Abstract
Cone and cone-rod dystrophies (CD and CRD, respectively) are degenerative retinal diseases that predominantly affect the cone photoreceptors. The underlying disease gene is not known in approximately 75% of autosomal recessive cases. Variants in NMNAT1 cause a severe, early-onset retinal dystrophy called Leber congenital amaurosis (LCA). We report two patients where clinical phenotyping indicated diagnoses of CD and CRD, respectively. NMNAT1 variants were identified, with Case 1 showing an extremely rare homozygous variant c.[271G > A] p.(Glu91Lys) and Case 2 compound heterozygous variants c.[53 A > G];[769G > A] p.(Asn18Ser);(Glu257Lys). The detailed variant analysis, in combination with the observation of an associated macular atrophy phenotype, indicated that these variants were disease-causing. This report demonstrates that the variants in NMNAT1 may cause CD or CRD associated with macular atrophy. Genetic investigations of the patients with CD or CRD should include NMNAT1 in the genes examined.
Introduction
Cone and cone-rod dystrophies (CD and CRD, respectively) are inherited retinal dystrophies (IRD), which predominantly or initially affect the cone photoreceptors. The patients with CD usually become symptomatic in the first or second decades of life presenting with reduced visual acuity, reduced colour vision, photophobia and variable nystagmus [1, 2]. Visual acuity loss is progressive and some patients later develop rod photoreceptor dysfunction, conferring a subsequent diagnosis of CRD [1]. The most common mode of inheritance is autosomal recessive, although autosomal dominant and X-linked recessive forms exist. Disease-causing variants are known in at least 30 genes, and 75% of CD and CRD are estimated to be caused by genes yet to be identified [1, 3].
Homozygous and compound heterozygous variants in NMNAT1 (nicotinamide nucleotide adenylyltransferase 1) are described as causative in Leber congenital amaurosis (LCA) [4–7]. LCA is an early-onset form of IRD manifesting in the first year of life with vision impairment due to severe disease of both rod and cone photoreceptors [3]. NMNAT1 encodes an enzyme involved in nicotinamide adenine dinucleotide (NAD) synthesis [8]. In this study, we report cases of CD and CRD due to either homozygous or compound heterozygous NMNAT1 variants, identifying new phenotypes associated with this gene.
Materials and methods
Subjects
Case 1 was a 26-year-old female of Indian ethnicity who presented with LCA. There was no known family history of IRD or consanguinity. At the age of 4 years, ophthalmic investigations undertaken in India reported “bull’s eye” macular changes and a provisional diagnosis of CD. At the age of 25 years, a change in diagnosis to LCA was made. Upon relocation to Australia, referral for LCA gene panel testing resulted in identification of a homozygous NMNAT1 variant, c.[271G > A] p.(Glu91Lys), reported as an “inconclusive” result (Casey Eye Institute, Molecular Diagnostics Laboratory, Oregon, USA). This prompted detailed ophthalmic and genetics review.
Case 2 was a 14-year-old female of Caucasian background with a diagnosis of CRD. There was no known family history of IRD or consanguinity. Genomic studies and detailed ophthalmic review were undertaken.
The Human Research Ethics Committee of the Sydney Children’s Hospital Network, Sydney, Australia, approved the study, and informed written consent was obtained.
Ophthalmic investigations
Ophthalmic examinations included ultra-wide-field fundus autofluorescence (Optos plc Dunfermline Scotland) and macular optical coherence tomography (OCT) (Spectralis, Heidelberg Engineering, Germany). Full-field electroretinograms (ERGs) (maximum flash intensity 12 cd*s/m2) (Espion, Diagnosys, Lowell, MA) were recorded according to the International Society for Clinical Electrophysiology of Vision (ISCEV) standards.
