Abstract
Aspergillus fumigatus is an important fungal pathogen of humans. Inhaled conidia of A. fumigatus adhere to pulmonary epithelial cells, causing opportunistic infection. However, little is known about the molecular mechanism of the adherence of resting conidia. Fungal molecules adhesive to host cells are presumed to be displayed on the conidial surface during conidial formation as a result of changes in gene expression. Therefore, we exhaustively searched for adhesion molecules by comparing the phenotypes and the gene expression profiles of A. fumigatus strains that have conidia showing either high or low adherence to human pulmonary A549 cells. Morphological observation suggested that strains that produce conidia of reduced size, hydrophobicity, or number show decreased adherence to A549 cells. K‐means cluster analyses of gene expression revealed 31 genes that were differentially expressed in the high‐adherence strains during conidial formation. We knocked out three of these genes and showed that the conidia of AFUA_4G01030 (encoding a hypothetical protein) and AFUA_4G08805 (encoding a haemolysin‐like protein) knockout strains had significantly reduced adherence to host cells. Furthermore, the conidia of these knockout strains had lower hydrophobicity and fewer surface spikes compared to the control strain. We suggest that the selectively expressed gene products, including those we identified experimentally, have composite synergistic roles in the adhesion of conidia to pulmonary epithelial cells.
Keywords: fungi (aspergillus), infection, microbial‐cell interaction, molecular genetic, transcriptomics, virulence
1. INTRODUCTION
The filamentous fungus Aspergillus fumigatus is one of the most common causative agents of invasive fungal infection in immune‐compromised patients and is associated with alarmingly high mortality rates. The main route of infection caused by the fungus is via inhalation of conidia. Inhaled conidia come in contact with airway epithelial cells or pulmonary macrophages, where the conidia adhere before initiating germination and hyphal growth (Croft, Culibrk, Moore, & Tebbutt, 2016; Gomez, Hackett, Moore, Knight, & Tebbutt, 2010; McCormick, Loeffler, & Ebel, 2010; Wasylnka & Moore, 2002, 2003), leading (ultimately) to aspergillosis. The interaction of conidia with respiratory epithelium cells is important to our understanding of aspergillosis and for finding drug targets for antifungal therapy.
In our in vivo experiments, conidia adhering to mouse lung epithelial cells were observed at 2 hr after infection, but no conidia were found in mouse lung alveoli or bronchi at 5 hr (data not shown). The time course of in vitro adhesion and disappearance are similar to those reported for in vivo infection of humans by the fungus. This result indicates that conidial adherence to host cells is one of the first steps in infection by Aspergillus. However, little is known of the molecular mechanisms underlying the adherence of A. fumigatus conidia to host pulmonary epithelial cells (Sheppard, 2011). In this study, we sought factors that facilitate the adherence of A. fumigatus resting conidia to host pulmonary epithelial cells.
For swollen conidia and germlings, it is known that polysaccharides (e.g., galactofuranose and galactosaminogalactan) in the conidial cell walls are required for adherence to the host‐cell surface, extracellular matrix (ECM), and a variety of other substrates (Gravelat et al., 2013; Lamarre et al., 2009; Loussert et al., 2010; Sheppard, 2011). However, these polysaccharides are exposed on the outside of conidia only after swelling, and cellular adhesion is attenuated but not abolished in the null mutants of related genes (e.g., ugm1 and uge3 deletion mutants; Gravelat et al., 2013; Lamarre et al., 2009; Loussert et al., 2010; Sheppard, 2011). Also, it should be noted that most previous adherence studies have focused on hyphae, not on resting conidia. However, it is resting conidia that first come in contact with host airway epithelial cells after inhalation. We therefore hypothesized that resting conidia may possess additional adhesion factors.
In the previous study by Oosthuizen et al. (2011), human bronchial epithelial cells were coincubated with A. fumigatus conidia for 6 hr, followed by genome‐wide transcriptomic analysis using human and fungal microarrays. Those authors observed an up‐regulation of fungal genes involved in iron acquisition, vacuolar acidification, and formate dehydrogenase activities. Some of these genes were also up‐regulated in conidia exposed to human neutrophils (Sugui et al., 2008) and considered important for conidial resistance to macrophages and neutrophils. However, we postulated that it is important to compare the alteration of transcript levels during conidial formation rather than after coincubation of conidia with host cells, because some of the adhesion molecules on the surface of resting conidia may be preferentially produced during conidial formation.
Although there have been reports based on quantitative shotgun proteomic analysis during germination of conidia (Asif et al., 2006; Cagas, Jain, Li, & Perlin, 2011; Suh et al., 2012), only limited results, including data from our studies (Hagiwara et al., 2016; Hagiwara, Suzuki, Kamei, Gonoi, & Kawamoto, 2014), are available for comparative transcriptomic analysis during A. fumigatus conidial formation. To provide a better understanding of the mechanism for adherence of A. fumigatus conidia to host cells, we conducted comparative transcriptomic analysis during conidial formation using A. fumigatus strains of independent origins that exhibited either high or low adherence to A549 human pulmonary cells, a cell line that is widely used as a model for analysing the interaction of A. fumigatus with alveolar epithelial cells.
Our analysis identified 31 genes as new candidate for conidial adhesion factors; we showed that knockouts of two of three of these genes resulted in reduced conidial adherence to host cells in vitro.
2. RESULTS
2.1. Adhesion efficiency of conidia to A549 cells differs among fungal strains
A. fumigatus strains isolated from aspergillosis patients and soil are stored at the Medical Mycology Research Center, Chiba University. We chose 30 independently isolated strains from those stocks (Table S1) and studied the efficiency of adhesion of their resting conidia to A549 human lung adenocarcinoma epithelial cells. We found that conidia from different strains showed variable adherence to A549 cells (Figure 1). These adherence efficiencies did not appear to correlate with the origin of the strains, whether from environmental samples or clinical conditions (e.g., pulmonary aspergillosis, chronic necrotizing pulmonary aspergillosis, and invasive pulmonary aspergillosis; Figure S1A). We selected four highly adhesive strains (adhesion efficiencies ranging between 62.4% and 77.2%) and four weakly adhesive strains (adhesion efficiencies ranging between 30.5% and 38.5%) for exploring adhesive factors in the following experiments. In this paper, we number the selected high‐ and low‐adherence strains from 1 to 4 and 5 to 8, respectively (see Figure S1B, upper panel). The low‐adherence strains adhered to the host cells at levels similar to those seen with Aspergillus oryzae and Aspergillus nidulans, aspergilli that do not or seldom (respectively) cause disseminated infection (see Figure 1). A time course of conidial adhesion to A549 cells for the eight strains is shown in Figure S1B (lower panel). As described previously (DeHart, Agwu, Julian, & Washburn, 1997), conidia rapidly attached to confluent monolayers of A549 cells, with the numbers of adhering conidia plateauing after approximately 1 hr.
Figure 1.
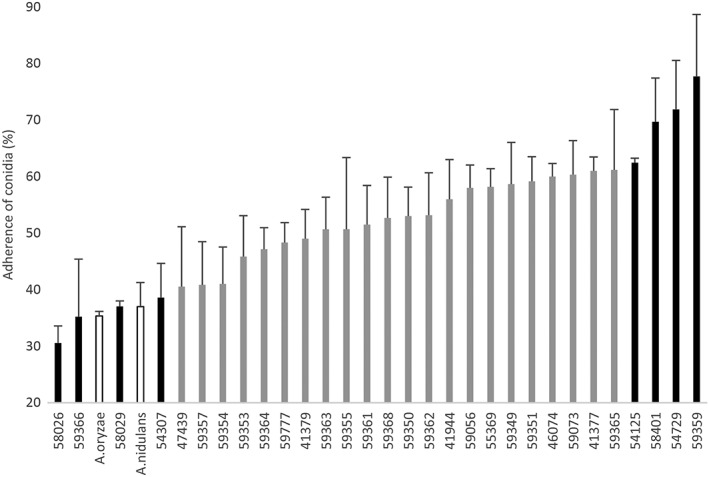
Adherence of conidia from different Aspergillus strains to A549 pulmonary epithelial cells. The columns indicate mean adherence values of three independent measurements for each strain. Black bars indicate high‐ and low‐adherence strains used in the experiments. White bars indicate non‐fumigatus strains. Strain numbers (IFM Nos.) of the Medical Mycology Research Center, Chiba University, are indicated on the abscissa. Percent values are mean ± SD of three independent measurements. The high‐adherence strains (IFM 59359, 54125, 58401, and 54729) and low‐adherence strains (IFM 59366, 58026, 58029, and 54307) are, respectively, called Strain 1 thru 4 and 5 thru 8 in this paper (Figure S1B)
It has been reported that A. fumigatus conidia adhere to the ECM of host pulmonary cells (Bromley & Donaldson, 1996; Gil, Penalver, Lopez‐Ribot, O'Connor, & Martinez, 1996; Yang, Jaeckisch, & Mitchell, 2000). Therefore, we first assessed the adhesion efficiencies of the eight selected strains to plastic dishes that were uncoated or had been coated with individual ECM components (collagen, fibronectin, and laminin). We observed nominal but non‐significant (as assessed by non‐parametric analysis) positive correlations among the efficiencies of adhesion to A549 cells and ECM‐coated plastic dishes, as well as to noncoated dishes (Figure 2a–d). We also found, for each A. fumigatus strain, that the efficiencies of conidial adhesion to the ECM were several times lower than those to A549 cells. Moreover, adherence of conidia to A549 and H292 cells (NCI‐H292 human lung mucoepidermoid carcinoma cells; American Type Culture Collection # CRL‐1848) was significantly correlated (Figure S1C). These results suggested the existence of additional host cell components, other than the ECM, that are shared between host epithelial cells and are recognised by conidial cell wall factors.
Figure 2.
