Abstract
Introduction
A large number of genes involved in autosomal recessive forms of intellectual disability (ID) were identified over the past few years through whole-exome sequencing (WES) or whole-genome sequencing in consanguineous families. Disease-associated variants in TRAPPC9 were reported in eight multiplex consanguineous sibships from different ethnic backgrounds, and led to the delineation of the phenotype. Affected patients have microcephaly, obesity, normal motor development, severe ID, and language impairment and brain anomalies.
Patients
We report six new patients recruited through a national collaborative network.
Results
In the two patients heterozygous for a copy-number variation (CNV), the phenotype was clinically relevant with regard to the literature, which prompted to sequence the second allele, leading to identification of disease-associated variants in both. The third patient was homozygote for an intragenic TRAPPC9 CNV. The phenotype of the patients reported was concordant with the literature. Recent reports emphasized the role of CNVs in the etiology of rare recessive disorders.
Conclusion
This study demonstrates that CNVs significantly contribute to the mutational spectrum of TRAPPC9 gene, and also confirms the interest of combining WES with CNV analysis to provide a molecular diagnosis to patients with rare Mendelian disorders.
Introduction
A large number of genes involved in autosomal recessive forms of intellectual disability (ID) have been identified over the past few years through whole-exome sequencing (WES) or whole-genome sequencing (WGS) in consanguineous families. Disease-associated variants in TRAPPC9 gene have been reported in eight multiplex consanguineous sibships from different ethnic backgrounds leading to the delineation of the phenotype [1–8]. All patients carried homozygous loss-of-function variants, either frameshift or nonsense. Through an exhaustive literature review, we also found a single individual born to consanguineous parents carrying a homozygous intragenic deletion of NIBP [9], the former name of TRAPPC9. Here we report six new patients whom we recruited through a national collaborative network for developmental anomalies (Filière AnDDI-Rares). Three of these patients were diagnosed after the identification of an intragenic copy-number variation (CNV) detected by array-comparative genomic hybridization (aCGH) in the TRAPPC9 gene, including two patients who were compound heterozygotes for an intragenic CNV and a disease-associated variant on the second allele.
Patients and methods
Informed consent was obtained from all patients and their parents.
Clinical reports
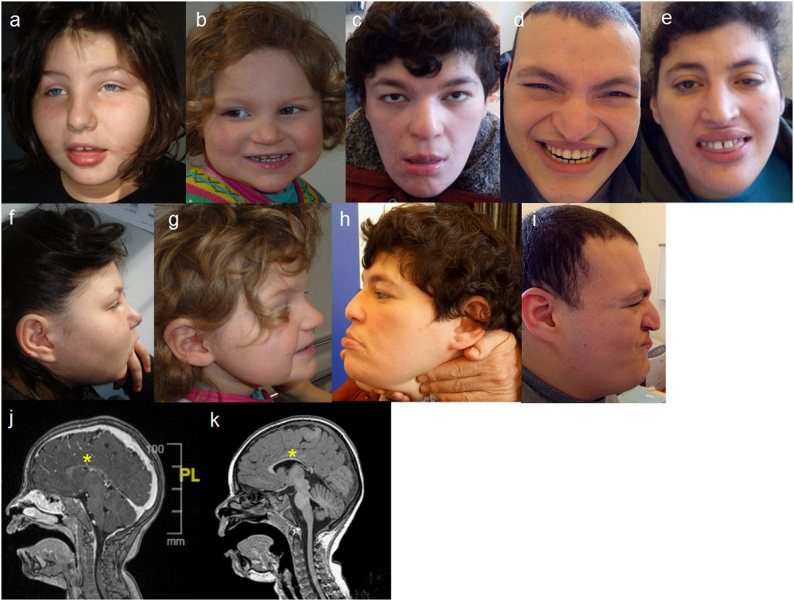
The phenotype of the patients is illustrated in Fig. 1.
