Abstract
Little is known about the link between gastric microbiota and the epidemiology of gastric cancer. In order to determine the epidemiologic and clinical relevance of gastric microbiota, we used 16S ribosomal RNA gene sequencing analysis to characterize the composition and structure of the gastric microbial community of 80 paired samples (non-malignant and matched tumor tissues) from gastric cardia adenocarcinoma (GCA) patients in Shanxi, China. We also used PICRUSt to predict microbial functional profiles. Compared to patients without family history of upper gastrointestinal (UGI) cancer in the non-malignant gastric tissue microbiota, patients with family history of UGI cancer had higher Helicobacter pylori (Hp) relative abundance (median: 0.83 vs. 0.38, P=0.01) and lower alpha diversity (median observed species: 51 vs. 85, P=0.01). Patients with higher (versus lower) tumor grade had higher Hp relative abundance (0.73 vs. 0.18, P=0.03), lower alpha diversity (observed species, 66 vs. 89, p=0.01), altered beta diversity (weighted UniFrac, P=0.002), and significant alterations in relative abundance of five KEGG functional modules in non-malignant gastric tissue microbiota. Patients without metastases had higher relative abundance of Lactobacillales than patients with metastases (0.05 vs. 0.01, P=0.04) in non-malignant gastric tissue microbiota. These associations were observed in non-malignant tissues but not in tumor tissues. In conclusion, this study showed a link of gastric microbiota to a major gastric cancer risk factor and clinical features in GCA patients from Shanxi, China. Studies with both healthy controls and gastric cardia and noncardia cancer cases across different populations are needed to further examine the association between gastric cancer and the microbiota.
Keywords: H. pylori, 16S rRNA gene, KEGG modules, microbiome, gastric cancer
Introduction
Gastric cancer is the fifth most common cancer in the world and the third leading cause of cancer death1. Gastic cancer incidence varies widely by populations with rates high in Asia, Eastern Europe, and Central and South America, and low in North America and Africa2. Gastric cancer may arise in cardia, proximal to the esophagus, or in the noncardia, including the fundus, body, or pylorus of the stomach. Chronic colonization of Helicobacter pylori (Hp) is known to increase the risk of noncardia cancer3. The association between Hp colonization and gastric cardia cancer varies by populations. In Western countries, there is a neutral or even negative association between cardia cancer and Hp colonization. In Eastern populations namely China, Japan and Korea, there is strong evidence of a higher risk of cardia cancer among subjects with Hp colonization4.
Shanxi Province has among the highest rates of gastric cancer in China, most of which are in the gastric cardia (gastric cardia adenocarcinoma, GCA). In addition to Hp infection, other known risk factors for GCA in Shanxi, China include family history of upper gastrointestinal (UGI) cancer, and some dietary exposures (including increased consumption of scalding hot foods and decreased consumption of fresh vegetables and fruits)5, 6. Tobacco smoking and alcohol consumption, which are important risk factors for esophageal and/or gastric cancer in western populations, have little or no relation to GCA in the population of Shanxi, China6.
Chronic infection by Hp is the major risk factor of gastric noncardia cancer as well as GCA in Eastern populations. However, although half of the world population is infected with Hp, only a small proportion (<3%) develops gastric cancer7. This has motivated studies to search for other potential risk factors including non-Hp gastric bacteria8, 9. The presence of non-Hp bacteria in the human stomach has been consistently indicated by both culture-based and culture-free studies (reviewed in9). The human stomach hosts a diverse and complex microbial community primarily composed of Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, and Fusobacteria. The role of the non-Hp gastric microbiota in human health is unclear. Recent studies in a transgenic mouse model suggested that the non-Hp gastric microbiota may play a role in gastric carcinogenesis, therefore potentially serving as biomarkers10, 11. However, studies of the diverse gastric microbiota in humans are scarce and limited. Little is known about its relationship with demographic or clinical features.
In this study, we analyzed the gastric microbiota of both non-malignant and tumor tissues from GCA patients in Shanxi, China, and examined its associations with epidemiologic and clinical characteristics in their paired non-malignant and tumor tissues.
