Summary
Marine phototroph and heterotroph interactions are vital in maintaining the nutrient balance in the oceans as essential nutrients need to be rapidly cycled before sinking to aphotic layers. The aim of this study was to highlight the molecular mechanisms that drive these interactions. For this, we generated a detailed exoproteomic time‐course analysis of a 100‐day co‐culture between the model marine picocyanobacterium Synechococcus sp. WH7803 and the Roseobacter strain Ruegeria pomeroyi DSS‐3, both in nutrient‐enriched and natural oligotrophic seawater. The proteomic data showed a transition between the initial growth phase and stable‐state phase that, in the case of the heterotroph, was caused by a switch in motility attributed to organic matter availability. The phototroph adapted to seawater oligotrophy by reducing its selective leakiness, increasing the acquisition of essential nutrients and secreting conserved proteins of unknown function. We also report a surprisingly high abundance of extracellular superoxide dismutase produced by Synechococcus and a dynamic secretion of potential hydrolytic enzyme candidates used by the heterotroph to cleave organic groups and hydrolase polymeric organic matter produced by the cyanobacterium. The time course dataset we present here will become a reference for understanding the molecular processes underpinning marine phototroph‐heterotroph interactions.
Introduction
The ocean is the Earth's largest biome covering 70% of the world's surface. Marine systems play a major role in global climate regulation, not only due to their ability to store and transport heat, but also because of the constant atmosphere—ocean exchange of CO2. Oceans are major carbon reservoirs and are known to buffer anthropogenic carbon emissions by drawing CO2 from the atmosphere and burying it in the deep ocean, a process known as the biological carbon pump (Jiao and Zheng, 2011; De La Rocha and Passow, 2014). This is a process whereby CO2 in the upper ocean is fixed by photosynthetic primary producers to form organic matter that is then transported to the deeper ocean by sedimenting particulate organic matter (POM) and the drawdown of dissolved organic matter (DOM) through mixing and dwelling (Jiao et al., 2010). Marine microbes are also major drivers of other global biogeochemical cycles such as those of nitrogen and sulphur (Karl and Church, 2014).
Marine picocyanobacteria, mainly belonging to the genera Prochlorococcus and Synechococcus, are numerically the world's dominant photosynthetic primary producers and a major component of marine phytoplankton. Despite being outnumbered by the heterotrophic community, these cyanobacteria are responsible for half of the marine primary production and play a key role in sustaining marine food webs (Falkowski, 2012). Hence, phytoplankton are at the base of the marine food chain feeding the ecosystem with DOM and POM that is released via cell lysis (e.g., cell death, inefficient grazing and viral lysis) or other cellular processes (e.g., outer membrane vesicles, active efflux processes or permeable membrane leakage) (Biller et al., 2014; Christie‐Oleza et al., 2015a, 2017; Grossowicz et al., 2017). Most of this organic matter will be used by the heterotrophic bacterioplankton as their main source of carbon and energy, returning inorganic nutrients to the phototrophic community (Christie‐Oleza et al., 2017). The Roseobacter group of the class Alphaproteobacteria is an important component of the microbial community within the marine euphotic layer, accounting for up to 20% of the sea surface bacterioplankton (Wagner‐Dobler and Biebl, 2006). This group of heterotrophs generally shows a positive correlation with the phytoplankton community and forms intimate associations with specific phytoplankton groups (Giebel et al., 2011; Morris et al., 2012). Interestingly, marine Roseobacter strains present a large genomic capability and metabolic versatility to use an array of organic substrates found within phytoplankton exudates and, hence, they are one of the first bacterioplankton groups to react to the input of organic matter produced, for example, during phytoplankton blooms (Newton et al., 2010; Romera‐Castillo et al., 2011; Christie‐Oleza et al., 2012a; Buchan et al., 2014; Landa et al., 2016; Simon et al., 2017). The strain Ruegeria pomeroyi DSS‐3 was the first roseobacterium to have its genome sequenced (Moran et al., 2004) and has served as a model organism to study biogeochemical, ecological and physiological strategies of this group of heterotrophic marine bacteria (Christie‐Oleza and Armengaud, 2015).
Around 70% of the DOM in the oceans is considered of low‐molecular weight (< 1 kDa) (Benner, 2002). The other 30% of DOM is of high molecular weight (> 1 kDa) and, curiously, is much less refractory and, hence, more readily degraded than the low‐molecular weight fraction (Decho and Gutierrez, 2017). Most of the high molecular weight DOM is in the form of biopolymers, and because biological membrane systems are only permeable to molecules smaller than 0.6 kDa (Weiss et al., 1991), exoenzymes or ectoenzymes play a key pivotal role in polymeric DOM hydrolysis and assimilation (Vetter and Deming, 1999) as these biopolymers must be hydrolyzed outside the cell before they can be taken up by most organisms (i.e., nongrazing organisms). The activities of secreted enzymes in marine microbes has generally been assessed through the use of fluorescently labeled substrates (Karner and Herndl, 1992; Martinez et al., 1996; D'Ambrosio et al., 2014; Arnosti, 2015), simple observation of bacterial growth on different polymeric substrates (e.g., Mitulla et al., 2016) or, more rarely, identifying and characterizing the actual enzymes involved in hydrolyzing the DOM (e.g., Hehemann et al., 2014; Xing et al., 2015). Interestingly, marine bacterial exoenzymes are proving highly distinct from their well‐characterized terrestrial counterparts (Michel and Czjzek, 2013) and are currently poorly identified.
The use of shotgun proteomics has become a powerful tool for detecting the array of proteins present in the extracellular medium of an organism under different experimental conditions (Armengaud et al., 2012) and, hence, a reliable high throughput method for identifying key secreted enzymes and proteins involved in cell‐to‐cell and cell–environment interactions. We previously analyzed the exoproteome of various Roseobacter strains grown in rich media and showed a diversity of trophic strategies within this clade, that is, through the abundant detection of secreted nutrient transporters, mobility proteins, adhesion‐like proteins or toxins (Christie‐Oleza et al., 2012b). Nevertheless, Roseobacter strains induced a completely different array of secreted proteins when grown in the presence of a more realistic source of organic matter, that is, DOM and POM produced by marine Synechococcus, with an increase in proteins involved in motility, microbial interactions, hydrolytic activities and capsid proteins of the genetic transfer agent (GTA) encoded in the genome and a strong decrease in the secretion of toxin‐like proteins (Christie‐Oleza et al., 2015b).
Synechococcus species generate large amounts of organic matter and requires the presence of a specialized heterotrophic community to remineralise the leaked photosynthate and obtain a constant feed‐back of inorganic nutrients (Christie‐Oleza et al., 2017). Based on this basic principle of phototroph–heterotroph interaction, illuminated R. pomeroyi–Synechococcus sp. co‐cultures are able to survive for extended time periods both in mineral enriched media and natural oligotrophic seawater (Christie‐Oleza et al., 2017). In this study, we analyzed for the first time the exoproteome of R. pomeroyi–Synechococcus co‐cultures under an extended time period (i.e., 100 days) both in nutrient‐enriched and natural oligotrophic seawater in order to generate a unique time course dataset that would highlight the molecular mechanisms involved in this dependent microbial interaction over time. We therefore aimed to (i) determine protein pattern shifts over the 100‐day time course and observe whether culture stability is reflected by a consistent exoproteome; (ii) identify culture stages and frame the trophic strategy of each microbe at each time point; and (iii) identify the secreted hydrolytic enzymes produced by the heterotroph and evaluate their variations over time.
Results and discussion
Correlation between culture growth and exoproteomes
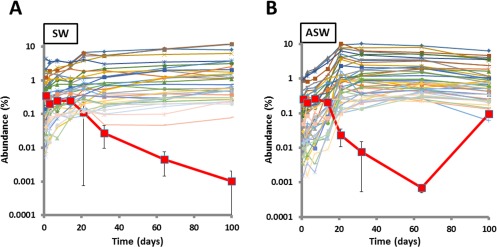
In both nutrient‐rich and oligotrophic media, the sustained survival of the co‐culture comes as a consequence of nutrient cycling between the phototroph and heterotroph (Christie‐Oleza et al., 2017). Here, the growth of Synechococcus sp. WH7803 and R. pomeroyi DSS‐3 during the 100 days co‐culture in both natural oligotrophic seawater (SW) and enriched artificial seawater (ASW) (Fig. 1) showed the expected trends as previously reported (Christie‐Oleza et al., 2017). While Synechococcus sp. WH7803 reaches high cell densities in ASW (> 109 cells ml−1) and growth is limited by light, in natural SW the cyanobacterium is limited by the availability of inorganic nutrients and cell densities reached 105 cells ml−1, numbers that are similar to those observed in natural marine ecosystems (Parsons et al., 2012). The heterotroph, R. pomeroyi DSS‐3, is limited by the availability of organic carbon and vitamins regenerating essential nutrients for the phototroph. Synechococcus–R. pomeroyi co‐cultures can persist over 200 days (Christie‐Oleza et al., 2017), but a sTable state is reached after just 21 days in natural SW (Fig. 1). However, in rich ASW medium, Synechococcus sp. WH7803 only enters the sTable state after 100 days when cell densities drop 10‐fold and stabilise at 108 cells ml−1 (Christie‐Oleza et al., 2017).
Figure 1.
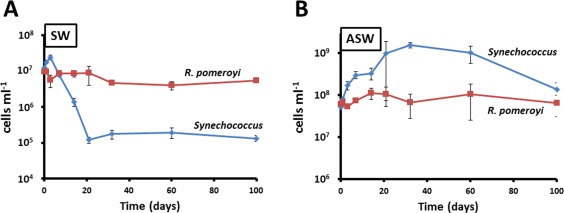
Growth curves of R. pomeroyi DSS‐3 and Synechococcus WH7803 over the 100‐day time course experiment in natural SW (A) and ASW medium (B). The average value of triplicate cultures (n = 3) is shown in panels (error bars show standard deviation).
