Abstract
DNA damage and alterations in global DNA methylation status are associated with multiple human diseases and are frequently correlated with clinically relevant information. Therefore, assessing DNA damage and epigenetic modifications, including DNA methylation, is critical for predicting human exposure risk of pharmacological and biological agents. We previously developed a higher-throughput platform for the single cell gel electrophoresis (comet) assay, CometChip, to assess DNA damage and genotoxic potential. Here, we utilized the methylation-dependent endonuclease, McrBC, to develop a modified alkaline comet assay, “EpiComet”, which allows single platform evaluation of genotoxicity and global DNA methylation [5-methylcytosine (5-mC)] status of single cell populations under user-defined conditions. Further, we leveraged the CometChip platform to create an EpiComet-Chip system capable of performing quantification across simultaneous exposure protocols to enable unprecedented speed and simplicity. This system detected global methylation alterations in response to exposures which included chemotherapeutic and environmental agents. Using EpiComet-Chip on 63 matched samples; we correctly identified single sample hypermethylation (≥1.5-fold) at 87% (20/23), hypomethylation (≥1.25-fold) at 100% (9/9), with a 4% (2/54) false negative rate (FNR) and 10% (4/40) false positive rate (FPR). Using a more stringent threshold to define hypermethylation (≥1.75-fold) allowed us to correctly identify 94% of hypermethylation (17/18), but increased our FPR to 16% (7/45). The successful application of this novel technology will aid hazard identification and risk characterization of FDA-regulated products, while providing utility for investigating epigenetic modes of action of agents in target organs, since the assay is amenable to cultured cells or nucleated cells from any tissue.
Keywords: Genotoxicity, Epigenetics, Global Methylation, Comet Assay, Methods, Platform Technology
Introduction
DNA methylation is an epigenetic modification that provides a stable gene silencing mechanism that, along with histone modification, plays an important role in regulating gene expression and maintaining genome stability (Bird 2002). DNA methylation primarily occurs by the covalent modification of cytosine residues in CpG dinucleotides, to yield 5-methylcytosine (5-mC), and in general, DNA global methylation status refers to the overall content of 5-mC in the genome, expressed as a percentage (5-mC%) (Waggoner 2007). Several normal biological processes (Reik 2007), environmental exposures (Feil and Fraga 2011; Benayoun et al. 2015) and pharmaceutical compounds (Csoka and Szyf 2009) have been shown to alter DNA methylation status in a variety of reproducible and predictable patterns. For example, treatment with cisplatin (Nyce 1989; Nyce et al. 1993), nalidixic acid (Nyce 1989), hydroxyurea (Nyce 1989) and higher dose methotrexate (MTX) (Nyce 1989; Nyce et al. 1993) have individually been shown to increase global methylation; whereas 5-azacytidine (5-Aza) (Christman 2002), procainamide (Cornacchia et al. 1988; Lee et al. 2005), 4,6-dioxoheltanoic acid (SA) (Wentzel et al. 2010; Lewies et al. 2014), tamoxifen (Wu et al. 2005; Tryndyak et al. 2006), valproic acid (Detich et al. 2003; Cribbs et al. 2015) and hydralazine (Cornacchia et al. 1988) exposures have each conversely resulted in decreased global methylation in defined physiological contexts.
Given that specific alterations in DNA methylation status and patterning have been strongly associated with clinical phenotypes and diseases such as cancer (Ordway et al. 2006; Esteller 2007; Baylin and Jones 2011; Rakyan et al. 2011; Pogribny and Beland 2013; McLean et al. 2014) and autoimmune disorders (Hu et al. 2008; Zhou and Lu 2008; Portela and Esteller 2010), it is of great importance to assess and characterize changes in DNA methylation status that may occur as a result of preventable human exposures. In fact, several chemotherapeutic agents designed to demethylate DNA have been shown to have clinical efficacy (Jones 2014; Yang et al. 2014; Chiappinelli et al. 2015; Licht 2015; Roulois et al. 2015; Paluch et al. 2016). Further, there is strong associative evidence that DNA methylation status and patterning can also be used in several contexts as biomarkers for guiding diagnosis and the treatment of the resulting disorders (Laird 2003; Wu et al. 2005; Bock 2009; Laird 2010; Heyn and Esteller 2012), again highlighting a critical need for rapid, cost-effective strategies to assess DNA methylation. Although enormous progress has been made in DNA methylation assays (Blueprint Consortium 2016) and in using DNA methylation markers in cancer diagnosis and prognosis (Bock 2009), the development of a sensitive and fast method for assessing DNA methylation that can utilize cells isolated from any tissue will be extremely useful and may provide valuable opportunities to enhance human risk assessment strategies.
In addition to DNA methylation modifying agents, it is similarly difficult for people to avoid exposure to exogenous chemicals (such as subsets of pharmaceuticals and food additives) which can also cause DNA damage. The resulting DNA damage may be mechanistically involved in the development of diseases, including cancer. Thus, a wide range of genetic toxicology assays that detect the early biological effects of DNA damaging agents are used for risk assessment and making regulatory decisions. One of these assays, the single cell gel electrophoresis (comet) assay, was recently validated for regulatory use (Azqueta and Dusinska 2015; Frotschl 2015) and an OECD Test Guideline for the in vivo assay (TG489) was approved in 2014 (OECD 2014). For the traditional comet assay, mammalian cells are embedded in agarose, lysed, incubated at high pH to denature DNA, and then subjected to electrophoresis. Due to the fact that damaged DNA migrates more readily than supercoiled, undamaged DNA, when the DNA is stained and analyzed under a microscope, the migration of damaged DNA away from the nucleoid creates a comet-like appearance. One advantage of the comet assay is that it can be used on both dividing and non-dividing cells. Therefore, the comet assay provides a way of detecting early DNA damage in cells cultured in vitro or cells isolated from tissues of animals treated in vivo.
Currently, no single platforms exist that are capable of evaluating, at the cellular level, the genotoxicity (DNA damage) and changes in global DNA methylation status (5-mC%) that occur as a result of exogenous exposures. We, and others, have previously described the use of the standard alkaline comet assay in vitro, on several human and nonhuman cell lines, as well as in vivo using animal models, as a powerful approach to assess genotoxicity (Collins 2004; Azqueta et al. 2009; Ding et al. 2011; Weingeist et al. 2013; Ding et al. 2014; Ge et al. 2014; Manjanatha et al. 2014; Azqueta and Dusinska 2015; Ge et al. 2015; Agnihothram et al. 2016; Ding et al. 2016). The comet assay is amenable to several modifications, including the addition of a subsequent restriction digest step after lysis in the traditional protocol. A variety of DNA lesions can be detected using the comet assay and slight modifications to the traditional procedure. For example, when run under alkaline conditions, the alkaline comet assay detects single-strand breaks (SSBs), double-strand breaks (DSBs), abasic sites (AB sites) and alkali labile sites (ALSs) (Epe et al. 1993). In contrast, under neutral conditions, the comet assay largely detects DSBs, but also detects SSBs to a level that does not allow for distinction between the two forms of damage using the assay. Of note, there is a specialized version of the neutral comet assay that incorporates a prolonged protease digestion step that occurs at a high temperature, which has been reported to selectively detect DSBs (personal communication with Andrew R. Collins and Gunnar Brunborg). In addition to the alkaline and neutral standard comet assay protocols, a modified comet assay approach has been developed to enable detection of base damage. Using the traditional alkaline comet assay, base damage cannot be detected because base lesions do not affect DNA migration. However, if after lysis, the resulting DNA is digested using a DNA glycosylase (e.g., formamidopyrimidine-DNA glycosylase (Fpg), endonuclease III (Endo III), or human 8-hydroxyguanine DNA-glycosylase (hOGG1)), undetectable base damage will be converted into detectable abasic sites and single strand breaks (Collins 2004; Smith et al. 2006; Azqueta et al. 2009; Ding et al. 2011; Ding et al. 2014; Manjanatha et al. 2014). Here, using an analogous approach, we introduced the restriction enzyme McrBC to design and validate a novel, high sensitivity version of the comet assay, “EpiComet”, capable of assessing global methylation status. The addition of McrBC induces “de novo” DNA damage at a majority of the 5-mC present in the DNA, thus the EpiComet assay is predicted to convert undetectable 5-mC into single strand breaks that can be quantified using the comet assay approach and system.
We have previously developed a higher-throughput platform for the comet assay, CometChip (Weingeist et al. 2013; Ge et al. 2014; Ge et al. 2015). Specifically, for the CometChip, cells are arrayed in microwells within agarose, and then analyzed for single strand breaks based upon the principle that damaged DNA (e.g., increased single strand breaks) migrates more readily than undamaged DNA when electrophoresed in agarose. The CometChip approach uses the same parameters as the traditional comet assay. Consistent with previous findings for CometChip alone (versus traditional throughput comet assay) (Ge et al. 2014; Ge et al. 2015), the use of EpiComet-Chip did not alter our results versus direct comparison to EpiComet alone (i.e. EpiComet technology not performed on CometChip platform). Key advantages to the CometChip are the higher throughput (only a few images per sample due to shared focal plane and automated image analysis) and increased sensitivity (reduced comet to comet variation). Here, instead of using the CometChip platform to solely analyze DNA damage, we leveraged the same platform in combination with EpiComet. This merged approach enables simultaneous analysis of DNA damage and global methylation levels under numerous exposure conditions with unprecedented speed and simplicity, via technology we have termed “EpiComet-Chip”.