Materials and Methods
Study subjects and sample collection
A total of 80 paired gastric tissue samples (non-malignant and paired tumor tissues) collected from GCA patients at Shanxi Cancer Hospital in Taiyuan, Shanxi Province, China between 1996 and 2001 were selected. Samples were selected based on DNA amount and quality appropriate for 16S rRNA gene amplification and sequence analysis. This study was approved by the Institutional Review Boards of the Shanxi Cancer Hospital and the National Cancer Institute (NCI) and all subjects provided written informed consent prior to participation. Cases were histologically confirmed as adenocarcinomas of the gastric cardia by pathologists at both the Shanxi Cancer Hospital and the NCI. The gastric tissues obtained during surgical resections were snap frozen in liquid nitrogen and stored at -130°C until used. At NCI, frozen tumor tissues were examined under the microscope following eosin staining to confirm that at least 50% of the cells were tumor before samples were selected for nucleic acid extraction. Frozen non-malignant tissues were similarly examined to confirm that they were free of tumor before selection for nucleic acid extraction. Total DNA was extracted using the AllPrep RNA/DNA/Protein Mini Kit from Qiagen. Epidemiologic data were collected via questionnaire, and clinical data were abstracted from medical records. No cases had prior therapy for cancer before their surgery.
Alcohol and pickled vegetable consumption were categorized based on intake frequencies in the questionnaire. Family history of UGI cancer refers to any UGI cancer in a first, second, or third degree relative. Tumor grade and stage were determined by pathology review as previouslyprevious described12. Tumor grade was categorized into Grade 1 (well differentiated), Grade 2 (moderately differentiated, Grade 3 (poorly differentiated), or Grade 4 (undifferentiated) based on primary tumor morphology. Tumor stage was categorized as I (invades mucosa or submucosa), II (invades mucularis mucosa), III (invades adventitia), or IV (invades adjacent structures) based on the extent of invasion of the primary tumor. Metastasis was determined by pathologic examination of lymph nodes removed during surgery (any positive nodes constituted metastasis) and radiographic. Survival was determined as days from gastric cancer surgery to death or the date of last contact.
16S rRNA gene amplification, sequencing and analysis
Sequence reads were processed to remove low quality, short, or chimera sequence reads in Quantitative Insights into Microbial Ecology (QIIME 1.8.0)13. The remaining reads with at least 97% identity were clustered into species-level Operational Taxonomy Units (OTUs) in QIIME. The command pick_open_reference_otus.py with the usearch61 clustering algorithm and other default settings were used to cluster sequences into OTUs. The OTUs were assigned to taxa (e.g. genus, family, phylum) based on Greengenes reference version 13_8 (ftp://greengenes.microbio.me/greengenes_release/gg_13_5/gg_13_8_otus.tar.gz). Each taxon’s proportion was its relative abundance. OTUs with only one read and samples with <1000 reads were excluded from the analysis. As a result of these quality filtering criteria, three non-malignant samples were excluded.
Alpha diversity measures the diversity of OTUs within a sample. The total number of OTUs, also known as richness, is a measurement of diversity that does not consider the relative abundance of the particular OTUs. Other measurements of alpha diversity, including Shannon’s Index14 and Phylogenetic Diversity (PD_whole_tree15), take the relative abundance of the particular OTUs into account. The PD_whole_tree measurements incorporate information about the phylogenetic relationship of OTUs. The phylogenetic tree of OTUs was prepared in QIIME based on the neighbor-joining method. Because the total number of OTUs depends on the depth (total number) of sequences, to compare alpha diversity across samples, we rarefied the OTU table to 1000 reads per sample (random sampling). Each alpha diversity measurement was the average over 20 such rarefied tables.
Beta diversity measures similarities between microbial communities of two samples. We estimated beta diversity with UniFrac, which measures the phylogenetic similarity of two communities based on the degree to which they share branch length on a bacterial tree of life16. The phylogenetic tree used for these measurements is the same as the tree used for PD_whole tree. Weighted UniFrac considers the taxa relative abundance whereas unweighted UniFrac does not.
For taxonomic analysis, OTUs were grouped into higher taxonomic levels including phylum, class, order, family, and genus. The taxa relative abundance was estimated as the proportion of OTUs assigned to a taxon. The taxa relative abundance was calculated based on the unrarefied OTU table.
Prediction of microbial function
We used Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt)1.0.017 to predict virtual metagenomes for each sample by using the 16S rRNA gene sequence data and a database of reference genomes, the Kyoto Encyclopedia of Genes and Genomes (KEGG) database18. Based on the predicted metagenomes, the relative abundance of KEGG genes, pathways, or modules within a sample was calculated.
