Abstract
Several studies have addressed cytokine gene polymorphisms and their possible associations with periodontitis. We examined the association between salivary anti‐ and pro‐inflammatory mediator polymorphisms and initial periodontitis in Finnish adolescents, taking into account the effect of smoking. Salivary samples of 93 clinically examined adolescents from Eastern Finland were analyzed. Their oral health and smoking habits were recorded. Periodontal probing depth (PPD), and bleeding on probing (BOP) at four sites per tooth, root calculus (RC), and visible plaque index (VPI) were recorded from the index teeth. Salivary MMP‐8 median values were assessed. The sites with ≥4 mm PD were categorized as follows: PPD1 = one or more ≥4 mm pocket, PPD2 = two or more ≥4 mm pockets, and PPD3 = three or more ≥4 mm pockets. Genomic DNA was extracted from 300 μl of the saliva samples by genomic QIAamp® DNA Blood Mini Kit and genotyped for polymorphisms. Genetic variants for genotyping were selected from the following genes of interest: S100A8, FCGR2A, FCGR2B, IL10, MMP8, MMP3, MMP13, VDR, TLR4, MMP2, MPO, ELANE, IL1A, IL1B, IL1RN, CD28, MMP9, DDX39B, NFKBIL1, LTA, TNF, SOD2, IL6, TLR4, TIMP1, and SYN1. After false discovery rate control (FDR), polymorphisms in MMP3 (rs679620, rs520540, rs639752), CD28 (rs3116496), and VDR (rs2228570) associated (FDR q < 0.05) with deepened periodontal pockets. Smoking did not affect the results. Genetic polymorphisms of pro‐inflammatory mediators MMP3, CD28, and VDR seem to link to initial periodontitis.
Keywords: adolescent, genetic, periodontitis, polymorphism
Periodontitis is a complex chronic low‐grade infection‐induced tissue destruction inflammatory disease involving environmental factors such as smoking. The potential periodontal pathogens activate matrix metalloproteinases (MMPs), pro‐inflammatory cytokines, and their regulators together with enhanced inflammatory burden (Sorsa et al., 2016). The pathophysiology of periodontitis is associated with variations in multiple disease modifying genes. The environmental factors may thus interact with genetics (Laine, Crielaard, & Loos, 2012), and smoking as a confounding factor needs to be taken into account when assessing the effect of immune system and host response in periodontitis patients.
Several studies have been published regarding cytokine gene polymorphisms and their possible associations with periodontitis pathogenesis, when attempting to explain the complex nature of this disease. MMP polymorphisms have been of interest because of the essential role of MMPs in periodontal tissue remodeling and destruction. MMP‐8 (neutrophil collagenase) is the most abundant and important biomarker in periodontitis reflecting the initiation and progression of the disease. Increased MMP‐8 levels reflect the severity and course of periodontitis (Sorsa et al., 2016; Sorsa, Tjäderhane, & Salo, 2004; Sorsa et al., 2006). In smokers, salivary MMP‐8 concentrations are usually lower than in non‐smokers, even in adolescents (Liede et al., 1999; Heikkinen et al., 2010).
The roles of MMP‐2, MMP‐8, MMP‐9, and MMP‐12 have been analyzed with respect to the genetic polymorphisms; they may contribute to the susceptibility to periodontitis. MMP3 and tissue inhibitor of MMPs (TIMP1) SNPs have also been considered as possible candidates in the pathogenesis of periodontitis (Letra et al., 2012; Emingil et al., 2014). However, regarding matrix metallopeptidase 9 (MMP9) gene polymorphism, the results are ambivalent. Gürkan et al. (2008) reported that carrier status of MMP9 ‐1562 T allele was lower in healthy Turkish subjects than in chronic periodontitis group (Gürkan et al., 2008). However, MMP9 ‐1562C/T was not related to the susceptibility to chronic periodontitis according to Holla, Fassmann, Muzík, Vanek, and Vasku (2006) and de Souza et al. (2005). Interleukins (IL) are pro‐inflammatory cytokines expressed mostly by leukocytes. Interleukin‐6 (IL‐6) is secreted by various cell types and has been shown to be important pro‐ and anti‐inflammatory mediator involved also in the pathogenesis of periodontitis by regulating the expression of IL‐1 and tumor necrosis factor alpha (TNF‐α) (Opal & DePalo, 2000). It also stimulates osteoclast differentiation as well as enhances B‐ and T‐cell growth, differentiation, and bone resorption (Hughes, Turner, Belibasakis, & Martuscelli, 2006). An association between the carriage rates of the IL‐1 , Alpha (IL1A‐889, rs1800587), and periodontitis has been demonstrated (Laine, Loos, & Crielaard, 2010). Other salivary inflammatory biomarker candidate genes for periodontitis are considered, such as myeloperoxidase (MPO‐463G/A) (Erciyas, Pehlivan, Sever, & Orbak, 2010) and calcium‐binding protein A8 (S100A8) (Sun et al., 2011), Vitamin D (1,25‐Dihydroxyvitamin D3) receptor (VDR) (Chen, Li, Zhang, & Wang, 2012 ; Laine et al., 2012 ), toll‐like receptor 2, 4 (Folwaczny, Glas, Török, Limbersky, & Folwaczny, 2004; Schröder & Schumann, 2005), and lymphotoxin alpha (LTA) (Vasconcelos et al., 2012). However, studies on these cytokine and inflammatory mediator polymorphisms are rare, and the results are contradictory.
In Finland, 48% of the 30‐ to 34‐year‐olds have periodontitis (Suominen‐Taipale et al., 2008). According to the thesis by Heikkinen (2011) and Heikkinen et al. (2008), 56% of one birth cohort aged 15‐ to 16‐year‐old adolescents had more than one ≥4 mm periodontal pockets (Heikkinen, 2011; Heikkinen et al., 2008). Taking into account attachment loss and bleeding on probing, 10% of the subjects had initial periodontitis, and most of them were smokers (Heikkinen, 2011; Heikkinen et al., 2008). Heikkinen et al. (2011, 2010, 2012 also observed that smokers (25% of all participants) frequently harbored more periodontal bacteria than non‐smokers (66%), and smoking significantly decreased the values of the salivary biomarkers MMP‐8 and polymorphonuclear leukocytes (PMN) elastase in boys (Heikkinen et al., 2010; Heikkinen, 2011; Heikkinen et al., 2012).
The aim of the present study was to examine the association between the polymorphisms in salivary anti‐ and pro‐inflammatory mediators and initial periodontitis. We hypothesized that the salivary anti‐ and pro‐inflammatory mediator polymorphisms are associated with initial periodontitis.