Materials and Methods
Cell Line Sourcing and Tissue Culture Methods
HeLa-S3 (ATTC Number CCL-2.2), HepG2 (ATCC Number HB-8065), MCF-7 (ATCC Number HTB-22) and TK-6 (ATCC Number CRL-8015), were all obtained directly for these studies from the American Type Culture Collection (ATCC.org), and subsequently were expanded, grown and cryopreserved at low passage number in our laboratory at the National Center for Toxicological Research. Cells were cultured to the exact specifications and cell culture media conditions that were recommended by ATCC for each cell line until they were exposed to the various treatments, as described in our report. Further, cells were kept in culture no longer than eight passages in our laboratory and no longer than one month in active culture to help preserve the integrity of the cellular identity of each line.
Exposures/Treatments of Cultured Cells
Cell lines in culture (as described above) were subdivided into treatment and control groups, with each treatment (each independent dose, each independent compound) performed in biological triplicate. Further, each set of biological triplicates was performed using a minimum of three experimental replicates. [Numerous pilot studies were performed to determine an effective dose and timeframe (between 18–72 hours (h)) in which we were able to quantify the modification in global methylation status for each compound tested]. All compounds were directly obtained from Sigma-Aldrich (St. Louis, MO, USA) and diluted immediately before use (i.e. freshly prepared for each experimental replicate) into stock concentrations using an appropriate diluent for the compound solubility and stability profiles (such as sterile water, sterile Dulbecco's Phosphate-Buffered Saline (DPBS), sterile dimethyl sulfoxide (DMSO), or filtered tissue culture media), which was rapidly diluted into a final concentration in fresh tissue culture media that was ultimately added to wells of confluent cells in culture for adherent cells, or mixed at an appropriate cellular density of cells for suspension cultures. Care was taken to use a concentration for our fresh stock preparations that minimized the amount of DMSO, a known oxidizing agent, added for all cellular treatments to less than 1%.
Alkaline Single Cell Gel Electrophoresis (Comet) Assay
The alkaline comet assay was performed as previously described. Briefly, 50 µl of single cell suspensions [in phosphate buffer saline (PBS)] derived from each treatment condition were mixed with 450 µl 0.8% (dilution in PBS) low melting-point agarose (LMPA) at 37°C, and 100 µl of this suspension was applied to microscope slides (Fisher Scientific, St. Louis, MO, USA) previously thin-coated with 1% agarose, or were added to CometChip assays as previously described (Weingeist et al. 2013; Ge et al. 2014; Ge et al. 2015). After solidification of the LMPA/embedded cell mixture at 4°C, the slides were placed in freshly prepared lysis buffer (2.5 M NaCl, 0.1 M ethylenediaminetetraacetic acid (EDTA), 10 mM Tris, 10% DMSO and 1% Triton X-100, pH 10.0) at 4°C in the dark for 3 h. The slides then were washed once in neutralization buffer (0.4 M Tris, pH 7.5) in the dark for 5 minutes (min) at 4°C and then transferred into chilled alkaline (unwinding/electrophoresis) solution (300 mM NaOH, 1 mM EDTA, pH > 13) in the dark for 30 min to unwind DNA. Immediately following unwinding, electrophoresis was performed in the same solution at 4°C in the dark for 30 min at 0.8 V/cm and ~300 mA. After removing the slides from the electrophoresis chamber, the slides were washed three times (5 min each) with neutralization buffer, and the slides were then fixed with ice cold ethanol (100%) and dried for 30 min. To visualize nucleic acids and facilitate scoring, the slides were stained with SYBR Gold (Invitrogen, Carlsbad, CA, USA) (1:10,000 dilutions in TBE buffer). For glass slide preparations, a minimum of three slides were scored from each treatment/sampling time; and at least 150 representative cells/nucleoids were selected randomly from each slide and scored using a system comprised of a Nikon 501 fluorescent microscope and Comet IV digital imaging software (Perceptive Instruments, Wiltshire, UK). For CometChip preparations, either built-for-use custom MATLAB (MathWorks, Natick, MA, USA) software (Weingeist et al. 2013; Ge et al. 2014; Watson et al. 2014; Ge et al. 2015) or Comet IV digital software was used to manually score individual cells/comets. For all CometChip preparations, a minimum of three “wells” were seeded with previously treated cells, allowing for the scoring of approximately 300 cells. Although, both software options yielded similar results and both were used for individual sets of experiments, we exclusively used one version of software for data collection and analysis methods for any and all experiments that were directly compared for statistical analyses in this report. Independent of the software package used, the percentage of DNA in tail (% Tail DNA), defined as the fraction of DNA in the tail divided by the total amount of DNA associated with a cell multiplied by 100, (i.e. 100 − [(head optical intensity/total optical intensity) × 100]), was used as the parameter for DNA damage analysis as previously described (Manjanatha et al. 2014).
EpiComet: DNA Methylation-Sensitive Modified Comet Assay
For the EpiComet assay, the standard procedure for the alkaline comet assay and alkaline CometChip assay was followed (as above) through the lysis steps of the protocol. After the lysis, the standard procedure was modified to allow for the determination of global DNA methylation status. Briefly, similar to the principles and procedure for the modified oxidative comet assay (Collins and Dusinska 2002; Ding et al. 2016), slides or CometChip preparations were washed in a unique Wash Buffer (50mM NaCl, 10mM Tris-HCl, 10mM MgCl2, 1mM DTT, pH 7.9) and briefly allowed to equilibrate with Wash Buffer at room temperature. Subsquently, samples were incubated at 37°C in a preheated damp chamber for 105 min by layering either the following: A) Control Treatment Buffer (Wash Buffer (50mM NaCl, 10mM Tris-HCl, 10mM MgCl2, 1mM DTT, pH 7.9) plus 100 µg/ml BSA and 1 mM GTP (required for McrBC enzymatic activity)), or B) Methylation-Specific Buffer (Control Treatment Buffer (50mM NaCl, 10mM Tris-HCl, 10mM MgCl2, 1mM DTT, pH 7.9, 100 µg/ml BSA, 1 mM GTP) plus 0.035U/µl McrBC (New England Biolabs, Ipswich, MA, USA)) and covering the enzyme solution with cover slips. The slides were then transferred into a chilled alkaline solution (as above for alkaline comet assay) and allowed to remain in the solution for 40 min in the dark to unwind DNA. After unwinding, electrophoresis was performed in the same solution at 4°C in the dark for 50 min at 0.8 V/cm and ~300 mA. The slides and plates were then washed with neutralizing buffer (as above) to neutralize the remaining alkali and remove detergent, and dried with ice cold 100% ethanol. Before scoring for comets, the slides were stained with SYBR Gold (Invitrogen, Carlsbad, CA) (1:10,000 dilution in TBE buffer) and ultimately scored and analyzed by the methods described above for the standard alkaline comet assay. Global DNA methylation is interpreted by measuring and subtracting the mean % Tail DNA from buffer-only slides (representative of endogenous strand breaks) from the mean % Tail DNA obtained after incubation with the methylation sensitive restriction enzyme. An increase or decrease in the % Tail DNA after enzyme treatment is indicative of DNA hypermethylation or hypomethylation, respectively. Results are expressed as either the normalized or absolute difference in the mean % Tail DNA after enzyme treatment, as indicated.
DNA Isolation for Confirmation Studies
DNA was isolated using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA) per the manufacturer's protocol, including an optional Proteinase K incubation, for DNA isolation from cells derived from tissue culture. Total DNA quality assessment and quantification was performed using the NanoDrop 2000 (Thermo Scientific, Wilmington, DE, USA), as described in the protocol from the manufacturer.
DNA Methylation Confirmation Study
To confirm the global DNA methylation results from the modified comet (EpiComet) assay, we used the MethylFlash Methylated DNA Quantification Assay (Epigentek, Farmingdale, NY, USA) on pooled cells removed from the single cell homogenate preparations that were obtained from the exact same treatment wells that were used for the EpiComet assay. This quantification of global DNA methylation was performed by isolating DNA (using the procedure described above), and then following the manufacturer's protocol, as in (Brown et al. 2014) and several other published manuscripts. Briefly, assays were performed with an equal amount of total DNA (typically 100 ng) used for all assay wells and the procedure involves first binding DNA to strip wells that are specifically treated to have a high DNA affinity, and subsequently detecting the methylated fraction of DNA using capture and detection antibodies. Methylated DNA quantification is conducted fluorometrically in a fluorescence microplate spectrophotometer at excitation 540 nm and emission at 590 nm. The amount of methylated DNA is proportional to the relative fluorescence units measured, which is calculated with the manufacturer's formula for the absolute quantification of 5-mC using a standard curve. Percentage (%) 5-mC is calculated by dividing this number by the total amount of DNA loaded into the assay.
Statistical Analysis
All statistical analysis was performed using either Microsoft Excel (Microsoft Corporation, Redmond, WA, USA) and/or SigmaPlot (Systat Software, Inc., San Jose, CA, USA).
