Abstract
Aims/hypothesis
MicroRNAs (miRNAs) are key regulators of gene expression and novel biomarkers for many diseases. We investigated the hypothesis that serum levels of some miRNAs would be associated with islet autoimmunity and/or progression to type 1 diabetes.
Methods
We measured levels of 93 miRNAs most commonly detected in serum. This retrospective cohort study included 150 autoantibody-positive and 150 autoantibody-negative family-matched siblings enrolled in the TrialNet Pathway to Prevention Study. This was a young cohort (mean age = 11 years), and most autoantibody-positive relatives were at high risk because they had multiple autoantibodies, with 39/150 (26%, progressors) developing type 1 diabetes within an average 8.7 months of follow-up. We analysed miRNA levels in relation to autoantibody status, future development of diabetes and OGTT C-peptide and glucose indices of disease progression.
Results
Fifteen miRNAs were differentially expressed when comparing autoantibody-positive/negative siblings (range −2.5 to 1.3-fold). but receiver operating characteristic (ROC) analysis indicated low specificity and sensitivity. Seven additional miRNAs were differentially expressed among autoantibody-positive relatives according to disease progression; ROC returned significant AUC values and identified miRNA cut-off levels associated with an increased risk of disease in both cross-sectional and survival analyses. Levels of several miRNAs showed significant correlations (r 0.22-0.55) with OGTT outcomes. miR-21-3p, miR-29a-3p and miR-424-5p had the most robust associations.
Conclusions/interpretation
Serum levels of selected miRNAs are associated with disease progression and confer additional risk of the development of type 1 diabetes in young autoantibody-positive relatives. Further studies, including longitudinal assessments, are warranted to further define miRNA biomarkers for prediction of disease risk and progression.
Keywords: Clinical science, Microarray, Prediction and prevention of type 1 diabetes
Introduction
Type 1 diabetes is a chronic autoimmune disease causing a severe loss of pancreatic beta cells and insulin deficiency [1]. Genetic and immunological markers identify individuals at increased risk of developing it. Autoantibodies to islet cell autoantigens GAD, insulinoma-associated antigen 2 (IA-2), insulin and zinc transporter 8 (ZnT8) are predictive biomarkers in natural history studies. Autoantibodies appear months to years before diagnosis, and the rate of progression varies among individuals. Additional biomarkers are needed to improve prediction, including (1) early biomarkers predicting the triggering of islet autoimmunity for primary prevention, and (2) biomarkers that improve the accuracy of risk stratification for prevention trials among autoantibody-positive relatives.
MicroRNAs (miRNAs) are non-coding RNAs that regulate gene expression [2], affecting all gene pathways by promoting the degradation and suppressing the translation of target mRNAs. In terms of type 1 diabetes, miRNAs regulate key functions of the immune system in multiple cell types, including lymphocytes and regulatory T cells [3]. miRNAs regulate insulin secretion (miR-375) [4], pancreatic development, beta cell differentiation (miR-375, miR-376, miR-7 and miR-9) [5, 6] and regeneration (miR-15a, miR-15b, miR-16 and miR-195) [7]. The miR-200 family has been linked to pancreatic beta cell survival and type 2 diabetes in mouse models [8]; miR-124 influences insulin secretion in type 2 diabetes [9]. Initial evidence links miRNAs to type 1 diabetes in studies of cellular miRNAs from individuals with diabetes [10-12]; miRNAs influence cytokine-mediated beta cell apoptosis, and their loss in beta cells enhances diabetes induced by multiple low doses of streptozotocin [13, 14]. Thus, mounting evidence links miRNAs to beta cell function and the pathogenesis of type 1 diabetes [15, 16].
miRNAs are present in the circulation, where they are stable and resistant to repeated ‘freeze–thaw’ cycles, extended storage, low/high pH and endogenous RNAse activity [17], possibly because they form complexes with ribonucleoproteins [18]. miRNAs can be assayed in either plasma or serum with identical results [17]. Circulating miRNAs are also packaged inside exosomes, which might transfer mRNAs and miRNAs between cells [19]. Thus, circulating miRNAs might reflect biological responses and mark disease states. Serum/plasma miRNAs are emerging as biomarkers of acute tissue injury and chronic diseases, including cancer and type 2 diabetes, in both individuals with diabetes and rodent models of type 2 diabetes and insulin resistance [20]. In the setting of type 1 diabetes, serum levels of miR-375, an islet cell miRNA, reflect beta cell destruction in mice [21] and islet transplant recipients [22], although this may be limited to acute damage and miR-375 is not exclusively of pancreatic beta cell origin. A few studies have reported perturbed serum miRNA levels in individuals with newly diagnosed type 1 diabetes compared with healthy participants [23, 24].
We hypothesised that levels of some of the circulating miRNAs might be associated with islet autoimmunity and/or disease progression. We measured levels of circulating miRNAs in samples obtained from non-diabetic relatives of individuals with type 1 diabetes, who were longitudinally characterised for autoantibody status and metabolic variables; a proportion of the relatives developed type 1 diabetes on follow-up.
Methods
Study participants
We studied archived serum samples from Type 1 Diabetes TrialNet, a consortium conducting multicentre trials. Samples were from 300 relatives enrolled in the TrialNet Pathway to Prevention Study [25], which screens relatives for autoantibodies and follows them longitudinally with periodic testing until clinical type 1 diabetes develops. All participants provided written informed consent. The ethical boards at all participating institutions approved the study and allowed ancillary studies of de-identified samples. We studied 150 family-matched sibling pairs, selected to be discordant for the presence vs absence of autoantibodies and on sample availability. Table 1 reports demographic characteristics and autoantibody profiles at screening; at the time the sample tested was drawn, none of the participants had type 1 diabetes and all were below age 18 years, with a mean age of 11.0 ± SD 3.7 years.
Table 1.
Demographic and clinical characteristics of 150 sibling pairs discordant for autoantibodies
| Participants | AAb− | AAb+ | AAb+ NP | AAb+ P | |
|---|---|---|---|---|---|
|
|
|||||
| Variable | n | 150 | 150 | 111 | 39 |
| Male/female | 73/77 | 75/75 | 55/56 | 20/19 | |
| Age (years)a | Mean ± SD | 11.1 ± 3.7 | 10.9 ± 3.7 | 10.8 ± 3.7 | 11.2 ± 3.5 |
| Range | 1-18 | 2-18 | 2-17 | 6-18 | |
| Age of onset of type 1 diabetes (years) | Mean ± SD | NA | NA | NA | 11.9 ± 3.7 |
| Range | NA | NA | NA | 6-20 | |
| Follow-up from screening (years)b | Mean ± SD | 2.1 ± 2.4 | 2.4 ± 2.4 | 2.3 ± 2.4 | 2.4 ± 2.5 |
| Range | 0-9 | 0-13 | 0-13 | 0.02-8.6 | |
| Interval between screening and miRNA sample (years) | Mean ± SD | 2.1 ± 2.4 | 1.8 ± 1.9 | 1.8 ± 1.9 | 1.7 ± 1.8 |
| Range | 0-9 | 0-8.8 | 0.8.8 | 0.01-7.4 | |
| Follow-up after miRNA sample (months)b | Mean ±SD | 6.4 ± 10.2 | 6.9 ± 11.2 | 6.0 ± 9.7 | 8.7 ± 12.5 |
| Range | 0-45.4 | 0-77.5 | 0-77.5 | 0-51 | |
| Autoantibody typea | GAD | 0 | 132 | 100 | 32 |
| IA2 | 0 | 92 | 66 | 26 | |
| ZnT8 | 0 | 46 | 36 | 10 | |
| Insulin | 0 | 100 | 74 | 26 | |
| Number of autoantibodiesa | 1 AAb | 0 | 12 (8%) | 8 (7%) | 4 (10%) |
| >1 AAb | 0 | 138 (92%) | 103 (93%) | 35 (90%) | |
At the time the sample tested was drawn
For progressors, follow-up ended with the development of type 1 diabetes
AAb, autoantibody; –, negative; +, positive; NP; non-progressor; P, progressor; NA, not applicable
Autoantibody assays
Relatives were screened for GAD, IA-2 and insulin autoantibodies with standardised radioimmunoassays [26] (performed by the TrialNet Autoantibody Core Laboratory, Barbara Davis Center, University of Colorado, USA). ZnT8 autoantibodies were assessed in individuals who were positive for the autoantibodies tested at screening.
