Abstract
Genetic factors likely influence individuals' concentrations of 25-hydroxyvitamin D [25(OH)D], a biomarker of vitamin D exposure previously linked to reduced risk of several chronic diseases. We conducted a genome-wide association study of serum 25(OH)D (assessed using liquid chromatography-tandem mass spectrometry) and 386,449 single nucleotide polymorphisms (SNPs). Our sample consisted of 1,829 participants randomly selected from the Sister Study, a cohort of women who had a sister with breast cancer but had never had breast cancer themselves. 19,741 SNPs were associated with 25(OH)D (p < 0.05). We re-assessed these hits in an independent sample of 1,534 participants who later developed breast cancer. After pooling, 32 SNPs had genome-wide significant associations (p < 5 × 10−8). These were located in or near GC, the vitamin D binding protein, or CYP2R1, a cytochrome P450 enzyme that hydroxylates vitamin D to form 25(OH)D. The top hit was rs4588, a missense GC polymorphism associated with a 3.5 ng/mL decrease in 25(OH)D per copy of the minor allele (95% confidence interval [CI]: −4.1, −3.0; p = 4.5 × 10−38). The strongest SNP near CYP2R1 was rs12794714, a synonymous variant (p = 3.8 × 10−12; β = 1.8 ng/mL decrease in 25(OH)D per minor allele [CI: −2.2, −1.3]). Serum 25(OH)D concentrations from samples collected from some participants 3–10 years after baseline (811 cases, 780 non-cases) were also strongly associated with both loci. These findings augment our understanding of genetic influences on 25(OH)D and the possible role of vitamin D binding proteins and cytochrome P450 enzymes in determining measured levels. These results may help to identify individuals genetically predisposed to vitamin D insufficiency.
Keywords: 25-dihydroxy vitamin D, genome-wide association study, single nucleotide polymorphism, vitamin D binding protein, CYP2R1
Introduction
While randomized clinical trials of have failed to establish direct links between vitamin D supplementation and health (Bjelakovic et al., 2011, 2014; Avenell et al., 2014), observational studies have demonstrated that high levels of 25-hydroxyvitamin D [25(OH)D], a vitamin D biomarker that can be measured in blood, are associated with lower mortality (Garland et al., 2014) and reduced risk of many chronic diseases, including heart disease and some cancers (Gandini et al., 2011; Autier et al., 2014; Zhang et al., 2017). Determinants of 25(OH)D include ultraviolet-B radiation, dietary supplements, and certain foods, including fish and fortified dairy products. Genetic factors are also thought to play a role in determining blood concentrations.
Vitamin D sufficiency is typically assessed by measuring serum or plasma concentrations of 25(OH)D, which is a stable precursor to the active form of vitamin D, 1,25(OH)2D. The Institute of Medicine guidelines suggest that 25(OH)D concentrations above 20 ng/mL are sufficient for bone health (Ross et al., 2011), but higher levels may provide additional health benefits (Garland and Gorham, 2016; Durazo-Arvizu et al., 2017; O'Brien et al., 2017b; Zhang et al., 2017). Previous genome-wide association studies (GWAS) have identified several single nucleotide polymorphisms (SNPs) associated with 25(OH)D concentrations. To date, eight such GWAS have been published (Benjamin et al., 2007; Ahn et al., 2010; Engelman et al., 2010; Wang et al., 2010; Lasky-Su et al., 2012; Anderson et al., 2014; Sapkota et al., 2016; Jiang et al., 2018), identifying a total of 17 SNPs in 7 chromosomal locations (in or near GC, NADSYN1/DHCR7, CYP2R1/RRAS2/PDE3B, CYP24A1, SSTR4/FOXA2, AMDHD1, and SEC23A) with association p-values < 5 × 10−8 (PheGen I: Phenotype-Genotype Integrator-National Center for Biotechnology Information, 20141; MacArthur et al., 2017).
Here, we introduce the first vitamin D GWAS to use the current gold-standard measure of 25(OH)D, liquid chromatography/tandem mass spectrometry (LC/MS). LC/MS has been shown to outperform other commonly-utilized methods (Farrell et al., 2012) and is capable of measuring concentrations of 3-epi-25(OH)D3, which is thought to play a similar biological role to 25(OH)D3 (Cashman et al., 2014). We conducted this work using serum collected from a random sample of women participating in a large prospective cohort study. We re-assessed the top SNPs in an independent set of serum samples from women in the same cohort who later developed breast cancer, and again in serum samples collected from some of the same cases (post-diagnosis) and non-cases 3–10 years after baseline. We also performed haplotype analyses of key genetic regions.
Materials and methods
This work was conducted using data from the Sister Study, a prospective cohort of 50,884 women who had a full or half-sister with a history of breast cancer, but who had never had breast cancer themselves at the time they enrolled in the study. Participants aged 35–74 and living in the United States or Puerto Rico joined the study between 2003 and 2009. They were visited in their homes by a trained examiner, who obtained written informed consent and collected the blood samples needed for 25(OH)D and genotype analyses. A subset of participants, including some who had developed breast cancer and some who had not, were asked to provide a second blood sample and other biospecimens in 2013–2014, 3–10 years after baseline. Further details on the study protocol, which also included extensive questionnaires and additional biospecimens, are available elsewhere (Sandler et al., 2017). Approval and oversight for the Sister Study is provided by the Institutional Review Boards of the National Institute of Environmental Health Sciences and the Copernicus group. Analyses were completed using data release 4.1 (July 2014).
Participants for the vitamin D and genetics sub-studies were selected using a case-cohort design that included a random sample from the full cohort (n = 1,829, including 67 women who went on to develop breast cancer) and all remaining women diagnosed with invasive breast cancer or ductal carcinoma in situ within 5 years of their baseline blood draw (n = 1,534 additional cases, for 1,601 total cases) (O'Brien et al., 2017b). This included 28 pairs of sisters, who were treated as independent observations despite their genetic similarities [between-sister R2 for 25(OH)D levels = 0.03]. Blood samples from these women were assayed for 533,631 SNPs using the Infinium OncoArray genotyping panel (Illumina Inc.) (Amos et al., 2016). This panel includes a full GWAS backbone, as well as ancestry informative markers (AIMs) and genes presumably or possibly linked to cancer or cancer-related factors. Serum samples were assessed for 25(OH)D concentrations using LC/MS at Heartland Assays, Inc. Measured concentrations were adjusted for batch effects and season of blood draw and thus approximate average annual 25(OH)D. Further details on both the SNP and 25(OH)D analysis can be found elsewhere (Amos et al., 2016; O'Brien et al., 2017a,b).
After excluding SNPs that did not meet quality control standards (n = 41,664, as described previously, Amos et al., 2016) or that had a minor allele frequency (MAF) less than 2% in the sub-cohort (n = 105,518), 386,449 SNPs remained. We calculated Hardy-Weinberg equilibrium p-values and examined our top hits for evidence of disequilibrium, but did not exclude SNPs based on these results. We regressed 25(OH)D on the number of copies of the minor allele for each of these SNPs using linear least-squares. The values of 25(OH)D looked normally distributed. We adjusted each model for age at blood draw (in years), self-reported race/ethnicity [Non-Hispanic White (n = 1,576), Black (n = 134), Hispanic (n = 81), or other (n = 38)] and genetic ancestry (proportion CEU, YRI, or CHB) (O'Brien et al., 2017a) and calculated the genomic control inflation factor (λ) to test for evidence of uncontrolled confounding due to population stratification. We also conducted sensitivity analyses adjusting for estimated total vitamin D intake at baseline (dietary plus supplement) and hours spent outdoors per year, as these both contribute to variations in measured 25(OH)D concentrations. Analyses were conducted using SAS (v9.3), R (v3.2.1), or PLINK (v1.07). Locus plots were made using LocusZoom (Pruim et al., 2011).
