Abstract
Background
Unravelling domestication processes is crucial for understanding how species respond to anthropogenic pressures, forecasting crop responses to future global changes and improving breeding programmes. Domestication processes for clonally propagated perennials differ markedly from those for seed-propagated annual crops, mostly due to long generation times, clonal propagation and recurrent admixture with local forms, leading to a limited number of generations of selection from wild ancestors. However, additional case studies are required to document this process more fully.
Scope
The olive is an iconic species in Mediterranean cultural history. Its multiple uses and omnipresence in traditional agrosystems have made this species an economic pillar and cornerstone of Mediterranean agriculture. However, major questions about the domestication history of the olive remain unanswered. New paleobotanical, archeological, historical and molecular data have recently accumulated for olive, making it timely to carry out a critical re-evaluation of the biogeography of wild olives and the history of their cultivation. We review here the chronological history of wild olives and discuss the questions that remain unanswered, or even unasked, about their domestication history in the Mediterranean Basin. We argue that more detailed ecological genomics studies of wild and cultivated olives are crucial to improve our understanding of olive domestication. Multidisciplinary research integrating genomics, metagenomics and community ecology will make it possible to decipher the evolutionary ecology of one of the most iconic domesticated fruit trees worldwide.
Conclusion
The olive is a relevant model for improving our knowledge of domestication processes in clonally propagated perennial crops, particularly those of the Mediterranean Basin. Future studies on the ecological and genomic shifts linked to domestication in olive and its associated community will provide insight into the phenotypic and molecular bases of crop adaptation to human uses.
Keywords: Adaptation, phylogeography, introgression, Oleaceae, pathogen, microbes
INTRODUCTION
The cultivated olive (Olea europaea L. subsp. europaea var. europaea; Box 1) is considered to be the most iconic tree of the Mediterranean Basin, with origins linked to the emergence of some of the most ancient civilizations, about six millennia ago (Loumou and Giourga, 2003; Kaniewski et al., 2012; Zohary et al., 2012). In classical times, olive cultivation expanded to new regions and intensified around the Mediterranean Basin and beyond (Infante-Amate et al., 2016). Today, hundreds of olive varieties are grown to produce high-quality fruit for oil and for table consumption (Bartolini et al., 2005), but debate about their origins continues (e.g. Díez et al., 2015; Besnard and Rubio de Casas, 2016). The relationships between cultivated olives and wild Mediterranean olives [Olea europaea subsp. europaea var. sylvestris (Mill.) Leh., or the so-called oleaster; Box 1] are also unclear. The multiple uses of cultivated and wild olive trees, as sources of food, wood and cattle fodder, explain the expansion of olive groves with the spread of human civilization. The dual role of olives as both wild elements of the Mediterranean vegetation and as a cultivated crop has posed challenges to researchers trying to decipher the domestication history of this species. It also remains difficult to distinguish between feral (escaped from cultivation) and genuinely wild Mediterranean olives, even with the recently developed use of genetic and phenotypic traits to assist identification (Box 1). Such a tenuous domestication syndrome is a key issue that has affected research carried out on domestication of the olive, but also of other Mediterranean woody crops such as grape and date palm (Zohary et al., 2012). For all these reasons, there has long been speculation about the origin and domestication history of olives, mostly based on botanical data (e.g. Newberry, 1937; Chevalier, 1948; Turrill, 1951).
Box 1. Ecology of wild olives and the domesticated status of cultivated olives
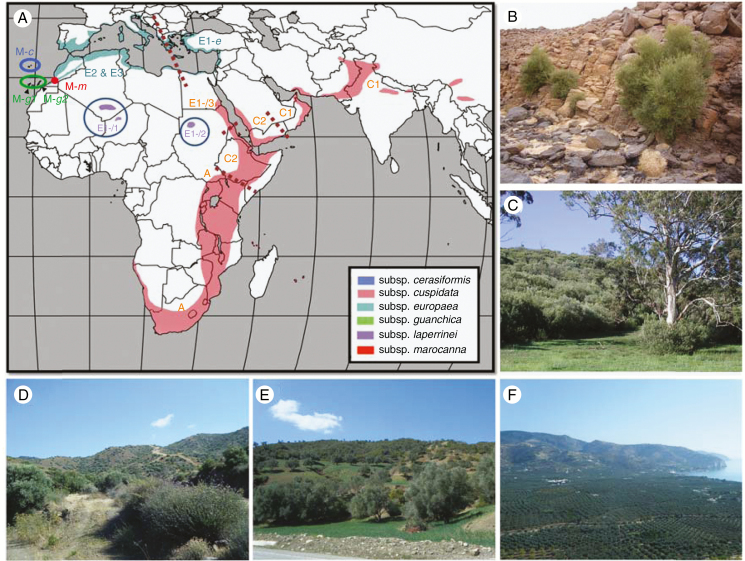
Six wild olive subspecies are currently recognized (Fig. B1A) and considered to be primary genetic resources for cultivated olive breeding (Zohary, 1994; Green, 2002). They are diploid, except for subspp. maroccana and cerasiformis, which are polyploid (6x and 4x, respectively; Besnard et al., 2008). In tropical and sub-tropical regions, non-Mediterranean olives (subspp. cuspidata and laperrinei) harbour small fruits (diameter generally <8 mm; Médail et al., 2001) and trees usually grow in mountainous areas (Fig. B1B). The African olive (subsp. cuspidata) can also invade anthropogenic habitats, as observed in Australia (Fig. B1C). The taxonomy of the olive complex is relatively well supported by genetic data (e.g. Rubio de Casas et al., 2006; Besnard et al., 2007). Each subspecies harbours specific plastid lineages/sub-lineages, with several lineages/sub-lineages detected within the four diploid subspecies. The 13 plastid lineages/sub-lineages are specified on the map (Fig. B1A).
Fig. B1.
The olive complex (Olea europaea L.). (A) Native distribution of wild olive relatives (according to Rubio de Casas et al., 2006). Six subspecies are currently recognized (Green, 2002). The plastid DNA data set used to define lineages (and sub-lineages) is available in Supplementary Data Table S2 and Fig. S2. Dotted lines indicate approximate limits of the distributions of two adjacent plastid lineages (indicative for putative secondary contacts). Note that lineages E2 and E3 are admixed in western Oleaster populations (subsp. europaea); (B–F) Various habitats with wild or cultivated olives in native and invasive ranges. (B) Ramets from the same stump of a Laperrine’s olive at Akerakar, south Algeria. Subspecies laperrinei persists in very dry habitats (mean annual rainfall <100 mm); (C) African olive invasion at Mt Annan, NSW, Australia (photo credit: Peter Cuneo). Subspecies cuspidata is highly invasive in east Australia, north New Zealand and Hawaii. It usually colonizes disturbed habitats, such as abandoned pastures in particular (Cuneo and Leishman, 2006); (D) scrubland dominated by oleasters at Lageia, Cyprus; (E) Traditional agrosystem with cultivated olives in northern Morocco (Chefchaouen, Rif). Annual crops (here, wheat) are usually cultivated between trees; (F) monoculture of olive trees near Mattinata, Puglia, Italy. A small number of genotypes (usually one or two major clones) are generally cultivated in such agrosystems.
Nuclear and plastid DNA data (e.g. Angiolillo et al., 1999; Besnard et al., 2007) show that the main wild progenitor of the cultivated olive (O. europaea subsp. europaea var. europaea) is the wild Mediterranean olive, also known as the oleaster (O. europaea subsp. europaea var. sylvestris; Fig. B1D). Two main oleaster genepools have been identified in the Western and Eastern Mediterranean Basin (Besnard et al., 2001b, 2013b; Breton et al., 2006; Belaj et al., 2007, 2011; Díez et al., 2015). Two specific plastid lineages (E2 and E3) are admixed in the Western Mediterranean Basin, and one lineage (E1) is highly diversified in the Eastern Mediterranean Basin (Besnard et al., 2013a). Nuclear DNA sequences have also revealed two or three divergent lineages of alleles in oleaster, with a weaker geographic structure than for chloroplast markers (Besnard and El Bakkali, 2014). This contrasting phylogeographic patterns between plastid and nuclear markers can be explained by differences in inheritance pattern (i.e. maternal for chloroplast markers and biparental for nuclear markers). However, the absence of a strong phylogeographic signal for nuclear markers probably reflects metapopulation dynamics, with recurrent breaks in gene flow and reconnections due to environmental changes (Rubio de Casas et al., 2006).
Oleaster and cultivated olive have overlapping distributions, ecological and morphological features, to the extent that the domesticated nature of olive is often questioned, as for many other perennials (Miller and Gross, 2011; Gaut et al., 2016; Gros-Balthazard et al., 2016). Historically, olive domestication processes have involved the selection of trees propagated by vegetative means and the repeated cultivation of spontaneously growing trees with favourable agronomic traits. Such mixtures of practices have continually shaped ecological, morphological and genetic differentiation between wild and cultivated trees since pre-historic times, through a long-standing process aiming to optimize fruit production (e.g. Terral et al., 2004a; Newton et al., 2014), and they continue to occur at various sites throughout the Mediterranean Basin. On dissection, cultivated olives and genuine oleaster fruits can be differentiated on the basis of ecological, morphological and genetic differences. Both cultivated and wild olive populations display significant genetic differentiation, despite the occurrence of numerous feral and admixed forms (escapees from cultivation), which considerably blur the pattern of genetic and phenotypic differentiation (e.g. Bronzini de Caraffa et al., 2002; Breton et al., 2006; Belaj et al., 2007, 2011; Hannachi et al., 2008; Besnard et al., 2013a, b; Dίez et al., 2015). This genetic differentiation is less strong in olive than in other fruit trees (Cornille et al., 2014; Gaut et al., 2015), possibly due to the particularly long generation time (>100 years for population turnover) of this species and recurrent cultivated–wild gene flow. Abrupt changes in the size and shape of olive stones during the Bronze Age (from round to elongate), with determinants other than environmental factors (Terral and Mengüal, 1999), also provide evidence for genetic changes in the exploited germplasm. Finally, quantitative trait analyses have revealed a panel of phenotypic and genetic differences between cultivated and wild olives for agronomic traits (Table 1). Furthermore, as genotypes can be maintained over long periods, somatic mutations may have also arisen (e.g. self-compatibility; Breton et al., 2014), generating phenotypic and genetic variability (Gaut et al., 2015).
The recent accumulation of paleobotanical, archeological, historical and molecular data (e.g. Terral et al., 2004a; Carrión et al., 2010; Kaniewski et al., 2012; Besnard et al., 2013b; Margaritis, 2013; Newton et al., 2014; Dίez et al., 2015; Rugini et al., 2016; see Supplementary Data Table S1 for current available genetic data) has made a crucial re-evaluation of the biogeography of wild olives and the history of their cultivation timely. Major issues in the domestication history of the olive remain to be resolved, and these points are dealt with in this review. Further investigations are required to elucidate the processes underlying the primary domestication and subsequent secondary diversification of olives. Besides, little is known about the selection by humans of agronomic or adaptive phenotypic traits in olive. Improving our understanding of the ecology of both wild and cultivated olives is also a critical step to deciphering their domestication history and assessing their probable future resilience to global changes (Holliday et al., 2017). Finally, a lot still needs to be learnt about the biotic and abiotic interactions of olives with their environment. The recent spread of major pathogens and pests remains also poorly explained (e.g. Verticillum, Phytophtora, olive fly; Martelli et al., 2002; Nardi et al., 2010). Xylella fastidiosa, a bacterium introduced from America and then disseminated by native vector insects in South-East Italian olive groves, is a prime example of the emergence of a pathogen, outbreaks of which pose a serious threat to both cultivated and wild olives (Saponari et al., 2014).
Here, we review the timeline of olive evolution in the Mediterranean Basin from its origin during the Quaternary until its domestication, diversification and selection for cultivation nowadays. In the light of new paleobotanical, archeological, historical and molecular data, we discuss the questions that remain unanswered, or even unasked, and we propose to study the ecological genomics of wild and cultivated olives to unravel the puzzle of olive domestication. In particular, we discuss perspectives of multidisciplinary researches and highlight recent technological advances in genomics, metabarcoding and ecological modelling that hold great promise for further documentation of the domestication process of the olive, as well as of other woody crops. These findings should contribute to the sustainable management of cultivated olive germplasm for future breeding programmes and for the conservation of wild olive populations, particularly in the current context of global change.
LATE NEOGENE PALEOGEOGRAPHICAL AND PLEISTOCENE CLIMATIC CHANGES HAVE SHAPED THE GENETIC DIVERSITY OF OLEASTER POPULATIONS
The natural distribution of wild olives (Olea europaea) extends from South Africa to South Asia, and encompasses the Saharan mountains, Macaronesia and Mediterranean countries (Green, 2002; Box 1). Paleobotanical data attest to the occurrence of Olea sp. in Europe during the Oligocene–Miocene boundary (gypsum flora of Aix-en-Provence, South-east France; de Saporta, 1873). This taxon became an important component of the vegetation of the Mediterranean region during the Early Miocene (Suc et al., 1984; Palamarev, 1989; Terral et al., 2004b), and was probably an ancestor of the oleaster (Box 1). Phylogenetic analyses have suggested that the most recent common ancestor of the six olive subspecies (Fig. 1) was present during the Late Miocene or Early Pliocene [about 4.0–8.3 million years ago (Mya); Besnard et al., 2009]. Gene flow between North African and tropical African olive populations has long been limited, due to successive phases of aridification of the Saharan region from the Late Miocene until the present (Schuster et al., 2006; Swezey, 2009), although conditions in this area were more humid from 11 800 to 4900 years before the present (BP) (Tierney et al., 2017). This limitation of gene flow may explain the early divergence of subsp. cuspidata from the other subspecies (Besnard et al., 2009). The divergence of the three extant plastid lineages of oleaster (E1, E2 and E3; Box 1; Fig. 1; Supplementary Data Fig. S1) has been dated to the Late Pliocene or Early Pleistocene (Besnard et al., 2013b), possibly around the time of the Messinian salinity crisis (Miocene/Pliocene). With the gradual establishment of the Mediterranean climate, thermophilic and xerophytic plant species (e.g. evergreen Quercus sp., Olea sp. and Pistacia sp.) emerged and rapidly replaced sub-tropical species (Suc et al., 1984).
