Abstract
Gain-of-function somatic mutations in the ubiquitin specific protease 8 (USP8) gene have recently been reported as a cause of pituitary adenomas in Cushing disease. Molecular diagnostic testing of tumor tissue may aid in the diagnosis of specimens obtained through therapeutic transsphenoidal surgery; however, for small tumors, availability of fresh tissue is limited, and contamination with normal tissue is frequent. We performed molecular testing of DNA isolated from single formalin-fixed and paraffin-embedded (FFPE) tissue sections of 42 pituitary adenomas from patients with Cushing disease (27 female patients and 15 male patients; mean age at surgery, 42.5 years; mean tumor size, 12.2 mm). By Sanger sequencing, we identified previously reported USP8 missense mutations in six tumors. Targeted next-generation sequencing (NGS) revealed known or previously undescribed missense mutations in three additional tumors (two with two different mutations each), with mutant allele frequencies as low as 3%. Of the nine tumors with USP8 mutations (mutation frequency, 21.4%), seven were from female patients (mutation frequency, 25.9%), and two were from male patients (mutation frequency, 13.3%). Mutant tumors were on average 11.4 mm in size, and patients with mutations were on average 43.9 years of age. The overall USP8 mutation frequency in our cohort was lower than in previously described cohorts, and we did not observe USP8 deletions that were frequent in other cohorts. We demonstrate that testing for USP8 variants can be performed from small amounts of FFPE tissue. NGS showed higher sensitivity for USP8 mutation detection than did Sanger sequencing. Assessment for USP8 mutations may complement histopathological diagnosis.
Keywords: FFPE, molecular diagnostic testing, ubiquitin specific protease
Sanger and targeted NGS of FFPE tissue from 42 corticotroph adenomas identified USP8 missense mutations in nine tumors; such tests may complement histopathological diagnosis.
Corticotropin (ACTH) is generated by cleavage of proopiomelanocortin in the anterior part of the pituitary gland in response to the release of hypothalamic corticotropin-releasing hormone. ACTH then increases cortisol secretion from the adrenal zona fasciculata. Excess production of ACTH by pituitary corticotroph adenomas causes Cushing disease, the most common form of endogenous Cushing syndrome [1, 2], characterized by the signs and symptoms of hypercortisolism, such as moon face, central obesity, diabetes, hypertension, and fragile skin. Over the past few years, molecular genetic studies have provided novel insight into the pathophysiology of these tumors. Whereas germline variants in tumor syndromes (including McCune-Albright syndrome, multiple endocrine neoplasia, Carney complex, and tuberous sclerosis) and mutations in classic proto-oncogenes and tumor suppressor genes are found in a very small fraction of pituitary adenomas [3, 4], somatic mutations in the USP8 gene, encoding the ubiquitin specific protease 8, were recently found to account for a substantial fraction of tumors.
Two groups performed exome and Sanger sequencing of corticotroph adenomas and independently reported somatic mutations in the USP8 gene as a cause of 35% (6 of 17) and 63% (75 of 120), respectively, of pituitary corticotroph adenomas [5, 6]. These findings were confirmed by targeted sequencing in several cohorts, with 35% (21 of 60) [7], 36% (48 of 134) [8], 17% (5 of 30) [9], and 31% (13 of 42) [10] USP8 mutation frequencies. Mutations were more common in female (43% to 68% mutation frequency) than in male (0% to 36% mutation frequency) patients [5–8], which is unexplained. Initially, age at peak incidence for female patients was suggested to range from 30 to 50 years [11], but a more recent study has shown similar prevalence among pediatric cases [10]. Apart from this association with sex, clinical and biochemical correlates (such as tumor size and hormonal parameters) of patients with somatic USP8 mutations have been conflicting [10].
The deubiquitinase activity of USP8 prevents the degradation of ubiquitinylated epidermal growth factor receptor (EGFR) in the lysosome [12]. Because EGFR signaling increases POMC transcription and secretion of ACTH [5], increased USP8 activity causes elevated ACTH production. Importantly, USP8 activity is regulated by the binding of 14-3-3 proteins [13], which may prevent proteolytic cleavage and activation. Recurrent gain-of-function mutations in USP8 cluster at the 14-3-3 binding site and impair the binding of 14-3-3 proteins. The resulting increased proteolytic cleavage of USP8 produces USP8 fragments with higher deubiquitinase activity [5]. This is thought to increase EGFR stability and EGFR-mediated ACTH secretion.
Prior studies have typically used fresh-frozen adenoma tissue for DNA extraction and sequencing [6–9]. However, fresh-frozen tissue is often not available, in particular for small tumors, for which the entire specimen is formalin-fixed and paraffin-embedded (FFPE) for diagnostic purposes [7]. Thus, such cohorts may be biased toward larger specimens. Here we investigate a cohort of 42 corticotroph adenomas from two centers in Germany and perform molecular genetic analysis from single sections of FFPE tissue.
1. Material and Methods
A. Patients and Clinical Data
We included 42 specimens from two cohorts of patients. Thirty-eight patients had undergone surgery for Cushing disease at the Johannes Wesling Hospital, Minden, Germany, between 2008 and 2014 (Table 1; these patients are labeled as HYP001 to HYP047, and missing numbers reflect duplicate samples or those for which no tumor tissue was identified on the slide), and four patients (labeled HYP050 to HYP055) were operated at the University Hospital Düsseldorf, Germany, between 2009 and 2014 (Table 1). Düsseldorf cases were selected in pathology on the basis of ACTH positivity, and clinical presentation was reviewed. Tumors with insufficient tumor tissue were excluded after assessment by an experienced neuropathologist.
Table 1.