1. MATERIAL AND METHODS
1.1. Study participants
Data were collected at the Kotka Health Center in Eastern Finland. The study was approved by the Ethical Committee of the Helsinki and Uusimaa Hospital District (Dnro 260/13/03/00/13). All the participants gave written informed consent. This study includes data from two Finnish adolescent birth cohorts collected at the Kotka Health Center in Eastern Finland in 2004–2005 and 2014–2015. Altogether, there were 94 participants for whom we had available saliva and DNA samples and who gave their approval for the saliva and DNA analyses (see Figure 1 for study flow of the participants).
Figure 1.
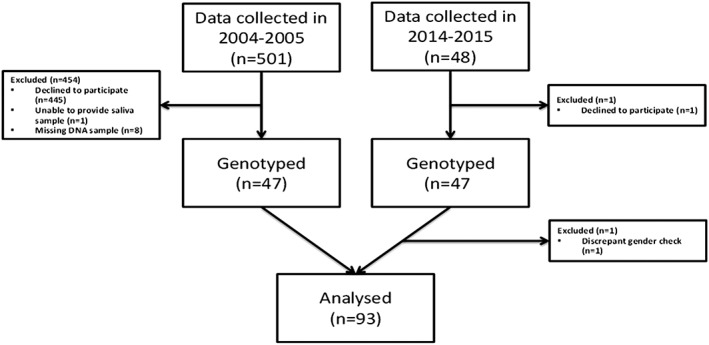
Study flow of the participants
Oral health of subjects was clinically examined in both cohorts. No periodontal therapy has been performed in the last recent year among these participants before this study, and after diagnostic procedures, they were treated by specialist in clinical periodontics. In short, periodontal parameters were recorded according to the WHO recommendations (World Health Organization, International Statistical Classification of Diseases and Related Health Problems, [Link]). Periodontal probing depth (PPD), bleeding on probing (BOP), and visible plaque index (VPI) were recorded at four sites per tooth, and all sites with ≥4 mm PD were recorded and categorized as follows: PPD1 = one or more ≥4 mm pocket, PPD2 = two or more ≥4 mm pockets, and PPD3 = three or more ≥4 mm pockets (Heikkinen et al., 2016). The cut‐point of the BOP value was 20 percentage meaning as gingivitis (Heikkinen, 2011; Heikkinen et al., 2008). Root calculus (RC) and VPI were recorded from index teeth.
Smoking habits (cigarettes per week indicated regular smoking) and pack‐years were recorded (Heikkinen et al., 2012). Stimulated salivary samples were collected and centrifuged, and the supernatants were used for the enzyme measurements, such as MMP‐8 analysis. MMP‐8 levels were analyzed by immunofluorometric assay (Medix Biochemica, Kauniainen, Finland). The interassay coefficient of variation (CV)% for this study was 6.3%, and detection limit for the assay was 0.08 μg/l (Heikkinen et al., 2010).
1.2. Candidate gene and SNP selection
Genetic variants for genotyping were selected from the following genes of interest: S100A8, Fc fragment of IgG, low affinity IIa, receptor (FCGR2A), Fc fragment of IgG, low affinity IIb, receptor (FCGR2B), prostaglandin‐endoperoxide synthase 2 (PTGS2), IL10, MMP8, MMP3, MMP13, VDR, tumor necrosis factor (TNF; ligand) superfamily, Member 11 (TNFSF11), toll‐like receptor 4 (TLR4), MMP2, MPO, elastase, neutrophil expressed (ELANE), IL‐1, IL1A, Beta (IL1B), interleukin 1 receptor antagonist (IL1RN), CD28 molecule (CD28), MMP9, DEAD (Asp‐Glu‐Ala‐Asp) box polypeptide 39B (DDX39B), nuclear factor of Kappa light polypeptide gene enhancer in B‐cells inhibitor‐like 1 (NFKBIL1), LTA, TNF, Superoxide Dismutase 2, Mitochondrial (SOD2), IL6, TIMP metallopeptidase inhibitor 1 (TIMP1), and Synapsin I (SYN1). The majority of SNPs were selected based on previous publications. Some of the originally selected SNPs failed the Sequenom iPLEX design because of technical reasons. For these genes (ELANE, IL1B, IL1RN, DDX39B), replacing SNPs were selected from the HapMap database (http://www.hapmap.org) by use of the tag SNP selection algorithm Tagger included in the Haploview 4.2 software (Nature, 2003; Barrett, Fry, Maller, & Daly, 2005). Taq SNP selection was used in order to select optimal SNPs covering most of the genetic variation in the genomic area of interest. Flanking areas of the size of 10 kb were included both upstream and downstream of the actual gene encoding regions. Taq SNPs were selected based on the criteria of minor allele frequency (MAF) >0.05 in European population and pair‐wise r2 threshold of 0.9. Based on these selection criteria, 74 SNPs were successfully included in three genotyping multiplexes. Three SNPs (rs3795391, rs2125685, and rs2544480) were excluded during the optimization phase of genotyping. Table 1 gives all the SNPs successfully genotyped in this study.
Table 1.