Statistical methods
To examine the association of alpha diversity or taxa relative abundance with epidemiologic and clinical variables (except survival), we used Wilcoxon rank-sum tests for categorical epidemiologic/clinical variables (e.g. tumor grade), and Spearman correlations for continuous epidemiologic/clinical variables (e.g. age). For variables (including family history of UGI cancer, and tumor grade) associated with microbiota measurements at the P<0.05 level based on either test, we used multiple linear regression models and stratified analyses to further evaluate the associations by taking potential confounders into account. To assess the relationship of survival to microbiota alpha diversity or taxa relative abundance, Kaplan-Meier (KM) plots were used to visualize survival difference by high (above median) versus low (below median) microbiota measurements;, log-rank tests were used to test for differences.
At each taxonomical/functional level, multiple taxa (e.g., genera) or functions (e.g. KEGG modules) were examined for associations with epidemiologic/clinical variables. To increase statistical power by reducing the number of tests, we excluded taxa/functions (those present in 10% of the samples or less). P values were Bonferroni-corrected for multiple comparisons using the R command, P.adjust. Bonferroni correction was not applied to the association with Hp because it is the only species-level taxon we examined.
Beta diversity is a matrix describing samples’ pairwise difference in microbial communities. We used the non-parametric permutation multivariate analysis of variance (MANOVA) to examine its association with epidemiologic/clinical variables (Adonis method in R Package Vegan19).
Quality control
We included two blank samples as negative controls to assess potential contamination, plus one vaginal and one stool sample as positive controls to evaluate DNA amplification and sequencing performance. The two positive control samples generated 2703 and 58201 reads, respectively, suggesting good performance of the experiment. The negative control samples had extremely low number of reads (41 and 43 reads/sample, respectively). Furthermore, the OTUs found in both blanks were extremely rare in the gastric samples, with accumulated relative abundance ranging of 0 to 0.006. Therefore, our results were unlikely to have been affected adversely by contamination.
The conventional DNA extraction method for microbiome studies often includes bead-beating as an extra cell lysis step to break the hard-to-break cell membranes of some species. To examine whether we missed taxa due to DNA extraction protocol we employed without the bead-beating step, we evaluated two gastric tissue samples using the DNA extraction protocol with the beat-beating step commonly used for microbiome studies20 and the protocol used in the study. We found 13 genera discovered by the extraction protocol with a bead-beating step that were not discovered by our DNA extraction method. However, these taxa were rare with the cumulated relative abundance of 0.006 and 0.037 for the two samples, respectively. Therefore, the DNA extraction method should not have adversely affected our findings, although we cannot exclude missing some rare taxa.
Results
After excluding samples with <1,000 reads, 77 non-malignant and 80 tumor tissue samples were included for analysis (mean, 16,293 reads). Characteristics of the 77 patients are shown in Table 1. Most patients were male (83%) and smoked cigarettes (74%). Daily alcohol use (14%), daily pickle vegetable consumption (29%), and family history of UGI cancer (21%) were less common. Most patients had tumors that were Stage III (92%), Grade 3 (63%), and had metastasized (70%).
Table 1.
Summary of the study subjects’ characteristics (n=77)
| Characteristics | Median (interquartile range)/number (percentage) |
|---|---|
| Age (years) | 62 (58–67) |
| Sex (Male) | 64 (83%) |
| Cigarette smoking status (Yes) | 57 (74%) |
| Alcohol intake | |
| Seldom/never | 59 (77%) |
| Weekly | 7 (9%) |
| Daily | 11 (14%) |
| Family history of UGI cancer1 | 16 (21%) |
| Pickled vegetable intake | |
| Never/less than monthly | 40 (52%) |
| Weekly | 15 (19%) |
| Daily | 22 (29%) |
| Tumor Grade2 | |
| 2 | 29 (38%) |
| 3 | 48 (62%) |
| Tumor stage | |
| II | 4 (5%) |
| III | 71 (92%) |
| IV | 2 (3%) |
| Metastasis (Yes) | 54 (70%) |
| Survival (days)3 | 496 (213–1410) |
Any UGI cancer in first, second, or third degree relatives.
Grade 2 is moderately differentiated, Grade 3 is poorly differentiated.
Days from gastric cancer surgery to death or date of last contact.