Clinical Data and Experimental Results in the Investigated Corticotroph Adenoma Patient Cohort
| HYP No. | Sex | Age (y) | Size (mm) | Repli-G | Sanger Result | NGS Result | EGFR IHC |
|---|---|---|---|---|---|---|---|
| Cushing disease | |||||||
| 002 | F | 15 | 5 | Yes | WT | p.P720R | − |
| p.S716Y | |||||||
| 005 | F | 14 | 5 | Yes | WT | p.S716F | − |
| p.S718P | |||||||
| 006 | F | 36 | 13 | Yes | p.S718P | NA | − |
| 019 | M | 50 | 3 | Yes | WT | p.P720R | − |
| 024a | F | 51 | 13 | Yes | p.P720Q | NA | − |
| 026 | F | 46 | 6 | Yes | p.P720R | NA | − |
| 029 | F | 36 | 11 | Yes | p.P720R | p.P720R | − |
| 039 | F | 66 | 18 | Yes | p.P720Q | NA | − |
| 051 | M | 81 | 29 | No | p.P720R | NA | − |
| 001 | M | 34 | 8 | Yes | WT | WT | − |
| 003b | M | 21 | 50 | Yes | WT | WT | (+) |
| 004 | F | 48 | Below detection | Yes | WT | WT | − |
| 007 | F | 25 | 4 | Yes | WT | WT | − |
| 009 | F | 61 | 20 | Yes | WT | WT | − |
| 011 | F | 42 | 6 | Yes | WT | WT | − |
| 013 | F | 45 | Below detection | Yes | WT | WT | − |
| 015 | F | 56 | 3 (ultrasonography) | Yes | WT | WT | − |
| 016 | F | 30 | 14 | Yes | WT | WT | (+) |
| 017b | F | 43 | NA | Yes | WT | WT | − |
| 018 | F | 65 | 3 | Yes | WT | WT | − |
| 020 | F | 52 | 8 | Yes | WT | WT | − |
| 021 | F | 24 | 6 | Yes | WT | WT | − |
| 022 | F | 52 | 3 (surgery) | Yes | WT | WT | − |
| 023 | M | 57 | 28 | Yes | WT | WT | − |
| 025 | F | 42 | 26 | Yes | WT | WT | − |
| 027 | M | 52 | 30 | Yes | WT | WT | − |
| 028c | F | 36 | 9 | Yes | WT | WT | − |
| 030b | M | 36 | NA | Yes | WT | WT | − |
| 032 | F | 54 | 8 | Yes | WT | WT | − |
| 033 | M | 49 | 3 | Yes | WT | WT | − |
| 036d | F | 35 | 5 | Yes | WT | WT | − |
| 037 | M | 20 | 3 | Yes | WT | WT | − |
| 038 | M | 35 | 4 | Yes | WT | WT | − |
| 041 | M | 40 | 4 | Yes | WT | WT | − |
| 043 | F | 29 | 3 | Yes | WT | WT | − |
| 044 | M | 38 | 9 (ultrasonography) | Yes | WT | WT | − |
| 045 | F | 47 | Below detection | Yes | WT | WT | − |
| 046 | F | 47 | 7 | Yes | WT | WT | − |
| 047a | M | 44 | 24 | Yes | WT | WT | − |
| 050 | F | 35 | 16 | No | WT | WT | − |
| 053 | M | 32 | 8 | No | WT | WT | − |
| 055a | M | 64 | 36 | No | WT | WT | − |
| Silent corticotroph adenomas | No | ||||||
| 056 | M | 59 | 13 | No | WT | WT | − |
| 063 | M | 57 | 15 | No | WT | WT | − |
Abbreviations: F, female; IHC, immunohistochemistry; M, male; Repli-G, whole-genome amplification of DNA; NA, not available; −, no positivity by immunohistochemistry; (+), possible very weak positivity by immunohistochemistry.
Atypical adenoma.
Studied tissue from recurrence, age and tumor size at first surgery are given.
Crooke cell adenoma.
Ectopic, sinus cavernosus (patient 2 in Knappe et al. [25]).
Cushing disease was diagnosed according to clinical signs and/or typical laboratory parameters [8]. Tumor size was determined on cross-sectional imaging. For tumors that were undetectable by cross-sectional imaging, size was determined by intraoperative ultrasonography or assessment during surgery (Table 1). The largest diameter was recorded. Informed consent was obtained from all patients. Transsphenoidal surgery was performed in most patients. Pathologic specimens were independently evaluated by at least two experienced neuropathologists, and pituitary ACTH cell adenoma was diagnosed in each instance. Two silent corticotroph adenomas from Düssseldorf (HYP056 and HYP063) were diagnosed on the basis of positive immunoreactivity for ACTH without clinical or biochemical evidence of hypercortisolism. Molecular and immunohistochemical analyses of FFPE tissue specimens were performed with approval of the institutional review board of the Medical Faculty, Heinrich Heine University Düsseldorf (study number 4938).
B. DNA Extraction, Polymerase Chain Reaction Amplification, and Sanger Sequencing
One fresh 10-µm-thick section of FFPE tumor tissue per sample mounted on a glass slide was scraped off and used for DNA isolation. In most cases, whole genome amplification was performed with the REPLI-g FFPE kit (Qiagen) from FFPE tissue directly, without prior DNA purification, according to the manufacturer's instructions. For large specimens, the QIAamp DNA FFPE Tissue kit (Qiagen) was used for DNA extraction.
Routine polymerase chain reaction (PCR) amplification of USP8 exon 14 was performed by using primers USP8_F and _R (Table 2) and the KAPA2G Fast Hot Start PCR kit (KapaBiosystems) following a step-down protocol. PCR products were verified by gel electrophoresis. For select cases, products were excised, and DNA was extracted by using the QIAquick Gel Extraction kit (Qiagen). PCR purification and direct bidirectional Sanger sequencing were performed at the Genewiz Sequencing Service facility (Takeley, United Kingdom). Results were analyzed with Sequencher version 4.7 software (Genecodes).
Table 2.
Primer Sequences
| Primer | Sequence 5′ > 3′ | Product Size (bp) |
|---|---|---|
| USP8_F | AGCCACAGATTCCTGCTGAG | 124 |
| USP8_R | ACTGTTGGAGTTACTGTTGGCT | |
| USP8big_Fa | AAAGCCAAGCCACAGATTCC | 234 |
| USP8big_Ra | TCCCTGACACTAACATACTGACA | |
| USP8bigadap_Fa | TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGAAAGCCAAGCCACAGATTCC | 301 |
| USP8bigadap_Ra | GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGTCCCTGACACTAACATACTGACA |
Used for NGS.