Successfully genotyped SNPs
| Marker | Chr. location (GRCh37) | Gene | Alleles (A1/A2) | A1A1 (n, %) | A1A2 (n, %) | A2A2 (n, %) | MAF, % | HWE p‐value |
|---|---|---|---|---|---|---|---|---|
| rs3806232 | 1:153364130 | S100A8 | T/C | 78 (85.7) | 10 (11.0) | 3 (3.3) | 8.8 | 0.02 |
| rs1801274 | 1:161479745 | FCGR2A | T/C | 23 (25.6) | 50 (55.6) | 17 (18.9) | 46.7 | 0.40 |
| rs5275 | 1:186643058 | PTGS2 | T/C | 39 (45.4) | 39 (45.4) | 8 (9.3) | 32.0 | 0.81 |
| rs689466 | 1:186650751 | PTGS2 | A/G | 59 (64.8) | 30 (33.0) | 2 (2.2) | 18.7 | 0.73 |
| rs1800872 | 1:206946407 | IL10 | C/A | 52 (56.5) | 36 (39.1) | 4 (4.4) | 23.9 | 0.58 |
| rs1800871 | 1:206946634 | IL10 | C/T | 52 (56.5) | 36 (39.1) | 4 (4.4) | 23.9 | 0.58 |
| rs1800896 | 1:206946897 | IL10 | A/G | 29 (31.5) | 46 (50.0) | 17 (18.5) | 43.5 | 1.00 |
| rs1800587 | 2:113542960 | IL1A | C/T | 48 (52.2) | 35 (38.0) | 9 (9.8) | 28.8 | 0.46 |
| rs3917365 | 2:113586469 | IL1B | C/T | 79 (86.8) | 11 (12.1) | 1 (1.1) | 7.1 | 0.37 |
| rs1143633 | 2:113590467 | IL1B | G/A | 45 (49.5) | 43 (47.3) | 3 (3.3) | 26.9 | 0.07 |
| rs16062 | 2:113591081 | IL1B | C/T | 92 (100) | 0.0 | |||
| rs1143627 | 2:113594387 | IL1B | T/C | 35 (38.5) | 43 (47.3) | 13 (14.3) | 37.9 | 1.00 |
| rs16944 | 2:113594867 | IL1B | G/A | 35 (39.3) | 42 (47.2) | 12 (13.5) | 37.1 | 1.00 |
| rs4251961 | 2:113874467 | IL1RN | T/C | 42 (46.2) | 40 (44.0) | 9 (9.9) | 31.9 | 1.00 |
| rs4251985 | 2:113877413 | IL1RN | G/T | 48 (52.8) | 29 (31.9) | 14 (15.4) | 31.3 | 0.02 |
| rs3213448 | 2:113879297 | IL1RN | G/A | 62 (67.4) | 26 (28.3) | 4 (4.4) | 18.5 | 0.50 |
| rs315935 | 2:113881365 | IL1RN | A/G | 85 (100) | 0.0 | |||
| rs4251998 | 2:113881962 | IL1RN | G/A | 88 (100) | 0.0 | |||
| rs3181051 | 2:113884673 | IL1RN | C/T | 87 (94.6) | 5 (5.4) | 0 | 2.7 | 1.00 |
| rs3087263 | 2:113885768 | IL1RN | G/A | 85 (93.4) | 5 (5.5) | 1 (1.1) | 3.9 | 0.11 |
| rs3087266 | 2:113889100 | IL1RN | C/T | 55 (60.4) | 28 (30.8) | 8 (8.8) | 24.2 | 0.15 |
| rs4252022 | 2:113890161 | IL1RN | G/A | 89 (98.9) | 1 (1.1) | 0 | 0.6 | 1.00 |
| rs315952 | 2:113890304 | IL1RN | T/C | 41 (44.6) | 32 (34.8) | 19 (20.7) | 38.0 | 0.01 |
| rs3116496 | 2:204594512 | CD28 | T/C | 66 (72.5) | 21 (23.1) | 4 (4.4) | 15.9 | 0.23 |
| rs160690 | 6:2799060 | DDX39B | G/T | 56 (61.5) | 30 (33) | 5 (5.5) | 22.0 | 0.76 |
| rs7766569 | 6:2807359 | DDX39B | A/G | 52 (61.2) | 32 (37.7) | 1 (1.2) | 20.0 | 0.17 |
| rs5004021 | 6:2807721 | DDX39B | C/T | 90 (98.9) | 1 (1.1) | 0 | 0.6 | 1.00 |
| rs12525298 | 6:2810889 | DDX39B | C/T | 53 (57.6) | 35 (38.0) | 4 (4.4) | 23.4 | 0.77 |
| rs186396 | 6:2811850 | DDX39B | A/G | 23 (25.3) | 57 (62.6) | 11 (12.1) | 43.4 | 0.01 |
| rs9405587 | 6:2812175 | DDX39B | T/G | 55 (59.8) | 33 (35.9) | 4 (4.4) | 22.3 | 1.00 |
| rs4084090 | 6:31218835 | Intergenic Chr. 6 | T/C | 86 (93.5) | 5 (5.4) | 1 (1.1) | 3.8 | 0.11 |
| rs2071591 | 6:31515799 | NFKBIL1 | C/T | 38 (42.2) | 44 (48.9) | 8 (8.9) | 33.3 | 0.48 |
| rs2857708 | 6:31533606 | LTA | G/A | 64 (69.6) | 27 (29.4) | 1 (1.1) | 15.8 | 0.46 |
| rs2009658 | 6:31538244 | LTA | C/G | 62 (67.4) | 29 (31.5) | 1 (1.1) | 16.9 | 0.45 |
| rs1800629 | 6:31543031 | TNF | G/A | 70 (76.1) | 22 (23.9) | 0 | 12.0 | 0.35 |
| rs2736189 | 6:31564728 | NCR3 | C/T | 62 (68.1) | 28 (30.8) | 1 (1.1) | 16.5 | 0.45 |
| rs4880 | 6:160113872 | SOD2 | T/C | 30 (32.6) | 47 (51.1) | 15 (16.3) | 41.9 | 0.83 |
| rs5746092 | 6:160114311 | SOD2 | C/G | 43 (46.7) | 41 (44.6) | 8 (8.7) | 31.0 | 0.81 |
| rs2758343 | 6:160114572 | SOD2 | G/T | 30 (32.6) | 48 (52.2) | 14 (15.2) | 41.3 | 0.53 |
| rs1800796 | 7:22766246 | IL6 | G/C | 90 (97.8) | 2 (2.2) | 0 | 1.1 | 1.00 |
| rs1800795 | 7:22766645 | IL6 | G/C | 34 (37.8) | 41 (45.6) | 15 (16.7) | 39.4 | 0.66 |
| rs11536889 | 9:120478131 | TLR4 | G/C | 78 (84.8) | 13 (14.1) | 1 (1.1) | 8.2 | 0.46 |
| rs3765620 | 11:102595492 | MMP8 | A/G | 36 (39.1) | 46 (50.0) | 10 (10.9) | 35.9 | 0.50 |
| rs1320632 | 11:102596063 | MMP8 | T/C | 81 (88.0) | 11 (12.0) | 0 | 6.0 | 1.00 |
| rs11225395 | 11:102596480 | MMP8 | G/A | 36 (39.1) | 46 (50.0) | 10 (10.9) | 35.9 | 0.50 |
| rs639752 | 11:102707339 | MMP3 | T/G | 34 (37.4) | 46 (50.6) | 11 (12.1) | 37.4 | 0.51 |
| rs650108 | 11:102708787 | MMP3 | G/A | 44 (47.8) | 40 (43.5) | 8 (8.7) | 30.4 | 1.00 |
| rs520540 | 11:102709425 | MMP3 | G/A | 35 (38.0) | 46 (50.0) | 11 (12.0) | 37.0 | 0.65 |
| rs679620 | 11:102713620 | MMP3 | G/A | 34 (40.5) | 41 (48.8) | 9 (10.7) | 35.1 | 0.63 |
| rs522616 | 11:102715048 | MMP3 | A/G | 51 (56.0) | 36 (39.6) | 4 (4.4) | 24.2 | 0.57 |
| rs2252070 | 11:102826539 | MMP13 | A/G | 25 (27.2) | 52 (56.5) | 15 (16.3) | 44.6 | 0.21 |
| rs7975232 | 12:48238837 | VDR | A/C | 22 (23.9) | 54 (58.7) | 16 (17.4) | 46.7 | 0.14 |
| rs1544410 | 12:48239835 | VDR | G/A | 47 (52.2) | 34 (37.8) | 9 (10.0) | 28.9 | 0.45 |