Alpha diversity measurements in the non-malignant tissue microbiota were analyzed for associations with the epidemiologic and clinical variables in Table 1. Family history of UGI cancer and tumor grade were associated with all three alpha diversity measurements based on Wilcoxon rank-sum tests (P<0.05) (Figure 1, Supprting information Table 1). Compared to patients without family history of UGI cancer, patients with family history of UGI cancer had lower alpha diversity. Compared to the patients with lower tumor grade (Grade 2), patients with advanced tumor grade (Grade 3) had lower alpha diversity. These results were confirmed by linear regression models which regressed both family history of UGI cancer and tumor grade against alpha diversity measures, as well as in stratification tests (Supporting information Table 1). No association was found between alpha diversity measurements and the other variables in Table 1.
Figure 1.
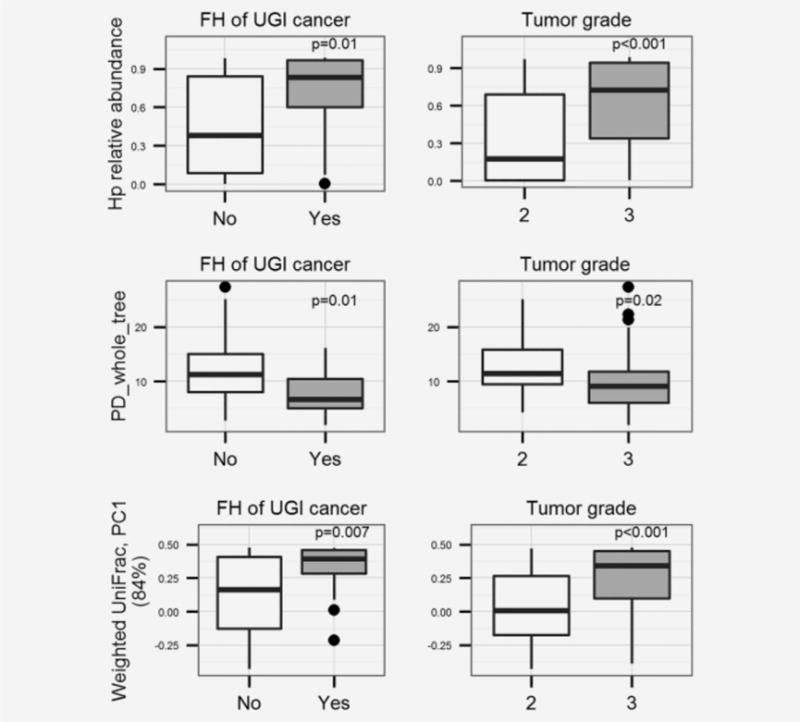
Non-malignant gastric tissue microbiota features associated with family history (FH) of UGI cancer and tumor grade, P values were based on Wilcoxon rank-sum tests. (a, b, c), comparison of Hp relative abundance, PD_whole_tree (an alpha diversity measurement) and first principal component (PC1, explain 84% variance) of unweighted UniFrac (a beta diversity measurement) by family history of UGI cancer. (d, e, f), comparison of Hp relative abundance, PD_whole_tree, and PC1 of unweighted UniFrac by tumor grade. The Boxes are interquartile range (IQR); median values are bands within the boxes; the lines outside the boxes are 1.5-times IQR. The dots are outliers.
Beta diversity measurements in the non-malignant tissue microbiota were also analyzed for association with epidemiologic/clinical variables. Multivariate MANOVA including both family history of UGI cancer and tumor grade as independent variables was applied. According to the weighted UniFrac distance, non-malignant tissue microbiota differed by tumor grade (P=0.002), and was marginally differerent by family history of UGI cancer (P=0.08). No associations were found with unweighted UniFrac distances, or between weighted/unweighted UniFrac and the other variables in Table 1.
Relative abundances of Hp and of all other taxa from genus to phylum levels in non-malignant tissue were analyzed for associations with variables in Table 1. Family history of UGI cancer, tumor grade, and metastasis were all associated with certain taxa based on Wilcoxon rank sum tests. Patients with family history of UGI cancer had higher Hp relative abundance than patients without family history of UGI cancer. Patients with lower tumor grade had lower Hp and higher Bacteroidetes relative abundance than patients with advanced tumor grade. These associations were confirmed in both multiple linear regression models and stratification tests (Supprting Information Table 1). Hp’s higher level taxa, including genus Helicobacter, family Helicobacteraceae, order Campylobacterales, and class Epsilonproteobacteria, were also associated with tumor grade (data not shown). In addition, metastasis status was also found to be associated with Lactobacillales relative abundance in non-malignant tissue microbiota (without vs. with metastases (median and interquartile): 0.05(0.01–0.08) vs. 0.01(0.00–0.04), Bonferroni-corrected P=0.04). No taxon was associated with the other variables in Table 1 after Bonferroni correction.