Metabolic evaluation
Autoantibody-positive relatives undergo OGTT metabolic evaluation every 6 months (after 2012, yearly for those with a single autoantibody). Samples for glucose and C-peptide measurements are obtained in the fasting state (−10 and 0 min), and at 30, 60, 90 and 120 min after ingestion of a 1.75 g/kg glucose dose (maximum 75 g of carbohydrate). Glucose and C-peptide AUC values are calculated using the trapezoidal method.
Serum collection and RNA extraction
Serum was obtained using the standardised protocol adopted by TrialNet (see electronic supplementary material [ESM] Methods, ‘Blood collection procedures’). Total RNA was extracted by mixing serum with phenol/guanidine-based lysis buffer, spiked with M2 bacteriophage RNA as a carrier, and isolated using a silica membrane separation-based method (miRNeasy, Qiagen, Germantown, MD, USA).
Analysis of miRNAs in serum
RNA was converted into cDNA using the miRCURY LNA Universal RT kit (Exiqon, Denmark). MiRNA levels were measured using the LNA-based quantitative (q)PCR method (Exiqon). The plates included interplate calibrators, in triplicate, a control primer set and negative/positive spike-in controls (ESM Methods, ‘Control miRNAs and assessment of hemolysis’). Plates were amplified on a Roche LightCycler 480. The assay cut-off was 38 cycles. Ct values were converted into a linear value (relative quantification [RQ]) to account for the exponential nature of PCR Ct values using the formula RQ= 2−(Global mean Ct − miRNA Ct). We assessed potential haemolysis comparing levels of miR-451a (highly expressed in erythrocytes) with those of miR-23a-3p (unaffected by haemolysis; ESM Methods, ‘Control miRNAs and assessment of hemolysis’); ΔCq (miR-23a-3p - miR-451a) values greater than 7–8 indicate a high risk of haemolysis, none of the samples studied had scores greater than 7, and the ΔCq scores for all samples studied (control and TrialNet) did not significantly differ (mean ± SD 5.32 ± 1.47 and 4.48 ± 1.51, respectively). We conducted preliminary investigations on local control samples to investigate whether age of the serum samples affected the detection of circulating miRNAs and to identify the miRNAs most frequently detected in serum (ESM Results and ESM Fig. 1); we then designed a custom panel inclusive of 93 miRNAs (ESM Table 1) that (and/or): (1) were most commonly detected in control samples (>80% of participants); (2) had a preliminary association with type 1 diabetes/islet autoimmunity from the literature and exploratory analyses of individuals with diabetes or their relatives from our institution (ESM Results and ESM Table 2); (3) had literature associations with other autoimmune diseases, insulin resistance, hyperglycaemia and beta cell function.
Data Analysis
miRNA data normalisation
We used the global mean approach for data normalisation [27] to minimise potential confounders when comparing data across groups, as other approaches (e.g. NormFinder) identified different miRNAs across comparisons.
Statistical analysis
For the primary comparison of miRNA levels by autoantibody status, we investigated and tested potential associations between selected miRNAs and islet autoimmunity under the hypothesis that there would be differential expression between autoantibody-positive and autoantibody-negative family-matched siblings. Power analysis via simulation was performed based on results from a pilot study of type 1 diabetes. For the individuals with diabetes and their relatives (data not shown), we evaluated power over a fold-change range of 1.5 to 3, with SDs between 1 and 1.23 (a conservative value, as it was below 1.23 for 95% of the pilot data). We employed a familial study design, which affords greater power, and assumed a correlation in expression level of 0.2 between siblings. Based on this power analysis, a comparison of 150 participants per group returned excellent power (0.89) to detect a 1.7-fold change among family-matched siblings discordant for autoantibody status.
Normalised data (RQ expression levels) were initially analysed, with an estimation and comparison of expression levels between groups. Linear-mixed models were constructed for the estimation and comparison of expression levels between the groups; this approach accounts for the correlation between members of the same family in variance estimates. Estimates of expression level and corresponding comparisons were made only when there were adequate numbers of observed miRNA expression levels for stable estimation. Sex and age were initially included in the models but were not found to be either significant or confounders in estimates of differential expression among autoantibody-positive and autoantibody-negative siblings. Correction for multiple comparisons was performed using empirical distributions generated via bootstrapped resampling with 1000 replications. Ranking of averaged bootstrapped p values is useful to identify a ‘shortlist’ of differentially expressed miRNA for further analysis, while accounting for small sample variability and family size error rates in multiple comparisons [28]. Pairs were kept intact for resampling to preserve the correlation structure of the observed data in generating the empirical distribution used in each replication and corresponding tests of significance for differentially expressed miRNAs. For each replication, analysis was performed on all miRNAs found to be differentially expressed in the original comparisons, and p values were averaged over all replications. CIs for p values were constructed to determine those which were significant based on bootstrap resampling. Levels of statistical significance were set at p<0.05 (uncorrected) and p<0.10 (corrected by bootstrapping).
Secondary analyses
We investigated potential associations of miRNA levels with disease progression and number of autoantibodies. We also assessed sensitivity and specificity for given miRNA levels, and correlations with OGTT indices.
The same methodology employed for the primary analysis was used to investigate differential expression of miRNAs according to future disease progression among autoantibody-positive participants.
We employed receiver operating characteristic (ROC) analysis to investigate the performance of those miRNAs with differential expression in the primary and secondary comparisons as a binary classifier system; this approach allows the identification of those miRNAs with better ability to discriminate groups and derive miRNA cut-off levels based on specificity and sensitivity. ROC curves and AUC values were generated using GraphPad Prism 7.0. For some analyses, groups were compared to estimate frequencies using Fisher’s exact test (GraphPad Prism 7.0); p values are two-sided.
Both univariate and multivariate analyses were undertaken. In univariate analysis, we used the Spearman test to assess correlations of miRNA levels with OGTT variables used to assess progression (glucose and C-peptide AUC, peak C-peptide, 30 minus 0 min C-peptide and type 1 diabetes Diagnostic Index60; Index60 was calculated as the log10 fasting C-peptide, 60 min C-peptide and 60 min glucose) [29, 30]; formulas for these indices and cut-offs were calculated using TrialNet data provided in mg dl−1 min−1 for glucose and ng ml−1 min−1 for C-peptide, respectively. Covariance analyses were performed for adjustments.
miRNA target prediction and pathway enrichment analysis
We investigate potential targets of selected miRNAs using miRWalk 2.0 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/, accessed 22 November 2016), an online bioinformatic program developed to identify predicted and experimentally verified miRNA–target interactions; miRWalk 2.0 documents miRNA-binding sites within the complete sequence of a gene and combines this information with a comparison of binding sites resulting from use of miRanda-rel2010, Targetscan 6.2, MirWalk 2.0 and RNA22 v2. Statistical significance of these predictions is reported after the Benjamini–Hochberg correction for multiple comparisons. Pathway analysis was performed using KEGG (the Kyoto Encyclopedia of Genes and Genomes, http://www.genome.jp/kegg/pathway.html) and WIKIPathways (http://wikipathways.org/index.php/WikiPathways); both sites were accessed on 22 November 2016).
Results
Selected miRNAs show modest association with islet autoimmunity
We compared miRNA levels in 150, family-matched, autoantibody-discordant sibling pairs. The groups were well matched for sex, age and length of follow-up (Table 1). Fifteen miRNAs (12 of which remained significant after bootstrapping) were differentially expressed between the two groups (Table 2). Mean differences in miRNA levels (fold change) between autoantibody-positive and autoantibody-negative siblings ranged from 0.40 (−2.5) to 1.33. Examples are shown as dot plots for miR-31-5p and miR-185-3p (ESM Fig. 2a); as is common in this type of investigation, we found statistically significant differences in mean expression levels even though individual levels might overlap to a large extent between groups. We used ROC to further investigate the utility of these miRNAs as risk biomarkers: 12/15 miRNAs had significant ROC p values (Table 2) but modest AUC values (range 0.56-0.66), indicating that these miRNAs have limited value for assessing risk and discriminating between groups.
