Abstract
G protein-coupled receptors (GPCRs) are the largest superfamily of transmembrane receptors and have vital signaling functions in various organs. Because of their critical roles in physiology and pathology, GPCRs are the most commonly used therapeutic target. It has been suggested that GPCRs undergo massive genetic variations such as genetic polymorphisms and DNA insertions or deletions. Among these genetic variations, non-synonymous natural variations change the amino acid sequence and could thus alter GPCR functions such as expression, localization, signaling, and ligand binding, which may be involved in disease development and altered responses to GPCR-targeting drugs. Despite the clinical importance of GPCRs, studies on the genotype-phenotype relationship of GPCR natural variants have been limited to a few GPCRs such as β-adrenergic receptors and opioid receptors. Comprehensive understanding of non-synonymous natural variations within GPCRs would help to predict the unknown genotype-phenotype relationship and yet-to-be-discovered natural variants. Here, we analyzed the non-synonymous natural variants of all non-olfactory GPCRs available from a public database, UniProt. The results suggest that non-synonymous natural variations occur extensively within the GPCR superfamily especially in the N-terminus and transmembrane domains. Within the transmembrane domains, natural variations observed more frequently in the conserved residues, which leads to disruption of the receptor function. Our analysis also suggests that only few non-synonymous natural variations have been studied in efforts to link the variations with functional consequences.
Keywords: G protein-coupled receptor, Non-synonymous natural variant, Structure
INTRODUCTION
G protein-coupled receptors (GPCRs) are the largest superfamily of transmembrane receptors, with approximately 800 genes identified in humans (Pierce et al., 2002). GPCRs transmit extracellular signals into the intracellular region resulting in interaction with various signaling proteins (Chun and Shim, 2015; Zhang et al., 2016; Zheng et al., 2016). These GPCR signaling regulate vital signaling pathways, which are involved in critical physiological and pathological functions in various organs in the sensory, metabolic, endocrine, neuromuscular, and central nervous systems (Pierce et al., 2002; Stoddard and Chun, 2015). Owing to their importance in physiology and pathology, approximately 30–40% of currently marketed drugs target GPCRs.
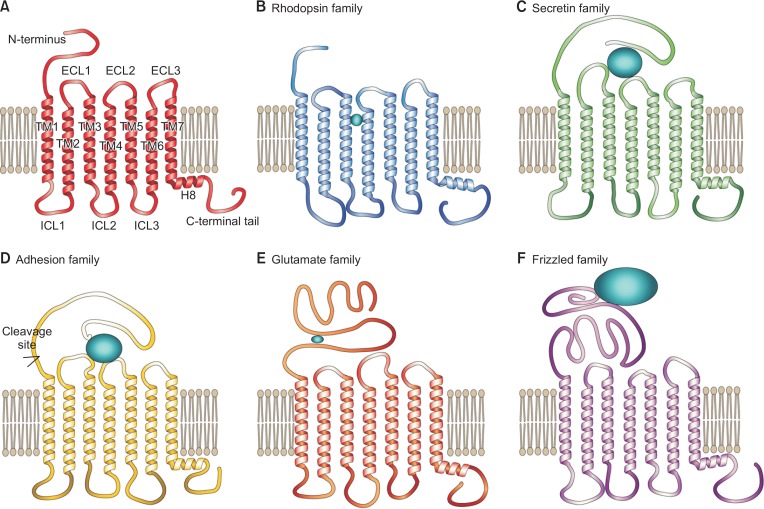
GPCRs are composed of seven transmembrane α-helical domains with an extracellular N-terminus and an intracellular C-terminus (Fig. 1A). Although GPCRs share a common overall structure, the sequence similarity is relatively limited, and the sequences and lengths of the N-termini, cytoplasmic loops, and C-termini vary among GPCRs (Tang and Insel, 2005). In 2003, human GPCRs were grouped into five families (Rhodopsin, Secretin, Adhesion, Glutamate, and Frizzled families) based on phylogenetic analysis (Fredriksson et al., 2003) (Fig. 1). Currently, a GPCR database, GPCRdb (http://gpcrbd.org/) (Munk et al., 2016), classified human GPCRs into 286 Rhodopsin (excluding olfactory receptors), 15 Secretin, 33 Adhesion, 22 Glutamate (including three taste 1 receptors), and 11 Frizzled families (Lagerstrom and Schioth, 2008; Munk et al., 2016). Twenty-five taste 2 receptors and six orphan receptors are not included in any of these families.
Fig. 1.
Description of the general GPCR structure and five GPCR families. (A) The structural domains of GPCRs. (B–F) Representations of the conserved and distinct features of the Rhodopsin (B), Secretin (C), Adhesion (D), Glutamate (E), and Frizzled (F) families.
Completion of the Human Genome Project revealed that many proteins have genetic variants such as nucleotide insertions, deletions, or exchanges. Although majority of the genetic variants are hypothesized to be functionally neutral (Kimura, 1968), some natural variants affect the function of the gene, usually involving non-synonymous or missense variants that change the amino acid sequence (Hecht et al., 2013). Genetic variants identified in GPCRs can influence receptor expression, proper folding, targeting within cell organelles, activation, function, desensitization, and ligand binding (Tang and Insel, 2005; Insel et al., 2007). Therefore, the genetic alterations in GPCRs are involved in the occurrence of genetic diseases and different drug responses (Thompson et al., 2014a, 2014b). It has been reported that there are extensive inter-patient variations in therapeutic responses to the GPCR-targeting drugs, that are caused by the genetic variations in GPCRs between patients (Drazen et al., 2000). Therefore, if we thoroughly understand the link between natural variants of GPCRs and the effects of these natural variants on disease development and drug responses, it would be possible to develop a rational scheme to selectively treat patients with genetically altered GPCRs.