Fig. 1.
a, f Patient 1 at 10 years and 6 months. b, g Patient 2 at 2 years and 4 months. c, h Patient 5 at 34 years. d, i Patient 6 at 32 years. e Patient 4 at 35 years. Common dysmorphic features are: full cheeks, prominent nasal bridge, short philtrum, everted inferior lip, and prominent median upper incisors. j, k Brain MRI in sagittal section from patient 1 (j) and patient 3 (k) showing thin corpus callosum (yellow stars) and white matter anomalies
Patients 1 and 2, unlike patients 3 and 4, were born from non-consanguineous parents. Nothing remarkable was noticed during pregnancy nor the first months of life. All patients were referred to hospitals at different ages for genetic examination in relation with syndromic ID, severe speech delay, and facial dysmorphism, associated with behavioral disorders leading to aCGH analysis. When performed, brain magnetic resonance imaging showed thin corpus callosum and white matter anomalies. Patients 5 and 6 are sister and brother of patient 4, respectively, and share the same phenotype.
More detailed information is available in Table 1 and in the Supplementary data.
Table 1.
Main clinical and radiological features associated with TRAPPC9 disease-associated variants in patients from the present study (columns 1–6) and those reported to date in literature[1–9] (columns 7 and 8)
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | Total patients + literature | ||
|---|---|---|---|---|---|---|---|---|
| Age, gender | 10 years, F | 6 years, F | 9 years, F | 35 years, F | 34 years, F | 32 years, M | N=34 | % |
| Relationship | Sibships | |||||||
| Consanguinity | No | No | Yes | Yes | Yes 31/34 | |||
| Geographic origin | Italy | France | Algeria | Tunisia | ||||
| Intellectual disability | +++ | +++ | +++ | +++ | +++ | +++ | 34/34 | 100% |
| Epilepsy | − | − | − | − | − | − | 3/27 | 11% |
| Microcephaly (<-2SD) | − | + | + | + | + | + | 26/31 | 84% |
| Behavioral disorder | + | + | + | + | + | + | 23/23 | 100% |
| Obesity/overweight | + | − | − | + | + | + | 11/22 | 50% |
| Stereotypic movements | + | + | + | + | + | − | 12/19 | 63% |
| Dysmorphic features | + | + | + | + | + | + | 21/30 | 70% |
| Full cheeks | + | + | + | − | + | + | 12/16 | 75% |
| Prominent nasal bridge | + | − | − | + | − | + | 10/16 | 63% |
| Prominent upper central incisors | + | − | − | + | nd | − | 4/10 | 40% |
| Thin superior lip | + | + | − | − | − | − | 5/13 | 38% |
| Short philtrum | + | + | + | + | − | + | 12/16 | 75% |
| Everted lower lip | + | + | + | + | + | + | 10/13 | 77% |
| Brain MRI | ||||||||
| Thin corpus callosum | + | + | + | + | + | + | 15/18 | 83% |
| White matter abnormalities | + | + | + | + | + | + | 17/17 | 100% |
| Status | Compound heterozygote | Compound heterozygote | Homozygote | Homozygotes | Homozygotes | |||
| CNV + point variation | CNV + point variation | CNVs | Point variations | |||||
All patients presented with ID, ranging from moderate to severe, behavioral disorders, and brain anomalies on magnetic resonance imaging (MRI). At the bottom, variant status. Over the 68 reported disease-associated alleles, 6 were CNVs (8.8%).
nd not determined, F female, M male
ACGH analyses
DNA was extracted by standard methods from peripheral blood lymphocytes for all patients and their parents.
In patients 1 and 2, aCGH was performed using a 4 × 180 K whole-genome oligonucleotide microarray following the manufacturer’s protocol (Agilent Technologies, Santa Clara, CA, USA). Results were interpreted with Cytogenomics software v3.0.1.1 (ADM2 method). A CNV was considered if at least three contiguous oligonucleotides presented an abnormal mean log ratio (>0.25 or <−0.25).
In patient 3, an Affymetrix Cytoscan HD array (Affymetrix, Santa Clara, CA, USA) was performed in accordance with the manufacturer’s protocol. CNVs >100 kb with more than 10 deviating probes were accessed with the Chromosome Analysis Suite software (ChAS). CNVs were confirmed by Sanger sequencing, quantitative PCR (qPCR) or fluorescence in situ hybridization (FISH) analysis (Supplementary data 4 and 5). Parental analysis was performed for each case when available. GRCh37 (hg19) was used as the reference sequence. CNVs are described using, respectively, ISCN 2016 and HGVS nomenclature.