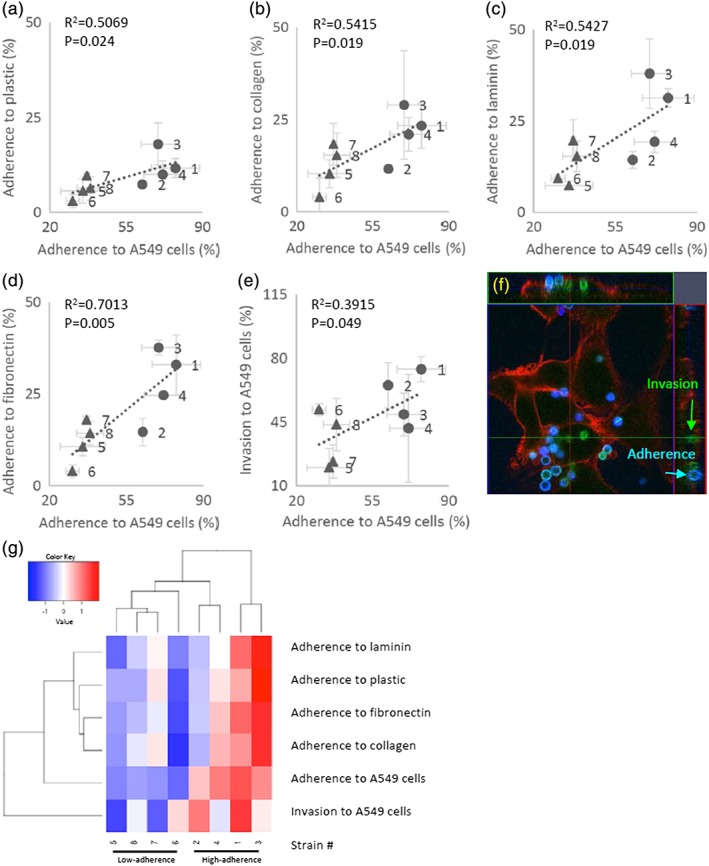
(a–d) Adhesion of Aspergillus fumigatus conidia to A549 cells (abscissa) versus extracellular matrices (ordinate), including collagen, fibronectin, and laminin, or uncoated‐plastic dishes in the high‐ (●) and low‐ (▲) adherence strains. (e) Invasion of conidia into A549 cells. Percent values are mean ± SD of three independent measurements. Regression lines, Spearman's non‐parametric rank correlation coefficients (R2), and p‐values (Student t‐test) are shown in each panel. (f) Green, conidia that invaded A549 cells at 5 hr after start of coincubation. Blue, conidia that adhered to A549 cells. Red, actin of A549 cells. (g) Heatmap of adherence to extracellular matrices and invasion data from the high‐adherence and low‐adherence strains. The heatmap was generated using the heatmap.2 function in the gplots package for the R programming language (R Core Team, 2014). The hierarchical clustering was conducted by Euclidian distance and complete linkage using variance scaling data
Some adherent conidia are internalised into bronchial or nasal epithelial cells, whereas other externally adherent conidia proceed to germinate while still on the host cell surface (Botterel et al., 2008; Gomez et al., 2010; Osherov, 2012). To investigate the relation between adherence and invasion by conidia, we compared adherence and invasion among the eight strains (Figure 2e,f). Two of the high‐adherence strains showed high invasion rates, and two of the low‐adherence strains showed low invasion rates. The remaining four strains exhibited intermediate invasion rates. These results suggested that adherence to host cells affects invasion, although not all high‐adherence strains have the trigger factors for invasion. The cluster dendrogram of adherence and invasion (Figure 2g) showed that the low‐adherence strains formed a distinct cluster, suggesting a weak (though not definitive) relationship between adhesion factors and conidial invasion.
2.2. Morphological properties of selected strains
To examine which conidial properties are associated with adherence to host cells, we compared the phenotypes of the high‐ and low‐adherence conidia. First, we found that the diameters of conidia of the low‐adherence strains were significantly smaller than those of the high‐adherence strains (Figure 3a). Conidia of the low‐adherence strains have nominally (but not significantly) lower hydrophobicity (Figure 3b), lighter colour (Figures 3c, S2A,B), and fewer spikes per unit surface area (Figures 3d, S2C,G) than those of the high‐adherence strains. Given that conidial hydrophobicity is associated with the surface‐displayed rodlet protein (Thau et al., 1994), we observed the rodlet layer using field emission‐scanning electron microscopy (Figure S2E,F). Strains that produced high‐hydrophobicity conidia (Strains 1–4, 7, and 8) and one strain that produced low‐hydrophobicity conidia (Strain 5) showed a rough surface, whereas a strain that produced other low‐hydrophobicity conidia (Strain 6) exhibited a smoother surface. Some of the low‐adherence strains had either thicker or thinner conidial cell walls in comparison to those of the high‐adherence strains (Figures 3e, S2D,F). In addition, the low‐adherence strains tended to show delayed conidiophore formation at 6 hr (Figure 3f), and two out of the four low‐adherence strains produced fewer conidia than the high‐adherence strains, even at 72 hr after the initiation of culturing (Figure 3g). From another perspective, the strains producing conidia with lower hydrophobicity, lighter colour, fewer spikes per unit surface area (excepting Strain 3), or irregular cell wall thickness exhibited lower adherence to A549 cells. Conidial adherence also was impaired in strains that exhibited delayed or decreased production of conidia.
Figure 3.
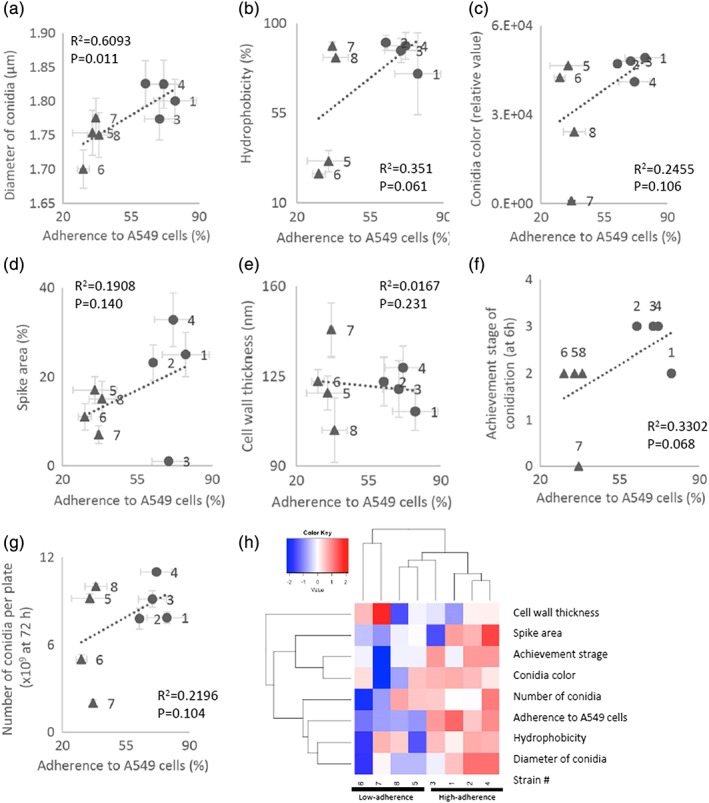
Comparison of conidial phenotypes in the high‐ and low‐adherence strains (7‐day‐old conidia). (a) Mean diameters of conidia. The mean diameters were measured using flow cytometry as described in Methods. (b) Hydrophobicity of conidia (ordinate) in the high‐ (●) and low‐ (▲) adherence‐strains. (c) Colony colours (ordinate) of the strains. Conidia (109) were collected into a glass capillary and photographed, and colour density was measured using image analysis software Image J (Schneider et al. 2012). (d) Ordinate: ratio (%) of areas occupied by spikes to the total conidial surface area. Scanning electron microscope images (Figure S2C) of conidia were processed using image analysis software, as described in Methods. (e) Ordinate: cell wall‐thickness of transmission electron microscopy images (Figure S2D) measured by image analysis software as described in Methods. (f) Correlation between asexual development in the high‐ (●) and low‐ (▲) adherence strains. Conidial formation stages were achieved at 12 hr after start of hyphal culture. Stage 0, mycelia before vesicle (conidiophore) formation; Stage 1, conidiophores and vesicle formation; Stage 2, phialide formation; Stage 3, almost all conidiophore have phialides and are initiating conidia formation; Stage 4, conidial maturation (also see Figure 3). (g) Number of conidia per plate (ordinate) at 72 hr after start of hyphal culture (see Methods). In Panels a–g, the abscissae show adherence of conidia to A549 cells. Each point represents a mean ± SD of three independent measurements. Regression lines, Spearman's non‐parametric rank correlation coefficients (R2), and p‐values (Student t‐test) are shown in each panel. (h) Heatmap of morphological property data from the high‐ and low‐adherence strains. The heatmap was generated using the heatmap.2 function in the gplots package for the R programming language (R Core Team, 2014). The hierarchical clustering was conducted by Euclidian distance and complete linkage using variance scaling data
A cluster dendrogram of all of these morphological properties showed that three of the high‐adherence strains grouped together as a single cluster. The remaining high‐adherence strain and two low‐adherence strains formed a second cluster, and the two remaining low‐adherence strains formed a third cluster (Figure 3h). Additionally, the dendrogram suggested that the property of “adherence to A549 cells” was associated with the diameter, hydrophobicity, and number of conidia produced by a given strain.
2.3. Principal component analysis (PCA) of gene expression patterns in high‐ and low‐adherence strains during conidial formation
Changes in gene expression during conidial formation were analysed using the RNA‐seq technology. To select distinctive conidiation stages for RNA‐seq analysis, we incubated mycelia of the eight A. fumigatus strains on potato dextrose agar (PDA) plates and used light microscopy to observe the cultures for specific developmental landmarks at 0, 6, 12, 24, and 48 hr. Figure 4a schematically summarises the developmental stages of typical A. fumigatus strains such as Af293 (the strain in which the genome was sequenced) as follows: A. fumigatus starts forming stipes and vesicles after 6 hr of incubation (Stage 1), starts forming phialides after approximately 12 hr (Stage 2), starts forming conidia after 24 hr (Stage 3), and achieves maturation of conidia (evidenced by the accumulation of a large number of dark conidia) by 48 hr of incubation (Stage 4). At every time point (0, 6, 12, 24, 48 hr), we compared gene expression patterns between the high‐ and low‐adherence strains.