There have been efforts to analyze the genetic variations in GPCR families with respect to their functional consequences and/or in relation to their distribution in the structural context. For example, GPCRdb or GPCR NaVa (http://nava.liacs.nl/) is devoted to provide graphically accessible information on GPCR mutations and their known functional consequences (Wahlestedt et al., 2004; Kazius et al., 2008). With these database resources, one can easily access the natural variant information of individual GPCRs, but understanding the bigger picture of the domain distribution of all known GPCR natural variants is not yet achieved. Comprehending the overall trend of domain distribution of GPCR natural variants would provide valuable information to prioritize the investigation of genotype-phenotype relationship and to predict yet-to-be-discovered natural variants. Currently, however, there are only few studies that analyzed the distribution of genetic variations of only a subset of GPCRs (Lee et al., 2003; Small et al., 2003; Wahlestedt et al., 2004). The present study analyzed the non-synonymous natural variants that occur in all human non-olfactory GPCRs from the five GPCR families (a total of 367 GPCRs) using UniProt (http://www.uniprot.org/) (Boutet et al., 2016) and GPCRdb databases. The positions of the natural variants were mapped onto the structural domains, and the occurrence patterns were analyzed.
MATERIALS AND METHODS
Database analysis
The list of human non-olfactory GPCRs was retrieved from GPCRdb (http://gpcrdb.org/). The classification of these GPCRs into five families and assignment of structural domains for all listed GPCRs were also based on GPCRdb. The non-synonymous natural variants of the listed human non-olfactory GPCRs were obtained from the UniProt database (http://uniprot.org/), and the positions of these variants were mapped on the structural domains based on the generic numbering system provided by GPCRdb.
Dopamine receptor type 1 mutant construct generation and cell lines
Dopamine receptor type 1 (D1R) cDNA was described in previous studies (Dearry et al., 1990; Cho et al., 2006). The DRY, WxP, NPxxY motifs were mutated to DHY, WxS, and NSxxY respectively via site-directed mutagenesis using PCR. Gαs-stably transfected HEK293 cell line was generated by selecting survived colonies after transfecting Gαs cDNA using Lipofectamine 2000 Reagent (ThermoFisher Scientific, Waltham, MA, USA) and G418 (Duchefa Biochemie, Haarlem, The Netherlands) antibiotic treatment.
cAMP assay
Gαs-stably transfected HEK293 cells were cultured in 12-well plate until the confluency reached 80%, and the cells were transfected with D1R wild type and mutant constructs. After 20–24 hrs later, about 3000 cells were seeded onto 96-well plate and were incubated for another 20–24 hrs in 5% CO2 at 37°C. The cells were pre-treated with 250 µM 3-isobutyl-1-methylxanthine (IBMX) (Sigma-Aldrich, St. Louis, MO, USA) and 200 µM Ro 20-1724 (Sigma-Aldrich) to prevent cAMP hydrolysis, and the D1R receptors were activated by 5 min incubation with 500 nM dopamine (Sigma-Aldrich) or buffer vehicle. cAMP levels were measured using cAMP-GloTM Assay kit (Promega, Fitchburg, WI, USA) according to the manufacturer’s manual. The luminescence levels were detected by Synergy HTX (BioTek Instruments, Winooski, VT, USA).
Immunofluorescence
D1R-expressing cells were washed with ice-cold PBS and fixed with 4% formaldehyde in PBS for 15 min. After washing out formaldehyde with PBS, cells were permeabilized with 0.25% Triton-X 100 in PBS for 5 min. After washing with PBS, cells were blocked with 10% BSA in PBS for 1 hr and incubated with FLAG M2 primary antibody (Sigma-Aldrich) for 2 hrs followed by Rhodamine Red-X secondary antibody (Invitrogen) for 1.5 hr. The immunofluorescence images were taken by Zeiss LSM 700 confocal microscope (Zeiss, Oberkochen, Germany).
RESULTS
Distribution of GPCR non-synonymous natural variants within GPCR families
Human GPCRs are categorized into five families based on phylogenetic analysis (Fredriksson et al., 2003). Each family shares seven transmembrane (TM) domains with varying N-termini and ligand-binding sites (Fig. 1B–1F) (Lagerstrom and Schioth, 2008). Several previous studies have analyzed the natural variants that occur in GPCRs (Rana et al., 2001; Small et al., 2002; Lee et al., 2003; Small et al., 2003; Wahlestedt et al., 2004). However, these studies analyzed only a subset of GPCRs such as monoamine or peptide-binding GPCRs (Rana et al., 2001; Lee et al., 2003). In the present study, non-synonymous natural variants of all human non-olfactory GPCRs (except twenty five taste 2 and six un-categorized orphan receptors) were obtained from the UniProt database; UniProt annotates non-synonymous natural variants, including polymorphisms; variations between strains, isolates, or cultivars; disease-associated mutations; and RNA editing events, whereas mutations that induce major changes such as frame-shifts or premature stops are not annotated. Non-synonymous natural variants were found in approximately 72% (207 out of 286) of the Rhodopsin family, 80% (12 out of 15) of the Secretin family, 79% (26 out of 33) of the Adhesion family, 82% (18 out of 22) of the Glutamate family, and 55% (6 out of 11) of the Frizzled family GPCRs showing that natural variants occur in majority of the GPCRs in all GPCR families.