Primary analyses used the baseline 25(OH)D measurements from the randomly selected sub-cohort. For replication analyses, we also analyzed associations in the set of women who later developed breast cancer, assessing the relationship between 25(OH)D and any SNPs for which the p-value had been < 0.05 in the sub-cohort. We excluded the 67 cases selected into the sub-cohort. In analyses pooling the sub-cohort and cases we defined genome-wide significance as p < 5 × 10−8. The pooled analysis included additional adjustment for future case status.
We also examined the association between these SNPs and 25(OH)D concentrations in blood samples collected 3–10 years after enrollment. In 2013, a total of 3,762 women who were originally selected for participation in the case-cohort sample were asked to provide secondary biospecimens. We collected second blood samples for 1,227 women who had been diagnosed with breast cancer while on study [assaying 811 for 25(OH)D] and 1,203 who had remained breast cancer-free [assaying 780 for 25(OH)D]. Measurements were based on LC/MS (Heartland Assays, Inc.).
We also conducted haplotype analyses of the top two regions using baseline data from the sub-cohort and cases. These were conducted separately in Non-Hispanic whites and African-Americans due to concerns about population stratification and racial differences in linkage disequilibrium (Stram and Seshan, 2012). We used expectation-maximization software (“hapassoc” package in R; Burkett et al., 2004, 2006) to estimate haplotype frequencies and the association between 25(OH)D and each copy of the index haplotype, relative to the most common haplotype in non-Hispanic whites. We assessed all haplotypes with estimated frequency of at least 2%, pooling the rare haplotypes into a single category. Models were adjusted for age at blood draw, ancestry, and future case status.
Results
Women in the sub-cohort were 55.3 years of age, on average, when they joined the study and had average serum 25(OH)D concentrations of 31.8 ng/mL (Table 1). Most participants were non-Hispanic white (86%). Women who later developed breast cancer were 57.4 years of age at baseline and had average 25(OH)D concentration of 31.0 ng/mL. On average, 7.8 years passed between baseline and follow-up blood collection. At that time, serum 25(OH)D concentrations were 40.4 ng/mL in non-cases and 43.5 ng/mL in post-diagnosis cases. Non-Hispanic white women comprised 89% the follow-up group.
Table 1.
Description of study sample.
| Baseline | Second blood draw 3–10 years after baseline (“Sisters Changing Lives”) | |||
|---|---|---|---|---|
| Sub-cohort (n = 1,829)b | Cases (n = 1,534)c,d | Non-cases (n = 780) | Cases (n = 811)d | |
| Age at blood draw; mean (std) | 55.3 (8.9) | 57.4 (8.9) | 63.1 (8.7) | 65.5 (8.8) |
| 25(OH)D (ng/mL)a; mean (std) | 31.8 (10.5) | 31.0 (10.1) | 40.4 (13.7) | 43.5 (14.3) |
| Self-reported Race/Ethnicity; n (%) | ||||
| Non-hispanic white | 1576 (86) | 1307 (85) | 693 (89) | 722 (89) |
| African-American | 134 (7) | 120 (8) | 46 (6) | 38 (5) |
| Hispanic | 81 (4) | 59 (4) | 27 (3) | 27 (3) |
| Other | 38 (2) | 47 (3) | 14 (2) | 23 (3) |
Adjusted for batch and season of blood draw.
Includes 67 women who went on to develop breast cancer.
Excludes cases selected as part of the random sub-cohort.
1 case missing self-reported race/ethnicity.
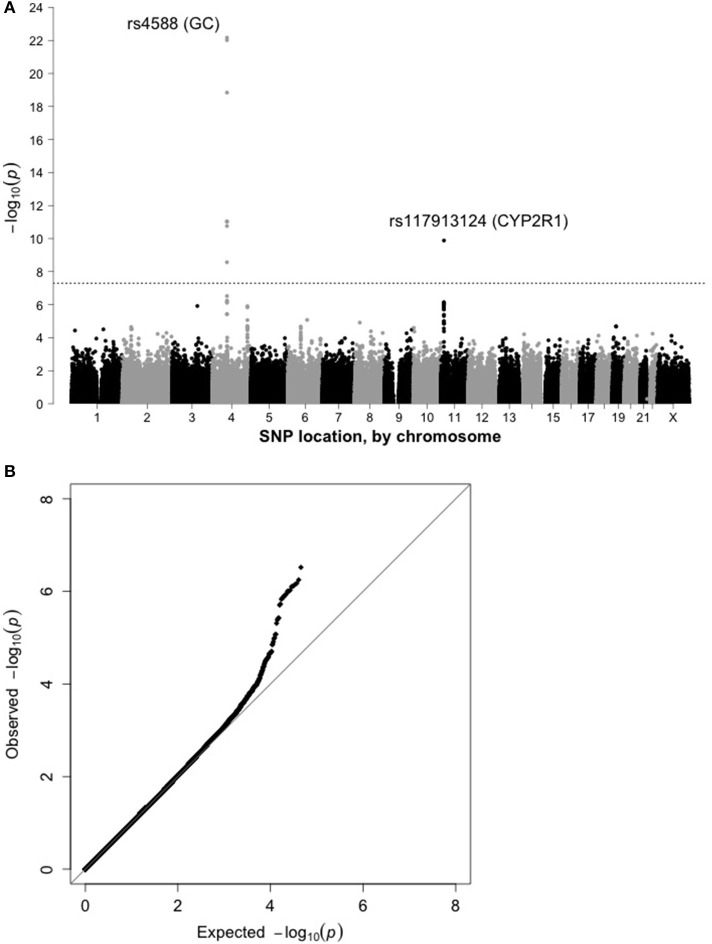
Results from preliminary analyses conducted within the sub-cohort are shown as a Manhattan plot (Figure 1A) and quantile-quantile plot (Figure 1B). There was no evidence of residual confounding due to population stratification (λ = 1.007). 19,741 SNPs were associated with 25(OH)D at p < 0.05. The top hit was rs4588 (p = 6.8 × 10−23; Table 2), located in the vitamin D binding protein (VDBP) gene (GC) on chromosome 4. SNPs in or near CYP2R1, which encodes a cytochrome P450 vitamin D hydroxylase, also showed evidence of an association with 25(OH)D, with rs117913124 having the lowest p-value in the sub-cohort (p = 1.3 × 10−10).
Figure 1.
(A) Manhattan plot for 25(OH)D in the sub-cohort (n = 1,829). (B) Quantile-quantile plot for 25(OH)D in the sub-cohort (n = 1,829).
Table 2.
Single nucleotide polymorphism associated with serum 25(OH)D levels at p < 5 × 10−8 in the Sister Study (2003–2009).