Exoproteomes tend to reflect microbial adaptive strategies (Christie‐Oleza et al., 2012bb), and as expected, the variations observed in the exoproteomes presented here corresponded nicely with the different growth‐phase physiologies observed in SW and ASW grown co‐cultures as shown by the principal component analyses (PCA) using the normalized exoproteomic data obtained from Synechococcus sp. WH7803 and R. pomeroyi over the time course experiment. The sum of the two first principal components represented 82% and 93% of the variability within the exoproteome of Synechococcus sp. WH7803 and R. pomeroyi over the 100 days, respectively (Fig. 2).
Figure 2.
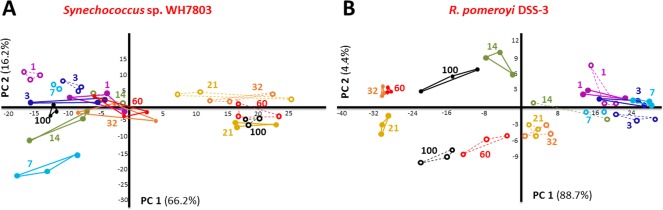
PCA of the normalized exoproteomes of Synechococcus sp. WH7803 (A) and R. pomeroyi DSS‐3 (B) when grown in co‐culture in ASW medium (solid circles and lines) and natural SW (open circles and dashed lines). Numbers refer to the culture day that the samples were collected.
PCA of the exoproteome of Synechococcus sp. WH7803
In natural SW, the exoproteome of Synechococcus showed a distinct shift between time points 1–14 and 21–100 days incubation (Fig. 2A), which coincides with the co‐culture entering the sTable state equilibrium observed at day 21 (Fig. 1). Interestingly, the exoproteome of ASW‐grown Synechococcus sp. WH7803 remained similar over the 100 days experiment, except for time point 21 which marks the transition from the exponential to the sTable state phase (Fig. 1). Curiously, time point 21 grouped with the sTable state time points of Synechococcus grown in oligotrophic SW (i.e., 21–100 days; Fig. 2A), suggesting a transitory peak of starvation in ASW media.
PCA of the exoproteome of R. pomeroyi
The heterotroph showed a similar transition over time in both nutrient‐poor and ‐rich media (Fig. 2B), mainly marked by the shift from exponential and sTable state cultures. Nevertheless, the slight divergence between SW and ASW exoproteomes was mainly resolved by the principal component 2 (Fig. 2B), suggesting a distinctness between nutrient‐rich and oligotrophic culture conditions.
Although the transitions that were observed over time are likely caused by the acclimation of the microbial populations to the varying conditions of the culture, genetic modifications (as demonstrated in Zambrano et al., 1993) cannot be ruled out and, hence, further research is needed to determine if genetic evolution also occurs during these long‐term co‐cultures.
The exoproteome of Synechococcus over the 100‐day time course
Just 187 and 221 of the 681 polypeptides detected in the co‐culture's exoproteome belonging to Synechococcus already represented 95% of the total protein abundance in SW and ASW, respectively. These proteins were grouped into functional categories (Supporting Information Table S4) and represented in Figure 3A.
Figure 3.
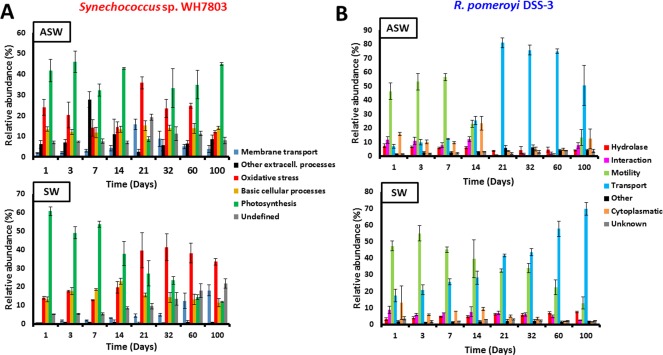
Functional category abundance of protein found in the exoproteomes of Synechococcus sp. WH7803 (A) and R. pomeroyi DSS‐3 (B) when grown in co‐culture in ASW medium and natural SW. The average value of triplicate cultures analyses (n = 3) are shown (error bars show standard deviation).
Photosynthesis and basic cellular processes
Synechococcus sp. release considerable amounts of organic matter not only in the form of carbohydrates (Biersmith and Benner, 1998; Bertlisson et al., 2005) but also in the form of protein (Christie‐Oleza et al., 2015a; 2017). Here, proteins involved in photosynthesis are among the most abundant categories found in the exoproteome of Synechococcus sp. WH7803, representing around 32% of the exoproteome (Table 1). Interestingly, this percentage is strikingly similar to the amount of photosynthetic proteins observed in cellular proteomic analyses (Table 1; Christie‐Oleza et al., 2017). While this may suggest that the exoproteome of Synechococcus is made entirely of proteins generated from cell lysis, other indicators such as the amount of cytoplasmic or ribosomal proteins are not in agreement. While ribosomal proteins in Synechococcus cells represent between 5.5% and 6.5% of the total cellular proteome, only between 1.0% and 1.6% of relative abundance of these proteins are found in the extracellular fraction (Table 1), suggesting that between 18% and 25% of the exoproteome may well come from cell lysis. When using cytoplasmic proteins as an indicator, the average percentage of predicted cell lysis increases to 50% (Table 1), but with bursts that coincide with the cell lysis expected from the growth curves. For example, the drastic reduction of Synechococcus cell abundance observed at days 7 and 14 under SW conditions (Fig. 1A) coincides with an increase in detection of cytoplasmic proteins in the exoproteome (i.e., 25–30%), suggesting that, during these time points, between 65% and 77% of the extracellular proteins may come from cell lysis. Nevertheless, at other time points cell lysis remains stably between 40% and 50%. It is interesting to note that some of these predicted cytoplasmic proteins are more abundant in the exoproteome than in the cellular fraction suggesting one of the following three possibilities: (i) they are more sTable in the extracellular milieu than other cytoplasmic proteins and, therefore, over time they are enriched in the exoproteome; (ii) they might actually be actively secreted; and (iii) they are ‘selectively leaked’ as suggested by others (Grossowicz et al., 2017). One of these proteins is the nucleotide‐binding protein SynWH7803_1823, a protein predicted to be involved in the temporal control of bacteriophage gene transcription, which represents 2.4% of Synechococcus exoproteome (almost 12% of the predicted cytoplasmic proteins in this fraction), whereas it only represents 0.1% in the cellular proteome. Proteins involved in photosynthesis are another example of unequal accumulation in the extracellular milieu or ‘selective leakiness’, and different elements of the photosynthetic apparatus are differentially partitioned, such as those from the phycobilisome (Table 1). Phycobilisomes should be localized on the cytoplasmic side of the thylakoidal membrane, but they seem to be enriched in the exoproteome of Synechococcus. Interestingly, the presence of these proteins from the photosynthetic antenna shows a strong decrease in the natural SW exoproteome over time (from 55% to 5% of the exoproteome; Fig. 3A and Supporting Information Table S4), suggesting a reduction in leakiness or a decrease in the production of cellular phycobilisomes as a consequence of nutrient stress. Protein trafficking in cyanobacteria remains poorly characterized (Schneider, 2014) and the ‘selective leakage’ observed here, such as of predicted cytoplasmic proteins or phycobilisomes, requires further research.
Table 1.
Relative abundance of ribosomal proteins and proteins form the photosynthetic apparatus detected in Synechococcus sp. WH7803 proteome datasets.
| Cellular fractiona | Exoproteome fractionb | |||||
|---|---|---|---|---|---|---|
| SW | ASW | SW | ASW | |||
| Synechococcus | ||||||
| Cytoplasmic proteinsc | 38.5 ± 1.1 | 33.1 ± 1.0 | 19.4 ± 4.9 | 50% | 16.6 ± 2.1 | 50% |
| Ribosomal proteins | 6.5 ± 0.9 | 5.5 ± 1.0 | 1.6 ± 0.8 | 25% | 1.0 ± 0.6 | 18% |
| Photosynthetic apparatus | 35.9 ± 1.3 | 39.4 ± 1.8 | 31.1 ± 19.8 | 87% | 32.2 ± 11.6 | 82% |
| Phycobilisomes | 29.8 ± 1.1 | 28.4 ± 1.6 | 28.2 ± 19.2 | 95% | 29.2 ± 11.7 | 103% |
| Other elements | 6.1 ± 0.4 | 11.1 ± 0.3 | 2.9 ± 0.8 | 48% | 2.9 ± 0.7 | 26% |
| R. pomeroyi | ||||||
| Cytoplasmic proteinsc | 57.6 ± 1.3 | n.a. | 6.0 ± 2.5 | 10% | 10.4 ± 6.4 | 18% |
| Ribosomal proteins | 8.0 ± 0.4 | n.a. | 0.2 ± 0.2 | 3% | 0.8 ± 0.7 | 10% |
n.a., not applicable due to the low number of proteins detected for R. pomeroyi in this analysis.
Data obtained from cellular proteomes published in Christie‐Oleza and colleagues (2017). Standard deviation from triplicate samples is shown.
This study. Standard deviation from triplicate experiments from all eight sampled time points is shown. Percentages were calculated by dividing the protein relative abundance obtained from exoproteome analyses by those obtained in cellular fractions.
Cytoplasmic proteins were predicted using the prediction server PSORTb.