Distribution of GPCR non-synonymous natural variants within structural domains
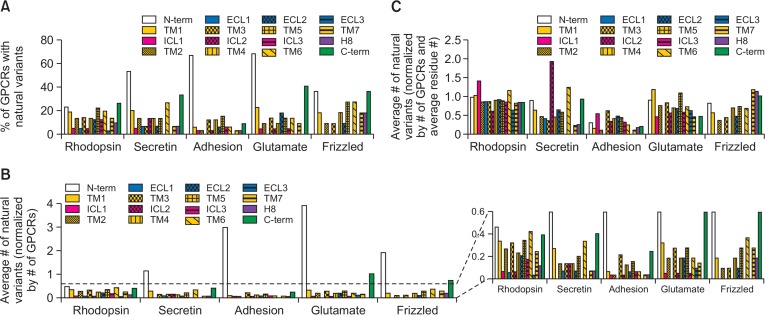
The positions of natural variants were mapped and assigned to 16 structural domains (i.e., one N-terminus, one C-terminal tail, seven TM domains, three intracellular loop regions, three extracellular loop regions, and 1 helix 8) for all 367 GPCRs (Table 1, Fig. 2). In all GPCR families, relatively more GPCRs have non-synonymous natural variants in the N-terminus, C-terminal tail, and TM domains than in the extracellular or intracellular loops (Table 1, Fig. 2A). Especially, the N-terminus contained the highest average number of non-synonymous natural variants in all GPCR families (Fig. 2B). The percentage of GPCRs with non-synonymous natural variants at the N-terminus or the average number of non-synonymous natural variants at the N-terminus were relatively smaller in the Rhodopsin family than in the other GPCR families (Fig. 2A, 2B). This is probably due to the shorter average N-terminal length of the Rhodopsin family compared to that of other GPCR families (Table 1). It has been proven that the average number of non-synonymous natural variants at the N-terminus becomes similar to that at the other domains for all GPCR families when it was normalized by the average residue numbers of the N-terminal region (Fig. 2C).
Table 1.
Summary of non-synonymous natural variants
| Family | N-term | TM1 | ICL1 | TM2 | ECL1 | TM3 | ICL2 | TM4 | ECL2 | TM5 | ICL3 | TM6 | ECL3 | TM7 | H8 | C-term |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rhodopsin (286) | ||||||||||||||||
| Average residue # | 47 ± 73.4 | 32 ± 2.6 | 4 ± 8.5 | 31 ± 10.7 | 6 ± 9.6 | 37 ± 9.6 | 10 ± 7.0 | 25 ± 6.0 | 22 ± 13.2 | 39 ± 12.4 | 20 ± 35.9 | 36 ± 2.7 | 4 ± 18.3 | 29 ± 6.7 | 13 ± 2.5 | 47 ± 43.6 |
| Natural variants | 132 | 96 | 18 | 76 | 15 | 91 | 18 | 66 | 58 | 98 | 50 | 120 | 8 | 69 | 32 | 112 |
| GPCRs with natural variants | 65 | 53 | 14 | 38 | 13 | 40 | 13 | 39 | 36 | 63 | 35 | 56 | 8 | 39 | 27 | 74 |
| Secretin (15) | ||||||||||||||||
| Average residue # | 127 ± 19.3 | 42 ± 0.0 | 6 ± 4.0 | 28 ± 1.0 | 16 ± 5.2 | 36 ± 2.5 | 7 ± 0.3 | 29 ± 0.0 | 10 ± 1.6 | 34 ± 1.5 | 8 ± 1.0 | 27 ± 1.8 | 7 ± 0.3 | 28 ± 1.8 | 26 ± 0.0 | 50 ± 27.3 |
| Natural variants | 17 | 4 | 0 | 2 | 1 | 2 | 2 | 2 | 1 | 3 | 0 | 5 | 0 | 1 | 1 | 6 |
| GPCRs with natural variants | 8 | 3 | 0 | 2 | 1 | 1 | 2 | 2 | 1 | 2 | 0 | 4 | 0 | 1 | 1 | 5 |
| Adhesion (33) | ||||||||||||||||
| Average residue # | 983 ± 1,098.1 | 38 ± 0.0 | 6 ± 1.5 | 27 ± 1.2 | 7 ± 4.0 | 34 ± 0.9 | 9 ± 2.9 | 29 ± 0.0 | 13 ± 11.4 | 34 ± 0.0 | 19 ± 16.2 | 25 ± 0.7 | 5 ± 3.8 | 27 ± 3.8 | 16 ± 2.9 | 148 ± 149.0 |
| Natural variants | 98 | 2 | 1 | 1 | 0 | 7 | 1 | 4 | 2 | 5 | 2 | 2 | 0 | 1 | 1 | 8 |
| GPCRs with natural variants | 22 | 2 | 1 | 1 | 0 | 4 | 1 | 4 | 2 | 5 | 2 | 2 | 0 | 1 | 1 | 3 |
| Glutamate (22) | ||||||||||||||||
| Average residue # | 436 ± 222.8 | 27 ± 0.0 | 10 ± 0.8 | 24 ± 1.6 | 11 ± 22.2 | 33 ± 0.0 | 8 ± 21.4 | 26 ± 0.0 | 27 ± 6.6 | 25 ± 0.0 | 8 ± 7.7 | 25 ± 0.0 | 15 ± 22.6 | 29 ± 0.0 | 12 ± 0.0 | 210 ± 361.7 |
| Natural variants | 86 | 7 | 1 | 4 | 0 | 6 | 1 | 4 | 4 | 6 | 1 | 4 | 2 | 3 | 0 | 22 |
| GPCRs with natural variants | 15 | 5 | 1 | 2 | 0 | 3 | 1 | 2 | 4 | 3 | 1 | 3 | 2 | 2 | 0 | 9 |
| Frizzled (11) | ||||||||||||||||
| Average residue # | 233 ± 33.8 | 32 ± 0.0 | 6 ± 0.0 | 24 ± 0.3 | 30 ± 10.5 | 36 ± 0.0 | 4 ± 0.3 | 26 ± 0.0 | 19 ± 0.0 | 37 ± 0.0 | 8 ± 0.8 | 53 ± 3.4 | 12 ± 4.6 | 23 ± 0.0 | 16 ± 0.0 | 76 ± 76.1 |
| Natural variants | 21 | 2 | 0 | 1 | 0 | 1 | 0 | 2 | 1 | 3 | 0 | 4 | 0 | 3 | 2 | 8 |
| GPCRs with natural variants | 4 | 2 | 0 | 1 | 0 | 1 | 0 | 2 | 1 | 3 | 0 | 3 | 0 | 2 | 2 | 4 |
Fig. 2.