| SNP-rare allele | Locationa | MAF, sub-cohort | HWE p-value, subcohort | P-value, sub-cohort | P-value, cases | βb (95% CI), pooled samplec | P-value, pooled samplec | P-value in follow-up studyc,d |
|---|---|---|---|---|---|---|---|---|
| CHROMOSOME 4, GC | ||||||||
| rs4588-A | 72618323 | 0.27 | 0.55 | 6.8 × 10−23 | 8.9 × 10−17 | −3.5 (−4.1, −3.0) | 4.5 × 10−38 | 5.8 × 10−10 |
| rs2282679-C | 72608383 | 0.26 | 0.28 | 9.6 × 10−23 | 3.1 × 10−16 | −3.5 (−4.0, −3.0) | 1.9 × 10−37 | 9.0 × 10−10 |
| rs1155563-G | 72643488 | 0.27 | 0.05 | 1.4 × 10−19 | 2.9 × 10−11 | −3.0 (−3.5, −2.5) | 9.6 × 10−29 | 6.6 × 10−7 |
| rs705120-A | 72614140 | 0.42 | 0.60 | 9.5 × 10−12 | 1.6 × 10−11 | −2.4 (−2.8, −1.9) | 4.5 × 10−22 | 6.2 × 10−5 |
| rs2201124-A | 72597009 | 0.31 | 0.70 | 9.0 × 10−12 | 3.5 × 10−7 | −2.2 (−2.8, −1.7) | 3.4 × 10−17 | 3.2 × 10−6 |
| rs4694105-A | 72592214 | 0.31 | 0.62 | 1.7 × 10−11 | 2.2 × 10−7 | −2.2 (−2.8, −1.7) | 3.5 × 10−17 | 4.7 × 10−6 |
| rs1526692-G | 72578724 | 0.40 | 0.31 | 2.7 × 10−9 | 6.8 × 10−8 | −2.0 (−2.5, −1.5) | 8.8 × 10−16 | 1.9 × 10−5 |
| rs6837549-A | 72596821 | 0.49 | 0.32 | 6.7 × 10−7 | 1.1 × 10−6 | 1.7 (1.2, 2.2) | 4.6 × 10−12 | 0.003 |
| rs13113067-A | 72739098 | 0.35 | 0.26 | 5.6 × 10−7 | 0.001 | −1.5 (−2.0, −1.0) | 3.8 × 10−9 | 0.14 |
| rs12639968-A | 72712872 | 0.18 | 0.53 | 7.7 × 10−7 | 0.001 | −1.8 (−2.4, −1.2) | 7.7 × 10−9 | 0.01 |
| rs962227-A | 72707517 | 0.29 | 0.17 | 3.0 × 10−7 | 0.005 | −1.5 (−2.1, −1.0) | 1.1 × 10−8 | 0.03 |
| rs10033936-G | 72743474 | 0.25 | 0.26 | 3.8 × 10−6 | 7.5 × 10−4 | −1.6 (−2.2, −1.0) | 2.2 × 10−8 | 0.09 |
| rs12512631-G | 72601331 | 0.35 | 0.72 | 3.7 × 10−6 | 0.004 | 1.4 (0.9, 1.9) | 3.8 × 10−8 | 0.003 |
| CHROMOSOME 11, CYP2R1 | ||||||||
| rs12794714-A | 14913575 | 0.42 | 0.18 | 7.9 × 10−7 | 9.4 × 10−7 | −1.8 (−2.2, −1.3) | 3.8 × 10−12 | 4.1 × 10−4 |
| rs201473898-A | 14893704 | 0.42 | 0.23 | 9.4 × 10−7 | 1.2 × 10−6 | −1.7 (−2.2, −1.2) | 6.5 × 10−12 | 3.5 × 10−4 |
| rs11023227-G | 14459087 | 0.36 | 0.27 | 7.1 × 10−7 | 2.2 × 10−6 | −1.7 (−2.2, −1.2) | 1.5 × 10−11 | 0.02 |
| rs10832275-C | 14478224 | 0.36 | 0.21 | 1.3 × 10−6 | 1.6 × 10−6 | −1.7 (−2.2, −1.2) | 1.6 × 10−11 | 0.02 |
| rs7121171-G | 14446420 | 0.36 | 0.16 | 9.9 × 10−7 | 2.3 × 10−6 | −1.7 (−2.2, −1.2) | 1.6 × 10−11 | 0.02 |
| rs12295888-G | 14450531 | 0.36 | 0.21 | 9.9 × 10−7 | 2.0 × 10−6 | −1.7 (−2.2, −1.2) | 1.7 × 10−11 | 0.02 |
| rs72261784-D | 14446060 | 0.36 | 0.22 | 1.4 × 10−6 | 2.2 × 10−6 | −1.7 (−2.2, −1.2) | 2.5 × 10−11 | 0.02 |
| rs11023223-G | 14457112 | 0.36 | 0.19 | 1.1 × 10−6 | 2.6 × 10−6 | −1.7 (−2.2, −1.2) | 2.5 × 10−11 | 0.02 |
| rs10832294-A | 14747427 | 0.41 | 0.27 | 4.1 × 10−6 | 1.5 × 10−6 | −1.7 (−2.2, −1.2) | 4.2 × 10−11 | 3.8 × 10−4 |
| rs10832268-A | 14465068 | 0.36 | 0.19 | 1.9 × 10−6 | 3.0 × 10−6 | −1.7 (−2.2, −1.2) | 4.7 × 10−11 | 0.01 |
| rs11023332-G | 14784110 | 0.42 | 0.31 | 1.0 × 10−5 | 6.6 × 10−7 | −1.7 (−2.2, −1.2) | 5.1 × 10−11 | 7.5 × 10−4 |
| rs10832269-A | 14465069 | 0.36 | 0.19 | 2.0 × 10−6 | 3.3 × 10−6 | −1.7 (−2.1, −1.2) | 5.6 × 10−11 | 0.01 |
| rs11023246-G | 14536956 | 0.36 | 0.29 | 4.9 × 10−6 | 1.7 × 10−6 | −1.7 (−2.2, −1.2) | 6.2 × 10−11 | 0.03 |
| rs117913124-A | 14900931 | 0.02 | 1.00 | 1.3 × 10−10 | 0.01 | −5.1 (−6.7, −3.6) | 1.2 × 10−10 | 4.2 × 10−6 |
| rs2305305-A | 14540942 | 0.36 | 0.26 | 1.1 × 10−5 | 1.8 × 10−6 | −1.6 (−2.1, −1.1) | 1.5 × 10−10 | 0.03 |
| rs10766188-G | 14660826 | 0.34 | 0.38 | 1.8 × 10−4 | 3.9 × 10−6 | −1.5 (−2.0, −1.0) | 6.1 × 10−9 | 0.003 |
| rs1993116-A | 14910234 | 0.38 | 0.69 | 1.4 × 10−5 | 1.3 × 10−4 | 1.4 (0.9, 1.9) | 9.7 × 10−9 | 6.3 × 10−4 |
| rs10741657-A | 14914878 | 0.37 | 0.73 | 4.2 × 10−5 | 2.2 × 10−4 | 1.4 (0.9, 1.9) | 4.7 × 10−8 | 0.002 |
| rs11023203-A | 14409815 | 0.33 | 0.34 | 2.8 × 10−5 | 3.3 × 10−4 | −1.4 (−1.9, −0.9) | 4.9 × 10−8 | 0.03 |
All models adjusted for age at blood draw, self-reported race/ethnicity, and estimated ancestry proportions; MAF, minor allele frequency; HWE, Hardy-Weinberg equilibrium.
GRch37/hg19.
Change in 25(OH)D (in ng/mL) per copy of the minor allele.
Adjusting for case status.
Serum 25(OH)D levels measured in blood collected 3–10 years after baseline for the “Sisters Changing Lives” follow-up study.
Bold values indicate SNP with lowest p-value for the region (based on pooled sample).
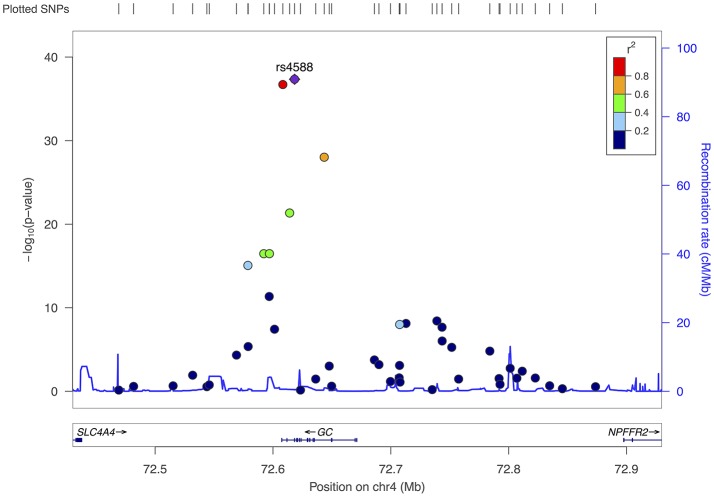
When we re-assessed the top 19,741 SNPs in the independent sample of women who later developed breast cancer, 1,121 were statistically-significant at p < 0.05. For the pooled sample, which included both cases and sub-cohort members, we identified 32 SNPs in two regions that were associated with 25(OH)D at p < 5 × 10−8 (Table 2). The top SNP for the GC region was again rs4588 (p = 4.5 × 10−38), with each copy of the variant A allele associated with an estimated 3.5 ng/mL decrease in 25(OH)D (95% confidence interval [CI]: −4.1, −3.0). This SNP also had a strong association in the smaller follow-up sample (p = 5.8 × 10−10). Twelve other SNPs in the region surrounding rs4588 were also strongly associated with 25(OH)D in the pooled baseline sample (Table 2 and Figure 2) and, with a few exceptions, also in the follow-up sample.