C. Targeted Next-Generation Sequencing and Data Analysis
Samples were prepared for ultradeep sequencing of USP8 exon 14 on the MiSeq sequencer (Illumina) according to the manufacturer's protocols. For barcoding of samples, the Nextera Index kit (Illumina) was used. The MiSeq run was performed with the MiSeq Reagent Nano kit version 2 for 300 cycles (Illumina), which comprises 150-bp paired-end reads. Fastq files were loaded as pairs in the CLC Genomic Workbench version 9.5.1 (Qiagen) and analyzed by merging the overlapping pairs, mapping them to the USP8 reference sequence, and applying the low-frequency variant detection tool. For a second evaluation, the data were reanalyzed according to the PCR Amplicon workflow guide of the Illumina MiSeq Reporter Software. A list of variants was generated with VariantCaller and sample sheet settings for detecting somatic variants. Called single nucleotide polymorphisms were assessed with the Integrative Genomics Viewer version 2.3.88 (Broad Institute). Protein-changing variants with minor allele frequency >2% within the previously described hotspot area (amino acids 713 to 735 in NP_005145.3) [11] were extracted.
D. Immunohistochemistry
Routine EGFR immunostaining was performed at the Department of Pathology and Neuropathology, University Hospital Essen [Zytomed clone 2.1E1, mouse monoclonal, 1:1000, Research Resource Identifier (RRID): AB_2721106] for samples from Minden or at the Department of Neuropathology, Heinrich Heine University Düsseldorf (pronase antigen retrieval, #M7239, DAKO, 1:300, RRID: AB_2721108) for samples from Düsseldorf. Antibody reactivity was confirmed by the use of a positive control. Images were recorded on a CTR500 microscope with a DFC 380 camera (both Leica).
E. Statistical Analysis
Statistical analyses were performed by using the Prism 7 program (GraphPad). Data are shown as mean ± standard error of the mean, unless otherwise indicated. Normality was assessed by the D’Agostino and Pearson normality test. Comparisons between two groups were performed by using the indicated statistical tests, and P values < 0.05 were considered to indicate significant differences.
2. Results
A. Characterization of the Patient Cohort
Among the 42 patients with Cushing disease, 15 were male and 27 were female. Tumors were typically excised by transnasal transsphenoidal surgery; patient 3 underwent right fronto-orbital craniotomy. Pathologic evaluation revealed pituitary adenomas in all cases. At the time of surgery, patients were age 42.5 ± 14.4 [mean ± standard deviation (SD); range, 14 to 81; median, 42.5] years. With the exception of two patients, all patients were adults (age >18 years). The tumor size was 12.2 ± 11.2 (mean ± SD; range, 3 to 50; median, 8) mm (tumors below detection are not included in analysis). Notably, 16 tumors were very small (≤5 mm, including the three that were undetectable by imaging); 26 tumors were microadenomas (<10 mm), and 14 were macroadenomas (≥10 mm). Data on tumor size at initial presentation were unavailable for two patients operated for recurrence. Clinical data are summarized in Table 1.
B. Sanger Sequencing Reveals USP8 Variants in 6 of 42 Cases
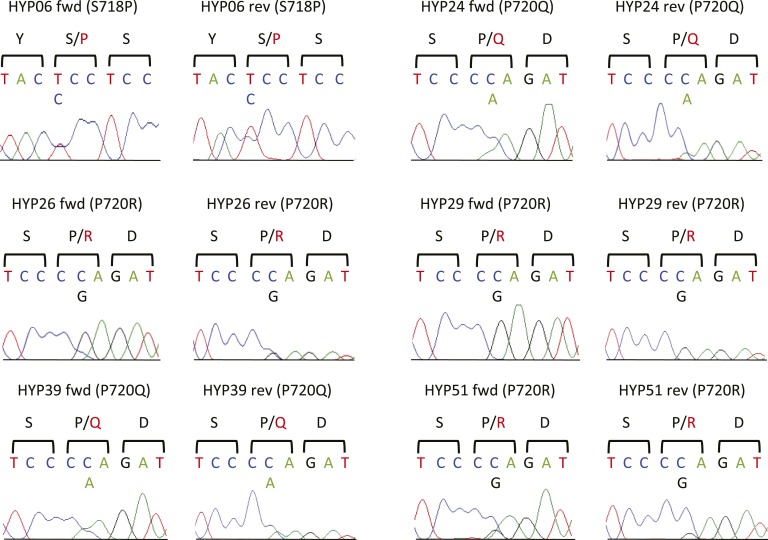
To determine the frequency of mutations in the USP8 gene, we initially extracted DNA from FFPE tissue and performed targeted PCR and direct Sanger sequencing of exon 14 of USP8, containing the previously described 14-3-3 binding site affected by gain-of-function variants [5]. We identified previously described variants in tumors of six patients (14.3% mutation frequency; three with p.P720R, one with p.S718P, two with p.P720Q variants) (Fig. 1; Table 3). All occurred in tumors >5 mm. In HYP029, only variant p.P720R and no wild-type (WT) sequence was detected, whereas all other variants appeared to be heterozygous.
Figure 1.
USP8 variants in corticotroph adenomas identified by Sanger sequencing. Sanger sequences of pituitary adenomas demonstrate USP8 variants in six tumors. Encoded protein sequences are shown in one-letter code, with mutant amino acids in red. The variant in HYP29 appears to be homozygous or hemizygous. Reverse sequence is shown as reverse complement. Fwd, forward; rev, reverse.
Table 3.
Identified USP8 Variants
| rs No. or COSMIC No. | Chromosomal Position | Nucleotide Change | Amino Acid Change |
|---|---|---|---|
| rs672601311 | 15:50490450 | C>G | P720R |
| rs672601307 | 15:50490443 | C>T | S718P |
| Identified in this study | 15:50490438 | C>A | S716Y |
| COSM416905 | 15:50490450 | C>A | P720Q |
| rs753615462 | 15:50490438 | C>T | S716F |
Chromosomal position refers to GRCh38.p7. Amino acid change refers to NP_005145.3.