Occurrence of non-synonymous natural variations in GPCR families. (A) Percentage of GPCRs with non-synonymous natural variants in the specified structural domains among all GPCRs of each family. (B) The reported average numbers of non-synonymous natural variations within the specified structural domains, which are normalized by the total number of GPCRs in each family. (C) The reported average numbers of non-synonymous natural variations within the specified structural domains, which are normalized by the total number of GPCRs in each family and average residue number of each structural domain.
Higher average number of non-synonymous natural variants was observed within the TM domains (TMs 1–7) than in the intracellular or extracellular loops (ICLs 1–3 and ECLs 1–3); moreover, no non-synonymous natural variant was observed in ICL1, ICL3, and ECL3 of the Secretin family; ECL1 and ECL3 of the Adhesion family; ECL1 of the Glutamate family; and ICL1, ECL1, and ECL3 of the Fizzled family (Table 1, Fig. 2A, 2B). When normalized by the average residue numbers, the occurrence of non-synonymous natural variants became similar throughout the TM, ICL, and ECL domains in all GPCR families except ICL2 of the Secretin family. The normalized average number of natural variants in ICL2 of the Secretin family was two to four times higher than that of the other TM, ICL, and ECL domains of the same family (Fig. 2C), which suggests that ICL2 of the Secretin family has higher frequency of non-synonymous natural variations than ICL2 of other GPCR families.
In the present study, we divided the C-terminus into two domains, helix 8 (H8) and C-terminal tail (Fig. 1A). The H8 domain is an α-helical domain (approximately 12–26 residues) located parallel to the plasma membrane (Fig. 1A). The percentage of GPCRs with non-synonymous natural variants at the H8 domain and the average number of non-synonymous natural variants at the H8 domain are smaller than the TM α-helical domains (Fig. 2A, 2B). The C-terminal tail contains a relatively larger number of non-synonymous natural variants than the other intracellular domains (Fig. 2A, 2B), although this number also becomes relatively similar to that of the other intracellular domains after normalization by average residue number (Fig. 2C).
Distribution of GPCR non-synonymous natural variants within helical domains and functional consequences
Non-synonymous natural variants of GPCRs can severely affect their functions such as membrane expression, activation, and drug interaction. As we describe in the present study, a high degree of genetic variation has been observed in the human GPCRs. However, the functional effects of the natural variants have been studied in only 218 (from 41 GPCRs) out of 1,440 non-synonymous natural variants (from 269 GPCRs) (Table 2).
Table 2.
Non-synonymous natural variants with known functional consequences
| Family | Total # of natural variants | # of natural variants with known functional consequences | % of natural variants with known functional consequences | Total # of GPCRs with natural variants | # of GPCRs with natural variants with known functional consequences | % of GPCRs with natural variants with known functional consequences |
|---|---|---|---|---|---|---|
| Rhodopsin | 1,059 | 155 | 14.6 | 207 | 31 | 15.0 |
| Secretin | 47 | 4 | 8.5 | 12 | 2 | 16.7 |
| Adhesion | 135 | 17 | 12.6 | 26 | 3 | 11.5 |
| Glutamate | 151 | 34 | 22.5 | 18 | 2 | 11.1 |
| Frizzled | 48 | 8 | 16.7 | 6 | 3 | 50.0 |
| Total | 1,440 | 218 | 15.1 | 269 | 41 | 15.2 |
Although the TM domains share certain degree of structural similarity, the sequences vary significantly between different GPCRs, with only a few conserved residues. Ballesteros and Weinstein developed a sequence-based generic GPCR numbering system for the Rhodopsin family GPCRs (Ballesteros and Weinstein, 1995). Subsequently, similar systems have been developed for other GPCR families (Pin et al., 2003; Wootten et al., 2013; Wang et al., 2014). In these systems, the residue numbers are assigned relative to the most conserved position in each TM, which is number 50. For example, in the Rhodopsin family, residue number 3.50 indicates the conserved arginine residue in TM3, and residue number 3.49 indicates one residue before the conserved arginine in TM3. Recently, a modified system was developed based on the GPCR crystal structures (Isberg et al., 2015), and GPCRdb assigned residue numbers for all GPCRs (except the olfactory receptors) using this new numbering system.