Table 2.
miRNAs differentially expressed in autoantibody-discordant sibling pairs
| Comparison of expression levels
|
ROC
|
|||||
|---|---|---|---|---|---|---|
| miRNA | Fold change | p value | Bootstrapped fold change | Bootstrapped p value | AUC | p value |
|
|
|
|||||
| miR-16-5p | 1.29 | 0.0046 | 1.292 | 0.0384 | 0.58 | 0.0123 |
| miR-106a-5p | 1.21 | 0.0112 | 1.217 | 0.0534 | 0.58 | 0.016 |
| miR-19a-3p | 1.27 | 0.0058 | 1.254 | 0.0545 | 0.56 | 0.0476 |
| miR-524-5p | 0.66 | 0.0245 | 0.650 | 0.0555 | 0.58 | 0.0699 |
| miR-523-3p | 0.40 | 0.0031 | 0.437 | 0.0612 | 0.66 | 0.0057 |
| miR-143-3p | 1.25 | 0.0238 | 1.284 | 0.0615 | 0.55 | 0.1267 |
| miR-31-5p | 0.73 | 0.0152 | 0.733 | 0.0639 | 0.59 | 0.0044 |
| miR-25-3p | 1.26 | 0.0159 | 1.276 | 0.0640 | 0.57 | 0.0379 |
| miR-451a | 1.33 | 0.0141 | 1.328 | 0.0689 | 0.58 | 0.0132 |
| miR-20a-5p | 1.20 | 0.0224 | 1.207 | 0.0779 | 0.57 | 0.0278 |
| miR-619 | 0.73 | 0.0446 | 0.711 | 0.0980 | 0.60 | 0.0455 |
| miR-92a-3p | 1.16 | 0.0298 | 1.161 | 0.0995 | 0.57 | 0.0406 |
| miR-485-3p | 0.84 | 0.0291 | 0.856 | 0.1144 | 0.58 | 0.0173 |
| miR-15a-5p | 1.15 | 0.0430 | 1.152 | 0.1276 | 0.55 | 0.1195 |
| miR-185-3p | 0.62 | 0.0302 | 0.701 | 0.1779 | 0.63 | 0.0437 |
Ratio for autoantibody-positive/autoantibody-negative siblings of the miRNA RQ (fold change) and corresponding ROC AUC values. miRNAs are ordered by bootstrapped p value
Selected miRNAs are associated with disease progression
We explored whether miRNA levels differed between autoantibody-positive participants according to future disease progression (39 had developed disease on follow-up). Those who did not develop type 1 diabetes during follow-up are considered in this analysis to be non-progressors. There were no differences in sex and age when autoantibody-positive relatives were stratified by disease progression (Table 1). The samples available for miRNA testing were obtained on average 1.8 ± SD 1.9 years from the initial screening; thus, follow-up from the study sample for the 150 autoantibody-positive relatives was on average 6.9 ± 11.2 months and was not statistically different when comparing progressors with non-progressors (Table 1; p=0.067). Seven miRNAs were detected at higher levels in the progressors group (Table 3), and 6/7 had significant ROC p values with AUC values ranging between 0.60 and 0.79. In choosing ROC cut-off values, we accepted specificity as low as 0.80 to maximise sensitivity, given that this was a population of relatives at high risk of developing type 1 diabetes.
Table 3.
miRNAs differentially expressed in autoantibody-positive participants according to disease progression on follow-up
| Comparison of expression levels
|
ROC
|
||||||
|---|---|---|---|---|---|---|---|
| miRNA | Fold change | p value | AUC | p value | Cut-off | Sensitivity | Specificity |
|
|
|
||||||
| miR-150-5p | 1.31 | 0.0206 | 0.61 | 0.0405 | 7.315 | 0.41 | 0.80 |
| miR-342-3p | 1.32 | 0.0250 | 0.64 | 0.0109 | 3.265 | 0.48 | 0.80 |
| miR-424-5p | 1.39 | 0.0010 | 0.68 | 0.0011 | 3.135 | 0.43 | 0.81 |
| miR-577 | 1.40 | 0.0254 | 0.60 | 0.0757 | 0.065 | 0.39 | 0.81 |
| miR-29a-3p | 1.46 | 0.0024 | 0.68 | 0.0007 | 2.740 | 0.43 | 0.80 |
| miR-491-5p | 1.49 | 0.0467 | 0.61 | 0.0457 | 0.085 | 0.34 | 0.80 |
| miR-21-3p | 2.33 | 0.0005 | 0.79 | 0.0009 | 0.085 | 0.60 | 0.85 |
|
|
|
||||||
Ratio for progressors/non-progressors of miRNA RQ (fold change) and corresponding ROC AUC values. ROC p values remained significant after Bonferroni correction for multiple comparisons for miR-424-5p, miR-29a-3p and miR-21-3p. miRNAs are ordered by fold change
ROC curves for miR-21-3p, miR-29a-3p and miR-424-5p with AUC values of 0.79, 0.68 and 0.68, respectively are shown in Fig. 1a,c,e; their ROC p values remained significant after Bonferroni correction for multiple comparisons. Figure 1b,d,f illustrates levels of miR-21-3p, miR-29a-3p and miR-424-5p, respectively, comparing progressors with non-progressors, and then stratified by number of autoantibodies. Relatives with multiple autoantibodies (92% of this cohort) were more likely to become progressors if they had miR-21-3p, miR-29a-3p or miR-424-5p levels above the respective ROC cut-off values (Table 4). There were too few relatives with a single autoantibody for a meaningful analysis.
Fig. 1.
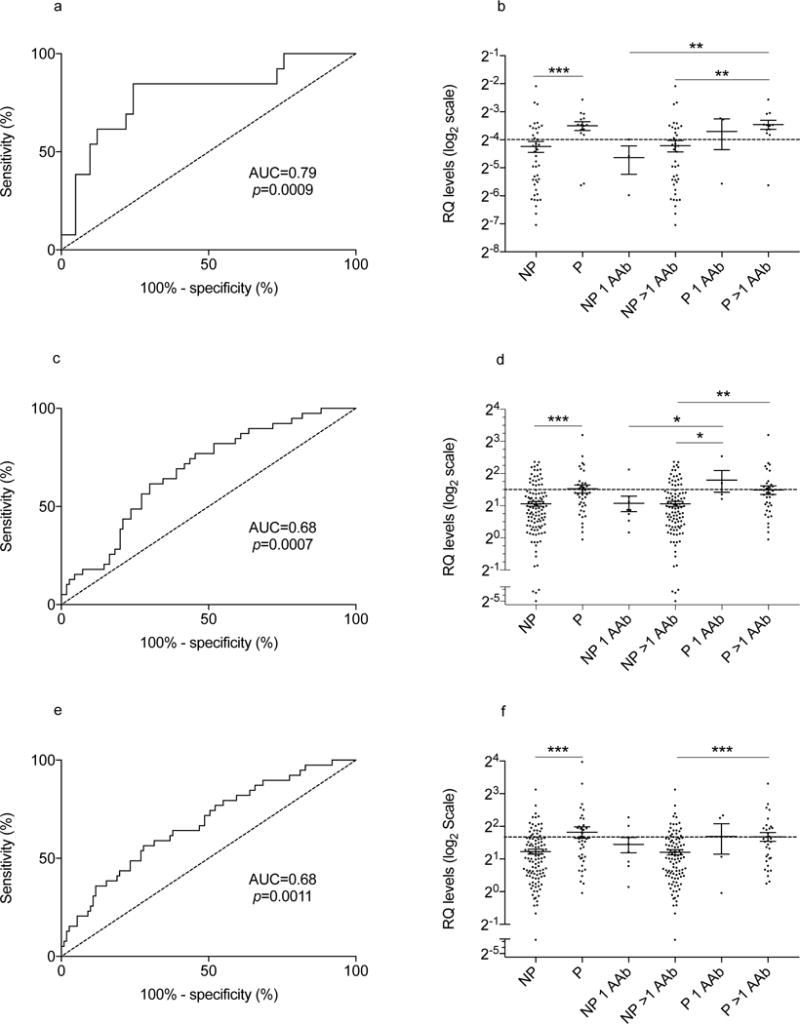
ROC curves and miR-21-3p, miR-29a-3p and miR-424-5p levels in autoantibody-positive participants. ROC curves (a, c, e) are shown for autoantibody-positive relatives according to progression to type 1 diabetes. RQ levels (b, d, f) plotted on a Log2 scale are shown according to disease progression (NP, non-progressor, P, progressor), and to disease progression stratified by autoantibody (AAb) positivity for single (1 AAb) or multiple (>1 AAb) autoantibodies
Table 4.