Results
Generation of the EpiComet and EpiComet-Chip Assays
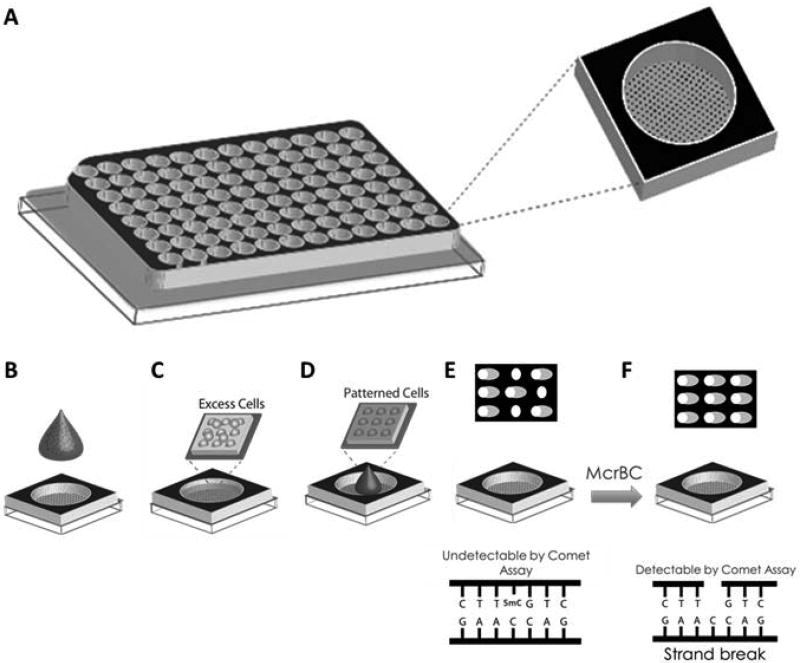
The major objective of this study was to develop a single platform capable of evaluating, at the cellular level, the genotoxicity (DNA damage) and changes in global DNA methylation status (5-mC%) that occur as a result of exogenous exposures. To accomplish this, we introduced the restriction enzyme McrBC to design and validate a novel, high sensitivity version of the comet assay, “EpiComet”, capable of assessing global methylation status. The addition of McrBC induces “de novo” DNA damage at a majority of the 5-mC present in the DNA, thus the EpiComet assay is predicted to convert undetectable 5-mC into single strand breaks that can be quantified using the comet assay approach and system (Figure 1E–F).
Figure 1. The Generation of the EpiComet and EpiComet-Chip Assays.
A, A bottomless 96-well plate is clamped onto an agarose slab that has an array of microwells created as described (Weingeist et al. 2013; Ge et al. 2015). After compressing and clamping, each well has within it ~300 microwells on the bottom surface. To perform the traditional CometChip assay, B, cells are placed above the microwells in agarose and loaded by gravity; C, excess cells are removed by sheer force; D, LMPA is added to the top to trap the cells in the microwells; and E, patterned cells are lysed, subjected to electrophoresis and comets are scored to quantitate DNA damage. For EpiComet-Chip, the procedures remains the same for A–E; however, immediately after lysis, we introduce F, an incubation step with McrBC or buffer alone, followed by electrophoresis and scoring of comets, allowing for the determination of global DNA methylation in addition to DNA damage.
To increase sensitivity and enable rapid throughput assessment, we also merged our EpiComet technology with our previously described platform, CometChip (a 96-well platform for measuring DNA damage in microarrayed cells, Figure 1A–E) (Weingeist et al. 2013; Ge et al. 2015). This combined approach, presented and defined here as the “EpiComet-Chip” single platform system (Figure 1), was utilized for several of our experiments to formally evaluate its potential in predicting exposure-mediated genotoxicity and global DNA methylation alterations at the cellular level. Consistent with previous findings for CometChip alone (versus traditional throughput comet assay) (Ge et al. 2014; Ge et al. 2015), the use of EpiComet-Chip did not alter our results versus direct comparison to EpiComet alone (i.e. EpiComet technology not performed on CometChip platform, data not shown).
Human Cell Culture Lines are Amenable to DNA Damage Detected by the Comet Assay
We, and others, have previously described the use of the standard alkaline comet assay in vitro, on several human and nonhuman cell lines, as well as in vivo using animal models, suggesting that we would not have difficulty performing the comet assay to assess for DNA damage in the immortalized human cells lines that were selected for this study (Collins 2004; Azqueta et al. 2009; Ding et al. 2011; Weingeist et al. 2013; Ding et al. 2014; Ge et al. 2014; Manjanatha et al. 2014; Azqueta and Dusinska 2015; Ge et al. 2015; Agnihothram et al. 2016; Ding et al. 2016). However, to formally test this hypothesis, we examined the ability of a known genotoxicant, methyl methanesulfonate (MMS) to induce dose-dependent induction of DNA damage that we could identify using the standard protocol for the alkaline comet assay. Specifically, we demonstrate statistically significant increases in MMS-induced DNA damage, as measured by increases in % Tail DNA, for human cell lines derived from breast (MCF-7), cervix (HeLa-S3), liver (HepG2), and spleen (TK-6), as shown in Supplemental Figure S1. Collectively, these data suggest that each of these cell lines is amenable to analysis of DNA damage via the comet assay and likely might have potential feasibility for probing global methylation status by using the EpiComet protocol presented here.
Global Methylation Status of Cultured Cells is Amenable to Interrogation with EpiComet
Although several previous studies have addressed the global methylation status of several cell lines (Vera et al. 2008), it has been demonstrated that global methylation status of immortalized cells lines of the same origin can widely vary in their quantified baseline levels of global methylation, with significant variation seen across quantification methods and individual laboratories. Therefore, we independently quantified the baseline 5-mC% of several key cell types using the MethylFlash Methylated DNA Quantification Assay. In particular, we examined MCF-7, TK-6, HeLa, and HepG2, which represent a broad range of well characterized human cell types. We found that our baseline level of global methylation for the MCF-7 cell line was 1.8 ± 0.56% (Mean 5-mC % ± Standard Error of the Mean (SEM)) (Figure S2); whereas this value was 1.6 ± 0.11% for HeLa-S3; 0.9 ± 0.28% for TK-6; and 1.5 ± 0.47 for HepG2; roughly consistent with similar values seen for each line in the literature (Vera et al. 2008). Given that global methylation values had the smallest SEM value in the HeLa-S3 line, we focused most of our subsequent experiments on this cell line.
Induction of Global Methylation Status Changes in Human Cell Lines
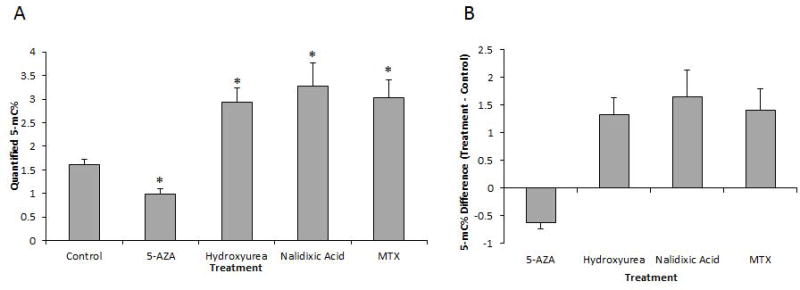
Many compounds are known to modify global methylation status, including chemotherapeutic, environmental and novel agents (Nyce et al. 1993; Zhao et al. 1997; Detich et al. 2003; Toffoli et al. 2003; Lee et al. 2005; Wu et al. 2005; Tryndyak et al. 2006; Zhou and Lu 2008). It is important to note that exposure-mediated changes in global methylation status can occur via both direct and indirect effects and that there are known agents that either increase DNA methylation (hypermethylate) or decrease global DNA methylation (hypomethylate). Given that our initial pilot experiments yielded a range of DNA methylation values (data not shown), we focused on a 48-hour (h) treatment exposure duration using the HeLa-S3 cell line and dosing of either control, 7.5 µM 5-Aza (an inhibitor of methyltransferases that is known to lead to hypomethylation), or one of the following exposures predicted to cause hypermethylation, albeit via different mechanisms: 100 µM hydroxyurea, 1 mM nalidixic acid or 0.25 µM MTX. As shown in Figure 2, we demonstrate that exposure to 5-Aza leads to a statistically significant reduction in global methylation status (5-mC%) in HeLa-S3 cells. Conversely, exposure to hydroxyurea results in a statistically significant increase in global methylation. Similarly, nalidixic acid and MTX dosing also both resulted in statistically significant increases in global methylation, as predicted.
Figure 2. Global Methylation Changes in Response to Various Exposure Conditions.
A, Quantification of mean 5-mC%, derived from a minimum of three independent experiments consisting of a 48h exposure to either control, 5 µM 5-Aza, 100 µM hydroxyurea, 1 mM nalidixic acid or 0.25 µM MTX, as indicated, with error bars reflecting SEM and * denoting statistical significance, defined here as p≤0.05. Baseline 5-mC% (in response to control exposure) was determined to be 1.61 ± 0.11% (Mean ± SEM). 5-Aza exposure led to hypomethylation, quantified at 0.99 ± 0.11%, *p=0.001. Conversely, exposures to the following led to hypermethylation: Hydroxyurea: 2.94 ± 0.30, *p=.005; Nalidixic acid: 3.27 ± 0.49%; *p=.005; MTX: 3.02 ± 0.39%; *p=.002. B, Derived value for global methylation change versus control, using the mean 5-mC% quantified in A. 5-Aza treatment resulted in a 0.62% decrease in global 5-mC% (39% decrease normalized to the control value). Conversely, the treatment with either hydroxyurea (1.33 increase in 5-mC%; 82% increase from control), nalidixic acid (1.66 increase in 5-mC%; 103% increase from control), and MTX (1.41 increase in 5-mC%; 87% increase from control) all resulted in increased global methylation.