Abbreviations: COSMIC number, catalog of somatic mutations in cancer database identifier, provided for variants without unique Single Nucleotide Polymorphism database identifier; Rs number, reference single-nucleotide polymorphism in Single Nucleotide Polymorphism database build 150.
C. Next-Generation Sequencing of USP8 Reveals Three Additional Cases With USP8 Variants
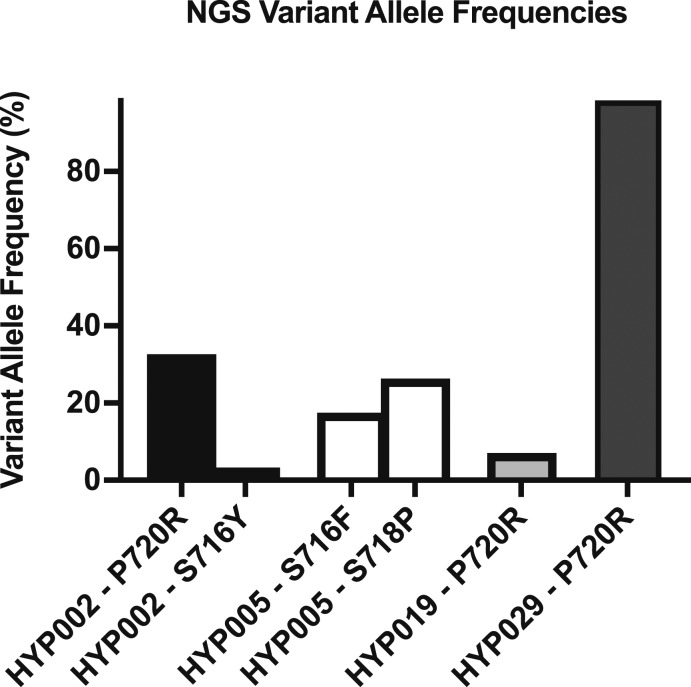
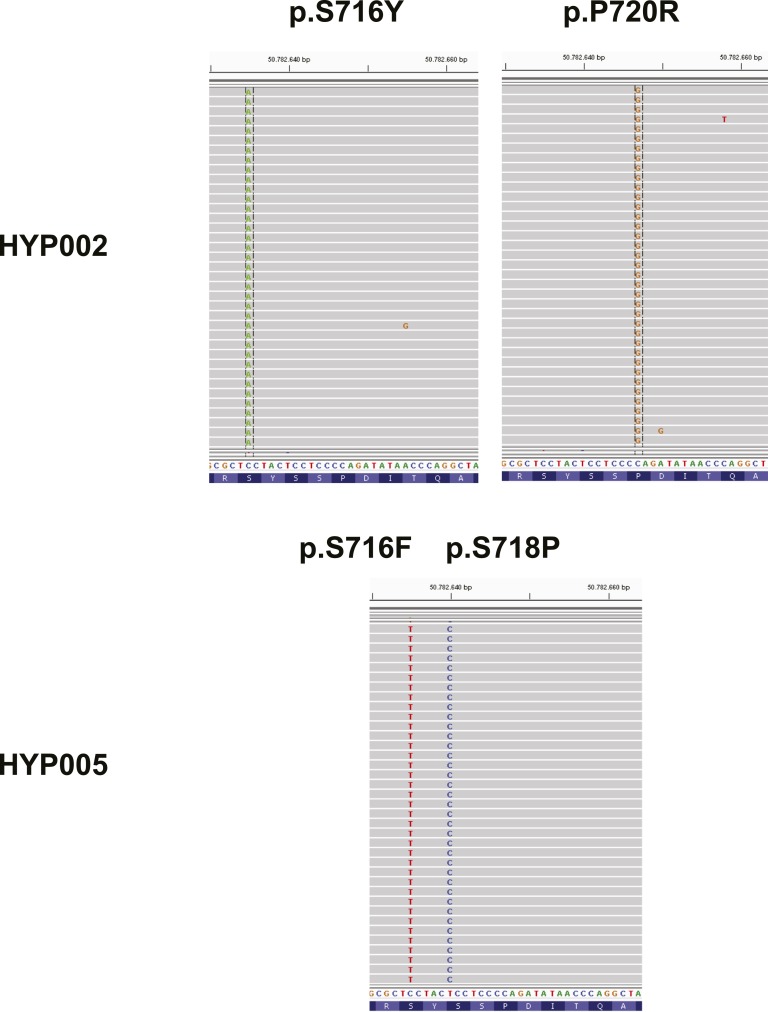
Because the prevalence of USP8 mutations detected by Sanger sequencing in our cohort was lower than in previous reports [5–8, 10], we performed targeted next-generation sequencing (NGS) of USP8 exon 14 on the Illumina MiSeq sequencer in samples without mutations detected by Sanger sequencing and sample HYP029 (positive control) (Fig. 2). In line with results of Sanger sequencing, the p.P720R variant in HYP029 showed an allele frequency of 98.5%, suggesting loss of heterozygosity or biallelic mutation. Further analysis revealed three additional samples with previously described USP8 variants. HYP019 with the p.P720R variant showed a low variant allele frequency of 7.09%, which is below the Sanger sequencing detection threshold. Two other tumors carried two different USP8 variants each. HYP005 carried the p.S716F variant previously described in skin cutaneous melanoma (ICGC MU4520263) (17.55% mutant allele frequency) and the p.S718P variant (26.33% mutant allele frequency) on the same allele. HYP002 carried the known p.P720R variant (32.67%) and the p.S716Y variant (3.30%, not found in public databases) on distinct alleles [HYP002 sequencing data have been submitted to the NCBI Sequence Read Archive under accession number SRR6448768 (https://www.ncbi.nlm.nih.gov/sra/?term=SRP128409)] (Fig. 3; Table 3). The three tumors in which variants were detected only by NGS were all ≤5 mm in size. No variants were detected in the two silent corticotroph adenoma controls, as previously reported [8].
Figure 2.