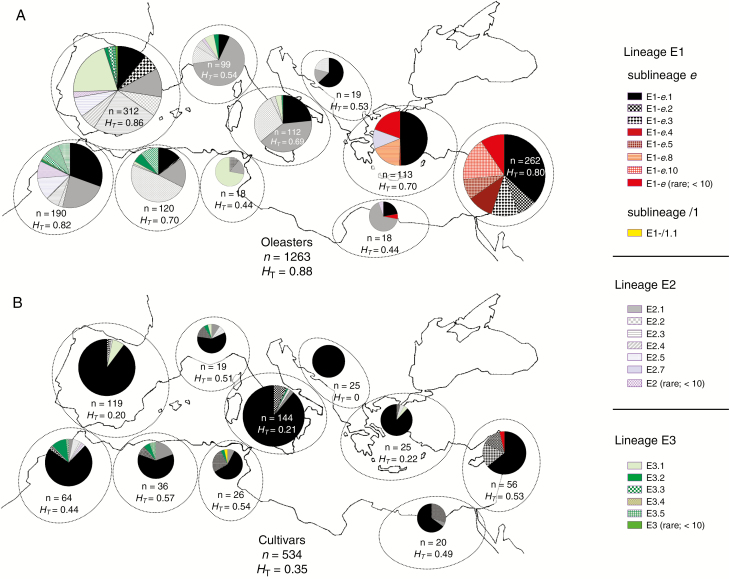
Fig. 1.
Distribution of chlorotypes (A) in oleaster populations, and (B) in the Mediterranean cultivated olive [based on data from Besnard et al. (2013b)]. Each chlorotype is represented by a specific motif, as defined in Supplementary Data Fig. S1. The number of accessions (n) and the total diversity (HT; Nei, 1987) of chloroplast DNA variation are given for each area and for the total sample. The size of pie charts is proportional to the number of individuals analysed per area. Chlorotypes primarily found in oleaster were mostly observed in the East (from the Peloponnese to the Levant; lineage E1) and westernmost part of the Mediterranean Basin (Iberian Peninsula and Morocco; lineages E2 and E3). Note that the geographic distribution of chlorotypes is clearly different between wild and cultivated olives, despite the extensive admixture of these two forms. In the western Mediterranean oleaster, lineage E1 is represented by only three chlorotypes (E1-e.1, E1-e.2 and E1-e.3) that were recently introduced into this area with the human-mediated spread of Levantine cultivated olives (Besnard et al., 2013b). For a detailed distribution of chlorotypes at the population level, see Besnard et al. (2013b).
The Pleistocene (from about 2.58 Mya to 11 700 BP) was also characterized by extreme climatic events, with cold and dry periods generally less favourable to thermophilic Mediterranean species such as the olive. These environmental shifts influenced the distribution of many species and left imprints in their genomes (Hewitt, 2004). The geographic distribution and genetic structure of wild olive populations are the result of a long process of recursive contraction and expansion in response to climatic shifts, and of the limited gene flow imposed by geographic distance and natural barriers, such as deserts and seas. Studies based on nuclear microsatellite markers have revealed the presence of two different oleaster genepools in the Western and Eastern Mediterranean Basin [i.e. ‘Wild East’ (WE) and ‘Wild West’ (WW), Fig. 2; Breton et al., 2006; Besnard et al., 2001b, 2013b; Díez et al., 2015]. In parallel, plastid markers have revealed strong differentiation between eastern and western oleaster populations (Fig. 1A). Before the spread of both oleaster and cultivated olives by humans, the E1 plastid lineage was probably restricted to the east, from the Levant to Greece, whereas the other two plastid lineages (E2 and E3) were specific to the western and central regions (Besnard et al., 2013b). A coalescent-based Bayesian approach indicated that the three Mediterranean plastid lineages diversified long before the Last Glacial Maximum, probably during the long MIS5 interglacial period (from 130 000 to 74 000 BP; Besnard et al., 2013b). Nuclear and plastid DNA analyses also revealed a natural zone of contact between eastern and western wild olive populations in the Peloponnese (Besnard et al., 2013a, b). Similar geographic patterns of genetic differentiation and diversification have been observed for two other shrubs found around the Mediterranean [Laurus nobilis and Myrtus communis (Rodríguez-Sánchez et al., 2009; Migliore et al., 2012)], suggesting that the climatic changes during the Late Pliocene and Pleistocene had similar effects on different Mediterranean species.
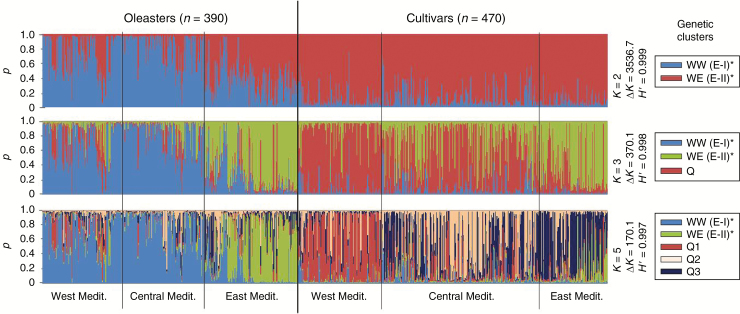
Fig. 2.
Bayesian inference of population structure (based on ten nuclear microsatellite loci) in the Mediterranean olive (including both cultivated and wild accessions; 860 individuals), for K = 2, 3 and 5 clusters [modified from Besnard et al. (2013a)], inferred with a model-based clustering method implemented in STRUCTURE v.2.3.4 (Pritchard et al., 2000). Q is the membership coefficient. H’ is the similarity coefficient between ten runs for each K, and ΔK is an ad-hoc measure described by Evanno et al. (2005). According to ΔK and H’, the most probable genetic structure model is K = 2 clusters (ΔK = 3536.7 and H’ = 0.99), with most wild accessions from the Western and Central Mediterranean Basin (cluster WW; * or E-I in the article by Besnard et al., 2013a) distinguished from cultivars and eastern wild accessions (cluster WE; or E-II); at K = 3, the western oleaster cluster remains but eastern cultivated olives and oleasters are distinguished from western and central Mediterranean cultivars (cluster Q); and at K = 5, a trend for the occurrence of a cultivar cluster in each pre-defined geographic zone [i.e. West (Q1), Central (Q2) and East (Q3)] is revealed, as reported by Haouane et al. (2011), Belaj et al. (2012) and Díez et al. (2012). Central Mediterranean cultivars revealed the highest level of admixture among the Q1, Q2 and Q3 genepools, consistent with the inferred admixed ancestry of most of its genotypes, whereas western and eastern cultivars were found to be more strongly assigned to their respective genepools. The five genetic clusters are named as described by Díez et al. (2015). In each group (oleaster or cultivars), the individuals are classified on the axis, from left to right, according to their geographic origin (from west to east).
Genetic markers have also facilitated the identification of hotspots of diversity in the wild olive and tracing of the origins of cultivated genotypes, through the characterization of nuclear and plastid DNA genepools, in particular (Box 1). Many plastid haplotypes (or chlorotypes) in oleaster are confined to small areas, but a few have spread across the Mediterranean Basin (Besnard et al., 2013b). Three regional diversity hotspots for plastid DNA have been identified in the Levant, the Aegean region and extending from southern Spain to Morocco (Gibraltar to the Anti-Atlas). The high degree of genetic diversity in these three areas suggests that they have served as long-term refugia for the oleaster, a hypothesis supported by paleobotanical data (Figueiral and Terral, 2002; Carrión et al., 2003; Terral et al., 2004b;Willner et al., 2009; Biltekin et al., 2015) and species distribution modelling (Besnard et al., 2013b). Barriers to dispersal (e.g. the Libyan Desert, Adriatic and Aegean Seas, and Rechinger’s Line in southern Turkey) and ecological factors (e.g. biotic interactions or abiotic factors) may also have played a crucial role in limiting long-distance dispersal, thereby maintaining strong genetic differentiation between distant geographic areas even during favourable, interglacial periods.
HUMAN-MEDIATED SPREAD AND FIRST USES OF OLEASTER DURING THE HOLOCENE
During the post-glacial period (about 11 700 to 8000 BP), wild olive populations recolonized the Mediterranean area from refugia, as shown by the fossil and sub-fossil records (Figueiral and Terral, 2002; Terral et al., 2004b; Carrión et al., 2010). It is thought that the expansion of olive populations was triggered by climate and then favoured by human activity, as habitats were cleared (e.g. green oak deforestation; Terral and Mengüal, 1999; Figueiral and Terral, 2002; Combourieu-Nebout et al., 2013). Indeed, the abundance of olive trees in Holocene palynological records steadily increased with human activity in both the Eastern and Western Mediterranean Basin (Carrión et al., 2010). Several authors have documented the human exploitation of oleaster since the Upper Paleolithic and Early Neolithic (Kislev et al., 1992; Terral, 2000; Terral et al., 2004a; Carrión et al., 2010; Kaniewski et al., 2012; Zohary et al., 2012). This early exploitation and further spread was probably linked to various uses, including fruit consumption, wood and cattle forage (Renfrew, 1972; Terral, 2000; Margaritis, 2013). Wild olive foliage is still frequently consumed by cattle in the Cádiz province (Spain), Corsica and Morocco, where this species remains a major component of the landscape (Box 1). Olive trees also provide livestock with shade in open habitats. Oleaster was integrated into early Mediterranean agrosystems, as a cornerstone species, but its early exploitation and spread around the Mediterranean Basin and beyond [e.g. Tassili n’Ajjer (Besnard et al., 2013a)] was considered by Renfrew (1972) and Margaritis (2013) to be a pre-domestication step. Indeed, the practice of pruning oleasters may have greatly favoured flowering and thus fruit production, a critical step for olive domestication, even before the selection and propagation of selected clones for their agronomic value.
EARLY OLIVE DOMESTICATION DURING PRE-HISTORIC TIMES AND THE REASONS FOR ITS SUCCESS
Olive domestication is characterized by vegetative propagation of the most valuable genotypes (Zohary et al., 2012), selected for their agronomic value (such as higher fruit set, larger fruits and higher oil content), for their ability to grow in anthropogenic environments and the ease with which they could be vegetatively propagated through cuttings or grafting. Olive cutting is relatively easy, but not the best way to propagate this species, because cuttings of cultivated varieties may have undesirable features (e.g. high sensitivity to soil pathogens). Grafting is considered to be a major innovation in the history of temperate and Mediterranean fruits and probably favoured the spread of these crops from the Middle East and Central Asia to Western Europe (Juniper and Maberly, 2006). The grafting of cultivated olive varieties onto local oleaster or ancient cultivars is a widespread practice (Díez et al., 2011; Barazani et al., 2014; Aumeeruddy-Thomas et al., 2017). Evidence of olive grafting is missing from the archeological record, but this practice is documented in texts and epigraphic data (Mudge et al., 2009). Olive grafting has been reported since Classical times, particularly in Ancient Greece and Ancient Rome, and is designed to provide productive varieties with more hardy roots (in Pease, 1933). In contrast to other fruit crops, such as grape or apple (Myles et al., 2011; Cornille et al., 2014; Warschefsky et al., 2016), the intentional breeding of rootstocks is poorly documented in olive (Barazani et al., 2014, 2017). Barazani et al. (2017) recently showed that scion/rootstock genotype combinations are not randomly distributed, suggesting that growers may have selected some combinations in the Levant area, possibly to improve oil quality or drought tolerance.
Elucidating ancient cultural shifts linked to the beginning of cultivated olive use and breeding is not a straightforward task (e.g. Galili et al., 1997; Terral et al., 2004a; Kaniewski et al., 2012; Zohary et al., 2012; Newton et al., 2014). Indeed, the multiple uses of olive trees have made it difficult to interpret archeological and sub-fossil data (i.e. pollen and charcoal abundance; Kaniewski et al., 2009). Olive domestication can be viewed as an ecological and historical process, that can be studied within a multidisciplinary framework, through the use of archeological, archeobotanical, historical and genetic approaches.
The archeological and archeobotanical evidence
Major human civilizations emerged in the Middle East during the Neolithic, and archeological remains have demonstrated the involvement of these civilizations in an olive oil trade during the Chalcolithic period (6000–5500 BP; Galili et al., 1997; Kaniewski et al., 2012; Zohary et al., 2012; Newton et al., 2014). Olive domestication is generally thought to have begun after this period (Liphschitz et al., 1991; Galili et al., 1997). Several studies have shown that olive cultivation was a regionally and temporally diverse process, possibly due to climatic fluctuations (e.g. Kaniewski et al., 2012; Langgut et al., 2016; Dighton et al., 2017). Olive domestication probably occurred in a diffuse manner in the Levant, making it difficult to track early origins based on archeobotanical data alone. In areas far outside the natural range of the oleaster, numerous finds of charred wood and stones associated with dates, pulses and cereals have provided indirect proof of technological developments in agronomy, such as seedling transplantation and crop irrigation (Neef, 1990; Newton et al., 2006; Zohary et al., 2012). Kislev et al. (1995) used traditional morphological stone characters to describe numerous Chalcolithic olive remains (dating from about 5600 BP) from a submerged site off the Carmel coast in Israel. Their findings provide evidence supporting the earlier existence of cultivation in localized regions. Kislev et al. (1995) showed that oleasters growing near the site were harvested by inhabitants, and concluded that the olive industry preceded olive domestication by several centuries.
Archeological and archeobiological records (i.e. changes in the size and shape of olive stones; Table 1) have also provided evidence of olive exploitation and management in the Central and Western Mediterranean during the Chalcolithic/Bronze Age (4500–4000 BP), an extensive process that was probably initiated during the Neolithic (e.g. Terral and Arnold-Simard, 1996; Terral, 2000; Terral et al., 2004a,Margaritis, 2013; Pagnoux, 2016; D’Auria et al., 2017). In the southern and eastern Iberian Peninsula, abrupt changes in the shape of charred olive stones recovered from Bronze Age settlements suggest the presence of domesticated olives two millennia before the arrival of the Phoenicians and Roman colonization, to which the introduction of new varieties into the region and the development of olive farming, respectively, are generally attributed (Brun, 2003, 2004; Terral et al., 2004a). A concomitant sudden increase in the numbers of olive stone remains was detected at Eastern and Western Mediterranean archeological sites, corresponding to the Late Bronze Age (about 3500–3000 BP; Terral et al., 2004a; Newton et al., 2014), but morphotypes typical of the Middle East did not appear in the western archeological record until 1000–1500 years later (Newton et al., 2014), attesting to the fundamental role played by East–West human migrations (e.g. (Chikhi et al., 2002) in the constitution of olive agrobiodiversity in the Western Mediterranean Basin.
Table 1.