Membrane transport
The periplasmic‐binding component of ABC transporters is commonly found in microbial exoproteomes (Christie‐Oleza and Armengaud, 2010; Johnson‐Rollings et al., 2014) and it has been suggested that some of these proteins could be intentionally translocated from the periplasm to the extracellular space (Giner‐Lamia et al., 2016). The detection of membrane transporters is a good indicator of (i) the nutrients that are targeted by each organism in a community (Christie‐Oleza et al., 2017) and (ii) an organisms' nutrient stress within the system (Saito et al., 2014). As expected, Synechococcus sp. WH7803 predominantly targets inorganic nutrients, mainly phosphate and metals such as iron (Supporting Information Table S4). Up to three different periplasmic substrate‐binding proteins for phosphate were detected in the ASW condition, although only one of them (i.e., SynWH7803_1045) was abundantly detected in both enriched and oligotrophic conditions. Interestingly, none of these periplasmic‐binding proteins co‐localizes in the genome with the other components of the ABC transporter for phosphate (i.e., permease and ATPase). Synechococcus shows a clear rise in P starvation over time under oligotrophic SW conditions as seen by the progressive increase in abundance of the periplasmic substrate binding protein for phosphate SynWH7803_1045 from 0.7% at day 1 to over 14% after the 100 day incubation (Supporting Information Table S4). A similar trend is observed for porins that are linked to P stress (SynWH7803_0993, 0.1–3.1%; SynWH7803_2236, below 0.1–0.7%) and an outer membrane efflux protein involved in protein secretion which needs further characterization (SynWH7803_2199, below 0.01–0.11%). In nutrient‐enriched ASW media, the cyanobacterium has a peak in all nutrient transport proteins (i.e., P and metals) at day 21, which then drops until day 100 (Fig. 3A), an trend that fits the variability observed in the PCA plot in Figure 2A.
Oxidative stress
Apart from nutrient recycling (Christie‐Oleza et al., 2017), the scavenging of reactive oxygen species (ROS) is considered a pivotal process in marine cyanobacteria – heterotroph interactions where the cyanobacterium, mostly lacking of catalase, relies on the heterotroph for depleting ROS (Hunken et al., 2008; Morris et al., 2011, 2008). Nevertheless, while catalase only catalyzes the decomposition of hydrogen peroxide, it is the enzyme superoxide dismutase (SOD) which helps deal with superoxide (O2–). SODs are found in all marine cyanobacteria although with different metal cofactors (Scanlan et al., 2009). While Ni‐containing SODs are prevalent in catalase‐lacking strains, Fe‐containing SODs prevail in catalase encoding cyanobacteria. Cellular proteomes of Synechococcus sp. WH7803 generally show below 1% abundance of this enzyme (Christie‐Oleza et al., 2017), but SOD has also been detected in the exoproteomes of different marine Synechococcus strains ranging between 0.7% and 3.4% (Christie‐Oleza et al., 2015a). SODs are possibly periplasmic‐located enzymes with a non‐canonical secretion system, but may also be ‘unavoidably leaked’ through the outer membrane, reinforcing the idea that ROS scavenging can be a public goods process as suggested by others (Morris et al., 2012). In this study, the abundance of the Fe‐containing SOD in the exoproteome of Synechococcus sp. WH7803 was surprisingly high throughout the entire time course experiment both in SW and ASW averaging 24% and 19% of total protein abundance, respectively (Supporting Information Table S4), and highlighting the relevance of this enzyme for dealing with ROS. The fact it was detected in both nutrient conditions is an indicator that its abundance is not an artefactual result and its variable abundance in ASW suggests it does not come as a consequence of its accumulation in the milieu. The second SOD encoded by Synechococcus sp. WH7803, the copper‐contain protein SynWH7803_0951, was also detected in both SW and ASW exoproteomes with an average detection of 0.19% and 0.03%, respectively.
Other extracellular processes
The variable abundance of structural type IV pili proteins in the exoproteome of Synechococcus sp. WH7803 is the most remarkable aspect within this functional category (Supporting Information Table S4). In a previous study, we already reported a high abundance of the pili proteins SynWH7803_1795 and SynWH7803_1796 (17.9% and 1.3%, respectively), which was exclusive to this strain (Christie‐Oleza et al., 2015a). The time course experiment has allowed us to assign the production of these proteins to nutrient‐rich conditions with a peak of SynWH7803_1795 detection at day 7 (25.7%) and an average detection of 8.1% over the 100 days, whereas the detection was low (< 0.1%) in natural SW conditions. The remarkable ecological and physiological roles of the pili need further characterization although a recent study in Synechococcus elongatus suggests this structure may be involved in cell buoyancy (Nagar et al., 2017). Interestingly, both alkaline phosphatases were detected in the exoproteome of Synechococcus sp. WH7803 but only in low abundance and peaked during day 21 in ASW (Supporting Information Table S4).
Secreted proteins of unknown function
Proteins of unknown function tend to predominate in the secreted fraction, highlighting the poor knowledge we currently have on microbe–environment interactions (Christie‐Oleza et al., 2015a). Datasets such as the one we present here are useful for flagging proteins of unknown function that are abundant and, hence, may have an important role and also to shed light on their possible function by monitoring their abundance over time. Interestingly, the five most abundant proteins in this category were common in both SW and ASW conditions (Supporting Information Table S4), but these showed interesting shifts in abundance over time (Fig. 4). The conserved proteins in bacteria or marine cyanobacteria SynWH7803_0982, SynWH7803_2169 and SynWH7803_1017 showed very similar behavior, with a progressive increase in oligotrophic SW and a peak in abundance at day 21 in ASW (Fig. 4), a time point when Synechococcus sp. WH7803 was nutrient‐starved as highlighted above. Hence, these proteins may have a role in response to nutrient stress. In contrast, protein SynWH7803_1824, which is unique to strain WH7803 and conserved only among Burkholderia, remained at a constant low abundance but became highly abundant in SW over time (Fig. 4). Finally, protein SynWH7803_1556 showed identical behavior in both culture conditions over time but with a lag of a few days in ASW.
Figure 4.
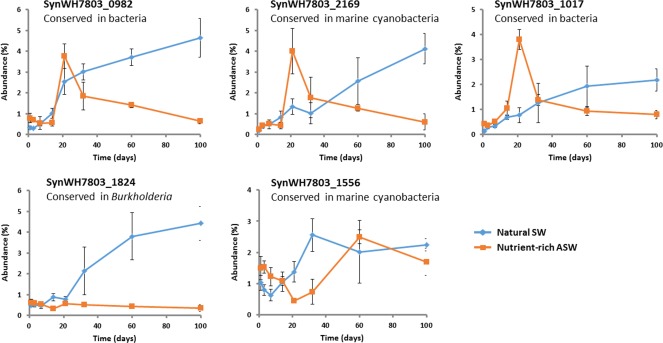
Variation of the five most abundant hypothetical proteins in the exoproteome of Synechococcus sp. WH7803 over time.
The average value of triplicate cultures analyses (n = 3) are shown (error bars show standard deviation).
The exoproteome of R. pomeroyi over time
From the 585 proteins detected in the exoproteome of the co‐culture belonging to R. pomeroyi, 184 and 222 represented over 95% of the total abundance in SW and ASW, respectively, and were grouped into functional categories (Supporting Information Table S5) and represented in Figure 3B.
Cytoplasmic proteins
As mentioned earlier, the detection of predicted cytoplasmic proteins and ribosomal proteins in the exoproteomes can be used as indicators of cell lysis. While the abundance of ribosomal proteins in cellular fractions of R. pomeroyi usually represents over 8% of the total proteome (Table 1), here we observed only 0.2% and 0.8% of these proteins in the exoproteomes of SW and ASW co‐cultures, respectively, suggesting that between 3% and 10% of the proteins detected in this study may come from cell lysis. Nevertheless, a higher cell lysis is suggested when predicted cytoplasmic proteins is used as an indicator (i.e., 10–18%; Table 1). There are several reasons that could explain this discrepancy: (i) that dying cells reduce their ribosomal protein content; (ii) the size of ribosomal complexes makes them less ‘leaky’ than other smaller cytoplasmic proteins; and (iii) the prediction of protein trafficking or selective leakiness has been understudied. For example, under both conditions (i.e., SW and ASW), the five most abundant cytoplasmic proteins in the exoproteome of R. pomeroyi already contributed to over 50% of the cytoplasmic category, being three of them common between both conditions (i.e., isocitrate dehydrogenase, a hypothetical protein with high similarity to a phosphoenolpyruvate mutase and imidazoleglycerol‐phosphate dehydratase; Supporting Information Table S5), hinting that these proteins, either moonlight (Jeffery, 2009), are dually secreted at least to the periplasm, or are highly sTable in the extracellular milieu and accumulate over time.