EpiComet Detects Global DNA Methylation Status Alterations Resulting from Exposure to DNA Methylation-Modifying Agents
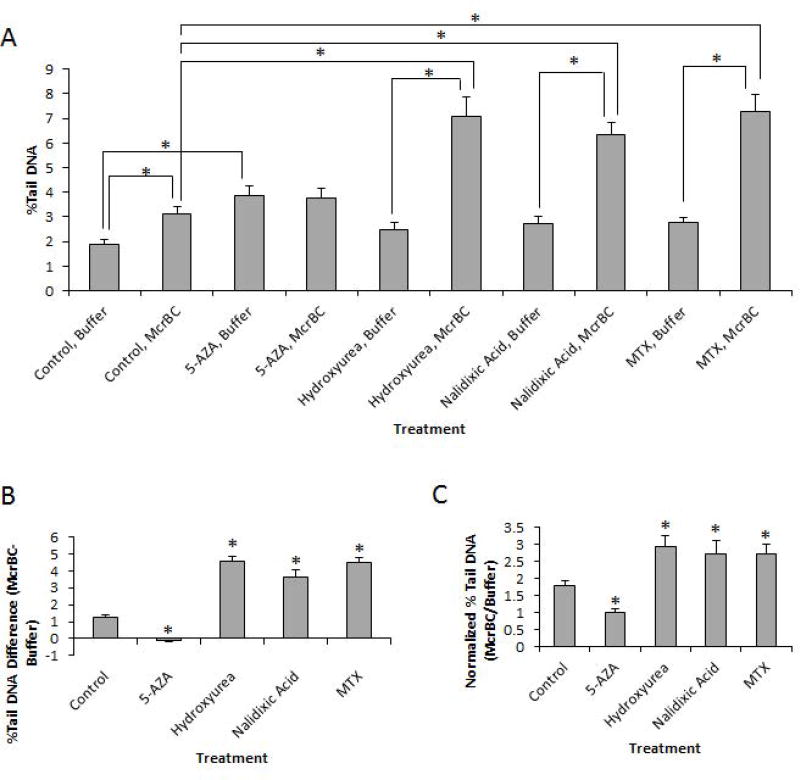
Given our 5-mC% quantification results above, we again performed 48h exposures in HeLa-S3 cells with either control, 5-Aza, hydroxyurea, nalidixic acid, or MTX, and sought to formally test whether the results we obtained using EpiComet from a single treatment could accurately detect hypomethylation or hypermethylation in response to specific exposure conditions. As shown in Figure 3, the incubation of control treated cells with McrBC resulted in a statistically significant increase in % Tail DNA versus buffer. This effect, as predicted, is likely to reflect the detection of baseline 5-mC% and the corresponding induced McrBC (methylation)-specific DNA damage at this baseline 5-mC. In contrast, exposure to the hypomethylating agent, 5-Aza, did not result in a significant difference in % Tail DNA for McrBC versus buffer; however, we did detect a statistically significant difference in 5-Aza/buffer versus Control/buffer (Figure 3A), consistent with literature reports of genotoxicity for the compound.
Figure 3. EpiComet Detects Changes in Global DNA Methylation Status with Exposure to DNA Methylation-Modifying Agents.
A, Determination of % Tail DNA by treatment protocol, derived from a minimum of three independent experiments consisting of a 48h exposure to either control, 5 µM 5-Aza, 100 µM hydroxyurea, 1 mM nalidixic acid or 0.25 µM MTX, as indicated, with error bars reflecting SEM and * denoting statistical significance, defined here as p≤0.05. The addition of McrBC to control treated cells resulted in a statistically significant increase in % Tail DNA versus buffer (Control/McrBC: 3.13 ± 0.29%; Control/buffer: 1.87 ± 0.22%; *p=2.71E-06), as predicted to reflect the detection of the induced McrBC(methylation)-specific DNA damage and baseline 5-mC%. In contrast, the predicted hypomethylating agent, 5-Aza, did not have a significant difference in % Tail DNA for McrBC versus buffer (5-Aza/McrBC: 3.86 ± 0.40%; 5-Aza/buffer: 3.76 ± 0.39%; p=0.725); however, there was a significant difference in 5-Aza/buffer versus Control-buffer (*p=7.49E-05), suggesting genotoxicity for the compound. The addition of each of the predicted hypermethylating agents (hydroxyurea, nalidixic acid and MTX) each resulted in a significant increase in % Tail DNA for the McrBC protocol versus buffer alone (Hydroxyurea/McrBC: 7.08 ± 0.80%; Hydroxyurea/buffer: 2.49 ± 0.27%; *p=2.11E-05. Nalidixic acid/McrBC: 6.36 ± 0.49%; Nalidixic acid/buffer: 2.73 ± 0.31%; *p=1.54E-05. MTX/McrBC: 7.29 ± 0.68%; MTX/buffer: 2.76 ± 0.20%; *p=2.12E-05). B, The absolute change in % Tail DNA that was observed with the addition of McrBC versus buffer alone for each condition (as calculated from the data in A), was as follows: Control: +1.26; 5-Aza: −0.09; Hydroxyurea: +4.59; Nalidixic acid: +3.63; MTX: +4.52. C, The normalized (to buffer alone) change in % Tail DNA was as follows: 1.79±0.13% (Mean ± SEM) for control treatment; 1.02 ± 0.08% with 5-Aza exposure; 2.95 ± 0.30% with hydroxyurea exposure; 2.71 ± 0.42% with nalidixic acid exposure; and 2.74 ± 0.26% with MTX exposure.
Cellular exposure to each of the hypermethylating agents (hydroxyurea, nalidixic acid and MTX) all resulted in a statistically significant increase in % Tail DNA for the McrBC protocol versus buffer alone, as also shown in Figure 3. In addition to the raw data, the absolute change in % Tail DNA that was observed with the addition of McrBC versus buffer alone for each condition is presented in Figure 3B for easier visualization. Further, in addition to direct subtraction of results obtained from experimental conditions to controls, we also normalized the individual treatments with McrBC to their exact treatment pair match with buffer alone to reduce several confounding variables, as shown in Figure 3C. These values were subsequently used to evaluate the strong correlation of normalized % Tail DNA to the value we independently obtained for global methylation status (5-mC%).
EpiComet Detects Global DNA Methylation Status
Although our EpiComet assay has the advantage of being able to use a single platform to examine both genotoxic endpoints and DNA methylation status, the ability to perform both of these functions adds a level of complexity to the system. Therefore, we sought to formally test if the results we obtained using EpiComet from a single treatment can accurately and directly detect global methylation status in a treatment agnostic manner. A continuum of hypomethylation, “normal” 5-mC and hypermethylation was obtained in HeLa-S3 cells in response to exposures to either control, 5-Aza, hydroxyurea, nalidixic acid or MTX. Unlike our previous experiment in which we formally addressed exposure-mediated effects, here we sought to determine if EpiComet was able to detect global DNA status using % Tail DNA as a surrogate for 5-mC% independent of the mechanism and treatment conditions by which we arrived at a final global methylation status.
Importantly, single cell homogenates from the exact same treatment well were collectively harvested, provided a unique identifier and divided into two equal cell populations. One half was used to quantitate DNA methylation using standard methods, as described above, and the other half was used to perform the EpiComet assay (subdivided again in either McrBC or buffer alone treatment groups) to obtain an absolute value for % Tail DNA, at which point the two values were subsequently directly compared as a matched pair. This matched pair comparison was used to derive values for the absolute difference and fold change in % Tail DNA between McrBC and buffer. It is important to note that all of these values are directly obtained from a single, specific cellular treatment well and singularly prepared single cell homogenate pools prior to being subjected to distinct protocol variations. This feature thus allows us to make direct comparisons without introducing an additional source of uncertainty that would be required to externally define a baseline % Tail DNA (and/or a baseline 5-mC%) level.
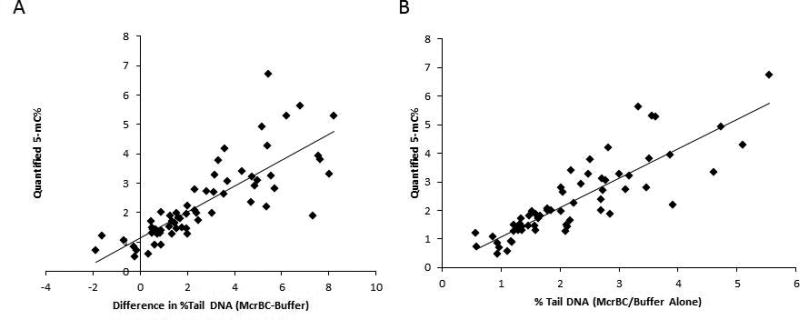
As shown in Figure 4A, independent of the method by which we used to hypo- or hyper- methylate global DNA, we were able to obtain a reasonably linear, direct correlation (r2 = 0.64) in which increases in the corrected difference of % Tail DNA (McrBC-buffer) are largely seen with increases in quantified 5-mC%. It should be noted that one should not simply look at non-corrected or non-normalized % Tail DNA numbers, (i.e. independent of exposure matching and knowledge of whether they are derived from McrBC or buffer), for two major reasons. First, it is known that several agents that alter DNA methylation status may also act in a genotoxic mechanism that can act independent of McrBC addition, as is the case in our study with some doses of 5-Aza in specific cell lines (Figure 3A). Therefore, there is a need to quantify % Tail DNA (Treatment) − % Tail DNA (Control) and examine only at exact same treatment conditions for direct comparisons, as one would normally do when performing a standard alkaline comet assay.
Figure 4. EpiComet and EpiComet-Chip % Tail DNA Single Cell Determinations Correlate to Quantified, Induced- DNA Methylation (5-mC%) Alterations.
A, The calculated difference in % Tail DNA for specific treatment isolates with McrBC minus buffer alone is plotted versus the quantified 5-mC% obtained for the same exact treatment. Notice the high level of correlation obtained in which increasing amounts of % Tail DNA correlate (r2 = 0.64) to increased values for global methylation. B, The normalized (to buffer alone) % Tail DNA for specific treatment isolates with McrBC addition is plotted versus the quantified 5-mC% obtained for the same exact treatment (using the same dataset that was used for A). Notice the higher correlation (r2 = 0.73) obtained for increasing amounts of normalized % Tail DNA with increased values for global methylation.
