Abstract
Langerhans cell histiocytosis (LCH) is characterized by a localized or systemic proliferation of Langerhans cells. BRAF mutations have been reported in 40–70% of cases and MAP2K1 mutations have been found in BRAF-negative cases, supporting the interpretation that LCH is a true neoplasm, at least in mutated cases. In a small subset of patients, LCH is detected incidentally in a biopsy specimen involved by lymphoma. These lesions are usually minute and rarely have been assessed for mutations. We assessed for BRAF and MAP2K1 mutations in 7 cases of LCH detected incidentally in biopsy specimens involved by lymphoma. We also performed immunohistochemical analysis for phosphorylated (p)-ERK. The study group included 4 men and 3 women (median age 54 years; range, 28–84). The biopsy specimens were lymph nodes (n=6) and chest wall (n=1). The lymphomas included 5 cases of classical Hodgkin lymphoma, 1 mantle cell lymphoma and 1 angioimmunoblastic T-cell lymphoma. All cases were negative for BRAF mutation by pyrosequencing, and MAP2K1 mutation by Sanger sequencing. Nevertheless, 3 of 7 (42.8%) cases were positive for p-ERK. We then performed mutation analysis of a panel of 134 commonly mutated genes (including BRAF and MAP2K1) by next-generation sequencing on 3 cases including the 2 cases positive for p-ERK by immunohistochemistry. No mutations were detected in any of the three cases assessed. Six patients were treated for lymphoma with chemotherapy; 2 also received radiotherapy, and 2 underwent stem cell transplantation. With a median follow-up of 21 months (range, 6–89), no patients developed recurrent LCH. We conclude that lymphoma-associated LCH is a clinically benign process that is not associated with BRAF or MAP2K1 mutations. Nevertheless, the expression of p-ERK in 2 cases suggests that the RAS-BRAF-MAP2K1-ERK pathway may be activated by non-mutational mechanisms, perhaps induced by the presence of lymphoma or lymphoma-microenvironment interaction.
Keywords: Langerhans cell histiocytosis, concurrent lymphoma, BRAF mutation, MAP2K1 mutation, p-ERK
Introduction
Langerhans cell histiocytosis (LCH) is a rare disease characterized histologically by a proliferation of Langerhans cells (LCs) admixed with eosinophils, lymphocytes, macrophages and multinucleated giant cells, with or without eosinophilic abscesses or necrosis.1 Virtually any anatomic site can be involved by LCH, and the disease may present as a localized lesion or as multiorgan disease. Prognosis is variable, correlating in part with extent of disease, but morphologic findings of LCH are identical in the localized or multifocal disease and are not an indicator of prognosis. Because of its heterogeneous clinical manifestations and benign morphology, the reactive or neoplastic nature of LCH was controversial for over 50 years.2 However, in 1994 two independent groups showed that a subset of LCH cases is monoclonal by using a human androgen receptor X-chromosome-inactivation assay (HUMARA).3,4
In 2010, Badalian-Very et al. described BRAF V600E mutation in 40–70% of cases of LCH5 supporting the interpretation that LCH is a neoplasm, at least in mutated cases. Similar findings also were reported by others.5–9 BRAF is a member of the rapid accelerating fibrosarcoma (RAF) kinase family that is activated by RAS and RAS-coupled receptor tyrosine kinases. RAS-RAF activated complexes transmit downstream signals to the mitogen activated protein kinase (MAPK) cascade including MEK/ERK kinases via protein phosphorylation.10–12 The RAS-RAF-MAPK pathway is a key regulator involved in cell proliferation, growth, differentiation and apoptosis by transmitting activating signals to several nuclear, cytoplasmic and cell membrane targets.10,11 Subsequently, a subset of BRAF wild-type LCH cases (30–50%) were shown to carry mutations in the MAPK-kinase 1 (MAP2K1 or MEK1) gene, further implicating the oncogenic MAPK pathway signaling in LCH pathogenesis.13,14 Most of these mutations cause constitutive activation of MAP2K1 and activation of the RAS-RAF-MAPK signaling cascade via ERK phosphorylation.13,14 BRAF V600E and MAP2K1 mutations are not exclusively seen in LCH and also have been reported with variable frequency in other hematopoietic neoplasms, for example, hairy cell leukemia as well as non-hematopoietic malignancies including melanoma, papillary thyroid carcinoma, colorectal carcinoma, and glioneuronal tumors.10–12,15–22
In a small subset of patients, LCH has been identified as an incidental finding in biopsy specimens involved by lymphoma. Classical Hodgkin lymphoma is the most common associated lymphoma,23–26 whereas only sporadic cases of other types of non-Hodgkin lymphoma associated with LCH are reported in the literature.23,25,27–30 The relationship between LCH and concurrent lymphoma remains unknown. To our knowledge, there are no previous studies that have assessed the status of BRAF or MAP2K1 in cases of LCH associated with concurrent lymphomas.
In this study, we identified 7 patients with concurrent LCH and lymphoma and assessed the LCH component for BRAF and MAP2K1 mutations.
Materials and Methods
Study group and immunohistochemistry
The archives of the Department of Hematopathology at The University of Texas MD Anderson Cancer Center from January 2000 to December 2015 were searched for cases diagnosed as LCH associated with lymphoma involving the same biopsy specimen. Cases with available paraffin-embedded tissue blocks or unstained slides were selected. Clinical and laboratory data were retrieved from the electronic medical record. This study was conducted under an Institutional Review Board-approved protocol.