Genomics and bioinformatics
Whole-genome sequencing (WGS) was performed on the Illumina HiSeq X Ten (Kinghorn Centre for Clinical Genomics, Garvan Medical Research Institute, Sydney, Australia). Alignment and variant calling used GATK best practice guidelines (https://software.broadinstitute.org/gatk/best-practices). Variants in disease genes associated with CD, CRD and other IRDs were examined using Ingenuity Variant Analysis (Qiagen, USA) and Alamut Visual v2.8.2 (Interactive Biosoftware, France), with analysis of allele frequencies in gnomAD (http://gnomad.broadinstitute.org/) and pathogenicity prediction by SIFT, PolyPhen-2, Align GVGD, Mutation Taster and conservations scores with PhyloP, as in our previous studies [9]. Prioritised variants were classified according to American College of Medical Genetics and Genomics (ACMG) guidelines [10]. The protein modelling tool HOPE was used to visualise amino acid substitutions [11]. Sanger sequencing was used to confirm and segregate the variants.
Results
Ophthalmic findings of CD in Case 1 and CRD in Case 2
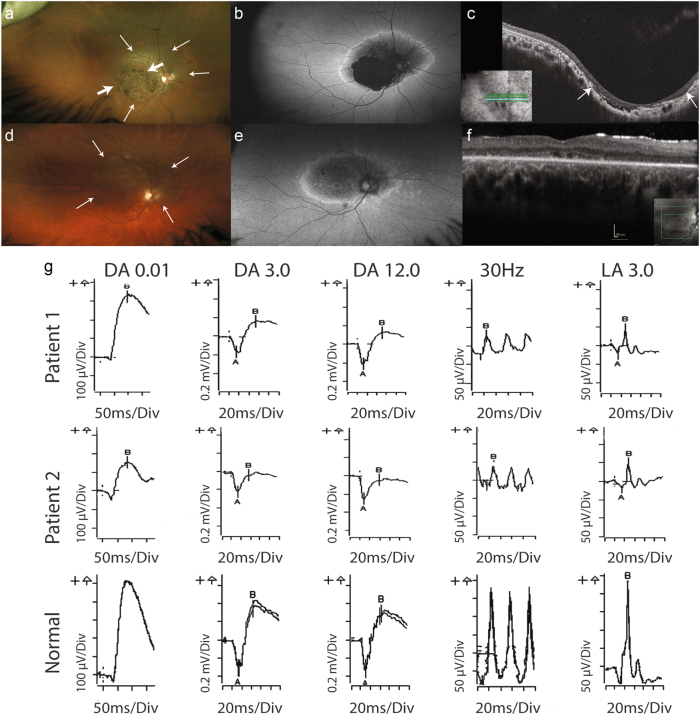
In Case 1, age 26 years, the best-corrected visual acuity was 6/45-1; N36, right eye and 6/120; N48, left eye. Refractive error was −3.00 / +2.75 × 45, right and −3.50 / +3.00 × 115, left. Fundal examination revealed bilateral coloboma-like macular atrophic changes with the retinal atrophy extending nasal to the optic disc. Fundus autofluorescence demonstrated macular hypoautofluorescence associated with the macular lesions, surrounded by a ring of hyperautofluorescence (Figs. 1a, b). Full-field ERG showed mildly reduced scotopic responses and decreased photopic responses. Specifically, there was a decrease in the amplitude of the 30 Hz flicker and the LA 3.0 (light-adapted single flash) (Fig. 1g). The loss of central vision and electrophysiology findings were consistent with CD.
Fig. 1.