Second, there is also a reasonable expectation that the McrBC and buffer treatment protocols could increase DNA damage (and thus % Tail DNA), due to the requirement of performing some of the steps at 37°C versus 4°C. Therefore, there is an additional need to examine % Tail DNA (Treatment X/McrBC) − % Tail DNA (Treatment X/Buffer). Thus, by analyzing the exact same tube under the exact same conditions to define a baseline value, we can account for and eliminate the need for introducing additional statistical methods to account for these known confounding variables. Further, by normalizing % Tail DNA for McrBC to % Tail DNA for buffer (i.e. % Tail DNA (McrBC) ÷ % Tail DNA (buffer)) for the same single exposure, we reduced many variables to demonstrate a high correlation (r2 = 0.73) of normalized % Tail DNA with quantified 5-mC%, as shown in Figure 4B.
EpiComet and EpiComet-Chip Accurately Predict Induced- DNA Methylation Alterations
Given that we were interested not only in creating an assay that correctly correlates % Tail DNA with absolute global methylation values (r2 = 0.73, Figure 4B), but rather an assay that can also act as a predictive test for hypo- and hyper-methylation that results from specific exposure conditions, we transformed our data by employing the following methods. First, using the EpiComet and EpiComet-Chip protocols as described above, we determined a mean value for the normalized % Tail DNA with control treatment and quantified the standard deviation (SD) in HeLa-S3 cells. Here, for % Tail DNA, we initially and arbitrarily defined Mean ± SD (as consistent with a “normal/baseline” level of methylation; less than Mean −SD as our prediction signal for hypomethylation; and greater than Mean + SD as our threshold for prediction of hypermethylation, and subsequently tested the ability of our EpiComet system to accurately predict the actual global methylation status.
Analogously, for our initial determination of “normal/baseline” methylation levels, we similarly used the Mean and SD values that we obtained for 5-mC% in control treated HeLa-S3 cells for 5-mC% for control treated cells. For this initial test with quantified 5-mC%, we therefore arbitrarily defined hypomethylation as values less than Mean −SD; normal for values in the range of Mean ± SD; and hypermethylation as greater than Mean + SD. Given that the quantification of 5-mC% occurred via a well-established and accepted protocol, we defined these data as the “truth situation” for which our EpiComet technology would attempt to predict.
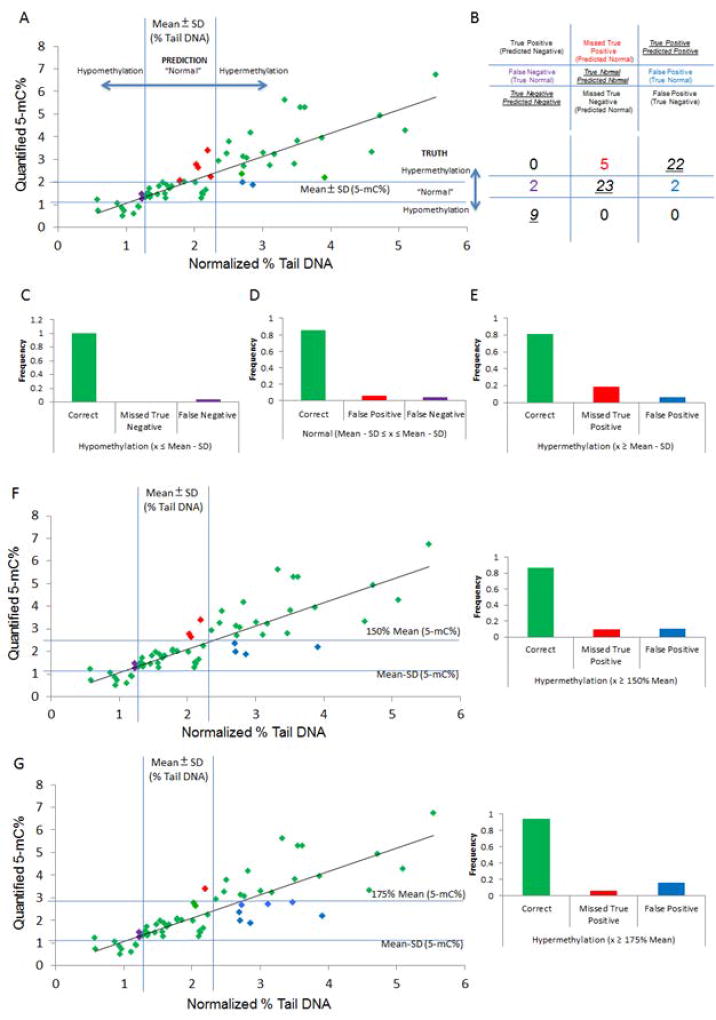
Using this approach, we directly examined the ability of our EpiComet readout to accurately predict global methylation status (Figure 5A–E), and subsequently subdivided status into smaller categories by relative methylation levels (Figure 5F–I). Overall, for 63 matched pairs, our 5-mC% quantification yielded nine treatments that caused hypomethylation, 27 treatments that resulted in normal methylation status, and an additional 27 that resulted in hypermethylation. Using our criteria for % Tail DNA, we accurately identified hypomethylation at a rate of 100% (Figure 5A–C); accurately identified normal methylation in 85% of cases (Figure 5A–B, D); accurately identified hypermethylation in 81% of instances (Figure 5A–B, E). Further, we had a 4% false negative rate (Figure 5A–B, D), 6% false positive rate (Figure 5A–B, D), and had no attribution of a case of actual hypermethylation/hypomethylation to the opposite category (Figure 5A–B and Supplemental Table S1 for annotated values).
Figure 5. EpiComet and EpiComet-Chip Accurately Predict Single Cell Induced- DNA Methylation Alterations.
A, Comparison of Normalized % Tail DNA plotted versus Quantified 5-mC%. We arbitrarily defined "normal/baseline" methylation values (x) to be in the range of Mean − SD ≤ x ≤ Mean + SD obtained from control treatment alone (i.e. Mean 5-mC% (1.61) ± SD (0.4), thus 1.21 – 2.01% for HeLa-S3 cells, denoted by lines on y-axis of graph). Note: Here, as opposed to the other figures, we are using standard deviation (SD) and not SEM to define our threshold parameters. We similarly arbitrarily defined "normal/baseline" % Tail DNA values to be in the range of Mean − SD ≤ x ≤ Mean + SD obtained from control treatment alone (i.e. Mean % Tail DNA (1.79) ± SD (0.5), thus 1.29 – 2.29% for HeLa-S3 cells, denoted by lines on x-axis of graph). B, Schematic and quantification of prediction accuracy for EpiComet prediction by category, using the threshold values defined in A. Note: Lines represent the four threshold values. C–E, Subdivision of prediction accuracy for EpiComet for (C) hypomethylation, (D) normal, and (E) hypermethylation using the threshold values defined in A. Using our criteria for % Tail DNA we accurately identified hypomethylation at a rate of 100% (9/9); accurately identified normal methylation in 85% of cases (23/27); accurately identified hypermethylation in 81% of instances ((22/27), missed true positives denoted by arrows); falsely attributed 2 normal methylation status samples as hypomethylation (4% false negative rate (2/54), denoted by asterisks), falsely attributed 2 normal methylation status samples as hypermethylation (6% false positive rate (2/36), denoted by stars), and had 0% (0/36) attribution of a case of actual hypermethylation/hypomethylation to the opposite category. F–G, Accuracy in predicting methylation with a less stringent upper threshold of 50% increase in methylation from control mean defined as "normal/baseline" (i.e. greater than 2.42 for 5-mC% scored as hypermethylation). Using this threshold, EpiComet accurately identified 87% (20/23) of the instances with a minimum of a 50% increase in methylation; however, our false positive rate was increased to 10%. H–I, Accuracy in predicting methylation with a less stringent upper threshold of 75% increase in methylation from control mean defined as "normal/baseline" (i.e. greater than 2.82 for 5-mC% scored as hypermethylation). Using this threshold, EpiComet accurately identified 94% (17/18), of instances with at least a 75% increase in methylation; however, our false positive rate was increased to 16%.
In nine instances, we obtained at least a 25% reduction in quantified 5-mC% versus the mean for control treatment. In all of these cases we accurately identified all instances as hypomethylation using EpiComet. In addition, we had 23 instances in which we observed at least a 50% increase in methylation versus the mean for control treatment, including a subset of 18 instances in which we quantified at least a 75% increase in methylation. EpiComet accurately identified 87% (Figure 5F–G) of the instances with a minimum of a 50% increase in methylation and 94% (Figure 5H–I) of instances with at least a 75% increase in methylation; however, our false positive rate was increased to 10% and 16%, respectively (from 6%), by altering our corresponding threshold values (Figure 5F–I and Supplemental Table S1).
To demonstrate that the utility of our approach was not limited to a single cell line, we also examined HepG2 and TK-6 in a limited sample set and found the same trends that were seen for HeLa-S3 as an initial proof of concept (data not shown). Although we originally hypothesized that our ability to detect a reduction in methylation might be largely limited, perhaps as a consequence of the low endogenous levels of DNA methylation in our cell lines; collectively our data suggest that the changes in global DNA methylation status of either single treatments or a group of exposures with DNA methylation-modifying agents correlate to a large degree to % Tail DNA determined using our novel EpiComet and EpiComet-Chip systems.