Routinely prepared hematoxylin-eosin stained slides for all cases were reviewed. Immunohistochemical studies were performed to confirm the diagnosis of lymphoma and LCH using antibodies specific for CD1a and CD21 (Leica Biosystem, Newcastle, UK); CD2, CD3, CD4, CD7, CD8, CD20, CD30, CD45/LCA, BCL-6, MUM1 and TIA-1 (DAKO, Carpinteria, CA); CD5 and cyclin D1 (Labvision/Neomarkers, Fremont, CA); CD10, CD23 and BCL-2 (Novocastra/Vision Biosystem, Benton Lane, Newcastle-upon-Tyne, UK); CD15 (Becton-Dickinson Biosciences, San Jose, CA); PAX-5 (Transduction Labs, San Diego, CA); CXCL-13 (R&D Systems, Minneapolis, MN); S-100 protein (BioGenex, Fremont, CA) and Langerin/CD207 (Novocastra Biosystem, Newcastle, UK). In situ hybridization for Epstein-Barr virus-encoded RNA (EBER) was also performed.
ERK phosphorylation analysis by immunohistochemistry
The highly specific antibody phospho-p44/42 MAPK (Thr202/Tyr204) (D13.14.4E) p-ERK (dilution 1:300, Cell Signaling, Danvers, MA) was used to assess for the presence of nuclear and cytoplasmic phosphorylated p44 and p42 MAPK (Erk1 and Erk2).
BRAF mutation analysis by immunohistochemistry
The VE-1 antibody (dilution 1:50, Spring Bioscience, Pleasanton, CA) was used to assess for cytoplasmic staining supportive of the presence of BRAF V600E mutation. The antibody is highly specific for this mutation as shown previously by Capper et al.31
BRAF mutation analysis by pyrosequencing
A polymerase chain reaction (PCR)-based pyrosequencing assay for BRAF mutation analysis was employed. This assay was developed at our institution and covers mutation hotspots in exons 11 (codon 468) and 15 (codons 595 to 600).32 This assay was chosen because all BRAF mutations in LCH identified previously have been clustered in exons 11 or 15 of the gene and alter the kinase domain of the protein.33 The LCH lesion was microdissected from fixed, paraffin-embedded tissue sections (10 μm thick) and DNA was extracted and PCR amplified using either a forward primer, 5′-TCCTGTATCCCTCTCAGGCATAAGGTAA-3′, and a reverse biotinylated primer, 5′-biotin-CGAACAGTGAATATTCCTTTGAT-3′ (for codon 468 of exon 11), or a forward primer, 5′-CATAATGCTTGCTCTGATAGGA-3′, and a reverse biotinylated primer, 5′-biotin-GGCCAAAAATTTAATCAGTGGA-3′ (for codons 595 to 600 of exon 15). PCR amplification was performed in duplicate on an ABI 2720 Thermocycler (Applied Biosystems, Grand Island, NY). The PCR products underwent electrophoresis on agarose gels to confirm successful amplification. Fifteen (15) μL of the PCR products were then sequenced in duplicate using primers 5′-TTGGATCTGGATCATTT-3′ (for exon 11) or 5′-GAAGACCTCACAGTAAAAATA-3′ (for exon 15) and the pyrosequencing PSQ96 HS System (Biotage AB, Uppsala, Sweden) according to the manufacturer’s instructions. The HL-60 cell line, negative for BRAF mutation, and A375, a melanoma cell line positive for BRAF mutation (exon 15 V600E), were used as negative and positive controls, respectively.
MAP2K1 mutation analysis
We performed Sanger sequencing to assess for mutations in exons 2 and 3 of MAP2K1. DNA was subjected to PCR using a pair of M13-tagged forward primer, 5′-TGTAAAACGACGGCC AGTAGTATTGACTTGTGCTCCCCA-3′, and reverse primer, 5′-CAGGAAACAGCTATGACCTGGTCCCCAGGCTTCTAAGT-3′, for exon 2, and a pair of M13-tagged forward primer, 5′-TGTAAAACGACGGCCAGTCATAAAACCTCTCTTTCTTCCACC-3′, and reverse primer, 5′-CAG GAA ACA GCT ATG ACC CCA GAG CCA CCC AAC TCT TA-3′, for exon 3, in a 50 μL reaction containing 10 ng DNA, 0.03 U/μL Go Taq polymerase (Promega, Madison, WI), 0.2 μM each of forward and reverse primers, 2 mM MgCl2, 1 mM dNTP mix and 1x Go Taq buffer. The reaction mix was first heated to 95°C for 10 min, then subjected to 40 cycles of 95°C for 30 sec, 60°C for 30 sec, and 72°C for 30 sec, followed by 72°C for 7 min. The PCR products underwent electrophoresis to confirm successful amplification, were purified using Agencourt AMPure Kit (Beckman Coulter, Indianapolis, IN), and sequenced using the same primer sets as shown above.
Next-generation sequencing (NGS)
We performed amplicon-based NGS targeting the coding regions of a panel of 134 genes that are commonly mutated in hematopoietic neoplasms using Torrent Suite platform (Thermo Fisher Scientific, Waltham, MA) on DNA extracted from paraffin-embedded tissues. For 2 cases, we were able to retrieve tissues that were not involved by LCH to be used as a control. We used 250 ng of DNA to prepare the genomic library. The genes included in the panel are as follows: ABL1, ACVRL1, AKT1, ALK, APC, APEX1, AR, ARAF, ATM, ATP11B, BAP1, BCL2L1, BCL9, BIRC2, BIRC3, BRAF, BRCA1, BRCA2, BTK, CBL, CCND1, CCNE1, CD274, CD44, CDH1, CDK4, CDK6, CDKN2A, CHEK2, CSF1R, CSNK2A1, CTNNB1, DCUN1D1, DDR2, DNMT3A, EGFR, ERBB2, ERBB3, ERBB4, ESR1, EZH2, FBXW7, FGFR1, FGFR2, FGFR3, FGFR4, FLT3, FOXL2, GAS6, GATA2, GATA3, GNA11, GNAQ, GNAS, HNF1A, HRAS, IDH1, IDH2, IFITM1, IFITM3, IGF1R, IL6, JAK1, JAK2, JAK3, KDR, KIT, KNSTRN, KRAS, MAGOH, MAP2K1, MAP2K2, MAPK1, MAX, MCL1, MDM2, MDM4, MED12, MET, MLH1, MPL, MSH2, MTOR, MYC, MYCL, MYCN, MYD88, MYO18A, NF1, NF2, NFE2L2, NKX2-1, NKX2-8, NOTCH1, NPM1, NRAS, PAX5, PDCDILG2, PDGFRA, PIK3CA, PIK3R1, PNP, PPARG, PPP2R1A, PTCH1, PTEN, PTPN11, RAC1, RAF1, RB1, RET, RHEB, RHOA, RPS6KB1, SF3B1, SMAD4, SMARCB1, SMO, SOX2, SPOP, SRC, STAT3, STK11, TERT, TET2, TIAF1, TP53, TSC1, TSC2, U2AF1, VHL, WT1, XOP1, and ZNF217. Following successful library generation and purification, DNA was used for multiplex paired-end sequencing on the Ion Proton platform, and analyzed using the Torrent Suite and OncoSeek data pipeline.