Ophthalmic investigations demonstrating macular atrophy with cone dystrophy in Case 1 and cone-rod dystrophy in Case 2. Patient 1—right eye (a–c): a Wide-field colour-corrected fundus images illustrating the coloboma-like macular atrophy with thick arrows marking the boundaries, and surrounding retinal atrophy extending beyond the vascular arcades and nasal to the optic discs with thin arrows marking this change. b Wide-field fundus autofluorescence showing the complete loss of autofluorescence coinciding with the coloboma-like macular atrophy surrounded by blotchy hypoautofluorescence and the ring of hyperautofluorescence bounding the atrophic area. c Optical coherence tomography (OCT) images showing the excavation and loss of the photoreceptor layers including the ellipsoid layer at the site of the macular atrophy, which is demarcated by the arrows. Patient 2—right eye (d–f): d Wide-field colour- corrected fundus images illustrating the macular atrophy with surrounding retinal atrophy extending beyond the vascular arcades and nasal to the optic discs, with thin arrows marking the extent of the atrophy. e Wide-field fundus autofluorescence showing blotchy hypoautofluorescence in the posterior pole and the ring of hyperautofluorescence bounding the atrophic area. f OCT images showing the loss of the photoreceptor ellipsoid layer at the site of the macular atrophy indicated by the arrows. g Full-field electroretinogram (ERG) performed according to ISCEV standards. Trace results for the right eye in patients 1 and 2 compared with normal. Patient 1 shows cone photoreceptor dysfunction with reduction in the photopic ERG indicated by reduction in the light-adapted (LA) 30 Hz flicker and LA 3.0 stimulus tests. There is a mild reduction in the rod-mediated scotopic ERG under the following stimulus conditions: DA0.01, DA3.0 and DA12.0. Patient 2 shows cone photoreceptor dysfunction with a reduction in the photopic ERG indicated by reduction in the 30 Hz flicker and LA 3.0 stimulus tests. There is a moderate reduction in the rod-mediated scotopic ERG under the following stimulus conditions: DA0.01, DA3.0 and DA12.0
In Case 2, examination at age 7 years showed that the visual acuity was 6/90 bilaterally. Refraction was +0.50 / +1.00 × 10 right eye and +1.50 / +1.00 × 130 left eye. Granular atrophic lesions involving the macula and atrophy nasal to the optic disc were present in both eyes. At age 14 years, visual acuity was 6/120 bilaterally. Fundi continued to show the atrophic changes, which were hypoautofluorescent and surrounded by a ring of hyperautofluorescence (Figs. 1d, e). The lesions were strikingly similar to those observed in Case 1 (Figs. 1a, b). In both cases, OCT showed the loss of the photoreceptor layer in the region of the macular atrophy (Figs. 1c, f). Full-field ERG indicated CRD, with decreased photopic and scotopic responses (Fig. 1g).
Variants in NMNAT1 in both cases
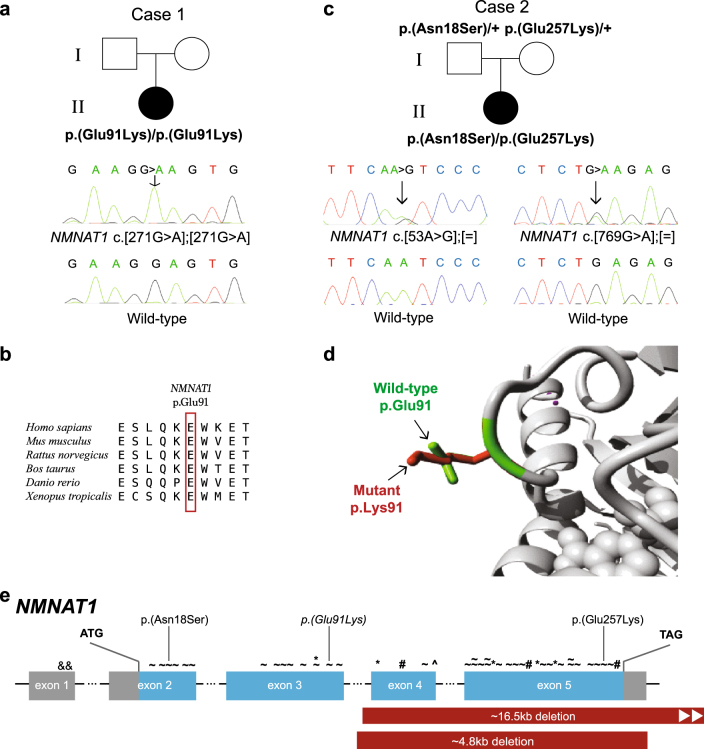
In Case 1, Sanger sequencing confirmed the homozygous NMNAT1 variant c.[271G > A] p.(Glu91Lys) (ClinVar accession ID: SCV000599466) (Fig. 2a). In silico assessment of the p.Glu91 showed conservation across various species (Fig. 2b) and a moderate score on PhyloP analysis, and there was a report where a different amino acid substitution at p.Glu91 segregated in an LCA family [12]. This variant was observed only once in gnomAD and had inconclusive in silico pathogenicity prediction (Table 1), although visualisation of the substituted amino acid indicated that the residue differed in both size and charge from the wild-type (Fig. 2d), and that the difference in ionic charge would disturb the residue interactions [11]. ACMG criteria indicated that this was a variant likely to affect function (Table 1).