Discussion
Assays that determine DNA damage and DNA methylation are extremely important in predicting the carcinogenicity of drugs, physical and biological agents, and environmental exposures. The comet assay is a sensitive and simple technique traditionally used to detect DNA strand breaks in single cells, which can be modified to detect a variety of DNA lesions. In this report we describe how we established unique tools and novel methods to combine the comet assay with restriction enzymes to create EpiComet and EpiComet-Chip for evaluating global DNA methylation status in individual cells in a high-throughput, cost effective manner to address this regulatory challenge. An analogous approach to the use of glycosylases to reveal DNA base damage is the use of enzymes that cleave DNA at sites of epigenetic structural changes to base structure. Methylation dependent restriction enzymes, such as HpaII, MspI, and more recently, McrBC, have been used extensively for a variety of epigenetic analyses (Pogribny et al. 1999; Fujiwara and Ito 2002; Ordway et al. 2006; Wentzel et al. 2010). In addition, HpaII and MspI have been previously used in conjunction with the comet assay, although with limited sensitivity (Wentzel et al. 2010). The limited sensitivity is likely due to the intrinsic properties of the enzymes. HpaII and its isoschizomer MspI recognize the same tetranucleotide sequence (5’-CCGG-3’) but display differential sensitivity to DNA methylation, in that neither enzyme will cleave when the external cytosine (in the sequence CCGG) is methylated, and MspI (unlike HpaII) can cleave the sequence when the internal cytosine residue is methylated (McClelland et al. 1994). Therefore, by using both MspI and HpaII independently, one can directly compare the ratio of enzymatic cuts for MspI/HpaII as a proxy for methylation status.
In contrast to the isoschizomer approach, McrBC possesses desirable characteristics that allow for the use of a single restriction digest for cleavage of a majority of the methylcytosines present in the DNA, across a wide range of global methylation statuses. Specifically, McrBC cleaves DNA containing 5-methylcytosine, 5-hydroxymethylcytosine or N4-methylcytosine on one or both strands (Gowher et al. 2000) and will not cut unmethylated DNA (Sutherland et al. 1992). McrBC recognizes two half-sites on DNA of the form (G/A)mC; these half-sites can be separated by up to 3 kb, with an optimal separation is 55–103 bp (recognition site-5’…PumC (N-40-3000) PumC…3’) (Gowher et al. 2000; Zhou et al. 2002). This short consensus sequence of McrBC, (PumC) allows the enzyme to recognize and cut a large proportion of the methylcytosines present in DNA. Further, McrBC is also capable of cleaving DNA which is not heavily methylated, as a low level of cleavage occurs even when the PumC elements are as far as 3 kb apart (Burman et al. 1999). Due to these desirable characteristics, we utilized McrBC to design and validate a novel, high sensitivity version of the comet assay, “EpiComet”. Specifically, after lysis, DNA is incubated with McrBC, which cleaves the DNA at sites of 5-mC, thus increasing DNA migration when analyzed using the comet assay approach. Thus, the EpiComet converts undetectable 5-mC into single strand breaks that can be quantified using the comet assay approach.
Our initial pilot studies related to exposure-mediated changes to global methylation status involved performing an exploratory series of exposure dose-response and exposure duration-response experiments with a variety of chemotherapeutic, environmental and novel agents of regulatory interest. These experiments exploited the properties of the following compounds shown in various previous contexts to either increase global methylation: cisplatin (Nyce 1989; Nyce et al. 1993), nalidixic acid (Nyce 1989), hydroxyurea (Nyce 1989) and higher dose MTX (Nyce 1989; Nyce et al. 1993); or decrease global methylation: 5-Aza (Christman 2002), procainamide (Cornacchia et al. 1988; Lee et al. 2005), 4,6-dioxoheltanoic acid (SA) (Wentzel et al. 2010; Lewies et al. 2014), tamoxifen (Wu et al. 2005; Tryndyak et al. 2006), valproic acid (Detich et al. 2003; Cribbs et al. 2015) and hydralazine (Cornacchia et al. 1988). Further, these pilot experiments allowed us to define appropriate dose ranges and duration of exposures for each agent/cell line pair.
Prior to our experiments, it was predicted that changes in global methylation might require timeframes between 16–96h and therefore we collected the mock and compound treated cells at three time points: 24h, 48h, or 72h after the initial dosing was performed. One limitation in our pilot studies was that the dynamics of methylation were not explored in a true full continuum, as our quantitation methods required the terminal use of the cellular material for analyses. In addition to beginning with uncertainty on which exposure duration to treat our cell lines, the effective dose in which we would anticipate to see an effect was also largely unknown. Whenever feasible, we set our middle dose concentration in a range of 4 doses (0, low, medium and high concentration) to reflect an expected value published in the field. Several initial preliminary findings were intriguing, but beyond the scope of this report, as several values may not reflect a true effect, as a consequence of limited experimental replicates for any one of the pilot treatments in the experimental design. Specifically, our experimental design was exploratory in nature and yielded many results with wide standard deviations in which conclusions could not be accurately made without substantial added effort. While the majority of our exposures behaved in the predicted manner in modifying DNA methylation, future studies should analyze preliminary findings which included several results that displayed cell line-dependent differences, did not produce a predicted detectable change in DNA methylation, produced biphasic trends loosely correlated with duration or exposure concentration, and/or did not provide consistent directional data that mirrored expectations from a large volume of previous experiments published in the literature.
As a consequence of these findings, we determined to focus on a subset of cell lines (mainly HeLa-S3) and exposure conditions (specific doses of four compounds) to perform our proof-of-principle studies and validate the EpiComet assay. Our goal was to develop and validate our assay in a manner that is consistent with accurately detecting differences in global methylation status in specific single cells that undergo exposure with chemical agents in the exact same treatment well. Therefore, we took advantage of the wide range of distinct sensitivities and dynamic responses of the distinct cell lines to exposure durations to examine if we could define the accuracy and limits for detectable differences in global methylation as determined using our study methods. Being able to determine the overall methylation status in a treatment agnostic manner provided us with great flexibility and has resulted in a versatile assay. In fact, this property of EpiComet allowed us to accurately and independently account for a unique failed experiment in which the lack of determined methylation-induction specifically could be attributed to a specific experiment in which an acute, limited dosing error had occurred (unpublished data).
Although the relationship between the EpiComet results and overall human exposure risk from any one agent is largely beyond the scope of this report, it is clear that having a relatively high throughput and low cost system that provides a rapid initial readout of global methylation status will have clear utility in a variety of human risk assessment protocols. Whereas EpiComet will have utility for identifying exposure conditions that globally hypermethylate and hypomethylate DNA above and below a user defined threshold, respectively, there exist several higher sensitivity (and more resource intensive) techniques that could subsequently be performed for a limited subset of exposures. In addition to having higher sensitivity, these additional testing procedures likely could characterize the localization and actual modifications that occur as a direct result to the exposure with an agent of interest in many instances. Further, EpiComet is unable to identify diverse epigenetic modifications, including changes in localized methylation patterns on a critical DNA region in the absence of overall changes in global methylation status. Therefore, in some settings EpiComet may not be the preferred initial screening method; however, given the existing OECD Test Guideline for the in vivo comet assay (TG489) for genotoxicity studies, the concomitant use of our EpiComet and EpiComet-Chip modification to this protocol should afford an opportunity to gain valuable epigenetic data with a relatively low input of additional laboratory or researcher resources required. Collectively, this suggests that the use of EpiComet may be warranted in most instances in which a standard comet assay normally would be performed.
Despite the fact that there are several methods by which to analyze global DNA methylation (Boch C 2016), there are no simple and cost effective methods available to use a single platform assay to examine for genotoxic induction of DNA damage and detect epigenetic modifications (such as global DNA methylation status) using a single platform, as described here using EpiComet. Overall, the successful application of this novel technology will aid hazard identification and risk characterization of FDA-regulated products, while providing utility for investigating epigenetic modes of action of agents in target organs, since the assay is amenable to cells in culture or nucleated cells from any tissue.
Supplementary Material
Supplemental Figure 1: Human Cell Lines are Amenable for Use in Alkaline Comet Assays. Quantification of mean percent (%) Tail DNA, derived from a minimum of three independent experiments and error bars reflecting SEM and * denoting statistical significance, defined here as p≤0.05. Background % Tail DNA levels were determined for the tested four human immortalized cells lines treated with control (vehicle) treatment as follows: HeLa-S3: 4.7 ± 1.3 (Mean ± SEM); HepG2: 8.0 ± 4.1; MCF-7: 12.5 ± 2.8; TK-6; 3.2 ± 0.7. Treatment with a 3h exposure to 10 µg/ml MMS, a known genotoxicant, induced DNA damage in all four cells lines as follows: HeLa-S3: 31.1 ± 0.5, Two-tailed Student's t test versus control: * p=0.004; HepG2: 28.9 ± 3.7, *p=0.05; MCF-7: 43.5 ± 0.8,*p=0.01; TK-6: 51.9 ± 0.3, *p=0.0002. A dose-dependent increase in DNA damage induction was seen for all lines except TK-6 with a 3h exposure to 20 µg/ml MMS, as follows: HeLa-S3: 51.9 ± 2.4, *p=0.006 (versus control), *p=0.01 (versus 10 µg/ml MMS); HepG2: 60.6 ± 0.8, *p=.001, *p=.027; MCF-7: 53.9 ± 1.9, *p=0.01, *p=0.01; TK-6: 50.1 ± 0.7, *p=0.001, p=0.08.
Supplemental Figure 2: Quantification of Background Levels of Global Methylation (5-mC%) in Human Cell Lines. Quantification of mean 5-mC%, derived from a minimum of three independent experiments and error bars reflecting SEM. Background 5-mC% levels were determined for the following four human immortalized cell lines as follows: HeLa-S3: 1.6 ± 0.11% (Mean ± SEM); TK-6: 0.9 ± 0.28%; HepG2: 1.5 ± 0.47; and MCF-7: 1.8 ± 0.56%.