Results
Clinical features
The study group included 7 patients, 4 men and 3 women, with a median age of 54 years (range, 28–84 years). The demographic, clinical, laboratory features at diagnosis, and pathology and molecular results are summarized in Table 1. One patient presented with mild leukocytosis (median white blood cell count, 5.6 ×103/μl; range, 4–13.2 ×103/μl; reference range, 4–11 ×103/μl); 3 patients had normocytic anemia (median hemoglobin, 13.3 g/dL; range, 9.8–16.2 g/dL; reference range, 14–18 g/dL for men and 12–16 g/dL for women), and 1 patient had thrombocytopenia (median platelet count, 158 ×103/μl; range, 41–368 ×103/μl; reference range, 140–440 ×103/μl). Serum lactate dehydrogenase was slightly elevated in 1 patient (median, 434 IU/L; range, 390–668 IU/L; reference range, 313–618 IU/L) and β2-microglobulin levels were elevated in 5 patients (median, 2.3 mg/L; range, 1.8–4.3 mg/L; reference range, 0.7–1.8 mg/L). Laboratory data were not available for 1 patient. The lymphomas include 5 cases of classical Hodgkin lymphoma and 1 case each of mantle cell lymphoma and angioimmunoblastic T-cell lymphoma. The cases of Hodgkin lymphoma were further classified as 3 nodular sclerosis, 1 mixed cellularity, and 1 recurrent disease. Concomitant LCH and lymphoma occurred in lymph nodes in 6 patients: 3 cervical, 1 axillary, 1 retroperitoneal and 1 inguinal. In one patient a biopsy was obtained from the chest wall.
TABLE 1.
Demographic, clinicopathologic and molecular features, treatment and outcome in patients with LCH and lymphomas
| Case | Age | Sex | WBC (x 103/μ l) |
Hgb (g/d L) |
Plt (x 103/ μl) |
LDH (IU/L ) |
B2M (mg/ L) |
Anatomic location |
Lymphoma subtype |
LCH distribution |
LCH component IHC |
pERK IHC |
BRAF V600E IHC |
BRAF V600E PCR |
MAP2K1 PCR |
Treatment | Follow up (months) |
Outcome |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 68 | F | N/A | N/A | N/A | N/A | N/A | Lymph node, cervical | AITL | Multiple small foci | CD1a + langerin + S100 + | Pos | Neg | Neg | Neg | N/A | N/A | N/A |
| 2 | 84 | F | 1 0.5 | 13.2 | 368 | 397 | 2 | Lymph node, cervical | CHL, mixed cellularity | Multiple small foci and scattered LCs | CD1a + langerin + S100 + | Neg | Neg | Neg | Neg | AVD XRT |
10 | CR |
| 3 | 28 | M | 6.6 | 16.2 | 154 | 465 | 2.8 | Chest wall | CHL, nodular sclerosis | Few small foci | CD1a + langerin + S100 + | Neg | Neg | Neg | Neg | ABVD | 67 | CR |
| 4 | 37 | M | 4.0 | 9.8 | 41 | 668 | 2.4 | Lymph node, axillary | CHL, nodular sclerosis | Multiple small foci, 2 large focus | CD1a + langerin + S100 + | Pos in subcap sular foci; neg in largest focus | Neg | Neg | Neg | ABVD, VDTPACE, IGEV, thalidomide, revlimid, XRT, auto-SCT | 21 | DOD (CHL) |
| 5 | 39 | F | 5.3 | 15.2 | 162 | 586 | 2.1 | Lymph node, cervical | CHL, nodular sclerosis | Multifocal, large clusters of LCs | CD1a + langerin + S100 + | Neg | Pos | Neg | Neg | ABVD, AVD, ICE 1st cycle, will be followed by SCT | 6 | Persistent disease in the mediastinum detected by imaging |
| 6 | 54 | M | 5.8 | 12.1 | 141 | 402 | 4.3 | Lymph node, retroperitone al | CHL, recurrent | Small cluster with central necrosis | CD1a + langerin + S100 + | Pos | Neg | Neg | Neg | ABVD, ICE, auto-SCT for CHL Azacitidine, decitabine, MUD-SCT for MDS |
21 | Therapy-relatedMDS |
| 7 | 61 | M | 13.2 | 13.4 | 344 | 390 | 1.8 | Lymph node, inguinal | MCL | Small cluster | CD1a + langerin + S100 + | Neg | Neg | Neg | Neg | Rituximab, hyper CVAD, methotrexate, cytarabine | 89 | CR |
Abbreviations: LCH, Langerhans cell histiocytosis; WBC, white blood cells; Hgb, hemoglobin; Plt, platelets; LDH, lactate dehydrogenase; B2M, β-2 microglobulin; IHC, immunohistochemistry; pERK, phosphorylated ERK; PCR, polymerase chain reaction; F, female; M, male; N/A, not available; AITL, angioimmunoblastic T-cell lymphoma; CHL, classical Hodgkin lymphoma; MCL, mantle cell lymphoma; LCs, Langerhans cells; Pos, positive; Neg, negative; AVD, doxorubicin, vinblastine and dacarbazine; XRT, radiotherapy; ABVD, doxorubicin, bleomycin, vinblastine and dacarbazine; VDTPACE, dexamethasone, thalidomide, cisplatin, doxorubicin, cyclophosphamide and etoposide; IGEV, ifosfamide, gemcitabine and vinorelbine; auto-SCT, autologous stem cell transplant; ICE, ifosfamide, carboplatin and etoposide; MUD-SCT, matched unrelated donor stem cell transplant; MDS, myelodysplastic syndrome; hyper CVAD, cyclophosphamide, vincristine, doxorubicin, and dexamethasone; CR, complete remission; DOD, died of disease
Morphologically, the lymphomas had pathologic and immunophenotypic findings that are typical of classical Hodgkin lymphoma (Figures 1 and 2), mantle cell lymphoma (Figure 3) and angioimmunoblastic T-cell lymphoma (Figure 4), and are not further discussed. In addition to the lymphomas, all biopsy specimens contained foci of LCH characterized by variable amounts of LCs with oval to folded nuclei with nuclear grooves, thin nuclear membranes, vesicular chromatin, inconspicuous nucleoli, and abundant pale to eosinophilic cytoplasm with ill-defined cell borders. The distribution of LCs was variable and ranged from a focal cluster of LCH to multiple foci of LCH intermingled intimately with lymphoma. Interspersed eosinophils, eosinophilic abscesses and areas of necrosis were observed variably (Figures 1–4). No cytologic atypia or increased mitotic activity was found in the LCH component in any case. Immunohistochemical stains for CD1a, langerin and S-100 were positive in the LCH component in all cases (Figures 1–4).