| rs2228570 | 12:48272895 | VDR | C/T | 39 (42.4) | 40 (43.5) | 13 (14.1) | 35.9 | 0.65 |
| rs9533156 | 13:43147671 | TNFSF11 | C/T | 23 (26.1) | 44 (50.0) | 21 (23.9) | 48.9 | 1.00 |
| rs2277438 | 13:43155168 | TNFSF11 | A/G | 56 (61.5) | 31 (34.1) | 4 (4.4) | 21.4 | 1.00 |
| rs498670 | 13:91763380 | TLR4 | G/A | 30 (33.0) | 49 (53.9) | 12 (13.2) | 40.1 | 0.28 |
| rs243865 | 16:55511806 | MMP2 | C/T | 47 (51.1) | 38 (41.3) | 7 (7.6) | 28.3 | 1.00 |
| rs2759 | 17:56348106 | MPO | A/G | 85 (92.4) | 6 (6.5) | 1 (1.1) | 4.4 | 0.15 |
| rs11575868 | 17:56352883 | MPO | G/A | 79 (85.9) | 13 (14.1) | 0 | 7.1 | 1.00 |
| rs2856857 | 17:56357833 | MPO | C/T | 69 (75.0) | 21 (22.8) | 2 (2.2) | 13.6 | 0.67 |
| rs2243828 | 17:56358884 | MPO | A/G | 63 (69.2) | 24 (26.4) | 4 (4.4) | 17.6 | 0.46 |
| rs2243827 | 17:56358941 | MPO | G/T | 69 (75.8) | 20 (22.0) | 2 (2.2) | 13.2 | 0.65 |
| rs740021 | 19:852104 | ELANE | G/T | 88 (96.7) | 3 (3.3) | 0 | 1.7 | 1.00 |
| rs17223045 | 19:855587 | ELANE | C/T | 90 (98.9) | 1 (1.1) | 0 | 0.6 | 1.00 |
| rs17216656 | 19:856015 | ELANE | G/A | 92 (100) | 0.0 | |||
| rs17576 | 20:44640225 | MMP9 | A/G | 27 (30.0) | 48 (53.3) | 15 (16.7) | 43.3 | 0.52 |
| rs3787268 | 20:44641731 | MMP9 | G/A | 55 (60.4) | 34 (37.4) | 2 (2.2) | 20.9 | 0.34 |
| rs1062849 | X:47445999 | TIMP1 | C/T | 92 (100) | 0.0 | |||
| rs6520279 | X:47448096 | SYN1 | T/C | 50 (54.4) | 17 (18.5) | 25 (27.2) | 36.4 | 0.00 |
| rs5906435 | X:47448410 | SYN1 | C/T | 54 (58.7) | 16 (17.4) | 22 (23.9) | 32.6 | 0.00 |
Chromosomal location (assembly Feb. 2009 (GRCh37/hg19), associated gene, alleles, genotype counts and frequencies, minor allele frequencies (MAF) and Hardy–Weinberg equilibrium (HWE) p‐values are given for each SNP.
1.3. DNA extraction and genotyping
All saliva samples in both cohorts were immediately frozen at −20°C and kept at −70°C until DNA extraction in both cohorts. DNA was extracted from 300 μl of the saliva using a genomic QIAamp® DNA Blood Mini Kit. Genotyping was performed using the Agena MassARRAY SNP genotyping system and iPLEX Gold assays (Agena Bioscience, San Diego, CA, USA). Allele discrimination is based on primer extension with single mass‐modified nucleotides followed by Matrix‐assisted laser desorption/ionization Time‐of‐Flight (MALDI‐TOF) mass spectrometry. All reactions were designed in multiplexes of up to 35 SNPs, by use of Assay Design v2.0 software (Agena Bioscience). Genotyping reactions were performed on 20 ng of dried genomic DNA in 384‐well plate format according to the manufacturer's recommendations. Concentrations of the extension primers were adjusted according to their mass and varied between 7 and 24.6 μM. The data was collected using the MassARRAY Compact System (Agena Bioscience), and the genotypes were identified using Typer 4 software (Agena Bioscience). For quality control reasons, the genotype calls were also checked manually and corrected when necessary. Genotyping quality was examined by a detailed QC procedure consisting of success rate checks, duplicated samples, gender check for X‐chromosomal markers, water controls, and Hardy–Weinberg Equilibrium (HWE) testing.
Genotyping success rates ranged from 90.3% to 98.9%. All duplicate samples gave concordant results. One subject was excluded because of discrepant gender check results. Table 1 gives the genotype and allele frequencies and HWE p‐values. Five SNPs turned out to be non‐polymorphic (rs16062, rs315935, rs4251998, rs17216656, and rs1062849). Three SNPs (rs5004021, rs4252022, and rs17223045) with MAF < 0.01 were excluded. Rs5906435 and rs6520279 are X‐chromosomal explaining their deviation from the HWE. All other SNPs were in HWE (p > 0.01). Finally, 93 subjects and 63 SNPs were included in the statistical analysis.
1.4. Statistical analysis
The association between dichotomic outcome variables (salivary MMP‐8 > 163 μg/l, median value, BOP > 20%, RC > median, PPD1, PPD2, and PPD3) and SNPs were modeled using logistic regression model. For continuous outcome (PPD total), linear regression model was used. We assumed additive effect of SNPs. For all outcomes, two models were calculated: unadjusted with SNP as only explanatory variable, and VPI, regular smoking (i.e., those who reported weekly smoking), and two separate occasions for birth cohorts adjusted model. All data analyses were carried out using R language (http://www.R‐project.org) with package “SNPassoc”(R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria, 2011). P values for each variable separately were corrected using false discovery rate (FDR) correction (Benjamini & Hochberg, 1995; Glickman, Rao, & Schultz, 2014). FDR q‐values less than 0.05 were considered significant. Population stratification or relatedness was not addressed.