Fig. 2.
NMNAT1 variants and gene structure. a, c Pedigrees of Cases 1 and 2, respectively, showing affected proband, unaffected parents and respective genotypes where available and confirmatory Sanger traces. Wild-type Sanger traces are shown for comparison. b Amino acid sequence conservation across different species d Ribbon diagram showing region of NMNAT1 affected by the variant in Case 1. Differences in size and structure are shown between wild-type p.Glu91 (green) and mutant p.Lys91 (red). e NMNAT1 gene structure with previously reported variants marked above the affected exon as follows: & non-coding, ~ missense, * nonsense, # frameshift, ^ canonical splice site point variants. Partial gene deletions are indicated by solid red bars. The variants identified in this study are indicated above, with the homozygous variant italicised
Table 1.
NMNAT1 (NM_022787.3) variants identified in association with cone and cone-rod dystrophy
| Case ID | Ophthalmic phenotype | Genomic coordinate | Nucleotide change | Predicted amino-acid change | Population allele frequency | In silico: Align GVGD class, SIFT, MutTaster, PolyPhen-2, PhyloP, splicing affect | ACMG classification (supporting evidence) |
|---|---|---|---|---|---|---|---|
| Case 1 | Cone dystrophy | Chr1:g.10035805G > A | c.[271G > A] homozygous | p.(Glu91Lys) | 0.000004 (1/245660) | C0, T, D, B, Mod, N/A | Likely affects function (PM1; PM2; PM5; PP2) |
| Case 2 | Cone-rod dystrophy | Chr1:g.10032184A > G | c.[53A > G] | p.(Asn18Ser) | 0.000018 (5/276912) | C0, T, D, B, Mod, N/A | Affects function (PS1; PM1; PM2; PM3; PP2) |
| Chr1:g.10042688G > A | c.[769G > A] | p.(Glu257Lys) | 0.0006968 (193/276988) | C0, T, D, B, Mod, N/A | Affects function (PS1; PM1; PM2; PM3; PP2) |
The variants described are located in the nicotinate-nucleotide adenylyltransferase protein domain and are heterozygous unless otherwise stated. In silico calculations were generated using Alamut Visual version 2.8.2 (Interactive Biosoftware, France). Reference sequence Genome Build GRCh37(hg19). Population allele frequencies sourced from gnomAD (http://gnomad.broadinstitute.org/). In silico definitions: C0 = amino acid substitution is unlikely to have a functional affect; D = deleterious/damaging; T = tolerated; B = benign; Mod = moderately conserved nucleotide; N/A = no affect predicted on splicing. ACMG classification assigned using in silico, pedigree information and the literature available. Further information regarding the evidence criteria can be found in the ACMG guidelines [10]
In Case 2, WGS analysis of known CD, CRD and other IRD genes showed compound heterozygous variants in NMNAT1 c.[53A > G];[769G > A] p.(Asn18Ser);(Glu257Lys) (ClinVar accession ID: SCV000599467), with no other variants of significance identified. Sanger sequencing confirmed these variants (Fig. 2c). Both variants were previously reported in LCA, but not in the same individual [5, 13]. In the population database gnomAD, these variants occurred with frequencies of 0.000018 (5/276912) and 0.0006968 (193/276988), respectively, in the heterozygous state, and were absent in the homozygous state. They scored as variants affecting function using ACMG criteria, despite their inconclusive in silico pathogenicity prediction (Table 1).
Discussion
The CD and CRDs display genetic and phenotypic heterogeneity rendering genetic diagnosis challenging. Coloboma-like macular atrophy is a rare phenotypic association in CD and CRD, with variants identified in ADAM9 and GUCA1A in only a few cases [14, 15]. Other IRDs associated with coloboma-like macular atrophy include North Carolina Macular dystrophy, early-onset retinitis pigmentosa and LCA [16–18]. In LCA, macular atrophy is described in association with variants in NMNAT1 and approximately a third of cases with AIPL1 variants [5, 6, 13, 19]. In both our cases, macular atrophy was present, but not included in referrals to the genetic clinic, highlighting the need for NMNAT1 to be recognised as a causative disease gene in CD and CRD, especially where genetic diagnosis is required.