Observed estimates of progression of type 1 diabetes among relatives with multiple autoantibodies and miRNA levels above the ROC cut-off
| Multiple autoantibodies
| |||||||
|---|---|---|---|---|---|---|---|
| miRNA | Progressors | Non-progressors | p value | RR | OR | PPV | NPV |
| miR-21-3p | 7/12 (58.3%) | 6/39 (15.4%) | 0.006 | 3.8 | 7.7 | 0.58 | 0.84 |
| miR-29a-3p | 15/35 (42.8%) | 21/102 (20.5%) | 0.01 | 2.1 | 2.9 | 0.42 | 0.79 |
| miR-424-5p | 15/35 (42.8%) | 19/103 (18.4%) | 0.006 | 2.3 | 3.3 | 0.44 | 0.80 |
PPV, positive predictive value; NPV, negative predictive value
We used Kaplan–Meier analysis to compare survival curves for disease progression according to the miRNA ROC cut-off levels listed in Table 3. There were no significant differences in sex between groups in each of the survival analyses. Figure 2 shows significantly different risks of development of type 1 diabetes for miR-21-3p, miR-150-5p, miR-29a-3p and miR-342-3p, respectively, with HRs estimates ranging from 2.2 to 3.4; curves were not significantly different for miR-424-5p, miR-491-5p and miR-577(ESM Fig. 3).
Fig. 2.
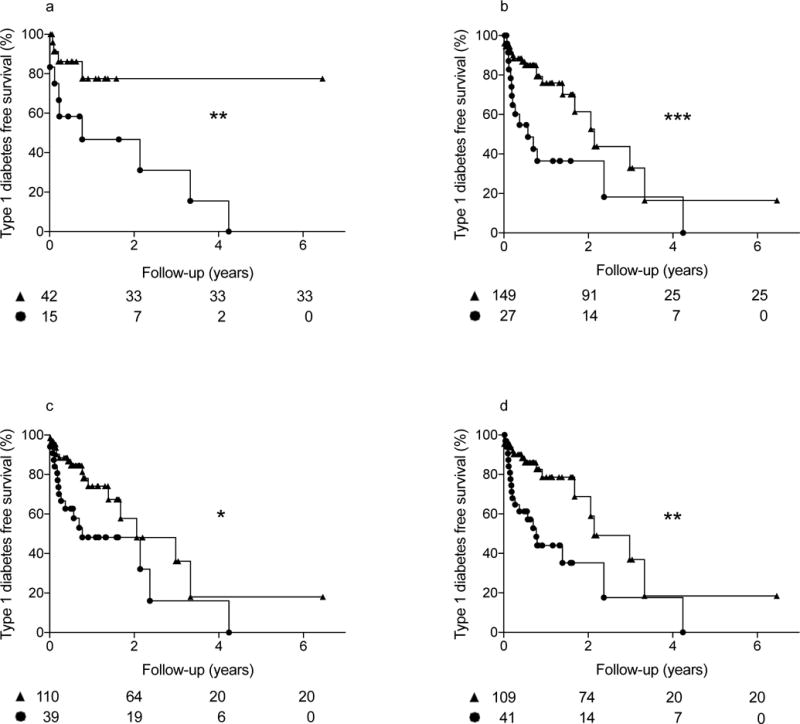
Kaplan–Maier survival curves for the development of type 1 diabetes among autoantibody-positive participants based on ROC cut-off levels of miRNAs associated with disease progression; circles indicate subjects with levels above the ROC cut-off, triangles identify subjects with levels below cut-off. Number of subjects at different time points for each group are shown below the X axis. Logrank Mantel–Cox test significance: *p<0.05, **p<0.01, ***p<0.001. HRs for panels a–d were 3.41 (95% CI 1.12, 10.37), 2.61 (95% CI 1.18, 5.79), 2.24 (95% CI 1.08, 4.64), and 2.82 (95% CI 1.36, 5.84), respectively. T1D, type 1 diabetes
For miR-150-5p, miR-29a-3p and miR-342-3p, the relatives with miRNA levels above the ROC cut-offs were younger than those with miRNA levels below the cut-offs (mean ± SD, respectively: miR-150-5p, 9.6 ± 3.6 vs 11.3 ± 3.6 years, p=0.0193; miR-29a-3p, 9.9 ± 3.1 vs 11/3 ± 3.8, p=0.0291; miR-324-3p, 9.7 ± 3.6 vs 11.4 ± 3.6, p= 0.0095). Given the higher prevalence of progressors in the groups with miRNA levels above the ROC cut-offs, these differences may reflect the known association of younger age with more rapid progression.
Association of miRNAs with metabolic indices of disease progression
TrialNet autoantibody-positive relatives are periodically evaluated by OGTT to detect signs of impaired glucose metabolism and insulin secretion. We analysed data from 96 relatives for whom the OGTT was performed on the same day the miRNA sample was drawn (Table 5 and ESM Table 3): 21 (21.8%) of these 96 participants developed type 1 diabetes on follow-up, within an average 7.0 ± SD 9.9 months; 19 such relatives had an abnormal OGTT when tested. Among the 75 diabetes-free relatives, 12 and 63 had an abnormal or normal OGTT, respectively. Table 6 lists Spearman correlations for miRNA levels with OGTT outcomes for the seven miRNAs associated with disease progression (Table 3), including correlations with fasting and AUC glucose and C-peptide levels, peak C-peptide, 30 minus 0 min C-peptide and Index60. Correlation data in Table 6 are illustrated in Figs 3-5 and ESM Fig. 4. Figures 3 and 4 show correlations between miR-424-5p, miR-29a-3p, miR-150-5p and miR-342-3p levels with C-peptide AUC (Fig. 3a,c,e,g), C-peptide peak values (Fig. 3b,d,f,h), Index60 (Fig. 4a,c,e,g) and 30 minus 0 min C-peptide values (Fig. 4b,d,f,h). Figure 5 shows correlations of glucose AUC with miR-424-5p (Fig. 5a), miR-21-3p (Fig. 5c) and miR-185-3p (Fig. 5e). Correlations of fasting C-peptide are plotted for with miR-424-5p (Fig. 5b), miR-150-5p (Fig. 5d) and miR-185-3p (Fig. 5f). On multiple regression analysis, the correlations reported in Table 6 were unaffected by sex, age and BMI except for correlations of miR-150p with fasting and 30 minus 0 min C-peptide, which were no longer significant after adjustment for BMI, and fasting C-peptide, which was no longer significant after adjustment for age and BMI. An additional nine miRNAs showed correlations with one or more of the above-mentioned OGTT-derived measures (ESM Table 4).
Table 5.
Demographic and clinical characteristics of 96 autoantibody-positive relatives with an OGTT performed on the same date the miRNA sample was drawn
| Participants | AAb+ | AAb+ NP | AAb+ P | |
|---|---|---|---|---|
|
|
||||
| Variable | n | 96 | 75 | 21 |
| Male/female | 58/38 | 45/30 | 13/8 | |
| Age (years)a | Mean ± SD | 11.2 ± 3.7 | 11.0 ± 3.8 | 11.7 ± 3.6 |
| Range | 3-18 | 3-18 | 7-18 | |
| Age of onset of type 1 diabetes (years) | Mean ± SD | NA | NA | 12.3 ± 3.8 |
| Range | NA | NA | 7-20 | |
| Follow-up/time to type 1 diabetes (months)b | Mean ± SD | 6.0 ± 8.8 | 5.5 ± 6.9 | 7.0 ± 9.9 |
| Range | 0-77.5 | 0-77.5 | 0-35.7 | |
| Autoantibody typea | GAD | 83 | 67 | 16 |
| IA-2 | 62 | 44 | 18 | |
| ZnT8 | 29 | 23 | 6 | |
| Insulin | 67 | 52 | 15 | |
| Number of autoantibodiesa | 1 AAb | 6 (6%) | 5 (7%) | 1 (5%) |
| >1 AAb | 90 (94%) | 70 (93%) | 20 (95%) | |
| With abnormal/normal OGTT | 31/65 | 12/63 | 19/2 | |
| OGTT fasting glucose (mmol/l) | Mean ± SD | 4.89 ± 0.53 | 4.89 ± 0.50 | 4.86 ± 0.60 |
| OGTT fasting C-peptide (nmol/l) | Mean ± SD | 0.49 ± 0.30 | 0.53 ± 0.30 | 0.37 ± 0.20 |
At the time the sample tested was drawn
From the time the sample tested was drawn
AAb, autoantibody; −, negative; +, positive; NP, non-progressor; P, progressor
Table 6.