Figure 4.
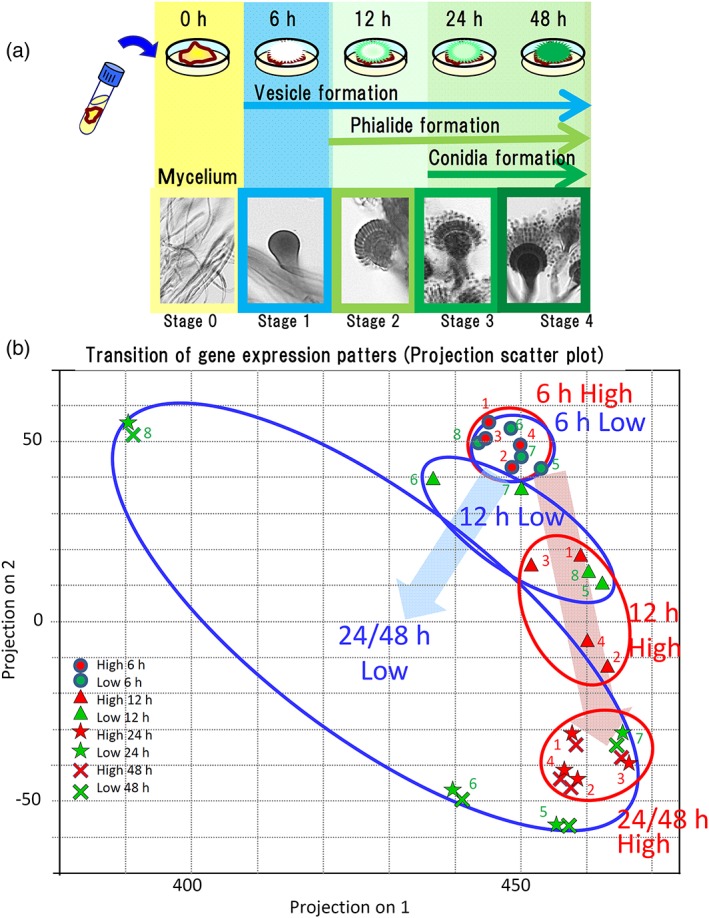
(a; top) A schematic drawing of the stages of conidial production in Aspergillus fumigatus. Note that time scale is not linear in the abscissa. (bottom) Photographs of the fungi at the corresponding stages. RNA‐seq analysis was conducted at 0, 1, 2, 24, and 48 hr. (b) Transition of gene expression patterns in the high‐ (red filled symbols) and low‐ (green) adherence strains during conidial formation. The pattern was examined through principal component analysis. Strain number designations (see Section 2) are shown next to the corresponding symbols. The principal components for the high‐ and low‐adherence strains at each time point are denoted with red and blue circles, respectively. Values at 0 hr (mycelia) are not shown
The eight strains expressed genes for late mycelial stages at Stage 0 (0 hr), showing diversity in PCA data without any apparent correlation to the strains' conidial adhesivities; therefore, we omitted these data in Figure 3b for clarity. We found no difference among the strains in transcript expression patterns during vesicle formation (Stage 1; 6 hr), but PCA analysis revealed evident differences between the high‐ and low‐adherence strains in expression patterns during conidial formation (12, 24, and 48 hr; Figure 4b). These results suggested that under‐ and over‐expression of the genes during conidial formation may be associated with the morphology and adherence of the conidia. Further analysis of gene expression revealed that more than 200 genes exhibited highly differential expression between the high‐ and low‐adherence strains during conidial formation; approximately one‐third of these genes encoded known cell‐wall‐biosynthesis‐associated functions (e.g., carbohydrate synthesis, cell membrane formation, and cell wall components; Figure S3). The PCA analysis also revealed that the expression of genes encoding proteins known to be central regulators of conidiation (brlA, abaA, and wetA; Yu, Mah, & Seo, 2006; Tao & Yu, 2011) remained at low levels for the first 12 hr in the strains of either adherence status, subsequently increasing by 24 hr only in the high‐adherence strains (Figure S4A).
The expression levels of genes that have been previously reported or predicted to encode adhesion‐related factors in swollen conidia, germlings, or hyphae (Figure S4B), as well as genes encoding cell‐wall components (Figure S4C), were compared between the high‐ and low‐adherence strains during conidial formation. Ugm1 (Lamarre et al., 2009), GAG Uge3 (Gravelat et al., 2013), LaeA (McDonagh et al., 2008), Asp f2 (Banerjee, Greenberger, Fink, & Kurup, 1998), Medusa (Gravelat et al., 2010), and Alp/Asp f13 (Crameri, 1998) have been reported to serve as adhesion‐related proteins in germling‐ and/or hyphal‐stage cells. However, the transcript levels of the corresponding genes did not differ significantly between the high‐ and low‐adherence strains during the early stage of conidial formation (Figure S4), suggesting that the expression of these adhesion‐associated fungal genes are up‐regulated at later stages of development (or are regulated post‐transcriptionally). In contrast, transcript levels of cspA (Levdansky et al., 2007; Levdansky, Kashi, Sharon, Shadkchan, & Osherov, 2010), alp2 (Reichard, Cole, Hill, Ruchel, & Monod, 2000), AFUA_1G13670 (EMBL‐EBI accession no. EAL90699.1, encoding a protein of unknown function; Suh et al., 2012), and AFUA_6G14470 (also encoding a protein of unknown function; Asif et al., 2006) were elevated during conidia formation in the high‐adherence strains (Figure S4B) but not in the low‐adherence strains. Notably, the protein encoded by AFUA_6G14470 previously was shown (via proteomic analysis) to accumulate in conidiating cells (Asif et al., 2006). These data suggested that the latter group of genes may be involved in conidial adhesion.
In addition, the expression levels of genes encoding cell wall components (or for the synthesis thereof) of resting conidia, including rodA (Thau et al., 1994), melanin‐synthesis‐related genes (pksP/alb1, arp1, abr1, and ayg1; Pihet et al., 2009), some carbohydrate‐synthesis‐related genes (e.g., rom2, Samantaray, Neubauer, Helmschrott, & Wagener, 2013; chsG, Mellado, Aufauvre‐Brown, Gow, & Holden, 1996), mnn9, and the gene encoding a subunit of galactomannan alpha‐1,6 mannosyltransferase (Gastebois, Clavaud, Aimanianda, & Latge, 2009) also increased in the high‐adherence strain during conidiation (Figure S4C). As shown in the supplemental data, we demonstrated a weak correlation between rodA expression level and conidial hydrophobicity and between the expression of melanin synthesis genes and conidial colour (Figure S4D). For instance, we observed that (during conidiation) the rodA transcript accumulated to high levels in Strains 2 and 4 and to a moderate level in Strains 1 and 3, and that all four of these strains yielded highly hydrophobic conidia. Additionally, all four of these strains expressed melanin synthesis genes at moderate levels and produced dark‐coloured conidia. In contrast, Strains 7 and 8 accumulated rodA transcripts to low levels but produced highly hydrophobic conidia; Strains 5 and 6 expressed several of the melanin synthesis genes only at low levels but still generated dark conidia. These results showed that rodA‐ and melanin‐synthesis‐encoding genes play an important role in causing the hydrophobicity or melanisation of conidia (respectively); nonetheless, a low R‐squared value and other results show that other genes are involved in these phenomena.
2.4. Search for genes whose expression specifically increases in high‐adherence strains during conidial formation (K‐means cluster analysis)
Expression patterns of each gene were classified into four clusters, a–d, as follows: (a) mainly expressed during the mycelial stage, (b) no significant changes during conidial formation, (c) expression levels increased during phialide formation and the early stage of conidial formation, and (d) expression levels increased during conidial formation and conidial maturation (Figure 5). The numbers of genes that were classified into each cluster are indicated in the lower panel of Figure 5. For all strains, the vast majority (over 98%) of the loci was assigned to cluster b, but 57, 77, and 60 sorted into clusters a, c, and d (respectively) in the high‐adherence strains. Expression patterns of most of the genes (98%) were shared in common among the test strains (i.e., did not differ significantly between the high‐ and low‐adherence strains), but 157 genes showed differential expression during conidiation when comparing the high‐ and low‐adherence strains. A subset of genes (n = 31), which were assigned to cluster “c” or “d” in the high‐adherence strains and to a distinct cluster in the low‐adherence strains (Figure 5, lower panel), were defined as genes that were specifically expressed in the high‐adherence strains during conidial formation and therefore were candidates for involvement in conidial adhesion (Table 1).
Figure 5.