A variety of homozygous and compound heterozygous variants in NMNAT1 have been identified in patients with LCA (Fig. 2e). NMNAT1 encodes an enzyme involved in NAD synthesis, playing a critical role in cellular metabolism essential for life since deficient mice cannot survive beyond early embryogenesis [8]. NMNAT1 variants may reflect altered rather than absent enzymatic activity [5]. The variants identified in this study had inconclusive in silico pathogenicity prediction and moderate conservation on PhyloP analysis, had heterozygous allele frequencies of 0.000004 to 0.0006968 in population databases and scored as likely to affect function and affecting function using ACMG criteria. This has been seen for other NMNAT1 variants, where comparison with frequencies in control databases has assisted in the designation of pathogenicity [7, 13]. Interestingly, the p.(Glu257Lys) variant, with a population allele frequency of 0.0006968, was not present in the homozygous state in the population database gnomAD, and has been seen in the homozygous state in an affected individual with LCA [5], but has also been reported in unaffected individuals in one family suggesting that this variant may be a hypomorphic allele [20].
The macular atrophy in our cases was very similar to that seen in LCA patients with NMNAT1 variants. Additionally, one patient described with LCA was reported with electrophysiological examination suggestive of CD [4]. Hence, despite the variations of in silico predictions for the identified NMNAT1 variants, the presence of these variants in combination with the striking macular atrophy phenotype strongly suggested that the variants in NMNAT1 were causative of the CD and CRD phenotypes in our cases. Our findings indicate that the homozygous variant in Case 1, initially reported as “inconclusive”, is significant and should not be ignored, and that the NMNAT1 variants in Case 2 are causative of the disease phenotype. This is further supported by the recent report of two siblings with a homozygous NMNAT1 variant c.[500A > G] p.(Asn167Ser) who had a clinical diagnosis of CRD with atrophic macular changes [21]. This affects clinical management, especially for adult cases where there may be a consanguineous union, heralding the need for investigation for the variant in the partner, since presence would indicate a 50% recurrence risk.
Our findings indicate that the disease-associated variants in NMNAT1 cause CD or CRD, where atrophic macular lesions may also be present. NMNAT1 should be included in the genes examined in CD and CRD patients.
Acknowledgements
We would like to thank the families for their willingness to participate in this study. This study was supported by the National Health and Medical Research Council of Australia (NHMRC Grant: 1099165 to R.V.J. and J.R.G.) and the Costco Organisation.
Compliance with ethical standards
Competing interests
The authors declare that they have no competing interests.
Footnotes
Benjamin M. Nash and Richard Symes contributed equally to this work.
References
- 1.Roosing S, Thiadens AA, Hoyng CB, Klaver CC, den Hollander AI, Cremers FP. Causes and consequences of inherited cone disorders. Prog Retin Eye Res. 2014;42:1–26. doi: 10.1016/j.preteyeres.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Michaelides M, Moore AT. The cone dysfunction syndromes. Br J Ophthalmol. 2004;88:291–7. doi: 10.1136/bjo.2003.027102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nash BM, Wright DC, Grigg JR, Bennetts B, Jamieson RV. Retinal dystrophies, genomic applications in diagnosis and prospects for therapy. Transl Pediatr. 2015;4:139–63. doi: 10.3978/j.issn.2224-4336.2015.04.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiang PW, Wang J, Chen Y, et al. Exome sequencing identifies NMNAT1 mutations as a cause of Leber congenital amaurosis. Nat Genet. 2012;44:972–4. doi: 10.1038/ng.2370. [DOI] [PubMed] [Google Scholar]