USP8 variants in corticotroph adenomas identified by NGS. Shown are variant allele frequencies of four tumors with USP8 variants detected by NGS. Frequencies were determined in the CLC Workbench program. A 98.5% mutant allele frequency in HYP029 confirms homozygosity or hemizygosity of the variant. HYP002 and HYP005 show two USP8 variants each.
Figure 3.
NGS reads in tumors with two concurrent USP8 variants. Analysis of mutant reads using Integrative Genomics Viewer demonstrates two variants on independent alleles or independent tumor subpopulations for HYP002 and two variants on the same allele for HYP005.
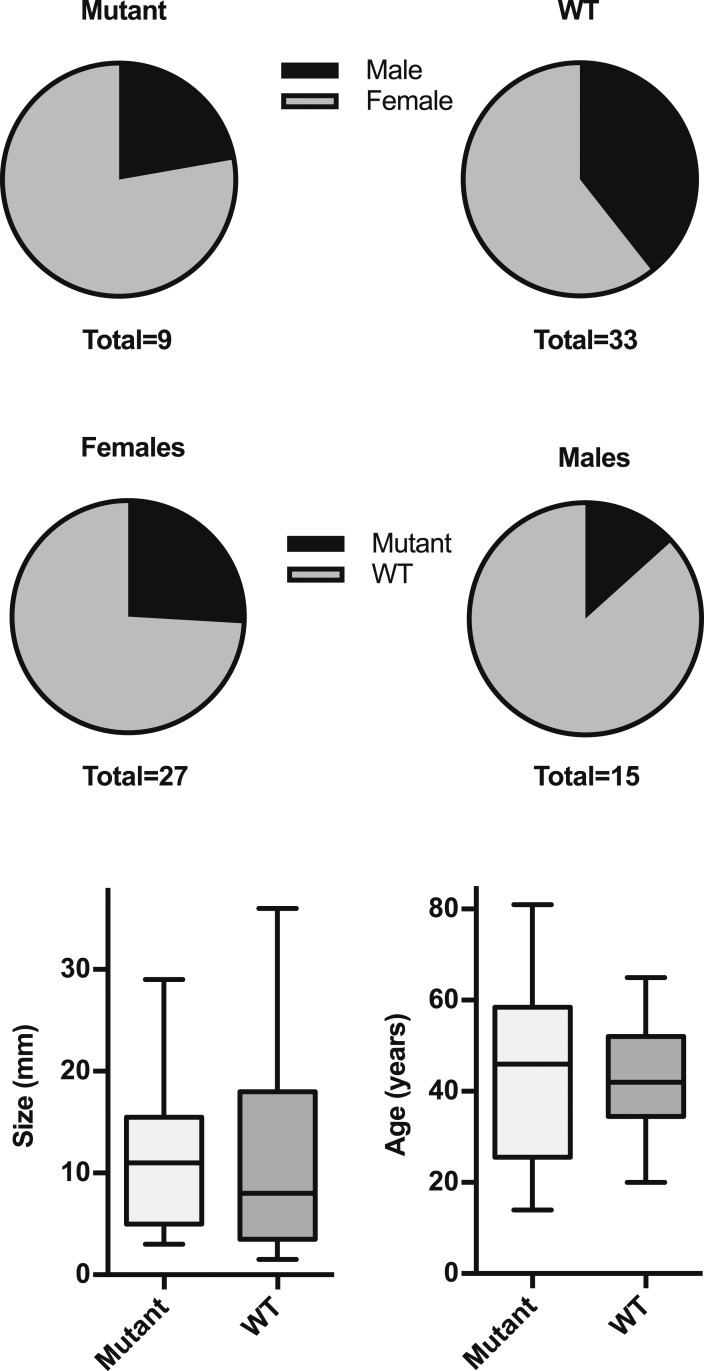
NGS analysis increased the number of tumors with somatic USP8 variants to nine and the total mutation frequency to 21.4%. Although female patients were overrepresented in the group with USP8 mutations (seven of nine mutant tumors were from female patients), there was no significant difference compared with the group without mutations (20 of 33 nonmutant tumors from female patients; P = 0.45, two-tailed Fisher’s exact test). The mutation frequency in female patients was 25.9%, and that in male patients was 13.3% (Fig. 4). There was no significant difference between mutant and WT tumors in terms of size [mutant tumors: 11.4 ± 8.2 (mean ± SD) mm; WT tumors: 12.1 ± 12.0 (mean ± SD) mm; P = 0.63 (two-tailed Mann-Whitney test)]. Similarly, patients with tumors bearing USP8 mutations did not significantly differ in age from those without such variants [mutant tumors: 43.9 ± 21.8 (mean ± SD) years; WT tumors: 42.1 ± 12.0 (mean ± SD) mm; P = 0.82 (unpaired two-tailed t test with Welch correction)]. One of the two pediatric patients (HYP005) carried two USP8 variants (Figs. 2 and 3).
Figure 4.
Clinical characteristics of individuals with USP8-mutant or WT tumors. Upper panels show sex distribution among mutant and WT tumors. Lower panel shows size and age in mutant and WT tumors (line, median; box, interquartile range; whiskers, 1.5x interquartile range). No significant differences were observed for either characteristic shown (see text for statistical analysis).