Figure 2.
Fine-mapping of region surrounding rs4588 (chromosome 4), all participants.
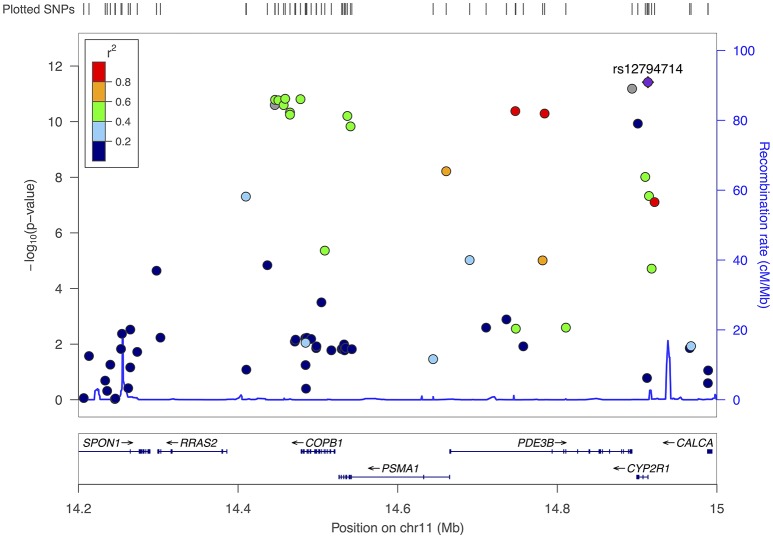
A more common CYP2R1 SNP, rs12794714, replaced rs117913124 as the top hit in the region for the pooled sample (p = 3.8 × 10−12 and MAF = 0.42 for rs12794714 vs. p = 1.2 × 10−10 and MAF = 0.02 for rs117913124), with each copy of the A allele associated with a 1.8 ng/mL decrease in 25(OH)D (95% CI: −2.2, −1.3). Seventeen other SNPs in this region also met criteria for genome-wide statistical significance in the pooled sample (Table 2). Though most of these were in moderate to high linkage disequilibrium with rs12794714, they spanned several gene regions, including COPB1, PSMA1, PDE3B, and RRAS2 (Figure 3). A second peak appeared ~454 kb away from rs12794714 at rs11023227 in COPB1, but the signal was not independent (original p = 1.5 × 10−11; p = 0.007 after adjusting for rs12794714; r2 = 0.33). All 19 of the genome-wide significant hits in this region were also associated with 25(OH)D concentrations from the follow-up visit (all p ≤ 0.03). None of the genome-wide significant hits showed evidence of Hardy-Weinberg disequilibrium.
Figure 3.
Fine-mapping plot for the region surrounding rs12794714 (chromosome 11), all participants.
Fifteen other SNPs that had false discovery rate q-values < 0.10 in the pooled sample are described in Supplementary Table 1. Three were from the chromosome 4 locus and four from the chromosome 11 locus (one on COPB1 and two on PDE3B). The remaining eight were independent signals, only two of which represented known genes (rs4951247 on ELK4 and rs360157 on MYO9B). The associations between these SNPs and 25(OH)D measured during follow-up were mostly consistent with those observed for baseline 25(OH)D.
When we additionally adjusted for both estimated vitamin D intake at baseline (measured using a food frequency questionnaire plus self-reported supplement use) and self-reported average hours spent outdoors per year, the results were very similar, with all but one of 32 SNPs reported here showing genome-wide significant associations (Supplementary Table 2). While the rankings shuffled around for the chromosome 11 locus, the only notable change was that while rs117913124 was the top ranked SNP for both the sub-cohort specific and pooled analysis, rs12794714 dropped down to 11th with the additional adjustments. There was little change to any of the effect estimates or p-values.
For haplotype analyses (Table 3), which were based on the pooled baseline sample, we selected SNPs with very small p-values (p < 1 × 10−10 for the chromosome 4 region and p < 5 × 10−8 for the chromosome 11 region) that were not in high linkage disequilibrium with any other, more strongly associated SNP (r2 < 0.80 in our sample). We excluded rs117913124 due to its low MAF. This resulted in a 6-SNP set including rs4588 and a 5-SNP set including rs12794714. For non-Hispanic whites, there were seven common (frequency >2%) haplotypes for both chromosomal sets. For chromosome 4, each copy of the “GCAAAG” haplotype (frequency = 21%) was associated with a 3.9 ng/mL decrease in 25(OH)D (95% CI: −4.6, −3.2; p = 9.8 × 10−30) in non-Hispanic whites, relative to the most common haplotype (AAGCCA, frequency = 43%). In African-Americans, only “GCACCA” (frequency = 9%) was significantly associated with 25(OH)D levels, with an estimated 4.6 ng/mL increase in 25(OH)D (95% CI: 1.2, 8.0; p = 0.008), per copy, relative to “AAGCCA” (frequency = 17%). For chromosome 11, “AGGGA” (frequency = 26%) was associated with an estimated 2.1 ng/mL decrease per copy in non-Hispanic whites (95% CI: −2.8, −1.5; p = 6.1 × 10−10), relative to the most common “GAAAG” haplotype (frequency = 34%). Haplotype distributions for the chromosome 11 locus were very different for African-Americans, with “GAAGG” being the most common haplotype (frequency = 53%) and no haplotypes showing a statistically-significant difference in 25(OH)D concentrations compared to “GAAAG” (frequency = 26%). Correlation matrices for SNPs in both of these regions are included as Supplementary Figures 1, 2.
Table 3.
Haplotype analysis.