Correlation of miRNA levels with OGTT outcomes
| Variable | miR-150-5p | miR-185-3p | miR-21-3p | miR-29a-3p | miR-342-3p | miR-424-5p | miR-491-5p |
|---|---|---|---|---|---|---|---|
|
|
|||||||
| n | 95 | 19 | 31 | 96 | 96 | 96 | 94 |
| Fasting glucose | −0.132 | 0.463* | 0.104 | −0.030 | −0.093 | −0.095 | 0.050 |
| AUC glucose | 0.068 | 0.498* | 0.432* | 0.157 | 0.065 | 0.280** | 0.097 |
| Fasting C-peptide | −0.299** | 0.552* | 0.042 | −0.134 | −0.199 | −0.224* | 0.096 |
| 30 minus 0 min C-peptide | −0.236* | −0.067 | −0.098 | −0.291** | −0.270** | −0.407*** | 0.045 |
| AUC C-peptide | −0.358*** | 0.149 | 0.040 | −0.225* | −0.276** | −0.373*** | 0.062 |
| Peak C-peptide | −0.347*** | 0.129 | 0.020 | −0.222* | −0.293** | −0.374*** | 0.047 |
| Index60 | 0.326** | 0.300 | 0.196 | 0.280** | 0.239* | 0.432*** | −0.002 |
Spearman correlations (r values) with OGTT outcomes for miRNAs associated with disease progression (see Table 3). Data are number (n) of participants with measurable miRNA levels
p<0.05,
p<0.01,
p<0.001
Fig. 3.
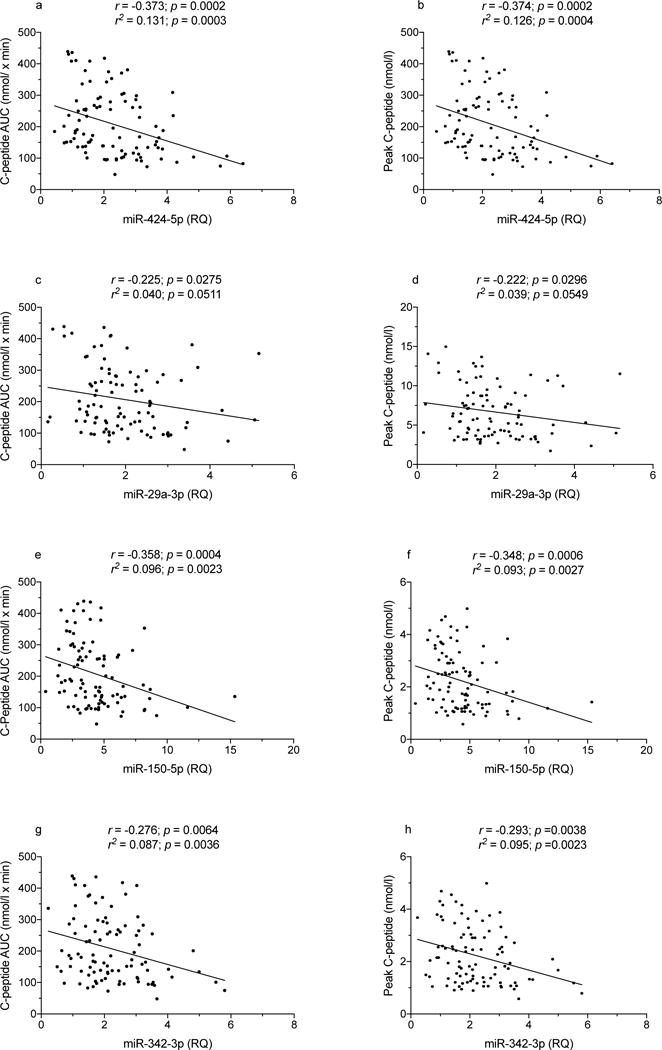
Spearman correlation of miR-424-5p, miR-29a-3p, miR-150-5p and miR-342-3p levels with C-peptide AUC and peak values in autoantibody-positive participants
Fig. 5.
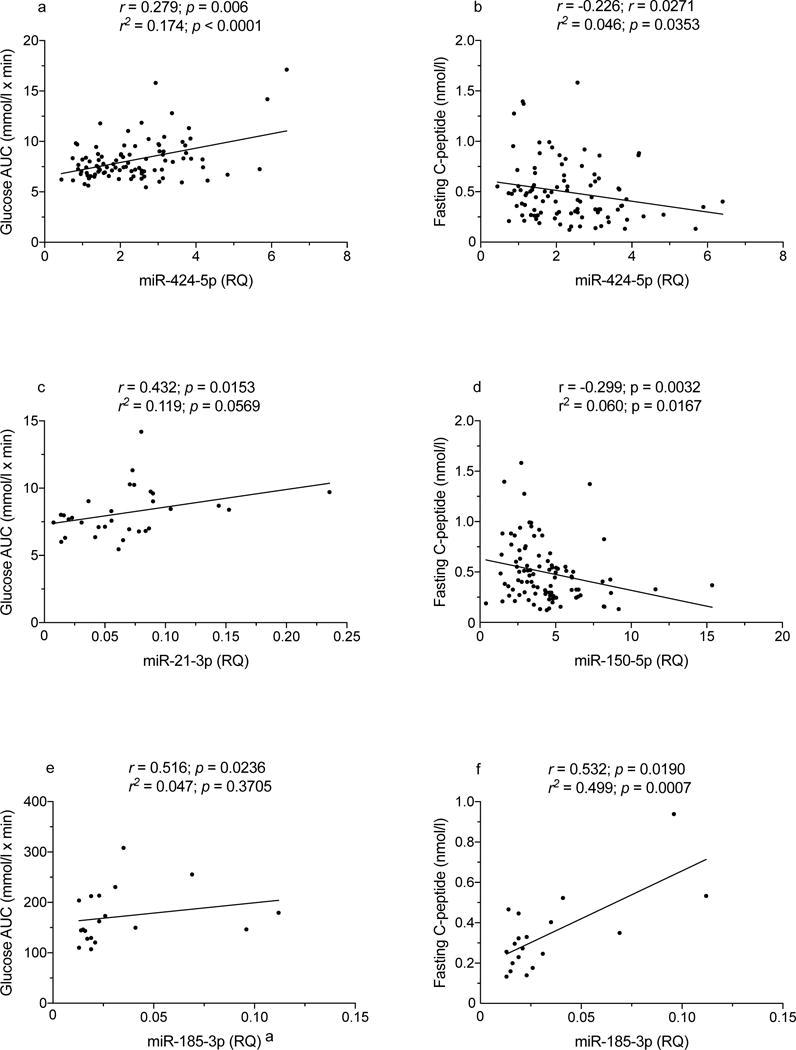
Spearman correlation of miR-424-5p, miR-21-3p, miR-150-5p and miR-185-3p levels with glucose AUC values in autoantibody-positive participants. (e) A single data point determined to be both a minor (inner) and a major (outer) outlier was removed from the analysis
Fig. 4.
