Abstract
Objectives
Deregulated production of IL-17 and IL-21 contributes to the pathogenesis of autoimmune disorders like SLE and RA. Production of IL-17 and IL-21 can be regulated by ROCK2, one of the two Rho kinases. Increased ROCK activation was previously observed in an SLE cohort. Here, we evaluated ROCK activity in a new SLE cohort, an RA cohort, and assessed the ability of distinct inhibitors of the ROCK pathway to suppress production of IL-17 and IL-21 by SLE T cells or human Th17 cells.
Methods
ROCK activity in PBMCs from 29 SLE patients, 31 RA patients, and 28 healthy controls was determined by ELISA. SLE T cells or in vitro-differentiated Th17 cells were treated with Y27632 (a pan-ROCK inhibitor), KD025 (a selective ROCK2 inhibitor), or simvastatin (which inhibits RhoA, a major ROCK activator). ROCK activity, IL-17, and IL-21 production were assessed. The transcriptional profile altered by ROCK inhibitors was evaluated by NanoString technology.
Results
ROCK activity levels were significantly higher in SLE and RA patients than healthy controls. Th17 cells exhibited high ROCK activity that was inhibited by Y276327, KD025, or simvastatin; each also decreased IL-17 and IL-21 production by purified SLE T cells or Th17 cells. Immune profiling revealed both overlapping and distinct effects of the different ROCK inhibitors.
Conclusions
ROCK activity is elevated in PBMCs from SLE and RA patients. Production of IL-17 and IL-21 by SLE T cells or Th17 cells can furthermore be inhibited by targeting the RhoA-ROCK pathway via both non-selective and selective approaches.
Keywords: Systemic Lupus Erythematosus, T cells, Autoimmunity, Rho Kinases
INTRODUCTION
Aberrant expansion and dysregulation of several TH effector subsets including Th17 and TFH cells can be associated with autoimmune disorders such as Systemic Lupus Erythematosus (SLE) and Rheumatoid Arthritis (RA).1–4 The pathogenicity of these cells has been linked to their ability to produce cytokines such as IL-17 and IL-21. Several transcription factors have been implicated in the regulation of Th17 differentiation.5 Notably, Th17 differentiation and the production of IL-17 and IL-21 are critically dependent on the presence of IRF4, a member of the IRF (interferon regulatory factor) family of transcription factors.6–8 The ability of IRF4 to drive high-levels of IL-17 and IL-21 production can be regulated via its phosphorylation by ROCK2.9 ROCK2 and the other member of the Rho kinase family, ROCK1, are two serine/threonine kinases whose activity is primarily controlled by binding of activated RhoA.10–13 The RhoA-ROCK pathway regulates multiple biological functions, including cytoskeletal reorganization, proliferation, and differentiation.10–13
While dysregulated activation of the RhoA-ROCK pathway in non-hematopoietic cells is known to contribute to cardiovascular, renal, and neurological disorders,14–16 recent work has also implicated aberrant ROCK activation in the pathogenesis of autoimmune disorders.17 In line with the ability of ROCK2 to control IRF4 function and the production of IL-17 and IL-21, ROCK2 is selectively activated in murine and human Th17 cells and aberrant ROCK2 activation has been observed in murine lupus T cells.9–18 Furthermore, administration of a pan-ROCK inhibitor, Fasudil, diminishes IL-17 and IL-21 production in vivo and ameliorates disease in spontaneous murine models of lupus.9–19 Consistent with these observations, SLE T cells, including T cells infiltrating the kidneys, exhibit increased phosphorylation of ERM proteins, a ROCK target, and the ROCK-mediated effects can be promoted by PP2A, a phosphatase expressed at higher levels in SLE T cells than T cells from healthy controls.20–21 An initial pilot study, furthermore, directly demonstrated enhanced ROCK activity in PBMCs from ≈60% of SLE patients suggesting that inhibition of this pathway represents a potential therapeutic target for SLE and potentially other autoimmune diseases like RA.22
The attractiveness of the RhoA-ROCK pathway as a therapeutic target for SLE is further strengthened by the availability of a wide array of ROCK inhibitors.23–27 Most ROCK inhibitors, such as the well-known Fasudil and Y27632, target the ATP-binding pocket of the ROCKs and, due to the high degree of homology in the kinase domain of ROCK1 and ROCK2, are thus non-isoform selective. Selective oral ROCK2 inhibitors, such as KD025 (formerly known as Slx-2119) that demonstrates 100-fold more selectivity toward ROCK2 than ROCK1, have, however, also been developed.18,28 Inhibition of ROCK activation is also a key mechanisms underlying the pleiotropic effects of statins since by inhibiting HMG-CoA reductase, statins interfere with RhoA activation and, consequently, decrease ROCK activity.29 Here, we systematically compared the ability of different classes of ROCK inhibitors to decrease IL-17 and IL-21 production by either SLE T cells or in vitro differentiated human Th17 cells. Additionally, we evaluated the broader transcriptional effects of these different inhibitors on human Th17 cells by NanoString technology.
METHODS
Human Subjects
SLE subjects
29 SLE patients followed at the Hospital for Special Surgery (HSS) were consented and enrolled. Inclusion criteria were: age 18-65 years and classification of SLE according to the revised 1997 ACR criteria.30 Exclusion criteria were: statin use, active infection by history, pregnancy, pulse glucocorticoids within 1 month, cyclophosphamide within 3 months, rituximab within 1 year, any anti-TNF therapy within 3 months. Clinical data including laboratory parameters, medications, complications, and serology were collected. Disease activity was assessed at time of sampling blood by the Systemic Lupus Erythematosus Disease Activity Index SELENA modification (SELENA-SLEDAI).31 5 of 29 SLE patients were taking ACE inhibitors or ARBs.
RA subjects
31 RA patients from HSS were consented and enrolled. Inclusion criteria: age >18 years and meeting ACR/EULAR 2010 RA classification criteria.32 Exclusion criteria were: active infection, pregnancy, recent surgery, history of Raynaud’s, or another autoimmune diagnosis. Clinical data including laboratory parameters, medications, complications, and serology were collected. Disease activity was assessed at time of sampling using the clinical disease activity index (CDAI).33 Radiographs of hands and wrists were also reviewed, if available, to determine presence or absence of erosions. RA patients were treated at the discretion of their doctor, and specific medications included one or more of the following: hydroxychloroquine (HCQ), leflunomide (LEF), methotrexate (MTX), etanercept (ETN), adalimumab (ADA), systemic corticosteroids (CS) and nonsteroidal anti-inflammatory drugs (NSAIDS). 4 of 31 RA patients were taking ACE inhibitors or ARBs.