The predicted KEGG pathways and modules in nonmalignant tissue microbiota were analyzed for associations with variables in Table 1 and the results are shown in Table 2. After Bonferroni correction, tumor grades differed in relative abundance of five KEGG modules, and marginally differed in six KEGG modules. No association was found with the other KEGG modules or pathways or other variables in Table 1.
Table 2.
KEGG modules whose relative abundance differed by tumor grade
| KEGG modules | Grade 2 (moderately differentiated) |
Grade 3 (poorly differentiated) |
P (Bonferroni-adjusted) |
|---|---|---|---|
| P<0.05 | |||
|
| |||
| Carbohydrate Metabolism | 0.158(0.142–0.166) | 0.142(0.137–0.153) | 0.02 |
| Metabolism | 0.042(0.033–0.048) | 0.034(0.032–0.039) | 0.04 |
| Transcription | 0.037(0.025–0.041) | 0.025(0.022–0.034) | 0.04 |
| Environmental Adaptation | 0.003(0.002–0.004) | 0.004(0.003–0.004) | 0.04 |
| Circulatory System | 0.001(0.000–0.003) | 0.002(0.001–0.003) | 0.05 |
|
| |||
| 0.05<P<0.1 | |||
|
| |||
| Enzyme Families | 0.032(0.029–0.033) | 0.029(0.028–0.031) | 0.08 |
| Digestive System | 0.001(0.000–0.001) | 0.000(0.000–0.001) | 0.06 |
| Folding Sorting and Degradation | 0.045(0.042–0.056) | 0.055(0.048–0.058) | 0.07 |
| Energy Metabolism | 0.098(0.089–0.116) | 0.116(0.101–0.121) | 0.08 |
| Cell Motility | 0.056(0.043–0.084) | 0.087(0.065–0.097) | 0.08 |
| Genetic Information Processing | 0.051(0.047–0.067) | 0.066(0.055–0.071) | 0.06 |
P-values were based on Wilcoxon rank-sum test with Bonferroni adjustment
Tumor tissue microbiota features, including alpha and beta diversity, and taxa relative abundance, were also examined for associations with the variables listed in Table 1, but no associations were found (data not shown).
Discussion
This study showed that gastric non-malignant tissue microbiota features were associated with a known gastric cancer risk factor (family history of UGI cancer) and clinical features (tumor grade and metastasis) in the GCA patients from Shanxi, China. These results suggested a potential role of gastric microbial communities in gastric cardia carcinogenesis and cancer progression in the population of Shanxi, China. To our knowledge, this is the first report documenting epidemiologic and clinical relevance for gastric microbiota in humans.
Gastric cancer is classified into two types based on the anatomic location within the stomach where the tumor develops: gastric cardia and noncardia cancer. The main gastric cancer type and risk factors vary by population. Shanxi, China has among the highest rates of gastric cancer, mainly GCA, in China. Other than Hp infection, the major known risk factor for GCA in this population is a family history of UGI cancer5, 6. In a previously-conducted case-control study in this same population, having a first degree relative with a family history of UGI cancer was associated with a 1.62-fold increased risk of GCA, while having two or more first degree relatives elevated GCA risk by 5.35-fold5. Beyond this study population, family history of UGI cancer has been consistently reported as a risk factor for gastric cancer including both GCA and gastric noncardia cancer in other ethnic populations and geographic regions. In the present study, we showed that family history of UGI cancer was also associated with features of the gastric microbiota. Interestingly, Hp, a well-known pathogen for gastritis, gastric ulcer and cancer, was the only taxon that differed between subjects by family history of UGI cancer9. A recent study also suggested a potential link between the gastric microbiota and gastric cancer risk8. With 20 samples from each of two Colombia populations with similar Hp prevalence but distinct gastric cancer risk (a 25-fold difference), the study showed that gastric microbiota composition differed between the populations. These results suggested that surveys of the gastric microbiota are needed to understand gastric cancer pathogenesis.