Motility
Ruegeria pomeroyi is a flagellum‐propelled motile bacterium. The extracellular flagellin filament and hook are rarely found in the cellular proteome as they are easily sheared off during sample processing and remain in the exoproteomic fraction. The flagellin was detected in very high abundance in the exoproteomes of both SW and ASW conditions (35% and 23%, respectively). During initial culture stages flagellin represented over 50% of the total protein abundance, but while its detection only reduced progressively over time in natural SW (down to 13% at day 100), it dropped to under 1% in ASW between time points 21 and 60 days to finally increase to 13% at day 100 (Fig. 3B). The sharp drop in motility between time points 21 and 60 days in ASW coincides with the maximum Synechococcus cell densities (Fig. 1) and, hence, highest photosynthate production and availability. Very low levels of flagellin were also detected in previous studies where R. pomeroyi was grown in carbon‐rich media (e.g., the flagellin was not detected in the exoproteome of this strain when grown in marine broth; Christie‐Oleza and Armengaud, 2010; Christie‐Oleza et al., 2012b), highlighting the ability of R. pomeroyi to switch between a motile–nonmotile lifestyle, which is dependent on nutrient availability. In order to prove this hypothesis, we visualized R. pomeroyi's motility under different nutrient conditions. No cell motility was observed when this strain was incubated in nutrient‐rich marine broth (containing 0.6%, wt/vol, of organic carbon, of which 0.5% is peptone and 0.1% yeast extract) or in carbon depleted media (i.e., ASW or ASW supplemented with ammonium and vitamins). Interestingly, R. pomeroyi did show notorious motility when incubated in filter‐sterilized ASW media obtained from a 2‐week old Synechococcus culture (Supporting Information Movie S1), which we predict contained ∼ 0.02% (wt/vol) of organic carbon (i.e., mainly protein) based on previous measurements (Christie‐Oleza et al., 2017). We then tested the motility of R. pomeroyi in the presence of varying concentrations of four different sources of organic matter (Fig. 5 and Supporting Information Movies S2 and S3). Glucose, yeast extract and peptone induced cell motility at concentrations as low as 0.005% (wt/vol). In contrast, higher substrate concentrations of yeast extract and peptone (i.e., 0.1%) caused a drop in motility (Fig. 5), suggesting a substrate repression in flagella biosynthesis as reported previously in other heterotrophic bacteria (Stella et al., 2008). These results are in agreement with the proteomic detection of flagellin when R. pomeroyi was grown in the presence of Synechococcus and demonstrates an adaptive life strategy of this strain in response to different levels of organic matter. Hence, R. pomeroyi remains immotile in total absence of a carbon source and very low concentrations of organic matter are enough to switch on its motile phenotype. The drop in flagella abundance observed in natural SW incubations (Fig. 3) is possibly caused by the lower concentrations of organic carbon produced by the phototroph under such nutrient‐deplete conditions. Previous measurements of organic matter in such oligotrophic conditions suggest concentrations as low as ∼ 0.0002% (wt/vol). While we did not test such low substrate concentrations in our controlled experiments (Fig. 5), these small amounts of substrate seem to be enough to induce a low level of flagella production and, hence, motility in R. pomeroyi. This is also supported by previous observations of 5.0–6.5% flagellin detected in an exoproteomic analysis where this heterotroph was incubated in natural coastal seawater (Christie‐Oleza et al 2012b). The abrupt decrease in flagellin abundance observed in nutrient‐enriched ASW conditions (Fig. 3) is more likely caused by elevated organic matter concentrations (i.e., > 0.1%), which represses the biosynthesis of this structure. This may well simulate a phytoplankton bloom where the heterotroph turns off its motility in order to remain within the high concentration patch.
Figure 5.
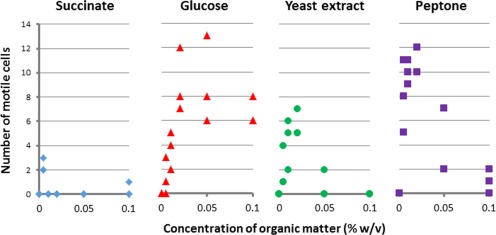
Motility of R. pomeroyi DSS‐3 in the presence of varying concentrations of different organic carbon sources.
Quantification of motility was based on the number of moving cells observed in 10‐s videos obtained from three different fields per condition.
Membrane transport
This is the most abundant category of proteins detected in the exoproteomes of R. pomeroyi (42% and 38% in ASW and SW, respectively), mostly made up of outer membrane transport systems and the periplasmic components of inner membrane transporters. For example, the periplasmic substrate‐binding component of ABC transporters made up 100% of the proteins detected for these transport systems, whereas the other components (i.e., permeases and ATP‐binding proteins) remained undetected (Supporting Information Table S5). It is interesting to note that the abundance of membrane transporters mirrors the one of cell motility and highlights how the cell invests resources in response to nutrient availability (Fig. 3B). This generalist marine heterotroph is adapted to scavenge a large variety of scarce nutrients in oligotrophic marine systems and to proliferate rapidly in nutrient‐rich patches of the ocean (Moran et al., 2004; Newton et al., 2010; Christie‐Oleza et al., 2012a, 2015b). The data we present here suggest that this life strategy is achieved by splitting resources between motility to search for nutrients when these are scarce and to rapidly take them up through the investment in membrane transporters when they are abundant. As expected, R. pomeroyi induces the production of a large array of specific membrane transporters for importing amino acids, amines and carbohydrates in the presence of Synechococcus, which is not surprising as these are the main components of the cyanobacterium photosynthate (Christie‐Oleza et al., 2017). Nevertheless, while all transporters showed an increase in abundance in accordance to the pattern shown by the ‘transport’ category in Figure 3B, the periplasmic component of the manganese ABC transporter showed a decrease in abundance in both SW and ASW conditions (Fig. 6). Although it is not clear which specific metal is transported by this transporter (i.e., manganese and zinc), R. pomeroyi clearly decreases its uptake possibly favoring the phototroph's high demand in metals for primary production.
Figure 6.
Abundance of R. pomeroyi DSS‐3 membrane transport proteins in natural SW (A) and ASW medium (B) over time.
Only transporter proteins with average abundance above 0.1% are represented. The average value of triplicate cultures analyses (n = 3) are shown. The periplasmic component of the manganese ABC transporter is highlighted in bold and, for convenience, error bars showing standard deviation were added for only this protein.
Interaction
The genetic transfer agent (GTA) and Repeats‐in‐ToXin (RTX‐like) proteins are the main contributors of this functional category. While GTAs are bacteriophage‐like particles produced by a large variety of bacteria which carry random fragments form the host's genome and that are thought to encourage horizontal gene transfer (Frost et al., 2005), RTX‐like proteins are elements that mainly play a direct role in cell‐to‐cell interactions ranging from toxicity to adhesion, although due to the enormous extant variability of these proteins, most are of unknown function (Linhartova et al., 2010). Interestingly, while RTX‐like proteins were highly produced in organic nutrient‐rich broths, for example, the RTX‐like protein PaxA represented over 50% of the proteins in the exoproteome when R. pomeroyi was grown in marine broth (Christie‐Oleza and Armengaud, 2010), this does not occur in the mineral media used in this study, where the heterotroph is constantly supplied with a smaller amount of photosynthate. In any case, RTX‐like proteins were still detected throughout the 100‐day experiment (1.7% and 0.5% in ASW and SW, respectively). GTA are commonly found in Roseobacter strains (Biers et al., 2008; Zhao et al., 2009). The high production of GTA by R. pomeroyi when grown in co‐culture with Synechococcus was highlighted previously (Christie‐Oleza et al., 2015b). However, from the time course data, it is interesting to note that the abundance of GTA elements (with an average protein abundance of 4.6% and 5.7% in ASW and SW, respectively) mimics that of the flagella, that is, dropping from 5% to 9% during days 1–14, to between 0.1% and 0.3% during time points 21–64 days in ASW (Supporting Information Table S5). This suggest a co‐regulation between the motile ‘scavenging‐for‐nutrients’ lifestyle mode and the burst of GTA production, possibly induced by the sensing of low nutrient availability. In fact, GTA production is known to be inhibited by high concentrations of phosphate (Westbye et al., 2013).
Hydrolytic enzymes for polymeric organic matter
One of the aims of this study was to highlight and monitor the production of potential secreted enzymes involved in the breakdown of phototrophic DOM over time. Marine Alphaproteobacteria are considered to have a very low extracellular hydrolytic potential due to the low number of enzymes that are encoded in their genomes (Barbeyron et al., 2016). In fact, previous exoproteomic analyses from a range of Roseobacter strains showed that different isolates produced a small but highly divergent, repertoire of exoenzymes suggesting that closely related species may be able to target different substrates and escape competition through the diversification of resources (Christie‐Oleza et al., 2015b). Here, the overall average abundance of this category was 5.3% and 5.5% for ASW and SW, respectively, but while the abundance of these proteins progressively dropped in ASW (7.4% at day 1 to 4.1% at day 100), an opposite trend was observed in SW with an increase from 3.3% to 7.6% during the 100‐day time course (Fig. 3B). Nineteen potential hydrolytic enzymes produced by R. pomeroyi were detected during this study (i.e., with an average detection > 0.1% in at least one of the culture conditions; Table 2). The two most abundantly detected hydrolases were AAV93776 (2.1% and 2.7% in ASW and SW, respectively) and AAV95476 (0.9% and 2.68% in ASW and SW, respectively), re‐annotated here as a possible pectate lyase and sialidase, respectively (Table 2). Hence, both proteins may be involved in polysaccharide hydrolysis, generating oligosaccharides and short chain sugars that can then be assimilated by the heterotroph. Despite being highly abundant, the pectate lyase‐like protein shows a drastic reduction in the ASW co‐culture between time points 21 and 60 coinciding with the period when Synechococcus is most abundant (Fig. 1). During this time period, Synechococcus is possibly at its maximum production of photosynthate and R. pomeroyi may fill its carbon and energy demands through the use of protein and other organic nitrogen compounds. Interestingly, it is between days 21 and 60 when R. pomeroyi shows an increased production of proteases (i.e., AAV95890 and AAV97448) and amidohydrolases (AAV97448 and AAV94260). Members of the Roseobacter group are known to preferentially target small nitrogen‐rich DOM (Bryson et al., 2016; Teira et al., 2017), and hence, it is not surprising that these organisms will specialize in using these compounds when they are present. While the abundance of organic matter in ASW allows R. pomeroyi to shift its targeted polymeric DOM, in SW it shows a more sTable production of hydrolases throughout the 100‐day co‐culture, with only the increase of the pectate lyase AAV93776 (from 1.4% to 3.6%) and the protease AAV95890 (from < 0.01% to 0.13%) over time. Ruegeria pomeroyi encodes and produces most enzymes required to breakdown the polymeric components within the cyanobacterial photosynthate (i.e., protein, polysaccharides, peptidoglycan and other sulfur and amide compounds), and hence, it is able to mineralize most of the primary produced organic matter, enabling the establishment of long term co‐cultures based on nutrient cycling described previously (Christie‐Oleza et al., 2017). Nevertheless, we acknowledge that the identification of most hydrolytic enzymes in Table 2 requires further experimentation to confirm their predicted function. For example, the most abundant enzyme detected, i.e., the pectate lyase‐like protein, was annotated as a hypothetical protein and only suggested here as a hydrolytic enzyme for polysaccharides based on a pectate lyase‐like domain it contains. Other pectate lyases have been detected in marine microbes, but these seem to have been acquired from a terrestrial origin (Hehemann et al., 2017). Nevertheless, this is not the norm and most marine microbes have a novel array of untapped hydrolytic enzymes that largely differ from their well‐known terrestrial counterparts (Hehemann et al., 2014), which require further characterization.