Figure 1. LCH and classical Hodgkin lymphoma, mixed cellularity (case 2).
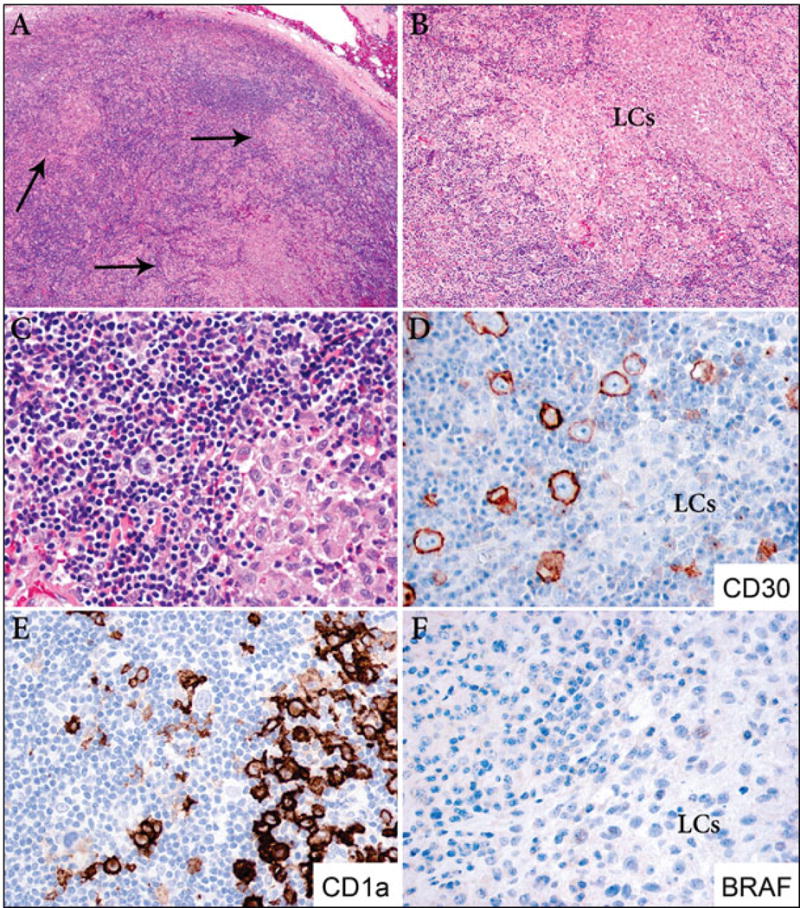
A. Low power image of a lymph node involved by classical Hodgkin lymphoma, mixed cellularity type, and multiple clusters of LCH of variable sizes (arrows) (40X). B. Largest ill-defined nodule of LCH (200X). C. Area of transition of classical Hodgkin lymphoma with a Reed-Sternberg cell (left) and LCH (right) (400X). D. Immunostain for CD30 highlights the Reed-Sternberg cells and is negative in the Langerhans cells (LCs) (400X). E. Immunostain for CD1a is positive in the LCs and is negative in the Reed-Sternberg cells (400X). F. BRAF is negative in both components (400X).
Figure 2. LCH and classical Hodgkin lymphoma, nodular sclerosis (case 5).