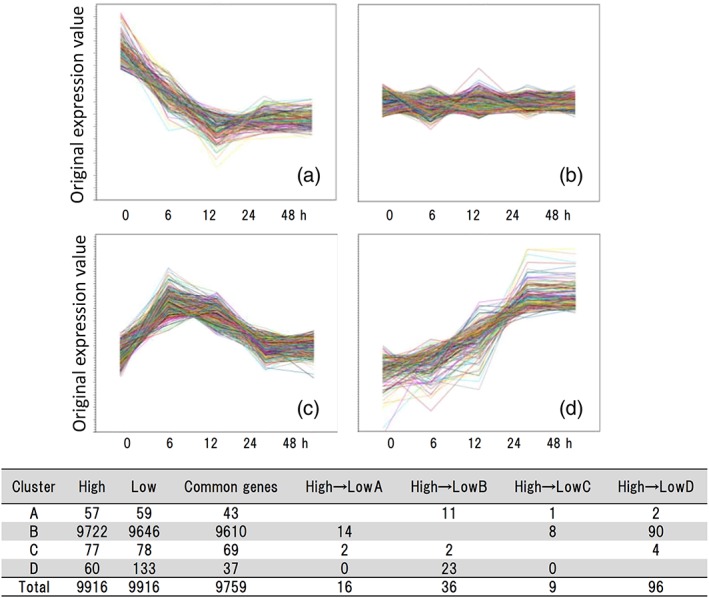
Diagrams of K‐means cluster analysis of gene expression levels during conidial formation. Genes were classified into four categories, (A–D), depending on changes in expression patterns, as follows: (A) mainly expressed in mycelium; (B) no significant changes were observed during conidiation; (c) genes with enhanced expression during conidiophore formation; (D) genes with enhanced expression during conidial formation. In A–D, the ordinate is expression level, and the abscissa is culture period. The diagrams show data for a representative experiment. The results of the parallel analysis conducted for all the strains are not shown. Table: Number of genes assigned to each cluster. High: High‐adherence strains. Low: Low‐adherence strains. For instance, “‘High A” means “the number of genes assigned to Cluster A.” At Line A, “High → Low B” means the number of genes assigned to Cluster A in high‐adherence strains and assigned to Cluster B in low‐adherence strains. The number of genes that were assigned to Cluster A in high‐adherence strains and assigned to Cluster A in low‐adherence strains is shown as “common genes’”
Table 1.
List of candidate genes for conidial adhesion factors
| Feature ID | Product | 12h(C) or 24h(D) means High/Low | |
|---|---|---|---|
| High C→Low A | AFUA_5G14680 | conserved hypothetical protein | 0.7 |
| AFUA_8G05985 | conserved hypothetical protein | 0.6 | |
| High C→Low B | AFUA_1G11190 | eukaryotic translation elongation factor 1 subunit Eef1‐beta, putative | 1.2 |
| AFUA_4G09310 | conserved hypothetical protein* | 6.2 | |
| High C→Low D | AFUA_1G02290 | conserved hypothetical protein | 1.5 |
| AFUA_1G17250 | conidial hydrophobin RodB* † | 3.8 | |
| AFUA_2G03720 | peptidyl‐prolyl cis‐trans isomerase | 1.2 | |
| AFUA_3G07430 | peptidyl‐prolyl cis‐trans isomerase/cyclophilin, putative | 1.3 | |
| High D→Low B | AFUA_1G04100 | conserved hypothetical protein | 8.7 |
| AFUA_1G10150 | conserved hypothetical protein | 6.3 | |
| AFUA_1G13670 | conserved hypothetical protein | 8.6 | |
| AFUA_2G00967 | Cupin domain protein | 5.0 | |
| AFUA_2G15290 | DUF636 domain protein | 1.7 | |
| AFUA_2G17580 | conidial pigment biosynthesis scytalone dehydratase Arp1 | 15.1 | |
| AFUA_3G07160 | class V chitinase, putative* | 9.8 | |
| AFUA_3G12900 | MFS transporter, putative | 3.0 | |
| AFUA_4G01030 | conserved hypothetical protein* † | 28.3 | |
| AFUA_4G02805 | Asp hemolysin‐like protein* † | 3.0 | |
| AFUA_4G05900 | conserved hypothetical protein | 5.0 | |
| AFUA_4G06370 | conserved hypothetical protein | 18.5 | |
| AFUA_4G09600 | GPI anchored protein, putative* | 62.4 | |
| AFUA_4G12470 | bZIP transcription factor CpcA | 2.6 | |
| AFUA_4G14530 | glutathione S‐transferase Ure2‐like, putative | 9.7 | |
| AFUA_5G00590 | hypothetical protein | 1.1 | |
| AFUA_5G02320 | conserved hypothetical protein | 3.7 | |
| AFUA_6G03210 | conidiation‐specific protein (Con‐10), putative* | 21.3 | |
| AFUA_6G03350 | GNAT family N‐acetyltransferase, putative | 2.2 | |
| AFUA_8G00510 | cytochrome P450 oxidoreductase OrdA‐like, putative | 3.6 | |
| AFUA_8G06180 | DOC family protein | 3.3 | |
| AFUA_8G06200 | conserved hypothetical protein* | 58.5 | |
| AFUA_8G07060 | hydrophobin, putative | 24.1 |
Genes for which deletion mutants were attempted.
Genes for which deletion mutants were obtained.
Twenty‐three of these selected genes were up‐regulated during conidiation in high‐adherence strains (categorised as “High D”), but induction was attenuated in low‐adherence strains (Figure 5 and Table 1). All 23 of these genes were categorised as “Low B” in low‐adherence strains, and no “High D” genes fell into either the “Low A” or “Low C” groups. Eight of these 23 High D/Low B genes encode function that are known (or thought) to be involved in conidial formation or cell wall localization in conidia (e.g., AFUA_4G02805, an Asp haemolysin‐like protein; AFUA_4G12470, the bZIP transcription factor CpcA; AFUA_3G07160, a putative class V chitinase; AFUA_8G07060, a putative hydrophobin; AFUA_2G17580, the scytalone dehydratase Arp1, which is involved in conidial pigment biosynthesis; AFUA_6G03210, homologous to the conidiation‐specific protein Con‐10; http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=330879). Six genes encode functions putatively involved in metabolism or other functions, and the rest (nine genes) encode hypothetical proteins. Eight genes were up‐regulated during vesicle and phialide formation in high‐adherence strains (categorised as High C) but not in low‐adherence strains (categorised as Low A, B, or D; Figure 5 and Table 1). Two of these genes showed attenuated induction in low‐adherence strains (High C to Low B: genes were categorised as Cluster c in high‐adherence strains and Cluster b in low‐adherence strains), and four of these genes showed delayed up‐regulation during conidiation in low‐adherence strains (High C to Low D).
2.5. Deletion mutant analysis of candidate adhesion‐associated genes AFUA_4G01030 (encoding a hypothetical protein), AFUA_4G02805 (encoding an Asp haemolysin‐like protein), and RodB (encoding a hydrophobin)
We attempted construction of deletion mutants for eight selected genes of the 31 genes listed in Table 1. These eight genes (indicated with an asterisk in Table 1) were selected to include the loci with the highest mean values of fold‐induction in High C (n = 2) and High D (n = 3), along with another three genes whose products (based on their protein structure) were expected to be localised to the cell surface. We succeeded in constructing deletion mutants for only three of these genes (marked as “‡” in Table 1), specifically AFUA_1G17250 (encoding the RodB hydrophobin), AFUA_4G02805 (encoding an Asp haemolysin‐like protein), and AFUA_4G01030 (encoding a hypothetical protein of unknown function). Compared to the conidia produced by the KU wild‐type strain, conidia produced by the AFUA_4G01030 and AFUA_4G02805 single‐mutant strains showed significantly reduced adherence to A549 cells, whereas conidia of the rodB deletion mutant did not (Figure 6a). Complementation of the AFUA_4G01030 and AFUA_4G02805 deletions with the respective genes restored conidial adherence to levels comparable to that of wild‐type conidia. Conidia of the AFUA_4G01030 and AFUA_4G02805 single‐mutant strains also showed reduced spike areas and hydrophobicities compared to wild‐type conidia, and, as above, complementation with the respective genes restored these two properties (Figures 6b–d). In addition, high‐resolution scanning electron microscope (SEM) showed that one of the mutants (⊿AFUA_4G01030) exhibited a smoother surface compared with that of the control KU strain (Figure S5D). Conidia of the AFUA_4G01030 and AFUA_4G02805 single‐mutant strains did not phenocopy other characteristics (changes in conidial diameter, colour, and cell wall thickness) that were observed in the low‐adherence strains (Figure S5).
Figure 6.
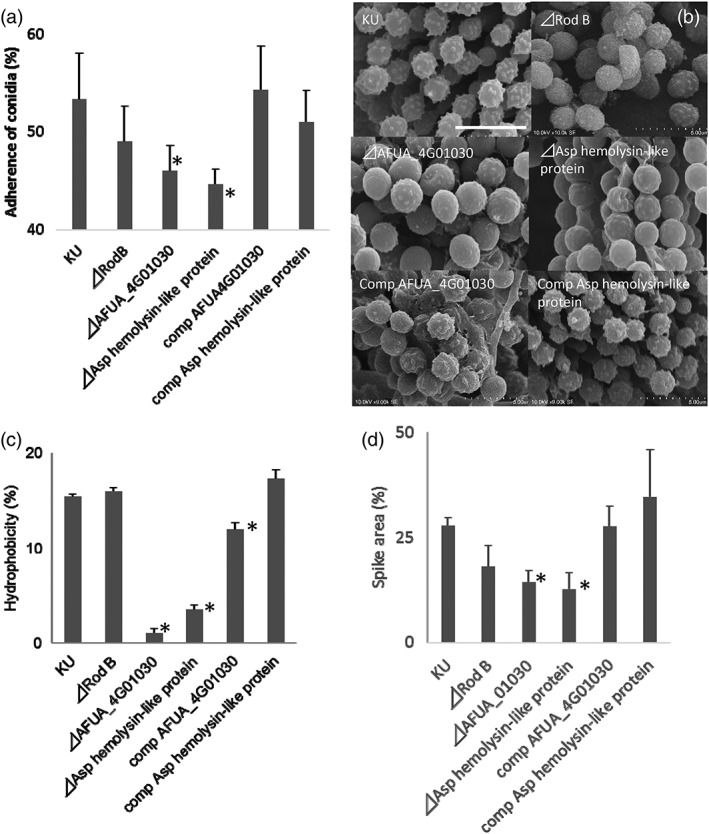
(a) Adhesion to A549 human lung adenocarcinoma epithelial cells of conidia produced by deletion mutants of AFUA_1G17250 (RodB), AFUA_4G01030, or AFUA_4G02805 (encoding an Asp haemolysin‐like protein). Adhesion data for the AFUA_4G01030 and AFUA_4G02805 (encoding an Asp haemolysin‐like protein) complemented strains (comp) are also shown to the right of the panel. Ordinate, adherence of conidia (%) to A549 cells (mean ± SD of three independent measurements). (b) Scanning electron microscope photographs showing conidial surfaces of the deletion mutants and the complemented strains. Scale bars: 5 μm. (c) Hydrophobicity of conidia in the deletion mutants and the complemented strains. (d) Proportion (%) of spike area divided by the total conidial surface area. Percent values are mean ± SD of three independent measurements. Comp, strains complemented with respect to the deleted genes. *: p < .05 in comparison to KU wt strain by Student t‐test in all panels
3. DISCUSSION
The adhesion of A. fumigatus conidia to host tissues is an essential event leading to persistence and infection of the host target organ (the lung). Several A. fumigatus molecules have been shown to serve as host cell adhesion factors in the swollen conidia, germling, and hyphal states (Al Abdallah et al., 2012; Sheppard, 2011). However, little is known about fungal factors involved in adherence to the host pulmonary epithelial cells in the initial stages after inhalation. In this study, A. fumigatus strains with high‐ and low‐adherence conidia in the dormant state were used to examine the morphological and transcriptomic differences between the two groups. The conidia of the four high‐adherence strains were similar in all characteristics studied except that conidia of one of the strains lacked spikes (Figure 3d). On the other hand, the conidia of the low‐adherence strains showed more variation in surface characteristics than did the high‐adherence strains (Figures 3 and S2). In addition, deletion mutants of genes encoding presumed adhesion factors AFUA_4G01030 (a protein of unknown function) and AFUA_4G02805 (an Asp haemolysin‐like protein) showed a reduced conidial spike areas and hydrophobicities but were not significantly altered in conidial cell diameter, colour, or cell wall thickness (Figures 6 and S5).