1.5. Comparison of allele frequencies between populations
Allele frequencies of the SNPs with statistically significant findings (rs679620, rs520540, rs639752, rs3116496, and rs2228570) were compared with allele frequencies given in the ExAC database (http://exac.broadinstitute.org/). Frequencies for SNP rs639752 were not found in the ExAC database, thus they were searched for HapMap‐CEU from dbSNP database (http://www.ncbi.nlm.nih.gov/SNP/). In addition, the frequencies were compared with American population (1000 genomes AMR (1000 Genomes Ad Mixed American, including populations: MXL, PUR, CLM, PEL) and HAPMAP‐MEX (Mexican ancestry in Los Angeles, California), obtained from dbSNP database (Table 2).
Table 2.
Comparison of allele frequencies between populations for SNPs with significant findings
| SNP | Allels (Major/minor) | MAF | ExAC | HapMap‐CEU | AMR | HapMap‐MEX | Letra et?al. (Brazil) # | Letra et?al. (US) # |
|---|---|---|---|---|---|---|---|---|
| rs679620 | G/A | 0.37 (A) | 0.40 | 0.59 | 0.32 | — | 0.48 | 0.41 |
| rs520540 | G/A | 0.37 (A) | 0.39 | 0.57 | 0.34 | — | 0.49 | 0.43 |
| rs639752 | T/G | 0.38 (G) | — | 0.57 | 0.34 | 0.35 | 0.48 | 0.51 |
| rs3116496 | T/C | 0.16 (C) | 0.14 | 0.17 | 0.11 | 0.13 | ||
| rs2228570 | A/G | 0.34 (A) | 0,36 | 0.41 | 0.48 | 0.52 |
MAF = minor allele frequency, minor allele given in parentheses; ExAC frequency given for European (Finnish) population; HapMap‐CEU = Utah residents with northern and western European ancestry from the CEPH collection; AMR = ad mixed American population; Mexican ancestry in Los Angeles, California.
Letra et?al. (2012), ref.no7.
2. RESULTS
Saliva MMP‐8 median values were calculated and appeared to be 163.1 μg/l. However, no statistical significant findings were observed between salivary MMP‐8 median values and SPNs, which were analyzed in this study (Table 1).
Of the subjects (n = 94), 18 were healthy (BOP < 20% and no ≥ 4 mm pockets), 15 subjects had gingivitis (BOP ≥ 20% and no ≥ 4 mm pockets), and 61 had one or more ≥4 mm pocket (PPD1). Fifty‐one subjects had two or more ≥4 mm pockets (PPD2) and 37 had three or more ≥4 mm pockets (PPD3). Table 3 gives the basic characteristics of the study population.
Table 3.
General characteristics of the participants with non‐missing DNA sample (n = 94)
| n | % | Missing (n) | ||||
|---|---|---|---|---|---|---|
| Male gender | 47 | 50 | ||||
| Female gender | 47 | 50 | ||||
| MMP‐8 > 163 μg/l | 30 | 52.6 | 37 | |||
| BOP > 20% | 54 | 57.4 | ||||
| RC > median 5.85 | 47 | 50 | ||||
| Smoking regularly # | 16 | 17 | ||||
| PPD1 | 61 | 64.9 | ||||
| PPD2 | 51 | 54.3 | ||||
| PPD3 | 37 | 39.4 | ||||
| Min. | Max. | Median | Mean | SD | Missing (n) | |
|---|---|---|---|---|---|---|
| Age | 15 | 17 | 15 | 15.51 | 0.76 | |
| MMP‐8 μg/l | 2.42 | 1147 | 163.1 | 176.4 | 165.9 | 37 |
| BOP% | 0.6 | 80.4 | 32.1 | 32.82 | 24.6 | |
| VPI % | 0 | 95.8 | 27.55 | 31.67 | 24.6 | |
| RC % | 0 | 79.2 | 5.85 | 9.52 | 12.6 | |
| PPD total | 0 | 32 | 2 | 4 | 5.8 |
Smoking weekly.
BOP = bleeding on probing; RC = root calculus; PPD1 = one or more ≥4mm pocket; PPD2 = two or more ≥4 mm pockets; PPD3 = three or more ≥4 mm pockets; VPI = visible plaque index.
The following results were obtained on the genetic polymorphism and periodontal disease clinical parameters and biomarkers. After false discovery rate control (FDR, q < 0.05) only for PPD3 (three or more ≥4 mm pockets) and MMP‐3 (rs679620, rs520540, and rs639752), PPD3 and CD28 (rs3116496) and PPD3 and VDR (rs2228570) were statistically significant (q = 0.04, 0.04, 0.04, and 0.04, respectively). Smoking or VPI did not affect these results. The significant results for the adjusted model are given in Table 4, and all association results (all phenotypes and both models) are given in the Supporting Information.
Table 4.
All significant (FDR q < 0.05) SNP associations between the analyzed SNPs and phenotypes of the VPI and weekly smoking adjusted model. Major alleles were used as reference
| Phenotype | Gene | SNP | Allele | OR | Lower | Upper | p‐value | FDR q‐value |
|---|---|---|---|---|---|---|---|---|
| PPD3 | MMP3 | rs679620 | A | 0.22 | 0.09 | 0.57 | 0.0006 | 0.04 |
| PPD3 | MMP3 | rs520540 | A | 0.31 | 0.14 | 0.69 | 0.0020 | 0.04 |
| PPD3 | MMP3 | rs639752 | G | 0.34 | 0.15 | 0.73 | 0.0036 | 0.04 |
| PPD3 | CD28 | rs3116496 | C | 0.20 | 0.06 | 0.64 | 0.0015 | 0.04 |
| PPD3 | Vitamin D receptor | rs2228570 | T | 2.85 | 1.39 | 5.83 | 0.0024 | 0.04 |
P‐value obtained from logistic regression corrected for multiple testing by false discovery rate (FDR).
PPD3 = three or more ≥4 mm pockets.
Allele frequencies of the five SNPs with statistically significant findings (rs679620, rs520540, rs639752, rs3116496, and rs2228570) were compared with allele frequencies in Finnish, European, and American populations (ExAC, HAPMAP‐CEU, 1000 genomes AMR and HAPMAP‐MEX). No major population specific allele differences were detected.