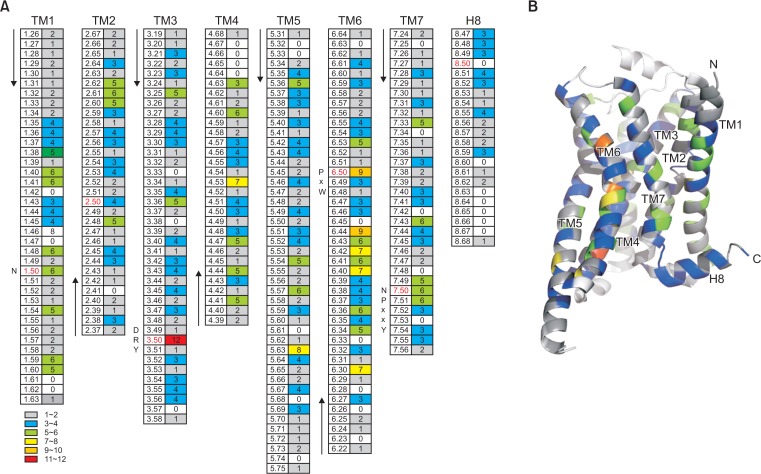
It has been reported that disease-causing non-synonymous polymorphisms of GPCRs occur more frequently within the TMs than non-disease-causing non-synonymous polymorphisms (Lee et al., 2003). Hence, to analyze the distribution pattern of the non-synonymous natural variants in these regions, we mapped all the non-synonymous natural variations in the TM and H8 domains based on the generic numbering system provided by GPCRdb (Fig. 3). In general, the non-synonymous natural variants are distributed throughout TM domains and H8 (Fig. 3). Overall, the occurrence of non-synonymous natural variants are not restricted to specific TM domains or residues. Nonetheless, we detected a few exceptional positions such as 3.50, 6.50, and 6.44, where more than nine non-synonymous natural variants were detected (Fig. 3).
Fig. 3.
Distribution of non-synonymous natural variants within the TM and H8 domains. (A) The positions of non-synonymous natural variants within the TM and H8 domains were assigned based on the generic numbering system provided by GPCRdb (left column), and the numbers of detected non-synonymous natural variants in these positions are indicated (right column). The conserved residues in each domain (x.50) are colored in red, and conserved motifs with functional significance (i.e., DR3.50Y, CP3.50xW, and NP3.50xxY) are indicated at their structural positions. (B) The frequencies of non-synonymous natural variations are highlighted using color-coding and mapped onto the crystal structure of a model GPCR (PDB 3SN6).
It is interesting to note that two of these exceptional residues are conserved residues with a residue number of x.50. More importantly, these conserved residues were found to have critical roles in the ligand-induced activation of GPCRs. In the Rhodopsin family, residue 3.50 is arginine (Fig. 4A, red stick), which is part of the D[E]R3.50Y motif; in X-ray crystal structures of the rhodopsin and a few other GPCRs, R3.50 in D[E]R3.50Y interacts with E6.30 forming an ionic lock in the inactive state, which is broken upon activation indicating that the ionic lock is an important factor for GPCR activation (Zhang et al., 2015). Among the 11 non-synonymous natural variations reported for residue R3.50, the conserved arginine was substituted with histidine in five cases, cysteine and leucine in two cases each, and glycine and tryptophan in one case each.
Fig. 4.
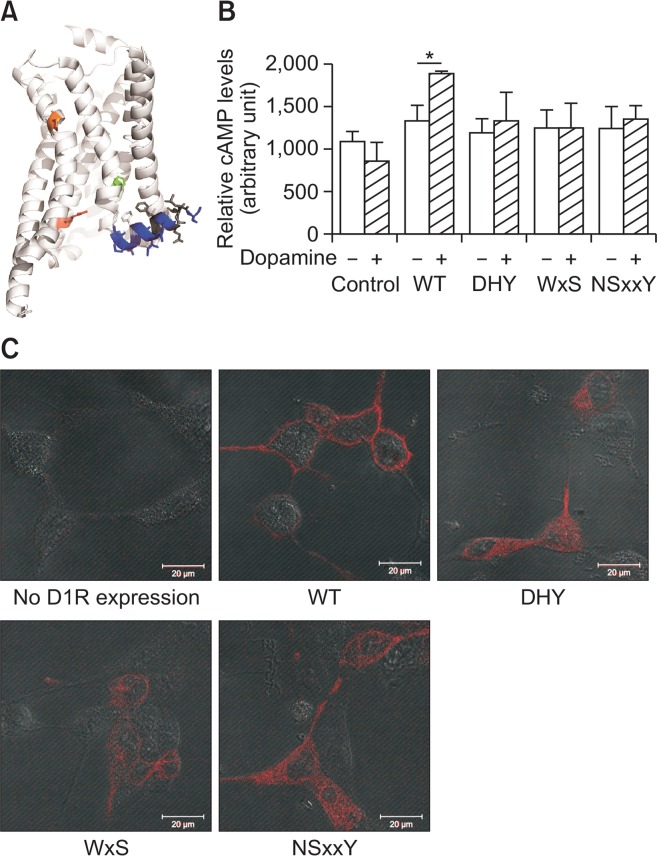
Positions of the conserved motifs and functional consequences of non-synonymous natural variations in the conserved motifs. (A) Positions of x.50 conserved residues within the DR3.50Y (red), CP6.50xW (orange), and NP7.50xxY (green) motifs are shown with sticks, and H8 domain is highlighted with sticks and color-coded based on Fig. 3. The figure is based on the crystal structure of a model GPCR (PDB 3SN6). (B) D1R variants-mediated G protein activation is analyzed by measuring agonist-induced cAMP generation. T-test was used for the statistical analysis, and *indicates p<0.05 from six experiments. (C) Cellular localization of D1R variants is analyzed by immunofluorescence assay.