Furthermore, among the high‐adherence strains, Strains 2 and 4 had complete properties (Figures 3 and S2) and sufficient expression of rodA and melanin biosynthetic genes (Figure S4D). Strain 3 lacked spikes (Figures 3, S2C,E), but the conidial surface was rough (Figure S2E) and showed high hydrophobicity while this strain exhibited moderate rodA expression (Figure S4D). Strain 1 showed low hydrophobicity while exhibiting moderate rodA expression (Figure S4D) and a rough surface (Figure S2E). The Day‐7 conidia of Strains 1 and 3 showed highly adherent behaviour, but this adherence was decreased on Day 15 (Figure S6), and the expression levels of melanin biosynthetic genes were low (Figure S4D). We presumed the decreasing adherence of conidia was caused by conidial aging with reduced reinforcement by melanin, hydrophobins, and/or other components. In the low‐adherence strains, Strain 7 in particular exhibited decreased conidial production (Figure 3g). Strain 7 showed delayed conidial formation and decreased conidial number, and the resulting conidia had fewer spikes and thicker cell walls (Figures 3 and S2), presumably as a result of a lack of cell wall components and decreased cell wall compaction. The expression levels of hydrophobin genes and melanin biosynthetic genes were also reduced in Strain 7 (Figure S4D). Conidia of Strains 5 and 6 showed low hydrophobicity and decreased expression of the rodA gene, and Strain 6 showed a smoother surface (Figure S2F). Conidia of Strain 8 appeared normal but had thin cell walls. The conidia of Strain 8 may lack some adhesin in the cell wall, although conidia of this strain did not exhibit apparent changes in the conidial surface. Each of the low‐adherence strains had distinct phenotypes and did not apparently share any one characteristic in common. Our experimental data suggested that morphological properties such as conidial size and hydrophobicity are related to the adhesivity of dormant conidia to host cells, such that a number of factors interact synergistically to determine the adhesivity of conidia.
Comparative transcriptomic analysis (using RNA‐seq) of multiple A. fumigatus strains identified 31 genes that were differentially expressed (comparing high‐ and low‐adherence strains) during conidiation; the products of these genes are candidates for fungal adherence factors. None of these proteins have been previously identified as adhesion‐associated factors in germinating conidia or hyphae. Strains deleted for each of three of these genes were constructed, and two of these single‐mutant strains (lacking AFUA_4G01030, which encodes a hypothetical protein of unknown function, or lacking AFUA_4G02805, which encodes an Asp‐haemolysin‐like protein) showed significant reductions in conidial adherence to A549 cells compared to the resting conidia of the wild‐type strain (Figure 6). The rodB deletion mutant, on the other hand, showed a nominal but non‐significant decrease in adherence compared to the parent strain. The Asp‐haemolysin‐like protein encoded by AFUA_4G02805 has been shown to bind human erythrocytes (Ebina, Ichinowatari, & Yokota, 1985), and the function of the AFUA_4G01030 protein remains unknown. Future studies on the conidial localization of these gene products and the effects of these mutations on in vivo pathogenicity are expected to be useful for evaluating the role of these factors.
The loci examined in the present study included genes whose products have been previously been implicated as global regulators of conidiation (Figure S4A), adhesion‐related factors (Figure S4B), or cell wall components (or enzymes responsible for the synthesis thereof; Figure S4C). The transcription of these genes was compared between the high‐ and low‐adherence strains. The expression levels of genes encoding global regulators of conidial formation (brlA, abaA, and wetA; Yu et al., 2006; Tao & Yu, 2011) were elevated in the high‐adherence strains but not in the low‐adherence strains, during conidial formation (Figure S4A). The expression levels of some adhesion‐related genes (previously characterised in swollen conidia, germlings, or hyphae) and cell wall component genes were increased during conidial formation in high‐adherence strains, and included the alp2, AFUA_1G13670, cspA, rodA, arp2, rom2, and mnn9 genes, among others. These genes also may be involved in adherence by resting conidia. The expression levels of other genes (including glfA/ugm1, uge3, laeA, asp f2, medusa, etc.) did not differ between high‐ and low‐adherence strains during conidiation. These genes presumably are either not involved in adherence or are involved in the adherence of other cell types (e.g., swollen conidia or hyphae), or their expression is regulated post‐transcriptionally. Figures S7 and S8 show, for the reader's information, that many (>90%) of these genes are expressed by the various high‐adherence strains to similar levels at each of the investigated time points. We hypothesise that the expression of some of these genes might be intricately linked to the adhesive properties of conidia.
We suggest, based on the comparative phenotypic and the transcriptomic analyses conducted in this study, that adherence of A. fumigatus resting conidia to host epithelial cells is mediated by a number of different fungal factors, including the products of the AFUA_4G01030 and AFUA_4G02805 (Asp haemolysin‐like protein) genes, as well as by previously studied factors (hydrophobins, melanin, and carbohydrates) whose synthesis is controlled by the central regulatory pathway of conidial formation (Tao & Yu, 2011; Yu et al., 2006). Additional gene disruption of the candidate genes identified in the present analysis is expected to reveal further details of the mechanisms of conidial adhesion to host cells.
4. EXPERIMENTAL PROCEDURES
4.1. Fungal strains and growth conditions
The Aspergillus strains used in this study are listed in Table S1. Species identification of all the isolates was molecularly confirmed by sequencing of the genes encoding beta‐tubulin, calmodulin, and actin. Fungi were grown on potato dextrose agar (Difco) at 37 °C in the dark for 7 days until conidia were fully mature. Conidia were harvested in phosphate‐buffered saline (PBS, pH 7.4) containing 0.1% Tween 20 and then rinsed twice with distilled water.
4.2. Cell culture
The A549 (Type II human pneumocyte) and H292 (NCI‐H292 human lung mucoepidermoid carcinoma) cell lines were obtained from American Type Culture Collection and maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (Gibco), 100 mg/L streptomycin (Sigma‐Aldrich, St. Louis, MO, USA) and 16 mg/L penicillin (Sigma). Cells were maintained at 37 °C in a humidified 5% CO2 incubator.
4.3. Conidial adhesion studies
The A549 cells were plated at 105 cells/well in 6‐well culture plates (BM Equipment Co., Ltd. Tokyo, Japan) and grown to confluence. For studying conidial adherence to the ECM, the 6‐well plates were precoated by adding PBS containing 0.05 mg/ml collagen Type I, fibronectin, or laminin (all from Sigma) to each well, incubating the plate at 37 °C for 1 hr followed by overnight incubation at 4 °C, and then rinsing the wells with PBS. The plastic wells, either covered with A549 cells grown to confluency, coated with the individual ECM components, or left uncoated, were overlaid with RPMI 1640 medium containing 100 conidia per well and incubated at 37 °C under 5% CO2. After 2 hr of incubation, the wells were rinsed three times with PBS, overlaid with Sabouraud's dextrose agar, and incubated at 37 °C. Fungal colonies that grew from the conidia that adhered to the cells (or to the wells) were counted after 24 hr of incubation. The results of preliminary studies assessing the effects of conidial age and conidial number are summarised in Figure S6. Conidial adherence levels were calculated by dividing the number of adherent conidia by the number of conidia added to the well and expressing the resulting value as a percentage. All experiments were repeated as three individual trials, each consisting of triplicate wells.
To count conidia internalised by the host cells, we prestained conidia with a fluorescent dye. Fungal conidia were suspended in RPMI 1640 medium containing 0.01 mg/ml fluorescein isothiocyanate isomer‐I and incubated at 37 °C for 20 min. Unbound fluorescein isothiocyanate was washed away with unsupplemented RPMI 1640 medium. The stained conidia (2 × 106/ml) were added to A549 cells that had been grown in 8‐well glass‐chamber slides (Nunc Lab‐Tek). After coincubation at 37 °C for 1 hr, the medium, including free conidia, was aspirated out of the well. Calcofluor white (0.5 mg/ml) was added to each well, and the slide was incubated at room temperature for 15 min to permit staining of conidia adhering to the host cell surface. Finally, cells were fixed overnight at 4 °C using 4% formaldehyde, and the A549 cells were counterstained with rhodamine phalloidin at room temperature for 30 min. The samples were visualised using a Zeiss LSM 5 fluorescence microscope, and micrographs were processed with the ZEN imaging software (Zeiss).