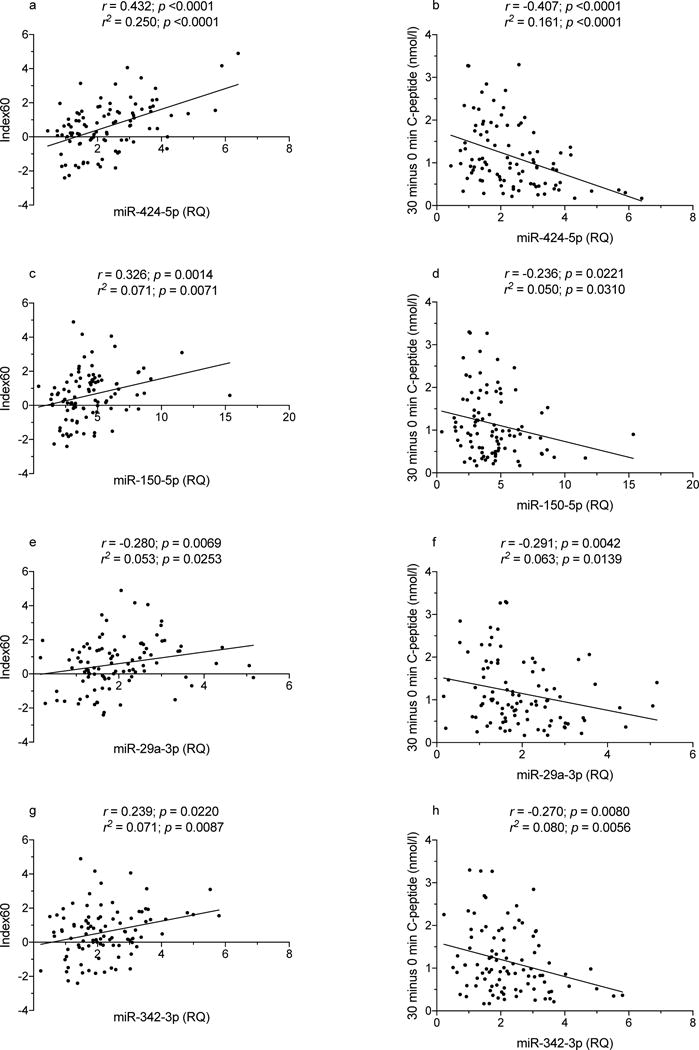
Spearman correlation of miR-424-5p, miR-150-5p, miR-29a-3p and miR-342-3p levels with Index60 and 30 minus 0 min C-peptide values in autoantibody-positive participants
miRNA target prediction and pathway enrichment analysis
We conducted an in silico investigation to determine which pathways and genes may be targeted by miR-21-3p, miR-29a-3p and miR-424-5p, since these showed the most robust associations with disease progression and correlations with OGTT outcomes; miRWalk 2.0 reported that miR-29a-3p and miR-424-5p gene targets were significantly enriched in the insulin signalling pathway (KEGG); these miRNAs are predicted to target 67 and 101 genes in the insulin signalling pathway, respectively (p=0.024 and p=2.4 × 10−10). miR-21-3p gene targets are also enriched in the insulin signalling pathway (WIKIPathways) with 75 predicted targets (p=0.00012).
Discussion
We investigated associations of circulating miRNAs with islet autoimmunity and progression to type 1 diabetes to determine their potential utility as biomarkers during the preclinical stages of the disease. To investigate the association with autoimmunity, we studied 150 autoantibody-positive relatives and compared them to their autoantibody-negative, family-matched siblings. This was a cohort of young individuals (mean age was 11 years); young age is associated with more rapid disease progression among autoantibody-positive relatives [31]. Moreover, among the autoantibody-positive participants, most (92%) had multiple autoantibodies, which confers higher disease risk than single autoantibody positivity [32]. The mean follow-up time from screening was 2.4 years, and the samples tested for miRNAs were obtained on average 1.8 years from screening; thus, our assessment of miRNAs was close to diagnosis for a subset of the participants studied (ESM Table 3). As none had clinical disease at testing, results are independent of symptomatic hyperglycaemia.
Fifteen miRNAs (Table 2) were differentially expressed in serum between the autoantibody-positive and autoantibody-negative siblings. The differences were modest and there was significant overlap in individuals’ levels between the groups. Accordingly, ROC curves returned modest AUC values. Thus, these miRNAs have limited value in predicting future risk of islet autoimmunity. However, we recognise that this was an initial, cross-sectional analysis and it is possible that more meaningful changes may be detected over time in longitudinal samples, for example samples spanning the transition from autoantibody negative to autoantibody positive.
Seven miRNAs were differentially expressed among autoantibody-positive relatives according to future development of diabetes (Table 3 and Figs 1 and 2); several of these miRNAs had a robust association with disease progression and correlation with OGTT indices of metabolic impairment (Table 6 and Figs 3-5). Similar correlations were reported in a cross-sectional study of individuals with type 1 or type 2 diabetes [16]. Three miRNAs (miR-21-3p, miR-424-5p and miR-29a-3p) showed the most robust associations and have the potential to be useful biomarkers. Levels above their respective ROC cut-off values conferred increased risk of future progression in relatives with multiple autoantibodies (Table 4), noting that generally it has been difficult to identify biomarkers that confer additional risk once the multiple autoantibody stage has been reached. Too few relatives with a single autoantibody were available to exclude that these miRNAs may not influence risk of progression at this earlier stage as well. Levels of several miRNAs were associated with risk of disease progression, including time to progression (Fig. 2); moreover, miRNA levels significantly correlated with OGTT indices of metabolic function, further supporting an association with disease processes and progression of diabetes, at least for an average period of 6 months, which was the follow-up time in this study. It is important that levels of certain miRNAs increased the risk of developing type 1 diabetes above the increased risk conferred by multiple autoantibodies, and this was observed in a young population with rapid progression, as shown by the steep survival curves (Fig. 2).
Published studies link these miRNAs to pathways relevant to type 1 diabetes. miR-21-3p targets histone deacetylase-8 mRNA [33]. Histone deacetylases are involved in inflammatory responses resulting in insulin resistance and beta cell failure, in particular those mediated by IL-1β, in both type 1 and type 2 diabetes [34], allelic variants of histone deacetylases are associated with susceptibility to type 1 diabetes [35], and inhibitors of these enzymes stimulate beta cell development, proliferation and differentiation [34]. miR-21-3p is produced in response to inflammatory stimuli and participates in paracrine signalling [36], which could explain its detection in the serum of autoantibody-positive relatives. The closely related miR-21-5p (which has the same stem–loop structure) has been linked to beta cell failure in vitro and in vivo [37, 38]; its manipulation prevents type 1 diabetes by blocking beta cell death [39], and its expression is altered in peripheral blood mononuclear cells (PBMCs) from individuals with type 1 diabetes [11] and serum from patients with type 1 or type 2 diabetes [16].
We previously reported miR-29a-3p levels enriched threefold in beta compared with alpha cells in human islets [40]. miR-29a-3p-deficient mice have dysregulated exocytosis, promoting diabetes after unfolded protein stress [41]. Increased glucose levels upregulated miR-29a-3p in rat and human islets, while miR-29a-3p inhibited glucose-stimulated insulin secretion and increased proliferation of INS-1E beta cells [42]. miR-29a-3p inhibits glucose-stimulated insulin secretion by targeting syntaxyn-1a, one of the two t-SNAREs (soluble NSF [N-ethylmaleimide sensitive fusion proteins] attachment protein receptor) involved in insulin exocytosis [43]. miR-29a-3p and miR-29b-3p affect insulin secretion by also silencing SLC16A1 mRNA in beta cells, which encodes for the monocarboxylate transporter-1 (MCT1) [44]. Levels of miR-29a-3p (as well as miR-29b-3p and miR-29c-3p) increase in the islets of NOD mice as islet autoimmunity progresses, and are upregulated in isolated mouse and human islets exposed to proinflammatory cytokines [14, 38]; overexpression of miR-29 miRNAs in MIN6 and dissociated islet cells impairs glucose-stimulated insulin secretion, related to decreased expression of the transcription factor Onecut2 (one cut homeobox 2) and the downstream elevation of an inhibitor of beta cell exocytosis, granuphilin [14]; overexpression of miR-29 miRNA also promotes apoptosis by decreasing the antiapoptotic protein Mcl1 (myeloid cell leukaemia sequence 1), a member of the Bcl2 family [14]. Taken together, changes in miR-29a-3p levels may reflect cytokine-mediated beta cell dysfunction occurring during the preclinical phases of type 1 diabetes. The elevated levels of miR-29a-3p we observed in autoantibody-positive relatives may reflect islet inflammation and the impairment of glucose-stimulated insulin secretion that becomes manifest as disease progresses, as suggested by the inverse correlation with the OGTT peak and AUC C-peptide values (Fig. 3); Nielsen et al. [23] have reported increased miR-29a-3p serum levels in individuals with type 1 diabetes.
Further interest in miR-21-3p and miR-29a-3p is spurred by the observation that their levels are altered in human islets on in vitro infection with a Coxsackie B virus (CBV5) [45]. Thus, increased levels of circulating miR-21-3p and miR-29a-3p in autoantibody-positive relatives may reflect enterovirus infections in the pancreatic islets [1]. Future studies should investigate how inflammatory stimuli and viruses influence the levels of miR-21-3p, miR-29a-3p and other miRNAs, as well as the downstream effects on gene expression and beta cell function, and possible correlations with serum markers of beta cell dysfunction (proinsulin/C-peptide ratio) [46] and death (e.g. levels of demethylated insulin gene DNA) [47].