Table 2.
Hydrolytic enzymes detected over time in the exoproteome of R. pomeroyi DSS‐3 when co‐cultured with Synechococcus sp. WH7803.
| Locus ID | Annotation (possible function) | Substrate | Day 1a | Day 3a | Day 7a | Day 14a | Day 21a | Day 32a | Day 60a | Day 100a | Average | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AAV93776 | Hypothetical SPO0459 (pectate lyase) | Polysaccharides | ASW | 4.94 | 4.05 | 3.23 | 3.08 | 0.04 | 0.02 | 0.08 | 1.63 | 2.13 |
| SW | 1.43 | 1.88 | 2.16 | 2.29 | 3.08 | 3.12 | 3.84 | 3.61 | 2.68 | |||
| AAV95476 | BNR/Asp‐box protein (sialidase) | Polysaccharides | ASW | 1.14 | 1.00 | 0.99 | 1.35 | 0.80 | 0.43 | 0.51 | 1.07 | 0.91 |
| SW | 1.07 | 1.08 | 1.31 | 1.22 | 1.77 | 1.59 | 1.49 | 1.25 | 1.35 | |||
| AAV95139 | Twin‐arginine pathway (phosphatase PhoX) | Phosphates | ASW | 0.58 | 0.56 | 0.40 | 0.33 | 0.16 | 0.88 | 0.65 | 0.15 | 0.46 |
| SW | 0.53 | 0.54 | 0.57 | 0.28 | 0.13 | 0.08 | 0.02 | < 0.01 | 0.27 | |||
| AAV96145 | Ser/Thr Phosphatase/nucleotidase (nucleotidase) | Nucleotides | ASW | 0.04 | 0.05 | 0.27 | 0.13 | 0.93 | 0.77 | 0.70 | 0.34 | 0.40 |
| SW | 0.06 | 0.18 | 0.23 | 0.44 | 0.69 | 0.58 | 1.02 | 1.49 | 0.59 | |||
| AAV95890 | Protease, S2 family (serine protease) | Proteins | ASW | 0.03 | 0.01 | 0.01 | 0.02 | 0.51 | 0.56 | 0.55 | 0.09 | 0.22 |
| SW | < 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.03 | 0.07 | 0.13 | 0.03 | |||
| AAV96272 | Metallo‐beta‐lactamase (alkyl sulfatase) | Others | ASW | 0.06 | 0.11 | 0.12 | 0.22 | 0.38 | 0.26 | 0.28 | 0.22 | 0.21 |
| SW | 0.03 | 0.10 | 0.23 | 0.28 | 0.35 | 0.37 | 0.38 | 0.43 | 0.27 | |||
| AAV93580 | Alkaline phosphatase (phosphatase PhoD) | Phosphates | ASW | 0.01 | 0.04 | 0.03 | 0.01 | < 0.01 | 0.43 | 0.78 | 0.01 | 0.17 |
| SW | 0.01 | 0.06 | 0.04 | 0.02 | 0.01 | 0.02 | 0.01 | < 0.01 | 0.02 | |||
| AAV95931 | LysM domain/M23/M37 peptidase (peptidase family M23) | Proteins | ASW | 0.12 | 0.28 | 0.19 | 0.15 | 0.11 | 0.18 | 0.11 | 0.10 | 0.16 |
| SW | 0.05 | 0.08 | 0.07 | 0.06 | 0.02 | 0.02 | 0.01 | 0.06 | 0.05 | |||
| AAV97448 | Amidohydrolase protein (amidohydrolase) | Others | ASW | 0.04 | 0.03 | 0.04 | 0.04 | 0.32 | 0.26 | 0.27 | 0.16 | 0.14 |
| SW | 0.02 | 0.07 | 0.11 | 0.15 | 0.15 | 0.08 | 0.18 | 0.40 | 0.14 | |||
| AAV95236 | Beta‐lactamase protein (carboxypeptidase) | Proteins | ASW | 0.01 | 0.05 | 0.24 | 0.36 | 0.03 | 0.05 | 0.05 | 0.09 | 0.11 |
| SW | < 0.01 | < 0.01 | < 0.01 | 0.01 | < 0.01 | < 0.01 | < 0.01 | < 0.01 | < 0.01 | |||
| AAV96841 | Peptidoglycan‐binding protein (glycoside hydrolase) | Peptidoglycan | ASW | 0.06 | 0.13 | 0.07 | 0.11 | 0.06 | 0.10 | 0.12 | 0.04 | 0.09 |
| SW | 0.02 | 0.09 | 0.07 | 0.06 | 0.03 | 0.01 | 0.01 | 0.03 | 0.04 | |||
| AAV94622 | Periplasmic serine protease (serine proteases) | Proteins | ASW | 0.04 | 0.05 | 0.04 | 0.11 | 0.06 | 0.06 | 0.04 | 0.03 | 0.05 |
| SW | 0.01 | 0.02 | 0.01 | 0.04 | 0.02 | 0.02 | 0.01 | < 0.01 | 0.01 | |||
| AAV95689 | Fumarylacetoacetate hydrolase (aromatic hydrolase) | Others | ASW | 0.02 | 0.02 | 0.05 | 0.04 | 0.09 | 0.07 | 0.06 | 0.03 | 0.05 |
| SW | 0.02 | 0.03 | 0.03 | 0.03 | 0.02 | 0.03 | 0.06 | 0.05 | 0.03 | |||
| AAV96493 | Glycosyl hydrolase, family 25 (acetylmuramidase) | Peptidoglycan | ASW | 0.08 | 0.10 | 0.10 | < 0.01 | < 0.01 | < 0.01 | < 0.01 | 0.01 | 0.04 |
| SW | 0.03 | < 0.01 | < 0.01 | < 0.01 | 0.01 | < 0.01 | < 0.01 | < 0.01 | 0.01 | |||
| AAV94066 | Cyclase family protein (aromatic hydrolase) | Others | ASW | < 0.01 | < 0.01 | < 0.01 | < 0.01 | 0.07 | 0.07 | 0.06 | 0.04 | 0.03 |
| SW | < 0.01 | < 0.01 | 0.01 | 0.01 | 0.01 | 0.02 | 0.04 | 0.07 | 0.02 | |||
| AAV93452 | Murein endopeptidase (murein endopeptidase) | Peptidoglycan | ASW | < 0.01 | 0.01 | 0.02 | 0.02 | 0.01 | 0.02 | 0.08 | 0.02 | 0.02 |
| SW | < 0.01 | < 0.01 | 0.02 | < 0.01 | < 0.01 | < 0.01 | < 0.01 | < 0.01 | 0.01 | |||
| AAV95558 | Hypothetical SPO2296 (lysozyme) | Peptidoglycan | ASW | 0.02 | < 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.04 | 0.03 | 0.02 |
| SW | 0.02 | < 0.01 | < 0.01 | < 0.01 | < 0.01 | 0.02 | < 0.01 | < 0.01 | 0.01 | |||
| AAV94260 | Amidohydrolase protein (amidohydrolase) | Others | ASW | < 0.01 | < 0.01 | < 0.01 | < 0.01 | 0.04 | 0.02 | 0.02 | 0.01 | 0.01 |
| SW | 0.01 | < 0.01 | < 0.01 | 0.01 | < 0.01 | < 0.01 | < 0.01 | < 0.01 | < 0.01 | |||
| AAV96597 | Peptidase, M16 family (peptidase M16) | Proteins | ASW | < 0.01 | < 0.01 | < 0.01 | < 0.01 | 0.03 | 0.03 | 0.02 | < 0.01 | 0.01 |
| SW | < 0.01 | < 0.01 | < 0.01 | 0.01 | < 0.01 | < 0.01 | < 0.01 | < 0.01 | < 0.01 |
The values shown are the average of relative abundance obtained from three biological replicate cultures. The full data can be obtained from Supporting Information Table S5.
Hydrolytic enzymes for phosphate
Both alkaline phosphatases PhoX and PhoD encoded by R. pomeroyi were found in the exoproteomes (Table 2). PhoX, and not PhoA, is prevalent in marine microbes and is thought to play an important role in marine oligotrophic systems (Sebastian and Ammerman, 2009, 2011). PhoX was the predominant alkaline phosphatase in our dataset (Table 2). Nevertheless, the less‐known PhoD, which was in very low abundance throughout most of the time course (abundance below 0.06%), peaked only at time points 32 and 60 (0.43% and 0.78%, respectively) under ASW conditions. While the substrates targeted by PhoX have been characterized (Sebastian and Ammerman, 2011), organic phosphate compounds targeted by PhoD remain unknown. Ruegeria pomeroyi also abundantly secretes the nucleotidase AAV96145 when grown in co‐culture with Synechococcus. It is worth noting the progressive increase in this nucleotidase especially under SW conditions (from 0.06% to 1.49%), which coincides with a strong decrease in PhoX abundance (Table 2). The ABC transporters for phosphate and organic phosphate (e.g., glycerol‐3‐phosphate) remain constant during the whole time course (Supporting Information Table S5), suggesting that R. pomeroyi's phosphorous starvation status does not vary over time, but the switch from PhoX to a nucleotidase indicates a variation in the organic phosphorous produced by Synechococcus. We hypothesize that Synechococcus may replace its phospholipids for sulfolipids (Van Mooy et al., 2009) and even initiate the accumulation of polyphosphates (Martin et al., 2014), which may become, together with nucleic acids, the main source of phosphorous in the system.