3. DISCUSSION
The main finding of this study was that three MMP3 SNPs (rs67620, rs520540, and rs639752) were associated as exposure agents with three or more at least 4‐mm deep periodontal pockets by the strict statistical analysis (FDR). Interestingly, the same SNPs have previously been significantly associated to chronic adult periodontitis (rs679620 in US Caucasians), rs520540 on a trend level in US Caucasians, and rs639752 in US Caucasians and Brazilians (Letra et al., 2012). Our results also showed that those with clinical signs of initial periodontitis were separated from the healthy ones using the same definitions as in our previous publications of the whole cohort (Heikkinen et al., 2010; Heikkinen, 2011; Heikkinen et al., 2008; Heikkinen et al., 2016).
Stromelysin‐1 (MMP‐3) is involved in periodontal extracellular matrix processing of the basal membrane as well as the MMP activation cascades (Sorsa et al., 2004; Uitto, Overall, & McCulloch, 2003). Fibroblasts, chondrocytes, and endothelial cells can produce MMP‐3 as a part of inflammatory tissue destruction cascades (Haerian, Adonogianaki, Mooney, Docherty, & Kinane, 1995; Alpagot, Bell, Lundergan, Chambers, & Rudin, 2001; Reynolds, Hembry, & Meikle, 1994). Increased MMP‐3 levels in gingival crevicular fluid reflecting the course and severity of periodontitis have also been reported by Toyman et al. (2015) in adult periodontitis patients (Toyman et al., 2015). Previously, Ding et al. (2015) reported in their meta‐analysis that MMP3 ‐1171 5A/6A polymorphism (rs35068180, not studied here) could be associated with decreased risk of periodontitis in an Asian population (Ding et al., 2015); however, Itagaki et al. (2004) pointed that MMP‐3 gene promoter polymorphisms did not influence the susceptibility to periodontitis in Japanese patients (Itagaki et al., 2004).
Another finding of this study was that CD28 SNP rs3116496 [also known as +17(T/C)] major allele T associated with PPD3 in the adolescents. Previously, the polymorphism did not show any association with periodontitis in adults (e Silva et al., 2013). However, among non‐smokers, they reported a higher frequency of the T− (CC) genotype in aggressive periodontitis compared with chronic periodontitis.
Furthermore, we observed that Vitamin D receptor SNP rs2228570 (also known as FokI) major allele C (also known as F allele) was protective to PPD3. In line, Naito et al. (2007) reported that heterozygous Ff individuals had a lower risk of severe chronic periodontitis than individuals without the F allele (Naito et al., 2007). On the contrary, Li et al. (2008) reported that F allele increased the susceptibility of aggressive periodontitis in Chinese (Li et al., 2008) and Park, Nam, and Choi (2006) reported that CC genotype associated with increased risk for generalized aggressive periodontitis in Koreans (Park et al., 2006). However, several studies have also reported no association between rs2228570 and periodontis (Wang, Zhang, & Chen, 2015; Wang et al., 2009; El Jilani et al., 2015; Tachi et al., 2003).
Laine et al. (2012) pointed in their review article that an association could be found between periodontitis and haplotypes in theIL4 and IL6 and VDR genes (Laine et al., 2012). Earlier, Kornman et al. (1997) observed that “severe periodontitis patients were accounted for by either smoking or the IL‐1 genotype” (Kornman et al., 1997). In our study, no such effects could be seen; in our study only 17% were regular smokers, and thus this potential confounding factor could not be detected. In this respect, cytokine gene polymorphisms are observed to be quite ambivalent.
However, as a limitation of this study of adolescents, the sample size was quite small. Majority of the participants were lost because in Finland it is difficult to obtain permission from adolescents and their parents for a genetic study. This study has several strengths such as comprehensive oral health examination, unique age group, and adolescents. Despite the strengths, the limited sample size needs to be acknowledged.
To conclude, our study was the first to investigate in an adolescent population the genetic background of pathogenesis of initial or early periodontitis. We found that the genetic polymorphisms in MMP3, CD28, or VDR gene seem to be important in this respect, otherwise than MMP8 gene, as might be assumed (Holla, Hrdlickova, Vokurka, & Fassmann, 2012). However, more investigations are needed in larger materials for final conclusion. Further, new point‐of‐care chair‐side diagnostic tools have been developed to conveniently and consistently identify those adolescents with elevated risk for ongoing active gingivitis and periodontitis to be guided to regular examination and treatments (Heikkinen et al., 2016). Nevertheless, our study hypothesis was confirmed by showing an association between genetic polymorphism and periodontal disease parameters and markers.
CONFLICT OF INTEREST
All authors certify that they have no affiliations with or involvement in any organization or entity with any financial interest or non‐financial interest in the subject matter or materials discussed in this manuscript.
Supporting information
Table S1 Supporting info item
ACKNOWLEDGMENTS
This work was supported by Helsinki University Hospital Research Foundation Grants TYH 2013353 and TYH 2014244 and TYH 2016251grants from the Finnish Dental Society Apollonia, and from the Karolinska Institutet, Stockholm, Sweden.
Heikkinen, A. M. , Kettunen, K. , Kovanen, L. , Haukka, J. , Elg, J. , Husu, H. , Tervahartiala, T. , Pussinen, P. , Meurman, J. , and Sorsa, T. Inflammatory mediator polymorphisms associate with initial periodontitis in adolescents. Clinical and Experimental Dental Research 2016;2:208–215. doi: 10.1002/cre2.40.