D. EGFR Expression Levels are Not Upregulated in USP8 Mutant Tumors
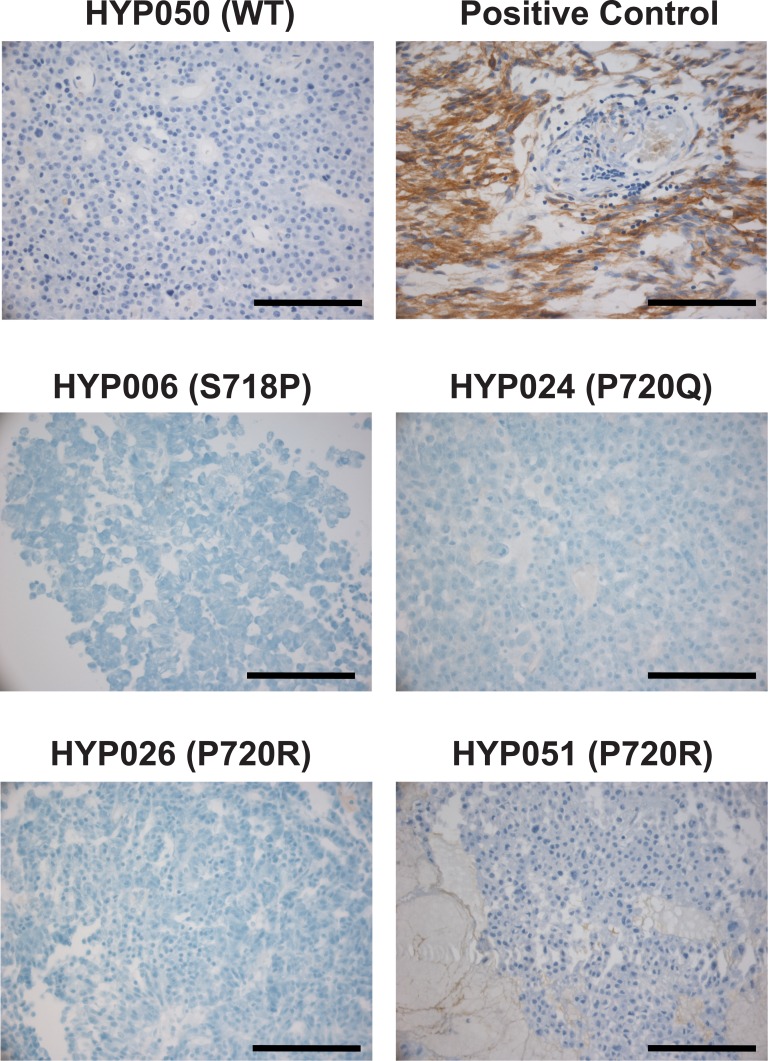
To assess EGFR expression levels in tumors, we performed immunohistochemistry with antibodies against EGFR. Interestingly, none of the mutation-positive corticotroph adenomas were EGFR-positive (Fig. 5); very weak EGFR positivity of connective tissue was occasionally observed.
Figure 5.
EGFR immunohistochemistry. HYP050 is shown as a representative EGFR-negative WT tumor. The positive control represents a glioblastoma. All mutant tumors stained negative for EGFR; representative pictures are shown. Some tumors showed very weak EGFR positivity of connective tissue, as demonstrated for HYP051. Scale bars, 100 µm.
3. Discussion
Our analysis of a cohort of 42 ACTH-producing pituitary adenomas in Cushing disease demonstrates the utility of a targeted NGS approach in detecting USP8 variants, specifically in small tumors with a size of ≤5 mm. When such small tumors are resected, in particular, fragmented tumors from transsphenoidal surgery, the distinction between normal pituitary tissue and adenoma tissue can be histologically challenging on hematoxylin-eosin–stained sections; several histochemical and immunohistochemical stains are typically performed [14]. Obtaining fresh tissue for DNA extraction and Sanger sequencing is not advised when small tumors are excised. Even with use of FFPE tissue for DNA extraction, separating tumor from normal tissue for DNA extraction can be challenging. Under these circumstances, targeted NGS can reveal low-frequency variants due to contamination with normal tissue or due to tumor heterogeneity. We show that this analysis can be performed by using DNA extracted from a single 10-µm-thick section of FFPE tissue, enabling a molecular diagnosis after routine histology. Such an analysis may complement clinical assessment, histology, and immunohistochemistry in the diagnosis of pituitary adenomas causing Cushing disease.
The USP8 mutation frequency in our cohort was lower than in most published cohorts. Only one smaller study (mostly on macroadenomas) reported a lower frequency [9], whereas most studies found USP8 mutations in about 35% of ACTH cell adenomas [5, 7, 8, 10], and a single study from Asia reported mutations in 62% of these tumors [6]. The higher frequency in the Asian cohort may be due to differences in patient ascertainment or to ethnic differences—mutations in the KCNJ5 potassium channel gene in adrenal aldosterone-producing adenomas are also more common in European women and more prevalent in most Asian cohorts [15, 16].
Many factors may explain the lower prevalence of USP8 mutations in our cohort. First, female patients were only about twofold overrepresented in our cohort (27 female and 15 male patients), whereas female patients were about three-to fivefold enriched compared with male patients in several published cohorts [6–8, 17]. The higher prevalence of mutations in female patients in these studies would therefore have contributed to a higher overall mutation prevalence. Whereas the mutation frequency in male patients was in a similar range as previously reported (although numbers were small) [8, 10], the mutation frequency in female patients was lower than previously reported (25% vs approximately 40%). The inclusion of a high number of very small tumors (38% of tumors were ≤5 mm in size) may have contributed to this divergence as well. We initially suspected that contamination with normal tissue may have led to a lower mutation detection rate in our cohort; however, mutation frequencies remained lower than in other cohorts even after NGS, which detected variants with allele frequencies as low as 3%. Surprisingly, we observed only missense mutations in our cohort, whereas one common deletion (p.S718del) accounted for >20% of mutations in other cohorts, with other rare deletions reported [6–8]. Manual assessment of Illumina reads in our cohort confirmed the absence of deletions. Of note, the p.S718del variant showed the highest deubiquitinase activity of all assessed variants in vitro [5]. Whether clinical correlations led to the exclusion of tumors with this variant from our cohort is unknown. Last, mutations that are present in only a fraction of the tumor can be missed when only limited tumor material is used for DNA extraction.
We note that several pituitary adenomas, both in previously published cohorts [5, 6] and in our cohort, carried two USP8 variants in a single tumor. The finding of an apparently homozygous or hemizygous USP8 variant in HYP029 supports this notion and may be due to loss of the WT allele or mitotic recombination, although we cannot exclude an FFPE artifact. Of note, all previously described variants were heterozygous [5–8]. The finding of a second USP8 variant with lower allele frequency in HYP002 demonstrates that tumors with USP8 variants are not necessarily clonal; independent hits may contribute to tumor heterogeneity [18, 19].