| Haplotype | Non-Hispanic whites (n = 2,883) | African-Americans (n = 254) | ||||
|---|---|---|---|---|---|---|
| Haplotype frequency | βa (95% CI), pooled samplee | P-value, pooled samplee | Haplotype frequency | βa (95% CI), pooled samplee | P-value, pooled samplee | |
| CHROMOSOME 4 (GC): rs1526692, rs6837549, rs2201124, rs705120, rs4588, rs1155563 | ||||||
| AAGCCAa | 0.43 | 0.0 | 0.17 | 0.0 | ||
| GCAAAGb | 0.21 | −3.9 (−4.6, −3.2) | 9.8 × 10−30 | 0.05 | 2.7 (−1.8, 7.1) | 0.24 |
| ACGACAc | 0.07 | −1.0 (−2.0, 0.0) | 0.05 | 0.10 | 1.1 (−2.5, 4.7) | 0.54 |
| GCACCAa | 0.07 | −0.7 (−1.7, 0.3) | 0.17 | 0.09 | 4.6 (1.2, 8.0) | 8.5 × 10−3 |
| GCGACAc | 0.07 | −1.3 (−2.4, −0.3) | 0.01 | 0.12 | 2.1 (−1.2, 5.5) | 0.21 |
| AAGAAGb | 0.03 | −4.6 (−6.2, −3.1) | 3.2 × 10−9 | – | – | – |
| ACGCCAa | 0.03 | −0.7 (−2.4, 0.9) | 0.40 | 0.16 | −1.0 (−5.2, 2.1) | 0.52 |
| GAGCCAa | – | – | – | 0.10 | −1.4 (−5.1, 2.3) | 0.47 |
| GAGACAc | – | – | – | 0.06 | −1.9 (−6.5, 2.7) | 0.43 |
| AAGACAc | – | – | – | 0.04 | 1.0 (−4.9, 6.9) | 0.74 |
| Rare pooledd | 0.08 | −2.3 (−3.3, −1.3) | 5.4 × 10−6 | 0.11 | −1.9 (−5.2, 1.4) | 0.26 |
| CHROMOSOME 11 (CYP2R1): rs11023203, rs11023227, rs10766188, rs1993116, rs12794714 | ||||||
| GAAAG | 0.34 | 0.0 | 0.26 | 0.0 | ||
| AGGGA | 0.26 | −2.1 (−2.8, −1.5) | 6.1 × 10−10 | 0.09 | −2.4 (−5.8, 1.0) | 0.17 |
| GAAGG | 0.15 | −0.7 (−1.5, 0.1) | 0.07 | 0.53 | −1.8 (−7.2, 3.7) | 0.53 |
| GAAGA | 0.06 | −1.4 (−2.5, −0.2) | 0.02 | 0.03 | −1.6 (−3.9, 0.6) | 0.15 |
| GGGGA | 0.05 | −2.0 (−3.2, −0.8) | 1.5 × 10−3 | – | – | – |
| GAGGA | 0.03 | −1.8 (−3.3, −0.3) | 0.02 | – | – | – |
| AGAGA | 0.03 | −2.9 (−4.6, −1.3) | 4.0 × 10−4 | – | – | – |
| Rare pooledd | 0.08 | −1.0 (−2.1, 0.0) | 0.05 | 0.11 | −0.8 (−3.9, 2.2) | 0.59 |
All models adjusted for age at blood draw, estimated ancestry proportions, and breast cancer case status.
Corresponds to the “Gc1s” vitamin D binding protein variant in Non-Hispanic whites (based on genotypes for rs705120 and rs4588).
Corresponds to the “Gc2” vitamin D binding protein variant in Non-Hispanic whites (based on genotypes for rs705120 and rs4588).
Corresponds to the “Gc1f” vitamin D binding protein variant in Non-Hispanic whites (based on genotypes for rs705120 and rs4588).
Rare haplotypes (frequency < 2%) pooled together.
Change in 25(OH)D (in ng/mL) per copy of the index haplotype relative to the most common haplotype (“AAGCCA” for chromosome 4, “GAAAG” for chromosome 11), controlling for all other haplotypes.
Bold values indicate SNP with lowest independently measured p-value (based on pooled sample).
Discussion
In this GWAS of vitamin D serum levels, we identified two regions strongly associated with serum 25(OH)D—one on chromosome 4 surrounding the GC gene and the second on chromosome 11 including SNPs from CYP2R1, COPB1, PSMA1, and PDE3B. The identified loci replicated in an independent sample of women selected because they later developed breast cancer and the SNPs were also strongly associated with 25(OH)D concentrations measured in second blood samples from the same participants collected 3–10 years after baseline. To our knowledge, this was the first GWAS to use the gold-standard LC/MS methods to measure total 25(OH)D and the first to examine haplotypes.
The GC gene encodes the VDBP, a member of the albumin family that stores and transports both 25(OH)D and the active form of vitamin D, 1,25(OH)2D (Speeckaert et al., 2006). It seems quite plausible that a variant that affects VDBP could directly impact measured serum 25(OH)D. CYP2R1 polymorphisms also have the capacity to directly impact 25(OH)D concentrations, as this gene encodes a cytochrome P450 enzyme responsible for hydroxylating vitamin D and converting it to 25(OH)D (Shinkyo et al., 2004).
Our findings augment the results of previous GWAS. Those that reported hits in GC (Ahn et al., 2010; Wang et al., 2010; Lasky-Su et al., 2012; Anderson et al., 2014; Jiang et al., 2018) observed the smallest p-values for rs1155563, rs17467825, rs2282679, and rs3755967. We did not assess rs17467825 or rs3755967, but rs2282679 and rs1155563 had the second and third smallest p-values in our study. However, they both are synonymous substitutions, while rs4588 is a missense substitution in an exon (exon 12; amino acid change Thr to Lys at position 436). Therefore, given their high correlations with rs4588 (r2 = 0.97 and 0.77, respectively), these previously identified SNPs may just be tags for rs4588, which may be a variant that causally impacts 25(OH)D concentrations. This possibility is supported by a number of candidate gene studies that have reported strong associations between rs4588 and measured 25(OH)D (Engelman et al., 2008; Lu et al., 2012; Perna et al., 2013; Robien et al., 2013; Li et al., 2014; Nissen et al., 2014). Of note, rs4588 was not associated with gene expression in quantitative trait loci analyses, but five other nearby genome-wide significant GC SNPs (rs1155563, rs13113067, rs12639968, rs962227, and rs10033936) were associated with expression of VDBP in stomach tissue2.
Another putative causal variant is rs7041, which also results in a missense substitution in exon 12 (amino acid change Asp to Glu, position 432). We did not measure this SNP, but it is highly correlated with rs705120 in whites (r2 = 0.97 in the CEU sample of 1,000 genomes) (Johnson et al., 2008; 1000 Genomes Project Consortium et al., 2015), and rs705120 had a p-value of 9.5 × 10−12 in our sample. Together, the genotypes for rs4588 and rs705120/rs7041 determine individual's VDBP variants (“AA” = Gc2, “AC” = Gc1f, or “CC” = Gc1s) (Powe et al., 2013). The combination of these variants determine an individual's phenotype, where each phenotype has a different glycosylation pattern and binding affinity (Braun et al., 1992; Abbas et al., 2008; Powe et al., 2013). In our sample, carriers of the Gc2-coding haplotypes had the lowest 25(OH)D concentrations (Table 3). This is consistent with the results of one previous study, which showed that individuals homozygous for Gc2-coding haplotypes (Gc2/Gc2 phenotype) had significantly lower serum concentrations of 25(OH)D than those with Gc1S/Gc1S, Gc1S/Gc1F, or Gc1s/Gc2 phenotypes (Sollid et al., 2016). The same study reported that individuals with the Gc2/Gc2 phenotype had lower VDBP than all other phenotypes.
However, the links between rs4588, rs705120/rs7041, GC haplotypes, and concentrations of VDBP and 25(OH)D remain uncertain, as other studies, including some with large samples of African-Americans, have not observed these same associations (Powe et al., 2013; Batai et al., 2014; Denburg et al., 2016; Yao et al., 2017). The role of the other SNPs included in this haplotype analysis is also unclear, though we note that any haplotypes that contained “A” alleles for both rs4588 and rs705120 also contained the “G” allele for rs1155563, a synonymous polymorphism identified in a previous GWAS (Anderson et al., 2014).
In our sample, African-Americans were less likely to carry the minor allele at rs4588 (MAF = 0.11 vs. 0.28 in non-Hispanic whites; Supplementary Table 3) and the SNP was not associated with 25(OH)D in African-Americans, though our sample size was quite small (data not shown). In general, most of the GWAS and candidate gene studies that observed positive associations between 25(OH)D and SNPs in GC were conducted in populations of European or Chinese descent (Engelman et al., 2008, 2010; Ahn et al., 2010; Wang et al., 2010; Jorde et al., 2012; Lasky-Su et al., 2012; Lu et al., 2012; Perna et al., 2013; Robien et al., 2013; Zhang et al., 2013; Anderson et al., 2014; Batai et al., 2014; Li et al., 2014; Nissen et al., 2014; Clendenen et al., 2015), with less consistent results seen for African-American populations (Engelman et al., 2008; Batai et al., 2014; Yao et al., 2017).