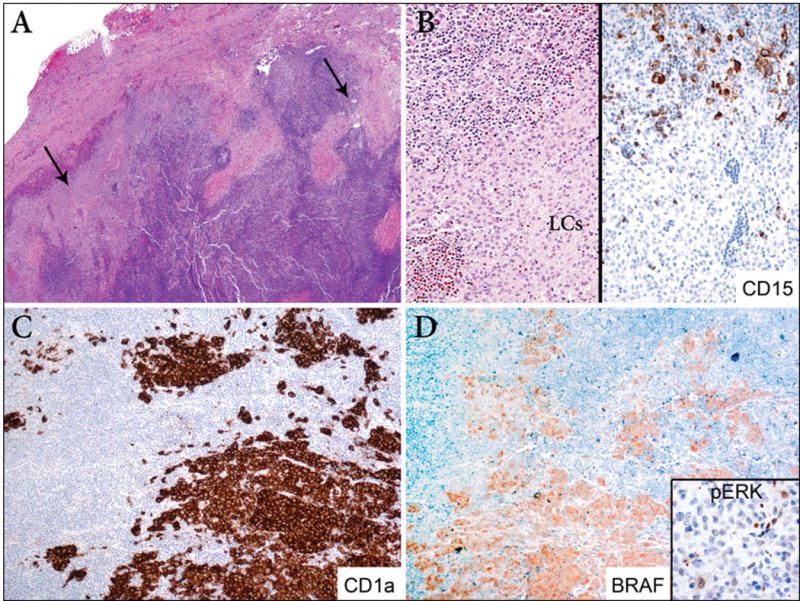
A. The lymph node capsule is thickened and contains multiple fibrous bands extending into the parenchyma that incompletely surround cellular areas. In addition, the subcapsular region contains large clusters of LCH (arrows) (40X). B. Area of transition between classical Hodgkin lymphoma (top) and a nodule of Langerhans cells (bottom) (200X). Immunostain for CD15 highlights the Reed-Sternberg cells (top) and is negative in the Langerhans cells (bottom) (200X). C. The widespread involvement of the lymph node by LCH is more evident with immunostain for CD1a (100X). D. Immunostain for BRAF is positive only in the LCH component with a weak cytoplasmic pattern of staining (100X). Despite a positive BRAF V600E immunohistochemical result, BRAF pyrosequencing demonstrated a wild type sequence in codons 596 (GGT) and 600 (GTG). This case is negative for pERK by immunohistochemistry (Inset, 100X).
Figure 3. LCH and mantle cell lymphoma (case 7).
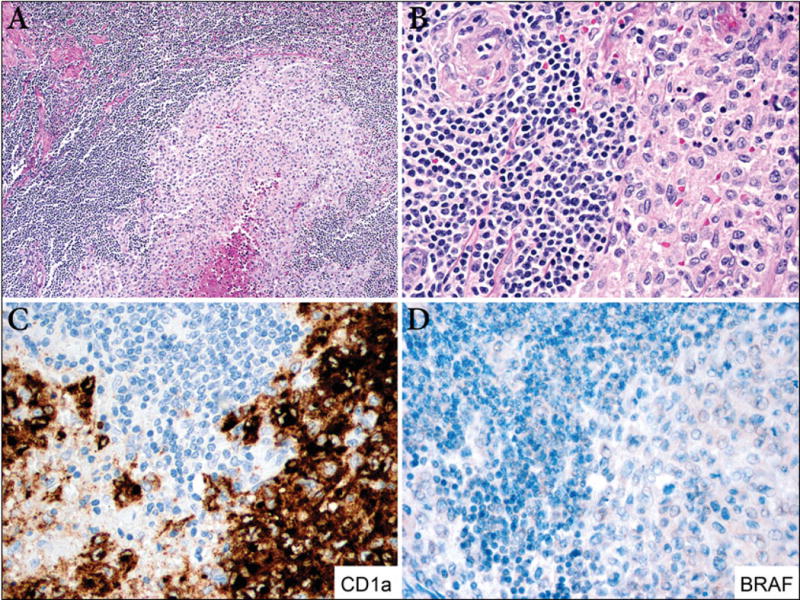
A. The lymph node is replaced by a monotonous proliferation of small lymphocytes with focal sclerosis. A single nodule of LCH with central necrosis is identified at the center of the lymph node (100X). B. Left, mantle cell lymphoma composed of small lymphocytes with cleaved nuclei and perivascular fibrosis. Right, LCH module with scattered apoptotic cells (400x). C. Immunostain for CD1a is positive in the Langerhans cells and is negative in mantle cell lymphoma cells (400X). D. BRAF is negative in both components (400X).
Figure 4. LCH and angioimmunoblastic T-cell lymphoma (AITL) (case 1).
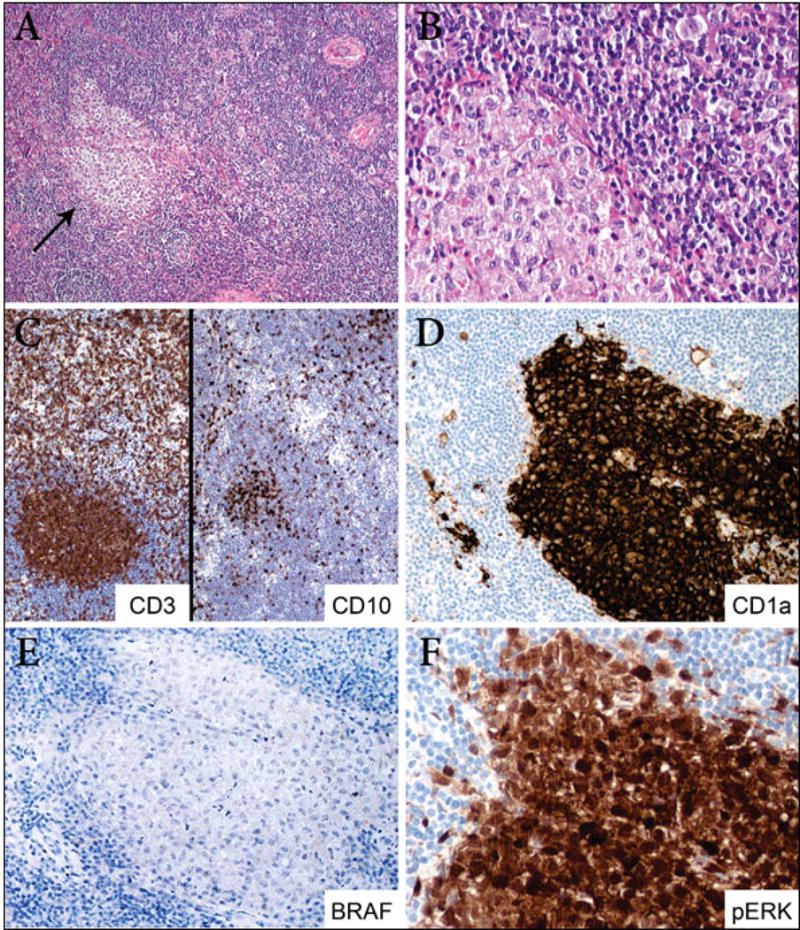
A. At low power, the lymph node is replaced by a diffuse lymphoproliferative process composed of small lymphocytes, clear cells and increased vascularity, characteristic of AITL. A well circumscribed pale nodule of LCH (arrow) is present within the lymphomatous process (100X). B. At higher magnification, the paler nodule (left) is composed of Langerhans cells and scattered eosinophils (400X). C. By immunohistochemistry, the lymphoma cells are positive for CD3 and CD10 (200X). D. The LCH nodule is positive for CD1a (200X). E. BRAF is negative in both AITL and LCH (200X). F) Immunostain with the anti-pERK antibody is positive in both the nucleus and cytoplasm of Langerhans cells (400X).