Healthy controls
24 sex-age-ethnically similar healthy controls were consented and enrolled at HSS. 4 healthy controls samples were provided by the Genotype and Phenotype Registry, a service of the Tissue Donation Program at The Feinstein Institute for Medical Research, Manhasset, NY, USA (supported by NIH grant 5RC2AR05092). Healthy controls were age >18 years. Exclusion criteria were: active infection, pregnancy, recent surgery, history of Raynaud’s, or another known diagnosis of an autoimmune disease. Of the healthy controls, 9 matched by age (±5 years), sex and ethnicity to the SLE patients, 13 matched to the RA patients and 6 matched to both SLE and RA.
The HSS Institutional Review Board approved the study, and all blood donors provided written informed consent.
Samples
PBMCs
Heparinized venous blood samples (10-20ml) were obtained by hospital phlebotomists and processed immediately. Peripheral blood mononuclear cells (PBMCs) were isolated from each subject, whole cell extracts obtained, and then frozen. At a later date extracts were thawed and ROCK assays performed on groups of samples.
T cell cultures
SLE CD4+ T cells were negatively selected by MACS (Miltenyi). Cells were plated at 2×105 cells/well in X-VIVO15 media (Lonza) and stimulated with soluble αCD28 (1 μg/ml) and plate-bound αCD3 (5 μg/ml; OKT3 clone) for 4 days. In separate experiments, cryopreserved human cord blood CD4+ T lymphocytes from different donors were obtained from Allcells, LLC. CD4+ T cell purity was >95%. Cells were thawed according to manufacturer’s instructions and cultured as described above. For Th17 differentiation, medium was supplemented with TGF-β1 (5 ng/ml), IL-1β (10 ng/ml), IL-6 (20 ng/ml), and IL-23 (50 ng/ml). Y27632 60-90μM (EMD), KD025 5μM (provided by Kadmon Corporation) and simvastatin 0.1-0.2μM (Calbiochem) were added at day 1. Supernatants were analyzed for IL-17 (Biolegend), IL-21 (eBioscience), CCL20 (R&D Systems), and IFN-γ (Biolegend) levels by ELISA.
ROCK Activity Assays and Western blotting
ROCK activity in extracts was measured using the 96-well ROCK Activity Assay Kit (Cell Biolabs, Inc.) as described.22 Active ROCK2 (1-4ng) served as a positive control. IRF4 phosphorylation was detected with a phospho-IRF4 Ab9 and total levels of IRF4 with a goat anti-IRF4 antibody (M-17; Santa Cruz Biotechnology, Inc.). Blots were also probed for either phospho-Stat3 (Y705; #9131; Cell Signaling) or total Stat3 (Cat: 610189; BD Transduction Labs).
RESULTS
Increased ROCK kinase activity in SLE and RA PBMCs compared to HC PBMCs
In an initial cross-sectional study, we had observed that ≈60% of SLE patients exhibited increased ROCK kinase activity as compared to healthy controls.22 To confirm these findings we investigated ROCK kinase activity in a second SLE cohort (Table 1). ROCK activity was significantly elevated in SLE PBMCs as compared to PBMCs from healthy controls (0.457 [IQR: 0.368-0.743] versus 0.278 [IQR: 0.230-0.332] respectively, p<0.0001, MW) (Figure 1A). PBMCs from ≈70% of SLE patients from this second cohort displayed ROCK activity levels that were 2SD above the mean of the ROCK activity levels observed in PBMCs from healthy controls. Thus SLE patients from two independent cohorts demonstrate increased ROCK activity.
Table 1.
Summary of demographic data and patient characteristics
| Parameter | SLE Patients N=29 |
RA Patients N=31 |
Healthy Controls N=28 |
|---|---|---|---|
| Mean age, years ± SD | 38 ± 11 | 59 ± 14 | 46 ± 15 |
| Females, N (%) | 27 (93) | 27 (87) | 25 (89) |
| Males, N (%) | 2 (7) | 4 (13) | 3 (11) |
| Ethnicity | |||
| African American, N (%) | 9 (31) | 4 (13) | 4 (14) |
| Asian, N (%) | 2 (7) | 6 (19) | 3 (11) |
| Caucasian, N (%) | 5 (21) | 17 (55) | 13 (46) |
| Hispanic, N (%) | 12 (41) | 4 (13) | 8 (29) |
| SLEDAI ± SD | 6 ± 4 | ||
| Nephritis, N (%) | 13 (45) | ||
| Mean ESR (mm/hr) ± SD | 28.0 ± 21.3 | ||
| Mean CRP (mg/dL) ±SD | 2.16 ± 3.92 | ||
| CDAI, Mean ± SD | 13.8 ± 7.52 |
Figure 1. Increased ROCK kinase activity in SLE and RA PBMCs compared to HC PBMCs.
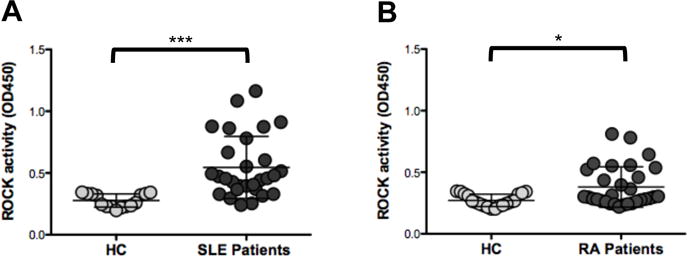
PBMCs were isolated from heparinized blood samples of 29 SLE patients (A) and 31 RA patients (B). PBMCs f rom healthy controls of similar ethnicity/race, sex and age ± 5 years (Table 1) for each of the disease group were also assayed. Whole cell extracts were prepared and ROCK activation was assessed by an ELISA-based ROCK kinase activity assay. Active ROCK2 (1-4ng) was used a positive control. Data were analyzed by Kruskal-Wallis test (χ2 = 26.25, p<0.0001) followed by the Dunns Multiple Comparison analysis (HC vs RA p= 0.087, RA vs SLE **p<0.01, HC vs SLE ***p<0.001).