Main characteristics usually used to distinguish between oleaster and cultivated olive in the Mediterranean Basin
| Criteria | Oleaster | Cultivated olive | Remarks | References |
|---|---|---|---|---|
| Ecology | Scrublands (generally in association with Pistacia lentsicus, Rhamnus, Rosmarinus officinalis, evergreen Quercus spp., Pinus spp.) | Agrosystems (crop fields, gardens) | Wild and feral olives may be encountered in similar habitats, but genuine oleaster populations may be restricted to remote areas. Non-cultivated olives locally frequent in the agrosystem (e.g. pastures in southern Spain or northern Morocco) | Alcántara and Rey (2003), Lumaret et al. (2004), Carrión et al. (2010), Zohary et al. (2012) |
| Fruit traits (size, shape and oil content) | Round to elongate, diameter <8 mm, less fleshy mesocarp and low oil content (about 5–15 % of wet matter). Bird dispersed. | Usually elongate, diameter 10–20 mm, fleshy mesocarp and high oil content (about 15–30 % of wet matter) | Abrupt morphological changes during olive domestication, as demonstrated by archeobotanical data. Feral olives or admixed forms between wild and cultivated olives may harbour intermediate traits. The genetic basis of fruits traits have been explored. | Terral et al. (2004a), Médail et al. (2001), Hannachi et al. (2008), Belaj et al. (2011), Zohary et al. (2012), Atienza et al. (2014), Pérez et al. (2014) |
| Tree architecture | Slow-growing, old trees often multistemmed, young trees with spinescent juvenile shoots | Human management promoting fast growth and more regular fruiting. Usually more vigorous trees than in the wild. Mostly vegetatively propagated (grafting or cuttings). | Architectural traits influenced by environmental conditions, but genetic factors strongly control growth and branching. Higher vigour of trees possibly due to heterosis | Zohary et al. (2012), Biton et al. (2012), Ben Sadok et al. (2013, 2015) |
The genetic evidence
The genome of current olive varieties bears witness to the origin of cultivated olives largely from oleaster populations in the Eastern Mediterranean region, consistent with a primary origin in the Middle East (e.g. Besnard et al., 2001b, 2013a; Lumaret et al., 2004; Baldoni et al., 2006; Breton et al., 2006; Dίez et al., 2015). Three chlorotypes, common to wild olives and belonging to lineage E1 (i.e. E1-e.1, E1-e.2 and E1-e.3), characterize about 90 % of cultivars (Fig. 1; Supplementary Data S1). These chlorotypes are now observed in feral olives throughout the Mediterranean Basin. The chlorotypes of the remaining 10 % of cultivars belong to the E2 and E3 lineages (i.e. E2.1, E2.2, E3.1 and E3.2). Based on the current distribution of E1 diversity in oleasters, the main cultivated olive chlorotypes (i.e. E1-e.1, E1-e.2 and E1-e.3) are thought to have originated along the current border between north-west Syria and south-east Turkey, close to an area inferred to be suitable for the long-term persistence of olive trees during the Late Pleistocene (Besnard et al., 2013b). In contrast, a recent study suggested that wild olives may have persisted over long time periods on the Mount Carmel Coast (north-west Israel), where early cultivation has been reported (Kislev et al., 1992), and trees from this area may have contributed to the cultivated olive genepool (Barazani et al., 2016). However, several genetic studies have shown that the oleaster populations of the Mount Carmel area are mostly feral (e.g. Besnard et al., 2001b; Lumaret et al., 2004; Díez et al., 2015). The presence of long-standing populations should be also supported by endemic chloroplast haplotypes, but chloroplast profiling has revealed the presence of the main cultivated haplotype only (E1-e.1; Besnard et al., 2013b), suggesting that these local wild forms probably escaped from cultivation.
Nuclear markers have also confirmed the strong eastern affiliation of the cultivated genepool, although a significant contribution of western oleasters has also been demonstrated in the Central and West Mediterranean Basin (Besnard et al., 2001b, 2013a; Lumaret et al., 2004; Breton et al., 2006; Díez et al., 2015). In addition, studies of the nuclear genetic diversity of cultivated olive have revealed a weak genetic structure in the Mediterranean Basin, mostly explained by geographic origin and different uses (i.e. oil or as whole fruits) of varieties (e.g. Claros et al., 2000; Belaj et al., 2001; Besnard et al., 2001a; Hagidimitriou et al., 2005; Owen et al., 2005; Marra et al., 2013; Linos et al., 2014; Yoruk and Taskin, 2014; Biton et al., 2015). These data, therefore, support multiple geographic origins of cultivars, but reflect a process of diversification in the Central and West Mediterranean Basin (Besnard et al., 2001b, 2013a; Díez et al., 2015). However, these studies based on nuclear markers are not entirely satisfactory, because they were based on limited oleaster samples or small numbers of loci. Additional analyses will be required to refine the domestication and diversification scenario for cultivated olives (Box 2). Additional sampling of genuinely wild olives will also be required to determine whether there were diffuse origins of domesticated olives in the Middle East (e.g. Barazani et al., 2016; Langgut et al., 2016; Dighton et al., 2017).
Finally, the local selection of cultivars by farmers may have played a key role during the recent diversification history of the cultivated olive, as described for other woody crops, such as grapes (Bowers et al., 1999; Myles et al., 2011; Bacilieri et al., 2013). Parentage analyses on cultivated genotypes from the same area (e.g. Marra et al., 2013; Dίez et al., 2015) revealed a narrow genetic basis of the current elite olive material in the West Mediterranean Basin. In particular, Dίez et al. (2015) used a large number of cultivars to demonstrate that most south Iberian varieties (group Q1) have a recent origin following a strong bottleneck (possibly during the Al-Andalus period; about 711 to 1492 AD). Using co-ancestry analyses, they identified two ancient genotypes that may have been the main progenitors of Q1. Thus, selection of the Q1 cultivar group (which includes some of the most important varieties worldwide, such as ‘Picual’, ‘Cornicabra’, ‘Hojiblanca’, ‘Manzanilla de Sevilla’ and ‘Picholine Marocaine’) was initially based on a very small number of genotypes.
Box 2. Single or multiple independent domestications: a challenging enigma.
New and older lines of evidence clearly indicate the existence of multiple centres of diversity for cultivated olive trees. The characterization with nuclear microsatellites of comprehensive samplings of modern olive cultivars by independent research teams revealed the existence of three main cultivated genepools (Figs B2A, B) corresponding to three geographic areas: the west, centre and east Mediterranean Basin (i.e. Q1, Q2 and Q3, respectively, according to Dίez et al., 2012, 2015; see also Haouane et al., 2011; Belaj et al., 2012; Besnard et al., 2013a). Other studies have reported regional structure, probably due to the selection of very closely related individuals locally (e.g. Khadari et al., 2003; Breton et al., 2006; Albertini et al., 2011; Muzzalupo et al. 2014). It remains unclear whether the centres of diversity result from one or multiple local domestication events. Dίez et al. (2015) stressed the relatively close association between the Q2 group and the WW wild genepool, with much greater plastid variation in Q2 than in Q1 and Q3 (Fig. B2C). The Q2 group may, therefore, represent a separate domesticated lineage originating from WW that subsequently admixed with cultivated eastern germplasm (Q3). Dίez et al. (2015) favoured this hypothesis of an independent domestication of Q2 because (1) multiple maternal plastid lineages may reflect multiple domestication events; (2) admixture estimates suggest a substantial proportion of the WW group (20 %) in Q2; and (3) Q2 cultivars bear some wild-like phenotypic characteristics, such as low endocarp weight and a smooth endocarp surface (Belaj et al., 2011; Klepo et al., 2013). This hypothesis remains to be explored, because cultivated olive diversification may also have occurred in the Central and Western Mediterranean Basin not as the result of local independent domestication, but as a consequence of admixture between local unselected pre-domesticated oleaster and cultivars introduced from the east (Besnard et al., 2001b, 2013a, b; Lumaret et al., 2004; Baldoni et al., 2006; Dίez et al., 2015). In accordance with this second hypothesis, Besnard et al. (2013a) showed, through nuclear microsatellite analysis, that most Mediterranean cultivars were assigned most strongly to the eastern oleaster genepool (cluster WE), whereas no cultivar was unambiguously assigned to the western genepool (WW), including those with plastid lineages originating from the Western Mediterranean Basin (Fig. 2). This finding supports the hypothesis that current elite cultivars either belong to the eastern genepool or are admixed. In addition, several teams have reported a significant excess of heterozygosity in cultivated olive (Dίez et al., 2011; Besnard et al., 2014), consistent with the hypothesis of admixture-mediated diversification of the crop (i.e. a single initial domestication followed by secondary domestication events). Biton et al. (2012) reported hybrid vigour in F1 olive progeny, suggesting that admixture between genepools may generate superior new genotypes (i.e. due to heterosis), particularly for agronomic traits (e.g. fruits weight or oil content).
Alternative scenarios of independent local domestications, primary domestication followed by secondary diversification, or this second scenario plus local domestication events can be formally tested (see propositions in Fig. B2C), by approximate Bayesian computation (ABC), for example (see Díez et al., 2015). Such investigations would require consensual sampling and genetic markers. In particular, caution is required when feral olives are considered, because these trees originate directly from cultivated olive and may be admixed with other spontaneously growing olives (Beghé et al., 2017). Their direct filiation to cultivars is a strong limitation on studies aiming to assign cultivated trees to wild genepools (e.g. Breton et al., 2006), and a large number of genetic markers and large samples of both cultivated and uncultivated olives will be required to distinguish feral trees (escapees from cultivation) from genuinely wild olives. Admixed populations should also be avoided to exclude recent gene flow between cultivated and wild olives.
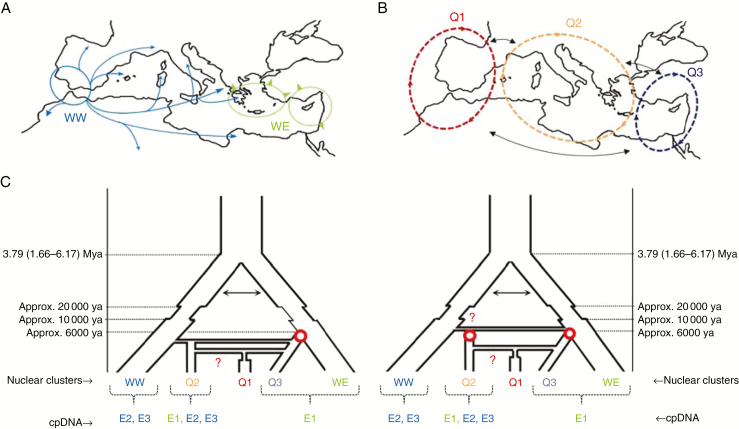
Fig. B2.
Debate about olive evolutionary history and domestication. (A and B) Distribution of wild and cultivated genepools in the Mediterranean Basin (Besnard et al., 2013a; Dίez et al., 2015). (A) WW and WE (E-I and E-II, respectively, in the article by Besnard et al., 2013a) correspond to the western and eastern Mediterranean oleaster populations that expanded extensively during the Holocene, with habitat clearance mediated by humans. Contact is thought to occur in the Central Mediterranean area (e.g. Peloponnese; Besnard et al., 2013a). (B) Q1, Q2 and Q3 correspond to the main genetic clusters of cultivated olive (Fig. 2; named according to Dίez et al., 2015). (C) Simplified scenarios of domestication. Two alternative scenarios of population divergence and admixture are proposed, but there are many other non-exclusive possibilities. In both cases, the divergence of eastern and western oleaster genepools is thought to have started during the Late Pliocene (i.e. based on molecular dating; Besnard et al., 2009, 2013b), with possible gene flow (as indicated by arrows). Population reduction followed by subsequent expansion is also thought to have occurred during the Last Glacial Maximum and the Holocene. On the left, a primary domestication event (red circle) is thought to have occurred in the Eastern Mediterranean Basin, leading to Q3, whereas Q2 and Q1 were derived from admixture events (horizontal connections) between Q3 and WW (for Q2) or Q3 and Q2 (for Q1). For Q1, the red question mark indicates a possible alternative admixture event involving local wild genepools (i.e. WW instead of Q2; see Dίez et al., 2015). A bottleneck is also indicated for Q1, as revealed by Dίez et al. (2015). On the right, two independent primary domestication events are considered, one for Q3 in the Eastern Mediterranean Basin, and one for Q2 in the Central Mediterranean Basin. As Q2 cultivars include a high proportion of E1 haplotypes (see Dίez et al., 2015), and a relatively strong genetic affinity with WE on the basis of nuclear loci (Fig. 2; Besnard et al., 2013a; Dίez et al., 2015), we presume that admixture between WE and WW occurred before domestication, but it would be difficult to distinguish between this scenario and early domestication from WW, followed by introgression from Q3, as suggested by Dίez et al. (2015). The origin of Q1 is identical in both scenarios. Note that feral olives are not shown here, but could be considered as additional populations. Indeed, numerous intermediate forms have probably escaped from cultivation and may have contributed to the diversification of cultivated olives.
BEYOND THE MEDITERRANEAN AND HYBRIDIZATIONS BETWEEN OLIVE SUBSPECIES: NEW OPPORTUNITIES FOR CROP DIVERSIFICATION?
Wild diploid olive subspecies are cross-compatible and can be considered as primary genetic resources for improving the cultivated olive genepool (e.g. Zohary, 1994; Besnard et al., 2008; Hannachi et al., 2009; Klepo et al., 2013; Cáceres et al., 2015), although some genomic incompatibilities may exist. As described above, wild olives are naturally distributed over three continents and grow in contrasting environments (Box 1; Médail et al., 2001; Green, 2002). They have a considerable potential for crop improvement. The introgression of diversity from wild species into cultivated olive might facilitate the introduction of various adaptive traits, such as pathogen and pest resistance and/or lead to the production of vigorous rootstocks resistant to extreme abiotic conditions (e.g. Lavee and Zohary, 2011; Warschefsky et al., 2014; Arias-Calderón et al., 2015; Trapero et al., 2015). Non-natural contacts between the cultivated olive and non-Mediterranean wild relatives have been mediated by humans through the diffusion of cultivars, and gene flow between different olive subspecies has occurred in both the native and introduced ranges of O. europaea (Besnard et al., 2013a, 2014).
Within its native range, the cultivated olive historically spread beyond the boundaries of the Mediterranean Basin, further into the Middle and Far East (from Iraq to south-wesr China), but also into numerous oases (e.g. Palmyra, Syria; Kharga, Egypt; Ziz Valley, Morocco; Kufra, Libya; and Erkowit, north-east Sudan), the Canary Islands and, possibly, the mountains of the Sahara (Besnard et al., 2013a; Noormohammadi et al., 2014; Mousavi et al., 2014, 2017; Hosseini-Mazinani et al., 2014; Zhan et al., 2015; Djamali et al., 2016). During these various introduction events, cultivated olives may have come into contact with the wild subspecies cuspidata (e.g. China, Iran, Arabia and north-east Sudan), guanchica (Canary Islands) or laperrinei (Saharan Mountains). Crop diversification through admixture with close relatives has been documented in other fruits trees over their worldwide range, particularly for grapes (Myles et al., 2011), apples (Cornille et al., 2012), almonds (Delplancke et al., 2013) and date palms (Hazzouri et al., 2015; Zehdi-Azouzi et al., 2015). There have been few studies of such a scenario in olives, but anecdotal cases of early-generation hybrids (BC1 or BC2) between Laperrine’s olive and Mediterranean olives have been reported (e.g. the cultivar ‘Dohkar’; Besnard et al., 2013a).