Clinical follow-up data were available for 6 patients. Six patients received chemotherapy for the specific lymphoma (detailed chemotherapy regimens are listed in Table 1); 2 received additional radiation therapy, 2 underwent stem cell transplantation, and 1 patient is on the list in anticipation of stem cell transplantation. No patient received treatment specific for LCH. Treatment and outcome information were not available for 1 patient who was lost to follow up. With a median follow-up of 21 months (range, 6–89 months), 3 patients remained in clinical remission for both LCH and lymphoma, one patient developed recurrent mediastinal lymphoma detected by imaging (biopsy not performed at the time of this study), one patient developed therapy-related myelodysplastic syndrome, and one patient died of Hodgkin lymphoma. No patient developed recurrent LCH.
BRAF and phosphorylated ERK immunohistochemical assessment
The LCH component was negative for BRAF V600E mutant protein by IHC in 6 of 7 cases. The case of LCH positive for BRAF (case 5) is shown in Figure 2D. This result was performed twice with identical results.
The LCH component was positive for p-ERK by IHC in 3 of 7 cases (cases 1, 4, and 6). P-ERK expression was observed in the nucleus and cytoplasm of LCs (Figure 4F). In case 4, 2 small subcapsular LCH nodules were positive for p-ERK whereas the largest focus found in the mid portion of the lymph node was negative (not shown). In all cases, stromal cells, fibroblasts, and endothelial cells were positive for p-ERK (internal controls).
BRAF and MAP2K1 mutation analysis
None of the 7 cases harbored a BRAF or MAP2K1 mutation by PCR/pyrosequencing and PCR/Sanger sequencing, respectively.
Next-generation sequencing results
To evaluate the discrepancy between immunohistochemistry and Sanger sequencing for BRAF mutation on one case, and to explore the mutational status of the three cases positive for p-ERK, we performed NGS analysis to assess mutation status of a panel of 134 genes that are commonly mutated in hematopoietic neoplasms, including ARAF, BRAF, ERBB1, ERBB2, ERBB3, KRAS, MAP2K1, NRAS, and PIK3CA, on 3 cases with DNA available. No mutations in any gene were detected in all 3 cases.
Discussion
We assessed the frequency of LCH-related mutations in biopsy specimens involved by LCH and lymphoma in 7 patients. This form of LCH, virtually always detected incidentally, is rare with most cases reported in the literature as case reports or small case series.23–30 Most of these studies were published before the discovery of BRAF and MAP2K1 mutations in LCH, and therefore, as far as we are aware, this is the first study to assess BRAF V600E and MAP2K1 mutation status in the LCH component in cases with concomitant LCH and lymphoma.
Using molecular methods, we did not identify BRAF V600E or MAP2K1 mutations in the LCH lesions in this cohort. Nevertheless, we detected p-ERK by IHC in 3 of 7 cases consistent with activation of this pathway. NGS analysis for a panel of 134 genes (including ARAF, BRAF, ERBB1, ERBB2, ERBB3, KRAS, MAP2K1, NRAS, and PIK3CA) on two of these three cases failed to detect any mutation. Our data do not support an explanation for the positive p-ERK result in these 3 cases. One possibility is that the sensitivity of the methods we employed is insufficient to detect mutations. In our opinion, this seems unlikely as the LCH lesions were microdissected and had well above 10% LCs in the specimen analyzed, the lower limit of sensitivity for the methods employed. We considered the possibility of non-V600E BRAF mutations, but the NGS results in two cases exclude this idea. It seems reasonable to suggest that the RAS/RAF/MAPK signaling pathway in some cases of incidental LCH may be activated by non-mutational mechanisms. Possibly, a local cytokine-mediated process imparting a growth advantage to LCs through activation of p-ERK might be involved, perhaps induced by the presence of lymphoma or a manifestation of lymphoma–tumor microenvrionmental interaction.
Using a commercially available monoclonal BRAF V600E specific antibody (VE-1), the results were negative in 6 cases of LCH, but was positive in 1 case (case 5). The reason for the discrepancy between the IHC result and molecular results in this case is uncertain. Methods do not appear to be an explanation as we used a pyrosequencing-based assay in all our cases to identify BRAF mutations similar to most published studies.8,34,35 This case was also assessed by NGS methods and was negative for mutations in a 134 gene panel that included BRAF and MAP2K1. The BRAF antibody has been shown to have a high sensitivity and specificity for the BRAF mutation and represents an excellent tool for screening tissue samples.35 It is possible, however, that the BRAF V600E antibody uncommonly may yield false positive results. In a large study of colorectal carcinomas performed at our institution, Estrella and colleagues showed that 7 of 323 (2.2%) carcinomas shown to be wild type for BRAF V600E mutation by sequencing analysis were diffusely positive using the VE-1 antibody.36 It is important to mention that the case of LCH positive for BRAF V600E by IHC was negative for p-ERK, indicating that that RAS/RAF/MAPK pathway was not active in this case.