- 5.Koenekoop RK, Wang H, Majewski J, et al. Mutations in NMNAT1 cause Leber congenital amaurosis and identify a new disease pathway for retinal degeneration. Nat Genet. 2012;44:1035–9. doi: 10.1038/ng.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perrault I, Hanein S, Zanlonghi X, et al. Mutations in NMNAT1 cause Leber congenital amaurosis with early-onset severe macular and optic atrophy. Nat Genet. 2012;44:975–7. doi: 10.1038/ng.2357. [DOI] [PubMed] [Google Scholar]
- 7.Falk MJ, Zhang Q, Nakamaru-Ogiso E, et al. NMNAT1 mutations cause Leber congenital amaurosis. Nat Genet. 2012;44:1040–5. doi: 10.1038/ng.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sasaki Y, Margolin Z, Borgo B, Havranek JJ, Milbrandt J. Characterization of Leber congenital amaurosis-associated NMNAT1 mutants. J Biol Chem. 2015;290:17228–38. doi: 10.1074/jbc.M115.637850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma AS, Grigg JR, Ho G, et al. Sporadic and familial congenital cataracts: mutational spectrum and new diagnoses using next-generation sequencing. Hum Mutat. 2016;37:371–84. doi: 10.1002/humu.22948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richards A, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association of Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venselaar H, Te Beek TA, Kuipers RK, Hekkelman ML, Vriend G. Protein structure analysis of mutations causing inheritable diseases. An e-science approach with life scientist friendly interfaces. BMC Bioinformatics. 2010;11:548. doi: 10.1186/1471-2105-11-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin X, Qu LH, Meng XH, Xu HW, Yin ZQ. Detecting genetic variations in hereditary retinal dystrophies with next-generation sequencing technology. Mol Vis. 2014;20:553–60. [PMC free article] [PubMed] [Google Scholar]
- 13.Siemiatkowska AM, van den Born LI, van Genderen MM, et al. Novel compound heterozygous NMNAT1 variants associated with Leber congenital amaurosis. Mol Vis. 2014;20:753–9. [PMC free article] [PubMed] [Google Scholar]
- 14.El-Haig WM, Jakobsson C, Favez T, Schorderet DF, Abouzeid H. Novel ADAM9 homozygous mutation in a consanguineous Egyptian family with severe cone-rod dystrophy and cataract. Br J Ophthalmol. 2014;98:1718–23. doi: 10.1136/bjophthalmol-2014-305231. [DOI] [PubMed] [Google Scholar]
- 15.Kamenarova K, Corton M, Garcia-Sandoval B, et al. Novel GUCA1A mutations suggesting possible mechanisms of pathogenesis in cone, cone-rod, and macular dystrophy patients. Biomed Res Int. 2013;2013:517570. doi: 10.1155/2013/517570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiernan DF, Shah RJ, Hariprasad SM, et al. Thirty-year follow-up of an African American family with macular dystrophy of the retina, locus 1 (North Carolina macular dystrophy) Ophthalmology. 2011;118:1435–43. doi: 10.1016/j.ophtha.2010.10.041. [DOI] [PubMed] [Google Scholar]
- 17.Ajmal M, Khan MI, Neveling K, et al. A missense mutation in the splicing factor gene DHX38 is associated with early-onset retinitis pigmentosa with macular coloboma. J Med Genet. 2014;51:444–8. doi: 10.1136/jmedgenet-2014-102316. [DOI] [PubMed] [Google Scholar]
- 18.Smith D, Oestreicher J, Musarella MA. Clinical spectrum of leber’s congenital amaurosis in the second to fourth decades of life. Ophthalmology. 1990;97:1156–61. doi: 10.1016/S0161-6420(90)32442-9. [DOI] [PubMed] [Google Scholar]
- 19.Aboshiha J, Dubis AM, van der Spuy J, et al. Preserved outer retina in AIPL1 Leber’s congenital amaurosis: implications for gene therapy. Ophthalmology. 2015;122:862–4. doi: 10.1016/j.ophtha.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 20.Siemiatkowska AM, Schuurs-Hoeijmakers JH, Bosch DG, et al. Nonpenetrance of the most frequent autosomal recessive leber congenital amaurosis mutation in NMNAT1. JAMA Ophthalmol. 2014;132:1002–4. doi: 10.1001/jamaophthalmol.2014.983. [DOI] [PubMed] [Google Scholar]
- 21.Khan AO, Budde BS, Nurnberg P, Kawalia A, Lenzner S, Bolz HJ. Genome-wide linkage and sequence analysis challenge CCDC66 as a human retinal dystrophy candidate gene and support a distinct NMNAT1-related fundus phenotype. Clin Genet. 2017. 10.1111/cge.13022 [DOI] [PubMed]