cfDNA‐extraction protocols.23 The Bioanalyzer used in this study specifically measures cfDNA instead of genomic DNA.
Previous studies showed that PCR‐based methods are able to detect minimal amounts of nucleic acids,24 on the other hand, the necessary bisulfate treatment for the used methylation‐specific PCR could result in degradation of DNA and therefore limited detection of very few copies of methylated DNA.17 In our study, 71‐91% of samples at the different timepoints had detectable levels of methylated GSTP1. For methylated APC, this was 56‐90% of the samples. This in is accordance with previous studies, in which a detection rate of 79‐95% of PCa cases for GSTP1 hypermethylation was reported and 27‐100% for APC hypermethylation.25
There were strong, positive correlations between PSA and marker responses after starting treatment (rs 0.305‐0.636), which were all statistically significant (P < 0.04). We evaluated the 11 cases in which the PSA level increased after starting treatment, to determine the prognostic value of the methylated markers in this group. There was a statistical difference in the survival distribution for the groups based on median baseline levels for GSTP1 copies (P = 0.004) and the combination of GSTP1+APC (P = 0.004).
Our results show that the cfDNA yield in samples as marker itself is a prognostic factor for overall survival. It would be very interesting to compare cfDNA level with CTC count. Especially because, De Bono et al11 showed that the number of CTC in peripheral blood has strong independent prognostic value in patients with metastatic CRPC when measured before start of treatment. Moreover, when measured during therapy an increase in CTC count was associated with reduced overall survival, especially shortly after introducing a new treatment.26 However, for CTC analysis the time frame between blood sampling, sample preparation and analysis needs to be as small as possible and it is more complicated than the methylation‐specific PCR. If cfDNA yield is equal or even better in predicting prognosis of CRPC patients, even stored blood plasma samples could be used for analysis. This could improve feasibility of using these markers in daily practice.
The limitations of this study: most importantly, the heterogeneity of the study population with patients receiving chemotherapy versus abiraterone acetate or enzalutamide. In addition, the number of PCa‐related deaths was relatively small and, therefore, the median overall survival difference could not be studied. Furthermore, there was little information available about the controls. In our analysis, several controls had detectable levels of GSTP1 methylation in the cfDNA (Table 2), however, in a previous study, no hypermethylation of GSTP1 was observed in healthy controls.16 However, also in PCa patients the detection rate of hypermethylation of GSTP1 was lower in this study with 42.3% versus 91% in our study.16
Despite its explorative character, this study indicates that pre‐treatment levels of cfDNA and epigenetic markers (GSTP1 and APC) are of prognostic value for CRPC patients. Moreover, measuring the variation of levels after starting treatment could potentially improve monitoring treatment response. Standardization and uniform standards for collection, processing and analysis are needed to study these markers. Besides, studies with matched control and patient groups are essential. Future research should include monitoring of disease progression with imaging and the radiological RECIST criteria.27 Moreover, it would be very interesting to collect samples at more timepoints short after starting treatment.
5. CONCLUSIONS
In conclusion, this study evaluated cfDNA concentrations in plasma cfDNA of CRPC patients and the levels of methylated GSTP1 and APC. The baseline value of the markers before starting treatment was prognostic for overall survival. Furthermore, the variation of levels after starting treatment are promising to improve personalized treatment of CRPC patients in the future.
CONFLICTS OF INTEREST
The authors would like to declare the following conflicts of interest: Smit, VanderSmissen, and Van Voorde are employees of MDxHealth. Mulders, Van Criekinge, and Schalken report consultancy with honoraria for MDxHealth. The other authors declare no potential conflicts of interest.
Supporting information
Additional Supporting Information may be found online in the supporting information tab for this article.
Table S1. Epigenetic markers in circulating cell‐free DNA as prognostic markers for survival of castration‐resistant prostate cancer patients.
ACKNOWLEDGMENTS
This research was funded by MDxHealth (Irvine, CA).