4.4. Flow cytometry
The relative size of the conidia was determined through flow cytometry. Freshly harvested conidia (105 cells/ml in PBS containing 0.1% Tween 80) were vortexed briefly, and then the sizes were analysed with side‐scattered light using an on‐chip flow cytometer (On‐Chip Flow, On‐Chip Biotechnologies Co., Ltd, Japan) and 0.5‐μm beads for calibration.
4.5. Hydrophobicity measurement
The conidia were harvested, washed twice, and suspended in PBS at an optical density at 540 nm (OD540) of 0.4. The conidial suspension was treated with excess xylene (2.5:1, v/v) and mixed vigorously for 2 min; conidia then were allowed to settle for 20 min. The OD540 of the aqueous phase was determined, and the relative hydrophobicity was calculated as described previously (Reeves et al., 2006).
4.6. Measurement of conidia colour
The conidia in PBS were harvested into glass tubes (3 mm in diameter, 3 cm in depth) at a concentration of 109 conidia/ml and then allowed to settle to the tube bottom for 10 hr at 4 °C. The settled cells were photographed, and the colour densities were measured using image analysis software “Image J” (Schneider, Rasband, & Eliceiri, 2012).
4.7. Electron microscopy
For transmission electron microscopy analysis, hyphae were cultured on PDA medium for 18 hr and prefixed with 2% (w/v) glutaraldehyde containing 0.1 M phosphate buffer (pH 7) at 4 °C for 10 hr, followed by staining with 1% osmium tetroxide in phosphate buffer at room temperature for an additional hour. After dehydration, tissues were embedded in epoxy resin. Ultrathin sections were stained with uranyl acetate and lead citrate and visualised under a JEM 1400 electron microscope (JEOL, Japan). The cell wall thickness was measured using SMile View software (JEOL, Japan).
For SEM analysis, hyphae were cultured on PDA medium at 37 °C for 48 hr, and agar blocks containing fungal cells were fixed as described for the transmission electron microscopy analysis. The blocks were then dehydrated by passage through a graded series of ethanol solutions, replaced with isoamyl acetate, and dried using the critical‐point method (EM CPD030; Leica, Germany). After sputter coating with platinum‐palladium (using an E102 Ion sputter; Hitachi, Japan), samples were visualised using an S‐3400 SEM (Hitachi, Japan). The proportion of spike area to the total surface area of each conidium was measured using Image J. For field emission‐scanning electron microscopy analysis, dehydrated samples were visualised on poly‐L‐lysine coated silicon wafers under CrossBeam 550 (Zeiss).
4.8. Ribonucleic acid sequencing
For the synchronised induction of asexual development, conidia (105 conidia/ml) were cultivated in 0.1% yeast extract ‐ 1% glucose minimum medium at 37 °C for 18 hr, and conidia‐free mycelia were harvested using Miracloth (Merck, Frankfurter, Germany), washed with distilled water, and transferred onto 0.1% yeast extract ‐ 1% glucose minimum medium agar plates. The plates were then incubated in the dark at 37 °C, with the start of incubation defined as 0 hr. The mycelia were harvested at time points of 0, 6, 12, 24, and 48 hr.
Total RNA was extracted from fungal bodies (approximately 1 g for each sample) using the RNeasy®Mini Kit (Qiagen) and treated with DNase I (TaKaRa, Japan). Polyadenylated mRNA was then extracted from the total RNA and fragmented using the TruSeq RNA Sample Preparation Kit v2‐Set A (Illumina). A 200‐ to 300‐nucleotide size selection was performed, and the resulting RNA was converted into an Illumina sequencing library according to the manufacturer's protocol. Libraries were sequenced on a Miseq sequencer (Illumina) as 25‐bp paired‐end reads. The CLC genomics workbench (CLC bio, Demark) was used to analyse the sequencing results.
4.9. Construction of disruption and complementation vectors
Gene disruption vectors were constructed as follows. Approximately 2‐kb fragments from upstream and downstream of the target gene were amplified by polymerase chain reaction (PCR) using the primer sets listed in Table S2 and A. fumigatus (Af293) genomic DNA as the template. The hygromycin resistance gene (hph) was amplified using a separate set of primers (Hyg‐F and Hyg‐R in Table S2) and pCB1004 (Sakai, Kinoshita, Shimizu, & Nihira, 2008) as the template (Zhu, Wang, Yang, Wang, & Cui, 2009). The amplified fragments were assembled by introducing the PCR products for each gene into the SmaI site of pUC19. For construction of complementation vectors, gene fragments with native promoters were amplified using the primer sets listed in Table S2 and A. fumigatus genomic DNA as the template. The amplified fragments were cloned (separately) into the SmaI site of pPTRI (TaKaRa). The sequences of the resulting constructs were confirmed by sequencing.
4.10. Transformation of A. fumigatus
A. fumigatus was transformed using the previously described protoplast‐polyethylene glycol method (Sakai et al., 2008), with the following modifications. After 3–7 days of cultivation on PDA, conidia of the KU strain (Afs35, aku70) were inoculated into 100 ml of potato dextrose broth and cultivated at 37 °C for 20 hr. The resulting mycelia were collected and converted to protoplasts by incubation in TF Solution 1 (1.2 M sorbitol, 21.8 mM MES) supplemented with 5 mg/ml Yatalase (Takara Bio, Shiga, Japan) and 5 mg/ml Trichoderma harzianum lysing enzyme (Sigma). After incubation at 30 °C for 2–3 hr, the resulting protoplasts were harvested and resuspended in TF Solution 2 (1.2 M Sorbitol, 50 mM CaCl2, 35 mM NaCl, 10 mM Tris–HCl, pH 7.5). The relevant plasmid vector was added to the protoplast suspension, and the mixture was held on ice for 30 min, followed by addition of 1 ml of TF Solution 3 (149 mM PEG4000, 50 mM CaCl2, 10 mM Tris–HCl, pH 7.5). After 20 min at room temperature, the mixture was centrifuged, and the resulting pellet was resuspended in TF Solution 2. The mixture was plated on a selection medium (0.8% PDA containing 200 mg/ml hygromycin B) and overlaid with 4.5 ml of soft agar of the same selection medium. The inoculated plates were incubated at 37 °C for 3–5 days. Homologous recombination was confirmed by PCR and sequencing.
Supporting information
Figure S1. A. Comparison of adherence to A549 cells by conidia derived from different sources. PA, pulmonary aspergillosis; CNPA, chronic necrotizing pulmonary aspergillosis; IPA, invasive pulmonary aspergillosis. Mean values are indicated by horizontal bars in the graph. No significant difference was observed among the clinical and environmental strains by ANOVA. B. Upper panel: the selected high‐ and low‐adherence strains are numbered from 1 to 4 and 5 to 8, respectively. Lower panel: Time course of adhesion of the selected eight strains. Mean ± SD. The experiments were repeated as three individual trials, each consisting of triplicate wells. C. Adherences to A549 cells and H292 (NCI‐H292 human lung mucoepidermoid carcinoma cells) exhibit significant correlation. % values are mean ± SD of three independent measurements. Regression line, Spearman's non‐parametric rank correlation coefficients (R2), and p‐values (Student t‐test) are shown in the panel.
Figure S2. A‐D. Conidial morphology of high‐ and low‐adherence strains. A, Colonies. B, Conidia harvested into glass tubes. C, SEM micrographs showing conidial surfaces. D, TEM micrographs showing cell wall thicknesses (7‐day‐old conidia).
Figure S2E‐F. FE‐SEM micrographs showing conidial surfaces of 7‐day‐old conidia.
Figure S2G‐H. G, SEM micrographs showing conidial surfaces of 2‐day‐old conidia. H, TEM micrographs showing cell‐wall thicknesses of 2‐day‐old conidia.
Figure S3. Number of genes in which expression levels (RPKM values, reads per kilobase of exon per million mapped sequence reads) significantly (p < 0.05 by Student's t‐test, n = 4) differed by more than three fold between the high‐ and low‐adherence strains. A and B, genes for which levels were more than three fold higher or lower, respectively, in the high adherence strains compared to levels in the low adherence strains. The abscissae indicate different time points in conidial formation (see Figure 3 for conidial stages). Putative functions of the genes are indicated with different colours for A and B
Figure S4A‐C. Comparison of expression levels of genes that are reportedly or presumably important for conidial formation. Black and grey lines of each panel represent the mean gene expression levels (RPKM values) in the high‐ and low‐adherence strains (respectively, mean ± SD of four isolates). The abscissae indicate different time points of conidial formation. The ordinates are RPKM values. S4A: Genes of central regulatory pathways that control the temporal and spatial expression of conidiation‐specific genes during conidiophore development and spore maturation. S4B (next page): Previously published putative adhesins and other related genes. S4C: Genes for cell‐wall components. *, p < 0.05 by Student's t‐test.
Figure S5. Morphological properties of deletion mutants. A. Diameter of conidia. B. Conidial colour. C. Cell wall thickness. Data are presented as mean ± SD of three independent measurements (n = 3). In A, B, and C, there was no significant difference among values of the mutant and wild ‐type strains by Student's t‐test. See Methods for the parameters.
Figure S5D. FE‐SEM micrographs showing conidial surfaces of 7‐day‐old conidia.
Figure S6 A. Effect on adhesion to A549 cells of delayed conidial maturation or conidial aging. To investigate whether defects in adherence reflected delayed maturation of the conidia of low‐adherence strains, we compared adherence of 7‐day‐old and 15‐day‐old conidia. Adherence of low‐adherence strains did not increase with an additional 8 days of maturation. However, the data did indicate that the adherence of some strains (1, 3, and 7) decreased at day 15, presumably as a result of conidial aging (Oliveira et al., 2015).