The cultivated olive has more recently been introduced into new regions, such as the New World, New Zealand and Australia. The introduction of multiple varieties, usually from different origins, has ensured the maintenance of substantial genetic diversity in the cultivated genepool in these regions. During crop diffusion, new genotypes (putatively more adapted to local conditions) were also selected, after uncontrolled crosses occurred between cultivars and feral olives (e.g. do Val et al., 2012; Mousavi et al., 2014; Beghé et al., 2015). The wild African olive (O. europaea subsp. cuspidata) was introduced into Australia, New Zealand and Hawaii (Box 1; Cuneo and Leishman, 2006; Besnard et al., 2014). Mediterranean and African subspecies have both become naturalized in south-east Australia (Cuneo and Leishman, 2006; Cornuault et al., 2015; (Cuneo and Leishman, 2006; Cornuault et al., 2015; Besnard and Cuneo, 2016). In this region, the selection of promising genotypes from naturalized olives has been reported (Sedgley, 2004), and the possibility of introgression into these trees of material from subspecies cuspidata should be investigated (see below).
TOWARD THE ECOLOGICAL GENOMICS OF OLIVE DOMESTICATION
Domestication can be viewed as a window on human history, but also as a model for studying short-term species evolution and adaptation to different ecological conditions (i.e. natural ecosystems vs. agrosystems; Holliday et al., 2017). Studies of the processes of adaptation in the context of domestication are also particularly relevant in terms of the crop community (pests, pathogens or symbionts), with frequent host shifts involving adaptation to new hosts following anthropogenic changes on the environment (Stukenbrock and McDonald, 2008; Xing et al., 2012; Gladieux et al., 2015). In this context, olive is a relevant model for deciphering the evolutionary processes involved in perennial tree species adaptation at different temporal and spatial scales. Both archeological and genetic evidence has demonstrated that olive domestication involved multiple spatial and temporal steps, and that this process is still ongoing (Besnard and Rubio de Casas, 2016; Díez and Gaut, 2016) as in several perennial species (e.g. Miller and Gross, 2011; Myles et al., 2011; Delplancke et al., 2013; Cornille et al., 2014; Hazzouri et al., 2015). However, a full understanding of this complex ecological process will require the elucidation of several aspects of the olive domestication. Indeed, while the major components of olive domestication are now relatively well established, several unanswered, or even unasked, questions remain to be addressed. (1) Can we detect a signature of selection associated with olive domestication, i.e. what are the traits and associated genes under selection in the cultivated olive? (2) Has there been any ecological shift between cultivated and wild olives? If so, with which ecological traits is it associated? (3) Was the associated community (e.g. pests, pathogens and symbionts) affected by olive domestication? Conversely, did the associated community play any role in olive domestication?
The answers to these questions lie in the analysis of signatures of adaptation at both the ecological and genomic levels, and neutral processes need to be taken into account through the accurate determination of olive demographic history (Box 3). Over the next few years, the flourishing of genomics methodologies has opened up new avenues of research for tackling these issues (e.g. McClure et al., 2014; Plomion et al., 2016; Holliday et al., 2017; Migicovsky and Myles, 2017; see Supplementary Data Table S1).
The demographic history of olive: a prerequisite for investigations of the ecological genomics of olive domestication
Archeobiological perspectives for tracking the domesticated lineages.
Analyses of sub-fossil or archeological records will continue to shed light on the history of domesticated olives, particularly their human-mediated spread, and recent advances in archeogenetics and archeogenomics have opened up new perspectives for such studies (e.g. Elbaum et al., 2006; Orlando et al., 2015). Complete plastomes, mitogenomes and a draft of the nuclear genome are now available for olive (Mariotti et al., 2010; Besnard et al., 2011; Cruz et al., 2016; Unver et al., 2017; Van de Paer et al., 2018). These genomic materials should facilitate investigations of targeted genes or genomic regions of ancient DNA under selection during olive domestication, through the use of baiting methods (Orlando et al., 2015). Such approaches have already been used in maize (da Fonseca et al., 2015; Ramos-Madrigal et al., 2016). For olive, single nucleotide polymorphisms (SNPs) have been used to track the spread of the main cultivar chlorotypes from the East to the West Mediterranean Basin (i.e. E1-e.1 and E1-e.2; Besnard et al., 2013b). The characterization of dated sub-fossils for such markers could help to clarify the timing of introduction of the Levantine cultivated lineage into different regions and to determine whether these introductions were linked to abrupt morphological changes in olive stones. These archeogenomic approaches may, however, encounter difficulties in the extraction of endogenous DNA from charred material, even with gene baiting methods (Nistelberger et al., 2016). This approach requires validation for organellar genomes, which should be the most amenable to such analyses due to their haploid nature, large numbers of copies per cell and available data concerning the distribution of their polymorphisms in current wild and cultivated olives.
The origin and parentage of cultivated clones.
Fine genetic characterization of ancient olive individuals on a regional scale may reveal a succession of principal cultivated genotypes (varieties) over time, with the replacement of some clones by others (e.g. new varieties grafted onto ancient ones), possibly due to past political or environmental changes. Monumental trees may, therefore, provide indications about the history of varieties, but it is essential to distinguish between rootstocks (wild or cultivated) and grafted genotypes (e.g. Dίez et al., 2011; Barazani et al., 2014, 2016; Chalak et al., 2015; Lazović et al., 2016). A high diversity would be expected for rootstocks, with mixtures of old varieties and oleaster, and the possibility of successive grafts onto the same tree (e.g. Baldoni et al., 2006; Dίez et al., 2011; Barazani et al., 2014, 2016).
The combination of pedigree reconstruction or parentage analyses with studies of historical records of variety use should also shed light on the process of variety diversification at the regional scale, as already demonstrated in grape (e.g. Sefc et al., 1998; Bowers et al., 1999; Myles et al., 2011). A few parentage analyses based on nuclear microsatellites have been reported for olive (e.g. Marra et al., 2013), but this approach is sensitive to genotyping errors and possible mutations of such highly variable loci (e.g. Barazani et al., 2014; Trujillo et al., 2014). The discriminating power of genetic markers may also be lower in genepools that have undergone strong bottlenecks (especially Q1; Dίez et al., 2015) and, in such cases, numerous unlinked loci (>30–50 microsatellites) may need to be used. With advances in sequencing technologies, thousands of SNPs are now available for the genotyping of olive varieties (e.g. Kaya et al., 2013; Biton et al., 2015; İpek et al., 2016). SNPs are less variable than microsatellites, but they are considered to be more reliable (due to their great stability, easiness of scoring and a better understanding of their mutation rates), and this should make it possible to reconstruct robust pedigrees in the near future. Among the most ancient varieties, it may be possible to identify the major progenitors of current cultivars (e.g. Dίez et al., 2015) and preferential crosses, as in grapes (Bowers et al., 1999).
The demographic history of wild and cultivated olives.
As stated above, the release of a draft genome for olive and recent advances in population genomics will make it possible to acquire large, affordable SNP data sets for further documentation of the timing and geography of olive domestication. Different hypotheses for single or multiple domestication events (Box 2) can be tested by computationally efficient population genomics methods developed for large SNP data sets (Box 3). The role of wild-to-crop and crop-to-crop gene flow in olive diversification also merits further investigation. In apples and grapes, wild-to-crop introgressions are key drivers of crop diversification (Myles et al., 2011; Cornille et al., 2012). Wild-to-crop introgressions were clearly involved in variety diversification in Mediterranean olives (Box 2). Introgressions from non-Mediterranean subspecies may also have played a role (in both the native and introduced ranges). This question has never been addressed by whole-genome approaches. The early generation of hybrids was detected with a few molecular markers (Besnard et al., 2013a; Besnard and El Bakkali, 2014), but the detection of more ancient footprints of introgression (>BC2) will require sequences or SNP data that are gradually becoming more accessible through the use of new sequencing technologies (Supplementary Data Table S1). Crop-to-crop gene flow (i.e. admixture between differentiated cultivated gene pools) may also have contributed to olive diversification. The contribution of the eastern and western wild genepools to the three main cultivated genepools could be assessed by genomic scans to characterize the regions along the chromosomes that have contributed to the genome architecture of cultivated olives (e.g. Scascitelli et al., 2010; Liu et al., 2015). The use of whole-genome sequencing data would also make it possible to determine how many and which parts (and genes) of the genome have been subject to introgression, and whether or not these regions were selected during domestication (Box 3, and see below). The answers to these questions will not only provide fundamental insight into the olive domestication process, but will also help us to understand the genomic processes underlying short-term species divergence in perennials.
Investigating the ecological genomics of olive domestication and its impact on the associated biotic community
Which traits and genes were under selection during olive domestication?
The deciphering of the olive domestication process requires investigations of the impact of human selection on the genome architecture of the olive, including, in particular, the genes and alleles controlling agronomic traits (Box 3). As in other perennial crops, no strong domestication syndrome has been described in olive, and some varieties have been vegetatively propagated over long periods. Selection may therefore not have occurred, or may have been limited to the first few generations after the wild ancestors. This situation may account for the tenuous nature of the domestication syndrome described in many fruit crops, in stark contrast to that of seed-propagated annuals (Glémin and Bataillon, 2009; McKey et al., 2010; Zohary et al., 2012; Gaut et al., 2016). However, there is evidence to suggest that human selection may have led to the adaptive divergence of cultivated perennial crops from their wild relatives (e.g. Cornille et al., 2014). In olive, human selection has resulted in phenotypic, ecological and genetic differences between oleaster and cultivated olives (Table 1). It remains unclear whether or not these differences are adaptive, and further studies are therefore required to determine the genomic basis of olive domestication in an ecological context.
Recently developed genomic approaches (Supplementary Data Table S1; Box 3) can detect footprints of positive selection on genes of agronomic importance (e.g. fruit size, organoleptic properties or tree architecture) and the signature of balancing selection for genes involved in resistance to pests and diseases (e.g. Verticillum, Phytophtora and olive fly). However, the detection of balancing selection on genes of advanced generations of admixed varieties remains challenging (e.g. Fijarczyk and Babik, 2015). Furthermore, the genes of the self-incompatibility (SI) system in olives may have been under strong selection during the process of cultivar selection (e.g. cultivation of compatible genotypes or selection of self-compatible mutants). An unusual sporophytic system with two compatibility groups has recently been described in the olive tribe (Saumitou-Laprade et al., 2017), but the genes and alleles involved have yet to be identified and their diversity has not been investigated in relation to cross-compatibility phenotypes (e.g. Breton et al., 2014).
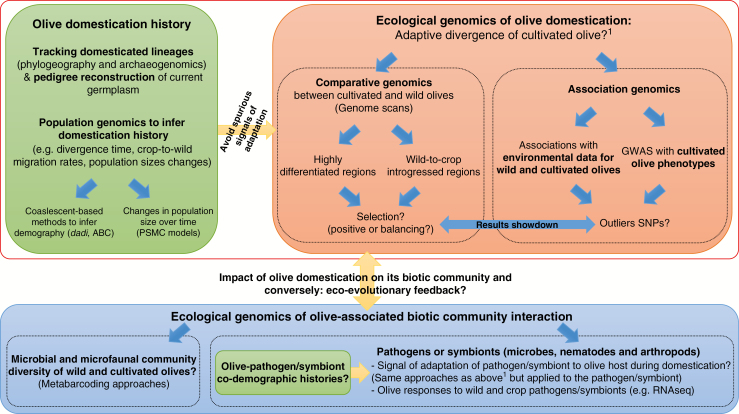
Box 3. Methodological prospects for the ecological genomics of olive domestication and of the olive–biotic community interaction
The recent release of a reference genome for olive and oleaster (Cruz et al., 2016; Unver et al., 2017), together with new population genetic frameworks designed to search for molecular signatures of evolutionary processes and to infer complex demographic histories, have made studies of the genomic consequences of olive domestication timely. Resequencing of the genomes of both wild and cultivated olives (as in date palm, peach and almond; Hazzouri et al., 2015; Velasco et al., 2016) can now be used for comparative and population genomics approaches, to infer demographic history and then to test for a precise genomic signature of adaptation during olive domestication. In parallel, the diversity and evolution of olive-associated biotic communities can also be investigated. The combination of these genomic approaches will provide us with a more precise picture of the ecological and genomic consequences of olive domestication and, more generally, of adaptation in perennials.
THE DEMOGRAPHIC HISTORY OF CULTIVATED AND WILD OLIVES
When testing for signals of adaptation, a lack of knowledge of the demography of the species studied can greatly skew estimates of allele frequency (Tiffin and Ross-Ibarra, 2014). It is therefore crucial to document and infer the demographic history of the model species. The history of olive cultivars can be reconstructed from sub-fossil data (archeogenomics) and current material (e.g. parentage analyses). Different hypotheses for single or multiple domestication events can then be tested with diffusion theory-based models of demographic history (dady; Gutenkunst et al., 2009), MSMC (Schiffels and Durbin, 2014) or by ABC (Beaumont, 2010), as recently reported for several tree species (Cornille et al., 2012; Delplancke et al., 2013; Besnard et al., 2014; Gerbault et al., 2014; Dίez et al., 2015; Mayol et al., 2015; Zhou et al., 2017). Genetic erosion during cultivar selection must also be assessed, with recently developed population genomics methods created for use with whole-genome sequences from hundreds of individuals. In particular, pairwise or multiple sequentially Markovian coalescent (PSMC or MSMC) models can be used to infer changes in population size over time from SNP frequency spectra (Liu and Fu, 2015). However, this method requires high-quality genome sequences (i.e. good sequencing coverage per site over the whole genome), to ensure that the estimated allele frequency spectra are accurate. The olive genome is relatively large (1.5–1.8 Gb; Cruz et al., 2016), so such methods will entail high sequencing costs, and the varieties used must therefore be selected with care (e.g. on the basis of genepool membership – Q1, Q2 and Q3 – or ancient vs. recent cultivars; Gross et al., 2014).
Fig. B3.
Methodological framework to address major questions on the impact of domestication on the evolution of olive and its associated biotic communities.
TESTING FOR SIGNALS OF ADAPTATION DURING OLIVE DOMESTICATION
Recently developed methods can be used to detect different types of selection. These methods include genome scans for signatures of positive selection (e.g. Hufford et al., 2012; Hoban et al., 2016; Booker et al., 2017), with the detection of hard or soft selective sweeps for candidate genes (e.g. Civáň et al., 2015; Akagi et al., 2016; Velasco et al., 2016; Hermisson and Pennings, 2017), and/or balanced selection for traits affected by heterosis or conferring parasite resistance (e.g. Bento et al., 2017). It will also be relevant to investigate whether the genes or genomic regions under positive selection detected have resulted from wild-to-crop gene flow.