Hendriks RJ, Dijkstra S, Smit FP, et al. Epigenetic markers in circulating cell‐free DNA as prognostic markers for survival of castration‐resistant prostate cancer patients. The Prostate. 2018;78: 336–342. https://doi.org/10.1002/pros.23477
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016; 66:7–30. [DOI] [PubMed] [Google Scholar]
- 2. Tannock IF, de Wit R, Berry WR, et al. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med. 2004; 351:1502–1512. [DOI] [PubMed] [Google Scholar]
- 3. de Bono JS, Oudard S, Ozguroglu M, et al. Prednisone plus cabazitaxel or mitoxantrone for metastatic castration‐resistant prostate cancer progressing after docetaxel treatment: a randomised open‐label trial. Lancet. 2010; 376:1147–1154. [DOI] [PubMed] [Google Scholar]
- 4. Fizazi K, Scher HI, Molina A, et al. Abiraterone acetate for treatment of metastatic castration‐resistant prostate cancer: final overall survival analysis of the COU‐AA‐301 randomised, double‐blind, placebo‐controlled phase 3 study. Lancet Oncol. 2012; 13:983–992. [DOI] [PubMed] [Google Scholar]
- 5. Ryan CJ, Smith MR, Fizazi K, et al. Abiraterone acetate plus prednisone versus placebo plus prednisone in chemotherapy‐naive men with metastatic castration‐resistant prostate cancer (COU‐AA‐302): final overall survival analysis of a randomised, double‐blind, placebo‐controlled phase 3 study. Lancet Oncol. 2015; 16:152–160. [DOI] [PubMed] [Google Scholar]
- 6. Scher HI, Fizazi K, Saad F, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012; 367:1187–1197. [DOI] [PubMed] [Google Scholar]
- 7. Beer TM, Armstrong AJ, Rathkopf DE, et al. Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med. 2014; 371:424–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Parker C, Nilsson S, Heinrich D, et al. Alpha emitter radium‐223 and survival in metastatic prostate cancer. N Engl J Med. 2013; 369:213–223. [DOI] [PubMed] [Google Scholar]
- 9. Onstenk W, de Klaver W, de Wit R, Lolkema M, Foekens J, Sleijfer S. The use of circulating tumor cells in guiding treatment decisions for patients with metastatic castration‐resistant prostate cancer. Cancer Treat Rev. 2016; 46:42–50. [DOI] [PubMed] [Google Scholar]
- 10. Hegemann M, Stenzl A, Bedke J, Chi KN, Black PC, Todenhofer T. Liquid biopsy: ready to guide therapy in advanced prostate cancer? BJU Int. 2016; 19. [DOI] [PubMed] [Google Scholar]
- 11. de Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration‐resistant prostate cancer. Clin Cancer Res. 2008; 14:6302–6309. [DOI] [PubMed] [Google Scholar]
- 12. Stroun M, Lyautey J, Lederrey C, Olson‐Sand A, Anker P. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin Chim Acta. 2001; 313:139–142. [DOI] [PubMed] [Google Scholar]
- 13. Jung K, Stephan C, Lewandowski M, et al. Increased cell‐free DNA in plasma of patients with metastatic spread in prostate cancer. Cancer Lett. 2004; 205:173–180. [DOI] [PubMed] [Google Scholar]
- 14. Wang BG, Huang HY, Chen YC, et al. Increased plasma DNA integrity in cancer patients. Cancer Res. 2003; 63:3966–3968. [PubMed] [Google Scholar]
- 15. Ellinger J, Muller SC, Stadler TC, Jung A, von Ruecker A, Bastian PJ. The role of cell‐free circulating DNA in the diagnosis and prognosis of prostate cancer. Urol Oncol. 2011; 29:124–129. [DOI] [PubMed] [Google Scholar]
- 16. Ellinger J, Haan K, Heukamp LC, et al. CpG island hypermethylation in cell‐free serum DNA identifies patients with localized prostate cancer. Prostate. 2008; 68:42–49. [DOI] [PubMed] [Google Scholar]
- 17. Bastian PJ, Palapattu GS, Lin X, et al. Preoperative serum DNA GSTP1 CpG island hypermethylation and the risk of early prostate‐specific antigen recurrence following radical prostatectomy. Clin Cancer Res. 2005; 11:4037–4043. [DOI] [PubMed] [Google Scholar]
- 18. Reibenwein J, Pils D, Horak P, et al. Promoter hypermethylation of GSTP1, AR, and 14‐3‐3sigma in serum of prostate cancer patients and its clinical relevance. Prostate. 2007; 67:427–432. [DOI] [PubMed] [Google Scholar]
- 19. Wang Y, Fan C, Yu J, Wang X. APC methylation predicts biochemical recurrence of patients with prostate cancer: a meta‐analysis. Int J Clin Exp Med. 2015; 8:15575–15580. [PMC free article] [PubMed] [Google Scholar]
- 20. Van Neste L, Bigley J, Toll A, et al. A tissue biopsy‐based epigenetic multiplex PCR assay for prostate cancer detection. BMC Urol. 2012; 12:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. El Messaoudi S, Rolet F, Mouliere F, Thierry AR. Circulating cell free DNA: Preanalytical considerations. Clin Chim Acta. 2013; 23: 222–230. [DOI] [PubMed] [Google Scholar]
- 22. Thijssen MA, Swinkels DW, Ruers TJ, de Kok JB. Difference between free circulating plasma and serum DNA in patients with colorectal liver metastases. Anticancer Res. 2002; 22:421–425. [PubMed] [Google Scholar]
- 23. Chiu RW, Poon LL, Lau TK, Leung TN, Wong EM, Lo YM. Effects of blood‐processing protocols on fetal and total DNA quantification in maternal plasma. Clin Chem. 2001; 47:1607–1613. [PubMed] [Google Scholar]
- 24. Bastian PJ, Palapattu GS, Yegnasubramanian S, et al. Prognostic value of preoperative serum cell‐free circulating DNA in men with prostate cancer undergoing radical prostatectomy. Clin Cancer Res. 2007; 13:5361–5367. [DOI] [PubMed] [Google Scholar]
- 25. Jeronimo C, Bastian PJ, Bjartell A, et al. Epigenetics in prostate cancer: biologic and clinical relevance. Eur Urol. 2011; 60:753–766. [DOI] [PubMed] [Google Scholar]
- 26. Scher HI, Jia X, de Bono JS, et al. Circulating tumour cells as prognostic markers in progressive, castration‐resistant prostate cancer: a reanalysis of IMMC38 trial data. Lancet Oncol. 2009; 10:233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009; 45:228–247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found online in the supporting information tab for this article.
Table S1. Epigenetic markers in circulating cell‐free DNA as prognostic markers for survival of castration‐resistant prostate cancer patients.