Conclusions
Microbial exoproteomes are good proxies to study the dynamic interactions of microbes with their environment. Here we generated a unique time course exoproteomic dataset of a 100‐day long Synechococcus sp. WH7803–R. pomeroyi DSS‐3 co‐culture incubated in both nutrient rich and natural oligotrophic seawater. The observed protein variations matched well with the culture's physiology over time and emphasizes the need of time course experiments such as the one we present here in order to obtain a comprehensive understanding of microbial interactions, as single time points may be misleading. This study has highlighted a number of interesting aspects in this phototroph–heterotroph system such as (i) the heterotroph's varying motility lifestyle depending on nutrient availability, (ii) the unexplained selective leakage of phycobilisomes to the milieu by Synechococcus, (iii) the specificity of nutrient acquisition though the production of an array of active membrane transport systems, (iv) the large production of SOD by Synechococcus to deal with ROS despite the presence of a heterotroph, (v) the varying abundance of a type IV pili structure produced by the phototroph, (vi) a list of uncharacterized hydrolytic enzymes secreted by R. pomeroyi to mineralise the polymeric organic matter generated by the cyanobacterium, (vii) a pattern of phosphatases that varies over time and that is believed to adapt to the pool of organic phosphorous present in the system and (viii) a list of relevant proteins of unknown function that require further research. Hence, the high‐resolution time course dataset we present here will become a reference for future characterization of specific molecular mechanisms involved in sustaining this microbial system.
Experimental procedures
Bacterial growth and experimental setup
Marine Synechococcus sp. WH7803 was grown in ASW (Wilson et al., 1996) at 22°C at a light intensity of 10 µmol photons m−2 s−1 with shaking (140 r.p.m.). Roseobacter strain R. pomeroyi DSS‐3 was grown in marine broth (Difco, France) at 28°C until stationary phase. Ruegeria pomeroyi and Synechococcus cells were harvested via centrifugation and washed twice in filter‐sterilized autoclaved seawater (SW, natural seawater collected from the Gulf Stream in the Gulf of Mexico; provided by Sigma, USA) prior to co‐inoculating both organisms in 100 ml ASW and SW at cell concentrations of ∼ 108 and 107 cell ml−1, respectively (Fig. 1). ASW and SW co‐cultures were incubated for up to 100 days in optimal conditions for Synechococcus (as described earlier). We allowed triplicate flasks for each one of the eight time points analyzed, i.e., days 1, 3, 7, 14, 21, 32, 60 and 100. At each time point, Synechococcus cell abundance was monitored by flow cytometry (BD FACScan), while viable heterotrophs were counted by colony forming units on marine agar (Difco, France) as previously suggested (Christie‐Oleza et al., 2017).
For motility visualization, R. pomeroyi was grown in 10 ml of marine broth for 40 h after which cells were washed in filter‐sterilized autoclaved SW, re‐suspended in 10 ml of SW and further incubated for 4 days to starve the cells. Then, 100 µl of starved cells were added to 100 µl of ASW supplemented with 2.5 mM of (NH4)2SO4 and 4× the standard concentration of f2‐media vitamin mix. Succinate, glucose, yeast extract (Merck, Germany) and bacto peptone (Merck, Germany) were added at final concentrations, 0.005%, 0.01%, 0.02%, 0.05% and 0.1% (wt/vol), and incubated for 4 h before imaging cell motility using concave microscope slides under a 100× objective of a light microscope (Nikon Eclipse Ti) equipped with a widefield camera (Andor Zyla sCMOS). For each one of the conditions, 10 s videos were recorded from three different fields containing approximately 60 ± 10 cells and the number of motile cells was counted.
Preparation of exoproteome samples
Triplicate 100 ml ASW and SW cultures were used for each time point. The exoproteomes contained in the culture milieu were collected after removing all cells via centrifugation at 4000 r.p.m. for 15 min at room temperature and further filtering the supernatant through 0.22 μm pore size filters (Millex‐GV; Millipore, Germany). A total of 40 and 80 ml of the supernatant of SW and ASW cultures, respectively, were used for trichloroacetic acid precipitation as described previously (Christie‐Oleza and Armengaud, 2010). The resulting protein pellets were dissolved in LDS loading buffer (Invitrogen, USA), and the equivalent of 20 ml of ASW cultures and 40 ml of SW cultures were loaded on a precast Tris‐Bis NuPAGE gel (Invitrogen, USA) using 1× MOPS solution (Invitrogen, USA) as the running buffer. SDS‐PAGE was performed for a short gel migration (5 mm). This allowed removing contaminants and purifying the polypeptides in the polyacrylamide gel.
Trypsin in‐gel proteolysis and nanoLC‐MS/MS analysis
Polyacrylamide gel bands containing the exoproteome were excised and standard in‐gel reduction with dithiothreitol and alkylation with iodoacetamide were performed prior to trypsin (Roche, Switzerland) proteolysis (Christie‐Oleza and Armengaud, 2010). The resulting tryptic peptide mixture was extracted using 5% formic acid in 25% acetonitrile and concentrated at 40°C in a speed‐vac. For mass spectrometry, the samples were resuspended in 2.5% acetonitrile containing 0.05% trifluoroacetic acid and filtered using a 0.22 μm cellulose acetate spin column 16,000g for 5 min in order to eliminate undissolved aggregates. Sample were analyzed by means of nanoLC‐ESI‐MS/MS using an Ultimate 3000 LC system (Dionex‐LC Packings) coupled to an Orbitrap Fusion mass spectrometer (Thermo Scientific, USA) using a 60 min LC separation on a 25 cm column and settings as previously described (Christie‐Oleza et al., 2015b).
Data analysis
Raw files were processed using the software package for shotgun proteomics MaxQuant version 1.5.5.1 (Cox and Mann, 2008) to identify and quantify protein using the UniProt databases of Synechococcus sp. WH7803 and R. pomeroyi DSS‐3. Samples were matched between runs. Other parameters were set by default. The peak intensities across the whole set of measurements were compared to obtain quantitative data for all the peptides in all the samples. Peak intensities and spectral counts assigned to each organism present in the co‐culture were compared (Supporting Information Table S1). The list of detected peptides and polypeptides is provided as Supporting Information Tables S2 and S3, respectively. The bioinformatic analysis pipeline was completed using the software Perseus version 1.5.5.3. Decoy and contaminants were removed. The relative protein abundance was obtained from the raw protein intensities from each sample after normalization to protein size and prior to converting to a logarithmic scale with base 2 (Murugaiyan et al., 2016). The missing values were imputed using the default parameters. The protein quantification and calculation of statistical significance were carried out using two‐sample Student's t test (P = 0.05) using a permutation‐based false discovery rate (q = 0.05). Protein categorization was based on KEGG annotations with manual curation using the Conserved Domain search tool from NCBI. Prediction of secreted proteins was carried out using the servers SignalP 4.1 (Petersen et al., 2011), SecretomeP 2.0 (Bendtsen et al., 2005), LipoP 1.1 (Juncker et al., 2003) and PSORTb (Yu et al., 2010).
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's web‐site.
Table S1. Spectral counts and mass intensities assigned to Synechococcus sp. WH7803 and R. pomeroyi DSS‐3 in each one of the mass spectrometry runs. Percentage assigned to each organism in the co‐culture is in brackets.
Table S2. List of peptides in the exoproteomes of Synechococcus sp. WH7803 and R. pomeroyi DSS‐3 co‐cultures detected by LC‐MS/MS.
Table S3. Raw list of polypeptides detected by LC–MS/MS in the exoproteomes of Synechococcus sp. WH7803 and R. pomeroyi DSS‐3 co‐cultures.
Table S4. (a) Protein categories and comparative exoproteomic analysis of Synechococcus sp. WH7803 proteins detected in SW co‐cultures with R. pomeroyi DSS‐3. (b) Protein categories and comparative exoproteomic analysis of Synechococcus sp. WH7803 proteins detected in ASW co‐cultures with R. pomeroyi DSS‐3.
Table S5. (a) Protein categories and comparative proteomic analysis of R. pomeroyi DSS‐3 proteins detected in SW co‐cultures with Synechococcus sp. WH7803. (b) Protein categories and comparative proteomic analysis of R. pomeroyi DSS‐3 proteins detected in ASW co‐cultures with Synechococcus sp. WH7803.
Movie S1. R. pomeroyi incubated in filter‐sterilized ASW obtained from a two‐week old Synechococcus culture.
Movie S2. R. pomeroyi incubated in ASW supplemented with ammonium and vitamins. Only Brownian motion is observed.
Movie S3. R. pomeroyi incubated in ASW supplemented with ammonium, vitamins and 0.02% peptone (wt/vol).
Acknowledgements
This work was supported by the NERC Independent Research Fellowship NE/K009044/1 and WISB, a BBSRC/EPSRC Synthetic Biology Research Centre (grant ref: BB/M017982/1) funded under the UK Research Councils' Synthetic Biology for Growth program. We also acknowledge the technical support from the WPH Proteomic Facility at the University of Warwick.