Variants with low allele frequency in particular could represent FFPE or whole-genome amplification artifacts, although the low-frequency variants in HYP002 and HYP019 did not match the C>T pattern caused by cytosine deamination in FFPE tissue [20]. We did not perform functional studies to assess whether the previously unidentified variant (p.S716Y) causes increased USP8 activity. Further studies would be required to assess the pathogenicity of this variant.
The percentage of EGFR-positive tumors was much lower in our cohort than in a published cohort, in which USP8 mutations were associated with higher EGFR expression [6]. Interestingly, this association did not replicate in another cohort [7]. Whereas USP8 variants increase EGFR signaling in cultured cells, such an effect has not been consistently shown in vivo, suggesting that the effect is small or temporary or that other factors [21] are involved in increased ACTH production and/or proliferation of USP8 mutation-positive tumors. The use of antibodies with different specificities or different staining protocols may in part account for inconsistent findings [17]; the antibodies used in our study are also used in clinical routine analysis, and a positive control was performed.
A limitation of our study is the incomplete retrospective availability of endocrine workup for some of the cases. This prevents a more detailed analysis of hormonal characteristics within our cohort. Nonetheless, we can demonstrate the utility of NGS in the diagnosis of small corticotroph adenomas. Importantly, many Cushing adenomas in large surgical series are very small, sometimes below the threshold for magnetic resonance imaging detection [22]. Molecular diagnosis by NGS could be particularly suitable for such cases. This could be beneficial for patients with recurrence [23] if USP8 mutations can be associated with response to specific therapies, such as those blocking EGFR or activating the somatostatin receptor [7, 24]. The presence of such associations is supported by the finding that USP8 knockdown or EGFR inhibition attenuates ACTH secretion in primary USP8-mutated tumor cells [7]. In conclusion, we suggest that NGS has diagnostic potential for pituitary Cushing adenomas.
Acknowledgments
We thank our patients for their invaluable contribution to this project and the Department of Pathology and Neuropathology, University Hospital Essen (Dr. K.W. Schmid, F.A. Saballs, I. Albertz, and G. Ladwig) for support. We also acknowledge members of the Düsseldorf and Minden Departments of Neurosurgery involved in patient care and Holger Willenberg (Rostock, Germany) for helpful discussions.
Financial Support: This work was supported by the Ministerium für Innovation, Wissenschaft und Forschung / Ministerium für Kultur und Wissenschaft des Landes Nordrhein-Westfalen, Germany (Junges Kolleg and Rückkehrprogramm, to U.I.S.). C.B.K.-T. is supported by the Max Eder Program of the German Cancer Aid (Deutsche Krebshilfe). Funding for the German Pituitary Tumor Registry from Novartis Pharma GmbH (Nuremberg, Germany), Novo Nordisk Pharma GmbH (Mainz, Germany), Pfizer Pharma GmbH (Berlin, Germany), and Ipsen Pharma GmbH (Berlin, Germany) (all W.S.) is gratefully acknowledged.
Author Contributions: U.I.S. designed the study; U.J.K. performed surgery, evaluated patients, and ascertained clinical information and selected Minden cases for analysis; C.B.K.-T. selected Düsseldorf cases for analysis; C.B.K.-T., A.C.R. G.R., and W.S. evaluated histological specimens, provided tissue specimens, and performed and analyzed immunohistochemistry; C.B., A.T., and H.E.K. extracted DNA and performed Sanger sequencing; C.B., A.T., and U.I.S. analyzed the data; A.T., S.S., and K. K. performed and analyzed NGS; U.I.S. wrote the manuscript, with all authors contributing to a critical revision.
Disclosure Summary: The authors have nothing to disclose.
Glossary
Abbreviations:
- ACTH
corticotropin
- EGFR
epidermal growth factor receptor
- FFPE
formalin-fixed and paraffin-embedded
- NGS
next-generation sequencing
- PCR
polymerase chain reaction
- RRID
Research Resource Identifier
- SD
standard deviation
- WT
wild-type
References and Notes
- 1. Orth DN. Cushing’s syndrome. N Engl J Med. 1995;332(12):791–803. [DOI] [PubMed] [Google Scholar]
- 2. Lacroix A, Feelders RA, Stratakis CA, Nieman LK. Cushing’s syndrome. Lancet. 2015;386(9996):913–927. [DOI] [PubMed] [Google Scholar]
- 3. Sbiera S, Deutschbein T, Weigand I, Reincke M, Fassnacht M, Allolio B. The new molecular landscape of Cushing’s disease. Trends Endocrinol Metab. 2015;26(10):573–583. [DOI] [PubMed] [Google Scholar]
- 4. Caimari F, Korbonits M. Novel genetic causes of pituitary adenomas. Clin Cancer Res. 2016;22(20):5030–5042. [DOI] [PubMed] [Google Scholar]
- 5. Reincke M, Sbiera S, Hayakawa A, Theodoropoulou M, Osswald A, Beuschlein F, Meitinger T, Mizuno-Yamasaki E, Kawaguchi K, Saeki Y, Tanaka K, Wieland T, Graf E, Saeger W, Ronchi CL, Allolio B, Buchfelder M, Strom TM, Fassnacht M, Komada M. Mutations in the deubiquitinase gene USP8 cause Cushing’s disease. Nat Genet. 2015;47(1):31–38. [DOI] [PubMed] [Google Scholar]