There is little experimental evidence to link miR-424-5p to type 1 diabetes; however, in silico statistically significant predictions suggest a role for miR-424-5p in regulating many genes in the insulin signalling pathway, including those coding for the insulin receptor and its substrates 1 and 2. We found statistically significant evidence that targets of miR-29a-3p and miR-21-3p are also enriched in insulin signalling. miR-150 was altered in PBMCs from individuals with type 1 diabetes [48]. miR-342 targets the PTPRN and PTPRN2 mRNAs [49], encoding for the IA-2 and IA-2β autoantigens; miR-342-5p is included in the imprinted 14q32 miRNA cluster, in which 32/56 miRNAs are predicted to target autoantigen mRNAs and could modulate their expression. MiR-342-5p suppresses Coxsackie virus B3 biosynthesis by targeting the 2C-coding region [50]. Among the miRNAs we found associated with islet autoimmunity, miR-25-3p was linked to type 1 diabetes by Nielsen et al. [23].
In closing, the analysis of a large, family-matched cohort of TrialNet relatives provides evidence that serum levels of certain miRNAs reflect islet autoimmunity and its progression; these miRNAs are candidates for further validation as novel biomarkers. Several of the miRNAs associated with progression correlated with OGTT indices of beta cell function. Longitudinal studies will help in better examining the association between these (and other) circulating miRNAs and disease progression, as levels could change over time. Further studies may identify more predictive miRNA combinations, or additional associations with other variables of disease progression (demethylated insulin gene DNA levels, gene expression profiles, or other immunological and metabolic readouts), which could be combined into composite biomarkers and risk scores.
Supplementary Material
Acknowledgments
We acknowledge the support of the Type 1 Diabetes TrialNet Pathway to Prevention Study Group, which identified study participants and provided samples and follow-up data for this study. The Type 1 Diabetes TrialNet Pathway to Prevention Study Group is a clinical trials network funded by the National Institutes of Health (NIH) through the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institute of Allergy and Infectious Diseases, and the Eunice Kennedy Shriver National Institute of Child Health and Human Development, through cooperative agreements U01 DK061010, U01 DK061034, U01 DK061042, U01 DK061058, U01 DK085465, U01 DK085453, U01 DK085461, U01 DK085466, U01 DK085499, U01 DK085504, U01 DK085509, U01 DK103180, U01 DK103153, U01 DK085476, U01 DK103266, U01 DK103282, U01 DK106984, U01 DK106994, U01 DK107013, U01 DK107014, UC4 DK106993, and the JDRF. The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the NIH or the JDRF.
The Sylvester Cancer Center Oncogenomics Core, University of Miami, USA supported the study; we acknowledge the support of T. Guettouche and S. Williams, who overviewed the services provided by the core.
Funding Specific funding for this study was provided by the JDRF (SRA-17-2011-293) and the Diabetes Research Institute Foundation, Hollywood, FL, USA. We thank S. Ahmed (JDRF) for programmatic support.
Abbreviations
- IA-2
Insulinoma-associated antigen 2
- miRNA
MicroRNA
- PBMC
Peripheral blood mononuclear cell
- ROC
Receiver operating characteristic
- RQ
Relative quantification
- ZnT8
Zinc transporter 8
Footnotes
Data availability According to the policies of the Type 1 Diabetes TrialNet (www.diabetestrialnet.org), data used for a publication are released by TrialNet to the National Institute of Diabetes and Digestive and Kidney Diseases Repository (www.niddkrepository.org) 12 months after the manuscript has appeared in print. This is to allow time for publication of related manuscripts using the same dataset.
Duality of interest The authors declare that there is no duality of interest associated with this manuscript.
Contribution statement JS, RLP, SMC and AP conceived the study or parts of the study. SMC was responsible for statistical design and the analysis plan. IVS and GA generated the data. IVS, GA, JS, RLP, SMC and AP analysed and the interpreted the data. All authors participated in preparation of the manuscript and approved the final version for publication. AP is the guarantor of the work.
References
- 1.Pugliese A. The multiple origins of type 1 diabetes. Diabet Med. 2013;30:135–146. doi: 10.1111/dme.12081. [DOI] [PubMed] [Google Scholar]
- 2.Yates LA, Norbury CJ, Gilbert RJ. The long and short of microRNA. Cell. 2013;153:516–519. doi: 10.1016/j.cell.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Cobb BS, Hertweck A, Smith J, et al. A role for Dicer in immune regulation. J Exp Med. 2006;203:2519–2527. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poy MN, Eliasson L, Krutzfeldt J, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 5.Correa-Medina M, Bravo-Egana V, Rosero S, et al. MicroRNA miR-7 is preferentially expressed in endocrine cells of the developing and adult human pancreas. Gene Expr Patterns. 2009;9:193–199. doi: 10.1016/j.gep.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 6.Joglekar MV, Joglekar VM, Hardikar AA. Expression of islet-specific microRNAs during human pancreatic development. Gene Expr Patterns. 2009;9:109–113. doi: 10.1016/j.gep.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Joglekar MV, Parekh VS, Mehta S, Bhonde RR, Hardikar AA. MicroRNA profiling of developing and regenerating pancreas reveal post-transcriptional regulation of neurogenin3. Dev Biol. 2007;311:603–612. doi: 10.1016/j.ydbio.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 8.Belgardt BF, Ahmed K, Spranger M, et al. The microRNA-200 family regulates pancreatic beta cell survival in type 2 diabetes. Nature Medicine. 2015;21:619–627. doi: 10.1038/nm.3862. [DOI] [PubMed] [Google Scholar]
- 9.Sebastiani G, Po A, Miele E, et al. MicroRNA-124a is hyperexpressed in type 2 diabetic human pancreatic islets and negatively regulates insulin secretion. Acta Diabetol. 2015;52:523–530. doi: 10.1007/s00592-014-0675-y. [DOI] [PubMed] [Google Scholar]
- 10.Sebastiani G, Grieco FA, Spagnuolo I, Galleri L, Cataldo D, Dotta F. Increased expression of microRNA miR-326 in type 1 diabetic patients with ongoing islet autoimmunity. Diabetes Metab Res Rev. 2011;27:862–866. doi: 10.1002/dmrr.1262. [DOI] [PubMed] [Google Scholar]
- 11.Salas-Perez F, Codner E, Valencia E, Pizarro C, Carrasco E, Perez-Bravo F. MicroRNAs miR-21a and miR-93 are down regulated in peripheral blood mononuclear cells (PBMCs) from patients with type 1 diabetes. Immunobiology. 2013;218:733–737. doi: 10.1016/j.imbio.2012.08.276. [DOI] [PubMed] [Google Scholar]
- 12.Hezova R, Slaby O, Faltejskova P, et al. microRNA-342, microRNA-191 and microRNA-510 are differentially expressed in T regulatory cells of type 1 diabetic patients. Cell Immunol. 2010;260:70–74. doi: 10.1016/j.cellimm.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 13.Mi QS, He HZ, Dong Z, Isales C, Zhou L. microRNA deficiency in pancreatic islet cells exacerbates streptozotocin-induced murine autoimmune diabetes. Cell Cycle. 2010;9:3127–3129. doi: 10.4161/cc.9.15.12596. [DOI] [PubMed] [Google Scholar]
- 14.Roggli E, Gattesco S, Caille D, et al. Changes in microRNA expression contribute to pancreatic beta-cell dysfunction in prediabetic NOD mice. Diabetes. 2012;61:1742–1751. doi: 10.2337/db11-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osmai M, Osmai Y, Bang-Berthelsen CH, et al. MicroRNAs as regulators of beta-cell function and dysfunction. Diabetes/metabolism research and reviews. 2016;32:334–349. doi: 10.1002/dmrr.2719. [DOI] [PubMed] [Google Scholar]
- 16.Seyhan AA, Nunez Lopez YO, Xie H, et al. Pancreas-enriched miRNAs are altered in the circulation of subjects with diabetes: a pilot cross-sectional study. Scientific reports. 2016;6:31479. doi: 10.1038/srep31479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR) Methods (San Diego, Calif) 2010;50:298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang K, Zhang S, Marzolf B, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A. 2009;106:4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 20.Guay C, Regazzi R. Circulating microRNAs as novel biomarkers for diabetes mellitus. Nat Rev Endocrinol. 2013;9:513–521. doi: 10.1038/nrendo.2013.86. [DOI] [PubMed] [Google Scholar]