The most frequent lymphoma associated with LCH in this case series was classical Hodgkin lymphoma, in accord with data reported in the literature.23–26 To our knowledge, we report for the first time an association of LCH with mantle cell lymphoma and angioimmunoblastic T-cell lymphoma. In keeping with the literature, none of the 6 patients with follow-up data developed recurrent LCH or systemic LCH. This behavior, the often focal nature of the LCH, and the absence of BRAF and MAP2K1 mutations suggests that LCH associated with lymphoma is a benign process, at least in most cases, and possibly reactive to the presence of lymphoma. This concept has been supported by others, who have suggested that focal LCH associated with a hematolymphoid tumor is better considered as LC hyperplasia.2,23–25,29 Christie et al. and others have suggested that these lesions be designated as “LC-like lesions”.29 Furthermore, some authors have suggested that this type of LCH or LC-like lesions may be driven by chemokine/cytokine or other stimuli produced by the associated neoplastic conditions.2,23–25,29 This hypothesis could be the explanation for why classical Hodgkin lymphoma, a lymphoma that is characterized by the production of a storm of chemokines and cytokines, is the most common lymphoma with associated incidental LCH. Although this suggestion is reasonable for the concurrent findings of these two lesions, we do not have sufficient data to address this idea adequately.
In conclusion, we did not detect BRAF V600E or MAP2K1 mutations in all 7 cases of LCH associated with lymphoma assessed. These data, combined with the incidental nature of LCH detection when associated with lymphoma, the small size of the LCH lesions, and the absence of systemic LCH or recurrence, suggests that LCH associated with lymphoma is benign, as has been suggested by others. Nevertheless, p-ERK was positive in 3 of 7 cases suggesting activation of the RAS/RAF/MAPK pathway via non-genetic mechanisms in a subset of cases. It seems possible that pathway activation in these lesions may be induced by the presence of lymphoma.
Acknowledgments
We thank Jawad manekia and Mohammad Mohammad for technical assistance in NGS study.
Abbreviations
- LC
Langerhans cells
- LCH
Langerhans cell histiocytosis
- MAPK
mitogen activated protein kinase
- NGS
next-generation sequencing
- PCR
polymerase chain reaction
Footnotes
Disclosure/Conflict of Interest
The authors have no conflicts of interest to disclose.
References
- 1.Bechan GI, Egeler RM, Arceci RJ. Biology of Langerhans cells and Langerhans cell histiocytosis. Int Rev Cytol. 2006;254:1–43. doi: 10.1016/S0074-7696(06)54001-X. [DOI] [PubMed] [Google Scholar]
- 2.Jaffe R. Langerhans cell histiocytosis and Langerhans cell sarcoma. In: Jaffe ES, Harris NL, Vardiman JW, Campo E, Arber DA, editors. Hematopathology. Philadelphia: Elsevier; 2011. pp. 811–826. [Google Scholar]
- 3.Willman CL, Busque L, Griffith BB, et al. Langerhans cell histiocytosis (histiocytosis X)–a clonal proliferative disease. N Engl J Med. 1994;331:154–160. doi: 10.1056/NEJM199407213310303. [DOI] [PubMed] [Google Scholar]
- 4.Yu RC, Chu C, Buluwela L, et al. Clonal proliferation of Langerhans cells in Langerhans cell histiocytosis. Lancet. 1994;343:767–768. doi: 10.1016/s0140-6736(94)91842-2. [DOI] [PubMed] [Google Scholar]
- 5.Badalian-Very G, Vergilio JA, Degar BA, et al. Recurrent BRAF mutations in Langerhans cell histiocytosis. Blood. 2010;116:1919–1923. doi: 10.1182/blood-2010-04-279083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haroche J, Charlotte F, Arnaud L, et al. High prevalence of BRAF V600E mutations in Erdheim-Chester disease but not in other non-Langerhans cell histiocytoses. Blood. 2012;120:2700–2703. doi: 10.1182/blood-2012-05-430140. [DOI] [PubMed] [Google Scholar]
- 7.Roden AC, Hu X, Kip S, et al. BRAF V600E expression in Langerhans cell histiocytosis: clinical and immunohistochemical study on 25 pulmonary and 54 extrapulmonary cases. Am J Surg Pathol. 2014;38:548–551. doi: 10.1097/PAS.0000000000000129. [DOI] [PubMed] [Google Scholar]
- 8.Sahm F, Capper D, Preusser M, et al. BRAFV600E mutant protein is expressed in cells of variable maturation in Langerhans cell histiocytosis. Blood. 2012;120:e28–34. doi: 10.1182/blood-2012-06-429597. [DOI] [PubMed] [Google Scholar]
- 9.Satoh T, Smith A, Sarde A, et al. B-RAF mutant alleles associated with Langerhans cell histiocytosis, a granulomatous pediatric disease. PLoS One. 2012;7:e33891. doi: 10.1371/journal.pone.0033891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montagut C, Settleman J. Targeting the RAF-MEK-ERK pathway in cancer therapy. Cancer Lett. 2009;283:125–134. doi: 10.1016/j.canlet.2009.01.022. [DOI] [PubMed] [Google Scholar]
- 11.Wellbrock C, Karasarides M, Marais R. The RAF proteins take centre stage. Nat Rev Mol Cell Biol. 2004;5:875–885. doi: 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
- 12.Young A, Lyons J, Miller AL, et al. Ras signaling and therapies. Adv Cancer Res. 2009;102:1–17. doi: 10.1016/S0065-230X(09)02001-6. [DOI] [PubMed] [Google Scholar]
- 13.Brown NA, Furtado LV, Betz BL, et al. High prevalence of somatic MAP2K1 mutations in BRAF V600E-negative Langerhans cell histiocytosis. Blood. 2014;124:1655–1658. doi: 10.1182/blood-2014-05-577361. [DOI] [PubMed] [Google Scholar]