As rs705120 is not correlated with rs7041 in African-Americans (r2 = 0.10 in YRI sample of 1,000 genomes), we cannot determine the VDBP variants of our African-American participants, but prior studies have reported that VDBP variant distributions differ markedly by race, with Gc1F being much more common in African-Americans than whites (Powe et al., 2013; Denburg et al., 2016). Our inability to capture phenotype-relevant haplotypes in African-Americans may explain why we observed no clear associations between GC haplotypes and 25(OH)D concentrations in this group. The positive association between 25(OH)D and “GCACCA” vs. the referent “AAGCCA” haplotype may indicate a more influential role of the first three SNPs (rs1526692, rs6837549, rs2201124) or other nearby correlated SNPs in African-American women, but we again note that our African-American-specific results are based on very small numbers. Larger studies of African-American are needed to help disentangle these complicated relationships.
The relationship between the SNPs on chromosome 11 and 25(OH)D is also complex. Our top hit for the region, rs12794714, is a synonymous substitution in exon 1 of CYP2R1. The rare SNP with the strongest association in the sub-cohort, rs117913124 (MAF = 0.02), also results in a synonymous substitution in CYP2R1 (exon 4) and in a recent whole genome sequencing analysis (Manousaki et al., 2017), this variant was strongly associated with vitamin D even after adjusting for more common, previously-identified SNPs in the same region. Earlier GWAS for serum 25(OH)D identified 5 genome-wide significant SNPs on chromosome 11 near CYP2R1 (rs10741657, rs2060793, rs11023332, rs12287212, rs1007392) (Ahn et al., 2010; Wang et al., 2010; Anderson et al., 2014; Jiang et al., 2018). Of these previous GWAS hits, we assessed rs10741657 and rs11023332 (p = 4.9 × 10−8 and p = 1.5 × 10−11, respectively, in the pooled sample). The former is close to rs12794714 (1.3 kb upstream), but is intergenic and the two are only loosely correlated (r2 = 0.40 in our sample). The latter is further away from rs12794714 (129 kb downstream), but is more strongly correlated (r2 = 0.90) and results in a silent substitution in PDE3B. None of the other previously identified hits is an obvious candidate for a causal association: rs1007392 is an intron variant of PDE3B, rs2060793 is located upstream of CYP2R1, and rs12287212 is intergenic. Though many candidate gene studies have assessed the association between 25(OH)D and rs12794714 or other highly correlated SNPs, there is no clear consensus as to the likely causal variant(s) (Ramos-Lopez et al., 2007; Zhang et al., 2012, 2013; Robien et al., 2013; Batai et al., 2014; Ordóñez-Mena et al., 2016). Previously documented ancestral heterogeneity in allele frequencies and measured associations contribute additional uncertainty (Batai et al., 2014; Elkum et al., 2014). Assessments of expression quantitative trait loci showed that the genome-wide significant SNPs in this region were only associated with expression of genes in the direct vicinity (CYP2R1, RRAS2, COPB1, CALCB, and PDE3B)2.
We did not see genome-wide significant association between 25(OH)D and any of the SNPs in DHCR7/NADSYN1 or SSTR4/FOXA2 identified in previous GWAS (Wang et al., 2010; Sapkota et al., 2016; Jiang et al., 2018), though one of the DHCR7/NADSYN1 hits, rs12785878, had a very low p-value (0.0007 in the sub-cohort and 5.9 × 10−5 in the pooled sample) with the minor allele associated with decreased 25(OH)D in both their report (Wang et al., 2010) and our sample (β = −1.1 ng/mL per allele, 95% CI: −1.5, −0.6 for our study). None of the previously reported SSTR4/FOXA2 SNPs showed evidence of an association in our data (all uncorrected p > 0.3), though we note that these SNPs were originally identified in a sample of Punjabi Sikhs (Sapkota et al., 2016). As none of the SNPs identified in a recent genome-wide meta-analysis of men and women of European descent (Jiang et al., 2018) were directly genotyped in our sample (rs17216707 in CYP24A1, rs10745742 in AMDHD1, or rs8018720 in SEC23A), we looked for signals in nearby SNPs, finding none with low p-values (all >0.05). This failure to replicate could be due to chance, lack of power in our study, which was much smaller than the meta-analysis, or to sex-specific differences in the association.
Although our sample size was somewhat limited, we were able to replicate our results in an independent sample of women who later developed breast cancer, and to examine associations between the top SNPs and 25(OH)D concentrations in the same individuals at a later time point. All 25(OH)D measures were based on LC/MS, the current gold standard because of its improved precision (Farrell et al., 2012) and its ability to capture 25(OH)D2, 25(OH)D3, and epi-25(OH)D3, where the latter is an epimer of 25(OH)D3 that is thought to have nearly identical functionality (Cashman et al., 2014).
These results may not be generalizable to women of all races, particularly our findings for the GC gene and VDBP variants. Additionally, because our sample included women who had a sister with breast cancer, our effect estimates could be inflated for SNPs that interact with one or more breast cancer-related variants in their influence on vitamin D. However, we saw little difference in our results when we assessed women who later became cases vs. the random sub-cohort and our top hits and their effect estimates are consistent with findings of previous studies.
In this sample of women enrolled in the Sister Study, SNPs in GC and CYP2R1 were strongly associated with serum 25(OH)D concentrations measured using LC/MS. Although these loci had been identified in earlier GWAS, these findings extend our understanding by pointing to possible roles for specific SNPs within these regions and further elucidating the importance of VDBP and cytochrome P450 enzymes in determining 25(OH)D concentrations. They may also help to identify individuals who are genetically predisposed to lower 25(OH)D and would most benefit from interventions to improve their circulating vitamin D levels.
Author contributions
KO conceived and designed the research project, analyzed the data, wrote the paper, and has primary responsibility for final content. DS developed the overall research plan, oversaw data collection, and conducted study oversight. She also contributed to writing the manuscript. MS performed statistical analyses and contributed to the writing of the manuscript. QH helped with the interpretation of the results and the writing of the manuscript. JT helped to develop the overall research plan and write the paper. CW oversaw the overall research plan, the study design and statistical analysis, and helped to write the paper.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Glossary
Abbreviations
- 25(OH)D
25-hydroxyvitamin D
- AIMs
ancestry informative markers
- CI
95% confidence interval
- GWAS
genome-wide association study
- LC/MS
liquid chromatography-mass spectrometry
- MAF
minor allele frequency
- SNP
single nucleotide polymorphism
- VDBP
vitamin D binding protein.
2Broad Institute The Genotype-Tissue Expression (GTEx) Project.
Funding. This work was supported by an Office of Dietary Supplement Research Scholars Program Grant (to KO) and the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences (project Z01-ES044005 to DS; Z01-ES102245 to CW; and Z01-ES049033 to JT).