Mechanisms underlying the link between gastric microbiota and family history of UGI cancer likely include shared genetic susceptibility and shared environment exposures (e.g. diet) among family members21. Host genetic factors, environmental exposures, and diet may play important roles in determining gastric microbiota composition22, 23. Patients with a family history of UGI cancer may have increased risk of GCA that is due, at least in part, to certain gastric microbiota features determined by genetic factors, environmental exposures, and/or diet. However, our study had a limited number of samples and was a case-only study so we were unable to examine associations between microbiota and GCA risk factors such as environmental exposures or diet. Large studies with non-cancer control populations for comparison are needed to evaluate such risk factors in relation to incident gastric cancer.
Typically, gastric cancer patients are thought to have gastric hypochlorhydria (pH>4) with consequent loss or burn out of Hp due to long term atrophic gastritis with loss of specialized glandular tissue and decreased acid secretion. This expectation is contradictory to our finding that Hp relative abundance was higher in cases with more advanced disease (i.e., higher tumor grade). Consistent with our findings, a recent study of 212 patients with chronic gastritis and 103 GCA cases in China evaluated using quantitative PCR showed that bacterial load in the gastric mucosa was higher in cancer cases than in patients with gastritis, and that the bacterial load correlated positively with the absolute quantity of Hp (R=0.38, P<0.001)24. This observation is consistent with a potential role for Hp in tumor progression. In addition to differences in Hp relative abundance, several predicted KEGG modules differed in relative abundance between patients by tumor grade. This suggested that change of Hp relative abundance may be responsible for the change in microbial function (e.g. reduced carbohydrate metabolism, increased environmental adaptation, or increased circulatory system function), which, in turn, may have favored tumor progression. In addition to tumor grade, the present study identified a link between gastric microbiota and metastasis. Together, the results suggest that gastric microbiota may play a role in GCA tumor progression, or that tumor progression may affect the microenvironment and microbiota of the surrounding area. Further studies are needed evaluate this issue.
Increased Bacteroidetes in our study was associated with lower tumor grade. Bacteroidetes the third most abundant phylum in the gastric microbiota, following Proteobacteria and Firmicutes. The most abundant Bacterioidetes genus in our samples was Prevotella, which is commonly detected in stomach and oral samples25, 26. Our results suggest that Bacteroidetes might be involved in protection against GCA tumor progression. Further studies are needed to investigate the potential role of non-Hp bacteria in tumor progression.
Another noteworthy taxon identified in the gastric microbiota here is Lactobacillales, which was negatively associated with metastasis. Lactobacillales bacteria are characterized by the formation of lactic acid as the sole or main end product of carbohydrate metabolism. They are commonly found in the human oral cavities, gastrointestinal tracts and vaginas26, as well as in decomposing plants or milk products. Lactobacillales is considered as a composition of healthy microbiota26 and may confer a health benefit on the host27–29. One of its genus lactobacillus is commonly used as probiotics. Here we showed that Lactobacillales might warrant further investigation regarding a possible involvement in protection against metastasis.
Increased function in carbohydrate metabolism, metabolism and transcription modules and decreased function in environmental adaptation and circulatory system were all associated with lower tumor grade. It would be interesting to investigate whether changes of these function could affect tumor progression by influencing production of certain products like short chain fat acid, which in turn might affect tumor progression through process like inflammation. However, we note that our functional results were based on prediction and, therefore, need to be interpreted with caution. The differences in function associated with tumor grade are mainly due to the difference in Hp relative abundance. The functional prediction may bias towards known genomes (e. g. Hp) and against unknown genomes. Studies with metagenomics sequences are needed to validate our findings.
Microbial communities have shown differences between tumor and matched non-malignant tissues in studies of other cancers such as colorectal and lung cancer patients30, 31. Consistent with this observation, we also showed that microbial profiles differed between gastric tumor and non-malignant tissues (manuscript submitted), and that the associations we observed between gastric microbiota features and GCA risk factors and clinical features in non-malignant gastric tissues were not also seen in tumor tissues.
To our knowledge, this is the first report to examine potential links between gastric microbiota and known GCA risk factors and clinical features. Our findings are strengthened by the fact that we used uniform procedures under sterile surgical conditions to obtain the tissue samples studied here, which minimized the risk of sample contamination at collection. Our study has limitations. As with all microbiota studies performed using 16S rRNA gene amplification and sequencing, we do not know whether the taxa we detected are actual members of the gastric microbiota, or if they are transient and/or dead bacteria. Other limitations include our limited sample size and the use of a DNA extraction protocol without a bead-beating step. In addition, we studied only gastric cardia cases, and we lacked healthy controls for comparison. Therefore, our results need to be confirmed, as they may not apply to healthy individuals, other populations with low Hp prevalence (e.g. Western populations), or cases of gastric cancer originating in the noncardia regions of the stomach.