- 21.Latreille M, Herrmanns K, Renwick N, et al. miR-375 gene dosage in pancreatic beta-cells: implications for regulation of beta-cell mass and biomarker development. Journal of molecular medicine (Berlin, Germany) 2015;93:1159–1169. doi: 10.1007/s00109-015-1296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanak MA, Takita M, Shahbazov R, et al. Evaluation of microRNA375 as a novel biomarker for graft damage in clinical islet transplantation. Transplantation. 2015;99:1568–1573. doi: 10.1097/TP.0000000000000625. [DOI] [PubMed] [Google Scholar]
- 23.Nielsen LB, Wang C, Sorensen K, et al. Circulating levels of microRNA from children with newly diagnosed type 1 diabetes and healthy controls: evidence that miR-25 associates to residual beta-cell function and glycaemic control during disease progression. Exp Diabetes Res. 2012;2012:896362. doi: 10.1155/2012/896362. Epub 2012 Jul 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchand L, Jalabert A, Meugnier E, et al. miRNA-375 a sensor of glucotoxicity is altered in the serum of children with newly diagnosed type 1 diabetes. Journal of diabetes research. 2016;2016:1869082. doi: 10.1155/2016/1869082. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahon JL, Sosenko JM, Rafkin-Mervis L, et al. The TrialNet Natural History Study of the development of type 1 Diabetes: objectives, design, and initial results. Pediatr Diabetes. 2009;10:97–104. doi: 10.1111/j.1399-5448.2008.00464.x. [DOI] [PubMed] [Google Scholar]
- 26.Lampasona V, Schlosser M, Mueller PW, et al. Diabetes antibody standardization program: first proficiency evaluation of assays for autoantibodies to zinc transporter 8. Clin Chem. 2011;57:1693–1702. doi: 10.1373/clinchem.2011.170662. [DOI] [PubMed] [Google Scholar]
- 27.Mestdagh P, Van Vlierberghe P, De Weer A, et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome biology. 2009;10:R64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mukherjee SN, Sykacek P, Roberts SJ, Gurr SJ. Sigkdd Explorations. 2003. Gene ranking using bootstrapped p-values; pp. 14–18. [Google Scholar]
- 29.Sosenko JM, Palmer JP, Rafkin LE, et al. Trends of earlier and later responses of C-peptide to oral glucose challenges with progression to type 1 diabetes in diabetes prevention trial-type 1 participants. Diabetes Care. 2010;33:620–625. doi: 10.2337/dc09-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sosenko JM, Skyler JS, DiMeglio LA, et al. A new approach for diagnosing type 1 diabetes in autoantibody-positive individuals based on prediction and natural history. Diabetes care. 2015;38:271–276. doi: 10.2337/dc14-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wherrett DK, Chiang JL, Delamater AM, et al. Defining pathways for development of disease-modifying therapies in children with type 1 diabetes: a consensus report. Diabetes care. 2015;38:1975–1985. doi: 10.2337/dc15-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ziegler AG, Rewers M, Simell O, et al. Seroconversion to multiple islet autoantibodies and risk of progression to diabetes in children. JAMA. 2013;309:2473–2479. doi: 10.1001/jama.2013.6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan M, Chen C, Gong W, et al. miR-21-3p regulates cardiac hypertrophic response by targeting histone deacetylase-8. Cardiovascular research. 2015;105:340–352. doi: 10.1093/cvr/cvu254. [DOI] [PubMed] [Google Scholar]
- 34.Christensen DP, Dahllof M, Lundh M, et al. Histone deacetylase (HDAC) inhibition as a novel treatment for diabetes mellitus. Molecular medicine (Cambridge, Mass) 2011;17:378–390. doi: 10.2119/molmed.2011.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nerup J, Pociot F. A genomewide scan for type 1-diabetes susceptibility in Scandinavian families: identification of new loci with evidence of interactions. Am J Hum Genet. 2001;69:1301–1313. doi: 10.1086/324341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bang C, Batkai S, Dangwal S, et al. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. Journal of clinical investigation. 2014;124:2136–2146. doi: 10.1172/JCI70577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roggli E, Britan A, Gattesco S, et al. Involvement of microRNAs in the cytotoxic effects exerted by proinflammatory cytokines on pancreatic beta-cells. Diabetes. 2010;59:978–986. doi: 10.2337/db09-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bravo-Egana V, Rosero S, Klein D, et al. Inflammation-mediated regulation of microRNA expression in transplanted pancreatic islets. Journal of transplantation. 2012;2012:723614. doi: 10.1155/2012/723614. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruan Q, Wang T, Kameswaran V, et al. The microRNA-21-PDCD4 axis prevents type 1 diabetes by blocking pancreatic beta cell death. Proc Natl Acad Sci U S A. 2011;108:12030–12035. doi: 10.1073/pnas.1101450108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klein D, Misawa R, Bravo-Egana V, et al. MicroRNA expression in alpha and beta cells of human pancreatic islets. PLoS One. 2013;8:e55064. doi: 10.1371/journal.pone.0055064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dooley J, Garcia-Perez JE, Sreenivasan J, et al. The microRNA-29 family dictates the balance between homeostatic and pathological glucose handling in diabetes and obesity. Diabetes. 2016;65:53–61. doi: 10.2337/db15-0770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bagge A, Clausen TR, Larsen S, et al. MicroRNA-29a is up-regulated in beta-cells by glucose and decreases glucose-stimulated insulin secretion. Biochemical and biophysical research communications. 2012;426:266–272. doi: 10.1016/j.bbrc.2012.08.082. [DOI] [PubMed] [Google Scholar]
- 43.Bagge A, Dahmcke CM, Dalgaard LT. Syntaxin-1a is a direct target of miR-29a in insulin-producing beta-cells. Hormone and metabolic research. 2013;45:463–466. doi: 10.1055/s-0032-1333238. [DOI] [PubMed] [Google Scholar]
- 44.Pullen TJ, da Silva Xavier G, Kelsey G, Rutter GA. miR-29a and miR-29b contribute to pancreatic beta-cell-specific silencing of monocarboxylate transporter 1 (Mct1) Molecular and cellular biology. 2011;31:3182–3194. doi: 10.1128/MCB.01433-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim KW, Ho A, Alshabee-Akil A, et al. Coxsackievirus B5 infection induces dysregulation of microRNAs predicted to target known type 1 diabetes risk genes in human pancreatic islets. Diabetes. 2016;65:996–1003. doi: 10.2337/db15-0956. [DOI] [PubMed] [Google Scholar]
- 46.Sims EK, Chaudhry Z, Watkins R, et al. Elevations in the fasting serum proinsulin-to-C-peptide ratio precede the onset of type 1 diabetes. Diabetes care. 2016;39:1519–1526. doi: 10.2337/dc15-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akirav EM, Lebastchi J, Galvan EM, et al. Detection of beta cell death in diabetes using differentially methylated circulating DNA. Proc Natl Acad Sci U S A. 2011;108:19018–19023. doi: 10.1073/pnas.1111008108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Estrella S, Garcia-Diaz DF, Codner E, Camacho-Guillen P, Perez-Bravo F. Expression of miR-22 and miR-150 in type 1 diabetes mellitus: Possible relationship with autoimmunity and clinical characteristics. Medicina clinica. 2016;147:245–247. doi: 10.1016/j.medcli.2016.05.016. [DOI] [PubMed] [Google Scholar]
- 49.Abuhatzira L, Xu H, Tahhan G, Boulougoura A, Schaffer AA, Notkins AL. Multiple microRNAs within the 14q32 cluster target the mRNAs of major type 1 diabetes autoantigens IA-2, IA-2beta, and GAD65. FASEB journal. 2015;29:4374–4383. doi: 10.1096/fj.15-273649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L, Qin Y, Tong L, et al. MiR-342-5p suppresses coxsackievirus B3 biosynthesis by targeting the 2C-coding region. Antiviral research. 2012;93:270–279. doi: 10.1016/j.antiviral.2011.12.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