Sequencing
TRAPPC9 sequencing was performed using Sanger sequencing for patients 1 and 2 on probands and parents. For patient 4, a WES was performed, using Agilent SureSelect Human All Exon V5 kit for the library preparation following manufacturer’s recommendations (Agilent Technologies) and a 75–100 bp paired-end sequencing on a HiSeq 2000 (Illumina, San Diego, CA, USA). Alignment against GRCh37/hg19 assembly was done with BWA_0.7.12 and Picard_1.136 software, variant calling with GATK_3.4-46 (Broad Institute, Boston, MA, USA), and annotation with SeattleSeqAnnotation138 software. Mean depth was nearly 100×, with 96.4% of target base >10× coverage. Variations should check the following criteria: phred score >30, impact on protein or splice site, frequency <1% in dbSNP 138 and absent in 70 local controls, affecting a gene implicated in human disease (OMIM). Sanger sequencing was used to confirm variations and to test siblings and parents. Reference sequence for TRAPPC9 gene was GenBank accession number NM_031466.5, reference for exon numbering was NG_016478.2.
Description of variants and phenotype
TRAPPC9 disease-associated variants and phenotype have been submitted to Decipher (www.decipher.sanger.ac.uk) for patients 1 and 3 (IDs 314942 and 349431, respectively), to PhenomeCentral (www.phenomecentral.org) for patient 2 (ID P004076) and to the gene variant database at www.LOVD.nl/TRAPPC9 for patients 4, 5, and 6 (ID #00105899).
Results
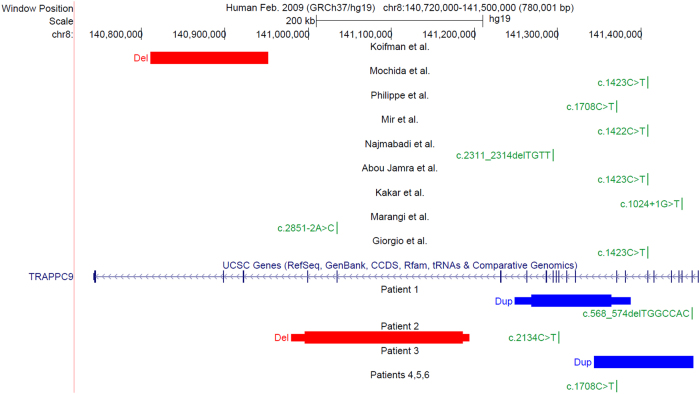
ACGH in patient 1 case showed a 119 kb intragenic duplication of in-frame exons 9–16 in TRAPPC9 (arr[GRCh37] 8q24.3(141268759_141364614)x3 (ISCN 2016)). This result was confirmed by qPCR and FISH. TRAPPC9 sequencing revealed a maternally inherited 7 bp deletion in exon 2 on the second allele (NM_031466.5:c.568_574delTGGCCAC; p.(Trp190Argfs*95) mat; alleles in HGVS format: chr8:g.[(141248399_141268759)_(141364614_141387694)dup]; [141461193_141461199delGTGGCCA] (GRCh37, NC_000008.10)). ACGH in patient 2 revealed a maternally inherited 189 kb intragenic deletion including out-of-frame exons 18 and 19 in TRAPPC9 (arr[GRCh37] 8q24.3 (140996534_141185717)x1 mat). TRAPPC9 sequencing showed a heterozygous nonsense variant in exon 12, paternally inherited (NM_031466.5:c.2134C>T;p.(Arg712*) pat; chr8:g.[(140980111_ 140996534)_(141185717_141193467)del];[141301106G>A] (GRCh37, NC_000008.10)). ACGH in patient 3 identified an homozygous 115 kb intragenic tandem duplication in TRAPPC9 gene, inherited from both parents, and encompassing out-of-frame exons 2–9 (arr[GRCh37] 8q24.3(141344339_141461062)x4 mat pat). Sequencing using long-range PCR define breakpoints and reveal a 61-nucleotide insertion (chr8:g.[141462486_141462487ins(61)ins141343433_141462486]; [141462486_141462487ins(61)ins141343433_141462486] (GRCh37, NC_000008.10)) (Supplementary data 5 for detailed inserted sequence). A homozygous nonsense variant in TRAPPC9 was identified in patient 4 (NM_031466.5:c.1708C>T; p.Arg570* mat pat; chr8:g.[141370230G>A];[141370230G>A] (GRCh37, NC_000008.10)), already reported as pathogenic in the ClinVar database (rs267607137). Among our point variants, this is the only one reported in ExAC database (http://exac.broadinstitute.org) with allele count 3/121,378 (frequency 2.472e−05) and no homozygote case. Parents were heterozygous carriers. The two affected siblings, patients 5 and 6, were homozygotes for the identified variant.