To assess whether elevated ROCK activation was specific for SLE or could also be observed in another autoimmune disease, we measured ROCK activity levels in PBMCs from 31 RA patients and 19 healthy controls of similar demographics with respect to age, gender, and race/ethnicity (Table 1). As compared to PBMCs from healthy controls, PBMCs from RA patients also expressed significantly higher median ROCK activity levels (0.299 [IQR: 0.264-0.522] versus 0.259 [IQR: 0.229-0.324] respectively, p<0.015, MW) (Figure 1B) although the extent of ROCK activation in RA PBMCs was not as great as that observed in some of the SLE PBMCs.
Effects of different ROCK inhibitors on the production of IL-17A and IL-21 by SLE T cells
Cytokine production by cord blood-derived Th17 cells can be decreased by Y27632, a nonselective ROCK inhibitor.22 To assess whether production of IL-17 and IL-21 by SLE T cells is also ROCK-dependent, we next purified CD4+ T cells from PBMCs of SLE patients and stimulated them ±Y27632 (Figure 2A–C). Given that ROCK activity can also be inhibited by statins via effects on RhoA, we also compared the capacity of Y27632 and simvastatin to decrease the production of IL-17A and IL-21 by SLE T cells (Figure 2A–C). Both Y27632 (at 90μM) and simvastatin (at 2μM) diminished the production of IL-17A and IL-21 by SLE T cells while no significant effects were observed on the production of IFN-γ (Figure 2A–C). Since simvastatin and Y27632 interfere with ROCK activation by different mechanisms, we also evaluated the effect of combining Y27632 and simvastatin (Figure 2A and Figure 2C). Addition of simvastatin to the lower dose of Y27632 (30μM) did not further increase the inhibition of IL-17A or IL-21 production (Figure 2A and Figure 2C). Thus both simvastatin and the nonselective ROCK inhibitor decrease the production of IL-17A and IL-21 by SLE T cells.
Figure 2. Decreased hIL-17A, hIL-21, and hIFN-γ production by SLE CD4+ T cells treated with ROCK inhibitors.
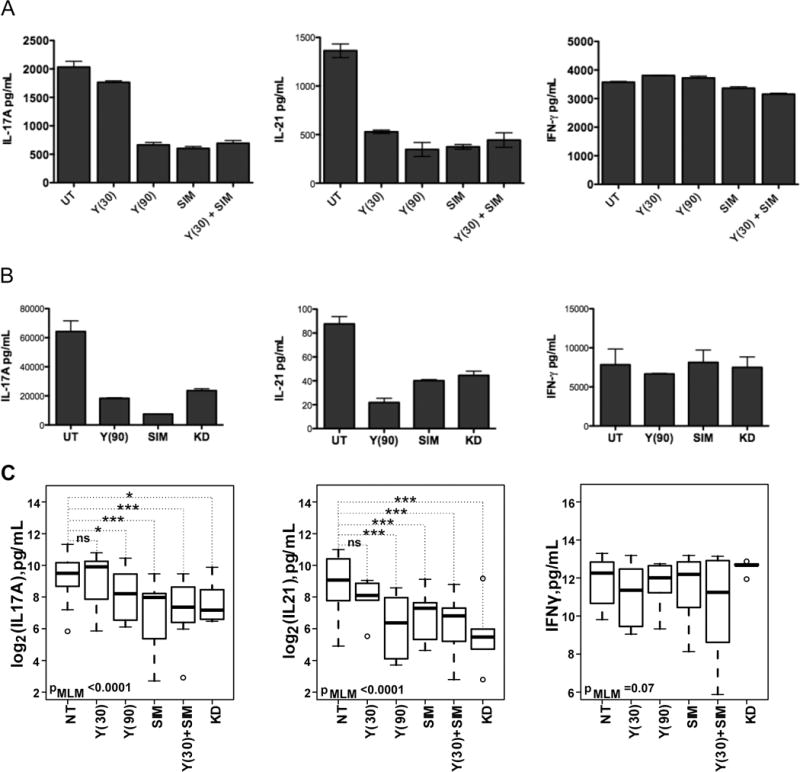
CD4+ T cells were isolated from SLE PBMCs and cultured with plate-bound anti-CD3 and anti-CD28 stimulation in X-VIVO15 media. One day later, CD4+ T cells were treated with (A) Y27632, simvastatin or Y27632 plus simvastatin or (B) Y27632 or simvastatin or KD025. Supernatants and cells were collected on day 4 and super atant levels of hIL-17, hIL-21 and hIFN-γ production were assessed. Shown is one representative experiment. (C) The inhibition of hIL-17A, hIL-21 and hCCL20 production by each inhibitor and the combination thereof was fitted to a mixed linear model. P-values from the likelihood ratio test comparing the null model with only the random effect of patients with the complete model that includes all treatments is shown on each panel. The p-values for individual treatment were calculated as described in the Materials and Methods Y(30): 30μM Y-27632, Y(90): 90μM Y-27632, SIM: 0.2μM simvastatin, KD: 5.0μM KD025.
In view of the recent availability of a selective ROCK2 inhibitor, KD025, we next compared the effects of KD025 to those of the pan-ROCK inhibitor, Y27632, and simvastatin on the production of IL-17A and IL-21. The ROCK2 selective inhibitor was also effective in diminishing the production of IL-17A (t=−2.57, p=0.014) and IL-21 (t=−4.08, p=0.0004). No effects on the production of IFN-γ were again observed (t=−0.27,p=0.7827). Thus, the production of IL-17A and IL-21 by SLE T cells is broadly amenable to inhibition by different ROCK inhibitors.
Effects of different ROCK inhibitors on cytokine production by differentiated Th17 cells
To evaluate whether different classes of ROCK inhibitors could also interfere with the production of IL-17A and IL-21 by non-autoimmune T cells differentiated under Th17 conditions we first compared the capacity of Y27632 and simvastatin to decrease the production of IL-17A, IL-21, and CCL20 by cord blood CD4+ T cells cultured under Th17-skewing conditions (Figure 3A and Figure 3C). Consistent with our prior results,22 Y27632 at 90μM was highly effective in inhibiting IL-17A production (t=−5.59, p<0.0001,MLM, Figure 3A–C). 60μM of Y27632 could also inhibit IL-17A production albeit to a lesser degree (t=−2.58, p=0.016,MLM, Figure 3A–C). As previously observed,22 IL-21 production was also sensitive to the inhibitory effects of Y27632 and both 60μM of Y27632 (t=−3.32, p=0.004) and 90μM of Y27632 (t=−4.96, p<0.0001) were highly effective in diminishing IL-21 levels (Figure 3A–C). The production of CCL20 was also decreased by both doses of Y27632 (60μM: t=−2.6, p=0.016; 90 μM: t=−4.49, p=0.0002) although this effect was less marked and the variance was larger compared to the inhibitory effects exerted on IL-21 production (Figure 3A–C).