The power to detect genes under positive selection associated with olive domestication will be enhanced by large-scale phenotyping efforts spanning multiple years in several environments and including replicates of wild and cultivated individuals in collections or common gardens (e.g. Belaj et al., 2012; El Bakkali et al., 2013; Ben Sadok et al., 2015; León et al., 2016). Over the last 10 years, segregating progenies have been used to identify genomic regions associated with agronomic traits [quantitative trait loci (QTLs) for olive oil quality, fruiting, growth and tree architecture], but the precise genes responsible for these trait variations remain unknown (e.g. Ben Sadok et al., 2013; Atienza et al., 2014; Pérez et al., 2014). Genome-wide association studies (GWAS) with large SNP datasets associated with phenotypic agronomic traits will make it possible to identify genes displaying signatures of recent positive selection associated with phenotypic agronomic traits (e.g. Wright et al., 2005; Migicovsky and Myles, 2017). These methods will require a consideration of olive history (e.g. cultivar filiation) and population structure (e.g. differentiated genepools or admixture; Segura et al., 2012). In particular, the high degree of genetic differentiation between eastern and western wild genepools and the complex diversification of the crop will have to be taken into account, to prevent spurious correlations related to historical effects (Tiffin and Ross-Ibarra, 2014). Finally, environmental association studies (Rellstab et al., 2015) would also make it possible to determine the contribution of local environmental factors to the adaptation of the cultivated olive, and to determine whether ecological differences led to the adaptive divergence between olive and oleaster.
WHAT ABOUT THE OLIVE-ASSOCIATED BIOTIC COMMUNITIES?
Olive domestication may have led to pathogens or symbionts becoming adapted to their crop hosts. This hypothesis can be tested by applying the same methods described above for olive (i.e. comparative and association genomics) to the associated populations (microbes, nematodes and arthropods). In parallel, phylogeographic methods coupled to ecological modelling comparing isolated, long-standing oleaster populations and their pathogens (e.g. olive fly) in the Mediterranean Basin would provide information about the co-demographic histories of the interacting species in the adaptation test. Microbial communities may also have played a crucial role in olive domestication, and olive adaptation to local conditions, with reciprocal effects, but these issues have yet to be investigated. A first step in this direction would involve investigations of the diversity of bacterial or fungal communities, and its differences between environments and between cultivated and wild olives.
Bridging the gap between genomics and ecology to unravel the olive domestication process.
The ecological context of domestication has been much less studied than its genomic basis (Milla et al., 2015). One reason for this may be the ease with which genomic data can be generated and obtained nowadays, relative to the burden of harvesting ecological data in the field. In recent years, access to large environmental databases (e.g. http://worldclim.org/), together with niche modelling, has facilitated the characterization of bioclimatic niches for many species (Guisan and Thuiller, 2005). The concerted advances in phenomics, genomics and ecological modelling have made it timely to bridge the gap between ecology and genomics to unravel olive domestication. In particular, the key role of climatic factors in the evolution of oleaster and cultivated olive during the Quaternary raises questions about the role of environmental conditions in shaping cultivated olive ecology, evolution and adaptation. Both in the wild (e.g. García-Verdugo et al., 2009; Granado-Yela et al., 2011; Rubio de Casas et al., 2011) and in common gardens (Rubio de Casas et al., 2011; Ben Sadok et al., 2015; León et al., 2016), strong environment effects have been demonstrated on many traits of wild olives and cross progeny. Little is known about the ecological and genomic basis of this phenotypic variation (e.g. Merilä and Hendry, 2014), but epigenetic determinants are likely involved (e.g. Platt et al., 2015). Genome-wide association studies with phenotypic traits should help us to understand the genomic basis underlying this variation and its association with olive domestication. Characterization of the bioclimatic niches of wild and cultivated olives will also facilitate investigations of the possible role of ecological shifts in tolerance to abiotic conditions in olive domestication (Milla et al., 2015). Environmental genome-wide association studies (Rellstab et al., 2015) should also shed light on whether local environmental factors were responsible for this variation and the local adaptation of cultivated olives (Box 3), and whether ecological differences have led to adaptive divergence between olive and oleaster. Ecological and genomic approaches comparing introduced non-native olives and local native olives should also reveal how olive trees respond to their local environment. Over the last two centuries, the invasion of South and East Australia by two olive subspecies has resulted in a realized niche shift (Cornuault et al., 2015). Investigations of ecological and genomic differentiation between native and invasive populations should reveal whether this shift is adaptive or the result of plasticity. Overall, these association and environmental studies will make it possible to predict the ability of olives to adapt to current global warming and aridification of the area around the Mediterranean Sea (Seager et al., 2014), which will be crucial to the survival of these trees in the face of current global changes.
The ecological genomics of the ‘olive–biotic community’ interaction.
Olives have intimate symbiotic or antagonistic relationships (e.g. symbionts or pathogens) with the diverse biotic community with which they are associated (microbes, nematodes and arthropods). The influences of the components of this system on each other and the effects of domestication on their interactions are so far understudied in olive (Box 3). Investigations of this issue will provide fundamental insight into the ecological processes and genomic basis of coevolution at the community level. This issue is not only of fundamental academic importance, it will facilitate the adoption of appropriate cropping practices for pest or disease control (e.g. Paredes et al., 2015; Decroocq et al., 2016). The efficacy of root endophyte bacteria (e.g. Pseudomonas fluorescens) as systemic biocontrol agents for a major fungal pathogen (Verticilium dahliae) has already been demonstrated in olive trees (Prieto et al., 2009; Aranda et al., 2011; Gómez-Lama Cabanás et al., 2014). Improvements in our understanding of olive ecology would help to unravel the history of such olive–microbe interaction and to predict its fate under future environmental and economic changes (e.g. Ponti et al., 2014; Seager et al., 2014).
Domestication may lead to significant changes in the evolutionary ecology of crop pathogens. For instance, crop domestication may involve the adaptation of pathogens to new hosts, or host switches from the wild to the crop (e.g. Gladieux et al., 2010; Cornille et al., 2014), potentially leading to an increase in pathogen virulence on the crop (Lê Van et al., 2012). In olive, human selection for genotypes with high growth and high fruit set rates may have had consequences for the defence responses of the cultivated olive (Massei and Hartley, 2000) and for gene dispersal (e.g. abundant pollen from a few clones, limited dispersal of large fruits; Alcántara and Rey, 2003; Perea and Gutiérrez-Galán, 2016), in turn affecting the interaction of olive trees with other organisms. Furthermore, due to the relentless modernization of orchards, ancestral cultivars are increasingly being discarded in favour of a few highly productive varieties (e.g. Khadari et al., 2008; Infante-Amate et al., 2016). With a limited number of clones cultivated at high density, pathogen outbreaks become more likely. A few studies have provided a glimpse of the genetic basis of olive–pathogen interactions (e.g. Corrado et al., 2012; Giampetruzzi et al., 2016), but whole-genome data are nevertheless required to identify the genomic variants involved in olive–parasite interactions. For instance, genes controlling the synthesis of secondary metabolites by the olive tree may be good candidates for involvement in olive–parasite interactions worthy of further investigation (e.g. Hashmi et al., 2015).
The microbial or microfaunal community associated with olive trees may also have been affected by olive domestication (Montes-Borrego et al., 2014; Ali et al., 2017). A high diversity of bacteria has been reported to be associated with the root systems of wild olives in Andalusia, with plant genotype- and site-specific communities and potential antagonism of pathogens (Aranda et al., 2011). However, too few such studies have been performed to date, and investigations of the structure of communities in different areas and under different conditions (e.g. oleasters vs. cultivars, or traditional vs. high-density orchards) are required to determine the impact of olive domestication and cultivation practices on diversity and susceptibility (e.g. Ali et al., 2017). Recent studies on nematodes (Palomares-Rius et al., 2012, 2015; Ali et al., 2017) and fungi (Montes-Borrego et al., 2014; Abdelfattah et al., 2015) associated with cultivated olives have also shown these communities to be influenced by environmental factors (e.g. soil parameters and cropping methods) and host genotype. Future investigation of this type may be facilitated by the development of DNA metabarcoding methods (e.g. Montes-Borrego et al., 2014; Abdelfattah et al., 2015) for describing the microbial communities associated with different cultivars. In addition, common garden studies carried out by interdisciplinary research consortia from around the world (multiple sites in different countries) should also provide valuable assessments of the effect of olive genotypes on associated communities.
Adaptive processes and demographic events must be distinguished in investigations of the ecological genomics of biotic interactions. Investigations of co-demographic histories between olives and the communities associated with them are therefore a useful approach for unravelling the impact of domestication on the olive and its biotic community. The arthropod, nematode and microbial communities associated with the oleaster and other Mediterranean shrubs may have a common phylogeography due to host specificity or constrained evolutionary histories in common habitats that have been fragmented, particularly during unfavourable periods, such as glaciations (e.g. Nieto Feliner, 2014). Recent studies have shown that the diversification of the olive fly is closely related to that of its host (Nardi et al., 2010; van Ash et al., 2015) and the endophytic bacterial communities of olive leaves have also revealed an East–West geographic structure of beta-diversity (Müller et al., 2015). Recently developed comparative methods (e.g. Satler and Carstens, 2016) coupled with ecological modelling for isolated, long-standing oleaster populations and other organisms (e.g. nematodes or olive fly) in the Mediterranean Basin will constitute a first step towards accurate investigations of the adaptive genomics of olive–biota interaction.
OVERALL CONCLUSIONS: TOWARD THE IMPROVEMENT OF SUSTAINABLE OLIVE AGRICULTURE
Over the last two decades, the contributions of several disciplines have improved our understanding of the processes of domestication, spread and diversification, for olive, as for other fruit trees and shrubs (e.g. Myles et al., 2011; Delplancke et al., 2013; Cornille et al., 2014; Hazzouri et al., 2015). The olive is a complex case study in fruit tree domestication. From methodological and analytical perspectives, deciphering the adaptive evolutionary processes involved in olive domestication remains challenging. More than ever, ecological and socio-economic issues relating to the future of the olive are fundamental in light of current global changes (climatic, societal, political, economic, etc.), particularly in the Mediterranean Basin, the cradle of olive domestication (e.g. Ponti et al., 2014). Interdisciplinary research is a key element for (1) understanding the functioning of oleaster communities and assessing their vulnerability/resilience in the face of increasing environmental constraints, predominantly of climatic and anthropogenic origin; (2) reconstituting the paleoanthropological, ecological and biogeographic history of the olive tree; (3) exploring agrobiodiversity and understanding the ecological plasticity of varieties of major interest; (4) developing strategies for the conservation of wild olive populations and development of local varieties; and (5) understanding the role of social, political and economic processes in the diversification and development of sustainable olive cultivation. A detailed knowledge of the origins of olive would have major implications for the management of genetic resources and the investigation of genetic factors involved in the variation of agronomic traits and, more generally, would provide insight into fruit tree domestication in the Mediterranean Basin.
The aim of our review was not to be all inclusive but rather to be selective with the up to date literature. Throughout it, we have shown, that unravelling the ecological genomics of olive domestication will also provide essential insight into the evolutionary processes involved in phenotypic and molecular evolution, and in adaptation and coevolution.
SUPPLEMENTARY DATA
Supplementary data are available at https://academic.oup.com/aob and consist of the following. Table S1: summary of the current genetic data used to infer histories of cultivated and wild olives, with associated methodologies, limitations and main conclusions, and the future genomic data required to test for the neutral and adaptive genomics of domestication in olive. Table S2: data matrix for the 147 plastid DNA haplotypes identified in the olive complex with 71 loci. Figure S1: reduced median networks of Mediterranean olive plastid DNA haplotypes. Fig. S2: median joining network of olive chlorotypes reconstructed with NETWORK.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the members of the EDB laboratory for fruitful discussions and, particularly, D. Kaniewski, J. Hackel, C. Van de Paer and J. Bruxaux and G. Volk. We also thank P. Cuneo, R. Rubio de Casas and E. Chapuis for helpful comments. G.B. is supported by TULIP (ANR-10-LABX-0041), CEBA (ANR-10-LABX-25-01) and PESTOLIVE (ARIMNet action KBBE 219262).