References
- Armengaud, J. , Christie‐Oleza, J.A. , Clair, G. , Malard, V. , and Duport, C. (2012) Exoproteomics: exploring the world around biological systems. Expert Rev Proteomics 9: 561–575. [DOI] [PubMed] [Google Scholar]
- Arnosti, C. (2015) Contrasting patterns of peptidase activities in seawater and sediments: an example from Arctic fjords of Svalbard. Mar Chem 168: 151–156. [Google Scholar]
- Barbeyron, T. , Thomas, F. , Barbe, V. , Teeling, H. , Schenowitz, C. , Dossat, C. , et al (2016) Habitat and taxon as driving forces of carbohydrate catabolism in marine heterotrophic bacteria: example of the model algae‐associated bacterium Zobellia galactanivorans DsijT. Environ Microbiol 18: 4610–4627. [DOI] [PubMed] [Google Scholar]
- Bendtsen, J.D. , Kiemer, L. , Fausbøll, A. , and Brunak, S. (2005) Non‐classical protein secretion in bacteria. BMC Microbiol 5: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benner, R. (2002). Chemical composition and reactivity In Biogeochemistry of Marine Dissolved Organic Matter. Hansell D.A., and Carlson C.A. (eds). Cambridge, MA, USA: Academic Press, pp. 56–90, Chapter 3. [Google Scholar]
- Bertlisson, S. , Berglund, O. , Pullin, M.J. , and Chisholm, S.W. (2005) Release of dissolved organic matter by Prochlorococcus . Vie Milieu 55: 225–232. [Google Scholar]
- Biers, E.J. , Wang, K. , Pennington, C. , Belas, R. , Chen, F. , and Moran, M.A. (2008) Occurrence and expression of gene transfer agent genes in marine bacterioplankton. Appl Environ Microbiol 74: 2933–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biersmith, A. , and Benner, R. (1998) Carbohydrates in phytoplankton and freshly produced dissolved organic matter. Mar Chem 63: 131–144. [Google Scholar]
- Biller, S.J. , Schubotz, F. , Roggensack, S.E. , Thompson, A.W. , Summons, R.E. , and Chisholm, S.W. (2014) Bacterial vesicles in marine ecosystems. Science 343: 183–186. [DOI] [PubMed] [Google Scholar]
- Buchan, A. , LeCleir, G.R. , Gulvik, C.A. , and González, J.M. (2014) Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat Rev Microbiol 12: 686–698. [DOI] [PubMed] [Google Scholar]
- Bryson, S. , Li, Z. , Pett‐Ridge, J. , Hettich, R.L. , Mayali, X. , Pan, C. , et al (2016) Proteomic sTable isotope probing reveals taxonomically distinct patterns in amino acid assimilation by coastal marine bacterioplankton. mSystems 1: e00027‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie‐Oleza, J.A. , and Armengaud, J. (2010) In‐depth analysis of exoproteomes from marine bacteria by shotgun liquid chromatography‐tandem mass spectrometry: the Ruegeria pomeroyi DSS‐3 case‐study. Mar Drugs 8: 2223–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie‐Oleza, J.A. , and Armengaud, J. (2015) Proteomics of the Roseobacter clade, a window to the marine microbiology landscape. Proteomics 15: 3928–3942. [DOI] [PubMed] [Google Scholar]
- Christie‐Oleza, J.A. , Armengaud, J. , Guerin, P. , and Scanlan, D.J. (2015a) Functional distinctness in the exoproteomes of marine Synechococcus . Environ Microbiol 17: 3781–3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie‐Oleza, J.A. , Fernandez, B. , Nogales, B. , Bosch, R. , and Armengaud, J. (2012a) Proteomic insights into the lifestyle of an environmentally relevant marine bacterium. ISME J 6: 124–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie‐Oleza, J.A. , Pina‐Villalonga, J.M. , Bosch, R. , Nogales, B. , and Armengaud, J. (2012b) Comparative proteogenomics of twelve Roseobacter exoproteomes reveals different adaptive strategies among these marine bacteria. Mol Cell Proteomics 11: M111.013110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie‐Oleza, J.A. , Scanlan, D.J. , and Armengaud, J. (2015b) “You produce while I clean up”, a strategy revealed by exoproteomics during Synechococcus–Roseobacter interactions. Proteomics 15: 3454–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie‐Oleza, J.A. , Sousoni, D. , Lloyd, M. , Armengaud, J. , and Scanlan, D.J. (2017) Nutrient recycling facilitates long‐term stability of marine microbial phototroph‐heterotroph interactions. Nat Microbiol 2: 17100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, J. , and Mann, M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.‐range mass accuracies and proteome‐wide protein quantification. Nat Biotechnol 26: 1367–1372. [DOI] [PubMed] [Google Scholar]
- D'Ambrosio, L. , Ziervogel, K. , MacGregor, B. , Teske, A. , and Arnosti, C. (2014) Composition and enzymatic function of particle‐associated and free‐living bacteria: a coastal/offshore comparison. ISME J 8: 2167–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decho, A.W. , and Gutierrez, T. (2017) Microbial extracellular polymeric substances (EPSs) in ocean systems. Front Microbiol 8: 922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Rocha, C.L. , and Passow, U. (2014) The biological pump In Treatise on Geochemistry. Second edition Holland H., and Turekian K. (eds). Oxford: Elsevier, pp. 93–122. [Google Scholar]
- Falkowski, P. (2012) Ocean science: the power of plankton. Nature 483: S17–S20. [DOI] [PubMed] [Google Scholar]
- Frost, L.S. , Leplae, R. , Summers, A.O. , and Toussaint, A. (2005) Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol 3: 722–732. [DOI] [PubMed] [Google Scholar]
- Giebel, H.A. , Kalhoefer, D. , Lemke, A. , Thole, S. , Gahl‐Janssen, R. , Simon, M. , and Brinkhoff, T. (2011) Distribution of Roseobacter RCA and SAR11 lineages in the North Sea and characteristics of an abundant RCA isolate. ISME J 5: 8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giner‐Lamia, J. , Pereira, S.B. , Bovea‐Marco, M. , Futschik, M.E. , Tamagnini, P. , and Oliveira, P. (2016) Extracellular proteins: novel key components of metal resistance in Cyanobacteria?. Front Microbiol 7: 878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossowicz, M. , Roth‐Rosenberg, D. , Aharonovich, D. , Silverman, J. , Follows, M.J. , and Sher, D. (2017) Prochlorococcus in the lab and in silico: the importance of representing exudation. Limnol Oceanogr 62: 818–835. [Google Scholar]
- Hehemann, J.H. , Boraston, A.B. , and Czjzek, M. (2014) A sweet new wave: structures and mechanisms of enzymes that digest polysaccharides from marine algae. Curr Opin Struct Biol 28: 77–86. [DOI] [PubMed] [Google Scholar]
- Hehemann, J.‐H. , Truong, L.V. , Unfried, F. , Welsch, N. , Kabisch, J. , Heiden, S.E. , et al (2017) Aquatic adaptation of a laterally acquired pectin degradation pathway in marine gammaproteobacteria. Environ Microbiol 19: 2320–2333. [DOI] [PubMed] [Google Scholar]
- Hunken, M. , Harder, J. , and Kirst, G.O. (2008) Epiphytic bacteria on the Antarctic ice diatom Amphiprora kufferathii Manguin cleave hydrogen peroxide produced during algal photosynthesis. Plant Biol (Stuttg) 10: 519–526. [DOI] [PubMed] [Google Scholar]
- Jeffery, C.J. (2009) Moonlighting proteins—an update. Mol Biosyst 5: 345–350. [DOI] [PubMed] [Google Scholar]
- Jiao, N. , Herndl, G.J. , Hansell, D.A. , Benner, R. , Kattner, G. , Wilhelm, S.W. , et al (2010) Microbial production of recalcitrant dissolved organic matter: long‐term carbon storage in the global ocean. Nat Rev Microbiol 8: 593–599. [DOI] [PubMed] [Google Scholar]
- Jiao, N. , and Zheng, Q. (2011) The microbial carbon pump: from genes to ecosystems. Appl Environ Microbiol 77: 7439–7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson‐Rollings, A.S. , Wright, H. , Masciandaro, G. , Macci, C. , Doni, S. , Calvo‐Bado, L.A. , et al (2014) Exploring the functional soil‐microbe interface and exoenzymes through soil metaexoproteomics. ISME J 8: 2148–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juncker, A.S. , Willenbrock, H. , Von Heijne, G. , Brunak, S. , Nielsen, H. , and Krogh, A. (2003) Prediction of lipoprotein signal peptides in Gram‐negative bacteria. Protein Sci 12: 1652–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl, D.M. , and Church, M.J. (2014) Microbial oceanography and the Hawaii Ocean Time‐series programme. Nat Rev Microbiol 12: 699–713. [DOI] [PubMed] [Google Scholar]
- Karner, M. , and Herndl, G.J. (1992) Extracellular enzymatic activity and secondary production in free‐living and marine‐snow‐associated bacteria. Mar Biol 113: 341–347. [Google Scholar]
- Landa, M. , Blain, S. , Christaki, U. , Monchy, S. , and Obernosterer, I. (2016) Shifts in bacterial community composition associated with increased carbon cycling in a mosaic of phytoplankton blooms. ISME J 10: 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhartova, I. , Bumba, L. , Masin, J. , Basler, M. , Osicka, R. , Kamanova, J. , et al (2010) RTX proteins: a highly diverse family secreted by a common mechanism. FEMS Microbiol Rev 34: 1076–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, P. , Dyhrman, S.T. , Lomas, M.W. , Poulton, N.J. , and Van Mooy, B.A. (2014) Accumulation and enhanced cycling of polyphosphate by Sargasso Sea plankton in response to low phosphorus. Proc Natl Acad Sci USA 111: 8089–8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, J. , Smith, D.C. , Steward, G.F. , and Azam, F. (1996) Variability in ectohydrolytic enzyme activities of pelagic marine bacteria and its significance for substrate processing in the sea. Aquat Microb Ecol 10: 223–230. [Google Scholar]