Residue 6.50 is another conserved residue in the CWxP6.50 motif in the Rhodopsin family (Fig. 4A, orange stick) and has a critical function in GPCR activation; W6.48 is known to be a rotamer toggle switch that undergoes a conformational change upon agonist binding to induce movement of TM6 (Zhang et al., 2015). Among the nine non-synonymous natural variants reported in P6.50, four were substitutions with serine, two with arginine and leucine each, and one with alanine. Although less frequent than residues 3.50 and 6.50, residue 7.50 (proline in the Rhodopsin family) (Fig. 4A, green stick) also showed six non-synonymous natural variations: three were substituted with serine, and the other three were substituted with leucine, histidine, or arginine. P7.50 is also a part of a conserved motif (NP7.50xxY) with functional significance (Zhang et al., 2015). It is interesting to note that there is a bias (approximately 45%) of substituted amino acids in these conserved regions; 5 out of 11 R3.50 variations involved a substitution with histidine, 4 out of 9 P6.50 variations involved a substitution with serine, and 3 out of 6 P7.50 variations involved a substitution with serine.
We have tested the functional consequences of the non-synonymous natural variants in these conserved residues by using dopamine receptor type 1 (D1R) as a model GPCR. D1R is a Rhodopsin family GPCR and has conserved D[E] R3.50Y, CWxP6.50, and NP7.50xxY motifs. Here, we mutated the conserved residues within D1R into the most frequently found non-synonymous natural variants: R3.50 into histidine, P6.50 and P7.50 into serine. All mutations in the conserved residues resulted in impairment of G protein activation (Fig. 4B). When the cellular expression was analyzed, the mutated D1R variants were localized not in the plasma membrane but inside of the cells (Fig. 4C), which suggests that the mutations in these conserved residues lead to mis-localization of the receptor and thus impaired signaling.
It has been shown that the H8 domain has a critical role in the expression/localization and activation of GPCRs (Ahn et al., 2010; Kuramasu et al., 2011; Kawasaki et al., 2015; Sounier et al., 2015; Zhu et al., 2015). The H8 domain is located beneath the plasma membrane, with one side of the H8 domain in contact with the plasma membrane and the other sides exposed to the cytosol (Fig. 1A). The residues that interact with the plasma membrane (residues 8.50, 8.54, and 8.57/8.58) are hydrophobic, with residue 8.50 being a conserved phenylalanine or leucine. Interestingly, residues that contact the plasma membrane show no or less frequent non-synonymous natural variant occurrence than the residues that are exposed to the cytosol (Fig. 3, 4A).
DISCUSSION
Approximately 30–40% of marketed drugs target GPCRs, and 6 out of the top 15 selling small-molecule drugs target GPCRs according to the 2015 annual report by Pharma-Compass. With the high-resolution X-ray crystal structures of various GPCRs being published since 2007, the structural understanding of GPCR activation mechanism has greatly improved, and the structure-based drug design for GPCR-targeting drugs is of great interest in the pharmaceutical industry (Shoichet and Kobilka, 2012). Given the pharmacological and pathological importance of GPCRs, it will be critical to understand their genotype-phenotype relationships, but only a few (approximately 15%) have been studied (Table 2). Moreover, it has been suggested that the variability of the GPCR superfamily is not well defined, as its false positive rate is high (68%) (Small et al., 2002). Therefore, it is necessary to validate the natural variants in the databases and investigate the physiological and/or pathological consequences of the variants. The current study, for the first time, analyzed the distribution of all known non-synonymous natural variations from all 367 non-olfactory GPCRs (except twenty five taste 2 and six un-categorized orphan receptors) in the structural context. Majority of GPCRs showed non-synonymous natural variations mainly in the N-terminus and TM regions throughout five GPCR families (Fig. 2).
Analysis of the positions of non-natural variations within the TM domains revealed their presence throughout the TM regions, and a few were more frequently detected in the conserved and functionally critical residues (x.50) (Fig. 3). Most of the non-synonymous natural variations in the conserved residues (Fig. 4A) have been detected in disease states such as Hirschsprung disease-2, X-linked nephrogenic diabetes insipidus, hyperthyroidism, hypogonadotropic hypogonadism, glucocorticoid deficiency 1, and retinitis pigmentosa according to the Uniprot database. The functional consequences and underlying mechanisms of these substitutions have not been fully understood, and only a few have been studied. For example, substitution of R3.50 with histidine in the gonadotropin-releasing hormone receptor or vasopressin V2 receptor has been reported to reduce receptor activity, whereas substitution with cysteine or leucine in the vasopressin V2 receptor increases receptor activity according to the UniProt database. Substitution of P7.50 with serine in the neuromedin-K receptor has been reported to impair receptor signaling. Here, we analyzed the functional consequences and underlying mechanisms of the most common non-synonymous natural variants in the conserved residues by using D1R as a model GPCR. We found that all the tested variants showed impaired cellular localization, which lead to impaired signaling (Fig. 4B, 4C). We suspect that other Rhodopsin family receptors would show similar functional consequences when the conserved residues are substituted with the same amino acids (i.e R3.50 into histidine, P6.50 and P7.50 into serine).