REFERENCES
- Alpagot, T. , Bell, C. , Lundergan, W. , Chambers, D. W. , & Rudin, R. (2001). Longitudinal evaluation of GCF MMP‐3 and TIMP‐1 levels as prognostic factors for progression of periodontitis. Journal of Clinical Periodontology, 28, 353–359. [DOI] [PubMed] [Google Scholar]
- Barrett, J. C. , Fry, B. , Maller, J. , & Daly, M. J. (2005). Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics, 15(21), 263–265. [DOI] [PubMed] [Google Scholar]
- Benjamini, Y. , & Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Royal Statistics Soc. Ser B (Methodological), 57, 289–300. [Google Scholar]
- Chen, L. L. , Li, H. , Zhang, P. P. , & Wang, S. M. (2012). Association between vitamin D receptor polymorphisms and periodontitis: A meta‐analysis. Periodontology 2000, 83, 1095–1103. [DOI] [PubMed] [Google Scholar]
- de Souza, A. P. , Trevilatto, P. C. , Scarel‐Caminaga, R. M. , de Brito, R. B Jr. , Barros, S. P. , & Line, S. R. (2005). Analysis of the MMP‐9 (C‐1562 T) and TIMP‐2 (G‐418C) gene promoter polymorphisms in patients with chronic periodontitis. Journal of Clinical Periodontology, 32, 207–211. [DOI] [PubMed] [Google Scholar]
- Ding, C. , Chen, X. , Zhang, P. T. , Huang, J. P. , Xu, Y. , Chen, N. , & Zhong, L. J. (2015). Matrix metalloproteinase‐3 ‐1171 5A/6A polymorphism (rs35068180) is associated with risk of periodontitis. Scientific Reports, 5, 11667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- e Silva, M. R. , Moreira, P. R. , da Costa, G. C. , Saraiva, A. M. , de Souza, P. E. , Amormino, S. A. , … Dutra, W. O. (2013). Association of CD28 and CTLA‐4 gene polymorphisms with aggressive periodontitis in Brazilians. Oral Diseases, 19, 568–576. [DOI] [PubMed] [Google Scholar]
- El Jilani, M. M. , Mohamed, A. A. , Zeglam, H. B. , Alhudiri, I. M. , Ramadan, A. M. , Saleh, S. S. , … Enattah, N. S. (2015). Association between vitamin D receptor gene polymorphisms and chronic periodontitis among Libyans. Libyan J Med, 19(10), 216–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emingil, G. , Han, B. , Gürkan, A. , Berdeli, A. , Tervahartiala, T. , Salo, T. , … Sorsa, T. (2014). Matrix metalloproteinase (MMP)‐8 and tissue inhibitor of MMP‐1 (TIMP‐1) gene polymorphisms in generalized aggressive periodontitis: Gingival crevicular fluid MMP8 and TIMP‐1 levels and outcome of periodontal therapy. Journal of Periodontology, 85, 1070–1080. [DOI] [PubMed] [Google Scholar]
- Erciyas, K. , Pehlivan, S. , Sever, T. , & Orbak, R. (2010). Genetic variation of myeloperoxidase gene contributes to aggressive periodontitis: a preliminary association study in Turkish population. Disease Markers, 28, 95–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folwaczny, M. , Glas, J. , Török, H. P. , Limbersky, O. , & Folwaczny, C. (2004). Toll‐like receptor (TLR) 2 and 4 mutations in periodontal disease. Clinical and Experimental Immunology, 135, 330–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman, M. E. , Rao, S. R. , & Schultz, M. R. (2014). False discovery rate control is a recommended alternative to Bonferroni‐type adjustments in health studies. Journal of Clinical Epidemiology, 67, 850–857. [DOI] [PubMed] [Google Scholar]
- Gürkan, A. , Emingil, G. , Saygan, B. H. (2008). Gene polymorphisms of matrix metalloproteinase‐2, ‐9 and ‐12 in periodontal health and severe chronic periodontitis. Archives of Oral Biology, 53, 337–345. [DOI] [PubMed] [Google Scholar]
- Haerian, A. , Adonogianaki, E. , Mooney, J. , Docherty, J. P. , & Kinane, D. F. (1995). Gingival crevicular stromelysin, collagenase and tissue inhibitor of metalloproteinases levels in healthy and diseased sites. Journal of Clinical Periodontology, 22, 505–509. [DOI] [PubMed] [Google Scholar]
- Heikkinen, A. M. (2011). Oral health, smoking and adolescence. University of Helsinki, Faculty of Medicine, Institute of Dentistry. http://urn.fi/URN:ISBN:978‐952‐10‐7250‐5.
- Heikkinen, A. M. , Pajukanta, R. , Pitkäniemi, J. , Broms, U. , Sorsa, T. , Koskenvuo, M. , & Meurman, J. H. (2008). The effect of smoking on periodontal health of 15‐ to 16‐year‐old adolescents. Journal of Periodontology, 79, 2042–2047. [DOI] [PubMed] [Google Scholar]
- Heikkinen, A. M. , Sorsa, T. , Pitkäniemi, J. , Tervahartiala, T. , Kari, K. , Broms, U. , … Meurman, J. H. (2010). Smoking affects diagnostic salivary periodontal disease biomarker levels in adolescents. Journal of Periodontology, 81, 1299–1307. [DOI] [PubMed] [Google Scholar]
- Heikkinen, A. M. , Pitkäniemi, J. , Kari, K. , Pajukanta, R. , Elonheimo, O. , Koskenvuo, M. , & Meurman, J. H. (2012). Effect of teenage smoking on the prevalence of periodontal bacteria. Clinical Oral Investigations, 16, 571–580. [DOI] [PubMed] [Google Scholar]
- Heikkinen, A. M. , Nwhator, S. O. , Rathnayake, N. , Mäntylä, P. , Vatanen, P. , & Sorsa, T. (2016). Pilot study on oral health status as assessed by an active matrix metalloproteinase‐8 chairside mouthrinse test in adolescents. Journal of Periodontology, 87, 36–40. [DOI] [PubMed] [Google Scholar]
- Holla, L. I. , Fassmann, A. , Muzík, J. , Vanek, J. , & Vasku, A. (2006). Functional polymorphisms in the matrix metalloproteinase‐9 gene in relation to severity of chronic periodontitis. Journal of Periodontology, 77, 1850–1855. [DOI] [PubMed] [Google Scholar]
- Holla, L. I. , Hrdlickova, B. , Vokurka, J. , & Fassmann, A. (2012). Matrix metalloproteinase 8 (MMP8) gene polymorphisms in chronic periodontitis. Archives of Oral Biology, 57, 188–196. [DOI] [PubMed] [Google Scholar]
- Hughes, F. J. , Turner, W. , Belibasakis, G. , & Martuscelli, G. (2006). Effects of growth factors and cytokines on osteoblast differentiation. Periodontology 2000, 41, 48–72. [DOI] [PubMed] [Google Scholar]
- Itagaki, M. , Kubota, T. , Tai, H. , Shimada, Y. , Morozumi, T. , & Yamazaki, K. (2004). Matrix metalloproteinase‐1 and ‐3 gene promoter polymorphisms in Japanese patients with periodontitis. Journal of Clinical Periodontology, 31, 764–769. [DOI] [PubMed] [Google Scholar]