- 14.Chakraborty R, Hampton OA, Shen X, et al. Mutually exclusive recurrent somatic mutations in MAP2K1 and BRAF support a central role for ERK activation in LCH pathogenesis. Blood. 2014;124:3007–3015. doi: 10.1182/blood-2014-05-577825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greaves WO, Verma S, Patel KP, et al. Frequency and spectrum of BRAF mutations in a retrospective, single-institution study of 1112 cases of melanoma. J Mol Diagn. 2013;15:220–226. doi: 10.1016/j.jmoldx.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rahman MA, Salajegheh A, Smith RA, et al. B-Raf mutation: a key player in molecular biology of cancer. Exp Mol Pathol. 2013;95:336–342. doi: 10.1016/j.yexmp.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 17.Tiacci E, Trifonov V, Schiavoni G, et al. BRAF mutations in hairy-cell leukemia. N Engl J Med. 2011;364:2305–2315. doi: 10.1056/NEJMoa1014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schindler G, Capper D, Meyer J, et al. Analysis of BRAF V600E mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol. 2011;121:397–405. doi: 10.1007/s00401-011-0802-6. [DOI] [PubMed] [Google Scholar]
- 19.Marks JL, Gong Y, Chitale D, et al. Novel MEK1 mutation identified by mutational analysis of epidermal growth factor receptor signaling pathway genes in lung adenocarcinoma. Cancer Res. 2008;68:5524–5528. doi: 10.1158/0008-5472.CAN-08-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murugan AK, Dong J, Xie J, Xing M. MEK1 mutations, but not ERK2 mutations, occur in melanomas and colon carcinomas, but none in thyroid carcinomas. Cell Cycle. 2009;8:2122–2124. doi: 10.4161/cc.8.13.8710. [DOI] [PubMed] [Google Scholar]
- 21.Wagle N, Emery C, Berger MF, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29:3085–3096. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waterfall JJ, Arons E, Walker RL, et al. High prevalence of MAP2K1 mutations in variant and IGHV4-34-expressing hairy-cell leukemias. Nat Genet. 2014;46:8–10. doi: 10.1038/ng.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burns BF, Colby TV, Dorfman RF. Langerhans’ cell granulomatosis (histiocytosis X) associated with malignant lymphomas. Am J Surg Pathol. 1983;7:529–533. doi: 10.1097/00000478-198309000-00003. [DOI] [PubMed] [Google Scholar]
- 24.Egeler RM, Neglia JP, Arico M, et al. The relation of Langerhans cell histiocytosis to acute leukemia, lymphomas, and other solid tumors. The LCH-Malignancy Study Group of the Histiocyte Society. Hematol Oncol Clin North Am. 1998;12:369–378. doi: 10.1016/s0889-8588(05)70516-5. [DOI] [PubMed] [Google Scholar]
- 25.Egeler RM, Neglia JP, Puccetti DM, et al. Association of Langerhans cell histiocytosis with malignant neoplasms. Cancer. 1993;71:865–873. doi: 10.1002/1097-0142(19930201)71:3<865::aid-cncr2820710334>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 26.Greaves WO, Bueso-Ramos C, Fayad L. Classical Hodgkin’s lymphoma associated with Langerhans cell histiocytosis: multiagent chemotherapy resulted in histologic resolution of both the classical Hodgkin’s lymphoma and Langerhans cell proliferation components. J Clin Oncol. 2011;29:e76–78. doi: 10.1200/JCO.2010.31.2413. [DOI] [PubMed] [Google Scholar]
- 27.Adu-Poku K, Thomas DW, Khan MK, et al. Langerhans cell histiocytosis in sequential discordant lymphoma. J Clin Pathol. 2005;58:104–106. doi: 10.1136/jcp.2003.015537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Almanaseer IY, Kosova L, Pellettiere EV. Composite lymphoma with immunoblastic features and Langerhans’ cell granulomatosis (histiocytosis X) Am J Clin Pathol. 1986;85:111–114. doi: 10.1093/ajcp/85.1.111. [DOI] [PubMed] [Google Scholar]
- 29.Christie LJ, Evans AT, Bray SE, et al. Lesions resembling Langerhans cell histiocytosis in association with other lymphoproliferative disorders: a reactive or neoplastic phenomenon? Hum Pathol. 2006;37:32–39. doi: 10.1016/j.humpath.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 30.Licci S, Boscaino A, De Palma M, et al. Concurrence of marginal zone B-cell lymphoma MALT-type and Langerhans cell histiocytosis in a thyroid gland with Hashimoto disease. Ann Hematol. 2008;87:855–857. doi: 10.1007/s00277-008-0489-5. [DOI] [PubMed] [Google Scholar]
- 31.Capper D, Preusser M, Habel A, et al. Assessment of BRAF V600E mutation status by immunohistochemistry with a mutation-specific monoclonal antibody. Acta Neuropathol. 2011;122:11–19. doi: 10.1007/s00401-011-0841-z. [DOI] [PubMed] [Google Scholar]
- 32.Verma S, Greaves WO, Ravandi F, et al. Rapid detection and quantitation of BRAF mutations in hairy cell leukemia using a sensitive pyrosequencing assay. Am J Clin Pathol. 2012;138:153–156. doi: 10.1309/AJCPL0OPXI9LZITV. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 34.Alayed K, Medeiros LJ, Patel KP, et al. BRAF and MAP2K1 mutations in Langerhans cell histiocytosis: a study of 50 cases. Hum Pathol. 2016;52:61–67. doi: 10.1016/j.humpath.2015.12.029. [DOI] [PubMed] [Google Scholar]
- 35.Jabbar KJ, Luthra R, Patel KP, et al. Comparison of next-generation sequencing mutation profiling with BRAF and IDH1 mutation-specific immunohistochemistry. Am J Surg Pathol. 2015;39:454–461. doi: 10.1097/PAS.0000000000000325. [DOI] [PubMed] [Google Scholar]
- 36.Estrella JS, Tetzlaff MT, Bassett RL, et al. Assessment of BRAF V600E in coloreactal carcinoma: tissue-specific discordance between immunohistochemistry and seqie3ncing. Mol Cancer Ther. 2015;14:2887–2895. doi: 10.1158/1535-7163.MCT-15-0615. [DOI] [PubMed] [Google Scholar]