Figure 3. ROCK inhibitors decrease hIL-17A, hIL-21 and hCCL20 production by in vitro differentiated Th17 cells.
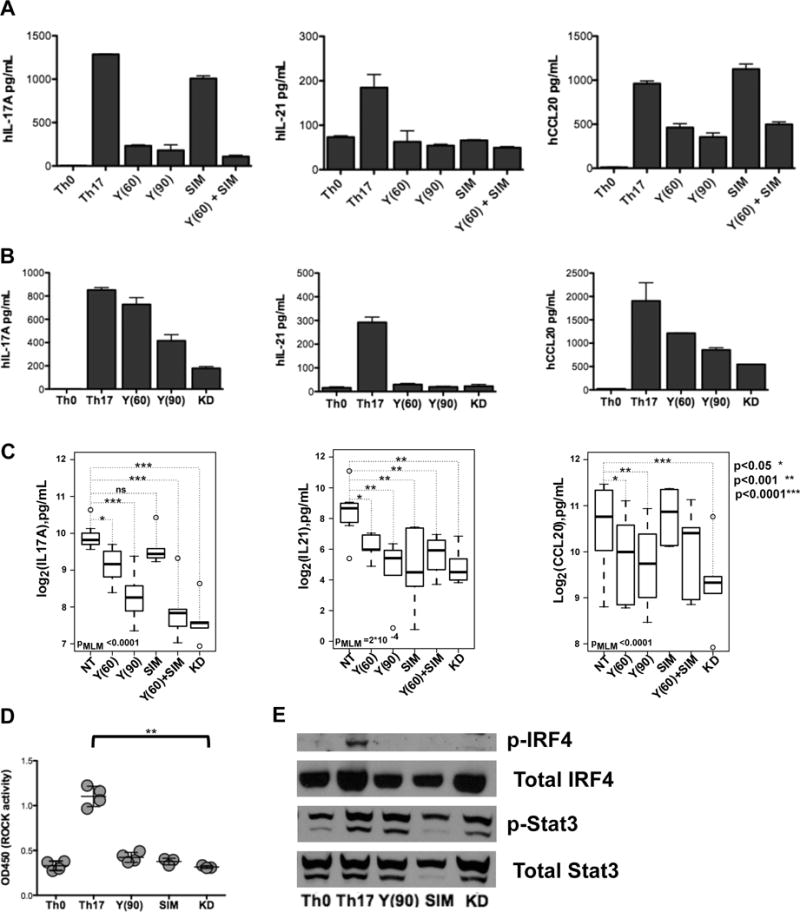
Human umbilical cord CD4+ T cells (106 cells/well) were cultured under Th0 or Th17-polarizing conditions for two days and then treated with (A) Y27632, simvastatin or Y27632 plus simvastatin or (B) Y27632 or KD025. Supernatants and cells were collected on day 4 and supernatant levels of hIL-17A, hIL-21 and hCCL20 were assessed. Shown is one representative experiment. (C) The inhibition of hIL-17A, hIL-21 and hCCL20 production by each inhibitor and the combination thereof was fitted to a mixed linear model. P-values from the likelihood ratio test comparing the null model with only the random effect of patients with the complete model that includes all treatments is shown on each panel. The p-values for individual treatment were calculated as described in the Materials and Methods. (D) Whole cell extracts were prepared and ROCK activity was measured. Shown are 3-4 independent experiments. Data were analyzed by Kruskal-Wallis test (χ2 = 13.48, p<0.009) followed by the Dunns Multiple Comparison analysis (**p<0.01). (E) Phosphorylation of Stat3 protein was assessed by western blotting, blots were stripped and reprobed for phosphorylation of IRF4 protein, then subsequently stripped and reprobed for total Stat3, and total IRF4 respectively. Y(60): 60μM Y-27632, Y(90): 90μM Y-27632, SIM: 0.1 μM simvastatin, KD: 5.0μM KD025.
In comparison to Y27632, simvastatin (0.1 μM) alone did not decrease IL-17A (t=−1.11, p=0.27) or CCL20 production (t=1.9, p=0.06) while the effect on IL-21 production was more variable (Figure 3A and 3C). Higher doses of simvastatin exerted toxic effects in this cell culture system and could not be evaluated (data not shown). As observed for SLE T cells, addition of simvastatin to the lower dose of Y27632 did not further increase the inhibition of CCL20 or IL-21 production but enhanced the ability of the lower dose of Y27632 to decrease IL-17A production (Figure 3C). We did not detect any significant inhibitory effects of either Y27632 or simvastatin on the levels of IRF4 mRNA indicating that the effects of the inhibitors are not due to decreased IRF4 transcription (Supplementary Figure 2A). Thus the nonselective ROCK inhibitor was more effective than simvastatin alone in modulating the cytokine profile of in vitro differentiated Th17 cells.
We next compared the effects of KD025 to those of the pan-ROCK inhibitor, Y27632, on cytokine production by Th17 cells. KD025 (5μM) was as effective as the higher dose of Y27632 in inhibiting the production of IL-17A (t=−7.1 p<0.0001), IL-21 (t=−3.9, p=0.0008), and CCL-20 (t=−6.58, p< 0.0001) and more effective than the lower dose of Y27632 at decreasing IL-17A and CCL-20 production (Figure 3B–C). When assessing IRF4 as a measure of cell viability and T cell stimulation, we did not observe any inhibitory effect on IRF4 gene expression in Th17 cells treated with either the nonselective ROCK inhibitor or the ROCK2 selective inhibitor (Supplementary Figure 2B). To confirm that the effects of the different inhibitors on cytokine production were linked to effects on ROCK activation, we performed an ELISA-based ROCK kinase assay (Figure 3D). Consistent with the finding that the higher levels of ROCK activity observed in Th17 cells than in Th0 cells are primarily due to ROCK2 activation,9 the ROCK2 selective inhibitor was as effective as the nonselective ROCK inhibitor or simvastatin at diminishing ROCK activity (Figure 3D). In line with these results, all three inhibitors diminished the phosphorylation of IRF4, a known target of ROCK2 (Figure 3E). Neither Y27632 nor KD025 decreased Stat3 phosphorylation (Figure 3E). Therefore, a selective ROCK2 inhibitor is as effective as a nonselective ROCK inhibitor at decreasing cytokine production by in vitro differentiated Th17 cells.