Results are displayed in Fig. 2.
Fig. 2.
UCSC Genome Browser view in GRCh37/hg19 showing TRAPPC9 gene (https://genome.ucsc.edu). Top, patients’ disease-associated variants from the literature[1–9]. Bottom, CNVs and point disease-associated variants found in the present study. HGVS names are based on NM_031466.5 and NG_016478.2 sequences. Except for patients 1 and 2 who are compound heterozygotes, all patient are homozygotes
Discussion
Our study contributes to a better delineation of the phenotype associated with TRAPPC9 disease-associated variants. Similarly to most genes involved in recently described autosomal recessive disorders, TRAPPC9 was identified through studies of consanguineous families. The first patients were reported as having non-syndromic ID with non-specific brain anomalies [1–5]. Further reports, including the patient from Koifman et al. [9] and the patients described here, demonstrated that patients carrying homozygous or compound heterozygous SNVs/CNVs displayed a clinically recognizable phenotype [7]. Neurological involvement includes severe ID predominating on speech with motor development within the normal range, and behavioral anomalies such as temper tantrums, aggressiveness, and stereotypies. Clinically, the facial appearance of young patients is remarkably similar: round face, prominent nasal bridge, full cheeks, short and upturned philtrum, everted lower lip, and prominent and widely spaced upper central incisors. In older patients, the face elongates. Post-natal microcephaly is variable but none of the patients had an occipito-frontal circumference above −1 SD. Truncal obesity is present in half of the patients. Interestingly, patient 4 and her siblings were initially suspected of Cohen syndrome based on the combination of microcephaly, severe ID, obesity, prominent central upper incisors, and myopia in the older.
The present report broadens the spectrum of TRAPPC9 mutational events, and demonstrates that CNVs significantly contribute to the mutational burden of this gene with 6 CNVs (8.8%) out of 68 disease-associated alleles (34 patients), including 3 deleted and 3 duplicated alleles. Recent studies have emphasized the role of CNVs in the etiology of recessive disorders [10]. Deletion of an allele, intragenic deletion, and intragenic duplication behave as loss-of-function variants. In genetically heterogeneous conditions, such as Usher syndrome [11], Cohen syndrome, genetics deafness, retinal dystrophies, and ciliopathies [12], diagnosis strategies include aCGH in addition to gene sequencing in order to detect genomic rearrangements. The proportion of large genomic rearrangements could reach up to one-third of all mutational events [13]. It is much more challenging, however, to determine the clinical relevance of a heterozygous CNV in a gene involved in a very rare recessive disorder. According to the criteria established by Miller et al. [14], a CNV inherited from a healthy parent is more likely to be benign. However, this CNV could uncover a recessive variant on the other chromosome. In theory, this hypothesis should be raised when a CNV affecting a gene involved in a recessive disorder is found. Some authors have recommended combining WES with CNV analysis to provide a molecular diagnosis to patients with rare Mendelian disorders [8]. However, it is likely that most individuals carrying a CNV on one allele are simply heterozygotes. When WES is not available in routine, confirmation of the diagnosis requires Sanger sequencing of a gene, which is time-consuming and expensive. It is then important to evaluate the clinical relevance of the CNV by comparing the patient’s phenotype with data from the literature. The description of the phenotype of TRAPPC9-associated ID illustrates the so-called forward phenotyping strategy [15]. In the near future, we expect that WES or WGS will allow detection of DNA point variants, CNVs, and structural variations with the same technique [16]. Various software are now available to perform these analyses from WGS data. Simultaneous analysis of the data can easily solve cases where compound heterozygosity is suspected. Nevertheless, a clinical description is essential to infer causality and direct further investigations.
Electronic supplementary material
Acknowledgements
We thank the families for their cooperation. We also thank all the lab technicians involved in this research. Affymetrix microarrays were processed in the Microarray Core Facility of the Institute in Regenerative Medicine and Biotherapy, CHRU-INSERM-UM Montpellier, http://irmb.chu-montpellier.fr.
Compliance with ethical standards
Conflict of Interest
The authors declare that they have no competing financial interests.