B. Suitability of experimental procedure for adherence of conidia. In the majority of the experiments, we employed an assay that used 100 conidia per well in a 6‐well plate. To confirm the suitability of that experimental technique, we repeated the measurement of adherence using a distinct procedure. For the confirmatory experiment, A549 cells were grown to full confluence in6‐well plates, and cells were overlaid with RPMI1640 containing 1000 conidia per well. After 2 h of incubation, the wells were rinsed three times with PBS (to remove non‐adherent conidia), and the A549 cells then were lysed with 0.1% Triton X100 (to free the conidia that had adhered to the A549 cells). The resulting suspensions, including adherent conidia, were mixed with SDA, plated into 9‐cm petri dishes, and incubated to allow fungal growth. The results showed that adherence assessed using 1000 conidia strongly correlated with adherence assessed using 100 conidia. Based on these experiments, we selected the procedure using 100 conidia, which was technically less cumbersome. The data are shown as mean ± SD of three independent measurements.
Figure S7. Venn diagrams of gene expression data at the different time points in the four high‐adherence strains. Genes with RPKM >5 were used in the analysis. The diagram shows that more than 90% of genes were commonly expressed among the four strains at each time point.
Figure S8. A. Venn diagram of genes expressed in high‐adherence strains at different stages of conidial formation. Numbers of prospective stage‐specific genes are encircled and labelled a to e in the diagram. a, Mycelial stage (stage 0 in Figure 3); b, stipe and vesicle formation (stage 1); c, phialide formation (stage 2); d, conidiation (stage 3); and e, conidial maturation (stage 4). Genes with RPKM >5 were used in the analysis. For distinguishing gene expressions between vesicle formation and conidial formation stages, gene expression values at 12 h were omitted in this analysis, B. Stacked numbers and proposed or hypothetical functions of the stagespecific genes.
Table S1. Aspergillus strains used in this study.
Table S2. PCR primers used for gene disruption and complementation
ACKNOWLEDGEMENTS
This study was supported by a Grant‐in‐Aid for Scientific Research (to A.T‐N, and to T.G.) from the Ministry of Education, Science, Sports, and Culture and partially supported by the following: a Cooperative Research Grant of NEKKEN (2010–2017); a Cooperative Research Program of Medical Mycology Research Center, Chiba University (12‐2); and the MEXT Special Budget for Research Projects: The Project on Controlling Aspergillosis and Related Emerging Mycoses. This work was also supported by the National BioResource Project − Pathogenic Microbes, funded by the Ministry of Education, Culture, Sports, Science, and Technology, Japan (http://www.nbrp.jp/). We declare that the authors have no conflict of interest.
Takahashi‐Nakaguchi A, Sakai K, Takahashi H, et al. Aspergillus fumigatus adhesion factors in dormant conidia revealed through comparative phenotypic and transcriptomic analyses. Cellular Microbiology. 2018;20:e12802 https://doi.org/10.1111/cmi.12802
REFERENCES
- Al Abdallah, Q. , Choe, S.‐I. , Campoli, P. , Baptista, S. , Gravelat, F. N. , Lee, M. J. , & Sheppard, D. C. (2012). A Conserved C‐Terminal Domain of the Aspergillus fumigatus Developmental Regulator MedA Is Required for Nuclear Localization, Adhesion and Virulence. PloS One, 7(11), e49959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asif, A. R. , Oellerich, M. , Amstrong, V. W. , Riemenschneider, B. , Monod, M. , & Reichard, U. (2006). Proteome of conidial surface associated proteins of aspergillus fumigatus reflecting potential vaccine candidates and allergens. Journal of Proteome Research, 5, 954–962. [DOI] [PubMed] [Google Scholar]
- Banerjee, B. , Greenberger, P. A. , Fink, J. N. , & Kurup, V. P. (1998). Immunological characterization of asp f 2, a major allergen from aspergillus fumigatus associated with allergic bronchopulmonary aspergillosis. Infection and Immunity, 66, 5175–5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botterel, F. , Gross, K. , Ibrahim‐Granet, O. , Khoufache, K. , Escabasse, V. , Coste, A. , … Bretagne, S. (2008). Phagocytosis of aspergillus fumigatus conidia by primary nasal epithelial cells in vitro. BMC Microbiology, 8, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromley, I. M. , & Donaldson, K. (1996). Binding of aspergillus fumigatus spores to lung epithelial cells and basement membrane proteins: Relevance to the asthmatic lung. Thorax, 51, 1203–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagas, S. E. , Jain, M. R. , Li, H. , & Perlin, D. S. (2011). The proteomic signature of aspergillus fumigatus during early development. Molecular & Cellular Proteomics, 10(M111), 010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2014). R: A language and environment for statistical computing In R Foundation for Statistical Computing ). Vienna: Austria. URL; http://www.r-project.org/. [Google Scholar]
- Crameri, R. (1998). Recombinant aspergillus fumigatus allergens: From the nucleotide sequences to clinical applications. International Archives of Allergy and Immunology, 115, 99–114. [DOI] [PubMed] [Google Scholar]
- Croft, C. A. , Culibrk, L. , Moore, M. M. , & Tebbutt, S. J. (2016). Interactions of aspergillus fumigatus conidia with airway epithelial cells: A critical review. Frontiers in Microbiology, 7, 472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeHart, D. J. , Agwu, D. E. , Julian, N. C. , & Washburn, R. G. (1997). Binding and germination of Aspergillus fumigatus conidia on cultured A549 pneumocytes. The Journal of Infectious Diseases, 175(1), 146–150. [DOI] [PubMed] [Google Scholar]
- Ebina, K. , Ichinowatari, S. , & Yokota, K. (1985). Studies on toxin of aspergillus fumigatus. XXII. Fashion of binding of asp‐hemolysin to human erythrocytes and asp‐hemolysin‐binding proteins of erythrocyte membranes. Microbiology and Immunology, 29, 91–101. [DOI] [PubMed] [Google Scholar]
- Gastebois, A. , Clavaud, C. , Aimanianda, V. , & Latge, J. P. (2009). Aspergillus fumigatus: Cell wall polysaccharides, their biosynthesis and organization. Future Microbiology, 4, 583–595. [DOI] [PubMed] [Google Scholar]
- Gil, M. L. , Penalver, M. C. , Lopez‐Ribot, J. L. , O'Connor, J. E. , & Martinez, J. P. (1996). Binding of extracellular matrix proteins to aspergillus fumigatus conidia. Infection and Immunity, 64, 5239–5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez, P. , Hackett, T. L. , Moore, M. M. , Knight, D. A. , & Tebbutt, S. J. (2010). Functional genomics of human bronchial epithelial cells directly interacting with conidia of aspergillus fumigatus. BMC Genomics, 11, 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravelat, F. N. , Beauvais, A. , Liu, H. , Lee, M. J. , Snarr, B. D. , Chen, D. , … Sheppard, D. C. (2013). Aspergillus galactosaminogalactan mediates adherence to host constituents and conceals hyphal beta‐glucan from the immune system. PLoS Pathogens, 9, e1003575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravelat, F. N. , Ejzykowicz, D. E. , Chiang, L. Y. , Chabot, J. C. , Urb, M. , Macdonald, K. D. , … Sheppard, D. C. (2010). Aspergillus fumigatus MedA governs adherence, host cell interactions and virulence. Cellular Microbiology, 12, 473–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara, D. , Suzuki, S. , Kamei, K. , Gonoi, T. , & Kawamoto, S. (2014). The role of AtfA and HOG MAPK pathway in stress tolerance in conidia of aspergillus fumigatus. Fungal Genetics and Biology, 73, 138–149. [DOI] [PubMed] [Google Scholar]
- Hagiwara, D. , Takahashi, H. , Kusuya, Y. , Kawamoto, S. , Kamei, K. , & Gonoi, T. (2016). Comparative transcriptome analysis revealing dormant conidia and germination associated genes in Aspergillus species: An essential role for AtfA in conidial dormancy. BMS genomics, 17, 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarre, C. , Beau, R. , Balloy, V. , Fontaine, T. , Wong Sak Hoi, J. , Guadagnini, S. , … Latgé, J. P. (2009). Galactofuranose attenuates cellular adhesion of aspergillus fumigatus. Cellular Microbiology, 11, 1612–1623. [DOI] [PubMed] [Google Scholar]
- Levdansky, E. , Kashi, O. , Sharon, H. , Shadkchan, Y. , & Osherov, N. (2010). The aspergillus fumigatus cspA gene encoding a repeat‐rich cell wall protein is important for normal conidial cell wall architecture and interaction with host cells. Eukaryotic Cell, 9, 1403–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levdansky, E. , Romano, J. , Shadkchan, Y. , Sharon, H. , Verstrepen, K. J. , Fink, G. R. , & Osherov, N. (2007). Coding tandem repeats generate diversity in aspergillus fumigatus genes. Eukaryotic Cell, 6, 1380–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loussert, C. , Schmitt, C. , Prevost, M. C. , Balloy, V. , Fadel, E. , Philippe, B. , … Beauvais, A. (2010). In vivo biofilm composition of aspergillus fumigatus. Cellular Microbiology, 12, 405–410. [DOI] [PubMed] [Google Scholar]
- McCormick, A. , Loeffler, J. , & Ebel, F. (2010). Aspergillus fumigatus: Contours of an opportunistic human pathogen. Cellular Microbiology, 12, 1535–1543. [DOI] [PubMed] [Google Scholar]
- McDonagh, A. , Fedorova, N. D. , Crabtree, J. , Yu, Y. , Kim, S. , Chen, D. , … Bignell, E. (2008). Sub‐telomere directed gene expression during initiation of invasive aspergillosis. PLoS Pathogens, 4, e1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellado, E. , Aufauvre‐Brown, A. , Gow, N. A. , & Holden, D. W. (1996). The aspergillus fumigatus chsC and chsG genes encode class III chitin synthases with different functions. Molecular Microbiology, 20, 667–679. [DOI] [PubMed] [Google Scholar]