In conclusion, we showed that features of the gastric microbiota are associated with a known risk factor and clinical features in GCA patients from a high-risk population in China. Additional studies with both healthy controls and gastric cancers of the cardia and noncardia from different populations are needed to further examine the association between gastric cancer and the microbiome.
Supplementary Material
Supplementary Table 1, Association of family history of UGI cancer, tumor grade, and non-malignant gastric tissue microbiota features by Wilcoxon rank-sum tests, multiple linear regression, and stratification analyses.
Novelty and Impact.
To our knowledge, this is the first study to link gastric microbiota to known cancer risk factors and clinical outcomes in gastric cardia cancer patients from China. This study shows that H. pylori relative abundance and microbiota diversity differ between patients with and without a family history of upper gastrointestinal tract cancer, a major risk factor. This study also shows associations between tumor grade, metastasis, and gastric microbiota features.
Abbreviations
- FH
family history
- UGI
upper gastrointestinal tract
- Hp
Helicobacter pylori
- GCA
gastric cardia adenocarcinoma
- KEGG
Kyoto encyclopedia of genes and genomes
- PD_whole_tree
Phylogenetic diversity
- MANOVA
Multivariate analysis of variance
- rRNA
ribosomal RNA
- PICRUSt
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States
References
- 1.Ferlay JSI, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11. Vol. 2013. Lyon, France: International Agency for Research on Cancer; 2013. [Google Scholar]
- 2.Carneiro F. Stomach cancer. In: Steward B, Wild CP, editors. World Cancer Report. Lyon: International Agency for Research on Cancer; 2014. pp. 383–91. [Google Scholar]
- 3.Cancer IAfRo. Infection with Helicobacter pylori. IARC Monogr Eval Carcinog Risks Hum. 1994;61:177–240. [PMC free article] [PubMed] [Google Scholar]
- 4.Cavaleiro-Pinto M, Peleteiro B, Lunet N, Barros H. Helicobacter pylori infection and gastric cardia cancer: systematic review and meta-analysis. Cancer causes & control: CCC. 2011;22:375–87. doi: 10.1007/s10552-010-9707-2. [DOI] [PubMed] [Google Scholar]
- 5.Gao Y, Hu N, Han X, Giffen C, Ding T, Goldstein A, Taylor P. Family history of cancer and risk for esophageal and gastric cancer in Shanxi, China. BMC Cancer. 2009;9:269. doi: 10.1186/1471-2407-9-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao Y, Hu N, Han XY, Ding T, Giffen C, Goldstein AM, Taylor PR. Risk factors for esophageal and gastric cancers in Shanxi Province, China: a case-control study. Cancer Epidemiol. 2011;35:e91–9. doi: 10.1016/j.canep.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wroblewski LE, Peek RM, Wilson KT. Helicobacter pylori and Gastric Cancer: Factors That Modulate Disease Risk. Clin Microbiol Rev. 2010;23:713–39. doi: 10.1128/CMR.00011-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang I, Woltemate S, Piazuelo MB, Bravo LE, Yepez MC, Romero-Gallo J, Delgado AG, Wilson KT, Peek RM, Correa P, Josenhans C, Fox JG, et al. Different gastric microbiota compositions in two human populations with high and low gastric cancer risk in Colombia. Sci Rep. 2016;6 doi: 10.1038/srep18594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheh A, Fox JG. The role of the gastrointestinal microbiome in Helicobacter pylori pathogenesis. Gut Microbes. 2013;4:505–31. doi: 10.4161/gmic.26205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lofgren JL, Whary MT, Ge ZM, Muthupalani S, Taylor NS, Mobley M, Potter A, Varro A, Eibach D, Suerbaum S, Wang TC, Fox JG. Lack of Commensal Flora in Helicobacter pylori-Infected INS-GAS Mice Reduces Gastritis and Delays Intraepithelial Neoplasia. Gastroenterology. 