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2018.00067/full#supplementary-material
References
- Abbas S., Linseisen J., Slanger T., Kropp S., Mutschelknauss E. J., Flesch-Janys D., et al. (2008). The Gc2 allele of the vitamin D binding protein is associated with a decreased postmenopausal breast cancer risk, independent of the vitamin D status. Cancer Epidemiol. Biomarkers Prev. 17, 1339–1343. 10.1158/1055-9965 [DOI] [PubMed] [Google Scholar]
- Ahn J., Yu K., Stolzenberg-Solomon R., Simon K. C., McCullough M. L., Gallicchio L., et al. (2010). Genome-wide association study of circulating vitamin D levels. Hum. Mol. Genet. 19, 2739–2745. 10.1093/hmg/ddq155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos C. I., Dennis J., Wang Z., Byun J., Schumacher F. R., Gayther S. A., et al. (2016). The OncoArray Consortium: a network for understanding the genetic architecture of common cancers. Cancer Epidemiol. Biomarkers Prev. 26, 126–135. 10.1158/1055-9965.EPI-16-0106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D., Holt B. J., Pennell C. E., Holt P. G., Hart P. H., Blackwell J. M. (2014). Genome-wide association study of vitamin D levels in children: replication in the Western Australian Pregnancy Cohort (Raine) study. Genes Immun. 15, 578–583. 10.1038/gene.2014.52 [DOI] [PubMed] [Google Scholar]
- Autier P., Boniol M., Pizot C., Mullie P. (2014). Vitamin D status and ill health: a systematic review. Lancet Diabetes Endocrinol. 2, 76–89. 10.1016/S2213-8587(13)70165-7 [DOI] [PubMed] [Google Scholar]
- Avenell A., Mak J., O' Connell D. (2014). Vitamin D and vitamin D analogues for preventing fractures in post-menopausal women and older men. Cochrane Database Syst. Rev. 14:CD000227 10.1002/14651858.CD000227.pub4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batai K., Murphy A. B., Shah E., Ruden M., Newsome J., Agate S., et al. (2014). Common vitamin D pathway gene variants reveal contrasting effects on serum vitamin D levels in African Americans and European Americans. Hum. Genet. 133, 1395–1405. 10.1007/s00439-014-1472-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin E. J., Dupuis J., Larson M. G., Lunetta K. L., Booth S. L., Govindaraju D. R., et al. (2007). Genome-wide association with select biomarker traits in the Framingham Heart Study. BMC Med. Genet. 8(Suppl. 1):S11. 10.1186/1471-2350-8-S1-S11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjelakovic G., Gluud L. L., Nikolova D., Whitfield K., Krstic G., Wetterslev J., et al. (2014). Vitamin D supplementation for prevention of cancer in adults. Cochrane Database Syst. Rev. 23:CD007469 10.1002/14651858.CD007469.pub2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjelakovic G., Gluud L., Nikolova D., Whitfield K., Wetterslev J., Simonetti R. G., et al. (2011). Vitamin D supplementation for prevention of mortality in adults. Cochrane Database Syst. Rev. 6:CD007470 10.1002/14651858.CD007470.pub2 [DOI] [PubMed] [Google Scholar]
- Braun A., Bichlmaier R., Cleve H. (1992). Molecular analysis of the gene for the human vitamin-D-binding protein (group-specific component): allelic differences of the common genetic GC types. Hum. Genet. 89, 401–406. [DOI] [PubMed] [Google Scholar]
- Burkett K., Graham J., McNeney B. (2006). Hapassoc: software for likelihood inference of trait associations with SNP haplotypes and other attributes. J. Stat. Softw. 16 10.18637/jss.v016.i02 [DOI] [PubMed] [Google Scholar]
- Burkett K., McNeney B., Graham J. (2004). A note on inference of trait associations with SNP haplotypes and other attributes in generalized linear models. Hum. Hered. 57, 200–206. 10.1159/000081447 [DOI] [PubMed] [Google Scholar]
- Cashman K. D., Kinsella M., Walton J., Flynn A., Hayes A., Lucey A. J., et al. (2014). The 3 epimer of 25-hydroxycholecalciferol is present in the circulation of the majority of adults in a nationally representative sample and has endogenous origins. J. Nutr. 144, 1050–1057. 10.3945/jn.114.192419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clendenen T. V., Ge W., Koenig K. L., Axelsson T., Liu M., Afanasyeva Y., et al. (2015). Genetic polymorphisms in vitamin D metabolism and signaling genes and risk of breast cancer: a Nested Case-Control Study. PLoS ONE 10:e0140478. 10.1371/journal.pone.0140478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denburg M. R., Hoofnagle A. N., Sayed S., Gupta J., de Boer I. H., Appel L. J., et al. (2016). Comparison of two ELISA methods and mass spectrometry for measurement of vitamin D-binding protein: implications for the assessment of bioavailable vitamin D concentrations across genotypes. J. Bone Miner. Res. 31, 1128–1136. 10.1002/jbmr.2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durazo-Arvizu R. A., Dawson-Hughes B., Kramer H., Cao G., Merkel J., Coates P. M., et al. (2017). The reverse J-shaped association between serum total 25-hydroxyvitamin D concentration and all-cause mortality: the impact of assay standardization. Am. J. Epidemiol. 185, 720–726. 10.1093/aje/kww244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkum N., Alkayal F., Noronha F., Ali M. M., Melhem M., Al-Arouj M., et al. (2014). Vitamin D insufficiency in Arabs and South Asians positively associates with polymorphisms in GC and CYP2R1 genes. PLoS ONE 9:e113102. 10.1371/journal.pone.0113102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman C. D., Fingerlin T. E., Langefeld C. D., Hicks P. J., Rich S. S., Wagenknecht L. E., et al. (2008). Genetic and environmental determinants of 25-hydroxyvitamin D and 1,25-dihydroxyvitamin D levels in hispanic and African Americans. J. Clin. Endocrinol. Metab. 93, 3381–3388. 10.1210/jc.2007-2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman C. D., Meyers K. J., Ziegler J. T., Taylor K. D., Palmer N. D., Haffner S. M., et al. (2010). Genome-wide association study of vitamin D concentrations in Hispanic Americans: the IRAS Family Study. J. Steroid Biochem. Mol. Biol. 122, 186–192. 10.1016/j.jsbmb.2010.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell C. J., Martin S., McWhinney B., Straub I., Williams P., Herrmann M. (2012). State-of-the-art vitamin D assays: a comparison of automated immunoassays with liquid chromatography-tandem mass spectrometry methods. Clin. Chem. 58, 531–542. 10.1373/clinchem.2011.172155 [DOI] [PubMed] [Google Scholar]
- Gandini S., Boniol M., Haukka J., Byrnes G., Cox B., Sneyd M. J., et al. (2011). Meta-analysis of observational studies of serum 25-hydroxyvitamin D levels and colorectal, breast and prostate cancer and colorectal adenoma. Int. J. Cancer 128, 1414–1424. 10.1002/ijc.25439 [DOI] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium. Auton A., Brooks L. D., Durbin R. M., Garrison E. P., Kang H. M., et al. (2015). A global reference for human genetic variation. Nature 526, 68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland C. F., Gorham E. D. (2016). Dose-response of serum 25-hydroxyvitamin D in association with risk of colorectal cancer: a meta-analysis. J. Steroid Biochem. Mol. Biol. 168, 1–8. 10.1016/j.jsbmb.2016.12.003 [DOI] [PubMed] [Google Scholar]