- Oosthuizen, J. L. , Gomez, P. , Ruan, J. , Hackett, T. L. , Moore, M. M. , Knight, D. A. , & Tebbutt, S. J. (2011). Dual organism transcriptomics of airway epithelial cells interacting with conidia of aspergillus fumigatus. PLoS One, 6, e20527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osherov, N. (2012). Interaction of the pathogenic mold aspergillus fumigatus with lung epithelial cells. Frontiers in Microbiology, 3, 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pihet, M. , Vandeputte, P. , Tronchin, G. , Renier, G. , Saulnier, P. , Georgeault, S. , … Bouchara, J. P. (2009). Melanin is an essential component for the integrity of the cell wall of aspergillus fumigatus conidia. BMC Microbiology, 9, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves, E. P. , Reiber, K. , Neville, C. , Scheibner, O. , Kavanagh, K. , & Doyle, S. (2006). A nonribosomal peptide synthetase (Pes1) confers protection against oxidative stress in aspergillus fumigatus. The FEBS Journal, 273, 3038–3053. [DOI] [PubMed] [Google Scholar]
- Reichard, U. , Cole, G. T. , Hill, T. W. , Ruchel, R. , & Monod, M. (2000). Molecular characterization and influence on fungal development of ALP2, a novel serine proteinase from aspergillus fumigatus. International Journal of Medical Microbiology, 290, 549–558. [DOI] [PubMed] [Google Scholar]
- Sakai, K. , Kinoshita, H. , Shimizu, T. , & Nihira, T. (2008). Construction of a citrinin gene cluster expression system in heterologous aspergillus oryzae. Journal of Bioscience and Bioengineering, 106, 466–472. [DOI] [PubMed] [Google Scholar]
- Samantaray, S. , Neubauer, M. , Helmschrott, C. , & Wagener, J. (2013). Role of the guanine nucleotide exchange factor Rom2 in cell wall integrity maintenance of aspergillus fumigatus. Eukaryotic Cell, 12, 288–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, C. A. , Rasband, W. S. , & Eliceiri, K. W. (2012). NIH image to ImageJ: 25 years of image analysis. Nature Methods, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard, D. C. (2011). Molecular mechanism of aspergillus fumigatus adherence to host constituents. Current Opinion in Microbiology, 14, 375–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugui, J. A. , Kim, H. S. , Zarember, K. A. , Chang, Y. C. , Gallin, J. I. , Nierman, W. C. , & Kwon‐Chung, K. J. (2008). Genes differentially expressed in conidia and hyphae of aspergillus fumigatus upon exposure to human neutrophils. PLoS One, 3, e2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh, M. J. , Fedorova, N. D. , Cagas, S. E. , Hastings, S. , Fleischmann, R. D. , Peterson, S. N. , … Momany, M. (2012). Development stage‐specific proteomic profiling uncovers small, lineage specific proteins most abundant in the aspergillus fumigatus conidial proteome. Proteome Science, 10, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, L. , & Yu, J. H. (2011). AbaA and WetA govern distinct stages of aspergillus fumigatus development. Microbiology, 157, 313–326. [DOI] [PubMed] [Google Scholar]
- Thau, N. , Monod, M. , Crestani, B. , Rolland, C. , Tronchin, G. , Latge, J. P. , & Paris, S. (1994). Rodletless mutants of aspergillus fumigatus. Infection and Immunity, 62, 4380–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylnka, J. A. , & Moore, M. M. (2002). Uptake of aspergillus fumigatus conidia by phagocytic and nonphagocytic cells in vitro: Quantitation using strains expressing green fluorescent protein. Infection and Immunity, 70, 3156–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylnka, J. A. , & Moore, M. M. (2003). Aspergillus fumigatus conidia survive and germinate in acidic organelles of A549 epithelial cells. Journal of Cell Science, 116, 1579–1587. [DOI] [PubMed] [Google Scholar]
- Yang, Z. , Jaeckisch, S. M. , & Mitchell, C. G. (2000). Enhanced binding of aspergillus fumigatus spores to A549 epithelial cells and extracellular matrix proteins by a component from the spore surface and inhibition by rat lung lavage fluid. Thorax, 55, 579–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J. H. , Mah, J. H. , & Seo, J. A. (2006). Growth and developmental control in the model and pathogenic aspergilli. Eukaryotic Cell, 5, 1577–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, T. , Wang, W. , Yang, X. , Wang, K. , & Cui, Z. (2009). Construction of two gateway vectors for gene expression in fungi. Plasmid, 62, 128–133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. A. Comparison of adherence to A549 cells by conidia derived from different sources. PA, pulmonary aspergillosis; CNPA, chronic necrotizing pulmonary aspergillosis; IPA, invasive pulmonary aspergillosis. Mean values are indicated by horizontal bars in the graph. No significant difference was observed among the clinical and environmental strains by ANOVA. B. Upper panel: the selected high‐ and low‐adherence strains are numbered from 1 to 4 and 5 to 8, respectively. Lower panel: Time course of adhesion of the selected eight strains. Mean ± SD. The experiments were repeated as three individual trials, each consisting of triplicate wells. C. Adherences to A549 cells and H292 (NCI‐H292 human lung mucoepidermoid carcinoma cells) exhibit significant correlation. % values are mean ± SD of three independent measurements. Regression line, Spearman's non‐parametric rank correlation coefficients (R2), and p‐values (Student t‐test) are shown in the panel.
Figure S2. A‐D. Conidial morphology of high‐ and low‐adherence strains. A, Colonies. B, Conidia harvested into glass tubes. C, SEM micrographs showing conidial surfaces. D, TEM micrographs showing cell wall thicknesses (7‐day‐old conidia).
Figure S2E‐F. FE‐SEM micrographs showing conidial surfaces of 7‐day‐old conidia.
Figure S2G‐H. G, SEM micrographs showing conidial surfaces of 2‐day‐old conidia. H, TEM micrographs showing cell‐wall thicknesses of 2‐day‐old conidia.
Figure S3. Number of genes in which expression levels (RPKM values, reads per kilobase of exon per million mapped sequence reads) significantly (p < 0.05 by Student's t‐test, n = 4) differed by more than three fold between the high‐ and low‐adherence strains. A and B, genes for which levels were more than three fold higher or lower, respectively, in the high adherence strains compared to levels in the low adherence strains. The abscissae indicate different time points in conidial formation (see Figure 3 for conidial stages). Putative functions of the genes are indicated with different colours for A and B
Figure S4A‐C. Comparison of expression levels of genes that are reportedly or presumably important for conidial formation. Black and grey lines of each panel represent the mean gene expression levels (RPKM values) in the high‐ and low‐adherence strains (respectively, mean ± SD of four isolates). The abscissae indicate different time points of conidial formation. The ordinates are RPKM values. S4A: Genes of central regulatory pathways that control the temporal and spatial expression of conidiation‐specific genes during conidiophore development and spore maturation. S4B (next page): Previously published putative adhesins and other related genes. S4C: Genes for cell‐wall components. *, p < 0.05 by Student's t‐test.
Figure S5. Morphological properties of deletion mutants. A. Diameter of conidia. B. Conidial colour. C. Cell wall thickness. Data are presented as mean ± SD of three independent measurements (n = 3). In A, B, and C, there was no significant difference among values of the mutant and wild ‐type strains by Student's t‐test. See Methods for the parameters.
Figure S5D. FE‐SEM micrographs showing conidial surfaces of 7‐day‐old conidia.
Figure S6 A. Effect on adhesion to A549 cells of delayed conidial maturation or conidial aging. To investigate whether defects in adherence reflected delayed maturation of the conidia of low‐adherence strains, we compared adherence of 7‐day‐old and 15‐day‐old conidia. Adherence of low‐adherence strains did not increase with an additional 8 days of maturation. However, the data did indicate that the adherence of some strains (1, 3, and 7) decreased at day 15, presumably as a result of conidial aging (Oliveira et al., 2015).
B. Suitability of experimental procedure for adherence of conidia. In the majority of the experiments, we employed an assay that used 100 conidia per well in a 6‐well plate. To confirm the suitability of that experimental technique, we repeated the measurement of adherence using a distinct procedure. For the confirmatory experiment, A549 cells were grown to full confluence in6‐well plates, and cells were overlaid with RPMI1640 containing 1000 conidia per well. After 2 h of incubation, the wells were rinsed three times with PBS (to remove non‐adherent conidia), and the A549 cells then were lysed with 0.1% Triton X100 (to free the conidia that had adhered to the A549 cells). The resulting suspensions, including adherent conidia, were mixed with SDA, plated into 9‐cm petri dishes, and incubated to allow fungal growth. The results showed that adherence assessed using 1000 conidia strongly correlated with adherence assessed using 100 conidia. Based on these experiments, we selected the procedure using 100 conidia, which was technically less cumbersome. The data are shown as mean ± SD of three independent measurements.
Figure S7. Venn diagrams of gene expression data at the different time points in the four high‐adherence strains. Genes with RPKM >5 were used in the analysis. The diagram shows that more than 90% of genes were commonly expressed among the four strains at each time point.
Figure S8. A. Venn diagram of genes expressed in high‐adherence strains at different stages of conidial formation. Numbers of prospective stage‐specific genes are encircled and labelled a to e in the diagram. a, Mycelial stage (stage 0 in Figure 3); b, stipe and vesicle formation (stage 1); c, phialide formation (stage 2); d, conidiation (stage 3); and e, conidial maturation (stage 4). Genes with RPKM >5 were used in the analysis. For distinguishing gene expressions between vesicle formation and conidial formation stages, gene expression values at 12 h were omitted in this analysis, B. Stacked numbers and proposed or hypothetical functions of the stagespecific genes.
Table S1. Aspergillus strains used in this study.
Table S2. PCR primers used for gene disruption and complementation