Acknowledgments
The data and views presented here are those of the authors alone and do not reflect or imply the formal views of the U.S. Food and Drug Administration or any other entity. This study was supported by the National Center for Toxicological Research and the United States Food and Drug Administration (Protocol E7574.01). The authors thank Drs. Volodymyr Tryndyak and Barbara Parsons for their critical review of this manuscript; in addition to Nicole Sonnert, Michelle Bishop and Sharon Shelton for technical support of experiments. TAT was supported in part by the Commissioner's Fellowship Program with the United States Food and Drug Administration. MCP was supported in part by (U.S.) National Institutes of Health (NIH) Grants [T32-ES007020], [P01-CA026731] and [P30-ES002109] (to MIT Center for Environmental Health Sciences). Many of the experiments described in this research utilized a HeLa cell line. Henrietta Lacks, and the HeLa cell line that was established from her tumor cells without her knowledge or consent in 1951, have made significant contributions to scientific progress and advances in human health. We are grateful to Henrietta Lacks, now deceased, and to her surviving family members for their contributions to biomedical research.
Footnotes
Statement of Author Contributions
Drs. Townsend and Manjanatha designed the study and submitted the protocol for internal approval. Drs. Engelward and Townsend prepared Figure 1 with input from Dr. Parrish. Dr. Townsend performed the experiments, analyzed the data, and prepared Figures 2–5 and all Supplemental Materials with intellectual input from all authors. Drs. Parrish and Engelward provided CometChip tools, reagents and critical intellectual insight regarding the high-throughput assay development. Dr. Townsend prepared the manuscript draft with important intellectual input from Drs. Parrish, Engelward and Manjanatha. All authors approved the final manuscript and had complete access to the study data.
References
- Agnihothram SS, Vermudez SA, Mullis L, Townsend TA, Manjanatha MG, Azevedo MP. Silicon Dioxide Impedes Antiviral Response and Causes Genotoxic Insult During Calicivirus Replication. J Nanosci Nanotechnol. 2016;16(7):7720–7730. doi: 10.1166/jnn.2016.12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azqueta A, Dusinska M. The use of the comet assay for the evaluation of the genotoxicity of nanomaterials. Front Genet. 2015;6:239. doi: 10.3389/fgene.2015.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azqueta A, Shaposhnikov S, Collins AR. DNA oxidation: investigating its key role in environmental mutagenesis with the comet assay. Mutat Res. 2009;674(1–2):101–108. doi: 10.1016/j.mrgentox.2008.10.013. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11(10):726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benayoun BA, Pollina EA, Brunet A. Epigenetic regulation of ageing: linking environmental inputs to genomic stability. Nat Rev Mol Cell Biol. 2015;16(10):593–610. doi: 10.1038/nrm4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Blueprint Consortium T. Quantitative comparison of DNA methylation assays for biomarker development and clinical applications. Nat Biotech. 2016;34(7):726–737. doi: 10.1038/nbt.3605. [DOI] [PubMed] [Google Scholar]
- Boch CHF, Carmona FJ, Tierling S, Datlinger P, Assenov Y, Berdasco M, Bergmann AK, Booher K, Busato F, Campan M, Dahl C, Dahmcke CM, Diep D. Quantitative comparison of DNA methylation assays for biomarker development and clinical applications. Nat Biotechnol. 2016;34(7):726–737. doi: 10.1038/nbt.3605. [DOI] [PubMed] [Google Scholar]
- Bock C. Epigenetic biomarker development. Epigenomics. 2009;1(1):99–110. doi: 10.2217/epi.09.6. [DOI] [PubMed] [Google Scholar]
- Brown TC, Juhlin CC, Healy JM, Prasad ML, Korah R, Carling T. Frequent silencing of RASSF1A via promoter methylation in follicular thyroid hyperplasia: a potential early epigenetic susceptibility event in thyroid carcinogenesis. JAMA Surg. 2014;149(11):1146–1152. doi: 10.1001/jamasurg.2014.1694. [DOI] [PubMed] [Google Scholar]
- Burman RW, Yates PA, Green LD, Jacky PB, Turker MS, Popovich BW. Hypomethylation of an expanded FMR1 allele is not associated with a global DNA methylation defect. Am J Hum Genet. 1999;65(5):1375–1386. doi: 10.1086/302628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, Hein A, Rote NS, Cope LM, Snyder A, Makarov V, Budhu S, Slamon DJ, Wolchok JD, Pardoll DM, Beckmann MW, Zahnow CA, Merghoub T, Chan TA, Baylin SB, Strick R. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell. 2015;162(5):974–986. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christman JK. 5-Azacytidine and 5-aza-2'-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21(35):5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- Collins AR. The comet assay for DNA damage and repair: principles, applications, and limitations. Mol Biotechnol. 2004;26(3):249–261. doi: 10.1385/MB:26:3:249. [DOI] [PubMed] [Google Scholar]
- Collins AR, Dusinska M. Oxidation of cellular DNA measured with the comet assay. Methods Mol Biol. 2002;186:147–159. doi: 10.1385/1-59259-173-6:147. [DOI] [PubMed] [Google Scholar]
- Cornacchia E, Golbus J, Maybaum J, Strahler J, Hanash S, Richardson B. Hydralazine and procainamide inhibit T cell DNA methylation and induce autoreactivity. J Immunol. 1988;140(7):2197–2200. [PubMed] [Google Scholar]
- Cribbs A, Feldmann M, Oppermann U. Towards an understanding of the role of DNA methylation in rheumatoid arthritis: therapeutic and diagnostic implications. Ther Adv Musculoskelet Dis. 2015;7(5):206–219. doi: 10.1177/1759720X15598307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csoka AB, Szyf M. Epigenetic side-effects of common pharmaceuticals: a potential new field in medicine and pharmacology. Med Hypotheses. 2009;73(5):770–780. doi: 10.1016/j.mehy.2008.10.039. [DOI] [PubMed] [Google Scholar]
- Detich N, Bovenzi V, Szyf M. Valproate induces replication-independent active DNA demethylation. J Biol Chem. 2003;278(30):27586–27592. doi: 10.1074/jbc.M303740200. [DOI] [PubMed] [Google Scholar]
- Ding W, Bishop ME, Lyn-Cook LE, Davis KJ, Manjanatha MG. In Vivo Alkaline Comet Assay and Enzyme-modified Alkaline Comet Assay for Measuring DNA Strand Breaks and Oxidative DNA Damage in Rat Liver. J Vis Exp. 2016;(111) doi: 10.3791/53833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding W, Bishop ME, Pearce MG, Davis KJ, White GA, Lyn-Cook LE, Manjanatha MG. Sex-specific dose-response analysis of genotoxicity in cyproterone acetate-treated F344 rats. Mutat Res Genet Toxicol Environ Mutagen. 2014;774:1–7. doi: 10.1016/j.mrgentox.2014.08.005. [DOI] [PubMed] [Google Scholar]
- Ding W, Levy DD, Bishop ME, Lyn-Cook Lascelles E, Kulkarni R, Chang CW, Aidoo A, Manjanatha MG. Methyleugenol genotoxicity in the Fischer 344 rat using the comet assay and pathway-focused gene expression profiling. Toxicol Sci. 2011;123(1):103–112. doi: 10.1093/toxsci/kfr153. [DOI] [PubMed] [Google Scholar]
- Epe B, Pflaum M, Haring M, Hegler J, Rudiger H. Use of repair endonucleases to characterize DNA damage induced by reactive oxygen species in cellular and cell-free systems. Toxicol Lett. 1993;67(1–3):57–72. doi: 10.1016/0378-4274(93)90046-z. [DOI] [PubMed] [Google Scholar]
- Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum Mol Genet. 2007;16(1):R50–59. doi: 10.1093/hmg/ddm018. [DOI] [PubMed] [Google Scholar]
- Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet. 2011;13(2):97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- Frotschl R. Experiences with the in vivo and in vitro comet assay in regulatory testing. Mutagenesis. 2015;30(1):51–57. doi: 10.1093/mutage/geu069. [DOI] [PubMed] [Google Scholar]
- Fujiwara H, Ito M. Nonisotopic cytosine extension assay: a highly sensitive method to evaluate CpG island methylation in the whole genome. Anal Biochem. 2002;307(2):386–389. doi: 10.1016/s0003-2697(02)00053-2. [DOI] [PubMed] [Google Scholar]
- Ge J, Chow DN, Fessler JL, Weingeist DM, Wood DK, Engelward BP. Micropatterned comet assay enables high throughput and sensitive DNA damage quantification. Mutagenesis. 2015;30(1):11–19. doi: 10.1093/mutage/geu063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J, Prasongtanakij S, Wood DK, Weingeist DM, Fessler J, Navasummrit P, Ruchirawat M, Engelward BP. CometChip: a high-throughput 96-well platform for measuring DNA damage in microarrayed human cells. J Vis Exp. 2014;(92):e50607. doi: 10.3791/50607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowher H, Leismann O, Jeltsch A. DNA of Drosophila melanogaster contains 5-methylcytosine. EMBO J. 2000;19(24):6918–6923. doi: 10.1093/emboj/19.24.6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet. 2012;13(10):679–692. doi: 10.1038/nrg3270. [DOI] [PubMed] [Google Scholar]