LITERATURE CITED
- Abdelfattah A, Li Destri Nicosia MG, Cacciola SO, Droby S, Schena L. 2015. Metabarcoding analysis of fungal diversity in the phyllosphere and carposphere of olive (Olea europaea). PLoS One 10: e0131069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertini E, Torricelli R, Bitocchi E et al. 2011. Structure of genetic diversity in Olea europaea L. cultivars from central Italy. Molecular Breeding 27: 533–547. [Google Scholar]
- Akagi T, Hanada T, Yaegaki H, Gradziel TM, Tao R. 2016. Genome-wide view of genetic diversity reveals paths of selection and cultivar differentiation in peach domestication. DNA Research 23: 271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcántara JM, Rey PJ. 2003. Conflicting selection pressures on seed size: evolutionary ecology of fruit size in a bird-dispersed tree, Olea europaea. Journal of Evolutionary Biology 16: 1168–1176. [DOI] [PubMed] [Google Scholar]
- Ali N, Tavoillot J, Besnard G et al. 2017. How anthropogenic changes may affect soil-borne parasite diversity? Plant-parasitic nematode communities associated with olive trees in Morocco as a case study. BMC Ecology 17: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angiolillo A, Mencuccini M, Baldoni L. 1999. Olive genetic diversity assessed using amplified polymorphic fragment length polymorphisms. Theoretical and Applied Genetics 98: 411–421. [Google Scholar]
- Aranda S, Montes-Borrego M, Jiménez-Díaz RM, Landa BB. 2011. Microbial communities associated with the root system of wild olives (Olea europaea L. subsp. europaea var. sylvestris) are good reservoirs of bacteria with antagonistic potential against Verticillium dahliae. Plant and Soil 343: 329–345. [Google Scholar]
- Arias-Calderón R, Rodríguez-Jurado D, León L et al. 2015. Pre-breeding for resistance to Verticillium wilt in olive: fishing in the wild relative gene pool. Crop Protection 75: 25–33. [Google Scholar]
- van Asch B, Pereira-Castro I, Rei FT, da Costa LT. 2015. Marked genetic differentiation between western Iberian and Italic populations of the olive fly: southern France as an intermediate area. PLoS One 10: e0126702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atienza SG, de la Rosa R, León L, Martín A, Belaj A. 2014. Identification of QTL for agronomic traits of importance for olive breeding. Molecular Breeding 34: 725–737. [Google Scholar]
- Aumeeruddy-Thomas Y, Moukhli A, Haouane H, Khadari B. 2017. Ongoing domestication and diversification in grafted olive–oleaster agroecosystems in Northern Morocco. Regional Environmental Change 17:1315–1328. [Google Scholar]
- Bacilieri R, Lacombe T, Le Cunff L et al. 2013. Genetic structure in cultivated grapevines is linked to geography and human selection. BMC Plant Biology 13: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldoni L, Tosti N, Ricciolini C et al. 2006. Genetic structure of wild and cultivated olives in the Central Mediterranean Basin. Annals of Botany 98: 935–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barazani O, Westberg E, Hanin N et al. 2014. A comparative analysis of genetic variation in rootstocks and scions of old olive trees: a window into the history of olive cultivation practices and past genetic variation. BMC Plant Biology 14: 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barazani O, Keren-Keiserman A, Westberg E et al. 2016. Genetic variation of naturally growing olive trees in Israel: from abandoned groves to feral and wild?BMC Plant Biology 16: 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barazani O, Waitz Y, Tugendhaft Y et al. 2017. Testing the potential significance of different scion/rootstock genotype combinations on the ecology of old cultivated olive trees in the southeast Mediterranean area. BMC Ecology 17: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolini G, Prevost G, Messeri C, Carignani C. 2005. Olive germplasm: cultivars and world-wide collections FAO/Plant Production and Protection, Rome: Available at: http://www.apps3.fao.org/wiews/olive/oliv.jsp. [Google Scholar]
- Beaumont MA. 2010. Approximate Bayesian computation in evolution and ecology. Annual Review of Ecology, Evolution, and Systematics 41: 379–406. [Google Scholar]
- Beghé D, García-Molano JF, Fabbri A, Ganino T. 2015. Olive biodiversity in Colombia. A molecular study of local germplasm. Scientia Horticulturae 189: 122–131. [Google Scholar]
- Beghè D, Piotti A, Satovic Z, de la Rosa R, Belaj A. 2017. Pollen-mediated gene flow and fine-scale spatial genetic structure in Olea europaea subsp. europaea var. sylvestris. Annals of Botany 119: 671–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belaj A, Trujillo I, de la Rosa R, Rallo L. 2001. Polymorphism and discrimination capacity of randomly amplified polymorphic markers in an olive germplasm bank. Journal of the American Society for Horticultural Science 126: 64–71. [Google Scholar]
- Belaj A, Muñoz-Diez C, Baldoni L et al. 2007. Genetic diversity and population structure of wild olives from the north-western Mediterranean assessed by SSR markers. Annals of Botany 100: 449–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belaj A, Muñoz-Diez C, Baldoni L, Satovic Z, Barranco D. 2010. Genetic diversity and relationships of wild and cultivated olives at regional level in Spain. Scientia Horticulturae 124: 323–330. [Google Scholar]
- Belaj A, León L, Satovic Z, De La Rosa R. 2011. Variability of wild olives (Olea europaea subsp. europaea var. sylvestris) analyzed by agromorphological traits and SSR markers. Scientia Horticulturae 129: 561–569. [Google Scholar]
- Belaj A, del Carmen Dominguez-García M, Atienza SG et al. 2012. Developing a core collection of olive (Olea europaea L.) based on molecular markers (DArTs, SSRs, SNPs) and agronomic traits. Tree Genetics and Genomes 8: 365–378. [Google Scholar]
- Ben Sadok I, Moutier N, Garcia G et al. 2013. Genetic determinism of the vegetative and reproductive traits in an F1 olive tree progeny. Tree Genetics and Genomes 9: 205–221. [Google Scholar]
- Ben Sadok I, Martinez S, Moutier N et al. 2015. Plasticity in vegetative growth over contrasted growing sites of an F1 olive tree progeny during its juvenile phase. PLoS One 10: e0127539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento G, Routtu J, Bourgeois Y, Du Pasquier L, Ebert D. 2017. The genetic basis of resistance and matching-allele interactions of a host–parasite system: the Daphnia magna–Pasteuria ramosa model. PLoS Genetics 13: e1006596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard G, Hernández P, Khadari B, Dorado G, Savolainen V. 2011. Genomic profiling of plastid DNA variation in the Mediterranean olive tree. BMC Plant Biology 11: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard G, El Bakkali A. 2014. Sequence analysis of single-copy genes in two wild olive subspecies (Olea europaea L.): nucleotide diversity and potential use for testing admixture. Genome 57: 145–153. [DOI] [PubMed] [Google Scholar]
- Besnard G, Rubio de Casas R. 2016. Single vs multiple independent olive domestications: the jury is (still) out. New Phytologist 209: 466–470. [DOI] [PubMed] [Google Scholar]
- Besnard G, Baradat P, Bervillé A. 2001a. Genetic relationships in the olive (Olea europaea L.) reflect multilocal selection of cultivars. Theoretical and Applied Genetics 102: 251–258. [Google Scholar]
- Besnard G, Baradat P, Breton C, Khadari B, Bervillé A. 2001b. Olive domestication from structure of oleasters and cultivars using nuclear RAPDs and mitochondrial RFLPs. Genetics, Selection, Evolution 33: S251–S268. [Google Scholar]
- Besnard G, Rubio de Casas R, Vargas P. 2007. Plastid and nuclear DNA polymorphism reveals historical processes of isolation and reticulation in the olive tree complex (Olea europaea). Journal of Biogeography 34: 736–752. [Google Scholar]
- Besnard G, Garcia-Verdugo C, Rubio de Casas R et al. 2008. Polyploidy in the olive complex (Olea europaea L.): evidence from flow cytometry and nuclear microsatellite analyses. Annals of Botany 101: 25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard G, Rubio de Casas R, Christin PA, Vargas P. 2009. Phylogenetics of Olea (Oleaceae) based on plastid and nuclear ribosomal DNA sequences: tertiary climatic shifts and lineage differentiation times. Annals of Botany 104: 143–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard G, El Bakkali A, Haouane H et al. 2013a. Population genetics of Mediterranean and Saharan olives: geographic patterns of differentiation and evidence for early-generations of admixture. Annals of Botany 112: 1293–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard G, Khadari B, Navascués M et al. 2013b. The complex history of the olive tree: from Late Quaternary diversification of Mediterranean lineages to primary domestication in the northern Levant. Proceedings of the Royal Society B: Biological Sciences 280: 20122833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard G, Dupuy J, Larter M et al. 2014. History of the invasive African olive tree in Australia and Hawaii: evidence for sequential bottlenecks and hybridizations with the Mediterranean olive. Evolutionary Applications 7: 195–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biltekin D, Popescu SM, Suc JP et al. 2015. Anatolia: a long-time plant refuge area documented by pollen records over the last 23 million years. Review of Palaeobotany and Palynology 215: 1–22. [Google Scholar]
- Biton I, Doron-Faigenboim A, Jamwal M et al. 2015. Development of a large set of SNP markers for assessing phylogenetic relationships between the olive cultivars composing the Israeli olive germplasm collection. Molecular Breeding 35: 107. [Google Scholar]
- Besnard G, Cuneo P. 2016. An ecological and evolutionary perspective on the parallel invasion of two cross-compatible trees. AoB Plants 8: plw056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biton I, Shevtsov S, Ostersetzer O et al. 2012. Genetic relationships and hybrid vigour in olive (Olea europaea L.) by microsatellites. Plant Breeding 131: 767–774. [Google Scholar]
- Booker TR, Jackson BC, Keightley PD. 2017. Detecting positive selection in the genome. BMC Biology 15: 98. [DOI] [PMC free article] [PubMed]
- Bowers JE, Boursiquot JM, This P et al. 1999. Historical genetics: the parentage of Chardonnay, Gamay, and other wine grapes of Northeastern France. Science 285: 1562–1565. [DOI] [PubMed] [Google Scholar]
- Breton C, Tersac M, Bervillé A. 2006. Genetic diversity and gene flow between the wild olive (oleaster, Olea europaea L.) and the olive: several Plio-Pleistocene refuge zones in the Mediterranean basin suggested by simple sequence repeats analysis. Journal of Biogeography 34: 1916–1928. [Google Scholar]
- Breton CM, Farinelli D, Shafiq S, Heslop-Harrison JS, Sedgley M, Bervillé AJ. 2014. The self-incompatibility mating system of the olive (Olea europaea L.) functions with dominance between S-alleles. Tree Genetics and Genomes 10: 1055–1067. [Google Scholar]
- Bronzini de Caraffa V, Maury J, Gambotti C, Breton C, Bervillé A, Giannettini J. 2002. Mitochondrial DNA variation and RAPD mark oleasters, olive and feral olive from Western and Eastern Mediterranean. Theoretical and Applied Genetics 104: 1209–1216. [DOI] [PubMed] [Google Scholar]
- Brun JP. 2003. Archéologie du vin et de l’huile. De la Préhistoire à l’époque Hellénistique. Errance: Paris. [Google Scholar]
- Brun JP. 2004. Archéologie du vin et de l’huile dans l’Empire romain. Errance: Paris. [Google Scholar]
- Cáceres ME, Ceccarelli M, Pupilli F, Sarri V, Mencuccini M. 2015. Obtainment of inter-subspecific hybrids in olive (Olea europaea L.). Euphytica 201: 307–319. [Google Scholar]
- Carrión JS, Yll EI, Walker MJ, Legaz AJ, Chain C. 2003. Glacial refugia of temperate, Mediterranean and Ibero-North African flora in south-eastern Spain: new evidence from cave pollen at two Neanderthal man sites. Global Ecology and Biogeography 12: 119–129. [Google Scholar]
- Carrión Y, Ntinou M, Badal E. 2010. Olea europaea L. in the North Mediterranean basin during the Pleniglacial and the early-middle Holocene. Quaternary Science Reviews 29: 952–968. [Google Scholar]
- Chalak L, Haouane H, Essalouh L et al. 2015. Extent of the genetic diversity in Lebanese olives: a mixture of an ancient germplasm with recently introduced varieties. Genetic Resources and Crop Evolution 62: 621–633. [Google Scholar]
- Chevalier A. 1948. L’origine de l’Olivier cultivé et ses variations. Revue Internationale de Botanique Appliquée et d’Agriculture Tropicale 28: 1–25. [Google Scholar]
- Civáň P, Craig H, Cox CJ, Brown TA. 2015. Three geographically separate domestications of Asian rice. Nature Plants 1: 15164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikhi L, Nichols RA, Barbujani G, Beaumont MA. 2002. Y genetic data support the Neolithic demic diffusion model. Proceedings of the National Academy of Sciences, USA 99: 11008–11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros MG, Crespillo R, Aguilar ML, Canovas FM. 2000. DNA fingerprinting and classification of geographically related genotypes of olive tree (Olea europaea L.). Euphytica 116: 131–142. [Google Scholar]
- Combourieu-Nebout N, Peyron O, Bout-Roumazeilles V et al. 2013. Holocene vegetation and climate changes in the central Mediterranean inferred from a high-resolution marine pollen record (Adriatic Sea). Climate of the Past 9: 2023–2042. [Google Scholar]
- Cornille A, Gladieux P, Smulders MJM et al. 2012. New insight into the history of domesticated apple: secondary contribution of the European wild apple to the genome of cultivated varieties. PLoS Genetics 8: e1002703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornille A, Giraud T, Smulders MJ, Roldán-Ruiz I, Gladieux P. 2014. The domestication and evolutionary ecology of apples. Trends in Genetics 30: 57–65. [DOI] [PubMed] [Google Scholar]
- Cornuault J, Khimoun A, Cuneo P, Besnard G. 2015. Spatial segregation and realized niche shift during the parallel invasion of two olive subspecies in south-eastern Australia. Journal of Biogeography 42: 1930–1941. [Google Scholar]
- Corrado G, Alagna F, Rocco M et al. 2012. Molecular interactions between the olive and the fruit fly Bactrocera oleae. BMC Plant Biology 12: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz F, Julca I, Gómez-Garrido J et al. 2016. Genome sequence of the olive tree, Olea europaea. GigaScience 5: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuneo P, Leishman MR. 2006. African Olive (Olea europaea subsp. cuspidata) as an environmental weed in eastern Australia: a review. Cunninghamia 9: 545–577. [Google Scholar]
- D’Auria A, Buonincontri MP, Allevato E et al. 2017. Evidence of a short-lived episode of olive (Olea europaea L.) cultivation during the Early Bronze Age in western Mediterranean (southern Italy). The Holocene 27: 605–612. [Google Scholar]
- Decroocq S, Cornille A, Tricon D et al. 2016. New insights into the history of domesticated and wild apricot and its contribution to plum pox virus resistance. Molecular Ecology 25: 4712–4729. [DOI] [PubMed] [Google Scholar]
- Delplancke M, Alvarez N, Benoit L et al. 2013. Evolutionary history of almond tree domestication in the Mediterranean Basin. Molecular Ecology 22: 1092–1104. [DOI] [PubMed] [Google Scholar]
- Dίez CM, Gaut BS. 2016. The jury may be out, but it is important that it deliberates: a response to Besnard and Rubio de Casas about olive domestication. New Phytologist 209: 471–473. [DOI] [PubMed] [Google Scholar]
- Dίez CM, Trujillo I, Barrio E et al. 2011. Centennial olive trees as a reservoir of genetic diversity. Annals of Botany 108: 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dίez CM, Imperato A, Rallo L, Baranco D, Trujillo I. 2012. Worldwide core collection of olive cultivars based on simple sequence repeat and morphological markers. Crop Science 52: 211–221. [Google Scholar]