2011;140:210. doi: 10.1053/j.gastro.2010.09.048. + [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lertpiriyapong K, Whary MT, Muthupalani S, Lofgren JL, Gamazon ER, Feng Y, Ge ZM, Wang TC, Fox JG. Gastric colonisation with a restricted commensal microbiota replicates the promotion of neoplastic lesions by diverse intestinal microbiota in the Helicobacter pylori INS-GAS mouse model of gastric carcinogenesis. Gut. 2014;63:54–63. doi: 10.1136/gutjnl-2013-305178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang G, Hu N, Yang HH, Wang L, Su H, Wang C, Clifford R, Dawsey EM, Li JM, Ding T, Han XY, Giffen C, et al. Comparison of global gene expression of gastric cardia and noncardia cancers from a high-risk population in china. PLoS One. 2013;8:e63826. doi: 10.1371/journal.pone.0063826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu G, Fadrosh D, Goedert JJ, Ravel J, Goldstein AM. Nested PCR Biases in Interpreting Microbial Community Structure in 16S rRNA Gene Sequence Datasets. PloS one. 2015;10:e0132253. doi: 10.1371/journal.pone.0132253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shannon CE. The mathematical theory of communication. 1963. MD Comput. 1997;14:306–17. [PubMed] [Google Scholar]
- 15.Faith DP, Baker AM. Phylogenetic diversity (PD) and biodiversity conservation: some bioinformatics challenges. Evol Bioinform. 2006;2:121–8. [PMC free article] [PubMed] [Google Scholar]
- 16.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2011;5:169–72. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Thurber RLV, Knight R, Beiko RG, Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–21. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dixon P. VEGAN, a package of R functions for community ecology. Journal of Vegetation Science. 2003;14:927–30. [Google Scholar]
- 20.Flores R, Shi J, Fuhrman B, Xu X, Veenstra TD, Gail MH, Gajer P, Ravel J, Goedert JJ. Fecal microbial determinants of fecal and systemic estrogens and estrogen metabolites: a cross-sectional study. J Transl Med. 2012;10:253. doi: 10.1186/1479-5876-10-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yaghoobi M, Bijarchi R, Narod SA. Family history and the risk of gastric cancer. British journal of cancer. 2010;102:237–42. doi: 10.1038/sj.bjc.6605380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pena A. Genetic factors determining the host response to Helicobacter pylori. World journal of gastroenterology: WJG. 2000;6:624–5. doi: 10.3748/wjg.v6.i5.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. Diet-induced extinctions in the gut microbiota compound over generations. Nature. 2016;529:212–U08. doi: 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang LL, Zhou JH, Xin YN, Geng CX, Tian ZB, Yu XJ, Dong QJ. Bacterial overgrowth and diversification of microbiota in gastric cancer. Eur J Gastroenterol Hepatol. 2016;28:261–6. doi: 10.1097/MEG.0000000000000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fox J, Sheh A. The role of the gastrointestinal microbiome in Helicobacter pylori pathogenesis. Gut Microbes. 2013;4 doi: 10.4161/gmic.26205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.HMP C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanders ME, Guarner F, Guerrant R, Holt PR, Quigley EMM, Sartor RB, Sherman PM, Mayer EA. An update on the use and investigation of probiotics in health and disease. Gut. 2013;62:787–96. doi: 10.1136/gutjnl-2012-302504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Kasper LH. The role of microbiome in central nervous system disorders. Brain Behavior and Immunity. 2014;38:1–12. doi: 10.1016/j.bbi.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajkumar H, Kumar M, Das N, Kumar SN, Challa HR, Nagpal R. Effect of Probiotic Lactobacillus salivarius UBL S22 and Prebiotic Fructo-oligosaccharide on Serum Lipids, Inflammatory Markers, Insulin Sensitivity, and Gut Bacteria in Healthy Young Volunteers: A Randomized Controlled Single-Blind Pilot Study. J Cardiovasc Pharmacol Ther. 2015;20:289–98. doi: 10.1177/1074248414555004. [DOI] [PubMed] [Google Scholar]
- 30.Burns MB, Lynch J, Starr TK, Knights D, Blekhman R. Virulence genes are a signature of the microbiome in the colorectal tumor microenvironment. Genome Med. 2015;7:55. doi: 10.1186/s13073-015-0177-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu GQ, Gail MH, Consonni D, Carugno M, Humphrys M, Pesatori AC, Caporaso NE, Goedert JJ, Ravel J, Landi MT. Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol. 2016;17 doi: 10.1186/s13059-016-1021-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1, Association of family history of UGI cancer, tumor grade, and non-malignant gastric tissue microbiota features by Wilcoxon rank-sum tests, multiple linear regression, and stratification analyses.