- Kornman, K. S. , Crane, A. , Wang, H. Y. , di Giovine, F. S. , Newman, M. G. , Pirk, F. W. , … Duff, G. W. (1997). The interleukin‐1 genotype as a severity factor in adult periodontal disease. Journal of Clinical Periodontology, 24, 72–77. [DOI] [PubMed] [Google Scholar]
- Laine, M. L. , Loos, B. G. , & Crielaard, W. (2010). Gene polymorphisms in chronic periodontitis. Int J Dent, 2010, 324719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine, M. L. , Crielaard, W. , & Loos, B. G. (2012). Genetic susceptibility to periodontitis. Periodontology 2000, 58, 37–68. [DOI] [PubMed] [Google Scholar]
- Letra, A. , Silva, R. M. , Rylands, R. J. , Silveira, E. M. , de Souza, A. P. , Wendell, S. K. , … Vieira, A. R. (2012). MMP3 and TIMP1 variants contribute to chronic periodontitis and may be implicated in disease progression. Journal of Clinical Periodontology, 39, 707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. , Yang, M. H. , Zeng, C. A. , Wu, W. L. , Huang, X. F. , Ji, Y. , & Zeng, J. Q. (2008). Association of vitamin D receptor gene polymorphisms in Chinese patients with generalized aggressive periodontitis. Journal of Periodontal Research, 43, 360–363. [DOI] [PubMed] [Google Scholar]
- Liede, K. E. , Haukka, J. K. , Hietanen, J. H. , Mattila, M. H. , Rönkä, H. , & Sorsa, T. (1999). The association between smoking cessation and periodontal status and salivary proteinase levels. Journal of Periodontology, 70, 1361–1368. [DOI] [PubMed] [Google Scholar]
- Naito, M. , Miyaki, K. , Naito, T. , Zhang, L. , Hoshi, K. , Hara, A. , … Nakayama, T. (2007). Association between vitamin D receptor gene haplotypes and chronic periodontitis among Japanese men. International Journal of Medical Sciences, 22(4), 216–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap consortium (2003). The international HapMap project. Nature, 18(426), 789–796. HapMap database ( http://www.hapmap.org ) [DOI] [PubMed] [Google Scholar]
- Opal, S. M. , & DePalo, V. A. (2000). Anti‐inflammatory cytokines. Chest, 117, 1162–1172. [DOI] [PubMed] [Google Scholar]
- Park, K. S. , Nam, J. H. , & Choi, J. (2006). The short vitamin D receptor is associated with increased risk for generalized aggressive periodontitis. Journal of Clinical Periodontology, 33, 524–528. [DOI] [PubMed] [Google Scholar]
- R Development Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: 2011. http://www.R‐project.org/. [Google Scholar]
- Reynolds, J. J. , Hembry, R. M. , & Meikle, M. C. (1994). Connective tissue degradation in health and periodontal disease and the roles of matrix metalloproteinases and their natural inhibitors. Advances in Dental Research, 8, 312–319. [DOI] [PubMed] [Google Scholar]
- Schröder, N. W. , & Schumann, R. R. (2005). Single nucleotide polymorphisms of toll‐like receptors and susceptibility to infectious disease. The Lancet Infectious Diseases, 5, 156–164. [DOI] [PubMed] [Google Scholar]
- Sorsa, T. , Tjäderhane, L. , & Salo, T. (2004). Matrix metalloproteinases (MMPs) in oral diseases. Oral Diseases, 10, 311–318. [DOI] [PubMed] [Google Scholar]
- Sorsa, T. , Tjäderhane, L. , Konttinen, Y. T. , Lauhio, A. , Salo, T. , Lee, H. M. , … Mäntylä, J. (2006). Matrix metalloproteinases: Contribution to pathogenesis, diagnosis and treatment of periodontal inflammation. Annals of Medicine, 38, 306–321. [DOI] [PubMed] [Google Scholar]
- Sorsa, T. , Gursoy, U. K. , Nwhator, S. , Hernandez, M. , Tervahartiala, T. , Leppilahti, J. , … Mäntylä, P. (2016). Analysis of matrix metalloproteinases, especially MMP‐8, in gingival creviclular fluid, mouthrinse and saliva for monitoring periodontal diseases. Periodontology 2000, 70, 142–163. [DOI] [PubMed] [Google Scholar]
- Sun, X. , Meng, H. , Shi, D. , Xu, L. , Zhang, L. , Chen, Z. , … Lu, R. (2011). Analysis of plasma calprotectin and polymorphisms of S100A8 in patients with aggressive periodontitis. Performance Research, 46, 354–360. [DOI] [PubMed] [Google Scholar]
- Suominen‐Taipale, L. , Nordblad, A. , Vehkalahti, M. , & Aromaa, A. (2008). Oral health in the finnish adult population. Health 2000 Survey. Publications of the National Public Health Institute B 25 / 2008.
- Tachi, Y. , Shimpuku, H. , Nosaka, Y. , Kawamura, T. , Shinohara, M. , Ueda, M. , … Ohura, K. (2003). Vitamin D receptor gene polymorphism is associated with chronic periodontitis. Life Sciences, 73, 3313–3321. [DOI] [PubMed] [Google Scholar]
- Toyman, U. , Tüter, G. , Kurtiş, B. , Kıvrak, E. , Bozkurt, Ş. , Yücel, A. A. , & Serdar, M. (2015). Evaluation of gingival crevicular fluid levels of tissue plasminogen activator,plasminogen activator inhibitor 2, matrix metalloproteinase‐3 and interleukin 1‐β in patients with different periodontal diseases. Journal of Periodontal Research, 50, 44–51. [DOI] [PubMed] [Google Scholar]
- Uitto, V. J. , Overall, C. M. , & McCulloch, C. (2003). Proteolytic host cell enzymes in gingival crevice fluid. Periodontology 2000, 31, 77–104. [DOI] [PubMed] [Google Scholar]
- Vasconcelos, D. F. , da Silva, M. A. , Marques, M. R. , de Brito Júnior, R. B. , Vasconcelos, A. C. , & Barros, S. P. (2012). Lymphotoxin‐alpha gene polymorphism +252A/G (rs909253, A/G) is associated with susceptibility to chronic periodontitis: A pilot study. ISRN Dent, 2012, 617245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. , Zhao, H. , Xiao, L. , Xie, C. , Fan, W. , Sun, S. , … Zhang, J. (2009). Association between vitamin D receptor gene polymorphisms and severe chronic periodontitis in a Chinese population. Journal of Periodontology, 80, 603–608. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Zhang, T. L. , & Chen, D. (2015). Lack of association between the vitamin D receptor polymorphism rs2228570 and chronic periodontitis in a Han Chinese population. Genetics and Molecular Research, 14, 12299–122305. [DOI] [PubMed] [Google Scholar]
- World Health Organization, International Statistical Classification of Diseases and Related Health Problems , 10th Revision (2007). Available: http://www.who.int/classifications/apps/icd/icd10online/[2007, 2007, 09/14].
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Supporting info item