- Hu N, Qiu X, Luo Y, Yuan J, Li Y, Lei W, Zhang G, Zhou Y, Su Y, Lu Q. Abnormal histone modification patterns in lupus CD4+ T cells. J Rheumatol. 2008;35(5):804–810. [PubMed] [Google Scholar]
- Jones PA. At the tipping point for epigenetic therapies in cancer. J Clin Invest. 2014;124(1):14–16. doi: 10.1172/JCI74145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3(4):253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet. 2010;11(3):191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- Lee BH, Yegnasubramanian S, Lin X, Nelson WG. Procainamide is a specific inhibitor of DNA methyltransferase 1. J Biol Chem. 2005;280(49):40749–40756. doi: 10.1074/jbc.M505593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewies A, Van Dyk E, Wentzel JF, Pretorius PJ. Using a medium-throughput comet assay to evaluate the global DNA methylation status of single cells. Front Genet. 2014;5:215. doi: 10.3389/fgene.2014.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licht JD. DNA Methylation Inhibitors in Cancer Therapy: The Immunity Dimension. Cell. 2015;162(5):938–939. doi: 10.1016/j.cell.2015.08.005. [DOI] [PubMed] [Google Scholar]
- Manjanatha MG, Bishop ME, Pearce MG, Kulkarni R, Lyn-Cook LE, Ding W. Genotoxicity of doxorubicin in F344 rats by combining the comet assay, flow-cytometric peripheral blood micronucleus test, and pathway-focused gene expression profiling. Environ Mol Mutagen. 2014;55(1):24–34. doi: 10.1002/em.21822. [DOI] [PubMed] [Google Scholar]
- McClelland M, Nelson M, Raschke E. Effect of site-specific modification on restriction endonucleases and DNA modification methyltransferases. Nucleic Acids Res. 1994;22(17):3640–3659. doi: 10.1093/nar/22.17.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CM, Karemaker ID, van Leeuwen F. The emerging roles of DOT1L in leukemia and normal development. Leukemia. 2014;28(11):2131–2138. doi: 10.1038/leu.2014.169. [DOI] [PubMed] [Google Scholar]
- Nyce J. Drug-induced DNA hypermethylation and drug resistance in human tumors. Cancer Res. 1989;49(21):5829–5836. [PubMed] [Google Scholar]
- Nyce J, Leonard S, Canupp D, Schulz S, Wong S. Epigenetic mechanisms of drug resistance: drug-induced DNA hypermethylation and drug resistance. Proc Natl Acad Sci U S A. 1993;90(7):2960–2964. doi: 10.1073/pnas.90.7.2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OECD. Test No. 489: In Vivo Mammalian Alkaline Comet Assay. OECD Publishing; 2014. [Google Scholar]
- Ordway JM, Bedell JA, Citek RW, Nunberg A, Garrido A, Kendall R, Stevens JR, Cao D, Doerge RW, Korshunova Y, Holemon H, McPherson JD, Lakey N, Leon J, Martienssen RA, Jeddeloh JA. Comprehensive DNA methylation profiling in a human cancer genome identifies novel epigenetic targets. Carcinogenesis. 2006;27(12):2409–2423. doi: 10.1093/carcin/bgl161. [DOI] [PubMed] [Google Scholar]
- Paluch BE, Naqash AR, Brumberger Z, Nemeth MJ, Griffiths EA. Epigenetics: A primer for clinicians. Blood Rev. 2016;30(4):285–295. doi: 10.1016/j.blre.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogribny I, Yi P, James SJ. A sensitive new method for rapid detection of abnormal methylation patterns in global DNA and within CpG islands. Biochem Biophys Res Commun. 1999;262(3):624–628. doi: 10.1006/bbrc.1999.1187. [DOI] [PubMed] [Google Scholar]
- Pogribny IP, Beland FA. DNA methylome alterations in chemical carcinogenesis. Cancer Lett. 2013;334(1):39–45. doi: 10.1016/j.canlet.2012.09.010. [DOI] [PubMed] [Google Scholar]
- Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28(10):1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011;12(8):529–541. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447(7143):425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- Roulois D, Loo Yau H, Singhania R, Wang Y, Danesh A, Shen SY, Han H, Liang G, Jones PA, Pugh TJ, O'Brien C, De Carvalho DD. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell. 2015;162(5):961–973. doi: 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CC, O'Donovan MR, Martin EA. hOGG1 recognizes oxidative damage using the comet assay with greater specificity than FPG or ENDOIII. Mutagenesis. 2006;21(3):185–190. doi: 10.1093/mutage/gel019. [DOI] [PubMed] [Google Scholar]
- Sutherland E, Coe L, Raleigh EA. McrBC: a multisubunit GTP-dependent restriction endonuclease. J Mol Biol. 1992;225(2):327–348. doi: 10.1016/0022-2836(92)90925-a. [DOI] [PubMed] [Google Scholar]
- Toffoli G, Russo A, Innocenti F, Corona G, Tumolo S, Sartor F, Mini E, Boiocchi M. Effect of methylenetetrahydrofolate reductase 677C-->T polymorphism on toxicity and homocysteine plasma level after chronic methotrexate treatment of ovarian cancer patients. Int J Cancer. 2003;103(3):294–299. doi: 10.1002/ijc.10847. [DOI] [PubMed] [Google Scholar]
- Tryndyak VP, Muskhelishvili L, Kovalchuk O, Rodriguez-Juarez R, Montgomery B, Churchwell MI, Ross SA, Beland FA, Pogribny IP. Effect of long-term tamoxifen exposure on genotoxic and epigenetic changes in rat liver: implications for tamoxifen-induced hepatocarcinogenesis. Carcinogenesis. 2006;27(8):1713–1720. doi: 10.1093/carcin/bgl050. [DOI] [PubMed] [Google Scholar]
- Vera E, Canela A, Fraga MF, Esteller M, Blasco MA. Epigenetic regulation of telomeres in human cancer. Oncogene. 2008;27(54):6817–6833. doi: 10.1038/onc.2008.289. [DOI] [PubMed] [Google Scholar]
- Waggoner D. Mechanisms of disease: epigenesis. Semin Pediatr Neurol. 2007;14(1):7–14. doi: 10.1016/j.spen.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Watson C, Ge J, Cohen J, Pyrgiotakis G, Engelward BP, Demokritou P. High-throughput screening platform for engineered nanoparticle-mediated genotoxicity using CometChip technology. ACS Nano. 2014;8(3):2118–2133. doi: 10.1021/nn404871p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingeist DM, Ge J, Wood DK, Mutamba JT, Huang Q, Rowland EA, Yaffe MB, Floyd S, Engelward BP. Single-cell microarray enables high-throughput evaluation of DNA double-strand breaks and DNA repair inhibitors. Cell Cycle. 2013;12(6):907–915. doi: 10.4161/cc.23880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentzel JF, Gouws C, Huysamen C, Dyk E, Koekemoer G, Pretorius PJ. Assessing the DNA methylation status of single cells with the comet assay. Anal Biochem. 2010;400(2):190–194. doi: 10.1016/j.ab.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Wu H, Chen Y, Liang J, Shi B, Wu G, Zhang Y, Wang D, Li R, Yi X, Zhang H, Sun L, Shang Y. Hypomethylation-linked activation of PAX2 mediates tamoxifen-stimulated endometrial carcinogenesis. Nature. 2005;438(7070):981–987. doi: 10.1038/nature04225. [DOI] [PubMed] [Google Scholar]
- Yang X, Han H, De Carvalho DD, Lay FD, Jones PA, Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26(4):577–590. doi: 10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao CQ, Young MR, Diwan BA, Coogan TP, Waalkes MP. Association of arsenic-induced malignant transformation with DNA hypomethylation and aberrant gene expression. Proc Natl Acad Sci U S A. 1997;94(20):10907–10912. doi: 10.1073/pnas.94.20.10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Bui T, Auckland LD, Williams CG. Undermethylated DNA as a source of microsatellites from a conifer genome. Genome. 2002;45(1):91–99. doi: 10.1139/g01-119. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Lu Q. DNA methylation in T cells from idiopathic lupus and drug-induced lupus patients. Autoimmun Rev. 2008;7(5):376–383. doi: 10.1016/j.autrev.2008.03.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Human Cell Lines are Amenable for Use in Alkaline Comet Assays. Quantification of mean percent (%) Tail DNA, derived from a minimum of three independent experiments and error bars reflecting SEM and * denoting statistical significance, defined here as p≤0.05. Background % Tail DNA levels were determined for the tested four human immortalized cells lines treated with control (vehicle) treatment as follows: HeLa-S3: 4.7 ± 1.3 (Mean ± SEM); HepG2: 8.0 ± 4.1; MCF-7: 12.5 ± 2.8; TK-6; 3.2 ± 0.7. Treatment with a 3h exposure to 10 µg/ml MMS, a known genotoxicant, induced DNA damage in all four cells lines as follows: HeLa-S3: 31.1 ± 0.5, Two-tailed Student's t test versus control: * p=0.004; HepG2: 28.9 ± 3.7, *p=0.05; MCF-7: 43.5 ± 0.8,*p=0.01; TK-6: 51.9 ± 0.3, *p=0.0002. A dose-dependent increase in DNA damage induction was seen for all lines except TK-6 with a 3h exposure to 20 µg/ml MMS, as follows: HeLa-S3: 51.9 ± 2.4, *p=0.006 (versus control), *p=0.01 (versus 10 µg/ml MMS); HepG2: 60.6 ± 0.8, *p=.001, *p=.027; MCF-7: 53.9 ± 1.9, *p=0.01, *p=0.01; TK-6: 50.1 ± 0.7, *p=0.001, p=0.08.
Supplemental Figure 2: Quantification of Background Levels of Global Methylation (5-mC%) in Human Cell Lines. Quantification of mean 5-mC%, derived from a minimum of three independent experiments and error bars reflecting SEM. Background 5-mC% levels were determined for the following four human immortalized cell lines as follows: HeLa-S3: 1.6 ± 0.11% (Mean ± SEM); TK-6: 0.9 ± 0.28%; HepG2: 1.5 ± 0.47; and MCF-7: 1.8 ± 0.56%.