Immune profiling reveals both overlapping and distinct effects of different ROCK inhibitors
To gain a broader understanding of the regulatory effects of the different classes of ROCK inhibitors in Th17 cells, we employed the NanoString nCounter Technology to profile the expression of immune-relevant genes. Human immunology v.2 CodeSet contains probe sets recognizing 581 immuno-relevant target genes (http://www.nanostring.com/support/literature?cat=gene_lists). 225 genes passed initial screen for differential expression (Supplementary Methods) and were used for further analysis. Principal Component Analysis (R, FactoMineR)34 was used to investigate the underlying data structure of expression profiling data set containing 225 genes and 18 treatment conditions. The log transformed expression count values were standardized prior to the analysis. The first four principle components (PC) explained 39.1%, 16.5%, 13.5%, and 8.6% of the variance respectively (Figure 4A, Supplementary Figure 3A–C). Contribution of individual genes to each of the first three PCs was calculated as ratio of the squared factor score of a gene by the eigenvalue associated with that component using FactorMineR. Genes with the above average contribution for a given PC (PC1 - 112, PC2-73, PC2-71 gene, Supplementary Figure 3) were selected for further analysis. The PC1 separates groups of genes with similar regulation in response to either simvastatin or Y276327 (Figure 4B and Supplementary Figure 3A), indicating that these two compounds act on similar immune-related regulatory pathways. The PC2 effectively separates genes altered in KD025-treated cells (Figure 4B and Supplementary Figure 3B) and PC3 is primarily driven by the differences between Y276327 and other treatments (Supplementary Figure 3C).
Figure 4. KD025 and Y27632 regulate distinct subset of immune-relevant genes.
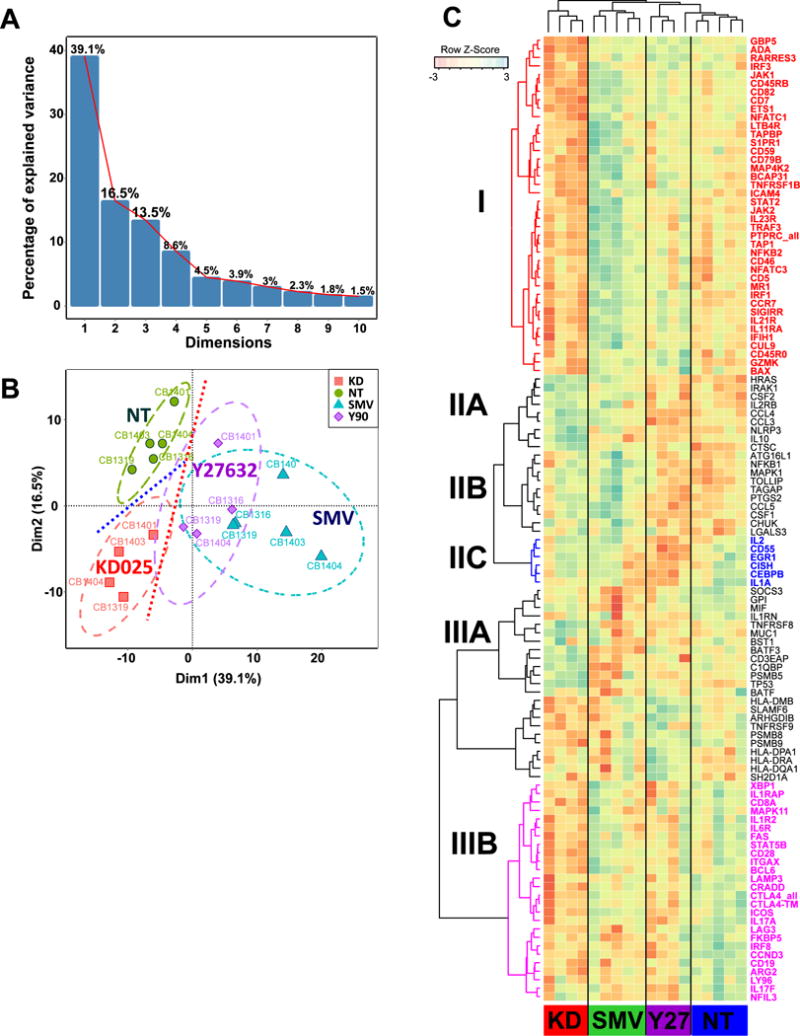
Following preliminary low-stringency NanoString screening for genes differentially expressed in Th17 cells treated with KD025, Y27632 we performed principal component analysis of independent cell samples (the sample coding is shown next to the data points) with gene expression as variables. (A) The first four principal components (PC) explain 77.7% of variance. (B) The first PC separates Y27632/SMV-treated Th17 cells from untreated (NT) and KD025-treated Th17 cells, whereas the second PC separates KD025-treated and NT cells. (C) Unsupervised hierarchical clustering were used to define the group of co-regulated genes I Th17 cells following treatments with Y27632, SMV or KD025 in comparison to not-treated cells. Genes specifically downregulated in KD025-treated cells (Cluster I) are shown in red, cluster IIC that contains a group of genes specifically downregulated by Y27632 is shown in blue, and cluster IIIB combines genes downregulated in Th17 cells treated with either Y27632 or KD025
Hierarchical clustering of genes differentially regulated by Y276327, KD025, and simvastatin in comparison with untreated cord blood-derived Th17 (p<0.05) revealed that all inhibitors downregulated a small number of genes, albeit to different extents (Fig. 4C, Cluster IIIB). In addition KD025 specifically inhibited a subset of genes (Cluster I), which was either largely unaffected or only weakly downregulated by Y27632 or simvastatin relative to untreated cells. Notably, KD025 downregulated the expression of both IL-23R and IL-21R (Cluster I). The gene profile affected by Y27632 and simvastatin was instead largely overlapping (Figure 4C and Supplementary Figure 3A) except for a small group of genes (Cluster IIC) that were primarily inhibited by Y27632 but not simvastatin or KD025 and that included IL-2. Finally, a small group of genes was upregulated by both KD025 and simvastatin (Cluster IIA and IIB) but not Y27632.