- Michel, G. , and Czjzek, M. (2013) Polysaccharide‐degrading enzymes from marine bacteria In Marine Enzymes for Biocatalysis: Sources, Biocatalytic Characteristics and Bioprocesses of Marine Enzymes. Trincone A. (ed.). Oxford: Elsevier, pp. 429–464. [Google Scholar]
- Mitulla, M. , Dinasquet, J. , Guillemette, R. , Simon, M. , Azam, F. , and Wietz, M. (2016) Response of bacterial communities from California coastal waters to alginate particles and an alginolytic Alteromonas macleodii strain. Environ Microbiol 18: 4369–4377. [DOI] [PubMed] [Google Scholar]
- Moran, M.A. , Buchan, A. , Gonzalez, J.M. , Heidelberg, J.F. , Whitman, W.B. , Kiene, R.P. , et al (2004) Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature 432: 910–913. [DOI] [PubMed] [Google Scholar]
- Morris, J.J. , Johnson, Z.I. , Szul, M.J. , Keller, M. , Zinser, E.R. , and Rodriguez‐Valera, F. (2011) Dependence of the cyanobacterium Prochlorococcus on hydrogen peroxide scavenging microbes for growth at the ocean's surface. PLoS One 6: e16805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, J.J. , Kirkegaard, R. , Szul, M.J. , Johnson, Z.I. , and Zinser, E.R. (2008) Facilitation of robust growth of Prochlorococcus colonies and dilute liquid cultures by “helper” heterotrophic bacteria. Appl Environ Microbiol 74: 4530–4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, J.J. , Lenski, R.E. , and Zinser, E.R. (2012) The Black Queen Hypothesis: evolution of dependencies through adaptive gene loss. MBio 3: e00036‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, R.M. , Frazar, C.D. , and Carlson, C.A. (2012) Basin‐scale patterns in the abundance of SAR11 subclades, marine Actinobacteria (OM1), members of the Roseobacter clade and OCS116 in the South Atlantic. Environ Microbiol 14: 1133–1144. [DOI] [PubMed] [Google Scholar]
- Murugaiyan, J. , Eravci, M. , Weise, C. , and Roesler, U. (2016) Label‐free quantitative proteomic analysis of harmless and pathogenic strains of infectious microalgae, Prototheca spp . Int J Mol Sci 18: E59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagar, E. , Zilberman, S. , Sendersky, E. , Simkovsky, R. , Shimoni, E. , Gershtein, D. , et al (2017) Type 4 pili are dispensable for biofilm development in the cyanobacterium Synechococcus elongatus . Environ Microbiol 19: 2862–2872. [DOI] [PubMed] [Google Scholar]
- Newton, R.J. , Griffin, L.E. , Bowles, K.M. , Meile, C. , Gifford, S. , Givens, C.E. , et al (2010) Genome characteristics of a generalist marine bacterial lineage. ISME J 4: 784–798. [DOI] [PubMed] [Google Scholar]
- Parsons, R.J. , Breitbart, M. , Lomas, M.W. , and Carlson, C.A. (2012) Ocean time‐series reveals recurring seasonal patterns of virioplankton dynamics in the northwestern Sargasso Sea. ISME J 6: 273–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, T.N. , Brunak, S. , von Heijne, G. , and Nielsen, H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786. [DOI] [PubMed] [Google Scholar]
- Romera‐Castillo, C. , Sarmento, H. , Alvarez‐Salgado, X.A. , Gasol, J.M. , and Marrasé, C. (2011) Net production and consumption of fluorescent colored dissolved organic matter by natural bacterial assemblages growing on marine phytoplankton exudates. Appl Environ Microbiol 77: 7490–7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, M.A. , McIlvin, M.R. , Moran, D.M. , Goepfert, T.J. , DiTullio, G.R. , Post, A.F. , et al (2014) Multiple nutrient stresses at intersecting Pacific Ocean biomes detected by protein biomarkers. Science 345: 1173–1177. [DOI] [PubMed] [Google Scholar]
- Scanlan, D.J. , Ostrowski, M. , Mazard, S. , Dufresne, A. , Garczarek, L. , Hess, W.R. , et al (2009) Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev 73: 249–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, D. (2014). Protein targeting, transport and translocation in cyanobacteria In The Cell Biology of Cyanobacteria. Flores E., and Herrero A. (eds). Cheshire, UK: Caister Academic Press, pp. 121–147. [Google Scholar]
- Sebastian, M. , and Ammerman, J.W. (2009) The alkaline phosphatase PhoX is more widely distributed in marine bacteria than the classical PhoA. ISME J 3: 563–572. [DOI] [PubMed] [Google Scholar]
- Sebastian, M. , and Ammerman, J.W. (2011) Role of the phosphatase PhoX in the phosphorus metabolism of the marine bacterium Ruegeria pomeroyi DSS‐3. Environ Microbiol Rep 3: 535–542. [DOI] [PubMed] [Google Scholar]
- Simon, M. , Scheuner, C. , Meier‐Kolthoff, J.P. , Brinkhoff, T. , Wagner‐Dobler, I. , Ulbrich, M. , et al (2017) Phylogenomics of Rhodobacteraceae reveals evolutionary adaptation to marine and non‐marine habitats. ISME J 11: 1483–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stella, N.A. , Kalivoda, E.J. , O'dee, D.M. , Nau, G.J. , and Shanks, R.M. (2008) Catabolite repression control of flagellum production by Serratia marcescens . Res Microbiol 159: 562–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teira, E. , Hernando‐Morales, V. , Guerrero‐Feijoo, E. , and Varela, M.M. (2017) Leucine, starch and bicarbonate utilization by specific bacterial groups in surface shelf waters off Galicia (NW Spain). Environ Microbiol 19: 2379–2390. [DOI] [PubMed] [Google Scholar]
- Van Mooy, B.A. , Fredricks, H.F. , Pedler, B.E. , Dyhrman, S.T. , Karl, D.M. , Koblizek, M. , et al (2009) Phytoplankton in the ocean use non‐phosphorus lipids in response to phosphorus scarcity. Nature 458: 69–72. [DOI] [PubMed] [Google Scholar]
- Vetter, Y.A. , and Deming, J.W. (1999) Growth rates of marine bacterial isolates on particulate organic substrates solubilized by freely released extracellular enzymes. Microb Ecol 37: 86–94. [DOI] [PubMed] [Google Scholar]
- Wagner‐Dobler, I. , and Biebl, H. (2006) Environmental biology of the marine Roseobacter lineage. Annu Rev Microbiol 60: 255–280. [DOI] [PubMed] [Google Scholar]
- Weiss, M.S. , Abele, U. , Weckesser, J. , Welte, W. , Schiltz, E. , and Schulz, G.E. (1991) Molecular architecture and electrostatic properties of a bacterial porin. Science 254: 1627–1630. [DOI] [PubMed] [Google Scholar]
- Westbye, A.B. , Leung, M.M. , Florizone, S.M. , Taylor, T.A. , Johnson, J.A. , Fogg, P.C. , et al (2013) Phosphate concentration and the putative sensor kinase protein CckA modulate cell lysis and release of the Rhodobacter capsulatus gene transfer agent. J Bacteriol 195: 5025–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, W.H. , Carr, N.G. , and Mann, N.H. (1996) The effect of phosphate status on the kinetics of Cyanophage infection in the oceanic cyanobacterium Synechococcus sp. WH7803. J Phycol 32: 506–516. [Google Scholar]
- Xing, P. , Hahnke, R.L. , Unfried, F. , Markert, S. , Huang, S. , Barbeyron, T. , et al (2015) Niches of two polysaccharide‐degrading Polaribacter isolates from the North Sea during a spring diatom bloom. ISME J 9: 1410–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, N.Y. , Wagner, J.R. , Laird, M.R. , Melli, G. , Rey, S. , Lo, R. , et al (2010) PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26: 1608–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrano, M.M. , Siegele, D.A. , Almirón, M. , Tormo, A. , and Kolter, R. (1993) Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science 259: 1757–1760. [DOI] [PubMed] [Google Scholar]
- Zhao, Y. , Wang, K. , Budinoff, C. , Buchan, A. , Lang, A. , Jiao, N. , et al (2009) Gene transfer agent (GTA) genes reveal diverse and dynamic Roseobacter and Rhodobacter populations in the Chesapeake Bay. ISME J 3: 364–373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's web‐site.
Table S1. Spectral counts and mass intensities assigned to Synechococcus sp. WH7803 and R. pomeroyi DSS‐3 in each one of the mass spectrometry runs. Percentage assigned to each organism in the co‐culture is in brackets.
Table S2. List of peptides in the exoproteomes of Synechococcus sp. WH7803 and R. pomeroyi DSS‐3 co‐cultures detected by LC‐MS/MS.
Table S3. Raw list of polypeptides detected by LC–MS/MS in the exoproteomes of Synechococcus sp. WH7803 and R. pomeroyi DSS‐3 co‐cultures.
Table S4. (a) Protein categories and comparative exoproteomic analysis of Synechococcus sp. WH7803 proteins detected in SW co‐cultures with R. pomeroyi DSS‐3. (b) Protein categories and comparative exoproteomic analysis of Synechococcus sp. WH7803 proteins detected in ASW co‐cultures with R. pomeroyi DSS‐3.
Table S5. (a) Protein categories and comparative proteomic analysis of R. pomeroyi DSS‐3 proteins detected in SW co‐cultures with Synechococcus sp. WH7803. (b) Protein categories and comparative proteomic analysis of R. pomeroyi DSS‐3 proteins detected in ASW co‐cultures with Synechococcus sp. WH7803.
Movie S1. R. pomeroyi incubated in filter‐sterilized ASW obtained from a two‐week old Synechococcus culture.
Movie S2. R. pomeroyi incubated in ASW supplemented with ammonium and vitamins. Only Brownian motion is observed.
Movie S3. R. pomeroyi incubated in ASW supplemented with ammonium, vitamins and 0.02% peptone (wt/vol).