- Garland C. F., Kim J. J., Mohr S. B., Gorham E. D., Grant W. B., Giovannucci E. L., et al. (2014). Meta-analysis of all-cause mortality according to serum 25-hydroxyvitamin D. Am. J. Public Health 104, 43–50. 10.2105/AJPH.2014.302034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., O'Reilly P. F., Aschard H., Hsu Y. H., Richards J. B., Dupuis J., et al. (2018). Genome-wide association study in 79,366 European-ancestry individuals informs the genetic architecture of 25-hydroxyvitamin D levels. Nat. Commun. 9:260. 10.1038/s41467-017-02662-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. D., Handsaker R. E., Pulit S. L., Nizzari M. M., O'Donnell C. J., De Bakker P. I. W. (2008). SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 24, 2938–2939. 10.1093/bioinformatics/btn564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorde R., Schirmer H., Wilsgaard T., Joakimsen R. M., Mathiesen E. B., Njølstad I., et al. (2012). Polymorphisms related to the serum 25-hydroxyvitamin D level and risk of myocardial infarction, diabetes, cancer and mortality. The Troms{ø} Study. PLoS ONE 7:e37295. 10.1371/journal.pone.0037295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasky-Su J., Lange N., Brehm J. M., Damask A., Soto-Quiros M., Avila L., et al. (2012). Genome-wide association analysis of circulating vitamin D levels in children with asthma. Hum. Genet. 131, 1495–1505. 10.1007/s00439-012-1185-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.-H., Yin X.-Y., Wu X.-H., Zhang L., Pan S.-Y., Zheng Z.-J., et al. (2014). Serum 25(OH)D and vitamin D status in relation to VDR, GC and CYP2R1 variants in Chinese. Endocr. J. 61, 133–141. 10.1507/endocrj.EJ13-0369 [DOI] [PubMed] [Google Scholar]
- Lu L. H., Sheng H., Li H., Gan W., Liu C., Zhu J., et al. (2012). Associations between common variants in GC and DHCR7/NADSYN1 and vitamin D concentration in Chinese Hans. Hum. Genet. 131, 505–512. 10.1007/s00439-011-1099-1 [DOI] [PubMed] [Google Scholar]
- MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., et al. (2017). The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 45, D896–D901. 10.1093/nar/gkw1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manousaki D., Dudding T., Haworth S., Hsu Y., Liu C., Medina-Gómez C., et al. (2017). Low frequency synonymous coding variation in CYP2R1 has large effects on vitamin D level and risk of multiple sclerosis. Am. J. Hum. Genet. 101, 1–12. 10.1016/j.ajhg.2017.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen J., Rasmussen L. B., Ravn-Haren G., Wreford Andersen E., Hansen B., Andersen R., et al. (2014). Common variants in CYP2R1 and GC genes predict vitamin D concentrations in healthy Danish children. PLoS ONE 9:e89907. 10.1371/journal.pone.0089907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien K. M., Sandler D. P., Kinyamu H. K., Taylor J. A., Weinberg C. R. (2017a). Single nucleotide polymorphisms in vitamin D-related genes may modify vitamin D-breast cancer associations. Cancer Epidemiol. Biomarkers Prev. 26, 1761–1771. 10.1158/1055-9965.EPI-17-0250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien K. M., Sandler D. P., Taylor J. A., Weinberg C. R. (2017b). Serum vitamin D and risk of breast cancer within five years. Environ. Health Perspect. 125, 1–9. 10.1289/EHP943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordóñez-Mena J. M., Maalmi H., Schöttker B., Saum K. U., Holleczek B., Wang T. J., et al. (2016). Genetic variants in the vitamin D pathway, 25(OH)D levels, and mortality in a large population-based cohort study. J. Clin. Endocrinol. Metab. 102, 470–477. 10.1210/jc.2016-2468 [DOI] [PubMed] [Google Scholar]
- Perna L., Felix J. F., Breitling L. P., Haug U., Raum E., Burwinkel B., et al. (2013). Genetic variations in the vitamin D binding protein and season-specific levels of vitamin D among older adults. Epidemiology 24, 104–109. 10.1097/EDE.0b013e318276c4b0 [DOI] [PubMed] [Google Scholar]
- Powe C. E., Evans M. K., Wenger J., Zonderman A. B., Berg A. H., Nalls M., et al. (2013). Vitamin D–binding protein and vitamin D status of Black Americans and White Americans. N. Engl. J. Med. 369, 1991–2000. 10.1056/NEJMoa1306357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim R. J., Welch R. P., Sanna S., Teslovich T. M., Chines P. S., Gliedt T. P., et al. (2011). LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 27, 2336–2337. 10.1093/bioinformatics/btq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Lopez E., Brück P., Jansen T., Herwig J., Badenhoop K., et al. (2007). CYP2R1 (vitamin D 25-hydroxylase) gene is associated with susceptibility to type 1 diabetes and vitamin D levels in Germans. Diabetes Metab. Res. Rev. 23, 631–636. 10.1002/dmrr.719 [DOI] [PubMed] [Google Scholar]
- Robien K., Butler L. M., Wang R., Beckman K. B., Walek D., Koh W. P., et al. (2013). Genetic and environmental predictors of serum 25-hydroxyvitamin D concentrations among middle-aged and elderly Chinese in Singapore. Br. J. Nutr. 109, 493–502. 10.1017/S0007114512001675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross A. C., Taylor C. L., Yaktine A. L., Del Valle H. B., Del Valle H. B. (2011). Dietary Reference Intakes: Calcium and Vitamin D. Washington, DC: The National Academies Press. [PubMed] [Google Scholar]
- Sandler D. P., Hodgson M. E., Deming-Halverson S. L., Juras P. J., D'Aloisio A. A., Suarez L., et al. (2017). The sister study: baseline methods and participant characteristics. Environ. Health Perspect. 125:127003. 10.1289/EHP1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapkota B. R., Hopkins R., Bjonnes A., Ralhan S., Wander G. S., Mehra N. K., et al. (2016). Genome-wide association study of 25(OH) vitamin D concentrations in Punjabi Sikhs: results of the Asian Indian diabetic heart study. J. Steroid Biochem. Mol. Biol. 158, 149–156. 10.1016/j.jsbmb.2015.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkyo R., Sakaki T., Kamakura M., Ohta M., Inouye K. (2004). Metabolism of vitamin D by human microsomal CYP2R1. Biochem. Biophys. Res. Commun. 324, 451–457. 10.1016/j.bbrc.2004.09.073 [DOI] [PubMed] [Google Scholar]
- Sollid S. T., Hutchinson M. Y., Berg V., Fuskevåg O. M., Figenschau Y., Thorsby P. M., et al. (2016). Effects of vitamin D binding protein phenotypes and vitamin D supplementation on serum total 25(OH)D and directly measured free 25(OH)D. Eur. J. Endocrinol. 174, 445–452. 10.1530/EJE-15-1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speeckaert M., Huang G., Delanghe J. R., Taes Y. E. (2006). Biological and clinical aspects of the vitamin D binding protein (Gc-globulin) and its polymorphism. Clin. Chim. Acta 372, 33–42. 10.1016/j.cca.2006.03.011 [DOI] [PubMed] [Google Scholar]
- Stram D. O., Seshan V.E. (2012). Multi-SNP haplotype analysis methods for association analysis, in Statistical Human Genetics. Methods in Molecular Biology (Methods and Protocols), eds Elston R. C., Satagopan J., Sun S. (New York, NY: Humana Press; ), 443–452. [DOI] [PubMed] [Google Scholar]
- Wang T. J., Zhang F., Richards J. B., Kestenbaum B., van Meurs J. B., Berry D., et al. (2010). Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet 376, 180–188. 10.1016/S0140-6736(10)60588-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S., Hong C. C., Bandera E. V., Zhu Q., Liu S., Cheng T. D., et al. (2017). Demographic, lifestyle, and genetic determinants of circulating concentrations of 25-hydroxyvitamin D and vitamin D–binding protein in African American and European American women. Am. J. Clin. Nutr. 105, 1362–1371. 10.3945/ajcn.116.143248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Li B., Gao X., Tian R., Pan Y., Jiang Y., et al. (2017). Serum 25-hydroxyvitamin D and the risk of cardiovascular disease: dose-response meta-analysis of prospective studies. Am. J. Clin. Nutr. 105, 810–819. 10.3945/ajcn.116.140392 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wang X., Liu Y., Qu H., Qu S., Wang W., et al. (2012). The GC, CYP2R1 and DHCR7 genes are associated with vitamin D levels in northeastern Han Chinese children. Swiss Med. Wkly. 142:w13636. 10.4414/smw.2012.13636 [DOI] [PubMed] [Google Scholar]
- Zhang Z., He J. W., Fu W. Z., Zhang C. Q., Zhang Z. L. (2013). An analysis of the association between the vitamin D pathway and serum 25-hydroxyvitamin D levels in a healthy Chinese population. J. Bone Miner. Res. 28, 1784–1792. 10.1002/jbmr.1926 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.