- Dίez CM, Trujillo I, Martinez-Uriroz N et al. 2015. Olive domestication and diversification in the Mediterranean basin. New Phytologist 206: 436–447. [DOI] [PubMed] [Google Scholar]
- Dighton A, Fairbairn A, Bourke S, Faith JT, Habgood P. 2017. Bronze Age olive domestication in the north Jordan valley: new morphological evidence for regional complexity in early arboricultural practice from Pella in Jordan. Vegetation History and Archaeobotany 26: 403–413. [Google Scholar]
- Djamali M, Jones MD, Migliore J et al. 2016. Olive cultivation in the heart of the Persian Achaemenid Empire: new insights into agricultural practices and environmental changes reflected in a late Holocene pollen record from Lake Parishan, SW Iran. Vegetation History and Archaeobotany 25: 255–269. [Google Scholar]
- El Bakkali A, Haouane H, Moukhli A et al. 2013. Construction of core collections suitable for association mapping to optimize use of Mediterranean olive (Olea europaea L.) genetic resources. PLoS One 8: e61265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaum R, Melamed-Bessudo C, Boaretto E et al. 2006. Ancient olive DNA in pits: preservation, amplification and sequence analysis. Journal of Archaeological Science 33: 77–88. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- Figueiral I, Terral JF. 2002. Late Quaternary refugia of Mediterranean taxa in the Portuguese Estremadura: charcoal based palaeovegetation and climatic reconstruction. Quaternary Science Reviews 21: 549–558. [Google Scholar]
- Fijarczyk A, Babik W. 2015. Detecting balancing selection in genomes: limits and prospects. Molecular Ecology 24: 3529–3545. [DOI] [PubMed] [Google Scholar]
- da Fonseca RR, Smith BD, Wales N et al. 2015. The origin and evolution of maize in the Southwestern United States. Nature Plants 1: 14003. [DOI] [PubMed] [Google Scholar]
- Galili E, Stanley DJ, Sharvit J, Weinstein-Evron M. 1997. Evidence for earliest olive-oil production in submerged settlements off the Carmel coast, Israel. Journal of Archaeological Science 24: 1141–1150. [Google Scholar]
- Garcίa-Verdugo C, Granado-Yela C, Manrique E, Rubio de Casas R, Balaguer L. 2009. Phenotypic plasticity and integration across the canopy of Olea europaea subsp. guanchica (Oleaceae) in populations with different wind exposures. American Journal of Botany 96: 1454–1461. [DOI] [PubMed] [Google Scholar]
- Gaut BS, Díez CM, Morrell PL. 2015. Genomics and the contrasting dynamics of annual and perennial domestication. Trends in Genetics 31: 709–719. [DOI] [PubMed] [Google Scholar]
- Gerbault P, Grimaldi IM, Allaby RG et al. 2014. Storytelling and story testing in domestication: can modeling help?Proceedings of the National Academy of Sciences, USA 111: 6159–6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giampetruzzi A, Morelli M, Saponari M et al. 2016. Transcriptome profiling of two olive cultivars in response to infection by the CoDiRO strain of Xylella fastidiosa subsp. pauca. BMC Genomics 17: 475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladieux P, Zhang XG, Roldan-Ruiz I et al. 2010. Evolution of the population structure of Venturia inaequalis, the apple scab fungus, associated with the domestication of its host. Molecular Ecology 19: 658–674. [DOI] [PubMed] [Google Scholar]
- Gladieux P, Feurtey A, Hood ME et al. 2015. The population biology of fungal invasions. Molecular Ecology 24: 1969–1986. [DOI] [PubMed] [Google Scholar]
- Glémin S, Bataillon T. 2009. A comparative view of the evolution of grasses under domestication. New Phytologist 183: 273–290. [DOI] [PubMed] [Google Scholar]
- Gómez-Lama Cabanás C, Schilirò E, Valverde-Corredor A, Mercado-Blanco J. 2014. The biocontrol endophytic bacterium Pseudomonas fluorescens PICF7 induces systemic defense responses in aerial tissues upon colonization of olive roots. Frontiers in Microbiology 5: 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granado-Yela C, García-Verdugo C, Carrillo K et al. 2011. Temporal matching among diurnal photosynthetic patterns within the crown of the evergreen sclerophyll Olea europaea L. Plant, Cell and Environment 34: 800–810. [DOI] [PubMed] [Google Scholar]
- Green PS. 2002. A revision of Olea L. (Oleaceae). Kew Bulletin 57: 91–140. [Google Scholar]
- Gros-Balthazard M, Newton C, Ivorra S, Pierre MH, Pintaud JC, Terral JF. 2016. The domestication syndrome in Phoenix dactylifera seeds: toward the identification of wild date palm populations. PLoS One 11: e0152394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross BL, Henk AD, Richards CM, Fazio G, Volk GM. 2014. Genetic diversity in Malus×domestica (Rosaceae) through time in response to domestication. American Journal of Botany 101: 1770–1779. [DOI] [PubMed] [Google Scholar]
- Guisan A, Thuiller W. 2005. Predicting species distribution: offering more than simple habitat models. Ecology Letters 8: 993–1009. [DOI] [PubMed] [Google Scholar]
- Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. 2009. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genetics 5: e1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagidimitriou M, Katsiotis A, Menexes G, Pontikis C, Loukas M. 2005. Genetic diversity of major Greek olive cultivars using molecular (AFLPs and RAPDs) markers and morphological traits. Journal of the American Society for Horticultural Science 130: 211–217. [Google Scholar]
- Hannachi H, Breton C, Msallem M et al. 2008. Differences between native and introduced cultivars as revealed by morphology of drupes, oil composition and SSR polymorphism; a case study in Tunisia. Scientia Horticulturae 116: 280–290. [Google Scholar]
- Hannachi H, Sommerlate H, Breton C et al. 2009. Oleaster (var. sylvestris) and subsp. cuspidata are suitable genetic resources for improvement of the olive (Olea europaea subsp. europaea var. europaea). Genetic Resources and Crop Evolution 56: 393–403. [Google Scholar]
- Haouane H, El Bakkali A, Moukhli A et al. 2011. Genetic structure and core collection of the World Olive Germplasm Bank of Marrakech: towards the optimised management and use of Mediterranean olive genetic resources. Genetica 139: 1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashmi MA, Khan A, Hanif M, Farooq U, Perveen S. 2015. Traditional uses, phytochemistry, and pharmacology of Olea europaea (Olive). Evidence-Based Complementary and Alternative Medicine 2015: 541591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazzouri KM, Flowers JM, Visser HJ et al. 2015. Whole genome re-sequencing of date palms yields insights into diversification of a fruit tree crop. Nature Communications 6: 8824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermisson J, Pennings PS. 2017. Soft sweeps and beyond: understanding the patterns and probabilities of selection footprints under rapid adaptation. Methods in Ecology and Evolution 8: 700–716. [Google Scholar]
- Hewitt GM. 2004. Genetic consequences of climatic oscillations in the Quaternary. Philosophical Transactions of the Royal Society B: Biological Sciences 359: 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoban S, Kelley JL, Lotterhos KE, et al. 2016. Finding the genomic basis of local adaptation: pitfalls, practical solutions, and future directions. The American Naturalist 188: 379–397. [DOI] [PMC free article] [PubMed]
- Holliday J, Aitken S, Cooke J et al. 2017. Advances in ecological genomics in forest trees and applications to genetic resources conservation and breeding. Molecular Ecology 26: 706–717. [DOI] [PubMed] [Google Scholar]
- Hosseini-Mazinani M, Mariotti R, Torkzaban B et al. 2014. High genetic diversity detected in olives beyond the boundaries of the Mediterranean Sea. PLoS One 9: e93146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufford MB, Xu X, van Heerwaarden J et al. 2012. Comparative population genomics of maize domestication and improvement. Nature Genetics 44: 808–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante-Amate J, Villa I, Aguilera E et al. 2016. The making of olive landscapes in the south of Spain. A history of continuous expansion and intensification. In: Agnoletti M, Emanueli F, eds. Environmental history. Biocultural diversity in Europe, Vol. 5 Cham: Springer, 157–179. [Google Scholar]
- İpek A, Yılmaz K, Sıkıcı P et al. 2016. SNP discovery by GBS in olive and the construction of a high-density genetic linkage map. Biochemical Genetics 54: 312–325. [DOI] [PubMed] [Google Scholar]
- Juniper BE, Maberly DJ. 2006. The story of the apple. Portland, OR: Timber Press. [Google Scholar]
- Kaniewski D, Van Campo E, Boiy T et al. 2012. Primary domestication and early uses of the emblematic olive tree: palaeobotanical, historical and molecular evidences from the Middle East. Biological Reviews 87: 885–899. [DOI] [PubMed] [Google Scholar]
- Kaniewski D, Paulissen E, Van Campo E, Bakker J, Van Lerberghe K, Waelkens M. 2009. Wild or cultivated Olea europaea L. in the eastern Mediterranean during the Middle–Late Holocene? A pollen-numerical approach. The Holocene 19: 1039–1047. [Google Scholar]
- Kaya HB, Cetin O, Kaya H et al. 2013. SNP discovery by Illumina-based transcriptome sequencing of the olive and the genetic characterization of Turkish olive genotypes revealed by AFLP, SSR and SNP markers. PLoS One 8: e73674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadari B, Breton C, Moutier N et al. 2003. The use of molecular markers for germplasm management in a French olive collection. Theoretical and Applied Genetics 106: 521–529. [DOI] [PubMed] [Google Scholar]
- Khadari B, Charafi J, Moukhli A, Ater M. 2008. Substantial genetic diversity in cultivated Moroccan olive despite a single major cultivar: a paradoxical situation evidenced by the use of SSR loci. Tree Genetics and Genomes 4: 213–221. [Google Scholar]
- Kislev ME. 1995. Wild olive stones at submerged Chalcolithic Kfar Samir, Haifa, Israel. Journal of the Israel Prehistoric Society 26: 134–195. [Google Scholar]
- Kislev ME, Nadel D, Carmi I. 1992. Epipalaeolithic (19,000 B.P.) cereal and fruit diet at Ohalo II, Sea of Galilee, Israel. Review of Palaeobotany and Palynology 73: 161–166. [Google Scholar]
- Klepo T, de la Rosa R, Satovic Z, León L, Belaj A. 2013. Utility of wild germplasm in olive breeding. Scientia Horticulturae 152: 92–101. [Google Scholar]
- Langgut D, Adams MJ, Finkelsetin I. 2016. Climate, settlement patterns and olive horticulture in the southern Levant during the Early Bronze and Intermediate Bronze Ages (c. 3600–1950 BC). Levant 48: 117–134. [Google Scholar]
- Lavee S, Zohary D. 2011. The potential of genetic diversity and the effect of geographically isolated resources in olive breeding. Israel Journal of Plant Sciences 59: 3–13. [Google Scholar]
- Lazović B, Adakalić M, Pucci C et al. 2016. Characterizing ancient and local olive germplasm from Montenegro. Scientia Horticulturae 209: 117–123. [Google Scholar]
- León L, Arias-Calderón R, de la Rosa R, Khadari B, Costes E. 2016. Optimal spatial and temporal replications for reducing environmental variation for oil content components and fruit morphology traits in olive breeding. Euphytica 207: 675–684. [Google Scholar]
- Lê Van A, Gladieux P, Lemaire C et al. 2012. Evolution of pathogenicity traits in the apple scab fungal pathogen in response to the domestication of its host. Evolutionary Applications 5: 694–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linos A, Nikoloudakis N, Katsiotis A, Hagidimitriou M. 2014. Genetic structure of the Greek olive germplasm revealed by RAPD, ISSR and SSR markers. Scientia Horticulturae 175: 33–43. [Google Scholar]
- Liphschitz N, Gophna R, Hartman M, Biger G. 1991. The beginning of olive (Olea europaea) cultivation in the Old World: a reassessment. Journal of Archaeological Science 18: 441–453. [Google Scholar]
- Liu KJ, Steinberg E, Yozzo A, Song Y, Kohn MH, Nakhleh L. 2015. Interspecific introgressive origin of genomic diversity in the house mouse. Proceedings of the National Academy of Sciences, USA 112: 199–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Fu YX. 2015. Exploring population size changes using SNP frequency spectra. Nature Genetics 47: 555–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loumou A, Giourga C. 2003. Olive groves: ‘the life and identity of the Mediterranean’. Agriculture and Human Values 20: 87–95. [Google Scholar]
- Lumaret R, Ouazzani N, Michaud H et al. 2004. Allozyme variation of oleaster populations (wild olive tree) (Olea europaea L.) in the Mediterranean basin. Heredity 92: 343–351. [DOI] [PubMed] [Google Scholar]
- McClure KA, Sawler J, Gardner KM, Money D, Myles S. 2014. Genomics: a potential panacea for the perennial problem. American Journal of Botany 101: 1780–1790. [DOI] [PubMed] [Google Scholar]
- McKey D, Elias M, Pujol B, Duputié A. 2010. The evolutionary ecology of clonally propagated domesticated plants. New Phytologist 186: 318–332. [DOI] [PubMed] [Google Scholar]
- Margaritis E. 2013. Distinguishing exploitation, domestication, cultivation and production: the olive in the third millennium Aegean. Antiquity 337: 746–757. [Google Scholar]
- Mariotti R, Cultrera NGM, Díez CM, Baldoni L, Rubini A. 2010. Identification of new polymorphic regions and differentiation of cultivated olives (Olea europaea L.) through plastome sequence comparison. BMC Plant Biology 10: 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra FP, Caruso T, Costa F et al. 2013. Genetic relationships, structure and parentage simulation among the olive tree (Olea europaea L. subsp. europaea) cultivated in Southern Italy revealed by SSR markers. Tree Genetics and Genomes 9: 961–973. [Google Scholar]
- Martelli GP, Salerno M, Savino V, Prota U. 2002. An appraisal of diseases and pathogens of olive. Acta Horticulturae 586: 701–708. [Google Scholar]
- Massei G, Hartley SE. 2000. Disarmed by domestication? Induced responses to browsing in wild and cultivated olive. Oecologia 122: 225–231. [DOI] [PubMed] [Google Scholar]
- Mayol M, Riba M, González-Martίnez C et al. 2015. Adapting through glacial cycles: insights from a long-lived tree (Taxus baccata). New Phytologist 208: 973–986. [DOI] [PubMed] [Google Scholar]
- Médail F, Quézel P, Besnard G, Khadari B. 2001. Systematics, ecology and phylogeographic significance of Olea europaea L. subsp. maroccana (Greuter & Burdet) P. Vargas et al., a relictual olive tree in south-west Morocco. Botanical Journal of the Linnean Society 137: 249–266. [Google Scholar]
- Merilä J, Hendry AP. 2014. Climate change, adaptation and phenotypic plasticity: the problem and the evidence. Evolutionary Applications 8: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migicovsky Z, Myles S. 2017. Exploiting wild relatives for genomics-assisted breeding of perennial crops. Frontiers in Plant Science 8: 460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliore J, Baumel A, Juin M, Médail F. 2012. From Mediterranean shores to central Saharan mountains: key phylogeographical insights from the genus Myrtus. Journal of Biogeography 39: 942–956. [Google Scholar]
- Milla R, Osborne CP, Turcotte MM, Violle C. 2015. Plant domestication through an ecological lens. Trends in Ecology and Evolution 30: 463–469. [DOI] [PubMed] [Google Scholar]
- Miller AJ, Gross BL. 2011. From forest to field: perennial fruit crop domestication. American Journal of Botany 98: 1389–1414. [DOI] [PubMed] [Google Scholar]
- Montes-Borrego M, Metsis M, Landa BB. 2014. Arbuscular mycorhizal fungi associated with the olive crop across the Andalusian landscape: factors driving community differentiation. PLoS One 9: e96397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi S, Hosseini-Mazinani M, Arzani K et al. 2014. Molecular and morphological characterization of Golestan (Iran) olive ecotypes provides evidence for the presence of promising genotypes. Genetic Resources and Crop Evolution 61: 775–785. [Google Scholar]