- 6. Ma ZY, Song ZJ, Chen JH, Wang YF, Li SQ, Zhou LF, Mao Y, Li YM, Hu RG, Zhang ZY, Ye HY, Shen M, Shou XF, Li ZQ, Peng H, Wang QZ, Zhou DZ, Qin XL, Ji J, Zheng J, Chen H, Wang Y, Geng DY, Tang WJ, Fu CW, Shi ZF, Zhang YC, Ye Z, He WQ, Zhang QL, Tang QS, Xie R, Shen JW, Wen ZJ, Zhou J, Wang T, Huang S, Qiu HJ, Qiao ND, Zhang Y, Pan L, Bao WM, Liu YC, Huang CX, Shi YY, Zhao Y. Recurrent gain-of-function USP8 mutations in Cushing’s disease. Cell Res. 2015;25(3):306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hayashi K, Inoshita N, Kawaguchi K, Ibrahim Ardisasmita A, Suzuki H, Fukuhara N, Okada M, Nishioka H, Takeuchi Y, Komada M, Takeshita A, Yamada S. The USP8 mutational status may predict drug susceptibility in corticotroph adenomas of Cushing’s disease. Eur J Endocrinol. 2016;174(2):213–226. [DOI] [PubMed] [Google Scholar]
- 8. Perez-Rivas LG, Theodoropoulou M, Ferraù F, Nusser C, Kawaguchi K, Stratakis CA, Faucz FR, Wildemberg LE, Assié G, Beschorner R, Dimopoulou C, Buchfelder M, Popovic V, Berr CM, Tóth M, Ardisasmita AI, Honegger J, Bertherat J, Gadelha MR, Beuschlein F, Stalla G, Komada M, Korbonits M, Reincke M. The Gene of the ubiquitin-specific protease 8 is frequently mutated in adenomas causing Cushing’s disease. J Clin Endocrinol Metab. 2015;100(7):E997–E1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de Araújo LJ, Lerario AM, de Castro M, Martins CS, Bronstein MD, Machado MC, Trarbach EB, Villares Fragoso MC. Transcriptome analysis showed a differential signature between invasive and non-invasive corticotrophinomas. Front Endocrinol (Lausanne). 2017;8:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Faucz FR, Tirosh A, Tatsi C, Berthon A, Hernández-Ramírez LC, Settas N, Angelousi A, Correa R, Papadakis GZ, Chittiboina P, Quezado M, Pankratz N, Lane J, Dimopoulos A, Mills JL, Lodish M, Stratakis CA. Somatic USP8 gene mutations are a common cause of pediatric Cushing disease. J Clin Endocrinol Metab. 2017;102(8):2836–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Perez-Rivas LG, Reincke M. Genetics of Cushing’s disease: an update. J Endocrinol Invest. 2016;39(1):29–35. [DOI] [PubMed] [Google Scholar]
- 12. Mizuno E, Iura T, Mukai A, Yoshimori T, Kitamura N, Komada M. Regulation of epidermal growth factor receptor down-regulation by UBPY-mediated deubiquitination at endosomes. Mol Biol Cell. 2005;16(11):5163–5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mizuno E, Kitamura N, Komada M. 14-3-3-dependent inhibition of the deubiquitinating activity of UBPY and its cancellation in the M phase. Exp Cell Res. 2007;313(16):3624–3634. [DOI] [PubMed] [Google Scholar]
- 14. Kleinschmidt-DeMasters BK, Lopes MB, Prayson RA. An algorithmic approach to sellar region masses. Arch Pathol Lab Med. 2015;139(3):356–372. [DOI] [PubMed] [Google Scholar]
- 15. Choi M, Scholl UI, Yue P, Björklund P, Zhao B, Nelson-Williams C, Ji W, Cho Y, Patel A, Men CJ, Lolis E, Wisgerhof MV, Geller DS, Mane S, Hellman P, Westin G, Åkerström G, Wang W, Carling T, Lifton RPK. K+ channel mutations in adrenal aldosterone-producing adenomas and hereditary hypertension. Science. 2011;331(6018):768–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lenzini L, Rossitto G, Maiolino G, Letizia C, Funder JW, Rossi GP. A Meta-analysis of somatic KCNJ5 K(+) channel mutations in 1636 patients with an aldosterone-producing adenoma. J Clin Endocrinol Metab. 2015;100(8):E1089–E1095. [DOI] [PubMed] [Google Scholar]
- 17. Theodoropoulou M, Reincke M, Fassnacht M, Komada M. Decoding the genetic basis of Cushing’s disease: USP8 in the spotlight. Eur J Endocrinol. 2015;173(4):M73–M83. [DOI] [PubMed] [Google Scholar]
- 18. Gicquel C, Le Bouc Y, Luton JP, Girard F, Bertagna X. Monoclonality of corticotroph macroadenomas in Cushing’s disease. J Clin Endocrinol Metab. 1992;75(2):472–475. [DOI] [PubMed] [Google Scholar]
- 19. Schulte HM, Oldfield EH, Allolio B, Katz DA, Berkman RA, Ali IU. Clonal composition of pituitary adenomas in patients with Cushing’s disease: determination by X-chromosome inactivation analysis. J Clin Endocrinol Metab. 1991;73(6):1302–1308. [DOI] [PubMed] [Google Scholar]
- 20. Do H, Dobrovic A. Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization. Clin Chem. 2015;61(1):64–71. [DOI] [PubMed] [Google Scholar]
- 21. Issaenko OA, Amerik AY. Chalcone-based small-molecule inhibitors attenuate malignant phenotype via targeting deubiquitinating enzymes. Cell Cycle. 2012;11(9):1804–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lüdecke DK, Flitsch J, Knappe UJ, Saeger W. Cushing’s disease: a surgical view. J Neurooncol. 2001;54(2):151–166. [DOI] [PubMed] [Google Scholar]
- 23. Chandler WF, Barkan AL, Hollon T, Sakharova A, Sack J, Brahma B, Schteingart DE. Outcome of transsphenoidal surgery for Cushing disease: a single-center experience over 32 years. Neurosurgery. 2016;78(2):216–223. [DOI] [PubMed] [Google Scholar]
- 24. Fukuoka H, Cooper O, Ben-Shlomo A, Mamelak A, Ren SG, Bruyette D, Melmed S. EGFR as a therapeutic target for human, canine, and mouse ACTH-secreting pituitary adenomas. J Clin Invest. 2011;121(12):4712–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Knappe UJ, Jaspers C, Buschsieweke D, Reinbold WD, Alomari A, Saeger W, Ehlenz K, Mann WA, Kann PH, Feldkamp J. Ectopic adrenocorticotropic hormone-secreting pituitary adenomas: an underestimated entity. Neurosurgery. 2017;80(4):525–533. [DOI] [PubMed] [Google Scholar]