Comparative analysis of regulatory pathways enriched in KD-responsive vs Y27-responsive genes (both up and down) using G:profiler35 further indicated that both inhibitors controlled a number of genes involved in the regulation of cytokine production (Supplementary Figure 4). In addition, Y27632 regulates a battery of unique genes that are involved in leukocyte differentiation and cell adhesion, viral response, and interactions between lymphoid and non-lymphoid cells. Furthermore, treatment with Y27632 is associated with induction of several elements of the interferon signaling pathway. KD025 instead altered the expression of genes involved in signal transduction downstream of multiple transmembrane receptors (Figure 4C and Supplementary Figure 4). Specifically, KD025 treatment downregulates several elements of the Jak-STAT pathway as well as several subunits of the immunoproteasome. Interestingly, Y27632 and KD025 potentially exert opposite effects on the same pathways. For example, while KD025 decreases mRNA levels for JAK1, JAK2, IRF3, and STAT5B, Y27632 upregulates the expression of these genes (Supplementary Figure 4).
DISCUSSION
The ROCKs are emerging as important regulators of T cell effector function via their ability to be activated under Th17 conditions and to regulate the production of IL-17 and IL-21 and initial studies revealed that ≈60% of SLE patients exhibit increased ROCK activity in their PBMCs.22 Here we have investigated ROCK activation in a new cohort of SLE patients and have confirmed that PBMCs from a majority of SLE patients exhibit higher levels of ROCK activity than healthy controls. Interestingly, PBMCs from RA patients were also found to exhibit higher ROCK activity levels than controls suggesting that dysregulated ROCK activation may represent a common pathogenic pathway in autoimmunity.
Given the potential of the ROCKs as therapeutic targets for SLE and other autoimmune diseases, here we directly compared the effectiveness of distinct classes of ROCK inhibitors for their effects on the production of IL-17 and IL-21 by T cells from SLE patients. Our results demonstrate that inhibition of ROCK-mediated cytokine production could be achieved by multiple therapeutic modalities. In line with prior observations demonstrating that the selective ROCK2 inhibitor, KD025, can down-regulate the production of IL-17 and IL-21 by RA T cells, KD025 decreased the production of these cytokines by SLE T cells, although the production of IFNγ appears to be ROCK2-dependent in RA but not in SLE T cells.18 The ability of KD025 to target IL-17A and IL-21 production in our study was, however, not accompanied by decreases in Stat3 phosphorylation or IRF4 expression,18,36 possibly due to the later timing of addition of the ROCK2 inhibitor to the cultures, which may have enabled the initial wave of signaling events to be protected from the inhibitory effects of KD025.
The increased ROCK activity observed in human Th17 cells as compared to Th0 cells was also effectively decreased not only by broad ROCK inhibitors such as Y27632 and simvastatin but also by the selective ROCK2 inhibitor, KD025, suggesting that the elevated ROCK activity in this T helper subset primarily reflects activation of the ROCK2 rather than the ROCK1 isoform. These results support previous murine and human studies, which have implicated ROCK2 as the major ROCK isoform involved in the differentiation of Th17 cells and TFH cells generated under Th17-cell skewing conditions and have demonstrated beneficial effects of ROCK inhibitors on the production of IL-17 and IL-21 and autoimmune pathogenesis9–19,36 and indicate that directly assessing cellular ROCK activity can be helpful in evaluating the efficacy of distinct ROCK inhibitors.
Interestingly, immune gene profiling using NanoString nCounter Technology uncovered broad differences amongst the distinct classes of ROCK inhibitors. In particular, the transcriptional profile altered by KD025 included inhibitory effects on the expression of the IL-23 and IL-21 receptors suggesting that this compound may not only target the production of key Th17 cytokines but also the cytokine responsiveness of Th17 cells. More studies will, however, be required to determine the functional relevance of these latter effects. In contrast to KD025, the immune profile modulated by Y27632 and simvastatin was largely overlapping. It remains to be determined whether the differences between the selective ROCK2 inhibitor (KD025) and the broader ROCK inhibitors (Y27632 and simvastatin) are due to the ability of the latter two compounds to concomitantly decrease ROCK1 activity and/or are due to additional ROCK-independent actions.
In summary, dysregulated ROCK activity is a consistent feature of PBMCs from autoimmune patients. Targeting of this pathway can be achieved by a number of both selective and nonselective inhibitors suggesting that this pathway could be amenable to a number of distinct therapeutic modalities. Whether ROCK2 selective inhibitors will provide a more advantageous risk/benefit profile than broader ROCK inhibitors will likely require a better understanding of the functional relevance of ROCK1-dependent pathways in immune cells and the development of tools to specifically monitor the activation of ROCK1- and/or ROCK2-specific pathways in individual patients.
Supplementary Material
Supplementary Figure 1. Standardized residuals versus fitted values plot for the cytokine levels produced by the Th17 cells from umbilical blood and SLE patients. Residuals values are uniformly distributed relative y=0 line indicating uniformity indicating homogeneity of variation across the range of cytokine concentration, consistent with a good concordance between experimental and fitted values. Non-homogeneity at the extreme low concentrations of cytokines, indicating potential deviation from the linearity during the concentration measurements.
Supplementary Figure 2. IRF4 gene expression in Th0 or Th17 cells treated with ROCK inhibitors. Human umbilical cord CD4+ T cells (106 cells/well) were cultured under Th0 or Th17-polarizing conditions for two days and then treated with (A) Y27632, simvastatin or Y27632 plus simvastatin or (B) Y27632 or KD025. Cells were collected on day 4 and mRNA levels of IRF4 were assessed. Shown is one representative experiment. Y(60): 60μM Y-27632, Y(90): 90μM Y-27632, SIM: 0.1 μM simvastatin, KD: 5.0μM KD025.
Supplementary Figure 3. Genes with the above average contributions to PC1, PC2 and PC3 are differentially sensitive to ROCK inhibitors and simvastatin in Th17 cells. Contribution of individual genes to the individual principal components (PC1-PC3) was evaluated using FactoMineR as described in the results. Log transformed expression values of genes with above average contribution were scaled and subjected to hierarchical clustering to define groups of potentially coregulated genes. (A) Genes with above average contribution to PC1 are typically altered (up or down) following treatment with either Y27632 or simvastatin relative to KD025 or non-treated Th17. (B) Top contributors to PC2 separate KD025-treated Th17 from the NT and (C) top contributors to PC3 are driven by Y27632-specific and KD025 specific genes. (D) Top contributing genes for PC1, PC2 and PC3 show little overlap, indicating that grouping based on PC contribution reflect functional or regulatory differences between top-contributors.