- Mousavi S, Mariotti R, Bagnoli F et al. 2017. The eastern part of the Fertile Crescent concealed an unexpected route of olive (Olea europaea L.) differentiation. Annals of Botany 119: 1305–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge K, Janick J, Scofield S, Goldschmidt E. 2009. A history of grafting. Horticultural Reviews 35: 437–493. [Google Scholar]
- Müller H, Berg C, Landa BB et al. 2015. Plant genotype-specific archaeal and bacterial endophytes but similar Bacillus antagonists colonize Mediterranean olive trees. Frontiers in Microbiology 6: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzzalupo I, Vendramin GG, Chiappetta A. 2014. Genetic biodiversity of Italian olives (Olea europaea) germplasm analyzed by SSR markers. Scientific World Journal 2014: 296590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myles S, Boyko AR, Owens CL et al. 2011. Genetic structure and domestication history of the grape. Proceedings of the National Academy of Sciences, USA 108: 3530–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardi F, Carapelli A, Boore JL et al. 2010. Domestication of olive fly through a multi-regional host shift to cultivated olives: comparative dating using complete mitochondrial genomes. Molecular Phylogenetics and Evolution 57: 678–686. [DOI] [PubMed] [Google Scholar]
- Neef R. 1990. Introduction, development and environmental implications of olive culture: the evidence from Jordan. In: Bottema S, Entjes-Nieborg G, van Zeist W, eds. Man’s role in the shaping of the Eastern Mediterranean landscape. Rotterdam: Balkema, 295–306. [Google Scholar]
- Nei M. 1987. Molecular evolutionary genetics. New York: Columbia University Press. [Google Scholar]
- Newberry PE. 1937. On some African species of the genus Olea and the original home of the cultivated olive tree. Proceedings of the Linnean Society of London 150: 3–16. [Google Scholar]
- Newton C, Terral JF, Ivorra S. 2006. The Egyptian olive (Olea europaea subsp. europaea) in the later first millennium BC: origins and history using the morphometric analysis of olive stones. Antiquity 80: 405–414. [Google Scholar]
- Newton C, Lorre C, Sauvage C, Ivorra S, Terral JF. 2014. On the origins and spread of Olea europaea L. (olive) domestication: evidence for shape variation of olive stones at Ugarit, Late Bronze Age, Syria: a window on the Mediterranean basin and on the westward diffusion of olive varieties. Vegetation History and Archaeobotany 23: 567–575. [Google Scholar]
- Nieto Feliner G. 2014. Patterns and processes in plant phylogeography in the Mediterranean basin. A review. Perspectives in Plant Ecology Evolution and Systematics 16: 265–278. [Google Scholar]
- Nistelberger HM, Smith O, Wales N, Star B, Boessenkool S. 2016. The efficacy of high-throughput sequencing and target enrichment on charred archaeobotanical remains. Scientific Reports 6: 37347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noormohammadi Z, Trujillo I, Belaj A, Ataei S, Hosseini-Mazinani M. 2014. Genetic structure of Iranian olive cultivars and their relationship with Mediterranean’s cultivars revealed by SSR markers. Scientia Horticulturae 178: 175–183. [Google Scholar]
- Orlando L, Gilbert MTP, Willerslev E. 2015. Reconstructing ancient genomes and epigenomes. Nature Reviews Genetics 16: 395–408. [DOI] [PubMed] [Google Scholar]
- Owen CA, Bita EC, Banilas G et al. 2005. AFLP reveals structural details of genetic diversity within cultivated olive germplasm from the Eastern Mediterranean. Theoretical and Applied Genetics 110: 1169–1176. [DOI] [PubMed] [Google Scholar]
- Pagnoux C. 2016. Emergence, développement et diversification de l’arboriculture en Grèce du Néolithique à l’époque romaine. Confrontation des données archéobotaniques, morphométriques, épigraphiques et littéraires. PhD, University of Paris 1 – Panthéon-Sorbonne, Paris, France. [Google Scholar]
- Palamarev E. 1989. Paleobotanical evidences of the Tertiary history and origin of the Mediterranean sclerophyll dendroflora. Plant Systematics and Evolution 162: 93–107. [Google Scholar]
- Palomares-Rius JE, Castillo P, Montes-Borrego M, Müller H, Landa BB. 2012. Nematode community populations in the rhizosphere of cultivated olive differs according to the plant genotype. Soil Biology and Biochemistry 45: 168–171. [Google Scholar]
- Palomares-Rius JE, Castillo P, Montes-Borrego M, Navas-Cortés JA, Landa BB. 2015. Soil properties and olive cultivar determine the structure and diversity of plant-parasitic nematode communities infesting olive orchards soils in southern Spain. PLoS One 10: e0116890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes D, Cayuela L, Gurr GM, Campos M. 2015. Is ground cover vegetation an effective biological control enhancement strategy against olive pests?PLoS One 10: e0117265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease AS. 1933. Notes on ancient grafting. Transactions of the American Philological Association 64: 66–76. [Google Scholar]
- Perea R, Gutiérrez-Galán A. 2016. Introducing cultivated trees into the wild: wood pigeons as dispersers of domestic olive seeds. Acta Oecologica 71: 73–79. [Google Scholar]
- Pérez AG, León L, Pascual M et al. 2014. Variability of virgin olive oil phenolic compounds in a segregating progeny from a single cross in Olea europaea L. and sensory and nutritional quality implications. PLoS One 9: e92898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt A, Gugger PF, Pellegrini M, Sork VL. 2015. Genome wide signature of local adaptation linked to variable CpG methylation in oak population. Molecular Ecology 15: 3823–3830. [DOI] [PubMed] [Google Scholar]
- Plomion C, Bastien C, Bogeat-Triboulot MB et al. 2016. Forest tree genomics: 10 achievements from the past 10 years and future prospects. Annals of Forest Science 73: 77–103. [Google Scholar]
- Ponti L, Gutierrez AP, Ruti PM, Dell’Aguila A. 2014. Fine-scale ecological and economic assessment of climate change on olive in the Mediterranean basin reveals winners and losers. Proceedings of the National Academy of Sciences, USA 111: 5598–5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto P, Navaro-Raya C, Valverde Corredor A et al. 2009. Colonization process of olive tissues by Verticillium dahliae and its in planta interaction with the biocontrol root endophyte Pseudomonas fluorescens PICF7. Microbial Biotechnology 2: 499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure from multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Madrigal J, Smith BD, Moreno-Mayar JV et al. 2016. Genome sequence of a 5,310-year-old maize cob provides insights into the early stages of maize domestication. Current Biology 26: 3195–3201. [DOI] [PubMed] [Google Scholar]
- Rellstab C, Gugerli F, Eckert AJ, Hancock AM, Holderegger R. 2015. A practical guide to environmental association analysis in landscape genomics. Molecular Ecology 24: 4348–4370. [DOI] [PubMed] [Google Scholar]
- Renfrew C. 1972. The emergence of civilisation. The Cyclades and the Aegean in the third millennium BC. London: Methuen. [Google Scholar]
- Rodríguez-Sánchez F, Guzmán B, Valido A, Vargas P, Arroyo J. 2009. Late Neogene history of the laurel tree (Laurus L., Lauraceae) based on phylogeographical analyses of Mediterranean and Macaronesian populations. Journal of Biogeography 36: 1270–1281. [Google Scholar]
- Rubio de Casas R, Besnard G, Schönswetter P, Balaguer L, Vargas P. 2006. Extensive gene flow blurs phylogeographic but not phylogenetic signal in Olea europaea L. Theoretical and Applied Genetics 113: 575–583. [DOI] [PubMed] [Google Scholar]
- Rubio de Casas R, Vargas P, Pérez-Corona E et al. 2011. Sun and shade leaves of Olea europaea respond differently to plant size, light availability and genetic variation. Functional Ecology 25: 802–812. [Google Scholar]
- Rugini E, Baldoni L, Muleo R, Sebastiani L. 2016. The olive tree genome. Cham: Springer International Publishing AG. [Google Scholar]
- Saponari M, Loconsole G, Cornara D et al. 2014. Infectivity and transmission of Xylella fastidiosa by Philaenus spumarius (Hemiptera: Aphrophoridae) in Apulia, Italy. Journal of Economic Entomology 107: 1316–1319. [DOI] [PubMed] [Google Scholar]
- de Saporta G. 1873. Etudes sur la végétation du sud-est de la France à l’époque Tertiaire. Révision de la flore des Gypses d’Aix. Annales des Sciences Naturelles de Paris 17 (Suppl. 1): 1–181. [Google Scholar]
- Satler JD, Carstens BC. 2016. Phylogeographic concordance factors quantify phylogeographic congruence among co-distributed species in the Sarracenia alata pitcher plant system. Evolution 70: 1080–1093. [DOI] [PubMed] [Google Scholar]
- Saumitou-Laprade P, Vernet P, Vekemans X et al. 2017. Elucidation of the genetic architecture of self-incompatibility in olive: evolutionary consequences and perspectives for orchard management. Evolutionary Applications 10: 867–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scascitelli M, Whitney KD, Randell RA et al. 2010. Genome scan of hybridizing sunflowers from Texas (Helianthus annuus and H. debilis) reveals asymmetric patterns of introgression and small islands of genomic differentiation. Molecular Ecology 19: 521–541. [DOI] [PubMed] [Google Scholar]
- Schaal BA, Olsen KM. 2000. Gene genealogies and population variation in plants. Proceedings of the National Academy of Sciences, USA 97: 7024–7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffels S, Durbin R. 2014. Inferring human population size and separation history from multiple genome sequences. Nature Genetics 46: 916–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M, Duringer P, Ghienne JF et al. 2006. The age of the Sahara desert. Science 311: 821. [DOI] [PubMed] [Google Scholar]
- Seager R, Liu H, Henderson N et al. 2014. Causes of increasing aridification of the Mediterranean region in response to rising greenhouse gases. Journal of Climate 27: 4655–4676. [Google Scholar]
- Sedgley M. 2004. Wild olive selection for quality oil production. Canberra, Australia: Rural Industries Research and Development Corporation, RIRDC Publication no 04/101. [Google Scholar]
- Sefc KM, Steinkellner H, Glossl J, Kampfer S, Regner F. 1998. Reconstruction of a grapevine pedigree by microsatellite analysis. Theoretical and Applied Genetics 97: 227–231. [Google Scholar]
- Segura V, Vilhjálmsson BJ, Platt A et al. 2012. An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nature Genetics 44: 825–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenbrock EH, McDonald BA. 2008. The origins of plant pathogens in agro-ecosystems. Annual Review of Phytopathology 46: 75–100. [DOI] [PubMed] [Google Scholar]
- Suc JP. 1984. Origin and evolution of the Mediterranean vegetation and climate in Europe. Nature 307: 429–432. [Google Scholar]
- Swezey CS. 2009. Cenozoic stratigraphy of the Sahara, Northern Africa. Journal of African Earth Sciences 53: 89–121. [Google Scholar]
- Terral JF. 2000. Exploitation and management of olive tree during Prehistoric times in Mediterranean France and Spain. Journal of Archaeological Science 27: 127–133. [Google Scholar]
- Terral JF, Arnold-Simard G. 1996. Beginnings of olive cultivation in Eastern Spain in relation to Holocene bioclimatic changes. Quaternary Research 46: 176–185. [Google Scholar]
- Terral JF, Mengüal X. 1999. Reconstruction of Holocene climate in Southern France and Eastern Spain using quantitative anatomy of olive wood and archaeological charcoal. Palaeogeography, Palaeoclimatology, Palaeoecology 153: 71–92. [Google Scholar]
- Terral JF, Alonso N, Buxò i Capdevila R et al. 2004a. Historical biogeography of olive domestication (Olea europaea L.) as revealed by geometrical morphometry applied to biological and archaeological material. Journal of Biogeography 31: 63–77. [Google Scholar]
- Terral JF, Badal E, Heinz C et al. 2004b. A hydraulic conductivity model points to post-Neogene survival of the Mediterranean olive. Ecology 85: 3158–3165. [Google Scholar]
- Tierney JE, Pausata FSR, de Menocal PB. 2017. Rainfall regimes of the Green Sahara. Science Advances 3: e1601503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiffin P, Ross-Ibarra J. 2014. Advances and limits of using population genetics to understand local adaptation. Trends in Ecology and Evolution 29: 673–680. [DOI] [PubMed] [Google Scholar]
- Trapero C, Rallo L, López-Escudero FJ, Barranco D, Díez CM. 2015. Variability and selection of verticillium wilt resistant genotypes in cultivated olive and in the Olea genus. Plant Pathology 64: 890–900. [Google Scholar]
- Trujillo I, Ojeda MA, Urdiroz NM et al. 2014. Identification of the Worldwide Olive Germplasm Bank of Córdoba (Spain) using SSR and morphological markers. Tree Genetics and Genomes 10: 141–155. [Google Scholar]
- Turrill WB. 1951. Wild and cultivated olives. Kew Bulletin 3: 437–442. [Google Scholar]
- do Val ADB, Ferreira JL, Vieira Neto J et al. 2012. Genetic diversity of Brazilian and introduced olive germplasms based on microsatellite markers. Genetics and Molecular Research 11: 556–571. [DOI] [PubMed] [Google Scholar]
- Unver T, Wu Z, Sterck L et al. 2017. Genome of wild olive and the evolution of oil biosynthesis. Proceedings of the National Academy of Sciences, USA114: E9413–E9422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Paer C, Bouchez O, Besnard G. 2018. Prospects on the evolutionary mitogenomics of plants: a case study on the olive family (Oleaceae). Molecular Ecology Resources, 18. doi:10.1111/1755-0998.12742. [DOI] [PubMed] [Google Scholar]
- Velasco D, Hough J, Aradhya M, Ross-Ibarra J. 2016. Evolutionary genomics of peach and almond domestication. Genes, Genomes, Genetics 6: 3985–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warschefsky EJ, Penmetsa RV, Cook DR, von Weitberg EJB. 2014. Back to the wilds: tapping evolutionary adaptations for resilient crops through systematic hybridization with crop wild relative. American Journal of Botany 101: 1791–1800. [DOI] [PubMed] [Google Scholar]
- Warschefsky EJ, Klein LL, Frank MH et al. 2016. Rootstocks: diversity, domestication, and impacts on shoot phenotypes. Trends in Plant Science 21: 418–437. [DOI] [PubMed] [Google Scholar]
- Willner W, Di Pietro R, Bergmeier E. 2009. Phytogeographical evidence for post-glacial dispersal limitation of European beech forest species. Ecography 32: 1011–1018. [Google Scholar]
- Wright SI, Bi IV, Schroeder SG et al. 2005. The effect of artificial selection on maize genome. Science 308: 1310–1314. [DOI] [PubMed] [Google Scholar]
- Xing X, Koch AM, Jones AM, Ragone D, Murch S, Hart MM. 2012. Mutualism breakdown in breadfruit domestication. Proceedings of the Royal Society B: Biological Sciences 279: 1122–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoruk B, Taskin V. 2014. Genetic diversity and relationships of wild and cultivated olives in Turkey. Plant Systematics and Evolution 300: 1247–1258. [Google Scholar]
- Zehdi-Azouzi S, Cherif E, Moussouni S et al. 2015. Genetic structure of the date palm (Phoenix dactylifera) in the Old World reveals a strong differentiation between eastern and western populations. Annals of Botany 116: 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan MM, Cheng ZZ, Su GC et al. 2015. Genetic relationships analysis of olive cultivars grown in China. Genetics and Molecular Research 14: 5958–5969. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Massonnet M, Sanjak JS, Cantu D, Gaut BS. 2017. The evolutionary genomics of grape (Vitis vinifera ssp. vinifera) domestication. Proceedings of the National Academy of Sciences, USA 114: 11715–11720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohary D. 1994. The wild genetic resources of the cultivated olive. Acta Horticulturae 356: 62–65. [Google Scholar]
- Zohary D, Hopf M, Weiss E. 2012. Domestication of plants in the Old World: the origin and spread of cultivated plants in Southwest Asia, Europe, and the Mediterranean Basin. Oxford: Oxford University Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.