The comprehensive analysis of non-synonymous natural variants in the current study would provide valuable insight for future research in GPCR non-synonymous natural variants. Firstly, for example, the current study suggests that ICL2 of the Secretin family has more frequency of non-synonymous natural variants than ICL2 of other GPCR families. Therefore, researchers can expect that non-synonymous natural variants would occur in ICL2 of the Secretin family although it is rare in other GPCR families. Secondly, although we have tested functional consequences of non-synonymous natural variants in a few conserved residues, we did not analyze comprehensive outlook of the genotype-phenotype relationship of GPCRs. This was mainly because of relatively poor database in the genotype-phenotype relationship of GPCRs. The current study provided the distribution of non-synonymous natural variants of GPCRs in the structural context. With aid of the extensive structural studies of GPCRs recent 10 years (Zhang et al., 2015), this data would provide information for prioritization of the GPCR genotype-phenotype relationship investigation.
Acknowledgments
This work was supported by the National Research Foundation of Korea funded by the Korean government (NFR-2015R1A1A1A05027473).
REFERENCES
- Ahn KH, Nishiyama A, Mierke DF, Kendall DA. Hydrophobic residues in helix 8 of cannabinoid receptor 1 are critical for structural and functional properties. Biochemistry. 2010;49:502–511. doi: 10.1021/bi901619r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros JA, Weinstein H. [19] Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 1995;25:366–428. doi: 10.1016/S1043-9471(05)80049-7. [DOI] [Google Scholar]
- Boutet E, Lieberherr D, Tognolli M, Schneider M, Bansal P, Bridge AJ, Poux S, Bougueleret L, Xenarios I. UniProtKB/Swiss-Prot, the manually annotated section of the UniProt KnowledgeBase: How to use the entry view. Methods Mol Biol. 2016;1374:23–54. doi: 10.1007/978-1-4939-3167-5_2. [DOI] [PubMed] [Google Scholar]
- Cho DI, Beom S, Van Tol HH, Caron MG, Kim KM. Characterization of the desensitization properties of five dopamine receptor subtypes and alternatively spliced variants of dopamine D2 and D4 receptors. Biochem Biophys Res Commun. 2006;350:634–640. doi: 10.1016/j.bbrc.2006.09.090. [DOI] [PubMed] [Google Scholar]
- Chun KS, Shim M. EP2 induces p38 phosphorylation via the activation of Src in HEK 293 cells. Biomol. Ther. (Seoul) 2015;23:539–548. doi: 10.4062/biomolther.2015.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearry A, Gingrich JA, Falardeau P, Fremeau RT, Jr, Bates MD, Caron MG. Molecular cloning and expression of the gene for a human D1 dopamine receptor. Nature. 1990;347:72–76. doi: 10.1038/347072a0. [DOI] [PubMed] [Google Scholar]
- Drazen JM, Silverman EK, Lee TH. Heterogeneity of therapeutic responses in asthma. Br Med Bull. 2000;56:1054–1070. doi: 10.1258/0007142001903535. [DOI] [PubMed] [Google Scholar]
- Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- Hecht M, Bromberg Y, Rost B. News from the protein mutability landscape. J Mol Biol. 2013;425:3937–3948. doi: 10.1016/j.jmb.2013.07.028. [DOI] [PubMed] [Google Scholar]
- Insel PA, Tang CM, Hahntow I, Michel MC. Impact of GPCRs in clinical medicine: monogenic diseases, genetic variants and drug targets. Biochim. Biophys. Acta. 2007;1768:994–1005. doi: 10.1016/j.bbamem.2006.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isberg V, de Graaf C, Bortolato A, Cherezov V, Katritch V, Marshall FH, Mordalski S, Pin JP, Stevens RC, Vriend G, Gloriam DE. Generic GPCR residue numbers - aligning topology maps minding the gaps. Trends Pharmacol Sci. 2015;36:22–31. doi: 10.1016/j.tips.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki T, Saka T, Mine S, Mizohata E, Inoue T, Matsumura H, Sato T. The N-terminal acidic residue of the cytosolic helix 8 of an odorant receptor is responsible for different response dynamics via G-protein. FEBS Lett. 2015;589:1136–1142. doi: 10.1016/j.febslet.2015.03.025. [DOI] [PubMed] [Google Scholar]
- Kazius J, Wurdinger K, van Iterson M, Kok J, Back T, Ijzerman AP. GPCR NaVa database: natural variants in human G protein-coupled receptors. Hum Mutat. 2008;29:39–44. doi: 10.1002/humu.20638. [DOI] [PubMed] [Google Scholar]
- Kimura M. Evolutionary rate at the molecular level. Nature. 1968;217:624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- Kuramasu A, Sukegawa J, Sato T, Sakurai E, Watanabe T, Yanagisawa T, Yanai K. The hydrophobic amino acids in putative helix 8 in carboxy-terminus of histamine H3 receptor are involved in receptor-G-protein coupling. Cell Signal. 2011;23:1843–1849. doi: 10.1016/j.cellsig.2011.06.021. [DOI] [PubMed] [Google Scholar]