Supplementary Figure 4. Y27632 and KD025 affect distinct regulatory pathways in Th17 cells. Comparative analysis of Y27632-upregulated (green) and KD025-sensitive (up- red and down-blue) gene lists were analyzed using compact comparison of gene annotations module35 of the g:Profiler, only differentially enriched pathways with the corrected p-values < 5*10−3 extracted from the Gene Onthology, KEGG and Reactome databases are shown. The number of sensitive genes whose products participate in specific pathways is shown inside the colored squares next to the p-values. The color intensity is inversely proportional to the magnitude of p-values. The hypergeometric test with Bonferroni multiple correction were used to evaluate the significance of observations.
Acknowledgments
Funding The research was supported by a grant from the NIH to JES and ABP (5R21AR62252-2), a grant from the NIH to ABP (1R01AI083440), a NIH Research Rheumatology Training grant to CR (T32AR007517), a American Heart Association grant to YC (11SDG5160006) and the Peter Jay Sharp Foundation and the David Z. Rosensweig Genomics Research Center.
Footnotes
Competing Interests ABP and JES received an Investigator-Initiated grant from Kadmon Corporation
References
- 1.Craft JE. Follicular helper T cells in immunity and systemic autoimmunity. Nature reviews Rheumatology. 2012;8(6):337–47. doi: 10.1038/nrrheum.2012.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crispin JC, Tsokos GC. Interleukin-17-producing T cells in lupus. Curr Opin Rheumatol. 2010;22(5):499–503. doi: 10.1097/BOR.0b013e32833c62b0. [DOI] [PubMed] [Google Scholar]
- 3.Pernis AB. Th17 cells in rheumatoid arthritis and systemic lupus erythematosus. J Intern Med. 2009;265(6):644–52. doi: 10.1111/j.1365-2796.2009.02099.x. [DOI] [PubMed] [Google Scholar]
- 4.Burmester GR, Feist E, Dorner T. Emerging cell and cytokine targets in rheumatoid arthritis. Nat Rev Rheumatol. 2014;10(2):77–88. doi: 10.1038/nrrheum.2013.168. [DOI] [PubMed] [Google Scholar]
- 5.Gaffen SL, Jain R, Garg AV, et al. The IL-23-IL-17 immune axis: from mechanisms to therapeutic testing. Nat Rev Immunol. 2014;14(9):585–600. doi: 10.1038/nri3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brustle A, Heink S, Huber M, et al. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nature immunology. 2007;8(9):958–66. doi: 10.1038/ni1500. [DOI] [PubMed] [Google Scholar]
- 7.Huber M, Brustle A, Reinhard K, et al. IRF4 is essential for IL-21-mediated induction, amplification, and stabilization of the Th17 phenotype. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(52):20846–51. doi: 10.1073/pnas.0809077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Q, Yang W, Gupta S, et al. IRF-4-binding protein inhibits interleukin-17 and interleukin-21 production by controlling the activity of IRF-4 transcription factor. Immunity. 2008;29(6):899–911. doi: 10.1016/j.immuni.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biswas PS, Gupta S, Chang E, et al. Phosphorylation of IRF4 by ROCK2 regulates IL-17 and IL-21 production and the development of autoimmunity in mice. J Clin Invest. 2010;120(9):3280–95. doi: 10.1172/JCI42856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amano M, Nakayama M, Kaibuchi K. Rho-kinase/ROCK: A key regulator of the cytoskeleton and cell polarity. Cytoskeleton. 2010;67(9):545–54. doi: 10.1002/cm.20472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schofield AV, Bernard O. Rho-associated coiled-coil kinase (ROCK) signaling and disease. Critical reviews in biochemistry and molecular biology. 2013;48(4):301–16. doi: 10.3109/10409238.2013.786671. [DOI] [PubMed] [Google Scholar]
- 12.Julian L, Olson MF. Rho-associated coiled-coil containing kinases (ROCK): structure, regulation, and functions. Small GTPases. 2014;5:e29846. doi: 10.4161/sgtp.29846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thumkeo D, Watanabe S, Narumiya S. Physiological roles of Rho and Rho effectors in mammals. Eur J Cell Biol. 2013;92(10–11):303–15. doi: 10.1016/j.ejcb.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Q, Gensch C, Liao JK. Rho-associated coiled-coil-forming kinases (ROCKs): potential targets for the treatment of atherosclerosis and vascular disease. Trends in pharmacological sciences. 2011;32(3):167–73. doi: 10.1016/j.tips.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komers R. Rho kinase inhibition in diabetic kidney disease. British journal of clinical pharmacology. 2013;76(4):551–9. doi: 10.1111/bcp.12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mueller BK, Mack H, Teusch N. Rho kinase, a promising drug target for neurological disorders. Nat Rev Drug Discov. 2005;4(5):387–98. doi: 10.1038/nrd1719. [DOI] [PubMed] [Google Scholar]
- 17.Pernis AB, Ricker E, Weng CH, et al. Rho Kinases in Autoimmune Diseases. Annu Rev Med. 2016;67:355–74. doi: 10.1146/annurev-med-051914-022120. [DOI] [PubMed] [Google Scholar]
- 18.Zanin-Zhorov A, Weiss JM, Nyuydzefe MS, et al. Selective oral ROCK2 inhibitor down-regulates IL-21 and IL-17 secretion in human T cells via STAT3-dependent mechanism. Proc Natl Acad Sci U S A. 2014;111(47):16814–9. doi: 10.1073/pnas.1414189111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stirzaker RA, Biswas PS, Gupta S, et al. Administration of fasudil, a ROCK inhibitor, attenuates disease in lupus-prone NZB/W F1 female mice. Lupus. 2012;21(6):656–61. doi: 10.1177/0961203312436862. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Harada T, Juang YT, et al. Phosphorylated ERM is responsible for increased T cell polarization, adhesion, and migration in patients with systemic lupus erythematosus. J Immunol. 2007;178(3):1938–47. doi: 10.4049/jimmunol.178.3.1938. [DOI] [PubMed] [Google Scholar]
- 21.Apostolidis SA, Rauen T, Hedrich CM, et al. Protein phosphatase 2A enables expression of interleukin 17 (IL-17) through chromatin remodeling. J Biol Chem. 2013;288(37):26775–84. doi: 10.1074/jbc.M113.483743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isgro J, Gupta S, Jacek E, et al. Enhanced rho-associated protein kinase activation in patients with systemic lupus erythematosus. Arthritis Rheum. 2013;65(6):1592–602. doi: 10.1002/art.37934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi J, Wei L. Rho kinases in cardiovascular physiology and pathophysiology: the effect of fasudil. Journal of cardiovascular pharmacology. 2013;62(4):341–54. doi: 10.1097/FJC.0b013e3182a3718f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen M, Liu A, Ouyang Y, et al. Fasudil and its analogs: a new powerful weapon in the long war against central nervous system disorders? Expert opinion on investigational drugs. 2013;22(4):537–50. doi: 10.1517/13543784.2013.778242. [DOI] [PubMed] [Google Scholar]