Footnotes
Electronic supplementary material
The online version of this article (10.1038/s41431-017-0018-x) contains supplementary material, which is available to authorized users.
References
- 1.Mochida GH, Mahajnah M, Hill AD, et al. A truncating mutation of TRAPPC9 is associated with autosomal-recessive intellectual disability and postnatal microcephaly. Am J Hum Genet. 2009;85:897–902. [DOI] [PMC free article] [PubMed]
- 2.Philippe O, Rio M, Carioux A, et al. Combination of linkage mapping and microarray-expression analysis identifies NF-κB signaling defect as a cause of autosomal-recessive mental retardation. Am J Hum Genet. 2009;85:903–8. [DOI] [PMC free article] [PubMed]
- 3.Mir A, Kaufman L, Noor A, et al. Identification of mutations in TRAPPC9, which encodes the NIK- and IKK-β-binding protein, in nonsyndromic autosomal-recessive mental retardation. Am J Hum Genet. 2009;85:909–15. [DOI] [PMC free article] [PubMed]
- 4.Najmabadi H, Motazacker MM, Garshasbi M, et al. Homozygosity mapping in consanguineous families reveals extreme heterogeneity of non-syndromic autosomal recessive mental retardation and identifies 8 novel gene loci. Hum Genet. 2007;121:43–8. [DOI] [PubMed]
- 5.Abou Jamra R, Wohlfart S, Zweier M, et al. Homozygosity mapping in 64 Syrian consanguineous families with non-specific intellectual disability reveals 11 novel loci and high heterogeneity. Eur J Hum Genet. 2011;19:1161–6. [DOI] [PMC free article] [PubMed]
- 6.Kakar N, Goebel I, Daud S, et al. A homozygous splice site mutation in TRAPPC9 causes intellectual disability and microcephaly. Eur J Med Genet. 2012;55:727–31. [DOI] [PubMed]
- 7.Marangi G, Leuzzi V, Manti F, et al. TRAPPC9-related autosomal recessive intellectual disability: report of a new mutation and clinical phenotype. Eur J Hum Genet. 2013;21:229–32. [DOI] [PMC free article] [PubMed]
- 8.Giorgio E, Ciolfi A, Biamino E, et al. Whole exome sequencing is necessary to clarify ID/DD cases with de novo copy number variants of uncertain significance: two proof-of-concept examples. Am J Med Genet A. 2016;170:1772–9. [DOI] [PubMed]
- 9.Koifman A, Feigenbaum A, Bi W, et al. A homozygous deletion of 8q24.3 including the NIBP gene associated with severe developmental delay, dysgenesis of the corpus callosum, and dysmorphic facial features. Am J Med Genet A. 2010;152A:1268–72. [DOI] [PubMed]
- 10.Boone PM, Campbell IM, Baggett BC, et al. Deletions of recessive disease genes: CNV contribution to carrier states and disease-causing alleles. Genome Res. 2013;23:1383–94. [DOI] [PMC free article] [PubMed]
- 11.Dad S, Rendtorff ND, Kann E, et al. Partial USH2A deletions contribute to Usher syndrome in Denmark. Eur J Hum Genet. 2015;23:1646–51. [DOI] [PMC free article] [PubMed]
- 12.Lindstrand A, Frangakis S, Carvalho CMB, et al. Copy-number variation contributes to the mutational load of Bardet-Biedl syndrome. Am J Hum Genet. 2016;99:318–36. [DOI] [PMC free article] [PubMed]
- 13.Rivera-Brugués N, Albrecht B, Wieczorek D, et al. Cohen syndrome diagnosis using whole genome arrays. J Med Genet. 2011;48:136–40. [DOI] [PubMed]
- 14.Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–64. [DOI] [PMC free article] [PubMed]
- 15.Amos JS, Huang L, Thevenon J, et al. Autosomal recessive mutations in THOC6 cause intellectual disability: syndrome delineation requiring forward and reverse phenotyping. Clin Genet. 2017;91:92–9. [DOI] [PubMed]
- 16.Pfundt, R, Del Rosario M, Vissers LELM, et al. Detection of clinically relevant copy-number variants by exome sequencing in a large cohort of genetic disorders. Genet Med Off J Am Coll Med Genet. 2016. 10.1038/gim.2016.163. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.