- Lagerstrom MC, Schioth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- Lee A, Rana BK, Schiffer HH, Schork NJ, Brann MR, Insel PA, Weiner DM. Distribution analysis of nonsynonymous polymorphisms within the G-protein-coupled receptor gene family. Genomics. 2003;81:245–248. doi: 10.1016/S0888-7543(03)00009-0. [DOI] [PubMed] [Google Scholar]
- Munk C, Isberg V, Mordalski S, Harpsoe K, Rataj K, Hauser AS, Kolb P, Bojarski AJ, Vriend G, Gloriam DE. GPCRdb: the G protein-coupled receptor database - an introduction. Br J Pharmacol. 2016;173:2195–2207. doi: 10.1111/bph.13509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- Pin JP, Galvez T, Prezeau L. Evolution, structure, and activation mechanism of family 3/C G-protein-coupled receptors. Pharmacol Ther. 2003;98:325–354. doi: 10.1016/S0163-7258(03)00038-X. [DOI] [PubMed] [Google Scholar]
- Rana BK, Shiina T, Insel PA. Genetic variations and polymorphisms of G protein-coupled receptors: functional and therapeutic implications. Annu Rev Pharmacol Toxicol. 2001;41:593–624. doi: 10.1146/annurev.pharmtox.41.1.593. [DOI] [PubMed] [Google Scholar]
- Shoichet BK, Kobilka BK. Structure-based drug screening for G-protein-coupled receptors. Trends Pharmacol Sci. 2012;33:268–272. doi: 10.1016/j.tips.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small KM, Seman CA, Castator A, Brown KM, Liggett SB. False positive non-synonymous polymorphisms of G-protein coupled receptor genes. FEBS Lett. 2002;516:253–256. doi: 10.1016/S0014-5793(02)02564-4. [DOI] [PubMed] [Google Scholar]
- Small KM, Tanguay DA, Nandabalan K, Zhan P, Stephens JC, Liggett SB. Gene and protein domain-specific patterns of genetic variability within the G-protein coupled receptor superfamily. Am. J. Pharmacogenomics. 2003;3:65–71. doi: 10.2165/00129785-200303010-00008. [DOI] [PubMed] [Google Scholar]
- Sounier R, Mas C, Steyaert J, Laeremans T, Manglik A, Huang W, Kobilka BK, Demene H, Granier S. Propagation of conformational changes during μ-opioid receptor activation. Nature. 2015;524:375–378. doi: 10.1038/nature14680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard NC, Chun J. Promising pharmacological directions in the world of lysophosphatidic acid signaling. Biomol. Ther. (Seoul) 2015;23:1–11. doi: 10.4062/biomolther.2014.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang CM, Insel PA. Genetic variation in G-protein-coupled receptors - consequences for G-protein-coupled receptors as drug targets. Expert Opin. Ther. Targets. 2005;9:1247–1265. doi: 10.1517/14728222.9.6.1247. [DOI] [PubMed] [Google Scholar]
- Thompson MD, Cole DE, Capra V, Siminovitch KA, Rovati GE, Burnham WM, Rana BK. Pharmacogenetics of the G protein-coupled receptors. Methods Mol Biol. 2014a;1175:189–242. doi: 10.1007/978-1-4939-0956-8_9. [DOI] [PubMed] [Google Scholar]
- Thompson MD, Hendy GN, Percy ME, Bichet DG, Cole DE. G protein-coupled receptor mutations and human genetic disease. Methods Mol Biol. 2014b;1175:153–187. doi: 10.1007/978-1-4939-0956-8_8. [DOI] [PubMed] [Google Scholar]
- Wahlestedt C, Brookes AJ, Mottagui-Tabar S. Lower rate of genomic variation identified in the trans-membrane domain of monoamine sub-class of Human G-Protein Coupled Receptors: the Human GPCR-DB Database. BMC Genomics. 2004;5:91. doi: 10.1186/1471-2164-5-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Wu H, Evron T, Vardy E, Han GW, Huang XP, Hufeisen SJ, Mangano TJ, Urban DJ, Katritch V, Cherezov V, Caron MG, Roth BL, Stevens RC. Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs. Nat Commun. 2014;5:4355. doi: 10.1038/ncomms5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootten D, Simms J, Miller LJ, Christopoulos A, Sexton PM. Polar transmembrane interactions drive formation of ligand-specific and signal pathway-biased family B G protein-coupled receptor conformations. Proc Natl Acad Sci USA. 2013;110:5211–5216. doi: 10.1073/pnas.1221585110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Zhao Q, Wu B. Structural Studies of G Protein-Coupled Receptors. Mol. Cells. 2015;38:836–842. doi: 10.14348/molcells.2015.0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Choi BG, Kim KM. Roles of dopamine D2 receptor subregions in interactions with β-Arrestin2. Biomol. Ther. (Seoul) 2016;24:517–522. doi: 10.4062/biomolther.2015.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Zhang X, Min C, Choi BG, Oh IJ, Kim KM. Functional regulation of dopamine D3 receptor through interaction with PICK1. Biomol. Ther. (Seoul) 2016;24:475–481. doi: 10.4062/biomolther.2016.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Zhang M, Davis JE, Wu WH, Surrao K, Wang H, Wu G. A single mutation in helix 8 enhances the angiotensin II type 1a receptor transport and signaling. Cell Signal. 2015;27:2371–2379. doi: 10.1016/j.cellsig.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]