- 25.Pan P, Shen M, Yu H, et al. Advances in the development of Rho-associated protein kinase (ROCK) inhibitors. Drug discovery today. 2013;18(23–24):1323–33. doi: 10.1016/j.drudis.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 26.Feng Y, LoGrasso PV. Rho kinase inhibitors: a patent review (2012 - 2013) Expert opinion on therapeutic patents. 2014;24(3):295–307. doi: 10.1517/13543776.2014.863279. [DOI] [PubMed] [Google Scholar]
- 27.Hahmann C, Schroeter T. Rho-kinase inhibitors as therapeutics: from pan inhibition to isoform selectivity. Cellular and molecular life sciences: CMLS. 2010;67(2):171–7. doi: 10.1007/s00018-009-0189-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JH, Zheng Y, von Bornstadt D, et al. Selective ROCK2 Inhibition In Focal Cerebral Ischemia. Annals of clinical and translational neurology. 2014;1(1):2–14. doi: 10.1002/acn3.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawada N, Liao JK. Rho/Rho-associated coiled-coil forming kinase pathway as therapeutic targets for statins in atherosclerosis. Antioxid Redox Signal. 2014;20(8):1251–67. doi: 10.1089/ars.2013.5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40(9):1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 31.Petri M, Kim MY, Kalunian KC, et al. Combined oral contraceptives in women with systemic lupus erythematosus. The New England journal of medicine. 2005;353(24):2550–8. doi: 10.1056/NEJMoa051135. [DOI] [PubMed] [Google Scholar]
- 32.Aletaha D, Neogi T, Silman AJ, et al. 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 2010;62(9):2569–81. doi: 10.1002/art.27584. [DOI] [PubMed] [Google Scholar]
- 33.Aletaha D, Smolen JS. The Simplified Disease Activity Index and Clinical Disease Activity Index to monitor patients in standard clinical care. Rheum Dis Clin North Am. 2009;35(4):759–72. viii. doi: 10.1016/j.rdc.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 34.Lê S, Josse J, Husson F. FactoMineR: An R Package for Multivariate Analysis. Journal of Statistical Software. 2008;25(1):1–18. [Google Scholar]
- 35.Reimand J, Arak T, Vilo J. g:Profiler–a web server for functional interpretation of gene lists (2011 update) Nucleic acids research. 2011;39:W307–15. doi: 10.1093/nar/gkr378. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weiss JM, Chen W, Nyuydzefe MS, et al. ROCK2 signaling is required to induce a subset of T follicular helper cells through opposing effects on STATs in autoimmune settings. Sci Signal. 2016;9(437):ra73. doi: 10.1126/scisignal.aad8953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aarts E, Verhage M, Veenvliet JV, et al. A solution to dependency: using multilevel analysis to accommodate nested data. Nature neuroscience. 2014;17(4):491–6. doi: 10.1038/nn.3648. [DOI] [PubMed] [Google Scholar]
- 38.J P, D B, S D, et al. nlme: Linear and Nonlinear Mixed Effects Models. 2016 R package version 31–126. http://CRAN.R-proiect.orq/packaqe=nlme.
- 39.Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22(12):1540–2. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Standardized residuals versus fitted values plot for the cytokine levels produced by the Th17 cells from umbilical blood and SLE patients. Residuals values are uniformly distributed relative y=0 line indicating uniformity indicating homogeneity of variation across the range of cytokine concentration, consistent with a good concordance between experimental and fitted values. Non-homogeneity at the extreme low concentrations of cytokines, indicating potential deviation from the linearity during the concentration measurements.
Supplementary Figure 2. IRF4 gene expression in Th0 or Th17 cells treated with ROCK inhibitors. Human umbilical cord CD4+ T cells (106 cells/well) were cultured under Th0 or Th17-polarizing conditions for two days and then treated with (A) Y27632, simvastatin or Y27632 plus simvastatin or (B) Y27632 or KD025. Cells were collected on day 4 and mRNA levels of IRF4 were assessed. Shown is one representative experiment. Y(60): 60μM Y-27632, Y(90): 90μM Y-27632, SIM: 0.1 μM simvastatin, KD: 5.0μM KD025.
Supplementary Figure 3. Genes with the above average contributions to PC1, PC2 and PC3 are differentially sensitive to ROCK inhibitors and simvastatin in Th17 cells. Contribution of individual genes to the individual principal components (PC1-PC3) was evaluated using FactoMineR as described in the results. Log transformed expression values of genes with above average contribution were scaled and subjected to hierarchical clustering to define groups of potentially coregulated genes. (A) Genes with above average contribution to PC1 are typically altered (up or down) following treatment with either Y27632 or simvastatin relative to KD025 or non-treated Th17. (B) Top contributors to PC2 separate KD025-treated Th17 from the NT and (C) top contributors to PC3 are driven by Y27632-specific and KD025 specific genes. (D) Top contributing genes for PC1, PC2 and PC3 show little overlap, indicating that grouping based on PC contribution reflect functional or regulatory differences between top-contributors.
Supplementary Figure 4. Y27632 and KD025 affect distinct regulatory pathways in Th17 cells. Comparative analysis of Y27632-upregulated (green) and KD025-sensitive (up- red and down-blue) gene lists were analyzed using compact comparison of gene annotations module35 of the g:Profiler, only differentially enriched pathways with the corrected p-values < 5*10−3 extracted from the Gene Onthology, KEGG and Reactome databases are shown. The number of sensitive genes whose products participate in specific pathways is shown inside the colored squares next to the p-values. The color intensity is inversely proportional to the magnitude of p-values. The hypergeometric test with Bonferroni multiple correction were used to evaluate the significance of observations.


